Abstract
Hedgehog (HH) ligands, classical morphogens that pattern embryonic tissues in all animals, are covalently coupled to two lipids-- a palmitoyl group at the N-terminus and a cholesteroyl group at the C-terminus. While the palmitoyl group binds and inactivates Patched 1 (PTCH1), the main receptor for HH ligands, the function of the cholesterol modification has remained mysterious. Using structural and biochemical studies, along with the re-assessment of prior cryo-electron microscopy structures, we find that the C-terminal cholesterol attached to Sonic Hedgehog (SHH) binds the first extracellular domain of PTCH1 and promotes its inactivation, thus triggering HH signalling. Molecular dynamics simulations show that this interaction leads to the closure of a tunnel through PTCH1 that serves as the putative conduit for sterol transport. Thus, SHH inactivates PTCH1 by grasping its extracellular domain with two lipidic pincers, the N-terminal palmitate and the C-terminal cholesterol, which are both inserted into the PTCH1 protein core.
INTRODUCTION
The Hedgehog (HH) signalling pathway is inextricably linked to cholesterol1. Secreted HH ligands are generated from a 45 kDa precursor that undergoes a self-cleavage reaction that leaves cholesterol covalently linked to the C-terminus through an ester bond (hereafter ShhNc for cholesteroylated ShhN)2. In a second step, a palmitoyl moiety is added through an amide bond at the N-terminus of ShhNc to produce the dually-lipidated mature ligand (hereafter pShhNc for palmitoylated and cholesteroylated ShhN)3. Separately, cholesterol plays an instructive role in target cells that respond to HH ligands by directly activating Smoothened (SMO), the G protein-coupled receptor (GPCR) that transduces HH signals across the membrane4,5. These two roles of cholesterol in ligand biogenesis and transmembrane signalling are linked by the 12-pass transmembrane (TM) protein PTCH1. In the absence of HH ligands, PTCH1 has been proposed to inhibit SMO by preventing its access to cholesterol4,5, likely through a transporter-like function6,7. HH ligands directly bind and inhibit PTCH1, thereby unleashing SMO activity8,9.
PTCH1 has homology to the Resistance-Nodulation-Division (RND)-family of ion-driven small molecule transporters and to the lysosomal cholesterol transporter Niemann Pick C1 (NPC1)10–12. PTCH1 contains a transmembrane domain (TMD) harbouring twelve TM helices, five of which form a sterol sensing domain (SSD) also found in NPC1 and other cholesterol-sensing proteins13, and a large extracellular region composed of two extracellular domains (ECDs): PTCH1ECD1 inserted between TM helices 1 and 2, and PTCH1ECD2 inserted between TM helices 7 and 8 (Fig 1a). Pioneering cryo-EM structures of an engineered PTCH1 (PTCH1TM, Fig. 1b) in complex with un-lipidated (ShhN) and dually-lipidated Shh (pShhNc) have confirmed that PTCH1 adopts the overall fold of NPC1 and revealed the molecular details of the interaction between PTCH1 and pShhNc14–16. One molecule of pShhNc binds to two molecules of PTCH1 in a 1:2 asymmetric complex: the N-terminal palmitate and subsequent 15 amino acid residues (aars) of pShhNc are inserted between the ECD1 and ECD2 of one PTCH1 molecule (hereafter PTCH1-molA) and the globular protein domain of the same pShhNc makes a high-affinity protein-protein interaction with the ECD1 of second PTCH1 molecule (hereafter PTCH1-molB)14,15. Extra features in the cryo-EM map, postulated to be consistent with sterol ligands, are found in the ECD1 and in the SSD of PTCH17,14–16. A tunnel extending through PTCH1 between these two sites has been proposed as a conduit for sterol transport7,15,16.
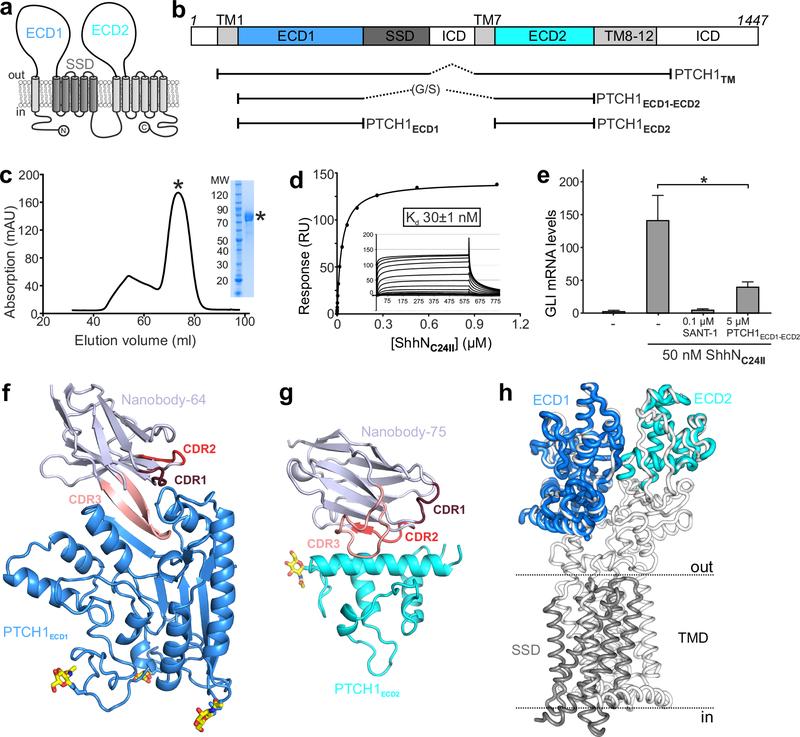
Figure 1 |. Structural and function characterization of PTCH1-nanobody interactions.
a, The pseudo-symmetric domain architecture of PTCH1: two 6-TM segments with extracellular domains (ECD1 and ECD2) interposed between the first two TM helices of each segment. The SSD, composed of TM helices 2–6, is marked in gray. b, Composition of the various protein constructs used in this study. c-e, Characterization of PTCH1ECD1-ECD2 used as an antigen to immunize Llamas. c, Typical SEC purification and corresponding SDS-PAGE of pooled fractions of PTCH1ECD1-ECD2. d, SPR equilibrium binding experiment between PTCH1ECD1-ECD2 (ligand) and non-lipidated ShhNC24II (analyte). This experiment was independently repeated 3 times with similar results. e, SHH signalling assay in mouse NIH-3T3 cells. Normalized Gli1 mRNA expression was used to assess SHH signalling activity by RT-qPCR after stimulation with purified ShhNC24II. Error bars denote SEM of 3 independent experiments. Statistical value is p=0.0387 determined by ordinary one-way ANOVA with Turkey’s multiple comparisons test. Sant-1 is a HH signalling inhibitor acting downstream of PTCH1. PTCH1ECD1-ECD2 acts as a ligand trap to inhibit Hh signalling. f-g, Cartoon representations of the high-resolution crystal structures of the PTCH1ECD1-NB64 (f) and PTCH1ECD2-NB75 (g) complexes. The complementary determining regions (CDRs) of the nanobodies are highlighted and N-linked glycans are shown in atomic colouring (carbon: yellow, oxygen: red, nitrogen: blue). h, Superposition of the PTCH1ECD1 and PTCH1ECD2 crystal structures on the previously determined cryo-EM PTCH1TM structure (PDB 6E1H16).
However, the modest resolution of these PTCH1 cryo-EM structures (at best 3.5Å) does not allow for the conclusive identification of the ligands, nor for visualization of the precise pose in which they interact with PTCH1. To address these limitations, we determined high-resolution crystal structures of the two soluble extracellular domains of PTCH1 in isolation and used them to reassess the previously solved cryo-EM structures.
RESULTS
Structures of the extracellular domains of PTCH1
To assess the integrity and ligand-binding activity of the PTCH1 extracellular domains (ECDs) when disconnected from the transmembrane domain (TMD), we linked the PTCH1ECD1 C-terminus to the PTCH1ECD2 N-terminus via a flexible glycine/serine (G/S)-rich linker (hereafter PTCH1ECD1-ECD2) (Fig. 1b). PTCH1ECD1-ECD2 was efficiently secreted by HEK293T cells, indicative of proper folding, showed high affinity (Kd ~ 30nM) binding to unlipidated ShhN and functioned as a ligand trap to inhibit SHH-triggered signalling (Fig. 1c–e). We raised camelid antibodies (nanobodies, NBs) against PTCH1ECD1-ECD2 and isolated two NBs (NB64 and NB75) that bound to PTCH1ECD1 and PTCH1ECD2, respectively, with nanomolar affinity (Supplementary Fig. 1a–f).
We determined crystal structures of the PTCH1ECD1-NB64 complex to 1.9 Å resolution using single-wavelength sulphur anomalous dispersion and the PTCH1ECD2-NB75 complex to 2.2 Å resolution by molecular replacement (Fig. 1f,g, Supplementary Fig. 2, Supplementary Table 1). The PTCH1ECD1-NB64 binding interface was formed predominantly by polar and van der Waals interaction between PTCH1ECD1 and the NB64 complementary determining regions (CDRs) 1 and 3, with a total buried surface area of 2570 Å (Supplementary Fig. 3). While CDR1 mainly interacts with the PTCH1ECD1 surface, CDR3 penetrates like an arrowhead into a cleft in PTCH1ECD1, with NB64-F102 forming a leading tip that reaches deep into PTCH1ECD1 (Fig. 1f, Supplementary Fig. 3e). This interaction generates a hybrid 3-stranded β-sheet structure with two strands from CDR3 and one from PTCH1ECD1. The main hydrophilic interactions are formed by six residues of the CDR3 loop (R101, F102, G104, S105, R106, R107) contributing to 13 hydrogen bonds and 5 salt bridges with PTCH1ECD1 (Fig. 1f, Supplementary Fig. 3d–e). In contrast, NB75 recognizes a flatter surface on PTCH1ECD2, using its CDR2 and CDR3 loops to cradle a helix (residues 844–865) in PTCH1ECD2 (Fig. 1g, Supplementary Fig. 3f).
PTCH1 ECD crystal structures compared to cryo-EM models
We compared the PTCH1ECD1-NB64 and PTCH1ECD2-NB75 crystal structures to a 3.5Å cryo-EM structure of PTCH1TM in a 2:1 complex with pShhNc (PDB Id. 6E1H16) (Fig. 1h). Structural superposition resulted in a r.m.s.d. of 0.95 Å for 271 of 301 equivalent Cα atoms for PTCH1ECD1 and a r.m.s.d. of 0.99 Å for 94 of 238 of equivalent Cα atoms for the PTCH1ECD2 domain, consistent with an identical overall fold. Taken together, our individual PTCH1ECD1 and PTCH1ECD2 structures cover ~70% of the PTCH1 extracellular region. Given that our crystal structures are higher in resolution by ~1.5Å, we re-refined the cryo-EM structure of the PTCH1TM-pShhNc complex using the deposited 3.5Å electron density (EMDB Id. EMD-895516). Several rounds of manual building and real space refinement led to an improvement of the cryo-EM model of the PTCH1TM-pShhNc complex (Fig. 2a; specific examples provided in Supplementary Fig. 4; overall structural comparison provided in Supplementary Fig. 5; metrics for model quality provided in Supplementary Table 2). This revised model identifies structural hot-spots for disease-associated mutations in PTCH1 that impair the stability of ECD1 and ECD2, likely impair the interaction of cholesterol with the SSD and disrupt a potential cation binding site at the center of the TMD that marks the path of cation flux (Supplementary Fig. 6 and 7).
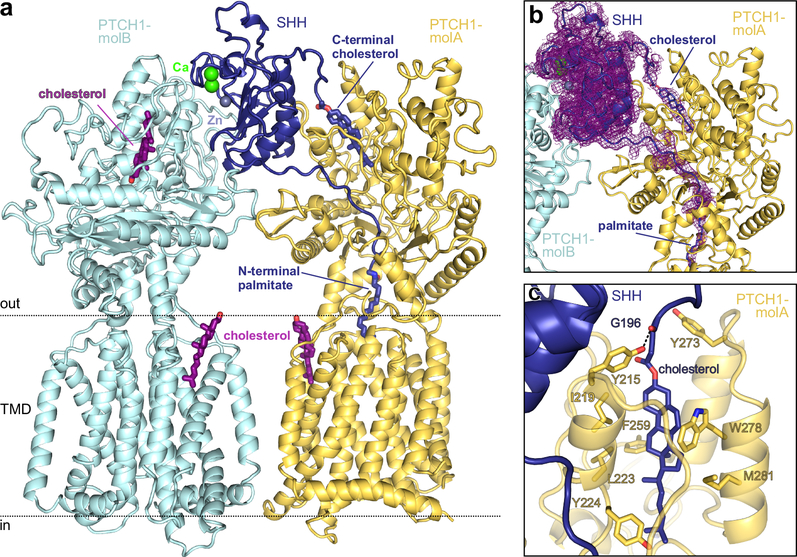
Figure 2 |. Structure of the PTCH1-pShhNc complex.
a, Rebuilt and improved model of the complex between one molecule of pShhNc and two molecules of PTCH1 (molA and molB). The high-affinity protein-protein interface between PTCH1-molB (cyan) and pShhNc is organized by the highly conserved calcium (green)- and zinc (grey)-binding sites of pShhNc (dark blue). The interface with PTCH1-molB (yellow) is composed of the cholesteryl- and palmitoyl- appendages of pShhNc that simultaneously penetrate the ECD. b, The previously deposited cryo-EM map of the SHH-PTCH1 complex (EMD-895516) mapped onto our improved model. Extra density is clearly discernible stretching from the C-terminus of pShhNc into ECD1 of PTCH1. c, Close-up view of the SHH-cholesterol molecule bound in the sterol-binding domain (SBD) of PTCH1, with residues in contact with cholesterol depicted in stick representation.
We assessed the identity and position of the sterol molecules located in the SSD and ECD1 (Supplementary Fig. 4d,e,i,h). These putative sterol ligands were initially identified in a different cryo-EM structure of PTCH1TM in complex with un-lipidated ShhN (PBD 6DMY), solved using PTCH1TM purified in the presence of cholesterol-hemisuccinate (HS)15, hence, this model was built with cholesterol-HS explicitly included in the two ligand binding sites. In contrast, the PTCH1TM-pShhNc complex (PDB 6E1H) used for our re-refinement16 used protein purified in the presence of the glyco-sterol detergent digitonin, but not cholesterol or cholesterol-HS. Despite the lack of cholesterol supplementation, we could clearly visualize cholesterol (not digitonin) bound both in the ECD1 and in the V-shaped cavity of the SSD that opens into the outer leaflet of the plasma membrane (Fig. 2a–c, Supplementary Fig. 6d–g).
This enhanced model allowed assignment of the cryo-EM density connecting the 3’-hydroxyl of cholesterol buried within PTCH1TM-ECD1 to the C-terminus of pShhNc (Fig. 2b), an observation that has been also noted in an independent study17. Notably, this region was mis-traced in the originally deposited structure (PDB 6E1H16), with PTCH1 residue R332 occupying the cholesterol density (Supplementary Fig. 4d,e). We were able to trace the missing C-terminal residues of pShhNc linked to a cholesterol molecule by an ester bond (Fig. 2b,c). The cholesterol in this binding site is encircled by three α-helices and lined with hydrophobic and aromatic residues (Fig. 2c). A hydrogen bond between Y215 in ECD1 and the backbone carbonyl of G196 in pShhNc positions cholesterol at the mouth of the hydrophobic pocket (Fig. 2c). Thus, our optimized model illuminates how pShhNc interacts in an unprecedented manner with PTCH1, simultaneously inserting both of its lipidic appendages, the palmitate and the cholesterol, into the ECD (Fig. 2a,b).
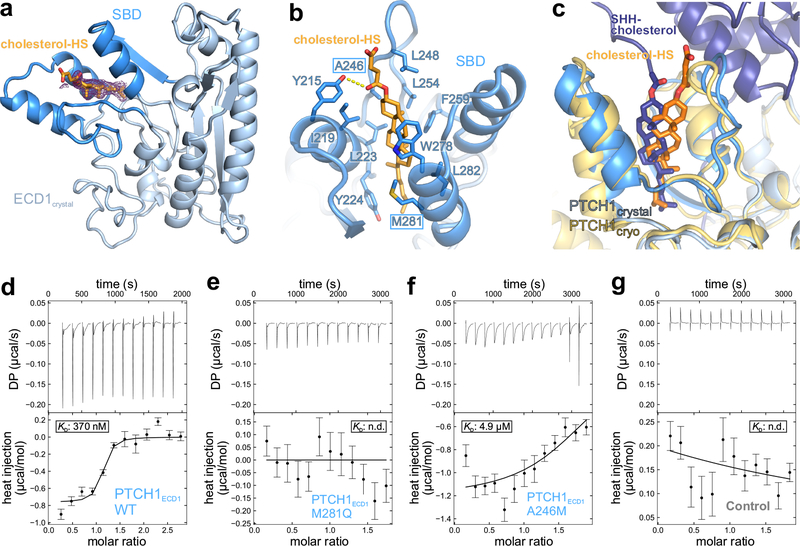
Structure of the PTCH1 ECD bound to cholesterol
To paint a portrait of the PTCH1ECD1-cholesterol interaction with finer strokes, we crystallised the PTCH1ECD1 in complex with cholesterol-HS and determined the structure to 1.9 Å resolution (Fig. 3a,b, Supplementary Fig. 2 h,i). The additional electron density contributed by cholesterol-HS was immediately apparent and occupies the same hydrophobic pocket formed by the three helices that accommodate the pShhNc-linked cholesterol in our optimized SHH-PTCH1TM model (Fig. 3c). Hereafter, the 3-helix motif in PTCH1ECD1 that binds to cholesterol is called the sterol binding domain (SBD). The binding pocket of the SBD is lined with aromatic (Y215, Y224, F259 and W278) and aliphatic (I219, L223, A246, L248, L254, M281 and L282) side-chains that make both π-π and van der Waals interactions with the sterol tetracyclic ring, positioned in the center of the pocket, and the iso-octyl chain, which extends towards the inner core of PTCH1ECD1 (Fig. 3b). While the TMDs and membrane-proximal portion of the ECDs are structurally similar across RND family members, the SBD of PTCH1, positioned at the apex of the ECD, is uniquely adapted to the cholesterol and pShhNc binding functions of PTCH1 (Supplementary Figure Fig. 8).
Figure 3 |. Structural and biophysical characterisation of the PTCH1 ECD-cholesterol complex.
a, Crystal structure of the PTCH1ECD1-cholesterol-hemisuccinate (cholesterol-HS) complex, with the 3-helix SBD coloured dark blue. The initial 2Fo-Fc map at 1.0 σ and 1.9 Å resolution before inclusion of cholesterol-HS is shown in wire representation. b, Close-up view of the sterol-binding site with cholesterol-HS depicted in orange sticks. c, Superposition of the PTCH1ECD1-cholesterol-HS crystal structure on the improved PTCH1-pShhNc cryo-EM structure. The entrance of the sterol-binding pocket is slightly rearranged, likely due to different steric constraints of the hemisuccinate group compared to the glycine ester linkage found in pShhNc. d-g, Raw ITC data (upper panel) and binding isotherms (lower panel) for titration of PEG-cholesterol into solutions containing PTCH1ECD1-WT (d), PTCH1ECD1-M281Q (e) and PTCH1ECD1-A246M (f). PEG200, unconjugated to cholesterol, was titrated into a PTCH1ECD1-WT solution as a control (g). Curves show the A+B ⇌AB binding model that was used for local fitting to the data. The Kd is shown where it could be calculated. Bars shown on the isotherm reflect errors associated with integration of the injection peaks in the corresponding thermograms (68% confidence around extrapolation of pre-and post-injection baselines).
To assess the relevance of the cholesterol-binding pocket in the SBD in solution, outside the context of a crystal lattice, we used isothermal titration calorimetry (ITC) to measure the interaction between PTCH1ECD1 and a cholesterol molecule modified with polyethylene glycol (PEG) to mimic the attached protein chain of ShhN (steroid 1 in Supplementary note). PEG-cholesterol bound with moderate affinity to wild-type PTCH1ECD1 (KD ~370 nM, Fig. 3d). To establish specificity, we mutated residues (A246 and M281, boxed in Fig. 3b) lining the SBD. Alteration of A246 to a bulkier methionine or M281 to a more polar glutamine did not substantially alter the expression levels or the thermostability of PTCH1ECD1 (Supplementary Fig. 9), but impaired its interaction with PEG-cholesterol (Fig. 3e–g).
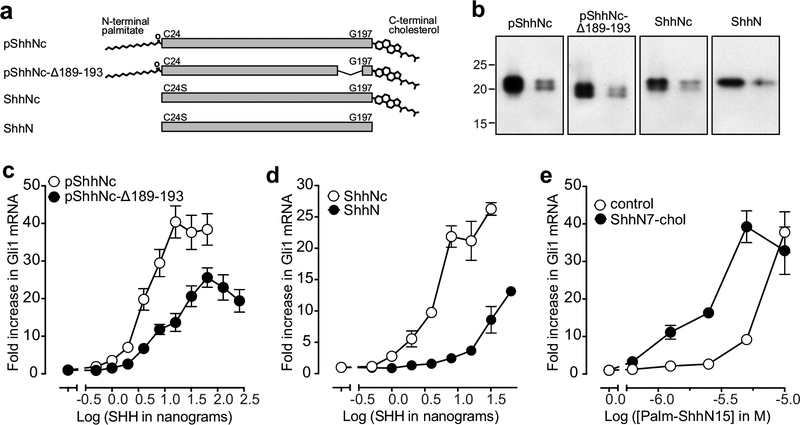
Inactivation of PTCH1 by the cholesterol attached to SHH
Does this newly uncovered interaction between the cholesterol attached to the C-terminus of HH ligands play a functional role in inactivating PTCH1 (and consequently activating signalling)? To address this question, we deleted five residues (N189-A193) in the extended peptide connector that links the globular domain of pShhNc to cholesterol, predicting that shortening this linkage would impair its ability to insert cholesterol into the ECD1 of PTCH1-molA while tightly bound to a second PTCH1-molB (Fig. 2a). Interestingly, the length of this peptide connector is evolutionarily conserved even though it does not form part of the ShhN globular core18. Indeed, pShhNc-Δ189–193 was less potent and less effective in activating HH signalling when added to NIH/3T3 cells, using mRNA levels of the immediate HH target gene Gli1 as a metric for signalling strength (Fig.4a–e, Supplementary Fig. 7c). To directly measure the effect of the cholesterol moiety on PTCH1 inactivation, separate from the known effect of the palmitate19, we produced two SHH ligands that both lacked the palmitate modification due to alteration of the palmitoylated Cys24 to Ser: ShhN is entirely un-lipidated and ShhNc is modified only with a cholesterol at the C-terminus (Fig. 4a). ShhNc is significantly more potent than ShhN in activating signalling, showing that the cholesterol modification of HH ligands can enhance their ability to inactivate PTCH1 and trigger HH signalling (Fig. 4d). Finally, a peptide composed of the C-terminal 7 aars of ShhN esterified to cholesterol20 (ShhN7-chol) significantly increased the signalling potency of an N-terminal 15 aars ShhN peptide bearing a palmitate (Palm-ShhN15) (Fig. 4e)19. The cholesterol esterified to ShhN7 cannot be directly acting on SMO because the interaction of SMO with cholesterol requires a free 3’-hydroxyl21. This cooperativity between the palmitate and cholesterol in PTCH1 inactivation, even when they are separated on different ShhN peptides, suggests that the sites occupied by cholesterol and palmitate (Fig. 2a,b) are allosterically linked. Thus, these lipids installed on all HH ligands can function as two arms of a pincer to coordinately inactivate PTCH1.
Figure 4 |. The cholesterol attached to pShhNc inactivates PTCH1.
a, Schematic of the various SHH proteins used in signalling assays. b, Two different amounts of each SHH variant from (h) were analyzed by immunoblotting. Full gels can be found on Supplementary Figure 7. c and d, Fold-increase in Gli1 mRNA (relative to the no SHH added condition) was used as a metric for signalling strength after treatment of NIH/3T3 cells with various concentrations of the indicated ligands. e, HH signalling strength at increasing concentrations of the N-terminal palmitoylated peptide (Palm-ShhN15) in the absence or presence (100 μM) of a cholesteroylated C-terminal peptide (ShhN7-chol). In c-e, the mean is depicted and error bars reflect standard deviation (n=4). Each experiment in c-e was repeated at least 3 times.
A role for ShhN-linked cholesterol in PTCH1 inactivation is supported by previous studies. Mouse embryos carrying an allele of Shh that cannot be cholesteroylated show defects in digit patterning caused by impaired HH signalling at longer distances from the ligand source22. In addition, ShhNc (lacking a palmitate and identical to the ligand used in our studies) induces more pronounced digit duplication compared to un-lipidated ShhN when expressed in the mesenchyme of the developing limb bud23. While these results support a role of cholesteroylation in long-distance ligand dispersal, they are also consistent with our finding that the cholesterol attached to HH ligands plays a direct role in PTCH1 inactivation. Such a role may become especially important at the low ligand concentrations found at longer distances from the ligand source.
The PTCH1 ECD1 can bind cholesterol in two inverted poses
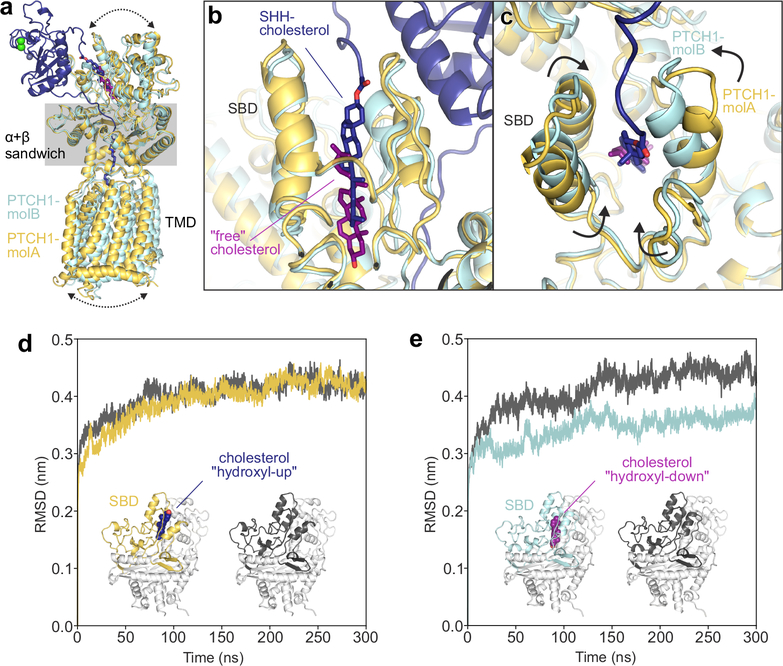
We uncovered a distinct mode of cholesterol interaction with the SBD when we compared PTCH1-molA to -molB by superimposing the α+β sandwich domains that link the ECD to the TMD (Fig. 5a). The interface between pShhNc and PTCH1-molA is driven by palmitate- and cholesterol contacts, while the interface between pShhNc and PTCH1-molB is composed of a protein-only contact organized by the two metal ion binding sites in ShhN (Fig. 2a). Comparison of the SBD in PTCH1-molA and PTCH1-molB revealed startlingly different orientations of the bound cholesterol molecules. In PTCH1-molA, the cholesterol attached to pShhNc is positioned with the ester bond located at the mouth of the pocket facing away from the TMD, an orientation we refer hereafter to as “hydroxyl-up” (Fig. 5b, blue). This “hydroxyl-up” pose resembles the cholesterol orientation seen in the crystal structure of the complex between PTCH1ECD1 and cholesterol-HS (Fig. 3c). However, in PTCH1-molB, a free cholesterol is found in an orientation inverted by 180-degrees, with the 3-hydroxy facing towards the TMD (hereafter “hydroxyl-down”, Fig. 5b, purple). Though we do not have a high-resolution crystal structure, several additional analyses support the interaction of PTCH1ECD1 with cholesterol in a “hydroxyl-down” orientation (Supplementary Figure 10).
Figure 5 |. The PTCH1 sterol-binding domain (SBD) can bind cholesterol in two opposite orientations.
a-c, Comparison of PTCH1-molA (yellow) and -molB (cyan) from the rebuilt 2:1 PTCH1-pShhNc complex. a, Superposition of molA and molB templated on the α+β sandwich domain. Differences in the relative positions of the TMD and the upper lobes of the ECD are indicated by arrows. b, Close-up of the SBDs from the superimposed structures. PTCH1-molA binds to the ShhN-attached cholesterol (dark blue) with the esterified cholesterol positioned at the SBD entrance (“hydroxyl-up”), while the cholesterol in PTCH1-molB is buried in the SBD core in an inverted orientation (“hydroxyl-down”). c, Conformational differences at the mouth of the SBD in PTCH1-molA and -molB. d and e, Cholesterol orientation-dependent differences in the stability of the PTCH1 upper lobe in molecular dynamics simulations. Average RMSD (compared to the starting structure) of the PTCH1ECD1 upper lobe across five independent atomistic simulations (300 ns each) of the entire PTCH1ECD from molA (d) and molB (e), with (yellow/cyan) and without (grey) bound cholesterol.
We speculate that the free cholesterol in the “hydroxyl-down” pose may represent an intermediate in the transport cycle of PTCH1. The deeper position of the cholesterol in PTCH1-molB allows a concerted claw-like closure of loops surrounding the pocket entrance (Fig. 5c). Thus, the SBD in PTCH1 can accommodate cholesterol in two entirely opposite orientations, including one orientation in which the cholesterol remains attached to ShhN. To understand the basis for this structural plasticity, we determined the high-resolution (1.95 Å) crystal structure of apo-PTCH1ECD1 (Supplementary Fig. 11a,b). Compared to the PTCH1ECD1-CHS-NB64 complex, the cholesterol-binding pocket is disordered and partially collapsed in apo-PTCH1ECD1, suggesting that the binding-pocket is not rigid but can adopt different conformations to accommodate cholesterol in both its free and ShhN-conjugated forms (Supplementary Fig. 11c–f).
Binding of cholesterol closes a transport tunnel in PTCH1
To understand the structural consequences of cholesterol binding to the SBD, we performed atomistic molecular dynamics (MD) simulations of extracted PTCH1ECD domain structures with cholesterol alone (not conjugated to ShhN) bound in either hydroxyl-up or hydroxyl-down orientations (Fig. 5d–e). Compared to simulations of PTCH1ECD lacking cholesterol altogether (apo-PTCH1ECD), RMSD changes in the upper lobe of PTCH1ECD-molA and apo-PTCH1ECD were similar during the course of the simulations, suggesting that cholesterol bound in the hydroxyl-up orientation did not change domain stability (Fig. 5d). In contrast, the upper lobe of PTCH1ECD-molB is significantly stabilized when cholesterol is bound more deeply in the hydroxyl-down orientation (Fig. 5e). Similar results are obtained when focusing on the bound cholesterol; the hydroxyl-up orientation shows much greater fluctuation as evidenced by a higher cholesterol atom RMSD over the course of the simulation compared to the hydroxyl-down orientation (Supplementary Fig. 12).
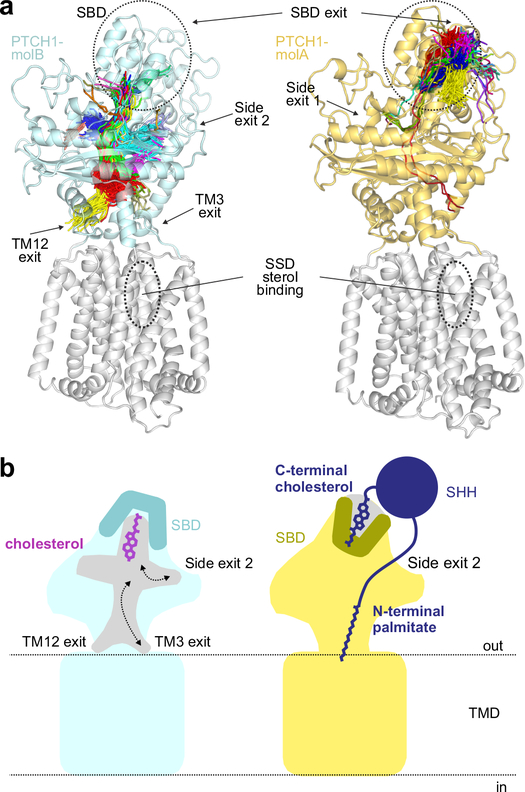
Previous analysis of PTCH1 identified a 150 Å tunnel, postulated to be the conduit for cholesterol transport, stretching from the SBD through the interior of the ECD to openings at TM3 (which is part of the SSD) or TM127,14,15. We explored the dynamic behaviour of tunnels in our simulations of PTCH1ECD (Fig. 6a). In PTCH1ECD-mol A, with cholesterol bound in the hydroxyl-up orientation, most tunnels were confined to the SBD (Fig. 6a, left panel), extending from the position of the SHH-cholesterol iso-octyl tail to the mouth of the SBD (Supplementary Table 3). In contrast, almost all the tunnels in PTCH1ECD-molB, which contains the deeper cholesterol molecule bound in the hydroxyl-down orientation, entered the ECD core (Fig. 6a, right panel). Notably, none of the tunnels in the simulations suggests efflux of this cholesterol out through the mouth of the SBD. Instead, the exit points of the tunnel were found in two locations: (1) just above TM3 and TM12, as noted previously7,14,15, or (2) at two symmetry-related positions in the middle of the ECD (labeled “side-exits” in Fig. 6a,b). Interestingly, side-exit 2 is where the palmitate conjugated to the N-terminus of pShhNc is inserted into the ECD in the model of PTCH1-molA14 (Fig. 2a). The direction in which PTCH1 transports cholesterol remains an unanswered question, with significant implications for the mechanism by which PTCH1 regulates SMO. Most prior studies speculate that PTCH1 transports cholesterol from the membrane out through the SBD to a membrane or protein acceptor6,7, but the structures and our simulations are also consistent with the reverse direction of transport (from the SBD to the membrane).
Figure 6 |. Tunnel analysis of PTCH1 structures.
a, Conduits that extend through the PTCH1ECD in molA (right, yellow) and molB (left, cyan) identified in a simulation from Fig. 4d,e are shown as lines drawn at the central point of each tunnel overlaid with the starting structure. Exit points of each tunnel are noted. The midpoint traces of the tunnels are clustered as indicated by the colours. b, Model showing the consequences of cholesterol bound in the two different orientations for PTCH1 function. When cholesterol is bound in the hydroxyl-down orientation (left, cyan) in molB, the mouth of the SBD pocket is closed; however, conduits through the ECD, at the side of the ECD or just above the TMD are open for cholesterol. When the cholesterol attached to pShhNc (right, molA) is bound in the SBD, the mouth of the pocket is open to accommodate the ShhN protein chain, but the conduits through the PTCH1 ECD are closed, thereby blocking the cholesterol transporter activity of PTCH1. The palmitoyl group further inserts into and plugs the side exit in the ECD.
DISCUSSION
The function of the cholesterol attached to HH ligands has remained a long-standing mystery in HH signalling. Prior work has focused on its role in ligand dispersal through tissues, though it seems to have opposite effects in Drosophila and mouse embryos, restricting ligands movement in the former and promoting ligand movement in the latter22,24. Our structural and biochemical work suggests a simple function for this cholesteroyl appendage - direct binding to the PTCH1-ECD1, which stabilizes a conformation of PTCH1 in which a continuous conduit for sterol transport through the ECD is closed.
It is striking that HH ligands make three entirely different interactions with their receptor PTCH1: one conventional protein-protein driven interaction (with PTCH1-molB, Fig. 2a), which mediates the high-affinity interaction between HH ligands and PTCH1, and two lipid-based interactions in which either their palmitoyl or cholesteroyl appendages are inserted into the PTCH1 ECD core (with PTCH1-molA, Fig. 2a). From a functional standpoint, which of these interactions are necessary or sufficient to inactivate PTCH1 and trigger signaling in target cells? ShhN, lacking both the palmitate and the cholesterol moieties, binds with high affinity to PTCH1, and it can inactivate PTCH1, albeit with considerably reduced potency, presumably through the interface seen between ShhN and PTCH1-molB3,23 (Fig. 2a). At the same time, a peptide composed of the N-terminal 22 amino acids of ShhN attached to palmitate, which can only engage PTCH1-molA (Fig. 2a), is also sufficient to inactivate PTCH1 at high, micromolar concentrations19. However, an isolated C-terminal ShhN peptide carrying the cholesterol moiety alone (ShhN7-chol, Fig.3e) did not inactivate PTCH1 at even at high concentrations (100 μM), though it synergized with the palmitate-modified peptide (suggesting a coordinated, pincer-like inactivation mechanism). Thus, under in vitro assay conditions where concentrations can be arbitrarily manipulated, none of these three interactions is essential for signalling. In addition, either the ShhN protein-based interaction (with PTCH1-molB) or the palmitate-based interaction (with PTCH1-molA) is sufficient to initiate signalling.
While the reason for this partially redundant, multi-partite interaction remains unclear, one possibility is that each of these interactions may play more or less important roles at different ligand concentrations. At high concentrations, the protein-protein interface or the palmitate interface may be sufficient to inactivate PTCH1 function, but at low ligand concentrations, distant from the ligand source, the cholesterol interface may also be required to maintain signalling activity. Each of these interactions likely activates PTCH1 in different ways: the cholesterol- and palmitate-based interfaces may inactivate the biochemical function of PTCH in cholesterol transport (Fig.6b), while the protein-protein interaction may function by promoting the endocytosis and clearance of PTCH1 from the plasma membrane and membrane of the primary cilium19. In this latter case, the protein-based interaction would control a distinct function of PTCH1 in sequestering HH ligands, shaping the gradient of HH ligands in a target tissue field25.
An important consideration is that the surface of ShhN that binds to PTCH-molB overlaps with the surface that binds to Heparan Sulfate Proteoglycans (HSPGs) as well as to other SHH co-receptors, including CDO, BOC, GAS1 and the inhibitor HHIP1,18,26–32 (Supplementary Fig. 13). Even the N-terminal peptide of ShhN, which is linked to palmitate, has a conserved Cardin-Weintraub motif that binds to the heparan sulphate chains of cell surface and extracellular HSPGs33. Hence, depending on the context, all three of these interfaces may not be simultaneously available for interaction with PTCH1.
We end by noting that our work supports a prescient hypothesis, proposed nearly a decade ago, for the multiple, seemingly unrelated roles for cholesterol in HH signalling34,35 The HH pathway may have evolved from a sterol-sensing and transport pathway: an ancestor of PTCH1 inhibited the function of a SMO-like sterol-responsive protein using its transporter-like activity to deplete sterols. Adaptation of this system to cell-cell communication in metazoans required the evolution of a ligand that is produced by one cell and regulates the activity of the pathway in a different cell. The simplest way to evolve such a ligand is to link the substrate for PTCH1 (cholesterol) to a protein chain that will sterically block transporter function-- the substrate, once attached to a protein chain, becomes an inhibitor. We predict that the cholesterol attached to ShhN was the original mechanism by which HH ligands blocked PTCH1 activity, with the palmitate and protein interfaces emerging later.
METHODS
Reagents.
Cholesterol hemisuccinate was obtained from Anatrace. Flp-In NIH/3T3 and HEK293T cells were certified stocks obtained directly from Thermo Fisher Scientific and ATCC, respectively. They were used at passages <10 without additional authentication. Incoming cell lines were confirmed to be negative for mycoplasma contamination. A rabbit monoclonal antibody (clone C9C5) against the N-terminal signaling domain of human SHH was obtained from Cell Signaling Technologies (cat#2207).
Constructs.
A synthetic and codon-optimised gene encoding full-length human Patched-1 (PTCH1) was obtained from Geneart (Grand Island, NY). Construct variants of PTCH1 (UniProt ID Q13635; PTCH1ECD1: 139–428, PTCH1ECD2: 772–1023, PTCH1ECD1-ECD2: (120–435)-GGSNGSGG-(789–1023), PTCH1TMD: (75–1185 including a deletion of intracellular loop 3 (Δ630–717)), either N- or C-terminally fused with a hexahistidine (His6)-tag, a BirA-recognition sequence or a 1D4 epitope tag36 were cloned into the pHLsec vector37. The two PTCH1ECD1 mutants (A246M and M281Q) designed to test for sterol binding were generated using a two-step overlap-extension PCR strategy. Nanobodies and ShhNC24II were cloned into bacterial expression plasmids pMESy45 (containing an N-terminal pelB leader sequence) and pET22b, respectively, with C-terminal His6-tag.
Expression and purification of secreted PTCH1 constructs.
Secreted PTCH1 constructs were produced by transient transfection in HEK293T cells in the presence of kifunensine. For protein biotinylation, PTCH1 constructs were co-transfected with a BirA expression plasmid and the media was supplemented with 100 μM biotin (Sigma). Conditioned media was collected three to five days’ post-transfection, clarified by centrifugation and filtering. For His6-tagged proteins, media was concentrated and diafiltrated into PBS pH 7.4 (Sigma) using a QuixStand™ benchtop system (GE Healthcare) connected to a 60 cm Xampler Cartridge (GE Healthcare) with a 10 kDa nominal MWCO. The proteins were incubated with TALON® beads for 1 hour at 20°C. Beads were washed with 20 mM HEPES pH 7.5, 500 mM NaCl, followed by washes in wash buffer containing 2 mM imidazole and 5 mM imidazole. The protein was eluted in wash buffer containing 250 mM imidazole. The buffers for PTCH1EDC2 cobalt-affinity chromatography contained an additional 1 mM β-mercaptoethanol. 1D4-tagged PTCH1 constructs were purified directly from media with Rho-1D4 antibody (University of British Columbia) coupled to CNBr-activated sepharose beads (GE Healthcare). Protein-bound beads were washed with 20 mM HEPES pH 7.5, 150 mM NaCl and eluted for 1–2 hours in 20 mM HEPES pH 7.5, 150 mM NaCl containing 500 μM “TETSQVAPA” peptide (Genscript). Samples were subjected to size exclusion chromatography (SEC) using a Superdex 200 16/60 column (GE Healthcare) equilibrated in 10 mM HEPES pH 7.5, 150 mM NaCl, for PTCH1ECD2 5mM DTT was included as well. Proteins were concentrated to 5–7 mg/ml using a 10 kDa MWCO filter (Millipore).
Expression and purification of transmembrane domain-containing PTCH1 constructs and amphipol exchange.
HEK293S-GnTI− (ATCC® CRL-3022TM) cells in suspension were cultivated in PEM (Invitrogen) supplemented with 1% FCS (Gibco), 1% NEAA (Gibco) and 1% L-Glutamine (Gibco) at 37°C, 8% CO2, 130 rpm. At a cell density of 2 × 10−6 cells mL−1, 0.8 L of cells were collected by centrifugation at 1.400 rpm and resuspended in 120 mL of FreeStyle 293 (Invitrogen) expression medium (1% NEAA, 1% L-Glutamine, 5 mM VPA (Sigma Aldrich)), containing 0.4 mg of plasmid DNA encoding C-terminally 1D4-tagged PTCH1 and 1.2 mL of PEI Max (Polysciences, 1 mg mL−1 in PBS). Cells were incubated at 37°C, 8% CO2 and 160 rpm for 4– 6 hours, after which 0.6 L of PEM was added. Proteins were expressed for 60– 72 hours until pelleted by centrifugation at 1100 x g for 4 min at RT. Proteins were solubilized in ice-cold membrane buffer (50 mM HEPES pH 7.5, 300 mM NaCl, 100 mM L-Arg, 10% glycerol) with a final detergent concentration of 1.1% DDM and 0.11% cholesterol-HS. The insoluble fraction was removed by centrifugation at 9300 x g for 30 min. The protein sample was diluted 2-fold with membrane buffer and incubated with Rho-1D4 antibody (University of British Columbia) coupled to CNBr-activated sepharose beads (GE Healthcare). Protein-bound beads were washed with 20 mM HEPES pH 7.5, 300 mM NaCl, 100 mM L-Arg, 10% glycerol, 0.03% DDM, 0.033% cholesterol-HS and eluted overnight in the same buffer containing 500 μM “TETSQVAPA” peptide (Genscript). The sample was subjected to SEC using a Superose 6 Increase 10/30 column (GE Healthcare) equilibrated in 10 mM HEPES pH 7.5, 300 mM NaCl, 5% glycerol, 0.03% DDM, 0.003% cholesterol-HS. All steps were carried out at 4 °C. For BLI experiments, protein samples were exchanged into amphipols (A8–35, Anatrace) at a mass ratio of 3:1 amphipol:protein and rotated at RT for 30 min. BioBeads (BIORAD), equilibrated in detergent-free buffer (10 mM HEPES pH7.5, 300 mM NaCl), were added to the protein-detergent-amphipol mixture at 10 mg per 100 μL and incubated overnight at 4°C to remove all detergent molecules.
Production of ShhNC24II.
The N-terminal signalling domain of SHH (residues 24–193, UniProt ID Q15465) was modified to enhance potency by replacing cysteine 24 with two isoleucine residues (ShhNC24II)38. ShhNC24II was cloned into pET22b and expressed in E. coli Rosetta 2(DE3)pLysS cells. Cultures were grown at 37 °C to an A600 of 0.6–0.8, cooled to RT, induced with 0.2 mM isopropyl-β-D-thiogalactopyranoside (IPTG) and grown overnight. The bacterial pellet was resuspended in 10 mM phosphate buffer pH 7.5, 500 mM NaCl, 1 mM β-mercaptoethanol and EDTA-free protease inhibitor cocktail (Roche). Cells were broken using sonication (Sonics Materials™, VCX 500) and fractionated by centrifugation (30,000 x g, 5 °C, 45 min). The supernatant was incubated with TALON® beads for 1 hour at RT. The resin was washed with 10 mM phosphate buffer pH 7.5, 500 mM NaCl, 1 mM β-mercaptoethanol (wash buffer) followed by a wash with wash buffer containing 10 mM imidazole. ShhNC24II was eluted in wash buffer containing 250 mM imidazole. The protein was further purified by SEC using a Superdex 75 16/60 column (GE Healthcare) equilibrated in 10 mM HEPES pH 7.5, 150 mM NaCl. ShhNC24II was concentrated to ~5 mg/ml using a 10 kDa MWCO filter (Millipore), snap-frozen and stored in aliquots at −80°C.
Production of SHH proteins by transient transfection of 293T cells.
All human SHH expression constructs were cloned into the pHLsec vector for expression in HEK293T cells2. The constructs encoded the following proteins: 1. Unlipidated ShhN: aars 1–197, Cys24Ser to abolish palmitoylation; 2. Cholesteroylated (but not palmitoylated) ShhNc: aars 1–462, Cys24Ser; 3. Palmitoylated and cholesterylated pShhNc: aars 1–462; 4. pShhcΔ189–193: aars 1–462 with a four amino acid deletion in residues 189–193. All SHH proteins were co-expressed with SCUBE2 to enhance secretion, as described previously39. HEK293T cell were transfected in 10cm plates with a mixture of constructs encoding a SHH variant and SCUBE2 using polyethylenimine (PEI, linear, MW ~25,000, Polysciences, Inc.). Twelve hours after transfection, the media was changed to serum-free Dulbecco’s modified Eagle’s medium (DMEM, Thermo Fisher). Media containing secreted SHH was collected after 36 hours, centrifuged at 18,000 x g, and then concentrated (Amicon-Ultra centrifugal filter device). The concentrations of each batch of the various SHH proteins were measured by quantitative immunoblotting (LI-COR Odyssey), in comparison to purified ShhN of known concentration produced in bacteria40.
Nanobody production.
Camelid single chain antibodies (nanobodies, NB) against the human PTCH1 ectodomain were produced using established protocols41. One animal was immunized repeatedly with PTCH1ECD1-ECD2. Subsequent to library generation, nanobodies were selected by phage display by trapping 1D4-tagged or directly adsorbed PTCH1ECD1-ECD2. E. coli strain WK6su− cultures containing the nanobody sequences in the pMES4y plasmid were grown at 37 °C to an optical density (A600) of 0.8, cooled to RT, induced by the addition of 1mM IPTG and grown over night at 22°C. Proteins were purified from the bacterial periplasm by IMAC chromatography. The cell pellet from 1 L of bacterial culture was resuspended in 23 mL of ice-cold TES buffer (0.2 M Tris-HCl pH 8.0, 0.5 mM EDTA, 0.5 M sucrose) and incubated on ice for 4 hours at 120 rpm. The lysate was diluted with 40 mL of 4-fold diluted TES buffer and incubated on ice for another 16 hours at 120 rpm. The insoluble fraction was removed by centrifugation at 12,000 x g for 30 min at 4°C. Ni-NTA resin (Qiagen) was added to the supernatant and incubated for 1 hour at 4°C. The resin was washed with 50 mM HEPES pH 7.5, 500 mM NaCl, followed by 50 mM HEPES pH 7.5, 200 mM NaCl, 10 mM imidazole. The protein was eluted in 20 mM HEPES pH 7.5, 150 mM NaCl, 250 mM imidazole. The sample was subsequently purified by SEC using a Superdex 75 16/60 column (GE Healthcare) equilibrated in 10 mM HEPES pH 7.5, 150 mM NaCl. Proteins were concentrated to 5 mg/ml, snap-frozen in liquid nitrogen and stored in aliquots at −80°C.
Crystallisation and data collection.
Prior to crystallisation, the proteins were concentrated to 5–7 mg/ml and treated with Endoglycosidase F1 for 1 hour at RT. For PTCH1ECD1 and PTCH1ECD2 nanobody complexes, the proteins were mixed in 1:1 PTCH1:NB molar ratio to a final concentration of 5–7 mg/ml. Crystallisation trials were set up using sitting-drop vapour diffusion utilising a Cartesian Technologies robot (100 nl protein sample plus 100 nl reservoir solution) in 96 well Greiner plates42. Crystals of the PTCH1ECD1-NB64 complex were obtained at 20°C by micro-seeding into a 2% dilution series (72% to 56%) of reservoir buffer containing 0.1 M MES/imidazole pH 6.5, 0.02M of each amino acid (L-glutamate, alanine, glycine, lysine, serine), 12.5% (v/v) MPD, 12.5% (w/v) PEG 1000, 12.5% (w/v) PEG 3350. For co-crystallisation experiments, the PTCH1ECD1-NB64 complex was mixed with a final concentration of 10 mM cholesterol hemisuccinate (HS). PTCH1ECD1-NB64-cholesterol-HS crystals were obtained at 20°C from mother liquor containing 0.1 M HEPES/MOPS pH 7.5, 0.03 M of each halide (sodium fluoride, sodium bromide, sodium iodide), 12.5% (v/v) MPD, 12.5% (w/v) PEG 1000, 12.5% (w/v) PEG 3350. Apo-PTCH1ECD1 crystallised at 5°C out of a mother liquor containing 0.1 M bicine/Trizma base pH 8.5, 0.03M of each ethylene glycol (diethyleneglycol, triethyleneglycol, tetraethyleneglycol and pentaethyleneglycol), 12.5% (v/v) MPD, 12.5% (w/v) PEG 1000, 12.5% (w/v) PEG 3350, and PTCH1ECD2-NB75 complex crystals were grown at 5°C from mother liquor containing 0.2 M sodium nitrate, 20% (w/v) PEG 3350. Prior to flash-freezing in liquid nitrogen, crystals were treated with 20% (v/v) ethylene glycol in mother liquor for cryo-protection. The cryo solution for the PTCH1ECD1-NB64-cholesterol-HS crystals contained an additional 10 mM cholesterol-HS. S-SAD data were collected at the Diamond Light Source, UK (DLS) long wave-length beamline I23 using an inverse beam operated at a wavelength of 2.7751 Å. Native data of the PTCH1ECD1-NB64 complex was collected at DLS beamline I04 (λ=0.97625 Å), the PTCH1ECD1-NB64-cholesterol-HS and PTCH1ECD2-NB75 complex data at DLS beamline I24 (λ=0.9686 Å), and the apo-PTCH1ECD1 data on DLS beamline I04 (λ=0.97950 Å). All data were collected at 100 K and processed and scaled with XIA243.
Structure solution and refinement.
We determined the structure of PTCH1ECD1-NB64 complex by Sulphur-SAD phasing. Using the SHELX suite44, we were able to identify the positions of 14 (out of 21) sulphur atoms observing an anomalous signal to 4 Å resolution. Phasing with the initial sub-structure was carried out in PHENIX.AUTOSOL45. This enabled determination of a partial PTCH1ECD1-NB64 complex model, which then was used as input for Molecular Replacement-SAD in PHASER46, also extending the resolution to 2.1 Å. Iterative rounds of manual model building in COOT47 and refinement in REFMAC48, PHENIX45 and AUTOBUSTER49 resulted in a well-ordered model with excellent stereochemistry (see Supplementary Table 1). The structures of the PTCH1ECD1-NB64-cholesterol-HS complex and apo-PTCH1ECD1 were determined by molecular replacement in PHASER46 using the PTCH1ECD1-NB64 structure as input model and refined with REFMAC48, and AUTOBUSTER49. The PTCH1ECD2-NB75 complex was determined by molecular replacement using PHASER with nanobody A26.8 VHH (PDB ID 4NBZ) and a truncated fragment of Patched (PDB ID 6DMY, residues K842-E970) as search models, followed by automatic model rebuilding using Phenix AutoBuild45 and iterative rounds of manual building in COOT47 and refinement in REFMAC48. Stereochemical properties were assessed with the MolProbity server (molprobity.biochem.duke.edu/). The crystallographic and Ramachandran statistics of the data collection and processing are shown in Supplementary Table 1.
Re-refinement and building of the 2:1 PTCH1:SHH complex.
The cryo-EM map of the PTCH1TM-pShhNc complex at 3.5Å resolution (EMDB code EMD-895550) was used to optimize the deposited model (PDB code 6E1H50), The PTCH1ECD regions were manually replaced with the higher resolution crystal structures of PTCH1ECD1-NB64 and PTCH1ECD2-NB75 (1.9 and 2.2 Å resolution, respectively). The rest of the initial model was manually adjusted using COOT47 by building and repositioning main-chains/side chains and adding molecules such as N-linked glycans and cholesterol molecules. The enhanced model was subjected to rigid-body and global-minimization used along NCS and secondary-structure restraints in ‘phenix.real_space_refine’ from PHENIX45.
Coarse grain molecular dynamics simulations.
Simulations were performed using the GROMACS 5.1.4 simulation package (www.manual.gromacs.org). PTCH1-molA and PTCH1-molB proteins were extracted from the rebuilt PTCH1-pShhNc (2:1) model. The missing TM6–7 loop (from residues 613 to 730) is too long to be modelled accurately using conventional modelling software and so LEU608-CYS613 and PRO725-TRP730 were modelled and residues 613 and 725 were directly connected using MODELLER 9.20 (salilab.org/modeller) to give an overall sequence (TM6)RRL-DIFCC//TKWT-LSS(TM7). 10 models were produced and the model with the lowest DOPE score was taken forward for each PTCH1 molecule. Default protonation states were used in simulations. Non-standard protonation states, as identified in our pKa computations, were not considered in our MD simulations. We acknowledge that these non-standard states may affect overall protein dynamical behaviour, and any subsequent simulations should be carried out considering such protonation states. As a resource, we have provided a list of such atypical states in Supplementary Table 4. PTCH1-molA and -molB were converted to coarse grain resolution using the MARTINI 2.2 forcefield with the ElNeDyn elastic network, a spring force constant of 500 kJ.mol−1 nm−2 and a cutoff of 0.9 nm (cgmartini.nl/index.php/force-field-parameters/particle-definitions). CG PTCH1 structures were embedded in a symmetric POPC:cholesterol (3:1) membrane and solvated with MARTINI water using insane.py in a 15 × 15 × 17 nm3 box (cgmartini.nl/index.php). The transmembrane region was centred in the bilayer and positioned along the z axis according to the atomistic bilayer position determined by MEMPROT MD (memprotmd.bioch.ox.ac.uk). The system was adjusted to approximately 0.15 M NaCl and two rounds of steepest descent energy minimisation were performed. The CG system was equilibrated for 25 ns using NPT ensemble with all protein beads restrained. A second 100 ns equilibration was subsequently performed using NPT ensemble and protein backbone beads restrained. The PTCH1-molA and PTCH1-molB systems were simulated for 5 × 10 μs each, with a 20 fs timestep. The MARTINI 2.2 forcefield was used to describe all components (cgmartini.nl/index.php). A v-rescale thermostat with a coupling constant τt = 1 ps was used to maintain temperature at 310 K. Pressure was maintained at 1 bar using the Parrinello-Rahman barostat, a coupling constant τp =12 ps and compressibility 3 × 10−4 bar−1. Long-range interactions were switched off at 1.1 nm and modelled using the reaction-field method. Lennard-Jones interactions were cutoff at 1.1 nm. The LINCS algorithm was used to constrain bonds to their equilibrium values.
Constructs used in atomistic molecular dynamics simulations.
PTCH1ECD1 crystal structures with cholesterol-HS bound (1.9 Å) or apo (1.9 Å) were used as initial structures for simulation. NB64 was removed and cholesterol was precisely positioned in the ECD1 sterol-binding pocket to replace cholesterol-HS where applicable. CHARMM-GUI pdb generator (www.charmm-gui.org) was used to rename cholesterol atoms to match those compatible with the CHARMM-36 forcefield and model disulphide bonds between C203-C226, C296-C304 and C234-C327. PTCH1ECD1 was centred in a 7 nm3 box, solvated using TIP4P water and neutralised using 0.15 M NaCl. One round of steepest descent energy minimisation was performed. In simulations of the full-length PTCH1ECD, residues A119-D436 and R772-G1023 were extracted from the rebuilt PTCH1-pShhNc (2:1) model with either the C-term SHH-cholesterol moiety bound (PTCH1ECD-molA, “hydroxyl-up” bound form), cholesterol bound (PTCH1ECD-molB, “hydroxyl-down” bound form) or devoid of cholesterol (PTCECD-mol A/B apo). CHARMM-GUI pdb generator (www.charmm-gui.org) was used to rename cholesterol atoms and model disulphide bonds between C234-C327 and C296–304 (PTCH1ECD-molA) and C203-C226, C234-C327 and C296-C304 (PTCH1ECD-molB) in agreement with the full length model. PTCH1ECD systems were centred in a 9 nm3 box, solvated using TIP4P water and adjusted to approximately 0.15 M NaCl. One round of steepest descent energy minimisation was performed. The final snapshot from CG simulations of PTCH1TM embedded in a POPC: cholesterol (3:1) membrane was backmapped to atomistic detail using a fragment based protocol51. The protein structure was backmapped according to the conformation of PTCH1-molA in the rebuilt model while lipids were backmapped to their end point positions. The backmapped PTCH1-lipid system was solvated with TIP4P water, adjusted to 0.15 M NaCl and subjected to three rounds of steepest descent energy minimisation.
Atomistic molecular dynamics simulations.
Simulations were performed using the GROMACS 2018 simulation package (www.manual.gromacs.org). In all simulations a 5 ns NVT equilibration was run followed by a 5 ns NPT equilibration with restraints applied to the protein in both steps. All atomistic simulations were run for 5 × 300 ns with the exception of the backmapped PTCH1TM in membrane, which was simulated for 1 × 100 ns. A 2fs timestep was used. The CHARMM-36 forcefield was used to describe all components. Periodic boundary conditions were applied. A Nosé-Hoover thermostat with a coupling constant τt = 0.5 ps was used to maintain temperature at 310 K. Pressure was maintained at 1 bar using the Parrinello-Rahman barostat, a coupling constant τp = 2 ps and compressibility 4.5 × 10−5 bar−1. A dispersion correction was not applied. Long-range electrostatics were modelled using the PME method and a 1.2 nm cut-off was applied to VDW interactions. The LINCS algorithm was used to constrain bonds to their equilibrium values. In simulations of the whole PTCH1 ECD restraints were applied to residues A119-N120, S435-D436, R772-D773 and I1022-G1023 to mimic the effect of the TMD.
Trajectory Analysis.
Analysis was performed using GROMACS tools (www.manual.gromacs.org) and VMD (www.ks.uiuc.edu/Research/vmd). VMD and pymol were used for visualization. A number of methods exist for tunnel identification and analysis52 and each of these has its strengths and limitations. In common with a number of recent studies53,54 we employed Caver Analyst 2.0 (www.caver.cz) which uses the same algorithm as Caver 3.0 (www.caver.cz/fil/download/manual/caver_userguide.pdf) but simplifies extension of tunnel analysis to trajectories. This increases the likelihood of identifying biologically relevant tunnels and transient tunnels which may not be apparent in a single static structure. A fitted and centred MD trajectory was loaded into the Caver Analyst 2.0 GUI which, like MOLE (mole.upol.cz), uses as Voronoi based algorithm for tunnel identification. Coordinates of cholesterol C17 were used as the start position for tunnel searching. The start point optimization maximum distance and desired radius were 5.0 Å. At each frame in the trajectory protein coordinates were used to construct a Voronoi diagram and tunnels were identified by following Voronoi vertices from the start point to the bulk solvent using a probe which specifies the minimum tunnel radius necessary for the vertex to be traversed. A probe radius of 2.0 Å was used and atoms were approximated using 8 balls to construct Voronoi diagrams. A shell radius of 3 Å and shell depth of 4 Å were used to define regions of the Voronoi diagram corresponding to bulk solvent. After tunnels have been identified at each frame in the trajectory an average-link hierarchical algorithm is used to cluster tunnels. The tree hierarchy of tunnel clusters was cut using a clustering threshold of 3.5.
Surface plasmon resonance (SPR).
SPR experiments were performed using a BIAcore T200 machine (GE Healthcare) at 25 °C in 10 mM HEPES, pH 7.5, 150 mM NaCl, 3 mM CaCl2, 0.005% (v/v) Polysorbate 20. The biotinylated PTCH1ECD1-ECD2 was immobilized via streptavidin onto a CM5 sensor Chip. PTCH1ECD1-ECD2 surface concentration was 820 response units. ShhNC24II (used as analyte) was dialyzed against SPR running buffer prior to use and a two-fold dilution series was prepared. The experiment was run at 5 μl/min with 600 s association and 400 s dissociation. The signal from the experimental flow cell was corrected by subtraction of a buffer and the reference signal from a control flow cell (streptavidin coated). All data were analyzed using SCRUBBER2 (Biologic) and GraphPad Prism Version 8 (GraphPad Software, San Diego USA). The dissociation constant (Kd) was obtained by nonlinear regression using a “one-site specific binding” model (Y=Bmax∙X/(Kd+X), where X is analyte concentration and Bmax is the maximum analyte binding).
Bio-layer Interference.
Multi-cycle kinetic measurements were carried out using an Octet RED384 instrument (Pall ForteBio) in 384-well plates at 25 °C and 1000 rpm. Streptavidin-coated biosensors (Pall ForteBio Dip and Read™) were hydrated in assay buffer for 10 min before being immobilised with biotinylated PTCH1 constructs at 0.1 mg/mL for 300 s. After recording a 60 s baseline step, the biosensors were exposed to an eight-point, two-fold dilution series of nanobody. Association and dissociation steps were carried out for 300 s respectively. Recorded data were double referenced by subtracting buffer interference on a PTCH1-loaded reference sensor and nanobody interaction on parallel biosensors with no PTCH1 immobilised. Data analysis was carried out using the Octet version V9.0 data analysis software provided by ForteBio.
Isothermal titration calorimetry.
Experiments were performed using a MicroCal PEAQ-ITC (Malvern) at 25 °C in 10 mM HEPES, pH 7.5, 150 mM NaCl, 3 % isopropanol, with a differential power of 10 μcal/s and stirring at 750 rpm. Experiments consisted of an initial test injection of 0.4 μl, followed 150–240 s later by 12 injections of 2 μl spread over x s, spaced 150 s apart. Due to the low solubility of cholesterol a PEGylated cholesterol PEG-CHL (see below) was used for affinity measurements. PTCH1ECD1 wildtype and mutants were dialysed against 0.15 M NaCl, 10 mM HEPES, 3% Isopropanol, final pH 7.5. Lyophilised PEG-CHL was resuspended in dialysis solution to a concentration of 1 mM. Protein concentrations were determined from the absorbance at 280 nm using calculated molar extinction coefficients. Cell concentrations of 40–70 μM PTCH1ECD1 proteins and syringe concentrations of 1–2.4 mM PEG-CHL were used for all experiments. Thermograms were integrated and corrected for heats of dilution using NITPIC (http://biophysics.swmed.edu/MBR/software.html). The resulting isotherms were analysed in SEDPHAT (http://www.analyticalultracentrifugation.com/sedphat/). Isotherms were fitted with the A+B ⇌AB model, where cell and syringe concentrations and baselines of each experiment were fitted locally. All figures were prepared using GUSSI (http://biophysics.swmed.edu/MBR/software.html).
UNcle stability.
The samples (9μL at 0.4mg/mL) were heated from 15 to 90 °C at a rate of 1 °C/min and tested in quadruplicate in 3 independent experiments on UNcle (Unchained Labs, US). The thermal unfolding was monitored by intrinsic fluorescence utilising excitation at 266nm with broad-spectrum emission detection. The raw spectral data were processed using the barycentric mean output. The barycentric mean of the fluorescence corresponds to the change in the center of the mass of the wavelength of the fluorescence emission. The barycentric mean as a function of temperature was used to calculate the melting temperatures. Data analysis was performed with the UNcle Analysis software and Graphpad Prism Version 8 (GraphPad Software, San Diego USA).
Hedgehog signalling assays using quantitative RT-PCR.
Gli1 mRNA is a commonly used metric for Hh signalling strength, because Gli1 is a direct Hh target gene. NIH/3T3 cells grown to confluency in DMEM with 10% Fetal Bovine Serum (FBS) were switched to media with 0.5% FBS (to induce ciliation) containing various SHH ligands for 24 hours. Gli1 mRNA levels were measured by quantitative Reverse Transcription PCR (qRT-PCR) using the Power SYBR Green Cells-to-CT kit (Thermo Fisher Scientific) as described previously55. Gli1 transcript levels relative to Gapdh were calculated using the ΔCt or ΔΔCt method. Primers used for PCR are as follows: Gli1 (forward primer: 5′-ccaagccaactttatgtcaggg-3′ and reverse primer: 5′-agcccgcttctttgttaatttga-3′); Gapdh (forward primer: 5′-agtggcaaagtggagatt-3′ and reverse primer: 5′-gtggagtcatactggaaca-3′). Each qRT–PCR experiment, which was repeated 3 times, included two biological replicates, each with two technical replicates.
Peptides.
All peptides were >90% pure as assessed by LC-MS. Palm-ShhN15 (Palmitate-CGPGRGFGKRRHPKK), a palmitoylated peptide containing the N-terminal 15 amino acids of human SHH was synthesized (Elim Biosciences) by solid-phase synthesis and purified by HPLC to >98% purity. ShhN7-chol (VAAKSGG-cholesterol), a cholesteroylated peptide containing the C-terminal 7 aars of human SHH, was kindly provided by Professor Herbert Waldmann. Its synthesis, purification (>90%) and characterization have been described in detail previously56.
Synthesis data of steroid 1.
For synthetic procedures, see Supplementary Note.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Herbert Waldmann and Sonja Sievers (Max Planck Institute of Molecular Physiology) for the kind gift of the cholesteroylated SHH peptide (ShhN7-chol), Nele Buys for the technical assistance during Nanobody discovery, Tom Walter and Karl Harlos (STRUBI) for help with crystallisation, and Oleg Fedorov (SGC) for help with the BLI measurements. CS was supported by grants from Cancer Research UK (C20724/A14414 and C20724/A26752) and a European Research Council grant (647278). DFC was supported by a grant from the National Institutes of Health (HL067773) and the Taylor Family Institute for Innovative Psychiatric Research. RR was supported by grants from the National Institutes of Health (GM118082 and GM106078), C.K. was supported by a Cancer Research UK studentship (C20724/A16135) and MK by a pre-doctoral fellowship from the National Science Foundation. M.S.P.S. was supported by the Wellcome Trust (208361/Z/17/Z and 102164/B/13/Z), BBSRC (BB/R00126X/1) and EPSRC (EP/L000253/1). This project made use of time on ARCHER granted via the UK High-End Computing Consortium for Biomolecular Simulation, HECBioSim (www.hecbiosim.ac.uk), supported by EPSRC (EP/R029407/1). This work benefited from access to the Nanobodies4Instruct centre (PID1129), we acknowledge the support and use of resources of Instruct-ERIC, part of the European Strategy Forum on Research Infrastructures (ESFRI). We acknowledge the Research Foundation - Flanders (FWO) for their support to the Nanobody discovery. The Wellcome Centre for Human Genetics, Oxford, is funded by Wellcome Trust Core Award 203852/Z/16/2.
Footnotes
Competing interests
The authors declare no competing interests.
DATA AVAILABILITY
Atomic coordinates and structure factors for PTCH1ECD1-NB64, PTCH1ECD1-NB64-cholesterol-hemisuccinate, apo-PTCH1ECD1 and PTCH1ECD2-NB75, as well as the coordinates for the revised PTCH1-SHH complex have been deposited in the Protein Data Bank (PDB) under accession numbers 6RTY, 6RTW, 6RTX, 6RVC and 6RVD.
MAIN MANUSCRIPT REFERENCES
- 1.Kong JH, Siebold C & Rohatgi R Biochemical mechanisms of vertebrate hedgehog signaling. Development 146, doi: 10.1242/dev.166892 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porter JA, Young KE & Beachy PA Cholesterol modification of hedgehog signaling proteins in animal development. Science 274, 255–259 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Pepinsky RB et al. Identification of a palmitic acid-modified form of human Sonic hedgehog. J Biol Chem 273, 14037–14045 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Luchetti G et al. Cholesterol activates the G-protein coupled receptor Smoothened to promote morphogenetic signaling. Elife 5, doi: 10.7554/eLife.20304 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang P et al. Cellular Cholesterol Directly Activates Smoothened in Hedgehog Signaling. Cell 166, 1176–1187 e1114, doi: 10.1016/j.cell.2016.08.003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bidet M et al. The hedgehog receptor patched is involved in cholesterol transport. PLoS One 6, e23834, doi: 10.1371/journal.pone.0023834 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y et al. Structural Basis for Cholesterol Transport-like Activity of the Hedgehog Receptor Patched. Cell 175, 1352–1364 e1314, doi: 10.1016/j.cell.2018.10.026 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marigo V, Davey RA, Zuo Y, Cunningham JM & Tabin CJ Biochemical evidence that patched is the Hedgehog receptor. Nature 384, 176–179 (1996). [DOI] [PubMed] [Google Scholar]
- 9.Stone DM et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature 384, 129–134 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Carstea ED et al. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science 277, 228–231 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Loftus SK et al. Murine model of Niemann-Pick C disease: mutation in a cholesterol homeostasis gene. Science 277, 232–235 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Tseng TT et al. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. Journal of molecular microbiology and biotechnology 1, 107–125 (1999). [PubMed] [Google Scholar]
- 13.Davies JP & Ioannou YA Topological analysis of Niemann-Pick C1 protein reveals that the membrane orientation of the putative sterol-sensing domain is identical to those of 3-hydroxy-3-methylglutaryl-CoA reductase and sterol regulatory element binding protein cleavage-activating protein. J Biol Chem 275, 24367–24374, doi: 10.1074/jbc.M002184200 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Qi X, Schmiege P, Coutavas E, Wang J & Li X Structures of human Patched and its complex with native palmitoylated sonic hedgehog. Nature 560, 128–132, doi: 10.1038/s41586-018-0308-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong X et al. Structural basis for the recognition of Sonic Hedgehog by human Patched1. Science 361, doi: 10.1126/science.aas8935 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Qi X, Schmiege P, Coutavas E & Li X Two Patched molecules engage distinct sites on Hedgehog yielding a signaling-competent complex. Science 362, doi: 10.1126/science.aas8843 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qian H et al. Inhibition of tetrameric Patched1 by Sonic Hedgehog through an asymmetric paradigm. Nature communications 10, 2320, doi: 10.1038/s41467-019-10234-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bishop B et al. Structural insights into hedgehog ligand sequestration by the human hedgehog-interacting protein HHIP. Nat Struct Mol Biol 16, 698–703 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tukachinsky H, Petrov K, Watanabe M & Salic A Mechanism of inhibition of the tumor suppressor Patched by Sonic Hedgehog. Proc Natl Acad Sci U S A 113, E5866–E5875, doi: 10.1073/pnas.1606719113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters C, Wolf A, Wagner M, Kuhlmann J & Waldmann H The cholesterol membrane anchor of the Hedgehog protein confers stable membrane association to lipid-modified proteins. Proc Natl Acad Sci U S A 101, 8531–8536, doi: 10.1073/pnas.0308449101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Byrne EF et al. Structural basis of Smoothened regulation by its extracellular domains. Nature 535, 517–522, doi: 10.1038/nature18934 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis PM et al. Cholesterol modification of sonic hedgehog is required for long-range signaling activity and effective modulation of signaling by Ptc1. Cell 105, 599–612 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Chen MH, Li YJ, Kawakami T, Xu SM & Chuang PT Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev 18, 641–659 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burke R et al. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell 99, 803–815 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Chen Y & Struhl G Dual roles for patched in sequestering and transducing Hedgehog. Cell 87, 553–563 (1996). [DOI] [PubMed] [Google Scholar]
- 26.McLellan JS et al. The mode of Hedgehog binding to Ihog homologues is not conserved across different phyla. Nature 455, 979–983 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bosanac I et al. The structure of SHH in complex with HHIP reveals a recognition role for the Shh pseudo active site in signaling. Nat Struct Mol Biol 16, 691–697 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Whalen DM, Malinauskas T, Gilbert RJ & Siebold C Structural insights into proteoglycan-shaped Hedgehog signaling. Proc Natl Acad Sci U S A 110, 16420–16425, doi: 10.1073/pnas.1310097110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kavran JM, Ward MD, Oladosu OO, Mulepati S & Leahy DJ All mammalian Hedgehog proteins interact with cell adhesion molecule, down-regulated by oncogenes (CDO) and brother of CDO (BOC) in a conserved manner. J Biol Chem 285, 24584–24590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chuang PT & McMahon AP Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature 397, 617–621, doi: 10.1038/17611 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Allen BL et al. Overlapping roles and collective requirement for the coreceptors GAS1, CDO, and BOC in SHH pathway function. Dev Cell 20, 775–787 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Izzi L et al. Boc and Gas1 each form distinct Shh receptor complexes with Ptch1 and are required for Shh-mediated cell proliferation. Dev Cell 20, 788–801 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rubin JB, Choi Y & Segal RA Cerebellar proteoglycans regulate sonic hedgehog responses during development. Development 129, 2223–2232 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Hausmann G, von Mering C & Basler K The hedgehog signaling pathway: where did it come from? PLoS Biol 7, e1000146, doi: 10.1371/journal.pbio.1000146 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bazan JF & de Sauvage FJ Structural ties between cholesterol transport and morphogen signaling. Cell 138, 1055–1056, doi: 10.1016/j.cell.2009.09.006 (2009). [DOI] [PubMed] [Google Scholar]
METHODS REFERENCES
- 36.Molday RS & MacKenzie D Monoclonal antibodies to rhodopsin: characterization, cross-reactivity, and application as structural probes. Biochemistry 22, 653–660 (1983). [DOI] [PubMed] [Google Scholar]
- 37.Aricescu AR, Lu W & Jones EY A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr 62, 1243–1250, doi: 10.1107/S0907444906029799 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Taylor FR et al. Enhanced potency of human Sonic hedgehog by hydrophobic modification. Biochemistry 40, 4359–4371 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Tukachinsky H, Kuzmickas RP, Jao CY, Liu J & Salic A Dispatched and scube mediate the efficient secretion of the cholesterol-modified hedgehog ligand. Cell Rep 2, 308–320, doi: 10.1016/j.celrep.2012.07.010 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whalen DM, Malinauskas T, Gilbert RJ & Siebold C Structural insights into proteoglycan-shaped Hedgehog signaling. Proc Natl Acad Sci U S A 110, 16420–16425, doi: 10.1073/pnas.1310097110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pardon E et al. A general protocol for the generation of Nanobodies for structural biology. Nat Protoc 9, 674–693, doi: 10.1038/nprot.2014.039 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walter TS et al. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr D Biol Crystallogr 61, 651–657, doi: 10.1107/S0907444905007808 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winter G xia2: an expert system for macromolecular crystallography data reduction. J Appl Crystallogr 43, 186–190, doi: 10.1107/S0021889809045701 (2010). [DOI] [Google Scholar]
- 44.Schneider TR & Sheldrick GM Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr 58, 1772–1779 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221, doi: 10.1107/S0907444909052925 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCoy AJ et al. Phaser crystallographic software. Journal of applied crystallography 40, 658–674, doi: 10.1107/S0021889807021206 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501, doi: 10.1107/S0907444910007493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murshudov GN et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67, 355–367, doi: 10.1107/S0907444911001314 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.BUSTER v. 2.10.2 (Global Phasing Ltd, Cambridge, United Kingdom, 2011). [Google Scholar]
- 50.Qi X, Schmiege P, Coutavas E & Li X Two Patched molecules engage distinct sites on Hedgehog yielding a signaling-competent complex. Science 362, doi: 10.1126/science.aas8843 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stansfeld PJ & Sansom MS From Coarse Grained to Atomistic: A Serial Multiscale Approach to Membrane Protein Simulations. Journal of chemical theory and computation 7, 1157–1166, doi: 10.1021/ct100569y (2011). [DOI] [PubMed] [Google Scholar]
- 52.Krone M et al. Visual Analysis of Biomolecular Cavities: State of the Art. Computer Graphics Forum 35, 527–551, doi: 10.1111/cgf.12928 (2016). [DOI] [Google Scholar]
- 53.Jussupow A, Di Luca A & Kaila VR I. How cardiolipin modulates the dynamics of respiratory complex I. Sci Adv 5, eaav1850, doi: 10.1126/sciadv.aav1850 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stock C et al. Cryo-EM structures of KdpFABC suggest a K(+) transport mechanism via two inter-subunit half-channels. Nature communications 9, 4971, doi: 10.1038/s41467-018-07319-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Byrne EF et al. Structural basis of Smoothened regulation by its extracellular domains. Nature 535, 517–522, doi: 10.1038/nature18934 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peters C, Wolf A, Wagner M, Kuhlmann J & Waldmann H The cholesterol membrane anchor of the Hedgehog protein confers stable membrane association to lipid-modified proteins. Proc Natl Acad Sci U S A 101, 8531–8536, doi: 10.1073/pnas.0308449101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.