Summary
Gene expression levels in clonal bacterial groups have been found to be spatially correlated. These correlations can partly be explained by the shared lineage history of nearby cells, however they could also arise from local cell-cell interactions. Here, we present a quantitative framework that allows us to disentangle the contributions of lineage-history, long-range spatial gradients, and local cell-cell interactions to spatial correlations in gene expression. We study pathways involved in toxin production, SOS stress response, and metabolism in Escherichia coli microcolonies and find for all pathways that shared lineage history is the main cause of spatial correlations in gene expression levels. However, long-range spatial gradients and local cell-cell interactions also contributed to spatial correlations in SOS-response, amino-acid biosynthesis, and overall metabolic activity. Together our data shows that the phenotype of a cell is influenced by its lineage history and population context, raising the question whether bacteria can arrange their activities in space to perform functions they cannot achieve alone.
Introduction
Many bacteria do not live in isolation, but instead are members of larger communities (Claessen et al, 2014). The microenvironment in these communities is partly determined by abiotic conditions, but it is also affected by the activities of a cell’s neighbors (Flemming et al, 2016; Stewart & Franklin, 2008). At the same time, cells adjust their phenotype by regulating gene expression based on the inferred state of the local microenvironment. The phenotype of a cell is thus likely influenced by its location in the community and by the identity and the activities of neighboring cells.
The functionality of the community as a whole depends on the combined activities of all of its members. Being part of a group can allow cells to specialize in performing different tasks (van Gestel et al, 2015a). Such interactions between different cell types can lead to new or improved functionality that goes beyond the sum of the activities of the individual cells (Claessen et al, 2014; van Vliet & Ackermann, 2015). In multispecies biofilms, most of this specialization is the result of genetic differences between member species. However, specialization can likewise occur in single species communities as the result of phenotypic variation among cells (van Gestel et al, 2015a; Ackermann, 2015; Mohr et al, 2013; Ackermann et al, 2008). One well studied example is in clonal Bacillus subtilis biofilms, where functionality depends on interactions between multiple different cell types (Lopez & Kolter, 2010; van Gestel et al, 2015a).
Interactions between different cell types can be sensitive to the spatial arrangements of the different types (Liu et al, 2016; Nadell et al, 2016). For example, the division of labor between nitrogen fixing and photosynthetic cells in multicellular cyanobacteria is likely more efficient due to the regular spacing of nitrogen fixing cells along the filaments (Muro-Pastor & Hess, 2012). Furthermore, recent work in B. subtilis colonies directly linked functionality at the group level to the spatial arrangement of two cell types that perform complementary functions (van Gestel et al, 2015b). More generally, we expect that correlations in the phenotypes of neighboring cells can be beneficial for a large number of activities (Ross-Gillespie & Kümmerli, 2014). Positive correlations in phenotypes (i.e. neighbors having similar phenotypes) can allow cells to coordinate their activities. This can for example be of benefit in the production of secreted effectors, by allowing a sufficient build-up in local effector concentrations. Negative correlations in phenotypes (i.e. neighbors having different phenotypes) can facilitate division of labor between cells. This can be of benefit in the context of anabolic pathways: cells can benefit from economies of scale by specializing on the biosynthesis of a subset of metabolites, while exchanging end products with neighbors specializing on complementary pathways (Johnson et al, 2012; Guantes et al, 2015).
Spatial correlations in phenotypes can be the result of several processes. In previous work it was found that phenotypes can be epigenetically inherited (Robert et al, 2010; Veening et al, 2008b; 2008a; Hormoz et al, 2015; Julou et al, 2013). Such epigenetic inheritance leads to positive correlations between the phenotypes of closely related cells (e.g. between sisters (Hormoz et al, 2015)). As neighboring cells tend to be closely related, epigenetic inheritance can lead to positive spatial correlations in phenotypes. Furthermore, neighboring cells are exposed to a similar combination of environmental gradients (Flemming et al, 2016; Stewart & Franklin, 2008). A shared gene regulatory response to these gradients can thus result in positive correlations in phenotypes. Finally, local cell-cell interactions can lead to a coupling in expression levels between neighbors (Risser et al, 2012; Bassler & Losick, 2006). These intercellular feedbacks can give rise to either positive or negative correlations in phenotypes. Spatial correlations in phenotypes will depend on the combined effects of epigenetic inheritance, spatial gradients, and intercellular feedbacks. However, we lack a quantitative understanding of the relative contributions of these processes to spatial correlations in phenotype.
Here we address the following question: to what extent are cellular activities correlated between neighboring cells? We are especially interested in disentangling the effects of epigenetic inheritance, spatial gradients, and intercellular feedbacks and quantifying their relative importance. As a model system, we used two-dimensional microcolonies of Escherichia coli. We followed the growth of a microcolony using time-lapse microscopy while tracking spatiotemporal gene expression patterns using transcriptional reporters. Subsequently, we quantified spatial correlations in gene expression and developed a novel quantitative approach to disentangle the effects of shared linage history, spatial gradients, and local interactions. We found strong spatial correlations for all studied pathways, which we could attribute to the effects of shared lineage history and, for some pathways, the effects of spatial gradients and local interactions.
Results
We used time-lapse microscopy to follow the growth of microcolonies of E. coli founded by a single cell. Gene expression was quantified using plasmid-based green fluorescent protein (GFP) transcriptional reporters. The mean fluorescent intensity of the transcriptional reporter is approximately proportional to the concentration of GFP and hence a proxy for the concentration of the protein encoded by the gene of interest; we refer to this quantity as protein level. We furthermore quantified the rate of change in the total fluorescent intensity over time, which is a proxy for the promoter activity (Kiviet et al, 2014, see Methods). We verified that our main findings can be reproduced using chromosomally integrated transcriptional reporters and are robust to changes in the bacterium used as model system (See Figure S10).
Neighboring cells have similar protein levels of Colicin Ib
We first investigated the spatial expression patterns of the bacteriocin colicin Ib (cib), which is a pore-forming toxin found in natural isolates of E. coli and Salmonella (Riley & Wertz, 2002; Cascales et al, 2007). In natural systems the extra cellular concentration of bacteriocin needs to reach a threshold concentration to inhibit the growth of nearby sensitive cells (Cascales et al, 2007). If neighboring cells coordinated their expression dynamics, it would be easier for them to reach this threshold concentration. We thus hypothesized that Colicin Ib protein levels should be positively correlated between neighboring cells.
cib transcription is co-repressed by the binding of LexA to the SOS-box (SOS DNA repair response) and by the binding of Fe2+-Fur complex to the iron box (Ferric uptake regulation) (Cascales et al, 2007; Nedialkova et al, 2014). cib transcriptional repression can be partly relieved by activation of the SOS-response in response to DNA damage, or by a shortage in ferrous iron (Fe2+) (Spriewald et al, 2015; Nedialkova et al, 2014). Here, we relieved repression by Fur by chelating free iron in the medium. We did not induce SOS response, so any cib expression is likely the result of SOS induction due to spontaneous occurring DNA damage (Pennington & Rosenberg, 2007).
Colicin Ib protein levels varied strongly between cells in a microcolony (median coefficient of variation=0.19, n=8, Figure 1A). This is consistent with previous reports of high variation in colicin expression levels (Silander et al, 2012). We do not expect any genetic variation between cells in the microcolony: when a microcolony is founded by a single cell there is a 88% chance that no mutations occur during 7 generations, giving rise to more than 100 cells (assuming a mutation rate of 10-3, per genome, per generation (Lee et al, 2012)). Likewise, we do not expect abiotic variation in the agar pads: in the absence of uptake or release of compounds by cells, diffusion should equalize any inhomogeneities across the colony within seconds (e.g. a molecule with a diffusion coefficient similar to that of glucose (D~600 μm2/s) diffuses across a microcolony (~13μm) in approximately 0.07 seconds). However, when cells excrete and take up nutrients and metabolites from the environment, they can create gradients on length scales comparable to the cell size, as long as the uptake rate (k) is comparable to the diffusion constant (length of gradient (Hiscock & Megason, 2015)). Phenotypic variation in the colony can thus be the result of cells adjusting their gene expression dynamics based on these local gradients. Furthermore, gene expression noise is likely an important factor leading to the observed phenotypic variation (Elowitz et al, 2002; Ozbudak et al, 2002; Kaern et al, 2005).
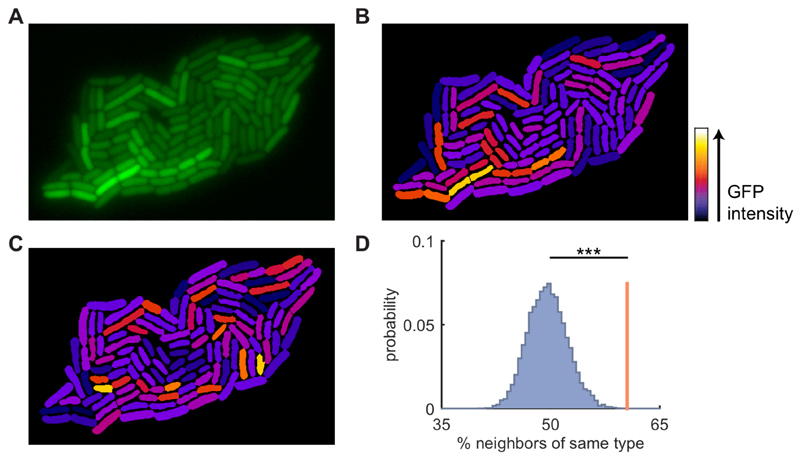
Figure 1. Neighboring cells have similar expression levels of colicin Ib.
A) Fluorescence image of an E. coli microcolony with GFP transcriptional reporter for colicin Ib (cib). B) Reconstructed image of the colony shown in A: cell shapes obtained from cell segmentation are uniformly colored based on their mean corrected intensity (see Figure S1). Note how neighboring cells tend to have similar intensities. C) Same as in B, but fluorescence intensities are randomly permuted among the cells. Note that the similarity between neighboring cells has been reduced compared to B. D) Cells are grouped into two clusters based on their intensity. The red line shows the average fraction of a cell’s neighbors that is of the same type. The blue distribution shows the same quantity obtained after randomly permuting the intensities among the cells (104 permutations). The observed similarity is significantly higher for the true data compared to the randomized data (p<1·10-4, randomization test). See also Figure S1, S2, & S3.
Visual inspection suggested that Colicin Ib protein levels were non-randomly distributed in the colony: neighboring cells appeared to have similar levels of Colicin Ib (Figure 1A). To quantitatively investigate the expression patterns, we first corrected the fluorescent images for optical artifacts (Figure 1B and S1, see Methods). Subsequently, we quantified the spatial correlation in Colicin Ib protein levels using a randomization test. We found that neighboring cells are significantly more similar to each other than can be expected by chance (Figure 1D, p=10-4, see Methods). We observed similar patterns for an additional 8 replicate microcolonies (Figure S2), confirming that Colicin Ib protein levels are spatially correlated.
Two main factors could contribute to the observed positive spatial correlation in Colicin Ib protein levels: shared lineage history and spatial proximity. However, these two factors are strongly correlated with each other. To disentangle their contributions we reconstructed the full, spatially resolved, lineage trees of the microcolonies (Figure 2A) and developed a statistical method to analyze these lineage trees.
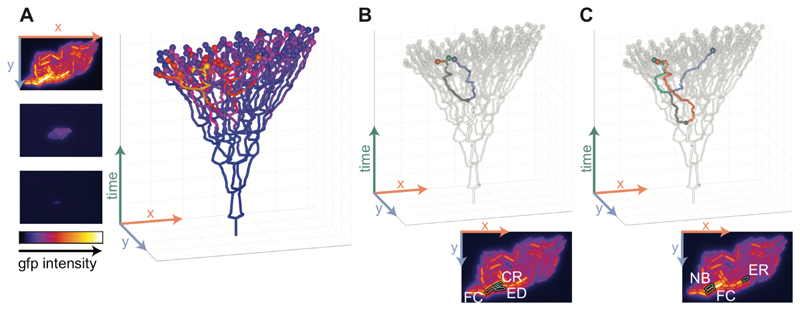
Figure 2. Reconstructing lineage trees to disentangle the effects of space and relatedness.
A) Left: frames from a time-lapse movie of a growing microcolony with a GFP reporter for cib. The images show GFP intensities using a heatmap representation for t=0,3,6h. Right: reconstructed lineage tree. Cells are plotted as a function of location (horizontal plane) and time (vertical axis). Branching points in the lineage tree mark cell division events. The spheres at the tip of the tree represent cells at the final time point with their color indicating the Colicin Ib level of the cell. B) Statistical test to quantify the effect of shared lineage history on similarity in expression levels. A focal cell (FC, red) is compared with its closest relative (CR, green) and with an equidistant cell (ED, blue), which is a cell that has the same distance to the focal cell as the closest relative, but that is less related. C) Statistical test to quantify the effect of spatial proximity on similarity in expression levels. A focal cell (FC, red) is compared with one of its neighbors (NB, green) and with an equally-related cell (ER, blue), which is a cell that has the same relatedness to the focal cell as the neighbor, but that is further away in space. B,C) The insets at the bottom show the positions of these cells in the GFP image for the last time point (see panel A).
Shared lineage history leads to spatial correlations in colicin Ib expression dynamics
First, we developed a test for the effect of shared lineage history on spatial correlations in phenotype. We disentangle the effect of relatedness from the effect of spatial proximity by analyzing a group of cells that differ in their relatedness, but that are identical in their spatial arrangement. Specifically, we select a focal cell, locate its closest relative (e.g. its sister), and then find a third cell (the equidistant cell) that has the same distance to the focal cell as the closest relative, but that is less related (Figure 2B). We then calculated the phenotypic difference (i.e., the difference in the Colicin Ib protein levels) between the focal cell and its equidistant cell (δED) and between the focal cell and its closest relative (δCR). To compare the magnitude of these phenotypic differences, we calculated the ratio δED/δCR that quantifies the effect of shared lineage history: values larger than 1 indicate that shared lineage history leads to similarity in phenotype (i.e. positive correlations), while values smaller than 1 indicate that shared lineage history leads to dissimilarity in phenotype (i.e. negative correlations).
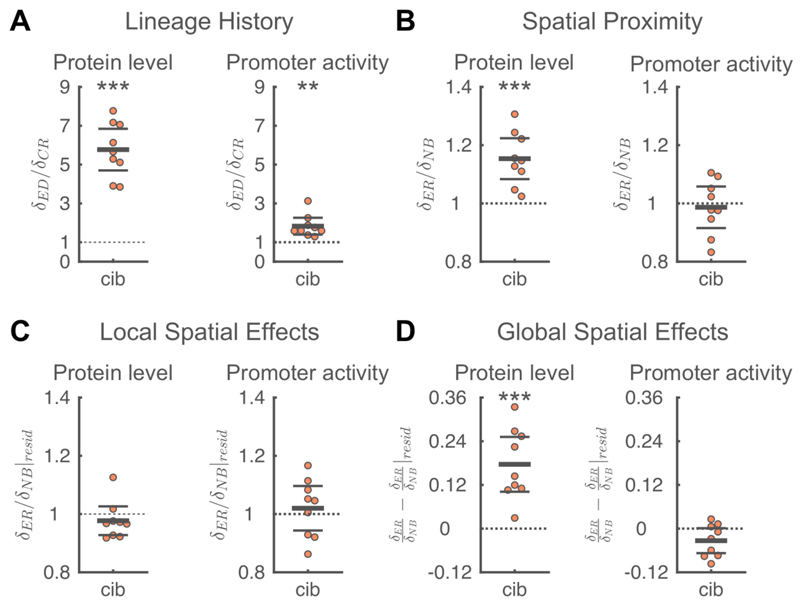
We found that shared lineage history leads to positive correlations in Colicin Ib protein levels: a cell is much more similar to its closest relative than to an equally distant (but less related) cell. Specifically, the phenotypic difference between a focal cell and the equidistant cell is on average 5.8 times higher than the phenotypic difference between the focal cell and its closest relative (Figure 3A, 〈δED/δCR〉=5.8, p<1·10-5, t-test, n=9). Additionally, we found that closely related cells are also similar with respect to their cib promoter activity (Figure 3A, 〈δED/δCR〉=1.8, p=0.002). While the similarity in protein level is to be expected due to protein inheritance, the similarity in promoter activity shows that closely related cells are also similar in their current activities.
Figure 3. Factors contributing to spatial correlations in colicin Ib expression dynamics.
A) Shared lineage history leads to similarity in Colicin Ib protein levels (left) and promoter activity (right). The phenotypic difference between a focal cell and an equidistant cell (δED) is significantly larger than the phenotypic difference between a focal cell and its closest relative (δCR), i.e. 〈δED/δCR〉 > 1. B) Spatial proximity leads to similarity in Colicin Ib levels but not in promoter activity. For Colicin Ib levels, the phenotypic difference between a focal cell and an equally-related cell (δED) is significantly larger than the phenotypic difference between the focal cell and one of its neighbors (δNB), i.e. 〈δER/δNB〉 > 1. C) Local spatial effects do not contribute to spatial correlations in Colicin Ib levels or promoter activity. Local spatial effects were calculated using the residuals of a linear regression of a cell’s phenotype to the distance of a cell to the colony edge. The difference in residuals between a focal cell and an equally-related cell (δER|resid) is not significantly different from the difference in residuals between the focal cell and one of its neighbors δNB|resid),i.e. δER/δNB|reside ≈ 1. D) Global spatial effects contribute to spatial correlations in Colicin Ib levels. Global spatial effects were calculated as the difference between the total effects of spatial proximity (panel B) and the local spatial effects (panel C). A-D) Each point corresponds to a microcolony with 117-138 (mean=128) cells; points are horizontally offset. Thick horizontal lines indicate mean, thin lines 95% confidence intervals. Dashed lines indicate the expected value under the null-hypothesis. Null hypothesis rejected with: *p<0.05, **p<0.01, ***p<0.001, t-test, n=9. The statistics are robust to the choice of the equally-related cell (Figure S4A), the size of the colony being analyzed (Figure S4B), and differences in the processing of fluorescent images (Figure S4C). Full distributions are shown in Figure S5. See also Figure S4-8
Spatial proximity leads to spatial correlations in Colicin Ib protein level
Can the observed spatial correlation in Colicin Ib protein levels be fully explained by the shared lineage history of neighbors, or are there additional factors that couple gene expression in neighboring cells? To answer this, we developed a test to quantify whether spatial proximity contributes to spatial correlations in phenotypes, after correcting for the effects of shared lineage history. To do so, we compare a group of cells that are identical in their relatedness, but that differ in how far they are apart in space. We then asked whether a cell is more similar to its neighbors in terms of colicin Ib expression than to cells that have the same relatedness, but that are further away in space.
Specifically, we select a focal cell, pick one of its neighbors, and then find a third cell (the equally-related cell) that has the same relatedness to the focal cell as the neighbor but that is further away in space (Figure 2C). For example, if the chosen neighbor is a first cousin of the focal cell, then we selected another cell that is also a first cousin of the focal cell but further away in space; this is the equally related cell. We then calculated the phenotypic difference between the focal cell and its equally-related cell (δER) and between the focal cell and its neighbor (δNB). To compare the magnitude of these phenotypic differences, we calculated the ratio δER/δNB that quantifies the effect of spatial proximity: values larger than 1 indicate that spatial proximity leads to similarity in phenotype (i.e. positive correlations), while values smaller than 1 indicate that spatial proximity leads to dissimilarity in phenotype (i.e. negative correlations). We verified that our test completely corrects for the effects of lineage history by applying it to a data set generated by a computer simulation of gene expression dynamics (Figure S6, see Methods for details).
We found that spatial proximity leads to significant similarity in Colicin Ib protein levels: the phenotypic difference between a focal cell and an equally-related cell is on average 15% larger than the phenotypic difference between the focal cell and its neighbor (Figure 3B, 〈δER/δNB〉=1.15, p=1·10-3). However, spatial proximity does not significantly affect promoter activity (Figure 3B, 〈δER/δNB〉=0.99, p=0.7).
Why did we observe that spatial proximity leads to similarity in protein levels, but not in promoter activities? One possible reason is that protein levels have a longer autocorrelation time than promoter activities. The amount of proteins inside a cell is the sum of protein production and protein inheritance and therefore depends on the transcriptional activity of both the cell itself and its ancestors. Contrarily, we determined the promoter activity as the amount of proteins produced during roughly one cell cycle. The overall (i.e. time-averaged) activities of cells could be spatially correlated even if they are not synchronized in time, for example due to transcriptional bursts or time-delays between the responses in neighboring cells. This would lead to an observed similarity over longer timescales (i.e. for protein levels) even when there is no similarity on shorter time scales (i.e. for promoter activity). Furthermore, promoter activities, which are calculated using temporal derivatives, are likely more affected by measurement noise than protein levels, which are directly measured. Weak correlations in promoter activity due to spatial proximity could thus be harder to detect.
In summary, our data shows that both shared lineage history and spatial proximity contribute to positive spatial correlations in Colicin Ib protein levels. Shared lineage history also contributes to positive correlations in cib promoter activity, but there is no evidence that spatial proximity also affects promoter activities.
Global spatial effects lead to correlations in Colicin Ib protein levels
Spatial proximity could lead to correlations in expression levels in two ways: by global and by local spatial effects. Global spatial effects refer to situations where expression dynamics vary systemically with the overall position of a cell in the microcolony. Specifically, we investigated whether expression dynamics correlated with the distance of a cell to the edge of the colony. Additionally, local (i.e. microscale) effects could lead to correlations in expression dynamics. Such local effects are most likely the result of interactions between neighboring cells. These interactions can be either the result of direct sharing of cellular components or be a consequence of intercellular feedbacks mediated though the local microenvironment. Both global and local effects could thus affect the phenotype of a cell through spatial variation in the environment. Although there is no fundamental difference between these two situations, we reserve the term global spatial effects for cases where the microenvironment varies on spatial scales that are (much) larger than the size of a cell and use local effects for cases where variation occurs on scales comparable to the size of a cell.
First, we analyzed whether global spatial effects contribute to spatial correlations in Colicin Ib protein levels. Visual inspection suggested that expression levels increased towards the center of the colonies. A linear regression of Colicin Ib protein levels with a cell’s distance to the colony edge confirmed this observation (mean r2=0.096, Figure S7). As neighboring cells are similar in their distance to the colony edge, the observed correlation between protein levels and the distance to the colony edge could thus lead to similar expression levels in neighboring cells. Cells at the colony edge could differ in phenotype from those in the center due to differences in the environment they experience (e.g. local density, nutrient availability, etc.) or due to differences in cell biology (e.g. cells at the colony edge tend to have older cell poles, Figure S7D,E).
To what extend does the observed global trend in Colicin Ib protein levels explain the effect of spatial proximity? To find out, we recalculated the effect of spatial proximity after correcting for the global spatial effects. We do this correction by subtracting the expected phenotype determined from the linear regression from the observed phenotype (Figure 3C). We then recalculated the effect of spatial proximity using the obtained residuals. The magnitude of global spatial effects is subsequently estimated as the difference between the effect of spatial proximity determined from the observed phenotype of a cell (δER/δNB, Figure 3B) with the effect of spatial proximity determined from the residuals of the linear regression (δER/δNB|resid, Figure 3C).
We found that the spatial correlations in Colicin Ib protein levels are strongly influenced by global spatial effects (Figure 3D, 〈δER/δNB − δER/δNB |resid〉=0.18, p=6·10-4). In fact, we no longer observe a significant effect of spatial proximity when we only analyze the local spatial effects (Figure 3C, 〈δER/δNB|resid〉=0.98,p=0.3). The spatial correlations in Colicin Ib protein levels are thus mainly the result of shared lineage history and shared overall position in the colony. Does this mean that intercellular feedbacks do not play any role in Colicin Ib expression patterns?
Direct cell-cell interactions in SOS response
We designed an experimental system where we could directly test if intercellular feedbacks affect Colicin Ib expression dynamics. The system consists of two strains: an inducible strain in which we can induce the expression of a target gene and a reporter strain that has a reporter for the same gene, but that does not respond to the inducing signal. If intercellular feedbacks were present, reporter cells neighboring inducible cells should have higher expression levels than isolated reporter cells.
We expect that cib expression is mainly the result of fluctuations in SOS response activity as we relieved transcriptional repression by Fur by chelating free iron in the medium (Nedialkova et al, 2014; Spriewald et al, 2015; Pennington & Rosenberg, 2007). We thus hypothesized that any interactions in colicin expression dynamics would most likely be the result of intercellular feedbacks in SOS response.
To test this hypothesis, we constructed a strain in which we could induce SOS response by expressing a nuclease inside the cell (Figure 4A, Methods). To measure SOS response levels, we used a transcriptional reporter for recA, which is a key component of the SOS response and has previously been used as a reporter for SOS induction levels (Friedman et al, 2005).
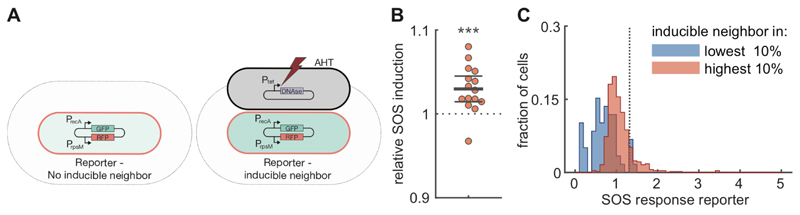
Figure 4. Direct cell-cell interactions in SOS response.
A) Test for direct interactions in SOS response. Cells with a transcriptional reporter for recA (pSV66-recA-rpsM, red cells) where grown together on agar pads with cells in which SOS response was induced by expressing the nuclease domain of colicin E2 (pSJB18, black cells). After 1h, the average SOS induction level was compared between reporter cells that do (right) and do-not (left) have inducible neighbors. The grey area indicates the region where cells are considered neighbors. Nuclease expression was induced by adding Anhydrotetracycline (AHT) to the agar pad. B). Cells neighboring inducible cells have higher SOS response levels. For each of 15 biological replicates, we measured the GFP intensity of a recA transcriptional reporter in cells with inducible neighbors (51-189 (mean=137) cells) and in cells with no direct inducible neighbors (359-713 (mean=575) cells). Each dot corresponds to a single biological replicate and shows the ratio between the mean GFP intensity in reporter cells next to inducible neighbors compared to the mean intensity in reporter cells without inducible neighbors. Points are horizontally offset, thick horizontal line indicates mean, thin lines 95% confidence intervals, over the 15 replicates. Reporter cells neighboring inducible cells have significantly higher levels of recA expression with a mean relative SOS induction of 1.030 (95% CI=1.015,1.045), p=9·10-4, t-test, n=15. C). Cells neighboring inducible cells with high levels of SOS response strongly upregulate their own stress response levels. Reporter cells (pUA66-recA) were mixed with inducible cells that also contained a recA transcriptional reporter (pSJB18 + pSV66-recA-rpsM) and grown together for 90 min on agar pads containing AHT. The distribution of SOS response levels is shown for reporter cells that are within 5μm of inducible cells with low levels (dimmest 10% of inducible cells, n=59 cells) of SOS response (blue) and for reporter cells that are within 5μm of inducible cells with high levels (brightest 10% of inducible cell, n=503 cells) of SOS response (red). The distributions were obtained by pooling the data of 4 biological replicates. The dashed vertical line indicates an SOS response level of 2 standard deviation above average. Distributions considering only direct neighbors are shown in Figure S9C.
We first tested for the presence of intercellular feedbacks by mixing recA reporter cells with cells with inducible SOS response. We grew the two strains together on agar pads and compared SOS induction levels in reporter cells that neighbored inducible cells with reporter cells without inducible neighbors. We found that in 14 out of 15 replicates SOS induction levels were higher when reporter cells neighbored an inducible cell (Figure 4B, mean relative induction=1.030, p=9·10-4, n=15). This suggests that cells can induce their neighbors’ SOS response with their own. However, from this data we cannot accurately estimate the strength of the induction, as only a small fraction (~11%) of the inducible cells actually has elevated levels of SOS response (Figure S9B).
We performed a second set of experiments where we added a recA reporter to the inducible cells, thus allowing us to determine which inducible cells had elevated levels of SOS response and how they influenced their neighbors. We then compared the SOS response levels in reporter cells neighboring inducible cells with high SOS response level (top 10%) to a control group consisting of reporter cell neighboring inducible cells with low SOS response levels (bottom 10%). When we only considered direct neighbors (cells touching), we found an increase of 26% in the mean SOS response level (Figure S9C). If we allowed for slightly longer-range interactions and considered cells to be neighbors if they are within 5μm of each other, we found an even higher effect size, with reporter cell close to highly induced cells having a 57% increase in their mean SOS response level compared to the control group (Figure 4C). Moreover, there was a three-fold increase in the fraction of reporter cells with very high SOS response levels (more than two standard deviations above average), from 5.1% in the control group to 15.1% in reporter cells close to highly induced cells (Figure 4C). Together, our data thus strongly suggests that there are intercellular feedbacks in SOS-response: cells appear to “sense” the SOS response level in their neighbors and respond by upregulating their own SOS response.
Despite finding evidence for direct intercellular feedbacks in SOS response, we did not observe any effect of spatial proximity on recA expression (Figure S8). RecA protein levels are spatially correlated (Figure S2), however this can be fully explained by the effects of shared lineage history (Figure S8A, 〈δED/δCR〉=5.9, p=1·10-4). We did not find a significant effect of spatial proximity (Figure S8B, 〈δER/δNB〉=1.07, p=0.2). One possible explanation for this is that the vast majority of cells in the microcolony had only very low SOS induction levels, making it unlikely that there could have been any transmission of the SOS response state.
Shared lineage history and spatial proximity lead to spatial correlations in anabolism
Next, we turn to our hypothesis that neighboring cells should be dissimilar in their expression levels of anabolic pathways. To test this hypothesis, we studied three pathways involved in amino acid biosynthesis in E. coli. Previous work using genetic consortia of complementary amino-acids autotrophs has shown that many amino acids can be exchanged through the environment (Mee et al, 2014; Wintermute & Silver, 2010). Furthermore, amino acid production costs appear to show economies of scale: the cost of producing an extra amino acid decreases with increasing production levels (Pande et al, 2013). A genetic consortia can thus grow faster if its members specialize on the production of complementary subset of amino-acids and exchange them with each other (Pande et al, 2013).
We hypothesized that genetically identical cells could phenotypically specialize on the production of different amino acids. A successful division of labor in amino-acid production would have two main requirements: i) efficient exchange of amino-acids between neighboring cells and ii) phenotypic variation in amino-acid production rates to would allow neighboring cells to specialize on different pathways (i.e. that would allow for symmetry breaking).
We used transcriptional reporters to follow the expression dynamics of PheL, the leader peptide of the pheLA operon that encodes for an enzyme involved tyrosine and phenylalanine biosynthesis; MetA, an enzyme involved in methionine biosynthesis; and TrpL, the leader peptide of the trpLEDCBA operon that encodes for enzymes involved in tryptophan biosynthesis. We chose trpL as previous work has shown that it has relatively high variation in expression levels (Silander et al, 2012). We chose pheL and metA as cells with knockout mutations in the biosynthesis pathways of phenylalanine and methionine tend to grow well when combined with a large number of complementary knockout strains in cross feeding cultures, suggesting that these two amino acids can efficiently be shared between cells (Mee et al, 2014; Wintermute & Silver, 2010).
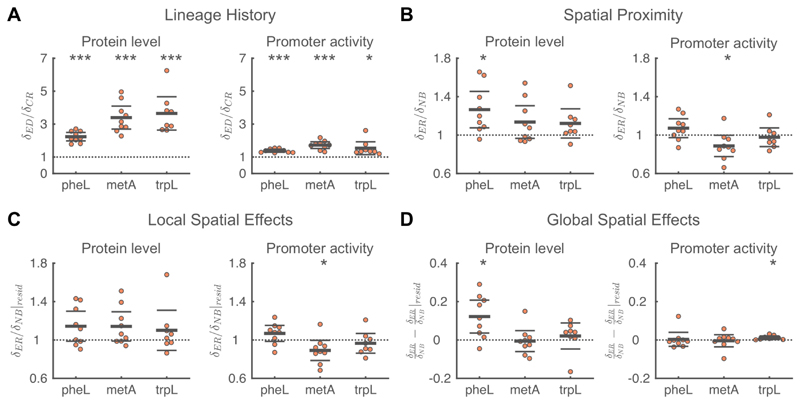
Contrary to our hypothesis, we observed strong positive spatial correlations in the protein levels of all three pathways (Figure S2). An important driver of these positive correlations is the effect of shared lineage history: for all three pathways we observe that closely related cells are similar in both protein levels and promoter activities (Figure 5A, 〈δED/δCR〉>1 for all pathways). However, even after correcting for lineage effects, we observed that protein levels are similar between neighboring cells. For all three pathways the phenotypic difference between a focal cell and its neighbor is smaller than the phenotypic difference between the focal cell and an equally related, but more distant, cell. However, this effect is only significant for pheL (Figure 5B, 〈δER/δNB〉>1 for all pathways). Based on protein levels we thus do not find any evidence for a division of labor in amino acid synthesis between neighboring cells.
Figure 5. Analyses of factors that contribute to spatial correlations in amino acid synthesis.
A) Shared lineage history leads to similarity in protein levels and promoter activity for all three pathways involved in amino acid synthesis. In all cases, the phenotypic difference between a focal cell and an equidistant cell (δED) is significantly larger than the phenotypic difference between a focal cell and its closest relative (δCR). B) Spatial proximity leads to similarity in PheL protein levels and dissimilarity in metA promoter activity. For PheL protein levels the phenotypic difference between a focal cell and an equally-related cell (δER) is significantly larger than the phenotypic difference between the focal cell and one of its neighbors (δNB). For metA promoter activities, neighboring cells are less similar than expected based on their relatedness. C) The dissimilarity in metA promoter activity is due to local spatial effects. For metA promoter activity, the difference in residuals between a focal cell and an equally-related cell (δER|resid) is significantly smaller than the difference in residuals between the focal cell and one of its neighbors (δNB|resid). D) Global spatial effects lead to similarity in PheL protein levels and trpL promoter activity. A-D) Each point corresponds to a microcolony with 117-138 (mean=128) cells, points are horizontally offset. Thick horizontal lines indicate mean, thin lines 95% confidence intervals. Dashed lines indicate the expected value under the null hypothesis. Null-hypothesis rejected with: *p<0.05, **p<0.01, ***p<0.001, t-test, n=9 (pheL, metA) or 8 (trpL). See also Figure S5,7.
Spatial dissimilarity in promoter activity of methionine biosynthesis
For metA, we observed that neighboring cells where significantly dissimilar in their promoter activities: a focal cell and its neighbor are more different in their promoter activity than the focal cell and an equally-related cell (Figure 5B, 〈δER/δNB〉=0.89, p=0.05). This difference does not change when we correct for global spatial effects by analyzing the residuals of a linear regression of metA promoter activity with the distance of a cell to the colony edge (Figure 5C, 〈δER/δNB|resid〉=0.89, p=0.04). This suggests that there are negative intercellular feedbacks affecting the expression of metA: if a cell transcribes metA at a high rate, its neighbors will tend to transcribe this gene at a lower rate. For pheA and trpL we found no evidence for similar negative feedback loops in promoter activity (Figure 5B).
For metA, we thus observed that neighboring cells where significantly dissimilar in their promoter activities, while their protein levels tended to be similar (though the latter effect was not significant, Figure 5B). This suggests that at any given time cells try to differentiate from their neighbors, giving rise to dissimilarity in promoter activities. However, as the identity of a cell’s neighbors continuously change with time (as a result of colony growth) this could prevent the establishment of dissimilarity in protein levels between neighbors.
Spatial similarity in overall metabolic state of cells
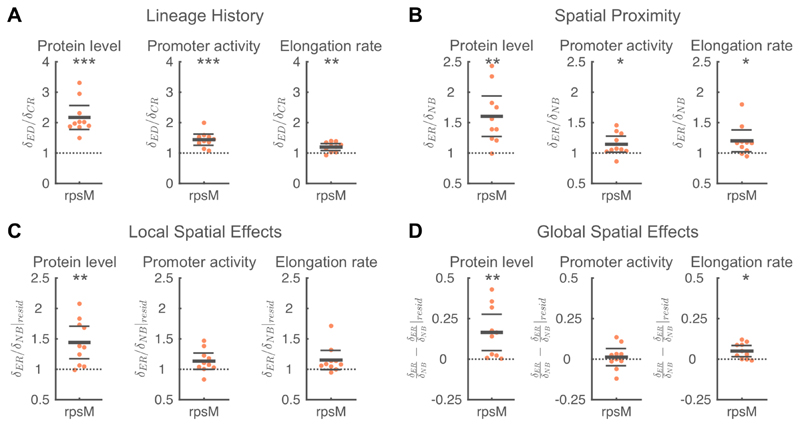
We observed positive spatial correlations in the protein levels of all three amino acid synthesis pathways we studied. This raises the question whether such positive correlations are a more general feature of a cell’s metabolism. To investigate this possibility, we simultaneously measured a cell’s elongation rate (i.e. growth rate) and the expression level of rpsM, which codes for the S13 ribosomal protein. Ribosome production levels have previously been shown to be strongly correlated to a cell’s growth rate and are thus expected to be a good proxy for a cell’s overall metabolic activity (Scott et al, 2010; 2014).
We observed significant positive spatial correlations in both RpsM protein levels and cell elongation rate (Figure S2). A large part of this correlation is again due to the effects of shared lineage history (Figure 6A). However, neighboring cells are more similar in the protein level and promoter activity of rpsM and in cell elongation rate than expected based on their relatedness (Figure 6B).
Figure 6. Analyses of factors that contribute to spatial correlations in metabolism.
A) Shared lineage history leads to similarity in RpsM protein levels (left), rpsM promoter activity (middle), and cell elongation rate (right). In all cases, the phenotypic difference between a focal cell and an equidistant cell (δED) is significantly larger than the phenotypic difference between a focal cell and its closest relative (δCR). B) Spatial proximity leads to similarity in RpsM protein levels, rpsM promoter activity, and cell elongation rate. In all cases, the phenotypic difference between a focal cell and an equally-related cell (δER) significantly is larger than the phenotypic difference between the focal cell and one of its neighbors (δNB). C) The similarity in RpsM protein levels is partly due to local spatial effects. For RpsM protein levels, the difference in residuals between a focal cell and an equally-related cell (δER|resid) is significantly larger than the difference in residuals between the focal cell and one of its neighbors (δNB|resid). D) Global spatial effects lead to similarity in RpsM protein levels and cell elongation rate. A-D) Each point corresponds to a microcolony with 117-138 (mean=128) cells, points are horizontally offset. Thick horizontal lines indicate mean, thin lines 95% confidence intervals. Dashed lines indicate the expected value under the null hypothesis. Null hypothesis rejected with: *p<0.05, **p<0.01, ***p<0.001, t-test, n=10. See Figure S10 for a chromosomal rpsM reporter in Salmonella Tm. See also Figure S5-7.
This similarity is due to both global and local spatial effects (Figure 6CD): RpsM protein levels and cell elongation rate are both (weakly) correlated with the distance of a cell to the edge of the colony (Figure S7). After correcting for these global effects, we still observe that spatial proximity tends to cause similarity in rpsM expression dynamics and cell growth rate, though the effect is only significant for RpsM protein levels (Figure 6C). Together these data show that metabolic activities are spatially correlated because of lineage history, local cell-cell interaction and global spatial gradients. For the latter, our analysis shows that cells in the colony center grow faster and had higher expression levels of rpsM, showing that nutrients are not limiting growth in the colony center (Figure S7).
Finally, we checked for the robustness of our results to the choice of model organism and the use of plasmid-based reporters by analyzing a transcriptional reporter for rpsM integrated in the chromosome of Salmonella Typhimurium. We find that our results for rpsM expression levels agree both qualitatively and quantitatively between a plasmid-based reporter in E. coli (Figure 6) and chromosomal reporter in Salmonella Tm. (Figure S10). The measurements for rpsM promoter activity in Salmonella Tm deviate from those measured in the plasmid-based system, potentially due to technical differences between the measurements: the lower signal intensity of the chromosomal construct resulted in a lower signal-to-noise ratio and required a reduced sampling frequency to reduce the effects of bleaching. Nonetheless, these results show that our main conclusion are robust to the genetic background of the model organism and suggest that our findings can be extended to other bacterial species.
Discussion
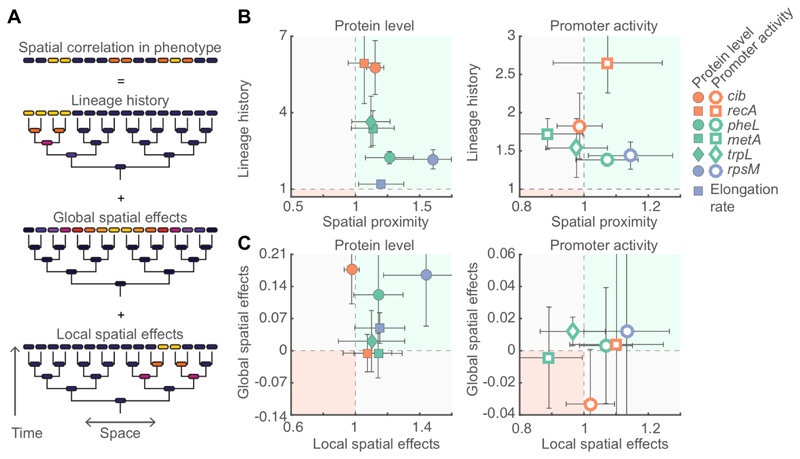
We found that lineage history effects contributed to positive correlations in gene expression in all pathways we studied, and global and local spatial effects affected a subset of the pathways (Figure 7). In general, we expect shared lineage history to always contribute to positive spatial correlations in gene expression as in most cases proteins are partitioned equally between daughter cells at cell division (Robert et al, 2010; Hormoz et al, 2015; Veening et al, 2008b; 2008a; Julou et al, 2013). Global spatial effects can also contribute to positive spatial correlations in gene expression and are likely caused by emergent spatial gradients that are the result of the uptake, release, and diffusion of metabolites during population growth (Julou et al, 2013; Stewart & Franklin, 2008; Flemming et al, 2016). Local interactions could lead to both positive and negative spatial correlations in gene expression. We found evidence for positive correlations in SOS stress response and RpsM expression levels and for negative correlations in metA promoter activity. These local interactions could be the result either of a direct exchange of cellular components between cells (e.g. via nanotubes (Dubey & Ben-Yehuda, 2011; Pande et al, 2015), pili (Hayes et al, 2010), vesicles (Schwechheimer & Kuehn, 2015), or membrane fusion (Ducret et al, 2013)) or indirectly through the microenvironment.
Figure 7. Causes of spatial correlations in phenotype.
A) Spatial correlations in phenotype are the consequence of shared lineage history, global spatial effects, and local spatial effects. B) For each pathway the relative importance of lineage history (〈δER/δNB〉) and spatial proximity (〈δED/δCR〉) is shown for protein level and cell elongation rate (left) and promoter activity (right). In most cases lineage history is the dominant factor (note the different scaling of the axis). C) For each pathway the relative importance of global spatial effects (〈δER/δNB − δER/δNB|resid〉) and local spatial effects (〈δER/δNB|resid〉) is shown for protein level and cell elongation rate (left) and promoter activity (right). B,C) Each point corresponds to the average value over 8-10 microcolonies; the data are identical to those shown in Figures 3, 5, and 6. Error bars indicate 95% confidence intervals. The green shaded region (upper right region) indicates that both factors contribute to similarity in phenotype; the red shaded region (bottom left) indicates that both factors contribute to dissimilarity in phenotype; in the other two regions (grey shading) the two factors have opposing effects.
Shared lineage history, global spatial effects, and positive intercellular feedbacks can all contribute to spatial correlations in gene expression. However, only intercellular feedbacks allow cells to directly coordinate their activities with their neighbors. In contrast, spatial correlations caused by lineage history effects or global spatial effects will largely be determined by the physics of cell growth (Nadell et al, 2013) and the physical and chemical properties of the environment (Stewart & Franklin, 2008). Intercellular feedbacks are thus the main mechanism that can allow for robust and consistent pattern formation irrespective of environmental conditions. Furthermore, intercellular feedbacks directly link the genotype of a cell to the spatial patterns of gene-expression at the colony level, potentially allowing for these patterns to evolve by natural selection.
Spatial correlations in expression levels can potentially have functional consequences. Positive correlations could allow cells to achieve a critical mass to collectively change their environment. This could be the explanation for the strong positive correlations we observe in the expression levels of the Colicin Ib toxin. Furthermore, positive correlations could allow for collective information processing where cells improve their inferences about the environment by pooling measurements between a group of neighboring cells (Hein et al, 2015; Simons, 2004; Berdahl et al, 2013; Popat et al, 2014). This could be beneficial in the context of stress response system: a cell’s survival chances might increase if it preemptively upregulates its stress response system if its neighbor is stressed, even if it is not yet exposed to any stressor itself. Consistent with this idea we found evidence for direct intercellular feedbacks in SOS response.
Negative spatial correlations in expression levels might allow for a division of labor strategy, however we did not find any evidence for them in this study. Although we did observe spatial dissimilarity in metA promoter activity, the stronger converging effect of lineage history resulted in an overall positive correlation in expression levels. Nonetheless, our data does not completely rule out the possibility of a phenotypic division of labor in amino acid biosynthesis: amino acid synthesis pathways are regulated by end product inhibition which cannot be measured using transcriptional reporters (Chubukov et al, 2014). Negative correlations in amino acid synthesis fluxes could thus be present without corresponding differences at the transcriptional level.
Our most important conclusion is that the phenotype of a cell depends to a large extend on the population context in which it grows. Our work thereby joins a growing number of recent studies showing that a large degree of phenotypic variation is not random, but rather determined by a cells lineage history, location in the population and the activities of its neighbors (Snijder & Pelkmans, 2011; Symmons & Raj, 2016). If we want to understand the activities of cells living in spatially structured assemblies, it is thus essential to learn more about the feedbacks between a cell and the surrounding population.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Simon van Vliet (simonvanvliet@gmail.com).
Experimental Model and Subject details
Strains and reporter plasmids
All experiments were done using E. coli MG1655, unless otherwise specified (see Table S1 for a list of strains and plasmids used in this study). Gene expression dynamics were followed using transcriptional reporters. The promoter region of the gene of interest was inserted in front of a gfpmut2 green fluorescent protein. For trpL, metA, pheL, and rpsM we used reporters based on the pUA66/pUA139 low copy number plasmids (Zaslaver et al, 2006). For recA we used a newly constructed dual reporter plasmid, pSV66-recA-rpsM, which is based on the low copy number plasmid pUA139 (Zaslaver et al, 2006). This plasmid contains a GFPmut2 transcriptional reporter for recA and an additional turboRFP (red fluorescent protein) transcriptional reporter for rpsM, allowing for the independent measurement of two promoters within the same cell (see Supplementary Text 1 and Table S2 for construction details). For cib we used the medium copy number plasmid pM1437 with a pBR322 background (Nedialkova et al, 2014; Spriewald et al, 2015). MG1655 does not naturally contain the cib operon in its chromosome. To measure Colicin Ib expression dynamics, we therefore transformed TB60 (MG1655 containing a chromosomal kanR cassette) with the p2-camR plasmid that is based on the natural occurring Salmonella pColB9 plasmid, which contains, among others, the cib operon (Stecher et al, 2012). We subsequently transformed the same strain with the pM1437 plasmid containing the cib transcriptional reporter. Additionally, we used the high copy number plasmids pGFP and pRFP with inducible green and red fluorescent proteins under control of the lac promoter to check for fluorescent bleed-through and halos (see Table S1). Finally, we used strain NF06, a S. Typhimurium SL1344 derivative with a GFP transcriptional reporter for rpsM inserted in the putPA locus on the chromosome.
Strain with inducible SOS response
To test for interactions in SOS response we constructed a plasmid, pSJB18, with which SOS response can be chemically induced. To do so, the nuclease domain of colicin E2 (colE2) was cloned downstream of the Ptet tetracycline inducible promoter of the pMG-Ptet vector (see Supplementary Text 1 and Table S2 for construction details). Upon induction, the nuclease activity results in DNA breaks, which in turn activates the cell’s SOS response. We made sure that the nuclease produced in a cell could not directly affect neighboring cells in two ways: i) pSJB18 does not contain the lysis gene that is part of the full colE2 operon; as colicins are released during cell lysis this greatly reduces the amount of extracellular nuclease (Cascales et al, 2007). ii) pSJB18 only contains the C-terminal nuclease domain of the colE2; the N-terminal and central domains that are required for Colicin E2 to enter a target cells (by mediating receptor binding and membrane translocation, respectively (Cascales et al, 2007)) were removed. A second plasmid, pSJB19, was constructed. This plasmid is identical to pSJB18, except that it also contain the coding sequence for the Colicin E2 immunity protein. Expression of this immunity protein inhibits the nuclease activity of Colicin E2 (see Supplementary File 2 for construction details).
We confirmed the functionality of the construct by co-transforming MG1655 with pSJB18 and pUA139-recA (Zaslaver et al, 2006). The latter contains a GFP transcriptional reporter for the SOS-response gene recA. Expression of the nuclease was induced by adding 100ng/ml of the non-toxic tetracycline analog anhydrotetracycline (AHT, Fluka, Buchs, Switzerland). Using flow cytometry and single-cell microscopy we confirmed that SOS-response activity (measured as recA expression levels) increased when the inducer was added (Figure S9).
Plasmid construction
Construction of pSJB18 and pSJB19
The pSJB18 plasmid is based on the pMG-Ptet plasmid (Neuenschwander et al, 2007) which contains the tetracycline inducible Ptet promoter. The colicin E2 nuclease domain (ceanuclease) was amplified from plasmid pColE2-p9 (Kerr et al, 2002; Hol et al, 2014) using primers ColE2_C-dom_tet_fwd and ColE2_tet_rev (see Table S2 for primer sequences). The forward primer includes an ATG start codon and Xbal restriction site, the reverse primer includes a Xhol restriction site. The amplicon was inserted into pMG-Ptet via Xbal/Xhol. The construct sequence was confirmed with sequencing.
We constructed a second plasmid, pSJB19, that is identical to pSJB18, except that it also includes the Colicin E2 immunity protein (cei). This immunity protein is co-expressed with the nuclease domain and inhibits the nuclease activity. The Colicin E2 nuclease domain (ceanuclease) was amplified together with the colicin E2 immunity protein (cei) from plasmid pColE2-p9 (Kerr et al, 2002; Hol et al, 2014) using primers ColE2_C-dom_tet_fwd and ImmE2_tet_rev (see Table S2). The forward primer includes an ATG start codon and Xbal restriction site, the reverse primer includes a Xhol restriction site. The amplicon was inserted into pMG-Ptet via Xbal/Xhol. The construct sequence was confirmed with sequencing.
Construction of pSV66-rpsM-rpsM dual-reporter
The pSV66 dual reporter is based on the low copy number pUA66/pUA139 (Zaslaver et al, 2006) reporter plasmid, but contains an additional transcriptional reporter based on the red fluorescent protein TurboRFP. The arrangement of the two reporters was based on the triple reporter plasmid pZS2-123 (Cox et al, 2010). Specifically, the gfmut2 and turborfp reporters are separated by a region containing multiple transcriptional terminators amplified from pZS2-123. The plasmid was constructed using a three-step Gibson assembly protocol (New England Biolabs, Ipswich, Massachusetts):
PrpsM-tRFP promoter-reporter construct. The promotor region of rpsM was amplified from plasmid pUA139-rpsM (Zaslaver et al, 2006) using primers Prpsm-fw/rv (see Table S2 for primer sequences). The coding sequence of turborfp was amplified from plasmid pTurboRFP (Hol et al, 2014) using rfp-fw/rv. The resulting products were joined using Gibson assembly.
PrpsM-turboRFP-terminator construct. The PrpsM-turboRFP construct from step 1 was amplified from the Gibson assembly product using primers Prpsm-fw and rfp-rv. The multiple terminator region was amplified from plasmid pZS2-123 (Cox et al, 2010) using primers ter-fw/rv. The resulting products were joined using Gibson assembly.
pSV66-rpsM-rpsM plasmid. The PrpsM-turboRFP-terminator construct from step 2 was amplified from the Gibson assembly product using primers ter-rv and Prfp-fw. The greater part of plasmid pUA139-rpsM ((Zaslaver et al, 2006), including the Prpsm-gfpmut2 reporter, origin of replication and kanamycin resistance cassette) was amplified using primers vector-fw/rv. The resulting products were joined using Gibson assembly.
We confirmed the sequence of the promoter regions, fluorescent proteins, terminator region and origin of replication using sequencing.
Construction of pSV66-recA-rpsM dual-reporter
To construct the dual reporter pSV66-recA-rpsM the promoter region in front of gfpmut2 of the pSV66-rpsM-rpsM was replaced with the promoter of recA using a one-step Gibson assembly process. The promoter for recA was amplified from the plasmid pUA139-recA (Zaslaver et al, 2006) using primer Pgfp-fw/rv and the backbone of pSV66-rpsM-rpsM was amplified with gfpVec-fw/rv. The two PCR products were then combined using Gibson assembly and the sequence of both the recA and rpsM promoter region, as well as the intermediate terminator region was confirmed using sequencing.
In all steps we used the Q5 high-fidelity DNA polymerase for DNA amplification and Gibson Assembly master mix for Gibson assembly (New England Biolabs, Ipswich, Massachusetts).
Media and growth conditions
In all cases cultures were started from a single colony taken from a LB-agar plate and grown overnight at 37°C in a shaker incubator. Subsequently the cultures were diluted 100 to 1000 fold and grown until mid-exponential phase. Cells containing reporters for cib, recA, and rpsM were grown in LB media (Sigma-Aldrich, Buchs, Switzerland, or Applichem, Darmstadt, Germany). For these reporters, microscopy was done on agar pads consisting of LB with 1.5% agar (Sigma-Aldrich or Applichem). Cells containing reporters for metA, pheL and trpL were grown overnight in M9 medium (47.76 mM Na2HPO4, 22.04 mM KH2PO4, 8.56 mM NaCl and 18.69 mM NH4Cl) supplemented with 1mM MgSO4, 0.1 mM CaCl2, 0.4% Glucose (all from Sigma-Aldrich), and 5% LB. Diluted cultures were grown in M9 medium supplemented with 1mM MgSO4, 0.1 mM CaCl2 and 0.4% Glucose. Microscopy was done on agar pads consisting of M9 salts with 1.5% agar and supplemented with 1mM MgSO4, 0.1 mM CaCl2 and 0.4% Glucose.
Plasmid maintenance was insured by adding the appropriate antibiotic to the culture medium and agar pads: 50μg/ml ampicillin (pM1437, pSJB18, pGFP, pRFP, Applichem), 50μg/ml kanamycin (pUA66, pUA139, pSV66, Sigma-Aldrich) and 15μg/ml chloramphenicol (p2-camR, Sigma-Aldrich). For experiments with the cib reporter we added additionally 0.1mM DTPA (diethylenetriaminepentaacetic acid, Fluka) to the medium of the diluted cultures and to the agar pads to chelate free iron. For the SOS interaction experiments 100ng/ml of anhydrotetracycline (AHT) was added to the agar pads to induce the nuclease in pSJB18. For the experiments done with a mixture of strains MG1655+pSJB18+pSV66-recA-rpsM and MG1655+pUA139-recA (Figure 4C) we additionally added 30μg/ml kanamycin to the agar pad. For the experiments done with a mixture of strains MG1655+pSJB18 and MG1655+pSV66-recA-rpsM (Figure 4B), no antibiotics were added to the agar pads as the two strains carry different resistance genes. As these experiments only lasted 1h, we expect plasmid loss to be negligible. For experiments using pGFP or pRFP we added 1mM IPTG (Isopropyl β-D-1-thiogalactopyranoside, Promega, Madison, Wisconsin) to the liquid cultures and agar pads to induce expression of the fluorescent proteins.
Method Details
Agar pad preparation
Agar pads were prepared by adding the appropriate supplements to molten aliquots of LB or M9 agar and adding 250μl of this mixture to the well of hollow-well microscope slides (Karl Hecht GmbH, Sondheim, Germany). The wells were sealed with a cover glass and dried at room temperature for 20 to 30min. Subsequently the cover glass was removed and the agar was cut into a square of approximately 5x5mm in the center of the well. 0.5 to 2μl of prepared cell suspension (see below) was added to the center of the pad and left to dry. Finally, the pad was sealed by adding a new cover glass. An air-tight seal was insured by adding a thin layer of lubricating grease (Glisseal, Borer, Zuchwil, Switzerland) between the two glass surfaces. The agar pad only occupies the central part of the well; the remaining area contains air to insure that sufficient oxygen is present for aerobic growth.
Before inoculation the optical density at 600nm (OD600) of the cultures was measured. The cultures were diluted to the desired OD600 (of 0.001 to 0.01) before adding 0.5 to 2μl of cells to the pad. For the SOS interaction experiment the two strains were first washed to remove antibiotics from the growth medium. Subsequently the strains were mixed in approximately a 1:1 ratio and added to the agar pad.
Microscopy
Time-lapse microscopy was done using fully-automated Olympus IX81 inverted microscopes (Olympus, Tokyo, Japan). Imaging was done using a 100X NA1.3 oil objective (Olympus) and either a F-View II CCD camera (for cib, Olympus Soft Imaging Solutions, Münster, Germany) or an ORCA-flash 4.0 v2 sCMOS camera (all other data, Hamamatsu, Hamamatsu, Japan). Fluorescent imaging was done using a X-Cite120 120 Watt high pressure metal halide arc lamp (Lumen Dynamics, Mississauga, Canada) and Chroma 49000 series fluorescent filter sets (N49002 for GFP and N49008 for RFP, Chroma, Bellows Falls, Vermont). Focus was maintained using the Olympus Z-drift compensation system and the entire setup was controlled with either the Olympus CellM or CellSens software. The sample was maintained at 37°C by a microscope incubator (Life imaging services, Basel, Switzerland). Images were taken every 3 (rpsM, elongation rate), 5 (cib) or 7.5 (recA, trpL, pheA, metA) minutes for several hours. We quantified the homogeneity of the illumination field and found that light intensities varied by less than 11% within the microcolony. Any potential negative effects of light exposure (bleaching, photo toxicity, etc.) are thus not expected to contribute to the observed spatial patterns of gene expression as they would affect all cells equally.
Quantification and Statistical Analysis
Selection and analysis of microcolonies
Fiji (Schindelin et al, 2012) was used for data visualization, image cropping and file-type conversions. All other processing was done using Matlab (version 2013 and newer, MathWorks, Natick, Massachusetts). For each reporter we analyzed 8 to 10 microcolonies, we decided on this sample size based on preliminary experiments with a colicin Ib reporter strain. Each micro colony is considered to be an independent biological replicate. From each agar pad we selected 1 to 6 (median=2) positions that contained an isolated micro colony of sufficient size (>128 cells before cell overlap occurs) and good optical quality. We manually determined the time range where cells were present in a single layer and cropped the images to only contain the area occupied by the colony.
Subsequently Schnitzcells 1.1 (Young et al, 2011) was used to segment and track cells. For cib, segmentation was done on phase contrast images. For trpL, metA, pheA, and rpsM segmentation was done on the GFP fluorescent images. The reporter plasmid for recA also contained a RFP reporter for rpsM, here segmentation was done on the RFP fluorescent images. As a last step custom Matlab code was used to extract fluorescent and geometrical properties of each cell (see fluorescent image processing and cell length determination below).
Fluorescent image processing
There are a number of optical artifacts that could cause neighboring cells to have similar fluorescent intensity levels, it is thus essential to correct for these artifacts before calculating the spatial similarity. Specifically, we applied the following corrections (see also Figure S1):
Shading correction. We corrected for inhomogeneities in the illumination field using a shading image. This image gives the normalized intensity of the incoming light for each pixel in the image. Subsequently we obtained the shading corrected image by dividing the intensity in each pixel of the captured fluorescent image by the intensity in the corresponding pixel of the shading image.
Deconvolution. Diffraction will cause the light of a point source to be spread across several pixels. Bright cells will thus generate a “halo” that increases the fluorescent intensity of its neighbors. We corrected for diffraction by deconvolving the shading corrected image with the experimentally measured point spread function (PSF) of the microscope (Kiviet et al, 2014). Deconvolution was done using the Matlab function “deconvlucy”, which uses the Lucy-Richardson method. We found that the accuracy of the deconvolution correction depends critically on the size of the PSF that is used (Figure S3B). When the size of the PSF is too small (e.g. 13x13 pixels) the halos are not completely removed, increasing fluorescent intensities in neighbors of bright cells. If the PSF is too large (e.g. 30x30 pixels) a “dark halo” artifact is formed, decreasing fluorescent intensities in neighbors of bright cells. We calibrated the required size of the PSF by mixing unlabeled wild type cells with cells carrying a high copy number plasmid with an inducible green fluorescent protein. We then selected the size of the PSF (i.e. 24x24 pixels) for which median fluorescent intensity in unlabeled cells neighboring GFP labeled cells are the same as for isolated unlabeled cells (Figure S3B,C). Furthermore, we confirmed that our statistical analysis of the effects of spatial proximity is robust to small changes in the size of the PSF (Figure S4C).
Background correction. We performed a background correction to compensate for temporal changes in the incoming light intensity. For each pixel the background corrected intensity (Icorr) was calculated as: Icorr = (I-Dark)/(Bg-Dark), where I is the pixel intensity after shading correction and deconvolution, Bg is the median intensity of all background pixels (i.e. all pixels that are not part of any segmented cell) and Dark is the median pixel intensity for the dark image (i.e. an image taken when no light reaches the camera taken with the same exposure settings).
Cell center intensity. As sell segmentation is imperfect, some pixels at the periphery will be misclassified. To increase robustness to such errors we calculated the mean fluorescent intensity only over the central area of the cell. The central area is found by eroding (i.e. shrinking) the cell segmentation mask on all sides with one quarter of the median cell width (the median cell width was determined over all cells in the microcolony). For most cells the intensity was thus be determined for the central 50%. If erosion removed all pixels in the cell mask, we progressively reduced the number of outer pixels we removed until at least a single row of pixels remained in the cell center.
Fluorescence bleed-through. The recA reporter strain contained a second RFP reporter for rpsM. We confirmed that there is no fluorescence bleed-through from the RFP to the GFP channel. To do so, we mixed unlabeled cells with cells carrying a high copy number plasmid with inducible red fluorescent protein. The distribution of fluorescent intensities in the GFP channel in unlabeled cells is identical to intensities in the GFP channel for the RFP labeled cells. This shows that emission from the red fluorescent protein do not affect measured intensities in the GFP channel (Figure S3A).
After performing all corrections, we obtained for each cell the mean fluorescent intensity, which is proportional to the concentration of GFP molecules in the cell and hence to the concentration of the gene of interest. Throughout the text we use protein level to refer to the mean fluorescent intensity.
Cell length determination
Cell length was determined following the procedure described in (Kiviet et al, 2014). In short: the cell centerline was determined by fitting the cells mask with a 3th degree polynomial (f(x)). To find the cell pole positions we calculated the silhouette proximity (sum of the squared distance to closest 25 pixels in cell mask) along the centerline. This measure is constant in the cell center, but increases sharply at the poles; the position of the cell poles was taken as the point along the centerline where the proximity silhouette reached 110% of the average value in the cell center. The cell length was subsequently calculated by numerical integration of where f′(x) is the derivative of f(x) and x0 and x1 are the positions of the cell pole.
Cell elongation rate
Cell elongation rates (r) were calculated for the microcolonies with a rpsM transcriptional reporter by fitting the exponential curve L(t) = L(0) · er·t to the cell length over time. The fitting was done using a linear fit on the log transformed cell lengths over a sliding time window of 7 time-points (21 minutes). When the time window exceeded the life time of a cell, it was extended by summing the cell lengths of the two daughter cells or by taking a fraction of L0/(L0 + L0,sister) of the mother cell length. Here, L0 and L0,sister are the lengths of a cell and its sister at their birth. This fraction takes the effects of asymmetries at cell division into account.
Promoter activity
Assuming that the rate of protein degradation is negligible, the promoter activity (PA) an be estimated as the rate of change in the total fluorescent intensity of a cell: The measurement of total fluorescent intensity as the summed intensity over all pixels in the cell mask is very sensitive to segmentation inaccuracies. To get a more accurate estimate of promoter activities we thus estimate the total fluorescent intensity by multiplying the mean fluorescent intensity of a cell, with its length, L(t); as the cell width is constant through the lifetime of a cell, this quantity is proportional to the total fluorescent intensity of a cell: The promoter activity is then estimated as the slope of a linear fit of this quantity over a window of 5 time points: When the time window exceeded the life time of a cell, it was extended by summing the total fluorescent intensities of the two daughter cells or by taking a fraction of the total intensity of the mother cell. Here, are the total fluorescent intensities of a cell and its sister at their birth. This fraction takes the effects of asymmetries at cell division into account.
Cell geometric and neighborhood properties
The cell width and its position were determined using the Matlab regionprops function and correspond to the minor-axis length of a fitted ellipse and the coordinates of the center of mass, respectively. The neighbors of a cell were found by expanding the cell mask in all directions with ¾ of the median cell width; all cells that overlap with this expanded area are classified as neighbors. The distance of a cell to the colony edge was determined as the minimum Euclidean distance between pixels inside the cell mask and pixels that are part of the colony boundary. Cell pole age was determined using the custom written Matlab code described in reference (Bergmiller & Ackermann, 2011).
Neighborhood similarity statistic
We quantitatively investigated the apparent non-randomness of expression patterns in the microcolonies using a randomization procedure. Cells in the colony were classified into two groups depending on whether their mean fluorescent intensity was above or below the median intensity in the microcolony. For each cell in the colony we calculated the fraction of neighboring cells that was classified in the same group and we computed the mean over all cells (red bar, Figure 1D and S2). We then randomly permuted intensities between cells in the colony and recalculated the mean fraction of neighbors classified in the same group. This procedure was repeated 104 times obtaining the distribution shown in Figure 1D. p-values were calculated by taking the fraction of randomized samples that have a higher mean fraction of neighbors of the same type than the non-randomized data.
Statistic for effect of shared lineage history
We tested for the effect of shared lineage history by quantifying how similar a cell is to its closest relative after correcting for the effects of spatial proximity. We compared the phenotypes within a group of three cells: a focal cell, its closest relative, and an equidistant cell. The closest relative will typically be a cell’s sister, however if the sister has already divided we selected one of its offspring (e.g. a niece of the focal cell) at random. The equidistant cell is a cell that directly neighbors the closest relative and that has a center-to-center distance to the focal cell that is the most similar to that of the closest relative.
We then calculated the difference in phenotype between the focal cell i and its closest relative (CRi): and between the focal cell i and its equidistant cell (EDi): where Xi, XCRi, and XEDi are the phenotypes (i.e. protein levels, promoter activities, or elongation rates) of the focal cell, closest relative, and equidistant cell, respectively. Finally, we calculated the effect of shared lineage history by taking the ratio of these two phenotypic distances: median where the median is taken over all cells in the colony.
Statistic for the effect of spatial proximity
We tested for the effect of spatial proximity by quantifying how similar a cell is to its neighbors after correcting for the effects of shared lineage history. We compared the phenotypes within a group of three cells: a focal cell, one of its neighbors, and an equally-related cell. We defined a cell’s neighbors as all cells that are directly adjacent (within ¾ cell width) to the focal cell (mean number of neighbors=5,95% range=[3,8]). For each neighbor we found a group of equally-related cells, these are cells that have the same relatedness to the focal cell as the neighbor, but that are further away in space (mean number of equally-related cells=20,95% range=[0,70]). From this group we selected the most distant equally-related cell, which is the equally-related cell with the largest Euclidean distance to the focal cell.
We then calculated the difference in phenotype between the focal cell i and its neighbor j: and between the focal cell and the most distant equally-related cell (ERi,j): where Xi, Xj, and XERi,j are the phenotypes (i.e. protein levels, promoter activities, or elongation rates) of focal cell i its neighbor j, and their most distant equally-related cell. Finally, we calculated the effect of spatial proximity by taking the ratio of these two phenotypic distances: median where the median is taken over all neighbor-focal cell pairs in the colony.
Our definition of equally-related cells insures that Were always calculated for pairs of cells that have been separated by the same amount of time. However, different sets of cells (i.e. sets based on different choices of focal and/or neighbor cell) do differ in their absolute degree of relatedness, e.g. for one focal cell both the neighbor and equally related cell could be first cousins, while for another focal cell both could by fourth cousins. This difference in the absolute degree of relatedness will likely affect the absolute values of but should not affect their ratio.
We also tested whether our choice of equally-related cell affected our conclusions (Figure S4A). We recalculated the statistics using the median phenotypic distance between the focal cell and all equally-related cells: is the phenotype of the kth equally-related cell of neighbor j of focal cell i and the median is taken over all equally-related cells. Subsequently we calculated the effect of spatial proximity as: median where the median is taken over all neighbor-focal cell pairs in the colony.
Statistic for local spatial effects
The phenotype of a cell can vary systematically within the colony, we corrected for such global effects using a linear regression of a cell’s phenotype (Xi) with its distance to the edge of the colony (di): Xi = α + β · di, where α and β are constants. The strength of local spatial effects could then be estimated by correcting the observed phenotype of a cell for the global trend by calculating the residuals of the regression: We then calculated the ratio of phenotypic differences as described above, where the phenotype of a cell (Xi) was replaced with the residual of the regression
Statistic for global spatial effects
The importance of global spatial effects was quantified by calculating to what extend the effect of spatial proximity is reduced when we correct for the systematic variation in phenotype. Specifically, we defined the global spatial effect as: whμere the first term describes the total effect of spatial proximity and the second term describes the local spatial effects.
Computational data set for validation of statistical method.
To validate our statistical tests, we used a computer to simulate protein levels under a null-model that only includes lineage history effects, but no spatial effects. Protein levels were computed using a stochastic model of unregulated gene expression:
where m represents the number of mRNA molecules and p the number of proteins in a cell. mRNA and protein levels were simulated on top of experimentally measured lineage trees using a Gillespie algorithm. Every cell produces mRNA and proteins according to the stochastic model above, and values for the protein levels were stored at the experimentally determined sampling times. Subsequently, p and m were distributed to the two daughter cells by sampling from a binomial distribution (thus incorporating lineage history effects) and the procedure was repeated until all cells were processed. For the first cell in the lineage tree p and m were set to the theoretically predicted steady state levels. We used the dataset of the rpsM reporter as source of the experimentally measure lineage trees and to parameterized our model. We estimated the parameters as follows: mRNA degradation rate (γm=0.1177min-1, (Chen et al, 2015)) and steady state RpsM protein number (pss =72767, (Schmidt et al, 2015)) were taken from the literature; the conversion factor from total GFP intensity to protein number was determined as where 〈Itot〉 is the measured average of the total fluorescence in the cells; the protein degradation rate γp was set to 0, as proteins were assumed to only be lost due to dilution at cell division; kp was estimated using the relation (Thattai & van Oudenaarden, 2001); and km was estimated from using the relation is the average measured growth rate of the cells. For each lineage tree, parameters were estimated separately and 1000 simulation were run. Subsequently we compared the values of our lineage history or spatial proximity statistic of the real data with the distribution of these statistics for the computed data set (Figure S6).
Data and Software Availability
Cell segmentation and tracking data, as well as the used Matlab code, has been deposited to the ETH Data Archive: http://dx.doi.org/10.5905/ethz-1007-77. A full data set containing the unprocessed microscopy data has been deposited to the Zenodo data archive: https://doi.org/10.5281/zenodo.268921.
Supplementary Material
Acknowledgements
We thank Daan Kiviet, Alejandra Manjarrez, Alejandra Rodriguez, Ben Roller, Susan Schlegel, and Clément Vulin for helpful comments and discussion on a previous version of this manuscript and Daan Kiviet and Roland Mathis for providing parts of the Matlab code used in this work. This work was supported by SNF grant nr. 31003A_149267 to MA and grants from the Deutsche Forschungsgemeinschaft, Priority Programme SB1617 to BS.
Footnotes
Author Contributions
Conception and design of study: SvV, MA
Engineering of genetic constructs: SvV, SS, BS
Data collection: SvV and ARW
Development of statistical analysis: SvV and ADC
Data analysis and interpretation: SvV with contributions of ARW and ADC
Drafting of the article: SvV with contributions of MA
Critical revision of the article: SvV, ADC, MA
Declaration of interest
The authors declare no competing interests.
References
- Ackermann M. A functional perspective on phenotypic heterogeneity in microorganisms. Nat Rev Microbiol. 2015;13:497–508. doi: 10.1038/nrmicro3491. [DOI] [PubMed] [Google Scholar]
- Ackermann M, Stecher B, Freed NE, Songhet P, Hardt W-D, Doebeli M. Self-destructive cooperation mediated by phenotypic noise. Nature. 2008;454:987–990. doi: 10.1038/nature07067. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Losick R. Bacterially speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Berdahl A, Torney CJ, Ioannou CC, Faria JJ, Couzin ID. Emergent sensing of complex environments by mobile animal groups. Science. 2013;339:574–576. doi: 10.1126/science.1225883. [DOI] [PubMed] [Google Scholar]
- Bergmiller T, Ackermann M. Pole age affects cell size and the timing of cell division in Methylobacterium extorquens AM1. J Bacteriol. 2011;193:5216–5221. doi: 10.1128/JB.00329-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Buchanan SK, Duché D, Kleanthous C, Lloubès R, Postle K, Riley M, Slatin S, Cavard D. Colicin biology. Microbiol Mol Biol Rev. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Shiroguchi K, Ge H, Xie XS. Genome-wide study of mRNA degradation and transcript elongation in Escherichia coli. Mol Syst Biol. 2015;11:781–781. doi: 10.15252/msb.20145794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubukov V, Gerosa L, Kochanowski K, Sauer U. Coordination of microbial metabolism. Nat Rev Microbiol. 2014;12:327–340. doi: 10.1038/nrmicro3238. [DOI] [PubMed] [Google Scholar]
- Claessen D, Rozen DE, Kuipers OP, Søgaard-Andersen L, van Wezel GP. Bacterial solutions to multicellularity: a tale of biofilms, filaments and fruiting bodies. Nat Rev Microbiol. 2014;12:115–124. doi: 10.1038/nrmicro3178. [DOI] [PubMed] [Google Scholar]
- Cox RS, Dunlop MJ, Elowitz MB. A synthetic three-color scaffold for monitoring genetic regulation and noise. Journal of Biological Engineering. 2010;4:10. doi: 10.1186/1754-1611-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey GP, Ben-Yehuda S. Intercellular nanotubes mediate bacterial communication. Cell. 2011;144:590–600. doi: 10.1016/j.cell.2011.01.015. [DOI] [PubMed] [Google Scholar]
- Ducret A, Fleuchot B, Bergam P, Mignot T, Greenberg P. Direct live imaging of cell–cell protein transfer by transient outer membrane fusion in Myxococcus xanthus. eLife Sciences. 2013;2:e00868. doi: 10.7554/eLife.00868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Flemming H-C, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- Friedman N, Vardi S, Ronen M, Alon U, Stavans J. Precise Temporal Modulation in the Response of the SOS DNA Repair Network in Individual Bacteria. Plos Biol. 2005;3:e238. doi: 10.1371/journal.pbio.0030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guantes R, Benedetti I, Silva-Rocha R, de Lorenzo V. Transcription factor levels enable metabolic diversification of single cells of environmental bacteria. ISME J. 2015;10:1122–1133. doi: 10.1038/ismej.2015.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautefort I, Proenca MJ, Hinton J. Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Applied and Environmental Microbiology. 2003;69:7480–7491. doi: 10.1128/AEM.69.12.7480-7491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes CS, Aoki SK, Low DA. Bacterial Contact-Dependent Delivery Systems. 2010;44:71–90. doi: 10.1146/annurev.genet.42.110807.091449. [DOI] [PubMed] [Google Scholar]
- Hein AM, Rosenthal SB, Hagstrom GI, Berdahl A, Torney CJ, Couzin ID. The evolution of distributed sensing and collective computation in animal populations. eLife Sciences. 2015;4:e10955. doi: 10.7554/eLife.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscock TW, Megason SG. Mathematically guided approaches to distinguish models of periodic patterning. Development. 2015;142:409–419. doi: 10.1242/dev.107441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hol F, Voges MJ, Dekker C, Keymer JE. Nutrient-responsive regulation determines biodiversity in a colicin-mediated bacterial community. BMC Biol. 2014;12:68. doi: 10.1186/s12915-014-0068-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormoz S, Desprat N, Shraiman BI. Inferring epigenetic dynamics from kin correlations. Proc Natl Acad Sci USA. 2015;112:E2281–9. doi: 10.1073/pnas.1504407112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DR, Goldschmidt F, Lilja EE, Ackermann M. Metabolic specialization and the assembly of microbial communities. ISME J. 2012;6:1985–1991. doi: 10.1038/ismej.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julou T, Mora T, Guillon L, Croquette V, Schalk IJ, Bensimon D, Desprat N. Cell-cell contacts confine public goods diffusion inside Pseudomonas aeruginosa clonal microcolonies. Proc Nat Acad Sci Usa. 2013;110:12577–12582. doi: 10.1073/pnas.1301428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- Kerr B, Riley MA, Feldman MW, Bohannan BJM. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- Kiviet DJ, Nghe P, Walker N, Boulineau S, Sunderlikova V, Tans SJ. Stochasticity of metabolism and growth at the single-cell level. Nature. 2014;514:376–379. doi: 10.1038/nature13582. [DOI] [PubMed] [Google Scholar]
- Lee H, Popodi E, Tang H, Foster PL. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc Natl Acad Sci USA. 2012;109:E2774–83. doi: 10.1073/pnas.1210309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Røder HL, Madsen JS, Bjarnsholt T, Sørensen SJ, Burmølle M. Interspecific Bacterial Interactions are Reflected in Multispecies Biofilm Spatial Organization. Front Microbiol. 2016;7:4258. doi: 10.3389/fmicb.2016.01366. Available at: http://journal.frontiersin.org/Article/10.3389/fmicb.2016.01366/abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Kolter R. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiology Reviews. 2010;34:134–149. doi: 10.1111/j.1574-6976.2009.00199.x. [DOI] [PubMed] [Google Scholar]
- Mee MT, Collins JJ, Church GM, Wang HH. Syntrophic exchange in synthetic microbial communities. Proc Natl Acad Sci USA. 2014;111:E2149–56. doi: 10.1073/pnas.1405641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr W, Vagner T, Kuypers MMM, Ackermann M, LaRoche J. Resolution of Conflicting Signals at the Single-Cell Level in the Regulation of Cyanobacterial Photosynthesis and Nitrogen Fixation. PLoS ONE. 2013;8:e66060. doi: 10.1371/journal.pone.0066060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro-Pastor AM, Hess WR. Heterocyst differentiation: from single mutants to global approaches. Trends Microbiol. 2012;20:548–557. doi: 10.1016/j.tim.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Nadell CD, Bucci V, Drescher K, Levin SA, Bassler BL, Xavier JB. Cutting through the complexity of cell collectives. Proc Biol Sci. 2013;280:20122770–20122770. doi: 10.1098/rspb.2012.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadell CD, Drescher K, Foster KR. Spatial structure, cooperation and competition in biofilms. Nat Rev Microbiol. 2016;14:589–600. doi: 10.1038/nrmicro.2016.84. [DOI] [PubMed] [Google Scholar]
- Nedialkova LP, Denzler R, Koeppel MB, Diehl M, Ring D, Wille T, Gerlach RG, Stecher B. Inflammation Fuels Colicin Ib-Dependent Competition of Salmonella Serovar Typhimurium and E. coli in Enterobacterial Blooms. PLoS Pathog. 2014;10:e1003844. doi: 10.1371/journal.ppat.1003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuenschwander M, Butz M, Heintz C, Kast P, Hilvert D. A simple selection strategy for evolving highly efficient enzymes. Nat Biotechnol. 2007;25:1145–1147. doi: 10.1038/nbt1341. [DOI] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- Pande S, Merker H, Bohl K, Reichelt M, Schuster S, de Figueiredo LF, Kaleta C, Kost C. Fitness and stability of obligate cross-feeding interactions that emerge upon gene loss in bacteria. ISME J. 2013 doi: 10.1038/ismej.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande S, Shitut S, Freund L, Westermann M, Bertels F, Colesie C, Bischofs IB, Kost C. Metabolic cross-feeding via intercellular nanotubes among bacteria. Nature Communications. 2015;6:6238. doi: 10.1038/ncomms7238. [DOI] [PubMed] [Google Scholar]
- Pennington JM, Rosenberg SM. Spontaneous DNA breakage in single living Escherichia coli cells. Nat Genet. 2007;39:797–802. doi: 10.1038/ng2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popat R, Cornforth DM, McNally L, Brown SP. Collective sensing and collective responses in quorum-sensing bacteria. Journal of the Royal Society Interface. 2014;12:20140882–20140882. doi: 10.1098/rsif.2014.0882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley MA, Wertz JE. Bacteriocins: Evolution, Ecology, and Application. Annu Rev Microbiol. 2002;56:117–137. doi: 10.1146/annurev.micro.56.012302.161024. [DOI] [PubMed] [Google Scholar]
- Risser DD, Wong FCY, Meeks JC. Biased inheritance of the protein PatN frees vegetative cells to initiate patterned heterocyst differentiation. Proc Natl Acad Sci USA. 2012;109:15342–15347. doi: 10.1073/pnas.1207530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert L, Paul G, Chen Y, Taddei F, Baigl D, Lindner AB. Pre-dispositions and epigenetic inheritance in the Escherichia coli lactose operon bistable switch. Mol Syst Biol. 2010;6:357. doi: 10.1038/msb.2010.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Gillespie A, Kümmerli R. Collective decision-making in microbes. Front Microbiol. 2014;5:54. doi: 10.3389/fmicb.2014.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Kochanowski K, Vedelaar S, Ahrné E, Volkmer B, Callipo L, Knoops K, Bauer M, Aebersold R, Heinemann ML. The quantitative and condition-dependent Escherichia coli proteome. Nat Biotechnol. 2015;34:104–110. doi: 10.1038/nbt.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol. 2015;13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T. Interdependence of cell growth and gene expression: origins and consequences. Science. 2010;330:1099–1102. doi: 10.1126/science.1192588. [DOI] [PubMed] [Google Scholar]
- Scott M, Klumpp S, Mateescu EM, Hwa T. Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol Syst Biol. 2014;10:747–747. doi: 10.15252/msb.20145379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silander OK, Nikolic N, Zaslaver A, Bren A, Kikoin I, Alon U, Ackermann M. A genome-wide analysis of promoter-mediated phenotypic noise in Escherichia coli. Plos Genet. 2012;8:e1002443. doi: 10.1371/journal.pgen.1002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons A. Many wrongs: the advantage of group navigation. Trends in Ecology & Evolution. 2004;19:453–455. doi: 10.1016/j.tree.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Snijder B, Pelkmans L. Origins of regulated cell-to-cell variability. Nat Rev Mol Cell Biol. 2011;12:119–125. doi: 10.1038/nrm3044. [DOI] [PubMed] [Google Scholar]
- Spriewald S, Glaser J, Beutler M, Koeppel MB, Stecher B. Reporters for Single-Cell Analysis of Colicin Ib Expression in Salmonella enterica Serovar Typhimurium. PLoS ONE. 2015;10:e0144647. doi: 10.1371/journal.pone.0144647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Denzler R, Maier L, Bernet F, Sanders MJ, Pickard DJ, Barthel M, Westendorf AM, Krogfelt KA, Walker AW, Ackermann M, et al. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc Natl Acad Sci USA. 2012;109:1269–1274. doi: 10.1073/pnas.1113246109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- Symmons O, Raj A. What's Luck Got to Do with It: Single Cells, Multiple Fates, and Biological Nondeterminism. Molecular Cell. 2016;62:788–802. doi: 10.1016/j.molcel.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thattai M, van Oudenaarden A. Intrinsic noise in gene regulatory networks. Proc Nat Acad Sci Usa. 2001;98:8614–8619. doi: 10.1073/pnas.151588598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gestel J, Vlamakis H, Kolter R. Division of Labor in Biofilms: the Ecology of Cell Differentiation. Microbiol Spectr. 2015a;3 doi: 10.1128/microbiolspec.MB-0002-2014. MB–0002–2014. [DOI] [PubMed] [Google Scholar]
- van Gestel J, Vlamakis H, Kolter R. From cell differentiation to cell collectives: Bacillus subtilis uses division of labor to migrate. Plos Biol. 2015b;13:e1002141. doi: 10.1371/journal.pbio.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet S, Ackermann M. Bacterial Ventures into Multicellularity: Collectivism through Individuality. Plos Biol. 2015;13:e1002162. doi: 10.1371/journal.pbio.1002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening J-W, Smits WK, Kuipers OP. Bistability, Epigenetics, and Bet-Hedging in Bacteria. Annu Rev Microbiol. 2008a;62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- Veening JW, Stewart EJ, Berngruber TW, Taddei F, Kuipers OP, Hamoen LW. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc Nat Acad Sci Usa. 2008b;105:4393–4398. doi: 10.1073/pnas.0700463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermute EH, Silver PA. Emergent cooperation in microbial metabolism. Mol Syst Biol. 2010;6:407. doi: 10.1038/msb.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JW, Locke JCW, Altinok A, Rosenfeld N, Bacarian T, Swain PS, Mjolsness E, Elowitz MB. Measuring single-cell gene expression dynamics in bacteria using fluorescence time-lapse microscopy. Nat Protoc. 2011;7:80–88. doi: 10.1038/nprot.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslaver A, Bren A, Ronen M, Itzkovitz S, Kikoin I, Shavit S, Liebermeister W, Surette MG, Alon U. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat Methods. 2006;3:623–628. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cell segmentation and tracking data, as well as the used Matlab code, has been deposited to the ETH Data Archive: http://dx.doi.org/10.5905/ethz-1007-77. A full data set containing the unprocessed microscopy data has been deposited to the Zenodo data archive: https://doi.org/10.5281/zenodo.268921.