Abstract
Genetic alterations in uterine myxoid leiomyosarcoma are unknown. We investigate the clinicopathologic features of 19 uterine tumors previously diagnosed as myxoid leiomyosarcomas in which tumoral RNA was subjected to targeted RNA sequencing. PLAG1, BCOR, BCORL1, HMGA2, and ALK break-apart fluorescence in situ hybridization (FISH) and BCOR, PLAG1, and ALK immunohistochemistry were performed in cases which failed or lacked fusions by sequencing. The diagnosis of myxoid leiomyosarcoma was confirmed in 15 cases after exclusion of 4 tumors with BCOR and ALK rearrangements. These 15 patients presented at a median age of 50 years with stage I (3), II (2), III (2), and IV (1), respectively; stage was unknown in 7 cases. Tumor size ranged from 10 to 24 cm. Matrix was myxoid in all tumors and also eosinophilic in 2. Cells were spindled, epithelioid, and both in 10, 2 and 3 tumors and showed mild, moderate, and severe nuclear atypia in 3, 8, and 4 tumors, respectively. Mitotic index ranged from <1 to 14/10 HPF, while tumor necrosis was present in 6 (40%). Novel TRPS1-PLAG1 or RAD51B-PLAG1 fusions were detected by sequencing in four tumors, three of which were also confirmed by FISH. Diffuse PLAG1 expression was seen in seven tumors, including four with PLAG1 rearrangement. No morphologic differences were seen among PLAG1 fusion-positive and fusion-negative tumors. No PLAG1, HMGA2, ALK, BCOR, or BCORL1 rearrangements were detected by FISH in 11 tumors. Based on sequencing and FISH results, PLAG1 rearrangements resulting in PLAG1 expression underpin ~25% of myxoid leiomyosarcomas and may serve as a useful diagnostic biomarker. Immunohistochemistry, targeted RNA sequencing, and/or FISH may distinguish myxoid leiomyosarcoma from its morphologic mimics.
Keywords: myxoid leiomyosarcoma, endometrial stromal sarcoma, inflammatory myofibroblastic tumor, PLAG1, BCOR, ALK, uterine sarcoma
INTRODUCTION
Myxoid leiomyosarcoma is a rare morphologic variant of malignant uterine smooth muscle neoplasia first described in 1982 because of an often deceptively bland appearance leading to misdiagnosis as benign. While clinical data is limited given its rarity, myxoid leiomyosarcoma appears to be more aggressive than usual leiomyosarcoma (1–4). Evaluation of myxoid uterine smooth muscle tumors is currently based on histologic features only with no information on the potential role of molecular alterations in tumor classification. In this study, we investigate the possible impact of molecular alterations in the distinction of myxoid leiomyosarcoma from other myxoid sarcomas.
Recognition of myxoid leiomyosarcoma is often difficult given the extensive histologic and immunophenotypic overlap with a variety of benign and malignant uterine mesenchymal tumors. An infiltrative pattern and lymphovascular invasion may help distinguish myxoid leiomyosarcoma from myxoid leiomyoma and leiomyoma with hydropic change. However, distinction from recently defined entities such as myxoid inflammatory myofibroblastic tumor (5–9) and endometrial stromal sarcoma with myxoid features, particularly those with BCOR genetic aberrations (10, 11) poses a significant challenge when using morphologic and/or immunohistochemical assessment alone.
The published literature on uterine myxoid leiomyosarcoma is currently limited to clinicopathologic studies, and genetic alterations involved in the pathogenesis of this rare tumor type are essentially unknown. Since gene rearrangements are frequently found among endometrial stromal sarcomas (12–14), inflammatory myofibroblastic tumors (5, 7, 9), and a subset of uterine leiomyomas (15, 16), we surmise that uterine myxoid leiomyosarcoma could potentially harbor fusions that may serve as diagnostic biomarkers. In this study, we performed targeted RNA sequencing and fluorescence in situ hybridization on a large cohort of uterine myxoid leiomyosarcomas to identify novel gene fusions in this rare tumor.
MATERIALS & METHODS
Case Selection
Nineteen malignant myxoid and at least focally spindled uterine tumors diagnosed as myxoid leiomyosarcoma were collected from three institutions. Ten, three, and two tumors were identified in the surgical pathology files of Memorial Sloan Kettering Cancer Center (New York, NY, USA), Massachusetts General Hospital (Boston, MA, USA), and Vancouver General Hospital (Vancouver, BC, Canada), respectively, by searching pathology databases using the following search terms: “myxoid leiomyosarcoma” and “leiomyosarcoma with myxoid features.” Four additional tumors originated from consultation files of two of the authors (EO and RHY) and the late Dr. Robert E. Scully. Available hematoxylin and eosin and immunohistochemical stained slides of each tumor were reviewed for independent diagnostic confirmation and detailed morphologic analysis by at least two gynecologic pathologists (JAS, SC, CHL, LNH, EO, RHY). Clinical data, including patient age, tumor stage, and clinical follow up were extracted from pathology reports and available clinical records. Archival formalin-fixed, paraffin-embedded tissue was retrieved. All tumors were subjected to the MSK-Solid Fusion assay (described below), immunohistochemistry for BCOR, PLAG1, and ALK (described below), and fluorescence in situ hybridization (FISH, described below). Any tumors found to harbor BCOR and ALK rearrangements were excluded from the study. The study was approved by the Institutional Review Board from respective institutions.
Next-Generation Sequencing
Samples were subjected to the MSK-Solid Fusion assay, a custom targeted RNA-based panel that utilizes the Archer Anchored Multiplex PCR (AMP™) technology to detect gene fusions and oncogenic isoforms in selected protein-coding exons of 62 genes(17). Tumor RNA was extracted from macrodissected formalin-fixed, paraffin-embedded tissue sections mounted on charged glass slides followed by cDNA synthesis and library preparation. Final targeted amplicons were sequenced on an Illumina Miseq instrument. The Archer™ analysis software V5.0 was used for data analysis.
Immunohistochemistry
Immunohistochemical stains using commercially available BCOR (C-10, 1:150, Santa Cruz, Dallas, TX), PLAG1 (3B7, Novus, Littleton, CO), and ALK (D5F3, Cell Signaling, Danvers, MA) antibodies were performed on 5-μm tumor tissue sections mounted on glass slides from all samples with available tumor tissue. All immunohistochemical assays were performed on Leica Bond-3 (Leica, Buffalo Grove, IL) automated stainer platforms employing heat-based antigen retrieval and a polymer-based secondary kit (Refine, Leica). Hematoxylin was used as a counterstain. A carrier-based multi-tissue block comprising of normal skin, colon, lung, testis, spleen, placenta, liver, kidney, and pancreas served as negative controls (18). Results were evaluated by two gynecologic pathologists (SC, JAS). Staining intensity (negative, weak, moderate, and strong) and estimated percentage of positive tumor cells were recorded. Only nuclear BCOR and PLAG1 and cytoplasmic ALK staining were considered positive. PLAG1 immunohistochemistry was specifically selected to study PLAG1 expression patterns in tumors harboring or lacking PLAG1 rearrangement by fluorescence in situ hybridization or targeted RNA sequencing.
Fluorescence In Situ Hybridization
Samples that were not sequenced due to low quality RNA, failed sequencing, or did not show gene fusions by targeted RNA sequencing were subjected to FISH to detect PLAG1, HMGA2, BCOR, , and ALK gene rearrangements. Custom probes were made by bacterial artificial chromosomes flanking PLAG1 (RP11–248B17, RP11–92A9, RP11–111I18, RP11–246A9, RP11–446E9, RP11–144E19), HMGA2 (RP11–937C6, RP11–167E10, RP11–662G15, RP11–230G5), BCOR (RP11–21D3, RP11–1105N2, RP11–37K20, RP11–973F20), and ALK (RP11–100C1, RP11–384J5, RP11–702A2, RP11–776G10) genes and obtained from BAC/PAC Resources (Children’s Hospital Oakland Research Institute, Oakland, CA, USA). BAC clones were labeled with nick translation and validated on normal metaphase chromosomes. Five-μm whole tumor tissue sections from formalin-fixed, paraffin-embedded tissue blocks were mounted on charged glass slides. Slides were deparaffinized, pretreated, and hybridized with denatured probes overnight, followed by post-hybridization washes and counterstaining with DAPI. Slides were examined on a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany) using Isis 5 software (Metasystems). Two hundred tumor nuclei were counted. Samples with >20% of tumor nuclei demonstrating break-apart signals were considered positive for gene rearrangement.
RESULTS
Clinicopathologic features
After re-review of available H&E slides as well as sequencing and FISH results, the diagnosis of myxoid leiomyosarcoma was confirmed in 15 tumors (Table 1, Figure 1); 4 were excluded based on the identification of BCOR and ALK rearrangements detected by targeted RNA sequencing (3 ZC3H7B-BCOR and 1 FN1-ALK fusion, data not shown). The median patient age was 50 (range, 28 to 74) years. Three, two, two, and one patients presented with FIGO stage I, II, III and IV disease, respectively; tumor stage was unknown in seven patients. Tumor size ranged from 10 to 24 (median, 16) cm. Tumor infiltrated the adjacent myometrium in five tumors and was circumscribed in one; tumor border could not be assessed in nine cases for which the adjacent myometrium was not present in available slides. Myxoid matrix was seen in all 15 tumors and was diffuse in 7 and multifocal in 6. Two tumors also showed eosinophilic matrix. Cells were spindled in 13 tumors, 3 of which also showed epithelioid features. Two tumors were composed entirely of epithelioid cells. Nuclear atypia was mild, moderate, and severe in three, eight, and four tumors, respectively. The mitotic index ranged from <1 to 14 per 10 HPF with five tumors having <2 mitoses per 10 HPFs. Delicate, thin-walled blood vessels were seen in 13 tumors, while thick-walled blood vessels were present in 2. Tumor cell necrosis was present in six tumors while lymphovascular invasion was absent in all 15 tumors.
Table 1.
Clinicopathologic characteristics
| PLAG1 fusion status | ||
|---|---|---|
| Positive n = 4 No. (%) |
Negative n = 11 No. (%) |
|
| Age, y | ||
| Median | 49 | 51 |
| Range | 28–74 | 44–65 |
| Stage | ||
| I | 2 | 1 |
| II | 1 | 1 |
| III | 0 | 2 |
| IV | 0 | 1 |
| Unknown | 1 | 6 |
| Tumor size, cm | ||
| Median | 17 | 15 |
| Range | 15–24 | 10–20 |
| Unknown | 1 | 5 |
| Tumor necrosis | ||
| Absent | 2 | 7 |
| Present | 2 | 4 |
| Nuclear atypia | ||
| Mild | 0 | 3 |
| Moderate | 4 | 4 |
| Severe | 0 | 4 |
| Mitotic index, #/10 HPF | ||
| <2 | 1 | 4 |
| 2–10 | 2 | 5 |
| >10 | 1 | 2 |
| Vascular invasion | ||
| Absent | 4 | 11 |
| Present | 0 | 0 |
Figure 1.
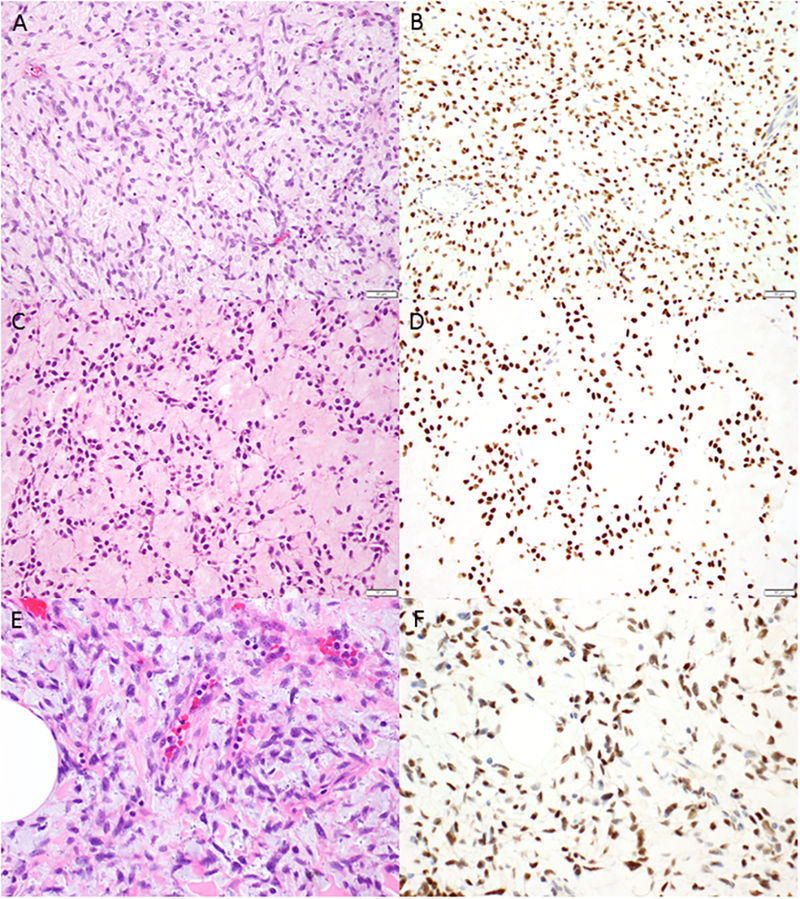
Myxoid leiomyosarcomas with PLAG1 rearrangement. Spindled cells with scant cytoplasm and ovoid nuclei (A, C, E) are embedded abundant amphophilic (A, E) or eosinophilic (C) myxoid matrix. Strong and diffuse nuclear PLAG1 expression is seen in all tumors with PLAG1 rearrangement (B, D, F).
Novel PLAG1 rearrangement defines a subset of uterine myxoid leiomyosarcoma
Targeted RNA sequencing was successful in eight tumors. Sequencing was attempted in four cases, but resulted in technical failure. Three tumors were not sequenced after RNA extraction due to low RNA quality. Rearrangements involving the PLAG1 gene were identified in four of eight (50%) tumors sequenced (cases 1–4) (Table 2). An in-frame fusion between exon 1 of the TRPS1 gene and exon 3 of the PLAG1 gene were detected in three tumors (cases 1–3) (Figure 2). One tumor (case 4) harbored an in-frame fusion between exon 9 of the RAD51B gene and exon 2 of the PLAG1 gene. PLAG1 rearrangement was also confirmed by FISH in three of these tumors for which additional material was available. FISH was successful in the four tumors that were negative for gene fusion by sequencing and the remaining seven that were subjected to sequencing, but resulted in technical failure, or were not subjected to sequencing due to suboptimal RNA quality. PLAG1 and HMGA2 rearrangements by FISH were not identified in any of these tumors (Table 2). After exclusion of tumors harboring BCOR and ALK fusions and based on both sequencing and FISH results, PLAG1 rearrangement was found in 4 of 15 (26%) myxoid leiomyosarcomas. No histologic differences were seen between fusion-positive and fusion-negative tumors.
Table 2.
Immunohistochemical and molecular characteristics
| Case | Immunohistochemistry | MSK-Solid Fusion Assay | Fluorescence in situ hybridization | Revised Diagnosis | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PLAG1 | ALK | BCOR | PLAG1 | HMGA2 | BCOR | BCORL1 | ALK | |||
| 1 | + (S >95%) | NP | NP | TRPS1-PLAG1 | NP | NP | NP | NP | NP | mLMS with PLAG1 fusion |
| 2 | + (S >95%) | NP | NP | TRPS1-PLAG1 | NP | NP | NP | NP | NP | mLMS with PLAG1 fusion |
| 3 | NP | NP | NP | TRPS1-PLAG1 | NP | NP | NP | NP | NP | mLMS with PLAG1 fusion |
| 4 | + (S >95%) | - | - | RAD51B-PLAG1 | NP | NP | NP | NP | NP | mLMS with PLAG1 fusion |
| 5 | + (M >95%) | - | + (W <5%) | - | - | - | - | - | - | mLMS |
| 6 | + (S >95%) | - | + (W <5%) | - | - | - | - | - | - | mLMS |
| 7 | NP | - | + (W 30%) | - | - | - | - | - | - | mLMS |
| 8 | + (W <5%) | - | + (M 50%) | - | - | - | - | - | - | mLMS |
| 9 | + (W <5%) | - | - | F | - | - | - | - | - | mLMS |
| 10 | - | - | - | F | - | - | - | - | - | mLMS |
| 11 | - | - | - | F | - | - | - | - | - | mLMS |
| 12 | + (W <5%) | - | - | F | - | - | - | - | - | mLMS |
| 13 | + (S >95%) | - | - | NP | - | - | - | - | - | mLMS |
| 14 | + (W <5%) | - | - | NP | - | - | - | - | - | mLMS |
| 15 | + (W 50%) | - | + (M 50%) | NP | - | - | - | - | - | mLMS |
F indicates failed; M, moderate; mLMS, myxoid leiomyosarcoma; NP, not performed; S, strong; W, weak.
Figure 2.
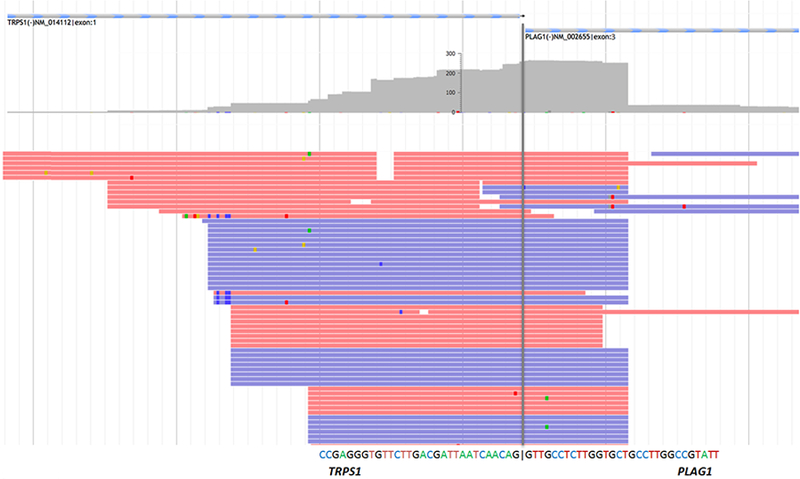
Detection of TRPS1-PLAG1 fusion in myxoid leiomyosarcoma. Schematic illustration of the TRPS1 Exon1 (NM_014112) and PLAG1 Exon3 (NM_002655) in-frame fusion. Red and blue lines represent bi-directional RNA sequencing reads supporting the fusion breakpoint, indicated by the grey line. The depth of coverage is shown above sequencing reads in grey.
Strong and diffuse PLAG1 immunoexpression is seen PLAG1-rearranged tumors
Strong nuclear PLAG1 expression in >95% of tumor cells was confirmed in three tumors (cases 1, 2, and 4) harboring PLAG1 rearrangement for which additional tumor tissue material was available (Figure 1). PLAG1 immunohistochemistry was not performed on the fourth tumor (case 3) harboring TRPS1-PLAG1 fusion for which all available tumor tissue sections were subjected to sequencing. Moderate to strong nuclear PLAG1 expression in >95% of tumor cells was also seen in 3 of 11 (27%) tumors that lacking gene fusions by targeted RNA sequencing and/or PLAG1, HMGA2, BCOR, and ALK rearrangements by FISH. One tumor showed weak PLAG1 expression in approximately 50% of tumor cells, while six tumors showed either weak staining in <5% of tumor cells or complete absence of immunoreactivity.
Fusions are not identified in the majority of bona fide myxoid leiomyosarcomas
No fusions were detected in the four tumors in which sequencing was successful after exclusion of endometrial stromal sarcoma and inflammatory myofibroblastic tumor. PLAG1, HMGA2, BCOR, and ALK rearrangements were also not detected in these four tumors and the seven cases that were subjected to sequencing, but resulted in technical failure, or were not sequenced due to low quality RNA.
DISCUSSION
In this study, we characterized the morphologic and genomic profiles of uterine myxoid leiomyosarcoma, a rare subtype of malignant smooth muscle neoplasia, to ascertain whether molecular features might help distinguish this entity from its morphologic mimics. By targeted RNA sequencing and FISH, we identified PLAG1 gene rearrangements resulting in PLAG1 overexpression in 26% of bona fide myxoid leiomyosarcomas after exclusion of tumors harboring BCOR and ALK rearrangements. PLAG1 rearrangement may serve as a useful biomarker in distinguishing a significant subset of myxoid leiomyosarcomas from other myxoid uterine mesenchymal tumors.
Uterine myxoid leiomyosarcomas is one of a variety of soft tissue and salivary gland tumors that often exhibit myxoid matrix and harbor rearrangement of the Pleomorphic Adenoma Gene 1 (PLAG1), a developmentally regulated zinc finger proto-oncogene located on chromosome 8q12. PLAG1 rearrangements have been previously reported in pleomorphic adenoma (19–22), carcinoma ex-pleomorphic adenoma (23–26), skin and soft tissue myoepithelioma (27, 28), and lipoblastoma (29–37). In pleomorphic adenomas, PLAG1 fusion occurs most frequently with CTNNB1, (20) followed by LIFR, TCEA1, and CHCHD7 (21, 38). HAS2, RAB2A, COL3A1, COL1A2, and RAD51L1 fusion partners have all been reported in lipoblastoma (29–32, 35). While LIFR-PLAG1 fusion has been detected in one soft tissue myoepithelioma (27), the fusion partner has not been identified in the vast majority of skin and soft tissue myoepitheliomas (27, 28). To date, TRPS1 and RAD51B fusion partners identified among uterine myxoid leiomyosarcomas have not been reported in other tumor types harboring PLAG1 rearrangement. The TPRS1 (Transcriptional Repressor GATA Binding 1) gene, located on chromosome 8q23.3, encodes a protein that represses GATA-mediated transcription, plays a central role in the control of cell cycle and cancer development, may be involved in the regulation of bone and cartilage growth (39–41). The RAD51B (RAD51 Paralog B) gene, located on chromosome 15q15.1, encodes a protein essential for DNA repair by homologous recombination (42, 43).
PLAG1 activation via translocation occurs as a result of promoter swapping with various fusion partner genes (20, 21, 29). Translocation breakpoints involve the 5’ untranslated regions of both PLAG1 and the fusion partner gene, resulting in the entire PLAG1 coding sequence being placed under transcriptional control of an active and ectopic promoter region. Strong and diffuse PLAG1 immunoexpression in all PLAG1-rearranged myxoid leiomyosarcomas tested provides evidence that PLAG1 rearrangement affects PLAG1 transcriptional up-regulation in these tumors. Diffuse PLAG1 expression in rare myxoid leiomyosarcomas lacking PLAG1 rearrangement may occur as a result of alternate mechanisms such as PLAG1 point mutations or HMGA1 rearrangement that may contribute to PLAG1 transcriptional up-regulation. Since there are no apparent differences in the clinicopathologic features of PLAG1 fusion-positive and fusion-negative myxoid leiomyosarcomas, PLAG1 immunohistochemistry in myxoid uterine sarcomas may be helpful in triaging tumors for fusion confirmation. Diffuse PLAG1 expression can also aid in distinguishing myxoid leiomyosarcomas from potential morphologic mimics such as ZC3H7B-BCOR high-grade endometrial stromal sarcoma.
Four tumors from our initial cohort were excluded from the study based on the identification of BCOR and ALK rearrangements by FISH and/or targeted RNA sequencing, emphasizing the extensive morphologic overlap among BCOR-rearranged high-grade endometrial stromal sarcoma, myxoid inflammatory myofibroblastic tumor, and myxoid leiomyosarcoma. In the most recent study describing the clinicopathologic features of ZC3H7B-BCOR high-grade endometrial stromal sarcomas, 4 of 17 (24%) of tumors were previously misdiagnosed as myxoid leiomyosarcomas (44). In the largest study of uterine myxoid leiomyosarcomas to date, 3 of 30 (10%) tumors may in fact represent ZC3H7B-BCOR high-grade endometrial stromal sarcomas based on the reported immunophenotype (4). Both myxoid leiomyosarcoma and ZC3H7B-BCOR high-grade endometrial stromal sarcoma are composed of spindle cells with at least focal myxoid matrix and collagen deposition and associated with large, thick-walled vessels (44). However, CD10 along with diffuse cyclin D1 and/or BCOR expression with absence of or only limited immunohistochemical evidence of myogenic differentiation (SMA and/or h-caldesmon positive, desmin negative) in any myxoid uterine sarcoma should raise suspicion for ZC3H7B-BCOR high-grade endometrial stromal sarcoma and prompt confirmation of the fusion. (11, 44). While h-caldesmon has been reported to be a sensitive marker of smooth muscle differentiation in the uterus and a useful tool in distinguishing smooth muscle tumors and endometrial stromal tumors, its utility mainly pertains to situations in which highly cellular leiomyoma and low-grade endometrial stromal sarcoma or endometrial stromal nodule is the differential diagnosis. We follow a similar practice as our soft tissue colleagues who utilize desmin with much more frequency than h-caldesmon and recommend that high-grade endometrial stromal sarcoma be considered in the differential diagnosis of any myxoid sarcoma lacking desmin positivity.
While inflammatory myofibroblastic tumors of the uterus are rare, they often exhibit spindled cells embedded in myxoid matrix that can be focal or extensive, mimicking myxoid leiomyosarcoma (5, 7). An inflammatory infiltrate is usually present, and ALK or ROS rearrangements are frequently found (5, 7). ALK immunohistochemistry is useful in distinguishing myxoid inflammatory myofibroblastic tumors from other uterine mesenchymal neoplasms with myxoid features. In one study, 4 of 30 (13%) of myxoid leiomyosarcomas expressed ALK, and 2 of them harbored an ALK rearrangement by FISH, leading to reclassification as inflammatory myofibroblastic tumor (4).
Frequent identification of genetic abnormalities driving oncogenesis, particularly gene fusions, among uterine mesenchymal tumors suggests that molecular methods are increasingly necessary in the routine diagnostic evaluation of these lesions. Gene rearrangements are detectable by FISH or sequencing of fusion transcripts. Because DNA-based sequencing requires primers flanking both target and fusion partners, this method allows detection of only known fusions. Targeted RNA sequencing methods based on the Anchored Multiplex PCR technology such as the one used in our study, however, enables detection of both known and novel fusions, providing an advantage in analytical sensitivity.
This study has important limitations. We were unable to identify gene fusions in the majority of myxoid leiomyosarcomas. High-quality RNA was difficult to obtain from samples older than three years due to RNA degradation over time. Many samples were individual contributions from different institutions; thus, variations in tissue processing may have also affected RNA quality. Many cases also lacked sufficient material for a more comprehensive immunohistochemical panel due to individual contributions as personal consultations from community practices. Limited clinical history also hinders our ability to draw conclusions regarding the behavior of PLAG1 fusion-positive sarcomas compared to fusion-negative tumors.
Despite these limitations, we identified PLAG1 gene rearrangements in a significant subset of uterine myxoid leiomyosarcomas. Detection of PLAG1 overexpression by immunohistochemistry can identify tumors likely harboring PLAG1 genetic abnormalities. Morphologic assessment of myxoid uterine mesenchymal tumors alone is insufficient in distinguishing myxoid leiomyosarcoma, ZC3H7B-BCOR endometrial stromal sarcoma, and myxoid inflammatory myofibroblastic tumor. Targeted RNA sequencing and FISH are helpful in identifying gene fusions that may aid in the classification of these tumors.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicts of interest to declare.
REFERENCES
- 1.King ME, Dickersin GR, Scully RE. Myxoid leiomyosarcoma of the uterus. A report of six cases. Am J Surg Pathol. 1982;6:589–98. [DOI] [PubMed] [Google Scholar]
- 2.Abeler VM, Royne O, Thoresen S, et al. Uterine sarcomas in Norway. A histopathological and prognostic survey of a total population from 1970 to 2000 including 419 patients. Histopathology. 2009;54:355–64. [DOI] [PubMed] [Google Scholar]
- 3.Burch DM, Tavassoli FA. Myxoid leiomyosarcoma of the uterus. Histopathology. 2011;59:1144–55. [DOI] [PubMed] [Google Scholar]
- 4.Parra-Herran C, Schoolmeester JK, Yuan L, et al. Myxoid leiomyosarcoma of the uterus: a clinicopathologic analysis of 30 cases and review of the literature with reappraisal of its distinction from other uterine myxoid mesenchymal neoplasms. Am J Surg Pathol. 2016;40:285–301. [DOI] [PubMed] [Google Scholar]
- 5.Bennett JA, Nardi V, Rouzbahman M, et al. Inflammatory myofibroblastic tumor of the uterus: a clinicopathological, immunohistochemical, and molecular analysis of 13 cases highlighting their broad morphologic spectrum. Mod Pathol. 2017;30:1489–503. [DOI] [PubMed] [Google Scholar]
- 6.Rabban JT, Zaloudek CJ, Shekitka KM, et al. Inflammatory myofibroblastic tumor of the uterus: a clinicopathologic study of 6 cases emphasizing distinction from aggressive mesenchymal tumors. Am J Surg Pathol. 2005;29:1348–55. [DOI] [PubMed] [Google Scholar]
- 7.Parra-Herran C, Quick CM, Howitt BE, et al. Inflammatory myofibroblastic tumor of the uterus: clinical and pathologic review of 10 cases including a subset with aggressive clinical course. Am J Surg Pathol. 2015;39:157–68. [DOI] [PubMed] [Google Scholar]
- 8.Pickett JL, Chou A, Andrici JA, et al. Inflammatory myofibroblastic tumors of the female genital tract are under-recognized: a low threshold for alk immunohistochemistry is required. Am J Surg Pathol. 2017;41:1433–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haimes JD, Stewart CJR, Kudlow BA, et al. Uterine inflammatory myofibroblastic tumors frequently harbor ALK fusions with IGFBP5 and THBS1. Am J Surg Pathol. 2017;41:773–80. [DOI] [PubMed] [Google Scholar]
- 10.Chiang S, Lee CH, Stewart CJR, et al. BCOR is a robust diagnostic immunohistochemical marker of genetically diverse high-grade endometrial stromal sarcoma, including tumors exhibiting variant morphology. Mod Pathol. 2017;30:1251–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoang LN, Aneja A, Conlon N, et al. Novel high-grade endometrial stromal sarcoma: a morphologic mimicker of myxoid leiomyosarcoma. Am J Surg Pathol. 2017;41:12–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiang S, Ali R, Melnyk N, et al. Frequency of known gene rearrangements in endometrial stromal tumors. Am J Surg Pathol. 2011;35:1364–72. [DOI] [PubMed] [Google Scholar]
- 13.Hoang L, Chiang S, Lee CH. Endometrial stromal sarcomas and related neoplasms: new developments and diagnostic considerations. Pathology. 2018;50:162–77. [DOI] [PubMed] [Google Scholar]
- 14.Lee CH, Nucci MR. Endometrial stromal sarcoma--the new genetic paradigm. Histopathology. 2015;67:1–19. [DOI] [PubMed] [Google Scholar]
- 15.Mehine M, Kaasinen E, Heinonen HR, et al. Integrated data analysis reveals uterine leiomyoma subtypes with distinct driver pathways and biomarkers. Proc Natl Acad Sci U S A. 2016;113:1315–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mehine M, Kaasinen E, Makinen N, et al. Characterization of uterine leiomyomas by whole-genome sequencing. N Engl J Med. 2013;369:43–53. [DOI] [PubMed] [Google Scholar]
- 17.Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20:1479–84. [DOI] [PubMed] [Google Scholar]
- 18.Frosina D, Jungbluth AA. A Novel technique for the generation of multitissue blocks using a carrier. Appl Immunohistochem Mol Morphol. 2016;24:668–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voz ML, Agten NS, Van de Ven WJ, et al. PLAG1, the main translocation target in pleomorphic adenoma of the salivary glands, is a positive regulator of IGF-II. Cancer Res. 2000;60:106–13. [PubMed] [Google Scholar]
- 20.Kas K, Voz ML, Roijer E, et al. Promoter swapping between the genes for a novel zinc finger protein and beta-catenin in pleiomorphic adenomas with t(3;8)(p21;q12) translocations. Nat Genet. 1997;15:170–4. [DOI] [PubMed] [Google Scholar]
- 21.Voz ML, Astrom AK, Kas K, et al. The recurrent translocation t(5;8)(p13;q12) in pleomorphic adenomas results in upregulation of PLAG1 gene expression under control of the LIFR promoter. Oncogene. 1998;16:1409–16. [DOI] [PubMed] [Google Scholar]
- 22.Matsuyama A, Hisaoka M, Nagao Y, et al. Aberrant PLAG1 expression in pleomorphic adenomas of the salivary gland: a molecular genetic and immunohistochemical study. Virchows Arch. 2011;458:583–92. [DOI] [PubMed] [Google Scholar]
- 23.Katabi N, Ghossein R, Ho A, et al. Consistent PLAG1 and HMGA2 abnormalities distinguish carcinoma ex-pleomorphic adenoma from its de novo counterparts. Hum Pathol. 2015;46:26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bahrami A, Perez-Ordonez B, Dalton JD, et al. An analysis of PLAG1 and HMGA2 rearrangements in salivary duct carcinoma and examination of the role of precursor lesions. Histopathology. 2013;63:250–62. [DOI] [PubMed] [Google Scholar]
- 25.Chiosea SI, Thompson LD, Weinreb I, et al. Subsets of salivary duct carcinoma defined by morphologic evidence of pleomorphic adenoma, PLAG1 or HMGA2 rearrangements, and common genetic alterations. Cancer. 2016;122:3136–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bahrami A, Dalton JD, Shivakumar B, et al. PLAG1 alteration in carcinoma ex pleomorphic adenoma: immunohistochemical and fluorescence in situ hybridization studies of 22 cases. Head Neck Pathol. 2012;6:328–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antonescu CR, Zhang L, Shao SY, et al. Frequent PLAG1 gene rearrangements in skin and soft tissue myoepithelioma with ductal differentiation. Genes Chromosomes Cancer. 2013;52:675–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bahrami A, Dalton JD, Krane JF, et al. A subset of cutaneous and soft tissue mixed tumors are genetically linked to their salivary gland counterpart. Genes Chromosomes Cancer. 2012;51:140–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hibbard MK, Kozakewich HP, Dal Cin P, et al. PLAG1 fusion oncogenes in lipoblastoma. Cancer Res. 2000;60:4869–72. [PubMed] [Google Scholar]
- 30.Warren M, Turpin BK, Mark M, et al. Undifferentiated myxoid lipoblastoma with PLAG1-HAS2 fusion in an infant; morphologically mimicking primitive myxoid mesenchymal tumor of infancy (PMMTI)--diagnostic importance of cytogenetic and molecular testing and literature review. Cancer Genet. 2016;209:21–9. [DOI] [PubMed] [Google Scholar]
- 31.Yoshida H, Miyachi M, Ouchi K, et al. Identification of COL3A1 and RAB2A as novel translocation partner genes of PLAG1 in lipoblastoma. Genes Chromosomes Cancer. 2014;53:606–11. [DOI] [PubMed] [Google Scholar]
- 32.Deen M, Ebrahim S, Schloff D, et al. A novel PLAG1-RAD51L1 gene fusion resulting from a t(8;14)(q12;q24) in a case of lipoblastoma. Cancer Genet. 2013;206:233–7. [DOI] [PubMed] [Google Scholar]
- 33.Bartuma H, Domanski HA, Von Steyern FV, et al. Cytogenetic and molecular cytogenetic findings in lipoblastoma. Cancer Genet Cytogenet. 2008;183:60–3. [DOI] [PubMed] [Google Scholar]
- 34.Brandal P, Bjerkehagen B, Heim S. Rearrangement of chromosomal region 8q11–13 in lipomatous tumours: correlation with lipoblastoma morphology. J Pathol. 2006;208:388–94. [DOI] [PubMed] [Google Scholar]
- 35.Morerio C, Rapella A, Rosanda C, et al. PLAG1-HAS2 fusion in lipoblastoma with masked 8q intrachromosomal rearrangement. Cancer Genet Cytogenet. 2005;156:183–4. [DOI] [PubMed] [Google Scholar]
- 36.Sciot R, De Wever I, Debiec-Rychter M. Lipoblastoma in a 23-year-old male: distinction from atypical lipomatous tumor using cytogenetic and fluorescence in-situ hybridization analysis. Virchows Arch. 2003;442:468–71. [DOI] [PubMed] [Google Scholar]
- 37.Gisselsson D, Hibbard MK, Dal Cin P, et al. PLAG1 alterations in lipoblastoma: involvement in varied mesenchymal cell types and evidence for alternative oncogenic mechanisms. Am J Pathol. 2001;159:955–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asp J, Persson F, Kost-Alimova M, et al. CHCHD7-PLAG1 and TCEA1-PLAG1 gene fusions resulting from cryptic, intrachromosomal 8q rearrangements in pleomorphic salivary gland adenomas. Genes Chromosomes Cancer. 2006;45:820–8. [DOI] [PubMed] [Google Scholar]
- 39.Wu L, Wang Y, Liu Y, et al. A central role for TRPS1 in the control of cell cycle and cancer development. Oncotarget. 2014;5:7677–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaiser FJ, Moroy T, Chang GT, et al. The RING finger protein RNF4, a co-regulator of transcription, interacts with the TRPS1 transcription factor. J Biol Chem. 2003;278:38780–5. [DOI] [PubMed] [Google Scholar]
- 41.Ludecke HJ, Schaper J, Meinecke P, et al. Genotypic and phenotypic spectrum in tricho-rhino-phalangeal syndrome types I and III. Am J Hum Genet. 2001;68:81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodrigue A, Coulombe Y, Jacquet K, et al. The RAD51 paralogs ensure cellular protection against mitotic defects and aneuploidy. J Cell Sci. 2013;126:348–59. [DOI] [PubMed] [Google Scholar]
- 43.Sigurdsson S, Van Komen S, Bussen W, et al. Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev. 2001;15:3308–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis N, Soslow RA, Delair DF, et al. ZC3H7B-BCOR high-grade endometrial stromal sarcomas: a report of 17 cases of a newly defined entity. Mod Pathol. 2017. [DOI] [PubMed] [Google Scholar]


