Abstract
Cleft lip with or without cleft palate (CL/P) is generally viewed as a complex trait with multiple genetic and environmental contributions. In 70% of cases, CL/P presents as an isolated feature and/or deemed non-syndromic. In the remaining 30%, CL/P is associated with multisystem phenotypes or clinically recognizable syndromes, many with a monogenic basis. Here we report the identification, via exome sequencing, of likely pathogenic variants in two genes that encode interacting proteins previously only linked to orofacial clefting in mouse models. A variant in GDF11 (encoding Growth Differentiation Factor 11), predicting a p.(Arg298Gln) substitution at the Furin protease cleavage site, was identified in one family that segregated with CL/P and both rib and vertebral hyper-segmentation, mirroring that seen in Gdf11 knockout mice. In the second family in which CL/P was the only phenotype, a mutation in FST (encoding the GDF11 antagonist, Follistatin) was identified that is predicted to result in a p.(Cys56Tyr) substitution in the region that binds GDF11. Functional assays demonstrated a significant impact of the specific mutated amino acids on FST and GDF11 function and, together with embryonic expression data, provide strong evidence for the importance of GDF11 and Follistatin in the regulation of human orofacial development.
Keywords: Cleft lip, cleft palate, follistatin, GDF11, vertebral hyper-segmentation
INTRODUCTION
Orofacial clefts (OFCs) collectively represent the second most common structural birth defect observed in humans (Dixon et al., 2011; Leslie & Marazita, 2013). OFCs can arise as a result of a principal genetic mutation or from the combined impact of genetic variants and environmental factors. In either situation, the precise spatiotemporally controlled growth and fusion of the embryonic facial prominences during the 6th (oronasal) and/or the 10th (secondary palate) weeks of gestation (Cox, 2004; Dixon et al., 2011) is sufficiently disrupted to result in the respective cleft type. The most common types of OFC presentations are clefts that involve the lip and/or alveolus (CL), clefts of both the lip/alveolus and secondary palate (CLP) and clefts of the secondary palate only (CP). Notably, CL often segregates in families with CLP and in such cases is referred to as cleft lip with or without cleft palate (CL/P), whereas it is uncommon to observe CP segregating with CL or CLP (Leslie & Marazita, 2013). This, and the temporal separation of primary and secondary palatal development, are consistent with the two processes being genetically distinct yet still sharing numerous genetic pathways and mechanisms (Cox, 2004). Severe clefting of the lip and alveolus can also physically restrict the secondary palatal shelves from contacting at the midline and thus in some cases the CP may be a secondary pathology (Cox, 2004). Although the most common forms of OFC are isolated and non-syndromic, 30% of cases are associated with multisystem phenotypes or clinically recognizable syndromes (Dixon et al., 2011; Leslie & Marazita, 2013).
Mice have proven to be useful models of human developmental disorders including OFCs. There are hundreds of reported mouse strains, including both gene targeted, spontaneously arisen and randomly mutagenized (for example, using N-ethyl-N-nitrosourea [ENU]) that exhibit OFCs as a component of the phenotype (Keteyian & Mishina, 2017). However, in contrast to humans, the vast majority involve only the secondary palate, with at most just a few dozen displaying CL. Nevertheless, mouse CP loci are frequently included in candidate gene lists for human OFCs (Gritli-Linde, 2012; Yu et al., 2017) and pathogenic variants have often been extensively characterized in mouse models well prior to the identification of the analogous human disorder.
We report the identification, via exome sequencing, of likely pathogenic variants in the genes GDF11 and FST, with both genes having been implicated previously in the pathogenesis of OFCs in mouse models. One family harboring a likely pathogenic FST variant presented with isolated CL/P whereas the other family, with a likely pathogenic GDF11 variant, represents a new orofacial clefting syndrome distinguished by vertebral and rib hyper-segmentation. Notably, the Gdf11 knockout mouse also exhibits both vertebral and rib anomalies, recapitulating the human phenotype. Moreover, GDF11 and FST encode physically interacting proteins: ligand and antagonist. Collectively, these results establish a previously unappreciated role for this regulatory pathway in human OFC pathogenesis.
METHODS
Patient recruitment and sample collection
The two families described in this report were part of a larger cohort of 72 families with multiple individuals affected with orofacial clefts (Cox et al., 2018) that were ascertained in the United States, Colombia and Australia. Each family was enrolled into the study under the respective institutional review board protocols: South Eastern Sydney Local Health District (HREC/13/POWH/203) Australia; Genetics of CL/P: a multicenter international consortium (PIROSTUDY10777) and the University of Iowa IRB (200109094). Informed consent was obtained prior to sample testing. DNA was extracted using standard protocols and sample quality control steps were taken including DNA quantification using Qubit (Invitrogen), co-efficient of inbreeding determinations, and XY genotyping to confirm the gender of the tested individuals.
Exome sequencing and genomic data generation
DNA samples from multiple affected individuals from each of the 72 families in the cohort were selected for exome sequencing as previously described (Cox et al., 2018). A Roche/Nimblegen SeqCap EZ v2.0 kit was used for enrichment capture and samples sequenced at the University of Washington’s Center for Mendelian Genomics (Seattle, WA) using an Illumina HiSeq 2500. The raw sequences which met internal quality standards were aligned to the human genome build hg19 (GRCh37) using BWA software (Li & Durbin, 2009). Single Nucleotide Variant (SNV) and Insertion / Deletion (indel) calls were performed using PICARD, SAMTOOLS, and GATK (DePristo et al., 2011; Li et al., 2009; Van der Auwera et al., 2013).
Bioinformatics and genomic data analysis
Individual-level single nucleotide variants (SNVs) and indels were joint-called using GATK (DePristo et al., 2011) into a single multi-sample VCF. Variants were annotated with the Ensembl Variant Effect Predictor (VEP) (McLaren et al., 2016). Variants were then filtered using the open-source GEMINI platform (Paila et al., 2013) according to a standardized set of criteria to identify rare and likely pathogenic variants (Cox et al., 2018). Given the pedigree structures of both families that are described in this report were consistent with autosomal dominant inheritance, shared heterozygous variants within each family were then prioritized using clinical and bioinformatic criteria, including population frequencies and in silico pathogenicity scores. Variants with allele frequencies >1% within the 1000 Genomes database (http://www.1000genomes.org/), the Seattle-based Exome Variant Server (EVS) database (http://snp.gs.washington.edu/EVS/), the Exome Aggregation Consortium (ExAC; http://www.exac.broadinstitute.org) and genome Aggregation Database (gnomAD; http://www.gnomad.broadinstitute.org) were excluded from further analysis. A conservative CADD threshold score (http://cadd.gs.washington.edu) of 15 was then used to prioritize variants for pathogenicity with other in silico scores including PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) and SIFT (http://sift.jcvi.org/) compared for consistency. Evolutionary conservation, effects on protein structure, absence in controls, the potential impact of the mutation on protein function, expression in mouse and human embryonic orofacial tissues, and links to biological pathways previously implicated in palatogenesis were then used to further prioritize candidate genes. Any variant deemed pathogenic and found in a gene with a mouse model that presented with OFC phenotypes was considered among the top candidates. We also assessed the full list of remaining candidate genes (sixteen for Family 4527, twenty for Family 22) using DOMINO, a variant effect prediction tool that predicts genes associated with dominant disorders based on linear discriminant analysis and independent of the mutational events. Top candidate variants in each family (seven in Family 4527, six in Family 22) were then assessed for segregation within each family using Sanger sequencing of PCR amplicons from all individuals for whom DNA was available.
3D protein modeling
Where possible, the effects of variants on protein function were inferred based on location in a conserved protein functional domain (PFAM database) with additional structural mutated protein analysis performed using the HOPE protein analysis server (http://www.cmbi.ru.nl/hope/). The GDF11-FST complex and location of the FST mutated residue was visualized using PyMOL based on the available PDB structural model (http://www.rcsb.org/pdb/home/home.do; 5JHW).
Expression analysis using SysFACE
The bioinformatics tool, SysFACE (Systems tool for craniofacial expression-based gene discovery), was used to analyze microarray-based genome-level gene expression profiles from various mouse embryonic orofacial tissues in the FaceBase (https://www.facebase.org) and NCBI GEO repositories (https://www.ncbi.nlm.nih.gov/geo/) (Lachke et al., 2012; Liu et al., 2017). The following expression datasets were analyzed using Lachke lab in-house scripts: Illumina HiSeq2500 RNA-seq data for E14.5 mouse posterior (FaceBase: FB00000768.01) and anterior palate tissue (FaceBase: FB00000769.01), Affymetrix 430 2.0 microarray data from the mouse embryonic orofacial tissues – mandible, maxilla and frontonasal tissue (at stages E10.5, E11.0, E11.5, E12.0, E12.5) (GEO: GSE7759) and E13.5 palate (FaceBase: FB00000468.01). The in-house scripts used for analysis are available upon request.
STRING-based protein-protein interaction network of GDF11
Known and predicted human protein-protein interactions (PPIs) for GDF11 were extracted from the STRING database (http://string-db.org) using an in-house Python script. The obtained high-confidence direct and nearest neighbor PPIs with interaction score > 0.07 were integrated into an open source tool, Cytoscape, for visualization of the GDF11 network.
Tissue collection and immunohistochemistry
Human fetal tissues were recovered under an approved protocol through the University of Washington (IRB# STUDY00000380). Specimens ranging in age between 57 and 70 days post conception, estimated by gestational ultrasound and/or fetal foot measurements (Shepard, 1975), were used as these represent the period during which the secondary palate fuses. Younger specimens spanning the time of primary palatal fusion were not available. Embryonic tissues were embedded in paraffin and 4-micron sections cut in the coronal plane. Sections were then stained with the following primary antibodies: anti-FST [H114] (1:100, Santa Cruz #sc30194) and anti-GDF11 (1:100, R&D Systems #MAB19581) followed by a biotinylated secondary antibody and ABC-HRP of the appropriate specificity as previously described (Tamasas & Cox, 2017).
Sanger sequencing
For candidate variants of interest, primers were designed for amplification of the respective genomic regions. Genomic DNA samples extracted from patients’ saliva or blood were used as template for PCR from available affected and unaffected relatives, with products sequenced on an Applied Biosystems Model 3730 (48-capillary) sequencer (Functional Biosciences, Inc., Madison, WI, USA). The chromatograms were analyzed and visualized by APE (http://biologylabs.utah.edu/jorgensen/wayned/ape/). Primer sequences for all variants are available upon request.
Plasmid constructs
The full coding sequence of human GDF11 was amplified by PCR using the following primers: 5’-CACCATGGTGCTCGCGGCCCCGCT-3’ and 5’-CGCTGTGGCTGCTCTTAA-3’. The product was then double-digested with NheI-HF and XbaI (New England Biolabs) and cloned into pcDNA3.1(+) (Life Technologies, Carlsbad, CA, USA). An alternate GDF11 expression construct was also made using the pRK5 vector (BD PharMingen) for experiments using HEK293T cells. Constructs for expression of wildtype human FST (isoform Fs288; in pcDNA3.1) (Cash et al., 2012a) and human Furin (in pcDNA4; Walker et al., 2017) were previously described. Site-directed mutagenesis was employed to generate the respective expression vectors carrying the GDF11:c.893G>A and FST:c.167G>A variants. Separately, four SMAD-binding elements (SBE, 5’-CAGACA-3’) were synthesized (IDT, Coralville, IA, USA) and cloned into pENTR/D-TOPO and then shuttled into a cFos-FFLuc plasmid (SBE-FFLuc) for the in vitro luminescence reporter activity assay.
Cell culture
The following cell lines were used in this study: a human embryonic oral epithelial cell line; GMSM-K (gift from Dr Daniel Grenier, Université Laval, Quebec, Canada); a human embryonic palatal mesenchyme cell line; HEPM (from ATCC, CRL-1486); and a derivative of the HEK293T epithelial cell line, HEK293-(CAGA)12, in which a SMAD3 binding element-linked luciferase reporter had been stably integrated (Cash et al., 2012a). GMSM-K cells were maintained in keratinocyte serum-free medium (Life Technologies) supplemented with EGF and bovine pituitary extract (Life Technologies). HEPM and HEK293 cells were maintained according to supplier guidelines. Cells were incubated at 37 °C in 5% CO2.
Luciferase reporter assays
For the dual luciferase activity assays in GMSM-K and HEPM cells, each SBE-FFLuc-based reporter construct was co-transfected with a Renilla Luciferase expression plasmid. Plasmids were electroporated into HEPM cells with Amaxa Basic Nucleofector II (Lonza) (Program: U-020), and electroporated into GMSM-K cells with Amaxa Cell Line Nucleofector Kit V (Lonza, Cologne, Germany) (Program: X-005). A Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) and 20/20n luminometer (Turner Biosystems, Sunnyvale, CA, USA) were employed to measure the luciferase activity at 72 hours post-transfection. Relative luciferase activities were calculated by the ratio between the value of firefly and Renilla luciferase activities. Three biological replicates were performed for each assay. All quantified results are presented as mean ±s.d. A Student’s t-test was used to determine statistical significance.
Luciferase assays to test for FST antagonism were performed using the FST isoform, Fs288, and the HEK293-(CAGA)12 reporter cell line as previously described (Cash et al., 2012b; Walker et al., 2018). In short, 2×104 HEK293-(CAGA)12 cells were seeded in a 96 well plate in growth media with serum for 18hrs. 50ng of Fs288 WT or Cys56Tyr mutant and 150ng of empty vector (for 200ng of total DNA) was then transfected using TransIT-LT1 reagent (Mirus Bio) in Opti-MEM media (Fisher 31985–070) according to the manufacturer’s protocol. 18hrs following DNA addition, the growth media was swapped out for serum free media containing 0.62nM of GDF11. 24hrs later, cells were lysed and luminesce measured. Data was plotted as fold relative to GDF11 activation and one-way Anova statistical analysis conducted within GraphPad Prism 5 software.
Western blotting
Westerns were conducted on samples from cells plated in a 6-well format. Briefly, 5×105 HEK293T cells were seeded in each well with growth media for 18 hours. After this time, 1.5ug of Fs288 WT or Cys56Tyr mutant vector DNA was transfected using TransIT-LT1 reagent (Mirus Bio). For GDF11, 1.5ug of WT or Arg198Gln construct DNA was transfected along with 1.5ug of Furin vector DNA to improve processing. 24hours post transfection, growth media was swapped for serum free media. Samples were collected and subject to standard western protocols. Primary antibodies against FST/Fs288 and GDF11 were purchased from R&D Biosystems (MAB66) and Abcam (ab124721), respectively. Goat anti-mouse (Millipore-Sigma; DC02L) and goat anti-rabbit (Santa Cruz; sc-2301) HRP-linked secondary antibodies, respectively, were used for detection. Western blot signal was captured using the C-DiGit blot scanner (LI-COR).
RESULTS
Clinical phenotype
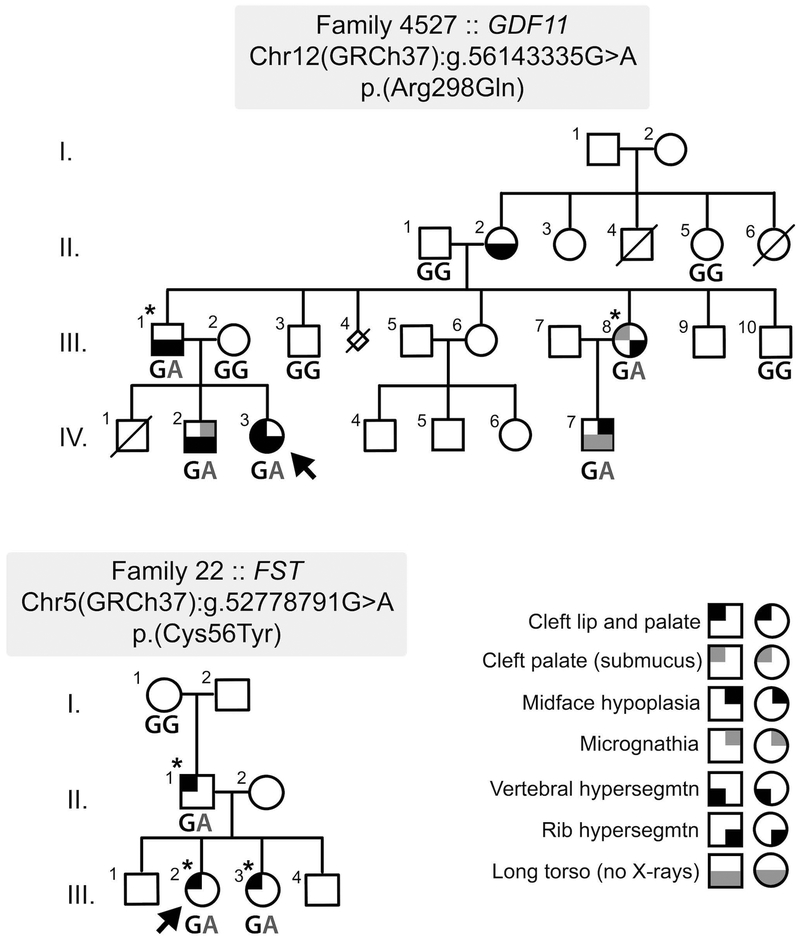
Family 4527 included six affected individuals each of whom manifested vertebral hyper-segmentation affecting different spinal levels. Multiple rib hyper-segmentation was also seen in three of these 6 individuals, while three presented with OFCs: two with unilateral CLP and one with a submucous CP. An additional member had midface hypoplasia (Table 1). Family 22 included three affected individuals: the female proband (CLP), her sister (CLP), and her father (CL). The proband’s two brothers and her paternal grandparents were unaffected (Figure 1).
Table 1:
Family 4527 Clinical Phenotype
| IV-2 Brother | IV-3 (Proband) | IV-7 Paternal Cousin | III-1 Father | III-7 Paternal Aunt | II-2 Paternal Grandmother | |
|---|---|---|---|---|---|---|
| Height | 60-97th percentile | 88th-94th percentile | >90th percentile | >99th percentile | >99th percentile | 95th percentile |
| Vertebrae | 8 Cervical | 7 Cervical | Long Torso, no x-rays | 8 Cervical | Rib hyper-segmentation | Thoracic hyper-segmentation |
| 14 Thoracic and 14 Ribs | 14 Thoracic and 14 Ribs | 14 Thoracic and 14 Ribs | ||||
| 6 Lumbar | 6 Lumbar | 6 Lumbar | ||||
| Ears | Darwinian tubercle | Bilateral supernumerary upper helix crus | Bilateral thickened Helices | L Pre-auricular fistula | NAD | |
| Thickened superior helix | L Pre-auricular fistula | L folded and thickened Superior helix | Bilateral dysmorphic pinnae | |||
| Folded superior helix | ||||||
| Orofacial | Overbite / micrognathia | Unilateral left cleft lip and palate | Midface hypoplasia | Unilateral Left Cleft Lip and Palate | Submucous cleft palate Velopharyngeal insuffic. Pharyngeal flap age 21 | NAD |
| Facial | Epicanthic folds | NAD | NAD | NAD | NAD | NAD |
| Blue sclerae | ||||||
| Upturned nasal tip | ||||||
| Prominent occiput | ||||||
| Widow's peak | ||||||
| Skeletal/Connective Tissue | Flat feet | NAD | NAD | NAD | NAD | NAD |
| Mild pectus excavatum | ||||||
| Winged scapulae | ||||||
| Hypermobile joints | ||||||
| Inguinal hernias | Inguinal hernias Treated 3 months | Inguinal hernia Treated 54 years | ||||
| Other | weight:height <1st percentile | VSD at birth, closed spontaneously | weight:height <1st percentile | Bilateral accessory nipples | Died 54 heart disease |
Figure 1: Pedigree structure of Families 4527 and 22 with GDF11 and FST variant segregation, respectively.
The proband in each pedigree is marked by an arrow. The individuals from each family that had exomes completed are asterisked.
Exome Sequencing
In Family 4527, the initial exome data analysis on the affected male 4527–2 (individual III-I; Figure 1, Table 1) and his affected sister 4527–13 (individual III-8; Figure 1, Table 1) detected 973 heterozygous rare variants in common with an allele frequency of <1% in control databases (gnomAD, ExAC, 1000 genomes and the Seattle EVS). Of these, 135 variants were unique to this family. Sixteen genes had one rare variant within the coding sequence, of which one was a truncating variant and five were predicted to be both damaging (Polyphen) and deleterious (SIFT) (Supp. Table S1). A missense variant in GDF11 (NC_000012.11:g.56143335G>A; NM_005811.4:c.[893G>A];[=]; NP_005802.1: p.(Arg298Gln)) was considered the top candidate as targeted knockout of its mouse ortholog results in orofacial and skeletal anomalies (McPherron et al., 1999). The perturbed arginine residue (Arg298) is highly conserved across multiple species. In silico analysis of the GDF11 variant uniformly predicted a deleterious effect on protein function: called deleterious by SIFT (0.001), probably damaging by Polyphen-2 (0.982), disease-causing by MutationTaster (p-value: 1), and deleterious by Provean (−3.2; threshold for consideration <−2.5). The variant was also classified in the top 0.03% of substitutions for deleteriousness by CADD with a score of 35 (Supp. Tables S1 and S5). Sanger sequencing in the 10 available family members (five affected and five unaffected) confirmed heterozygosity for the variant in each of the affected individuals, supporting an autosomal dominant mode of inheritance (Figure 1). The five unaffected family members examined were homozygous for the wildtype allele (G/G). As further support, GDF11 was also the top-ranked of the candidate genes from Family 4527 by DOMINO, being rated as “very likely dominant” (Supp. Table S2). Four other genes were classified as either “very likely dominant” (CHRM3), “likely dominant” (TNS3), or “either dominant or recessive” (PCDH8 and TRPM2). Segregation analysis of each of these additional four variants was then performed on available samples. Variants from HARS and TM2D3 were also included in the segregation analysis as they were noted as having a moderately small number (<100) of loss of function alleles listed in gnomAD, a feature of many other known CL/P genes. For these six additional candidate variants, none show perfect segregation with the clinical presentation, with each variant being present in 1 (CHRM3), 2 (TRPM2 and HARS), or 3 (TNS3, PCDH8 and TM2D3) unaffected individuals (Supp. Table S6A).
Exome sequencing was performed on three affected individuals in Family 22 (individuals II-1, III-2 and III-3) (Figure 1). These three individuals shared 793 heterozygous rare variants with an allele frequency of <1% in reference databases. Of these, 89 variants were unique to the family in this study and not present in any sequenced individuals from the additional 71 families in the CL/P cohort. Twenty genes had one rare variant within the coding sequence, of which three were truncating variants and seven were missense changes predicted to be both damaging (Polyphen) and deleterious (SIFT) (Supp. Table S3). A missense variant (NC_000005.9:g.52778791G>A; NM_013409.2 c.[167G>A];[=]; NP_037541.1: p.(Cys56Tyr)) identified in FST was considered as a strong candidate because a previous homozygous knockout of the mouse ortholog, Fst, had been reported to present with multiple anomalies including orofacial clefting and skeletal/rib defects, similar to the knockout of Gdf11 (Matzuk et al., 1995). The mutated cysteine (Cys56) is highly conserved across multiple species and this variant has not been described previously in over 245,000 alleles in gnomAD. The in silico analyses provided consistent support for pathogenicity including a PolyPhen2 score of 0.766 (possibly damaging), a SIFT score of 0 (deleterious) and a CADD score of 28.8 (see Supp. Tables S3 and S5). The heterozygous variant in FST was confirmed by Sanger sequencing in the three affected members of this family. DNA was only available for one of the two unaffected paternal grandparents, the grandmother, who was homozygous for the wildtype allele (G/G) (Figure 1). As with GDF11 in Family 4527, FST was also the top-ranked of the candidate genes from Family 22 by DOMINO, similarly being rated as “very likely dominant” (Supp. Table S4). Five other genes were classified as either “very likely dominant” (SPARC, HERC1, TENM2), “likely dominant” (RNF20), or “either dominant or recessive” (CRYBB2). Segregation analysis of the variants in SPARC, HERC1, TENM2 and RNF20 was then performed on samples from the three affected individuals (for validation) and the unaffected grandmother. The CRYBB2 variant was not pursued as pathogenic variants in this gene cause cataracts and are not associated with CL/P. As above, the EML4 variant was also included in the segregation analysis because it was noted to have <100 loss of function alleles listed in gnomAD. As expected, all five additional variants were confirmed in the three affected individuals, but notably the unaffected grandmother also carried each variant (Supp. Table S6B).
All variants reported here have been submitted to the Global Variome Shared LOVD database: https://databases.lovd.nl/shared/genes/.
Embryonic Craniofacial Expression of the GDF11/Gdf11 Network
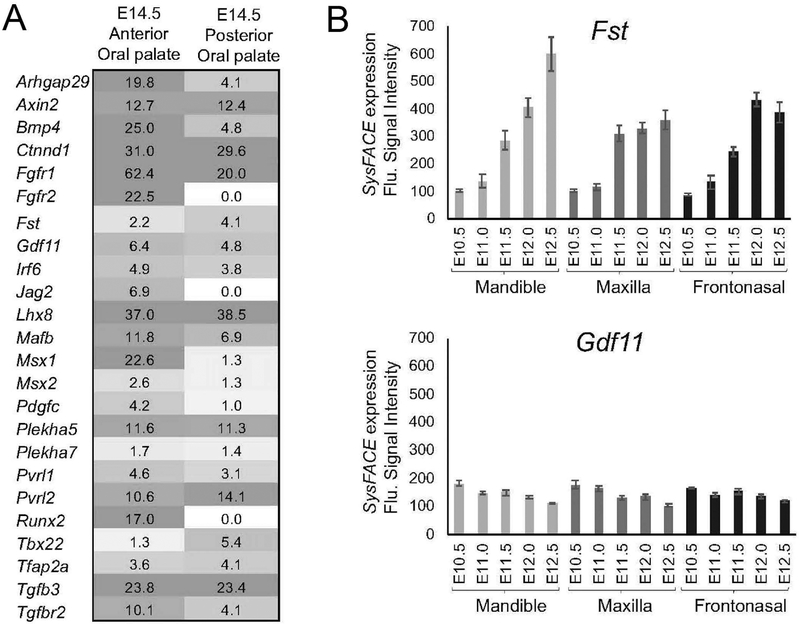
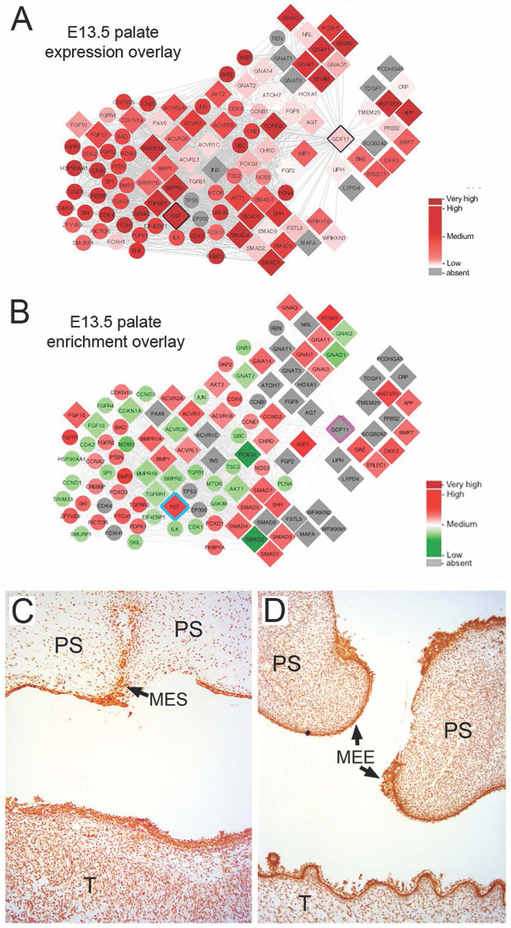
Previous studies have demonstrated that Gdf11 deficiency in mice causes cleft palate as well as skeletal anomalies as a result of dysregulation of anterior-posterior patterning of the axial skeleton (McPherron et al., 1999). Further, mouse Gdf11 and Fst compound mutants also exhibit patterning defects (McPherron et al., 1999). We therefore first analyzed Gdf11 and Fst expression in mouse anterior and posterior E14.5 palate tissue RNA-seq datasets, and found both genes to be expressed at similar levels and comparable to other known orofacial clefting-linked genes (e.g. Irf6, Pdgfc, Pvrl1, Tfap2a, etc.) (Figure 2A). Since tissue-enriched expression has been shown to be a good indicator of function in development of a given tissue (Kakrana et al., 2017; Lachke et al., 2012), Gdf11 and Fst expression was analyzed in further detail in various mouse embryonic facial tissue microarray datasets. Gdf11 is significantly expressed in maxillary, mandibular, and frontonasal tissues at E10.5 (Figure 2B). Next, using the human protein-protein interaction database (STRING), 66 potential GDF11 interactors (indicated by diamond shapes) and their nearest neighbors (secondary interactors, indicated by circles) were identified (Supp. Figure S1A). Using DAVID bioinformatics analysis, these interactors were enriched for the following GO (Gene Ontology) categories related to orofacial development: PI3K-Akt signaling pathway (hsa0415), TGFβ-signaling pathway (hsa04350), BMP signaling pathway (GO:0030509), and palate development (GO:0060021) (Supp. Figure S1B). Different tissue expression data were then overlaid on these candidates to identify the specific interactions within this network that are potentially relevant to GDF11 function in the affected facial tissues (Figure 3A,B; Supp. Figure S2). In addition to GDF11 and FST, several other direct protein-protein interactors of GDF11 (e.g. Erlec1, Pcsk5), were noted as significantly expressed in orofacial tissues (Figure 3A,B; Supp. Figure S2). Immunohistochemical staining of human palatal tissue sections confirmed the co-expression of GDF11 and FST both in the palatal epithelium (including the medial epithelial seam) and throughout the palatal and tongue mesenchyme (Figure 3C,D).
Figure 2: Expression of mouse Gdf11 and Fst in developing mouse facial tissues.
A. Gdf11 and Fst transcript expression was investigated in mouse E14.5 anterior and posterior palate RNA-seq datasets. Although their relative expression levels differ, both Gdf11 and Fst are significantly expressed in the anterior and posterior palate. Expression is given in Fragments per kilobase of transcripts per million mapped reads (FPKM). Further, Gdf11 and Fst expression levels are comparable to other known orofacial clefting-linked genes. B. Mouse mandible, maxilla, and frontonasal RNAseq datasets were analyzed using SysFACE for expression of Fst and Gdf11. Fst expression increases in these tissues as development progresses. Gdf11 is expressed at a constitutively low, but significant level in these facial tissues. Error bars represents standard error of mean (SEM).
Figure 3. Expression of GDF11 and FST in mouse and human palatal tissues.
Overlay of E13.5 mouse palatal (A) expression and (B) expression enrichment (tissue-specific expression compared to WB (whole embryonic tissue body)) from SysFACE on the Gdf11 protein-protein network. Gdf11 direct interactors, including FST, exhibit significant expression and enrichment in these tissues. For the expression overlays, differing intensities of red represent increasing transcript expression. For expression enrichment, differing intensities of red represent levels of enriched-expression, while differing intensities of green represent non-enriched expression. Immunohistochemical detection of human GDF11 (C) and FST (D) in 67–72d human embryonic secondary palate and tongue. Strong GDF11 and FST staining is seen in the palatal epithelia including the medial edge epithelia (MEE) and midline epithelial seam (MES) of the palatal shelves. Although less pronounced, specific staining is also seen throughout the palatal shelf (PS) and tongue (T) mesenchyme.
The p.Arg298Gln GDF11 Mutant is not Processed into its Active Form
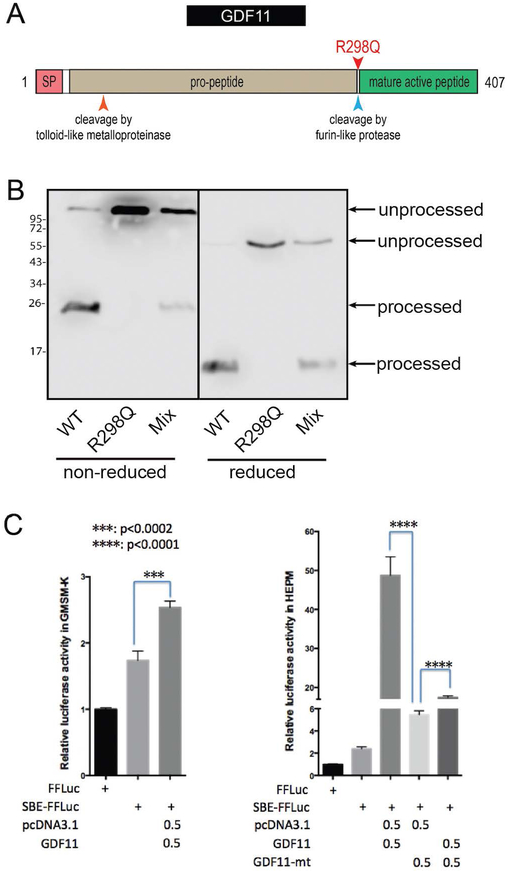
The GDF11 protein includes an N-terminal signal peptide, a Transforming Growth Factor-Beta-Related (IPR015615) prodomain, and a C-terminal mature peptide fragment that is released from the prodomain by Furin-type protease cleavage after the terminal Arginine residue of the RXXR recognition site (Figure 4A) (Walker et al., 2016). The GDF11 variant results in a substitution of the Arginine for Glutamine at the cleavage point between the prodomain and the mature peptide (see Figure 4A). Structural protein analysis using the HOPE protein analysis server (http://www.cmbi.ru.nl/hope/) showed that the wildtype arginine residue forms several internal hydrogen bonds. Substitution of this positively charged Arginine298 for the smaller neutral amino acid, glutamine, is predicted to abolish these hydrogen bonds. The change from the normal ionic interaction seen in the wildtype protein is predicted to impact recognition and subsequent cleavage of the site and thus generation of the active C-terminal mature signalling peptide.
Figure 4: The GDF11 p.Arg298Gln variant prevents cleavage of the prodomain and exhibits markedly reduced potential to activate SMAD-dependent reporter gene expression.
A. Schematic of the GDF11 protein. Pink = signal peptide (SP); beige = pro-peptide; green = mature active peptide. The mature active peptide is cleaved and released after Arg298 by furin-like proteases. B. Western blot of WT and R298Q GDF11 expression. A total of 1.5ug of WT GDF11 construct, R298Q mutant construct or an equal mix of each was transfected into HEK293T cells with 1.5ug of Furin vector DNA. 48 hours post transfection an anti-GDF11 western was conducted under non-reducing and reducing conditions. Unprocessed bands represent GDF11 covalently bound to prodomain while processed bands represent liberated GDF11 mature domain. C. Luciferase reporter assays in GMSM-K epithelial cells (left) and HEPM mesenchymal cells (right). FFLuc = enhancerless Firefly Luciferase construct; SBE-FFLuc = SMAD-binding element-Firefly Luciferase construct; pcDNA3.1 = empty vector control; GDF11 = GDF11 expression vector (in pcDNA3.1).
To test whether the Arginine298 to Glutamine substitution would indeed affect recognition and cleavage by the Furin convertase, we expressed the wildtype and p.Arg298Gln mutant GDF11 together with Furin in HEK293T cells. Western blotting demonstrated that all wildtype GDF11 was processed to its disulphide-linked dimeric form (Figure 4B). In contrast, the p.Arg298Gln mutant remained linked to its prodomain form, failing to be processed by Furin (Figure 4B) and thus is expected to remain inactive.
Previous studies have established that GDF11 activates the TGF-β-SMAD signaling pathway mainly through type-I activin receptors (ALK4 and ALK5) (Andersson et al., 2006). A SMAD-binding-element (SBE) reporter was therefore employed to assess the impact of the GDF11 variant when expressed in a human embryonic palatal mesenchyme (HEPM) cell line, GMSM-K oral epithelial cell line, and HEK293T epithelial cells. In all cell lines, wildtype GDF11 induced a significant increase in reporter activity over empty vector transfected controls (Figure 4C), with levels dependent on the cell line. In HEPM cells, in particular, wildtype GDF11 induced an approximate 50-fold increase of reporter activity over control levels. In contrast, the GDF11 p.Arg298Gln mutant failed to activate the SBE-reporter in HEPM cells. In fact, transfecting with both wildtype GDF11 and the p.Arg298Gln mutant resulted in a signal that was signficiantly lower than by wildtype GDF11 alone (Figure 4C).
The p.Cys56Tyr FST Mutant Disrupts the N-terminus and GDF11 Antagonism
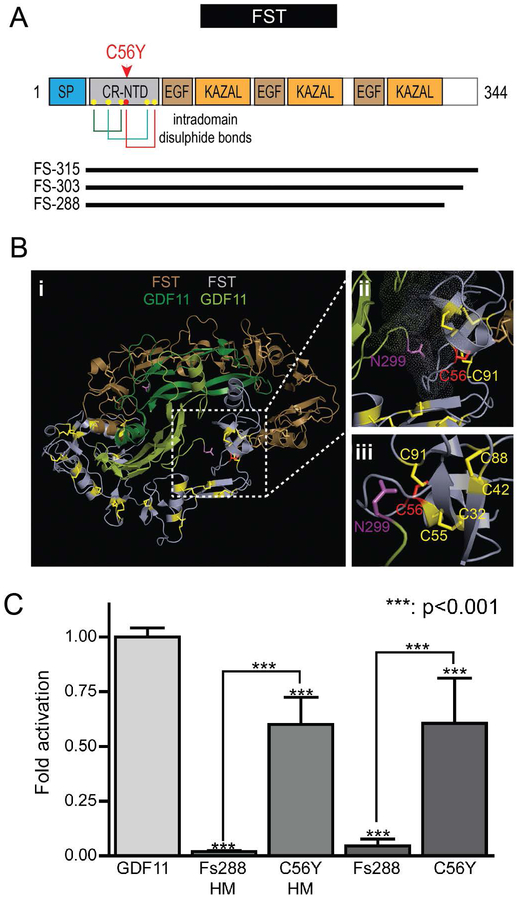
FST is comprised of multiple cysteine-rich domains, including a short signal sequence, a 63 residue N-terminal domain with six conserved cysteines, and three homologous C-terminal domains each with 10 conserved cysteines (Figure 5A). In both the N-terminal and C-terminal cysteine-rich domains, the cysteines form intradomain disulphide bridges to maintain a tight domain architecture (see Figure 5A,B) (Sidis et al., 2001). The identified FST variant involves Cysteine56, which resides in the 63 residue N-terminal domain. In the determined structure of the FST-GDF11 complex (PDB 5JHW) (Figure 5B), the N-terminal domain interacts with the hydrophobic pocket of GDF11 that is responsible for the interaction of the ligand with its receptors (Walker et al., 2017). Substitution of Cys56 with a hydrophilic tyrosine is predicted to prevent the formation of the appropriate disulphide bridge, and thus destabilize the N-terminus.
Figure 5: The FST p.Cys56Tyr variant disrupts the structure of the N-terminal domain and its ability to antagonize GDF11.
A. FST p.Cys56Tyr localizes to the cysteine-rich N-terminal domain (CR-NTD). Cysteine56 (red dot in A) is one of six cysteines (yellow dots) in the N-terminal domain of FST that form a stable structure mediated by three intradomain disulphide bonds (shown by colored lines): Cys32:Cys55, Cys56:Cys91, and Cys42:Cys88. The three main mature Follistatin isoforms are depicted below the schematic as black lines. B. (i) Visualization of the crystal structure of the GDF11:Follistatin [FS-288 isoform] complex (PDB: 5HJW). (ii) and (iii) Different views of Cys56 (red) showing its proximity to the N-terminus of mature GDF11 (Asn299) and the other cysteines (yellow) of the CR-NTD of FST. Mutation of Cys56 to tyrosine is predicted to impact proper folding of the CR-NTD and its ability to interact with GDF11 and related ligands. C. HEK293T (CAGA)12 cells were transfected with WT or C56Y Fs288 expression vectors with a C-terminal His-Myc tag (HM) or untagged then stimulated with exogenous GDF11. Activity is represented as fold activation normalized to activation by GDF11 on cells transfected with empty vector. One-way Anova analysis with a confidence interval of 99.9% conducted vs GDF11 (above each bar) or between WT and C56Y mutants (brackets). All experiments were conducted at least 3 times with experimental triplicate.
To simplify testing of the impact of the p.Cys56Tyr FST variant, we employed a similar SBE-reporter assay, but this time using a stable HEK293T cell line in which the SBE-luciferase reporter construct had been integrated into the genome. These HEK293-(CAGA)12 cells are sensitive to stimulation by GDF11 as judged by string reporter luminescence. Transfection of these cells with a construct expressing wildtype Fs288, as expected, completely inhibited the activation of the reporter following stimulation with wildtype GDF11. In contrast, expression of the p.Cys56Tyr mutant Fs288 in these cells had minimal impact on reporter activity following GDF11 stimulation (Figure 5C). We did observe that the mutant Fs288 was expressed at much lower levels than the wildtype Fs288 (Supp. Figure S3A) following transfection of equivalent amounts of vector. However, regardless of the amount of mutant Fs288 vector transfected, the mutant did not significantly inhibit the stimulation of reporter activity by GDF11 (Supp. Figure S3B).
DISCUSSION
We identified two families with CL/P that segregated for variants in GDF11 and FST, which encodes an antagonist of GDF11. We provide both genetic and biochemical evidence that the variants in these two genes are likely to represent the underlying cause of the orofacial cleft presentations in the respective families. Variants in GDF11 and FST have not been reported in association with any human monogenic disorder (OMIM, PubMed & Google Scholar review date 22 April 2019) nor is either variant reported in ClinVar or HGMD. The pathogenicity of each missense variant was assessed using the approach to variant classification outlined in Richards et al. (2015) and deemed to be likely pathogenic based on the following lines of evidence: well-established in vitro [GDF11] or in vivo [FST] functional studies supportive of a damaging effect on the gene product; the variants are located in well-established functional domains; each variant is absent from control populations; animal models for both GDF11 and FST recapitulate the essential morphological features of the disorder; and multiple lines of computational evidence support a deleterious effect on the gene or gene product. Of the candidate gene variants tested in each family, only the GDF11 and FST variants segregated completely with the clinical presentation.
GDF11 encodes a secreted protein that is a member of the bone morphogenetic protein family and the TGF-β superfamily. Most TGF-β superfamily proteins are synthesized as larger precursors that are cleaved at an RXXR motif by proprotein furin-type convertase endoproteases to generate an N-terminal prodomain and a C-terminal signaling domain. In line with this, GDF11 is initially produced as a 50kD precursor protein that undergoes proteolytic cleavage to generate an active 12.5kD C-terminal peptide that self-asociates through a disulphide bond to form a dimeric signaling molecule (mature GDF11) and a 37kD N-terminal peptide (GDF11 propeptide). The cleaved mature dimer (25kD) of GDF11 remains associated with the prodomain in a non-covalent latent complex which is subsequently activated in a tissue specific manner by BMP1/TLD-like proteinases via a further cleavage within the latent complex prodomain sequence (Ge et al., 2005). In GDF11, the final arginine of the RXXR motif, at which cleavage occurs, is Arginine298, the amino acid residue mutated in Family 4527. Consistent with a critical functional role for this residue, an arginine is present at this position in 245 out of 246 orthologs and paralogs of GDF11. Variants in the final arginine of this cleavage recognition site in other proteins (both naturally occurring and engineered) have been described as interfering with cleavage (de Zoeten et al., 2009; Ho et al., 2008) and to be disease causing (FGF23: p.(Arg179Gln) (ADHR Consortium, 2000)). Conversely, the creation of an RXXR domain is the basis of the engineered alpha-1-antitrypsin-Portland variant (α1-PDX), that functions as a suicide substrate inhibitor of Furin (Jean et al., 1998). The specific variant identified in Family 4527 is absent from the 119,616 GDF11 alleles reported in unrelated control individuals from ExAC v0.3 and over 245,000 alleles reported in gnomAD. There is only a single missense allele at the adjacent arginine p.Arg297Gln within the RXXR domain recorded in ExAC from a European sample but without phenotypic information.
Our functional data confirm that the p.Arginine298Glutamine substitution completely prevents cleavage by Furin (Figure 4B), suggesting the mutant GDF11 remains inactive. In line with this, GDF11 p.Arg298Gln showed minimal ability to activate a SMAD-sensitive reporter in comparison to wildtype GDF11. Given the autosomal dominant inheritance in family 4527 and that the phenotypic spectrum in affected individuals is intermediate to that of homozygous and heterozygous Gdf11 knockout mice (McPherron et al., 1999), we propose the phenotype in our family results from a dominant negative effect of the p.(Arg298Gln) variant. In support of this conclusion, when co-expressed, the mutant GDF11 protein appeared to interfere with the activity of wildtype GDF11 in the luciferase reporter assay (Figure 4C), although the mechanism for such action requires further investigation.
Follistatin (FST) is one of a number of extracellular antagonists of the TGF-beta family of growth factors, binding to BMP2, BMP4, BMP7, GDF8, GDF9, GDF11, activin A and activin B and neutralizing their activity. The crystal structure of the FST-GDF11 interaction shows two FST molecules symmetrically wrapping around the GDF11 dimer. This interaction masks the receptor binding sites on GDF11 (Walker et al., 2017). Homozygous Fst knockout mice exhibit cleft palate, while Keratin14-driven epithelial-specific conditional knockout of the gene shows tooth cusp morphology and delayed third molar development in mice, similar to that reported for the major CL/P gene, Irf6 (Blackburn et al., 2012; Chu et al., 2016). Although FST is widely expressed, it is reported to be most strongly expressed in follicles (from where it derived its name) and the skin. In our own analyses of developing facial tissues (mouse and human), we find that FST/Fst is also highly expressed in various embryonic facial tissues and is co-expressed with GDF11/Gdf11 in both the epithelial and mesenchymal populations. This is consistent with other reports for these two genes where they regulate, in an autocrine fashion, the differentiation of the respective progenitor cell populations (Gokoffski et al., 2011).
The identified FST p.(Cys56Tyr) variant alters one of six cysteine residues of the N-terminal domain (residue 27 of the mature protein following cleavage of the signal peptide). Cys56 is conserved across all follistatins and their homologs in all species, highlighting its likely functional importance. Using site-directed mutagenesis and biochemical interaction assays, Sidis et al (Sidis et al., 2001) reported that this N-terminal domain of FST is essential for its interaction with the related TGF-β family member, activin, and indeed substitution of both Cys55 and Cys56 with alanine was sufficient to abolish this interaction and its ability to regulate the bioactivity of activin. Similarly, substitution of hydrophobic tryptophan residues with hydrophilic residues in the vicinity of these cysteines also disrupted the interaction. Their data are consistent with the binding of FST to TGF-β family members involving a highly structured N-terminus (Keutmann et al., 2004; Sidis et al., 2001) that blocks the concave hydrophobic interface of these growth factors that normally mediates their interaction specifically with the type-I activin receptors (Walker et al., 2017; Walker et al., 2016). Indeed, consistent with this notion, we show in functional assays that, unlike wildtype FST which efficiently and completely antagonizes GDF11-stimulated reporter activity, the p.Cys56Tyr mutant protein retained minimal ability to inhibit GDF11-stimulated reporter activity and, by extension, would( not likely inhibit other TGFβ ligands, such as activin A (Figure 5C).
We have recently shown the importance of the p120-catenin-cadherin complex in Mendelian forms of non-syndromic orofacial clefting, having identified segregating pathogenic and likely pathogenic variants in multiple genes encoding interacting proteins of this complex (Cox et al., 2018). Despite only one variant having been identified for both GDF11 and FST in our cohort, the clinical phenotype, segregation, overlap with animal model phenotypes, tissue expression and the tested and predicted functional impact of the variants, provide compelling evidence of the involvement of the well-characterized GDF11-follistatin pathway in the molecular etiology of Mendelian forms of orofacial clefting. That said, it remains to be determined whether the orofacial clefting phenotype associated with inactivation of FST is simply due to changes in GDF11-mediated signaling or whether there is a more complex imbalance involving signaling via other TGF-β ligands regulated by FST, such as BMP4 or activin, that are also active in the developing facial tissue.
Finally, the unique association of orofacial clefting and vertebral/rib hypersegmentation with GDF11 warrants this presentation to be now classified as a novel syndromic form of orofacial clefting. We thus recommend that molecular testing for GDF11 should be considered in people with the combination of orofacial clefting and vertebral/rib hypersegmentation and that FST should be considered in orofacial gene panels for inclusion as a candidate gene for testing in individuals with non-syndromic CL/P.
Supplementary Material
ACKNOWLEDGEMENTS
Funding for this project included an Australian NHMRC Project grant # AU/1/BA51117 (TR, MLB, JZ, HvB, TCC, JCM, DAN, EK and ACL), the Laurel Foundation Endowment for Pediatric Craniofacial Research (TCC), NIH R01-DE014667 (ACL, KNK) and NIH R37-DE008559 (JCM), the March of Dimes Basil O’Connor award #FY98–0718 and research grant #6-FY01–616 (ACL, KNK), NIH R24-HD000836 (IAG, TCC), NIH R03-DE024776 (SAL) and NIH UM1-HG006493 (DAN, MJB), NIH R01-DE023575 (RAC), NIH R01-GM114640 (TBT), a Cleft Palate Foundation grant and American Association of Orthodontists Foundation Faculty Development Award and Start-up funds made available by the Ohio State University College of Dentistry (ACL) and start-up funds made available by the University of Iowa (LMU). RNA expression data obtained from FaceBase (www.facebase.org) were generated by projects U01DE020049 and U01DE020065. The FaceBase consortium and the FaceBase Coordinating Hub (1U01DE024449–01) are funded by the National Institute of Dental and Craniofacial Research. The authors have no conflicts of interest to declare.
Funding Information: Australian NHMRC Project grant # AU/1/BA51117, NIH R01-DE014667, NIH R37-DE008559, NIH R24-HD000836, NIH R03-DE024776, NIH UM1-HG006493, NIH R01-DE023575, NIH R01-GM114640, the March of Dimes Basil O’Connor award #FY98–0718 and research grant #6-FY01–616, the Laurel Foundation Endowment for Pediatric Craniofacial Research, a Cleft Palate Foundation grant, an AAOF Faculty Development Award, and start-up funds from Ohio State University College of Dentistry and the University of Iowa.
REFERENCES
- Andersson O, Reissmann E, & Ibanez CF (2006). Growth differentiation factor 11 signals through the transforming growth factor-beta receptor ALK5 to regionalize the anterior-posterior axis. EMBO Rep, 7(8), 831–837. doi: 10.1038/sj.embor.7400752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn J, Ohazama A, Kawasaki K, Otsuka-Tanaka Y, Liu B, Honda K, . . . Sharpe PT (2012). The role of Irf6 in tooth epithelial invagination. Dev Biol, 365(1), 61–70. doi: 10.1016/j.ydbio.2012.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu EY, Tamasas B, Fong H, Foster BL, LaCourse MR, Tran AB, . . . Cox TC (2016). Full spectrum of postnatal tooth phenotypes in a novel Irf6 cleft lip model. J Dent Res, 95(11), 1265–1273. doi: 10.1177/0022034516656787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ADHR Consortium. (2000). Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nat Genet, 26(3), 345–348. doi: 10.1038/81664 [DOI] [PubMed] [Google Scholar]
- Cash JN, Angerman EB, Kattamuri C, Nolan K, Zhao H, Sidis Y, Keutmann HT, & Thompson TB (2012a) Structure of myostatin·follistatin-like 3: N-terminal domains of follistatin-type molecules exhibit alternate modes of binding. J Biol Chem 287(2), 1043–1053. doi: 10.1074/jbc.M111.270801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash JN, Angerman EB, Keutmann HT, & Thompson TB (2012b) Characterization of follistatin-type domains and their contribution to myostatin and activin A antagonism. Mol Endocrinol 26(7), 1167–1178. doi: 10.1210/me.2012-1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LL, Cox TC, Moreno Uribe LM, Zhu Y, Richter CT, Nidey N, . . . Roscioli T (2018). Mutations in the Epithelial Cadherin-p120-Catenin Complex Cause Mendelian Non-Syndromic Cleft Lip with or without Cleft Palate. Am J Hum Genet, 102(6), 1143–1157. doi: 10.1016/j.ajhg.2018.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox TC (2004). Taking it to the max: the genetic and developmental mechanisms coordinating midfacial morphogenesis and dysmorphology. Clin Genet, 65(3), 163–176. [DOI] [PubMed] [Google Scholar]
- de Zoeten EF, Lee I, Wang L, Chen C, Ge G, Wells AD, . . . Ozkaynak E (2009). Foxp3 processing by proprotein convertases and control of regulatory T cell function. J Biol Chem, 284(9), 5709–5716. doi: 10.1074/jbc.M807322200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, . . . Daly MJ (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet, 43(5), 491–498. doi: 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MJ, Marazita ML, Beaty TH, & Murray JC (2011). Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet, 12(3), 167–178. doi: 10.1038/nrg2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge G, Hopkins DR, Ho WB, & Greenspan DS (2005). GDF11 forms a bone morphogenetic protein 1-activated latent complex that can modulate nerve growth factor-induced differentiation of PC12 cells. Mol Cell Biol, 25(14), 5846–5858. doi: 10.1128/MCB.25.14.5846-5858.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokoffski KK, Wu HH, Beites CL, Kim J, Kim EJ, Matzuk MM, . . . Calof AL (2011). Activin and GDF11 collaborate in feedback control of neuroepithelial stem cell proliferation and fate. Development, 138(19), 4131–4142. doi: 10.1242/dev.065870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritli-Linde A (2012). The mouse as a developmental model for cleft lip and palate research. Front Oral Biol, 16, 32–51. doi: 10.1159/000337523 [DOI] [PubMed] [Google Scholar]
- Ho AM, Marker PC, Peng H, Quintero AJ, Kingsley DM, & Huard J (2008). Dominant negative Bmp5 mutation reveals key role of BMPs in skeletal response to mechanical stimulation. BMC Dev Biol, 8, 35. doi: 10.1186/1471-213X-8-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean F, Stella K, Thomas L, Liu G, Xiang Y, Reason AJ, & Thomas G (1998). alpha1-Antitrypsin Portland, a bioengineered serpin highly selective for furin: application as an antipathogenic agent. Proc Natl Acad Sci U S A, 95(13), 7293–7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakrana A, Yang A, Anand D, Djordjevic D, Ramachandruni D, Singh A, . . . Lachke SA (2017). iSyTE 2.0: a database for expression-based gene discovery in the eye. Nucleic Acids Res. doi: 10.1093/nar/gkx837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keteyian AJ, & Mishina Y (2017). A Review of Orofacial Clefting and Current Genetic Mouse ModelsDesigning Strategies for Cleft Lip and Palate Care. In Almasri MA (Series Ed.): InTech. Retrieved from https://cdn.intechopen.com/pdfs/53696.pdf. doi: 10.5772/67052 [DOI] [Google Scholar]
- Keutmann HT, Schneyer AL, & Sidis Y (2004). The role of follistatin domains in follistatin biological action. Mol Endocrinol, 18(1), 228–240. doi: 10.1210/me.2003-0112 [DOI] [PubMed] [Google Scholar]
- Lachke SA, Ho JW, Kryukov GV, O’Connell DJ, Aboukhalil A, Bulyk ML, . . . Maas RL (2012). iSyTE: integrated Systems Tool for Eye gene discovery. Invest Ophthalmol Vis Sci, 53(3), 1617–1627. doi: 10.1167/iovs.11-8839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, & Marazita ML (2013). Genetics of cleft lip and cleft palate. Am J Med Genet C Semin Med Genet, 163C(4), 246–258. doi: 10.1002/ajmg.c.31381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, & Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25(14), 1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, . . . Genome Project Data Processing, S. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25(16), 2078–2079. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Busch T, Eliason S, Anand D, Bullard S, Gowans LJJ, . . . Butali A (2017). Exome sequencing provides additional evidence for the involvement of ARHGAP29 in Mendelian orofacial clefting and extends the phenotypic spectrum to isolated cleft palate. Birth Defects Res, 109(1), 27–37. doi: 10.1002/bdra.23596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk MM, Lu N, Vogel H, Sellheyer K, Roop DR, & Bradley A (1995). Multiple defects and perinatal death in mice deficient in follistatin. Nature, 374(6520), 360–363. doi: 10.1038/374360a0 [DOI] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, . . . Cunningham F (2016). The Ensembl Variant Effect Predictor. Genome Biol, 17(1), 122. doi: 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherron AC, Lawler AM, & Lee SJ (1999). Regulation of anterior/posterior patterning of the axial skeleton by growth/differentiation factor 11. Nat Genet, 22(3), 260–264. doi: 10.1038/10320 [DOI] [PubMed] [Google Scholar]
- Paila U, Chapman BA, Kirchner R, & Quinlan AR (2013). GEMINI: integrative exploration of genetic variation and genome annotations. PLoS Comput Biol, 9(7), e1003153. doi: 10.1371/journal.pcbi.1003153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard TH (1975). Normal and abnormal growth patterns In Gardner LI (Ed.), Endocrine and Genetic Diseases of Childhood (pp. pp. 1–6). Philadelphia: W. B. Saunders. [Google Scholar]
- Sidis Y, Schneyer AL, Sluss PM, Johnson LN, & Keutmann HT (2001). Follistatin: essential role for the N-terminal domain in activin binding and neutralization. J Biol Chem, 276(21), 17718–17726. doi: 10.1074/jbc.M100736200 [DOI] [PubMed] [Google Scholar]
- Tamasas B, & Cox TC (2017). Massively increased caries susceptibility in an Irf6 cleft lip/palate model. J Dent Res, 96(3), 315–322. doi: 10.1177/0022034516679376 [DOI] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, . . . DePristo MA (2013). From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics, 43, 11 10 11–33. doi: 10.1002/0471250953.bi1110s43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RG, Czepnik M, Goebel EJ, McCoy JC, Vujic A, Cho M, . . . Thompson TB (2017). Structural basis for potency differences between GDF8 and GDF11. BMC Biol, 15(1), 19. doi: 10.1186/s12915-017-0350-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RG, McCoy JC, Czepnik M, Mills MJ, Hagg A, Walton KL, . . . Thompson TB (2018). Molecular characterization of latent GDF8 reveals mechanisms of activation. Proc Natl Acad Sci USA, 115(5), E866–E875. doi: 10.1073/pnas.1714622115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RG, Poggioli T, Katsimpardi L, Buchanan SM, Oh J, Wattrus S, . . . Lee RT (2016). Biochemistry and Biology of GDF11 and Myostatin: Similarities, Differences, and Questions for Future Investigation. Circ Res, 118(7), 1125–1141; discussion 1142. doi: 10.1161/CIRCRESAHA.116.308391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Deng M, Naluai-Cecchini T, Glass IA, & Cox TC (2017). Differences in Oral Structure and Tissue Interactions during Mouse vs. Human Palatogenesis: Implications for the Translation of Findings from Mice. Front Physiol, 8, 154. doi: 10.3389/fphys.2017.00154 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.