Abstract
During meiosis, homologous chromosomes of a diploid cell are replicated and, without a second replication, are segregated during two nuclear divisions to produce four haploid cells (including discarded polar bodies in females of many species). Proper segregation of chromosomes at the first division requires in most species that homologous chromosomes be physically connected. Tension generated by connected chromosomes moving to opposite sides of the cell signals proper segregation. In the absence of the required connections, called crossovers, chromosomes often segregate randomly and produce aneuploid gametes and, thus, dead or disabled progeny. To be effective, crossovers must be properly distributed along chromosomes. Crossovers within or too near the centromere interfere with proper segregation; crossovers too near each other can ablate the required tension; and crossovers too concentrated in only one or a few regions would not re-assort most genetic characters important for evolution. Here, we discuss current knowledge of how the optimal distribution of crossovers is achieved in the fission yeast Schizosaccharomyces pombe, with reference to other well-studied species for comparison and illustration of the diversity of biology.
Keywords: meiosis, homologous recombination, DNA break hotspots, DNA break interference, crossover interference, crossover invariance, pericentric repression, Schizosaccharomyces pombe, linear element proteins
Introduction
To reproduce, sexual species alternate between having two copies of each chromosome in each cell to having only one copy of each chromosome. Cells of the former type (diploid) constitute the body of multicellular species, and cells of the latter type (haploid) are the gametes, such as eggs and sperm, that fuse to recreate diploid cells of progeny in the next generation. The process of making haploid cells from the diploid precursor cells is called meiosis and requires unique chromosome dynamics (Figures 1 and 2), which we divide into three stages – chromosome replication, DNA double-strand break (DSB) formation and repair, and chromosome segregation.
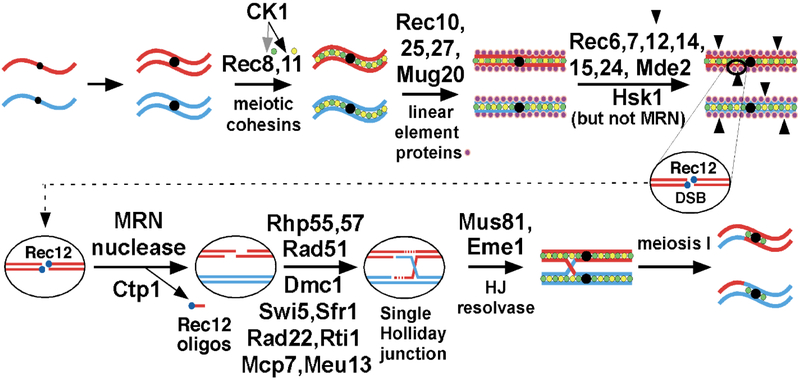
Figure 1. Pathway of meiotic recombination in S. pombe (modified from [93]).
Thick lines indicate the ds DNA of a chromatid, red for the chromosome from one parent and blue from the other parent; black dots indicate the centromeres. In ovals, thin lines indicate a single strand of DNA; dotted red lines indicate newly synthesized DNA. Accompanying replication, sister chromatid cohesins, containing the meiosis-specific subunits Rec8 and Rec11, are loaded onto chromosomes. In chromosomal arms casein kinase I (CK1; Hhp1 and Hhp2) phosphorylates Rec8, for its proteolysis and sister chromatid segregation, and Rec11, for its recruiting Rec10. Rec25, Rec27, and Mug20 direct Rec10 at high frequency to DSB hotspots. Rec10 binds Rec15, which with other indicated proteins activates Rec12 (Spo11 homolog) to make DSBs. The MRN complex and Ctp1 clip off Rec12 covalently bound to 5’ DNA ends and further resect the 5’ ends to produce long 3’-ended ss DNA tails. Rad51 and Dmc1 DNA strand-exchange proteins bind the tails and, with the additional proteins listed, form a displacement- (D-) loop with intact ds DNA. The D-loop (not shown) is cleaved to form a Holliday junction (HJ), which is resolved by the Mus81-Eme1 complex into crossover (shown) or non-crossover (gene conversion; not shown) products. HJ resolution is aided in an unknown way by Nse5 and Nse6, subunits of the Smc5-Smc6 complex [94]. Additional gene products required for meiotic recombination, but whose point of action remains unknown, include the following: mug1 (jnm1), mug5, pds5, rad54, rdh54, and rlp1; hop1 and mek1 [95], nse1 [96], pli1 [97], rec13, rec18, and rec21 [98], and rqh1 [99]. See [93] for references for genes not otherwise referenced here.
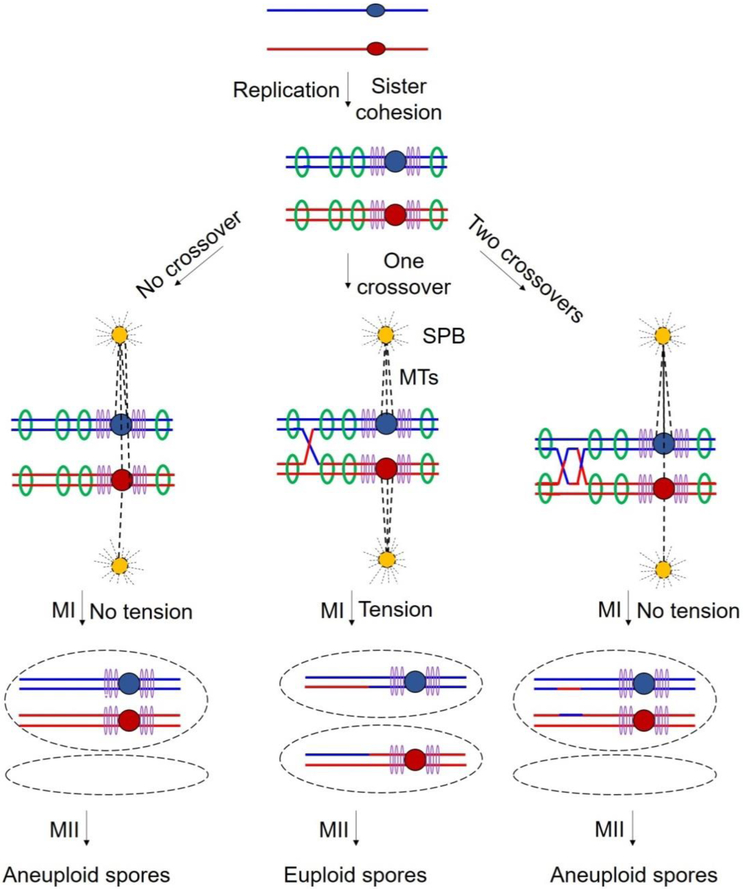
Figure 2. Crossovers are essential for proper segregation.
Each line indicates the ds DNA of a chromatid, red for the chromosome from one parent and blue from the other parent; central dots indicate the centromeres. After replication, sister chromatids of each parent are held together by cohesin complexes deposited at points across the chromosomal arms (green rings) and especially densely at the pericentric regions (purple rings). At MI, paired centromeres of each homolog attach to microtubules (MTs; dashed lines) originating from spindle pole bodies (SPBs) at opposite poles of the cell. Middle panels: Proper reductional division at meiosis I (MI) results from tension generated by centromeres being pulled to opposite poles by MTs and dependent on both cohesion between sister chromatids and crossovers between homologs. Left panels: Without crossovers, no tension is generated when MTs begin to pull the homologs to opposite poles. Right panels: Crossovers too close together may lack intervening cohesion and thus no tension being generated.
At the beginning of meiosis, the chromosomes are replicated to make two copies (sister chromatids) of each chromosome. Each species has a set number of pairs (N) of homologous chromosomes (homologs; one from each parent), so that after replication there are 4N total chromosomes. N varies from one (in a roundworm and an ant) to 630 (in a fern) and is 23 in humans [1-4]; thus, the total number of chromosomes at this point in the life cycle can be as high as 2520, resulting in a complex ballet of meiotic chromosomes. Here, we discuss meiosis in the fission yeast Schizosaccharomyces pombe, for which N is only three, but the general scheme applies to all species regardless of N.
During meiotic replication, the proteins (cohesins) that hold sister chromatids together are modified to promote the special pattern of chromosome segregation and exchange of chromosomal parts (recombination) unique to meiosis (Figures 1 and 2). In S. pombe the meiosis-specific Rec8 subunit of cohesin replaces the mitotic form Rad21; the Rec11 subunit replaces Psc3 but only in the chromosomal arms. The Rec8 and Rec11 cohesin subunits lead to the loading of another set of meiosis-specific proteins that form structures (linear elements, or LinEs) extending across the length of each chromosome [5, 6]. LinEs are like the synaptonemal complex of other species, but LinEs are less robust and appear to fall apart when the nucleus is disrupted: end-to-end structures are seen in live cells but fragments about 10% of the chromosomal lengths are seen in nuclear spreads [7]. Four LinE proteins (Rec10, Rec25, Rec27, and Mug20) have been identified and appear to form a multi-subunit complex non-randomly distributed along the chromosomes.
LinEs activate the formation of DSBs by Rec12 (homolog of Spo11 in other species) and its six partners (Figure 1). Rec12, probably acting as a dimer, remains covalently linked to each 5' end at a DSB. Endonucleolytic cutting by the MRN (Mre11-Rad50-Nbs1) complex acting with Ctp1 removes Rec12 attached to single-stranded (ss) DNA about 10 – 30 nucleotides long; these Rec12-oligos are degraded without serving any currently known function [8-10].
Repair of DSBs continues with further resection by MRN-Ctp1 and perhaps other nucleases, including exonuclease Exo1, to generate long ss DNA with a 3'-end on each side of the DSB. Aided by Rad51 and Dmc1, homologs of the bacterial RecA DNA strand-exchange protein, and other proteins, these ends invade intact double-stranded (ds) DNA and form a joint molecule, a single Holliday junction (HJ) [11]. HJs between sister chromatids are three or four times more frequent than HJs between homologs at DSB hotspots, described below; the opposite preference has been inferred for HJs in DSB-cold regions. In the budding yeast Saccharomyces cerevisiae, double HJs (two HJs roughly 1 kb apart and connecting the same pair of chromatids) are more frequent than single HJs [11]; they are more frequent between homologs than between sister chromatids at the several DSB hotspots tested [12]. The HJ form (single vs. double) and the chromatid preference for repair (intersister vs. interhomolog) in other species are unknown, to our knowledge.
Interhomolog (IH) HJs are resolved by the Mus81-Eme1 complex to form crossovers, in which DNA flanking the DNA strand-exchange region is from opposite parents on the two sides of the resolution point [13, 14]. In other species there are multiple mechanisms for HJ resolution into crossovers [15-19]. Crossovers are the structures important for directing proper chromosome segregation. Joint DNA molecules, including HJs, can also be resolved such that the flanking DNA comes from one chromatid or the other. If the joint is IH, DNA at the joint would contain a single strand from each parent, forming heteroduplex DNA, which can produce localized DNA exchanges, or gene conversions (GCs). GCs, like crossovers, are an important source of genetic variation, which is important for natural selection to propel evolution of the species. Thus, meiotic recombination plays a critical role in gamete formation (an essential role in species with large N), but its occurrence in the wrong place can be highly deleterious, as we describe below.
After DSBs are repaired, the chromosomes begin to segregate by being pulled by microtubules connected at one end to the fused sister centromeres via a huge protein structure, the kinetochore; the other microtubule end is attached to the spindle pole body (SPB; the centrosome in multicellular species) (Figure 2). Uniquely in the first meiotic division, sister centromeres remain connected because Rec8 cohesin in the pericentric region is protected from proteolytic cleavage by the meiosis- and pericentric-specific protein Sgo1 [20, 21]. As the microtubules shorten, by loss of tubulin subunits at the kinetochore attachment point, the connected sister centromeres of each homolog begin to move toward a SPB. If the paired centromeres of the two homologs move toward SPBs at opposite sides of the cell, as appropriate for homolog segregation, tension builds up but only if the homologs are properly connected. Tension requires both a crossover between homologs and cohesion between sister chromatids centromere-distal to the crossovers. Without a crossover or adequate cohesion, tension signaling proper segregation does not occur, and homologs segregate nearly at random. [A few species, such as males of the fruit fly Drosophila melanogaster, have an alternative mechanism directs proper segregation without any crossovers, and some species, including S. pombe, have a partially active system for this purpose [22].] If two crossovers involving the same two chromatids are too close, there is insufficient cohesion between the crossovers to generate the required tension (Figure 2). Thus, crossovers manifest spatial interference in most species. In addition, a crossover too near the centromere may interfere with proper kinetochore orientation. Mechanisms to promote crossover interference and crossover avoidance near the centromere are discussed below. Non-random distribution of crossovers helps assure successful reproduction and evolution of the species.
1. Keeping crossovers uniform
Classical genetic mapping indicates that meiotic crossovers occur across all of the genome (by definition), but comparison of genetic maps with physical (cytological or DNA) maps shows that this is not the case. Certain regions, such as the pericentric regions, have essentially no crossovers, and in certain species some regions have higher densities (crossovers per unit of DNA or microscopic length) than others. Certain mutations or natural variants create hotspots of crossing over or gene conversion [23, 24], indicating that genetic distance is not always proportional to physical distance. The basis for this non-uniformity is two-fold – non-random distribution of DSB formation and variation in partner choice (sister vs. homolog) for DSB repair. But in S. pombe and perhaps other species appropriate partner choice can counter the inherent non-uniformity of DSB hotspots to make a more uniform crossover landscape, as discussed below.
The discovery of DSBs as precursors to crossovers and high-resolution mapping of DSBs greatly advanced understanding of the first basis for crossover distribution. DSBs were first observed and mapped by Southern blot hybridization of DNA extracted from meiotic cells, and DSB hotspots corresponding to gene conversion hotspots were noted [25-29]. Enrichment of DNA covalently linked to Spo11 (Rec12 in S. pombe) and analyzed by microarray hybridization revealed hotspots and intervening cold regions across the genome [30, 31]. Sequencing of the Spo11 (or Rec12)-oligonucleotides released by the MRN-Ctp1 complex (MRX and Sae2 in S. cerevisiae) provided nearly single-nucleotide resolution and reliable distribution of DSBs across the genome in four species to date (Table 1; Figure 3) [32-35].
Table 1.
Chromosomal features of four well-studied speciesa.
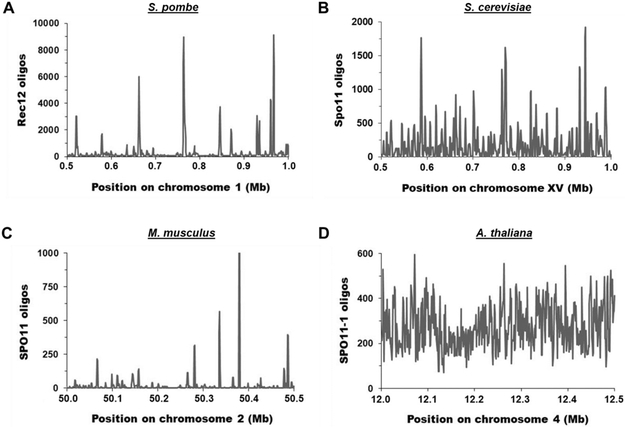
Figure 3. Distribution of Spo11 oligos (DSBs) across representative 0.5 Mb chromosomal arm regions in four species.
The number of Spo11 oligos per 1 kb bin is plotted for each representative genomic region. (A) S. pombe [33]. The mean and median values are 339.9 and 81.5, respectively. (B) S. cerevisiae [32]. The mean and median values are 177.1 and 76.5, respectively. (C) M. musculus [34]. The mean and median values are 19.5 and 2.1, respectively. (D) A. thaliana [35]. The mean and median values are 272.6 and 257.0, respectively.
Even setting aside the ultra-cold regions described below, DSBs are non-uniformly distributed across all genomes analyzed to date, but the degree of non-uniformity appears species-dependent. In S. pombe DSB hotspots range from having ~100 times more DSBs per kb of DNA than the genome median to being barely discernable above the genome median [33]. Considering hotspots ≥2.2-fold hotter than the genome mean, there are 603 hotspots across the 12.5 Mb genome (excluding the ultra-cold pericentric, mating-type, and ribosomal DNA regions, which have <5% as many DSBs/kb as the genome mean). These hotspots range from ~1 to 7 kb wide for strong hotspots and ~0.1 to 1 kb for weak hotspots; the mean hotspot width is 1.4 kb. The intervening cold regions are up to −250 kb wide (21 kb mean). Within these cold regions, DSBs appear approximately uniformly distributed and account for 28% of all DSBs. The three other species for which Spo11-oligo maps have been published appear to have considerable differences in the distribution of DSBs across the genome (Figure 3), in agreement with many other features of meiotic recombination differing among species. Quantitative comparisons, however, are complicated by necessity of arbitrary choice of parameters.
The molecular basis for DSB hotspots is best understood in S. pombe. The LinE proteins Rec25, Rec27, and Mug20 bind DSB hotspots with high specificity and are required for nearly all DSBs at nearly all hotspots [36]. They also bind Rec10, which physically interacts with Rec15, a member of the Rec12 complex that forms DSBs [37, 38]. Thus, the LinE proteins are determinants of DSB hotspots, but other features are also important. Transcription factors and their DNA binding sites are well-known determinants of hotspots, but chromatin structure, including histone modifications, also play a less-understood role [39-42].
The biological basis for different DSB distributions is not obvious; i.e., it is not clear why some species have considerably more uniformity of DSBs across the genome than do others. Many features of meiotic recombination, such as the proteins required, also differ among species, presumably a reflection of the ancient origin of meiotic recombination and its diverse evolution.
Comparison of the distribution of DSB hotspots and the S. pombe genetic map, with known physical distances between markers for assay of recombination, showed, surprisingly, their noncongruence and that a DSB hotspot need not be a crossover hotspot. Crossover frequency, measured as centiMorgans (cM) between markers, is about the same whether or not the interval assayed contains a strong DSB hotspot [43]. A 0.5 Mb region (Figure 3) containing ten genetic markers and six strong DSB hotspots (~10 to 100 times the genome median) was extensively analyzed. The mean crossover frequency of 0.10 cM/kb (range = 0.06 – 0.16 cM/kb) is about the same as that for the whole genome (1700 cM/12.5 Mb = 0.14 cM/kb) [44]. By contrast, a series of ade6 mutations, including hotspots of varying intensity, showed near linearity between DSB frequency and gene conversion frequency [29]. In addition, a DSB hotspot is a crossover hotspot if the markers used are sufficiently close together (~5 kb) [45].
The resolution to these seemingly contradictory results is the following [46]. DSBs at hotspots are repaired about 3 – 4 times more frequently with the sister chromatid than with the homolog. Since DSB repair with the sister does not yield a genetic recombinant, this feature reduces the recombinant frequency expected from the DSB frequency at a hotspot. DSBs in the cold region appear to be repaired preferentially, perhaps exclusively, with the homolog. Thus, in a large genetic interval cold-region DSBs are the major source of crossovers; the near-uniformity of cold-region DSBs leads to near uniformity of crossovers. In a short hotspot-containing interval, repair of hotspot DSBs is the major source of gene conversions, and their frequency is nearly proportional to the DSB frequency [29]. Even though repair of hotspot DSBs is preferentially with the sister, enough hotspot DSBs are repaired with the homolog to give a crossover hotspot if the interval is so short that the contribution of the more uniform cold-region DSBs is minimal.
The molecular basis for partner choice for DSB repair is unknown. Presumably, proteins more frequently bound to hotspots facilitate repair with the sister, or those bound to cold regions facilitate repair with the homolog. The S. pombe LinE proteins, determinants of hotspots, may also dictate partner choice; i.e., preferential repair with the sister. This proposal remains to be tested. The extent of differential partner choice among other species is unclear, but certain S. cerevisiae mutants, such as red1, hop1, and mek1, affect both DSB formation and partner choice [47, 12].
Why do DSB hotspots and countervailing partner choice exist? Early in the evolution of meiosis, mechanisms for DSB formation must have arisen and may have had DNA sequence-dependence, producing DSB hotspots. These would have allowed recombination between the groups of genes flanking a hotspot but not within the groups. Thus, evolution of the species would not have been as rapid as would occur with uniform recombination distribution. Dampening of hotspots, by directing repair with the sister, plus addition of DSBs between hotspots (cold-region DSBs) would have led to a more uniform recombination landscape and more nearly random assortment of alleles. This appears to be the case with present-day S. pombe. S. cerevisiae and the cruciferous plant Arabidopsis thaliana appear to have more uniform DSB distributions (Figure 3) and, consequently, recombination distribution. Other species, such as the mouse Mus musculus, may have not yet solved the problem and may be evolving more slowly (though this is hard to determine).
2. Keeping crossovers out
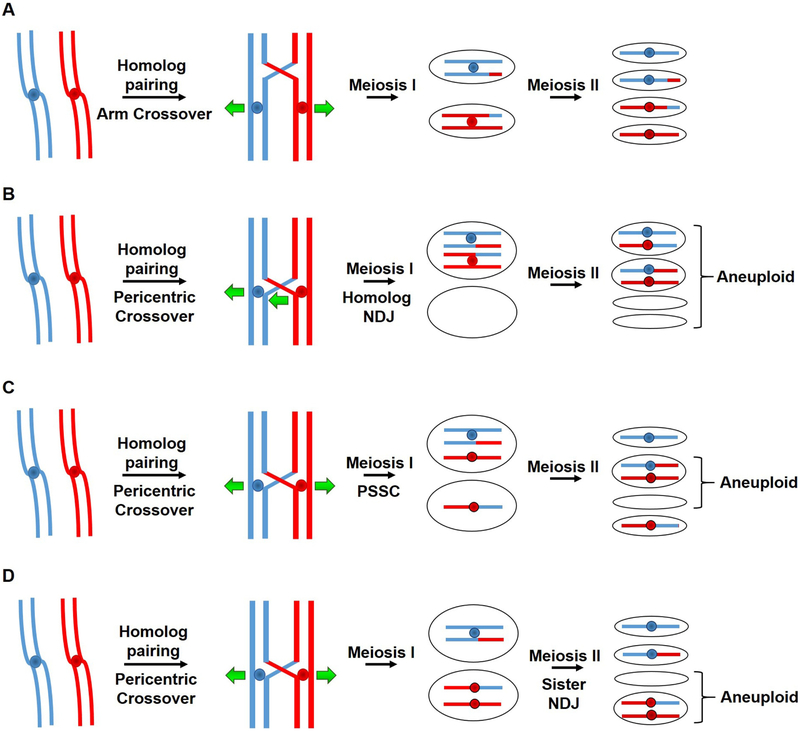
In contrast to maintaining a uniform distribution of crossovers across most of the meiotic chromosomes, active mechanisms exclude crossover formation in certain critical regions of the genome such as centromeres, telomeres, ribosomal DNA and the mating type locus in yeasts. Perhaps most important is repression of recombination near centromeres (pericentric regions). This conserved phenomenon, first reported in the early 1930s, is observed across many highly divergent species including humans [48]. Failure to prevent crossovers at centromeres has been associated with increased chromosome missegregation resulting in aneuploidy and consequent disability or inviability [49, 48]. Missegregation resulting in aneuploid or dead gametes can reflect non-disjunction (NDJ) of homologs at meiosis I [50, 51], precocious separation of sister chromatids (PSSC) at meiosis I, or NDJ of sisters at meiosis II [52] (Figure 4). The mechanism by which crossovers near the centromere cause these effects is unclear, but presumably crossovers constrain the chromosomes and kinetochores from assuming their proper configuration for segregation.
Figure 4. Positioning of crossovers on meiotic chromosomes dictates proper segregation and correct ploidy in gametes.
Each line indicates the ds DNA of a chromatid, red for the chromosome from one parent and blue from the other parent; central dots indicate the centromeres. Crossovers in the chromosomal arms (depicted as red-blue junctions) are needed for accurate chromosomal segregation during meiosis (A) (see Figure 2), but crossovers too close to the centromere are deleterious. Pericentric crossovers can lead to homolog non-disjunction (NDJ) at MI (B), precocious separation of sister chromatids (PSSC) when cohesion is lost between the sister chromatids at MI (C), or sister NDJ at MII (D). These missegregation events lead to aneuploid spores, either nullisomic (lacking a chromosome) or disomic (having an extra chromosome).
Molecular mechanisms of crossover repression at and around the centromeres were recently identified in two distantly related yeasts [53, 51]. Crossovers at such specific loci can be prevented by blocking DSB formation, favoring IS repair over IH repair, or both. In S. cerevisiae, the Ctf19 complex (part of the kinetochore) has an important role in suppressing DSB formation, as ctf19 mutants are ~5-fold derepressed for DSBs in the ~10 kb region around the centromere [53]. However, the increase in recombination frequency in ctf19 mutants is ~20-fold over wild-type, suggesting that DSB suppression is only part of the explanation for pericentric repression; to our knowledge, the mechanism of Ctf19's repression of DSB formation is unknown. The Ctf19 complex directs deposition of cohesins at the pericentromeres, which may bias DSB repair to be with the sister, as frequently occurs in mitotic DSB repair. The observed 20-fold increase in crossovers in Ctf19-component mutants may reflect more frequent DSB repair with the homolog in these mutants. This interpretation is supported by the observation that the rec8 cohesin mutant is derepressed for recombination but not for DSB formation and does not further increase DSB formation when a Ctf19-complex component is disrupted. Thus, in S. cerevisiae, pericentric crossovers are repressed by a combination of infrequent DSB formation and, presumably, biased DSB repair with the sister.
In S. pombe, however, repression of pericentric crossovers is predominantly, perhaps exclusively, controlled at the level of DSB formation. Heterochromatin, a type of condensed chromatin containing specific histone modifications that attract multiple proteins to repress gene expression, is abundant in pericentric regions and plays a vital role in meiotic crossover repression. Mutants altered in heterochromatin formation, such as clr4Δ or rik1Δ, which lack H3K9 methylation essential for heterochromatin, are highly derepressed for both pericentric DSBs and recombination (>100-fold) over the 35 – 100 kb pericentric regions [54]. Mutants lacking the downstream effector proteins, such as Swi6 or Chp2 (heterochromatin protein HP1 homologs), however, behave like wild type. Similarly, in D. melanogaster, Su(var) mutants lacking both Clr4 and HP1 homologs are modestly but significantly derepressed for pericentric crossovers (1.8 cM in wt vs. 2.5 cM in the double mutant; p < 0.01) and have ~2-fold more pericentric γH2Av foci than wt, presumably reflecting increased DSBs [55, 56]. Interestingly, non-crossovers are not susceptible to pericentric repression in D. melanogaster [57] and provide a mechanism other than IS exchange to repair potentially harmful pericentric DSBs. Mutants of A. thaliana lacking histone H3K9 or non-CG DNA methylation are also derepressed for meiotic pericentric DSBs (~1.2-fold) and recombination (~1.5-fold) [35, 58]. Thus, heterochromatin appears to be a wide-spread repressor of pericentric recombination, albeit with quantitative effects highly variable among species.
In S. pombe, pericentric recombination in clr4Δ depends on the meiosis-specific cohesin subunits Rec8 and Rec11 and the LinE protein Rec10 [54]. In the absence of Clr4, and hence methylated H3K9, the pericentric regions behave like chromosomal arms – recombination in both regions depends on all the tested proteins in the meiotic recombination pathway, including Rec8, Rec10, and Rec11 (Figure 1). Heterochromatin protein Swi6 is needed to deposit cohesins in the pericentric region in S. pombe [59], and it preferentially deposits cohesin complexes containing Rec8-Psc3, which does not promote DSB formation (Figure 5). In contrast, the chromosomal arms are bound by molecularly distinct cohesin complexes (containing Rec8-Rec11) that, via Rec10, activate the Rec12 complex to form DSBs (Figures 1 and 5). Hence, absence of Rec10 at the pericentric regions is a rate-limiting step that represses meiotic crossovers, which can be bypassed by forced Rec10 tethering to the pericentromeres [51]. Replacing the pericentromere-specific cohesin complex with that in the arms also successfully removes pericentric repression and allows robust recombination in that region. Therefore, the crucial role of cohesins in the repression of harmful pericentric crossovers appears to be a common underlying factor across both divergent yeast species, though by distinctly different molecular mechanisms. Since the S. pombe, but not S. cerevisiae, pericentric regions contain DNA repeats, IS repair in S. pombe could put the stability of the repeats at risk due to unequal sister chromatid exchanges, thereby warranting complete inhibition of DSB formation.
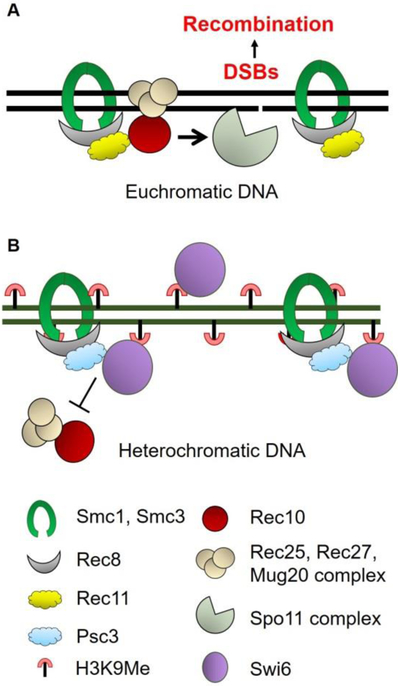
Figure 5. Composition of the cohesin complexes differentially controls distribution of DSB formation and recombination.
Each solid line indicates the ds DNA of a chromatid. In euchromatic DNA present largely in chromosomal arms, the meiosis-specific Rec8 and Rec11 subunits are part of the cohesin complex (with Smc1-Smc3). Rec11 activates Spo11- (Rec12)-dependent DSB formation by recruiting the LinE protein Rec10 (Figure 1). Other LinE proteins Rec25, Rec27 and Mug20 are recruited with Rec10 at DSB hotspots. Rec10 activates the Spo11 complex to induce DSB formation. In contrast, in heterochromatic DNA, especially at pericentric regions, the cohesin complex contains Rec8-Psc3, which does not bind Rec10 or activate DSB formation. Loading of Rec8-Psc3 cohesin complex is mediated by the HP1 homolog Swi6, which specifically binds to H3K9 methylated histones present in heterochromatin. Note that Swi6 has both a negative role (recruiting Psc3) and a potentially positive role (recruiting Rec8 via Psc3 recruitment); thus, swi6 mutants are not derepressed for pericentric recombination. The scheme shown is for S. pombe and is likely conserved even in mammals (see text).
The mechanism of pericentric repression in S. pombe is likely conserved even in mammals, which contain complex heterochromatic pericentromeres and a meiosis-specific homolog (STAG3) of Rec11 (apparently absent in S. cerevisiae). Localization of STAG3 preferentially to chromosomal arms appears to be similar in both S. pombe and mouse spermatocytes; like Rec11, STAG3 is phosphorylated during meiotic recombination and is required for fertility [60, 61]. Thus, blocking the first step of the pathway leading to meiotic DSBs and crossovers may be a common way of preventing harmful pericentric crossovers.
Other chromosomal regions with repetitive DNA containing heterochromatin are also at risk from recombination, which can produce deletions, duplications, translocations, and other rearrangements. These changes can adversely affect the functions of repetitive DNA. For example, the mating-type locus in S. pombe contains two silent gene copies of mat, flanked by homologous repeats and separated by the ~11 kb K region in which meiotic recombination is undetectable [62]. This region contains a ~4.3 kb segment that is homologous to part of the pericentromeric outer repeat sequences [63]. Removing the Clr4 partner protein Rik1 or Swi6, both of which are needed for formation of heterochromatin, allows abundant recombination in the K region [64, 65]. This suggests a conserved mechanism for repressing crossovers at this locus similar to that in pericentric regions, since the proteins known to bind to methylated H3K9 regions are similar.
DSB densities near telomeres are lower than the genome-wide average, although estimation of crossover density is challenging due to limited availability of markers. (As noted in Section 1, DSB frequencies and crossover frequencies are not always proportional.) In S. cerevisiae, Spo11-oligos, reflecting DSBs, are 3.5-times less frequent within 20 kb of telomeres [32]. This estimate is supported by analysis of single-stranded DNA intermediates from DSB sites in dmc1 mutant cells, which exhibit a 2 – 3 fold reduction in number of DSB hotspots within the same telomere-proximal intervals [66]. Similarly, in S. pombe, at least three of the four chromosomal ends analyzed (Chromosomes I and II) lack prominent DSB hotspots in the terminal 70 – 150 kb; a major DSB peak does occur, however, ~18 kb from the right end of Chromosome II [36]. In S. cerevisiae increased levels of DSBs were observed within 10 kb of the telomeres in sir2 mutants, which lack histone H4K16 deacetylase activity [67]. Increased acetylation in the absence of Sir2 may make the chromatin more “open” and conducive for Spo11 binding and activity. Overall, there is a reduction in DSB levels in the immediate vicinity of telomeres, and this could be correlated to the repression of crossovers as well.
In most species, rDNA genes are also tandemly repeated, and a fraction of the repeats are organized into heterochromatin. In S. cerevisiae the repressive chromatin mainly contains deacetylated histones, which are nucleated and spread by Sir1 – Sir4 proteins; in S. pombe, other fungi, plants and animals, rDNA contains methylated H3K9 histones, marks of heterochromatin [68, 69]. In S. cerevisiae meiotic recombination in rDNA gene clusters is repressed up to ~100-fold relative to other chromosomal intervals of similar size [70]. Mapping of DSBs in the rDNA regions in S. cerevisiae also confirmed the absence of strong hotspots in the ~1 Mb rDNA cluster region [66]. Similarly, in S. pombe, Rec12-oligos from rDNA are >100-times less frequent (per kb) than the genome mean [33]. This is consistent with the Spo11-oligo map for rDNA reported in S. cerevisiae (75-times less than the genome mean) [32]. Interestingly, in D. melanogaster at the rDNA region no significant accumulation of γ-H2av foci, a sign of DSBs, could be seen in the oocytes lacking the H3K9 methyltransferase SU(VAR)3-9 (Clr4 homolog) [56]. DSBs generated in this region may be repaired quickly and efficiently, or DSBs may not be generated if removal of SU(VAR)3-9 is not sufficient to modify the heterochromatin specifically at the rDNA, as it does at the centromeres (see above). Increased intrachromosomal rDNA recombination (up to 10-fold) occurs in S. cerevisiae sir2 mutants [71], which also have elevated DSB levels both within the rDNA and in genes flanking −150 kb of the rDNA cluster [67]. Absence of Pch2, a meiosis-specific protein with ATPase activity that localizes to the nucleolar organizer region harboring the rRNA genes, increases rDNA recombination 15-fold [72]. Pch2 may exclude the meiosis-specific protein Hop1 from the nucleolus, since Hop1 is observed in the nucleolus in pch2 and sir2 mutants but not in wild type. Redistribution of Hop1, which is required for homolog pairing during meiosis, efficient DSB formation, and crossing over [73-75], would then enable IH meiotic recombination in the rDNA.
Multiple mechanisms prevent the recruitment of pro-recombinogenic factors at heterochromatic loci. The most wide-spread mechanisms prevent DSB formation, the first fully committed step in homologous recombination. In S. pombe the earliest acting protein, Rec11, is blocked from recruitment to the pericentric region [51]. It is noteworthy that the process is blocked at the earliest step unique to recombination, as is common in intermediary metabolism. In one case (S. cerevisiae pericentric recombination) reducing IH repair during meiosis has been proposed to further lower pericentric crossover frequency. Loss of these regulatory pathways important for controlling crossover positioning can have detrimental effects on the outcome of meiosis and result in congenital birth defects and other genetic disorders.
3. Keeping crossovers apart
In addition to uniform distribution and region-specific exclusion of crossovers, the distance between two crossovers is also carefully regulated. As discussed above, both crossovers and sister chromatid cohesion are needed to generate tension between homologous chromosomes to ensure proper chromosome segregation in the first meiotic division. However, when crossovers are too close to each other, there may be insufficient cohesin between them (Figure 2, right panel) and hence not enough tension between segregating homologs, resulting in missegregation. Crossover interference, the occurrence of one crossover reducing the frequency of a second crossover nearby, reduces crossovers that are too close and thus detrimental.
Crossover interference was first reported in D. melanogaster by Sturtevant [76], who found there were often fewer double crossovers between three linked genetic markers than expected from independent occurrence of crossovers. He defined crossover interference as
where RD is the frequency of double events, and R1 and R2 are the frequencies of the individual events. I equals zero when the observed double crossover frequency is that expected from independence, indicating no interference. I equals 1 when no double crossovers are observed, indicating complete interference. I is negative when there are more double crossovers than expected, indicating coordinated crossover formation.
Crossover interference is reported in most model organisms but with different maximal strengths and appears in at least two species to be related to genetic distance rather than physical distance (DNA length). The roundworm Caenorhabditis elegans shows complete crossover interference - no double crossovers are observed on individual chromosomes (~50 cM or ~15 – 20 Mb) [77]. D. melanogaster and the bread mold Neurospora crassa both show different strengths of crossover interference in different size intervals [78]. For short intervals interference can be nearly complete (I ~1) in D. melanogaster and very strong (I ~0.8) in N. crassa [78]. Interference steadily decreases as interval sizes increase to ~35 – 50 cM (~15 – 20 Mb in D. melanogaster and ~0.6 – 0.9 Mb in N. crassa) [78]. In S. cerevisiae interference is detectable in intervals as long as −45 cM (−0.1 Mb) [79, 80], but there does not appear to be a simple relation between I and interval size; there may be chromosomal regional control of interference, but this has not to our knowledge been demonstrated. S. pombe has been reported to have no crossover interference from analysis of limited numbers of tetrads [81]. But more extensive analysis of random spores showed that S. pombe has weak but positive crossover interference (I = 0.26 ± 0.051) for the two short intervals (−11 cM or 69 kb) assayed [82].
Several models for crossover interference have been proposed, but the molecular basis of most of these models is unstated. King and Mortimer [83] proposed a “polymerization model” in which an unspecified factor polymerizes to each side of a crossover and ejects nearby crossover precursors. Foss et al. [78] proposed a “counting model” in which a crossover is made and then a set number of non-crossovers are made before another crossover is made; subsequent experiments disproved this model for S. cerevisiae [84]. Fujitani et al. [85] proposed a “diffusion model" in which a crossover precursor moves randomly along a chromosome before forming a crossover; it is inactivated, however, when it encounters another crossover precursor or a crossover. Kleckner et al. [86] proposed a “beam-film model” in which chromosomal stress is required to form a crossover, but formation of a crossover releases the stress locally, thereby preventing a second crossover nearby. Hultén [87] proposed a “chromosome oscillatory movement” model in which chromosomes, moved by their connections to the nuclear envelope and the kinetochores, adopt wave-like forms; crossovers form only at "nodes" at which homologs contact each other.
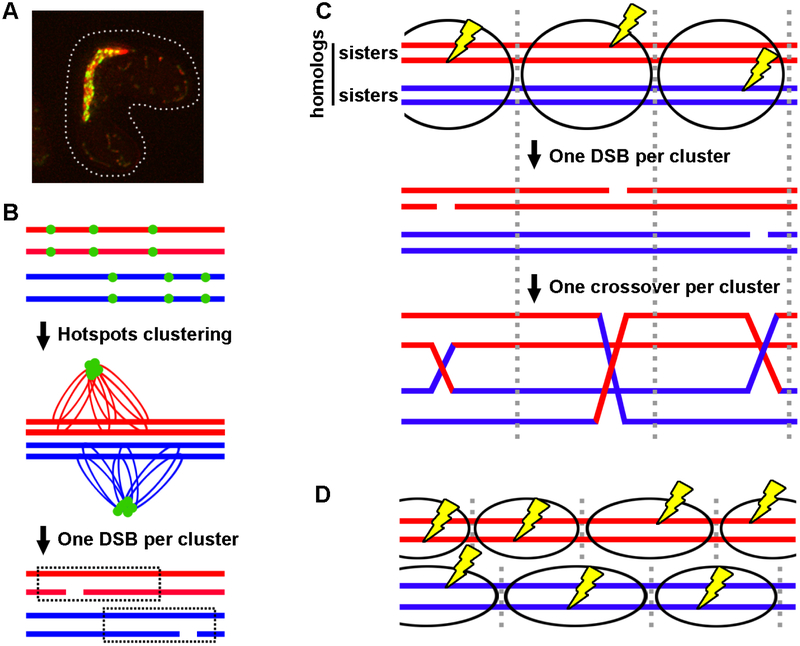
A new crossover interference model – the clustering model – was inspired by molecular and genetic findings in S. pombe (Figure 6) [82]. In this model, the linear element (LinE) proteins, which are DSB hotspots determinants [36], bind hotspots and form clusters of hotspots in −200 kb intervals; only one DSB is formed per cluster. Thus, DSB interference leads to crossover interference over −200 kb (−35 cM) regions. Clustering over only one homolog (two sister chromatids) allows independent DSBs on the two homologs, thereby accounting for relatively weak crossover interference in S. pombe; clustering over both homologs accounts for strong interference in other species (Figure 6C and 6D).
Figure 6. A molecular model for crossover interference – the clustering model.
(A) Live cell image of LinE foci at the "horsetail" stage of S. pombe meiosis. Multiple LinEs (represented by Rec25-GFP) concentrate and form linear or dot-like foci. DNA stained by Hoechst 33342 is red, and Rec25-GFP is green; overlap is yellow. (B-C) Model of crossover interference based on hotspot clustering and DSB interference. Each horizontal line is one sister chromatid (ds DNA molecule). Ovals indicate clusters, within which one DSB (yellow lightning bolt) occurs. (B) DSB hotspots are bound by LinEs (green dots) and form clusters of nearby hotspots on sister chromatids. Only one DSB and hence one crossover are formed per cluster. Independent cluster and DSB formation on separate homologs allows close double crossovers and thus low but positive crossover interference. (C) Clusters encompass all four chromatids in species with strong crossover interference. (D) Clusters encompass only two sister chromatids (one homolog) in species with weak crossover interference. Figure modified from [82].
The clustering model is supported by molecular and genetic evidence in S. pombe. LinE proteins (Rec25, Rec27, and Mug20) bind DSB hotspots and are required for DSB formation at most hotspots [36]. Genome-wide chromosome conformation capture (Hi-C) analysis coupled with immunoprecipitation (ChIA-PET) showed that LinE-bound DSB hotspots preferentially interact within −200 kb regions [82]. Cohesin, but not meiotic DSB formation, is needed for both LinE protein binding to hotspots and cluster formation, indicating that clusters are formed before DSB formation. DSB interference is strong for short intervals (~15 kb) but becomes negligible at −250 kb. In a tel1Δ mutant, lacking the ATM DNA damage-responsive protein kinase homolog, DSB interference is negative over the same range, and crossover interference is negative. Thus, Tel1 may be activated by the first DSB made in a cluster and prevent further DSB formation by phosphorylation of one or more components of the DSB-forming complex. DSB competition, the reduction of DSB formation at a hotspot by introduction of another hotspot nearby, extends over ~200 kb but only in cis (i.e., on one homolog), supporting the proposal that clusters form over only one homolog.
There are related observations in S. cerevisiae. DSB interference is positive in regions up to −150 kb but is negative over short regions (<10 kb) in tel1Δ mutants [88]. Crossover interference is reduced but not eliminated in tel1Δ mutants [89]. The S. cerevisiae "loop-cluster" model proposes that only one DSB is made per chromatin loop formed by chromosomal axis proteins [90]. DSB (or recombination) competition occurs in cis and in trans in S. cerevisiae [91, 92]; clusters, if they occur, may encompass both homologs and thereby give rise to stronger crossover interference than in S. pombe.
How DSB hotspot clusters are formed is not known. Presumably, some "machine" moves along the chromosome and forms clusters over a finite region, but the identity of the machine and what limits its action to a finite region are unknown. In S. pombe, the cohesin complex is needed for LinE protein binding to most DSB hotspots and formation of most DSBs and crossovers [36]; it may limit the region over which clusters form.
How do some species, such as S. pombe, show weaker crossover interference than others, such as D. melanogaster? Two possible explanations are based on molecular evidence from S. pombe. First, DSBs outside hotspots appear to be LinE-independent and thus may not cluster or manifest interference. Second, cluster formation involving only one homolog, not both homologs, accounts for DSB competition in cis but not in trans, as noted above, and would allow independent DSBs on homologs and thus some close double crossovers [82]. Strong crossover interference would result if all DSBs occur at hotspots clustered on both homologs (Figure 6C). S. pombe, with a high number of crossovers per chromosome (10 – 20), would have adequate cohesion and tension for proper chromosome segregation and high viability even with low crossover interference. Species with few crossovers per chromosome may need crossover interference to ensure adequate cohesion for proper segregation and fertility.
Acknowledgments
We thank Kyle Fowler, Ian Henderson, and Andrew Tock for generously providing data files; Sue Amundsen and Randy Hyppa for helpful comments on the manuscript; and Eric Alani, Scott Keeney, and Jeff Sekelsky for helpful discussions. Our research is supported by MIRA grant R35 GM118120 from the National Institutes of Health of the United States of America.
Abbreviations:
- ds
double-stranded
- ss
single-stranded
- DSB
DNA ds break
- HJ
Holliday junction
- IS
intersister
- IH
interhomolog
- LinE
linear element
- SPB
spindle pole body
- cM
centiMorgan
- PSSC
precocious separation of sister chromatids
- NDJ
non-disjunction
- MI
first meiotic nuclear division
- MII
second meiotic nuclear division
- MT
microtubules
- I
interference
- kb
kilobase pairs
- Mb
megabase pairs
- MRN
Mre11-Rad50-Nbs1 complex
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare no conflict of interest.
References
- [1].Powsner ER, Berman L, Number of chromosomes in the human cell, Nature, 188 (1960) 1045–1046. [DOI] [PubMed] [Google Scholar]
- [2].Sinha B, Srivastava D, Jha J, Occurrence of various cytotypes of ophioglossum reticulatum L. in a population from N. E. India, Caryologia, 32 (1979) 135–146. [Google Scholar]
- [3].Goday C, Pimpinelli S, Chromosome organization and heterochromatin elimination in parascaris, Science, 224 (1984) 411–413. [DOI] [PubMed] [Google Scholar]
- [4].Crosland MW, Crazier RH, Myrmecia pilosula, an ant with only one pair of chromosomes, Science, 231 (1986) 1278. [DOI] [PubMed] [Google Scholar]
- [5].Davis L, Rozalén AE, Moreno S, Smith GR, Martin-Castellanos C, Rec25 and Rec27, novel components of meiotic linear elements, link cohesin to DNA breakage and recombination in fission yeast, Current Biology, 18 (2008) 849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ding DG, Matsuda A, Okamasa K, Nagahama Y, Haraguchi T, Hiraoka Y, Meiotic cohesin-based chromosome structure is essential for homologous chromosome pairing in Schizosaccharomyces pombe, Chromosoma, 125 (2016) 205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bähler J, Wyler T, Loidl J, Kohli J, Unusual nuclear structures in meiotic prophase of fission yeast: a cytological analysis, J. Cell Biol, 121 (1993) 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Milman N, Higuchi E, Smith GR, Meiotic DNA double-strand break repair requires two nucleases, MRN and Ctp1, to produce a single size class of Rec12 (Spo11)-oligonucleotide complexes, Mol Cell Biol, 29 (2009) 5998–6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rothenberg M, Kohli J, Ludin K, Ctp1 and the MRN-complex are required for endonucleolytic Rec12 removal with release of a single class of oligonucleotides in fission yeast, PLoS Genet, 5 (2009) e1000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ma L, Milman N, Nambiar M, Smith GR, Two separable functions of Ctp1 in the early steps of meiotic DNA double-strand break repair, Nucl. Acids Res, 43 (2015) 7349–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR, Single Holliday junctions are intermediates of meiotic recombination, Cell, 127 (2006) 1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Humphryes N, Hochwagen A, A non-sister act: recombination template choice during meiosis, Experimental cell research, 329 (2014) 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Boddy MN, Gaillard P-HL, McDonald WH, Shanahan P, Yates JR, Russell P, Mus81-Eme1 are essential components of a Holliday junction resolvase, Cell, 107 (2001) 537–548. [DOI] [PubMed] [Google Scholar]
- [14].Smith GR, Boddy MN, Shanahan P, Russell P, Fission yeast Mus81•Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion, Genetics, 165 (2003) 2289–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].de los Santos T, Hunter N, Lee C, Larkin B, Loidl J, Hollingsworth NM, The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast, Genetics, 164 (2003) 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jessop L, Lichten M, Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis, Mol. Cell, 31 (2008) 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zakharyevich K, Tang S, Ma Y, Hunter N, Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase, Cell, 149 (2012) 334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Saito TT, Lui DY, Kim HM, Meyer K, Colaiacovo MP, Interplay between structure-specific endonucleases for crossover control during Caenorhabditis elegans meiosis, PLoS Genet, 9 (2013) e1003586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hatkevich T, Kohl KP, McMahan S, Hartmann MA, Williams AM, Sekelsky J, Bloom syndrome helicase promotes meiotic crossover patterning and homolog disjunction, Curr Biol, 27 (2017) 96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kitajima TS, Kawashima SA, Watanabe Y, The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis, Nature, 427 (2004) 510–517. [DOI] [PubMed] [Google Scholar]
- [21].Ishiguro T, Tanaka K, Sakuno T, Watanabe Y, Shugoshin-PP2A counteracts casein-kinase-1-dependent cleavage of Rec8 by separase, Nat Cell Biol, 12 (2010) 500–506. [DOI] [PubMed] [Google Scholar]
- [22].Davis L, Smith GR, Non-random homolog segregation at meiosis I in Schizosaccharomyces pombe mutants lacking recombination, Genetics, 163 (2003) 857–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Angel T, Austin B, Catcheside DG, Regulation of recombination at the his-3 locus in Neurospora crassa, Aust. J. Biol. Sci, 23 (1970) 1129–1240. [DOI] [PubMed] [Google Scholar]
- [24].Gutz H, Site specific induction of gene conversion in Schizosaccharomyces pombe, Genetics, 69 (1971) 317–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Game JC, Sitney KC, Cook VE, Mortimer RK, Use of a ring chromosome and pulsed-field gels to study interhomolog recombination, double-strand DNA breaks and sister-chromatid exchange in yeast, Genetics, 123 (1989) 695–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sun H, Treco D, Schultes NP, Szostak JW, Double-strand breaks at an initiation site for meiotic gene conversion, Nature, 338 (1989) 87–90. [DOI] [PubMed] [Google Scholar]
- [27].Cao L, Alani E, Kleckner N, A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae, Cell, 61 (1990) 1089–1101. [DOI] [PubMed] [Google Scholar]
- [28].Cervantes MD, Farah JA, Smith GR, Meiotic DNA breaks associated with recombination in S. pombe, Mol. Cell, 5 (2000) 883–888. [DOI] [PubMed] [Google Scholar]
- [29].Steiner WW, Schreckhise RW, Smith GR, Meiotic DNA breaks at the S. pombe recombination hotspot M26, Mol. Cell, 9 (2002) 847–855. [DOI] [PubMed] [Google Scholar]
- [30].Gerton JL, DeRisi J, Shroff R, Lichten M, Brown PO, Petes TD, Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae, Proc. Natl. Acad. Sci. USA, 97 (2000) 11383–11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cromie GA, Hyppa RW, Cam HE, Farah JA, Grewal SHIS, Smith GR, A discrete class of intergenic DNA dictates meiotic DNA break hotspots in fission yeast, PLoS Genetics 3(2007) e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, Tischfield SE, Zhu X, Neale MJ, Jasin M, Socci ND, Hochwagen A, Keeney S, A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation, Cell, 144 (2011) 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Fowler KR, Sasaki M, Milman N, Keeney S, Smith GR, Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome., Genome Research, 24 (2014) 1650–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lange J, Yamada S, Tischfield SE, Pan J, Kim S, Zhu X, Socci ND, Jasin M, Keeney S, The landscape of mouse meiotic double-strand break formation, processing, and repair, Cell, 167 (2016) 695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Choi K, Zhao X, Tock AJ, Lambing C, Underwood CJ, Hardcastle TJ, Serra H, Kim J, Cho HS, Kim J, Ziolkowski PA, Yelina NE, Hwang I, Martienssen RA, Henderson IR, Nucleosomes and DNA methylation shape meiotic DSB frequency in Arabidopsis thaliana transposons and gene regulatory regions, Genome Res, 28 (2018) 532–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Fowler KR, Gutiérrez-Velasco S, Martín-Castellanos C, Smith GR, Protein determinants of meiotic DNA break hotspots, Mol. Cell, 49 (2013) 983–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Estreicher A, Lorenz A, Loidl J, Mug20, a novel protein associated with linear elements in fission yeast meiosis, Current Genetics, 58 (2012) 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Miyoshi T, Ito M, Kugou K, Yamada S, Furuichi M, Oda A, Yamada T, Hirota K, Masai H, Ohta K, A central coupler for recombination initiation linking chromosome architecture to S phase checkpoint, Mol. Cell, 47 (2012) 722–733. [DOI] [PubMed] [Google Scholar]
- [39].White MA, Dominska M, Petes TD, Transcription factors are required for the meiotic recombination hotspot at the HIS4 locus in Saccharomyces cerevisiae, Proc. Natl. Acad. Sci. USA, 90 (1993) 6621–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kon N, Krawchuk MD, Warren BG, Smith GR, Wahls WP, Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe, Proc. Natl. Acad. Sci. USA, 94 (1997) 13756–13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hirota K, Mizuno K, Shibata T, Ohta K, Distinct chromatin modulators regulate the formation of accessible and repressive chromatin at the fission yeast recombination hotspot ade6-M26, Mol Biol Cell, 19 (2008) 1162–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B, PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice, Science, 327 (2010) 836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Young JA, Schreckhise RW, Steiner WW, Smith GR, Meiotic recombination remote from prominent DNA break sites in S. pombe, Mol. Cell, 9 (2002) 253–263. [DOI] [PubMed] [Google Scholar]
- [44].Egel R, Fission yeast in general genetics, in: Egel R (Ed.) The molecular biology of Schizosaccharomyces pombe: Genetics, Genomics and Beyond, Springer, Berlin, 2003, pp. 1–12. [Google Scholar]
- [45].Cromie GA, Rubio CA, Hyppa RW, Smith GR, A natural meiotic DNA break site in Schizosaccharomyces pombe is a hotspot of gene conversion, highly associated with crossing over, Genetics, 169 (2005) 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hyppa RW, Smith GR, Crossover invariance determined by partner choice for meiotic DNA break repair, Cell, 142 (2010) 243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kim KP, Weiner BM, Zhang L, Jordan A, Dekker J, Kleckner N, Sister cohesion and structural axis components mediate homolog bias of meiotic recombination, Cell, 143 (2010) 924–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Nambiar M, Smith GR, Repression of harmful meiotic recombination in centromeric regions, Seminars in Cell and Developmental Biology, 54 (2016) 188–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hassold T, Hunt P, To err (meiotically) is human: the genesis of human aneuploidy, Nat Rev Genet, 2 (2001) 280–291. [DOI] [PubMed] [Google Scholar]
- [50].Koehler KE, Hawley RS, Sherman S, Hassold T, Recombination and nondisjunction in humans and flies, Human Molecular Genetics, 5 (1996) 1495–1504. [DOI] [PubMed] [Google Scholar]
- [51].Nambiar M, Smith GR, Pericentromere-specific cohesin complex Ppevents meiotic pericentric DNA double-strand breaks and lethal crossovers, Mol Cell, 71 (2018) 540–553 e544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Rockmill B, Voelkel-Meiman K, Roeder GS, Centromere-proximal crossovers are associated with precocious separation of sister chromatids during meiosis in Saccharomyces cerevisiae, Genetics, 174 (2006) 1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Vincenten N, Kuhl LM, Lam I, Oke A, Kerr AR, Hochwagen A, Fung J, Keeney S, Vader G, Marston AL, The kinetochore prevents centromere-proximal crossover recombination during meiosis, eLife, 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ellermeier C, Higuchi EC, Phadnis N, Holm L, Geelhood JL, Thon G, Smith GR, RNAi and heterochromatin repress centromeric meiotic recombination, Proc. Natl. Acad. Sci. USA, 107 (2010) 8701–8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Westphal T, Reuter G, Recombinogenic effects of suppressors of position-effect variegation in Drosophila, Genetics, 160 (2002) 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Peng JC, Karpen GH, Heterochromatic genome stability requires regulators of histone H3 K9 methylation, PLoS Genet, 5 (2009) e1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Miller DE, Smith CB, Kazemi NY, Cockrell AJ, Arvanitakas AV, Blumenstiel JP, Jaspersen SL, Hawley RS, Whole-genome analysis of individual meiotic events in Drosophila melanogaster reveals that noncrossover gene conversions are insensitive to interference and the centromere effect, Genetics, 203 (2016) 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Underwood CJ, Choi K, Lambing C, Zhao X, Serra H, Borges F, Simorowski J, Ernst E, Jacob Y, Henderson IR, Martienssen RA, Epigenetic activation of meiotic recombination near Arabidopsis thaliana centromeres via loss of H3K9me2 and non-CG DNA methylation, Genome Res, 28 (2018) 519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y, Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast, Nat. Cell Biol, 4 (2002) 89–93. [DOI] [PubMed] [Google Scholar]
- [60].Prieto I, Suja JA, Pezzi N, Kremer L, Martinez AC, Rufas JS, Barbero JL, Mammalian STAG3 is a cohesin specific to sister chromatid arms in meiosis I, Nat. Cell Biol, 3 (2001) 761–766. [DOI] [PubMed] [Google Scholar]
- [61].Fukuda T, Pratto F, Schimenti JC, Turner JM, Camerini-Otero RD, Hoog C, Phosphorylation of chromosome core components may serve as axis marks for the status of chromosomal events during mammalian meiosis, PLoS Genet, 8 (2012) e1002485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Egel R, Two tightly linked silent cassettes in the mating-type region of Schizosaccharomyces pombe, Current Genetics, 8 (1984) 199–203. [DOI] [PubMed] [Google Scholar]
- [63].Grewal SI, Klar AJ, A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast, Genetics, 146 (1997) 1221–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Egel R, Willer M, Nielsen O, Unblocking of meiotic crossing-over between the silent mating-type cassettes of fission yeast, conditioned by the recessive, pleiotropic mutant rik1, Current Genetics, 15 (1989) 407–410. [Google Scholar]
- [65].Lorentz A, Heim L, Schmidt H, The switching gene swi6 affects recombination and gene expression in the mating-type region of Schizosaccharomyces pombe, Molecular and General Genetics, 233 (1992) 436–442. [DOI] [PubMed] [Google Scholar]
- [66].Blitzblau HG, Bell GW, Rodriguez J, Bell SP, Hochwagen A, Mapping of meiotic single-stranded DNA reveals double-stranded-break hotspots near centromeres and telomeres, Current Biology, 17 (2007) 2003–2012. [DOI] [PubMed] [Google Scholar]
- [67].Mieczkowski PA, Dominska M, Buck MJ, Lieb JD, Petes TD, Loss of a histone deacetylase dramatically alters the genomic distribution of Spo11p-catalyzed DNA breaks in Saccharomyces cerevisiae, Proc Natl Acad Sci U S A, 104 (2007) 3955–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Grewal SI, Rice JC, Regulation of heterochromatin by histone methylation and small RNAs, Curr Opin Cell Biol, 16 (2004) 230–238. [DOI] [PubMed] [Google Scholar]
- [69].Hickman MA, Froyd CA, Rusche LN, Reinventing heterochromatin in budding yeasts: Sir2 and the origin recognition complex take center stage, Eukaryot Cell, 10 (2011) 1183–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Petes TD, Botstein D, Simple Mendelian inheritance of the reiterated ribosomal DNA of yeast, Proc Natl Acad Sci U S A, 74 (1977) 5091–5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Gottlieb S, Esposito RE, A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA, Cell, 56 (1989) 771–776. [DOI] [PubMed] [Google Scholar]
- [72].San-Segundo PA, Roeder GS, Pch2 links chromatin silencing to meiotic checkpoint control, Cell, 97 (1999) 313–324. [DOI] [PubMed] [Google Scholar]
- [73].Hollingsworth NM, Byers B, HOP1: a yeast meiotic pairing gene, Genetics, 121 (1989) 445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Hollingsworth NM, Goetsch L, Byers B, The HOP1 gene encodes a meiosis-specific component of yeast chromosomes, Cell, 61 (1990) 73–84. [DOI] [PubMed] [Google Scholar]
- [75].Mao-Draayer Y, Galbraith AM, Pittman DL, Cool M, Malone RE, Analysis of meiotic recombination pathways in the yeast Saccharomyces cerevisiae, Genetics, 144 (1996) 71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Sturtevant AH, The behavior of the chromosomes as studied through linkage, Z. Indukt. Abstammungs. Vererbungsl, 13 (1915) 234–287. [Google Scholar]
- [77].Meneely PM, Farago AF, Kauffman TM, Crossover distribution and high interference for both the X chromosome and an autosome during oogenesis and spermatogenesis in Caenorhabditis elegans, Genetics, 162 (2002) 1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Foss E, Lande R, Stahl FW, Steinberg CM, Chiasma interference as a function of genetic distance, Genetics, 133 (1993) 681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Shinohara M, Sakai K, Shinohara A, Bishop DK, Crossover interference in Saccharomyces cerevisiae requires a TID1/RDH54- and DMC1-dependent pathway, Genetics, 163 (2003) 1273–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Shinohara M, Oh SD, Hunter N, Shinohara A, Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis, Nat Genet, 40 (2008) 299–309. [DOI] [PubMed] [Google Scholar]
- [81].Munz P, An analysis of interference in the fission yeast Schizosaccharomyces pombe, Genetics, 137 (1994) 701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Fowler KR, Hyppa RW, Cromie GA, Smith GR, Physical basis for long-distance communication along meiotic chromosomes, Proc Natl Acad Sci U S A, 115 (2018) E933–E9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].King JS, Mortimer RK, A polymerization model of chiasma interference and corresponding computer simulation, Genetics, 126 (1990) 1127–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Foss EJ, Stahl FW, A test of a counting model for chiasma interference, Genetics, 139 (1995) 1201–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Fujitani Y, Mori S, Kobayashi I, A reaction-diffusion model for interference in meiotic crossing over, Genetics, 161 (2002) 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Kleckner N, Zickler D, Jones GH, Dekker J, Padmore R, Henle J, Hutchinson J, A mechanical basis for chromosome function, Proc. Natl. Acad. Sci. USA, 101 (2004) 12592–12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Hulten MA, On the origin of crossover interference: A chromosome oscillatory movement (COM) model, Mol Cytogenet, 4 (2011) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Garcia V, Gray S, Allison RM, Cooper TJ, Neale MJ, Tel1(ATM)-mediated interference suppresses clustered meiotic double-strand-break formation, Nature, 520 (2015) 114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Anderson CM, Oke A, Yam P, Zhuge T, Fung JC, Reduced crossover interference and increased ZMM-independent recombination in the absence of Tel1/ATM, PLoS Genet, 11 (2015) e1005478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Cooper TJ, Garcia V, Neale MJ, Meiotic DSB patterning: A multifaceted process, Cell Cycle, 15 (2016) 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Wu TC, Lichten M, Factors that affect the location and frequency of meiosis-induced double-strand breaks in Saccharomyces cerevisiae, Genetics, 140 (1995) 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Fan QQ, Xu F, White MA, Petes TD, Competition between adjacent meiotic recombination hotspots in the yeast Saccharomyces cerevisiae, Genetics, 145 (1997) 661–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Cromie GA, Smith GR, Meiotic recombination in Schizosaccharomyces pombe: A paradigm for genetic and molecular analysis, in: Egel R, Lankenau D-H (Eds.) Recombination and meiosis: Models, means, and evolution, Springer-Verlag, Berlin, 2008, pp. 195–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Wehrkamp-Richter S, Hyppa RA, Prudden J, Smith GR, Boddy MN, Meiotic DNA joint molecule resolution depends on Nse5-Nse6 of the Smc5-Smc6 holocomplex, Nucl. Acids Res, 40 (2012) 9633–9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Latypov V, Rothenberg M, Lorenz A, Octobre G, Csutak O, Lehmann E, Loidl J, Kohli J, Roles of Hop1 and Mek1 in meiotic chromosome pairing and recombination partner choice in Schizosaccharomyces pombe, Mol Cell Biol, 30 (2010) 1570–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Pebernard S, McDonald WH, Pavlova Y, Yates JR 3rd, Boddy MN, Nse1, Nse2, and a novel subunit of the Smc5-Smc6 complex, Nse3, play a crucial role in meiosis, Mol Biol Cell, 15 (2004) 4866–4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Spirek M, Estreicher A, Csaszar E, Wells JL, McFarlane RJ, Watts FZ, Loidl J, SUMOylation is required for normal development of linear elements and wild-type meiotic recombination in Schizosaccharomyces pombe, Chromosoma, 119 (2010) 59–72. [DOI] [PubMed] [Google Scholar]
- [98].DeVeaux LC, Hoagland NA, Smith GR, Seventeen complementation groups of mutations decreasing meiotic recombination in Schizosaccharomyces pombe, Genetics, 130 (1992) 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Cromie GA, Hyppa RW, Smith GR, The fission yeast BLM homolog Rqh1 promotes meiotic recombination, Genetics, 179 (2008) 1157–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]