Abstract
Protein detection in complex biological fluids has wide-ranging significance across proteomics and molecular medicine. Existing detectors cannot readily distinguish between specific and nonspecific interactions in a heterogeneous solution. Here, we show that this daunting shortcoming can be overcome by using a protein bait-containing biological nanopore in mammalian serum. The capture and release events of a protein analyte by the tethered protein bait occur outside the nanopore and are accompanied by uniform current openings. Conversely, nonspecific pore penetrations by nontarget components of serum, which take place inside the nanopore, are featured by irregular current blockades. As a result of this unique peculiarity of the readout between specific protein captures and nonspecific pore-penetration events, our selective sensor can quantitatively sample proteins at single-molecule precision in a manner distinctive from those employed by prevailing methods. Because our sensor can be integrated into nanofluidic devices and coupled with high-throughput technologies, our approach will have a transformative impact in protein identification and quantification in clinical isolates for disease prognostics and diagnostics.
Keywords: FhuA, Ion channel, Protein-protein Interface, Protein dynamics, Stochastic sensing, Electrophysiology, Membrane Protein Engineering
Graphical Abstract:
A current challenge in personalized medicine is the ability to detect proteins in a specific, sensitive, and scalable fashion using biofluids.1, 2 Prevailing methods for protein analysis in vitro, such as biolayer interferometry, surface plasmon resonance, and isothermal titration calorimetry, are usually employed in homogeneous samples. Yet, these approaches cannot be used in a heterogeneous solution, because of nonspecific protein binding and deteriorated signal-to-noise ratio (SNR). In the past decade, significant progress has been made in developing alternative bioanalytical approaches for detecting proteins and their interactions in complex biofluids.3–8 Conventional techniques, such as mass spectrometry, enzyme-linked immunosorbent assay (ELISA), and Western Blot, have facilitated detection and exploration of proteins in biofluids. Here, we demonstrate accurate determinations of protein detection in mammalian serum at single-molecule precision using a modular pore-based nanostructure. By using conventional nanopore sensors, it is difficult to distinguish signals caused by specific interactions with the target protein from those caused by nonspecific pore penetrations of other constituents of a heterogeneous sample. This challenge is exacerbated in nanopore recordings, because both specific and nonspecific events appear as current blockades. Therefore, pore penetration by biofluid constituents enhances the susceptibility for sampling false capture events and precludes accurate protein detection.
In this work, we show that this persistent limitation can be overcome by detecting the protein analyte outside the nanopore by means of a tethered protein bait (Fig. 1a). The 455-residue single-polypeptide chain protein nanopore, also named t-FhuA,9, 10 is an extensive truncation of ferric hydroxamate uptake component A (FhuA)11 of E. coli. This is a robust monomeric β-barrel.12–15 The protein bait was engineered at the N terminus of t-FhuA via a flexible hexapeptide tether ((GGS)2). As a test case for the protein bait and protein analyte, we used 110-residue RNase barnase (Bn)16 and its 89-residue inhibitor barstar (Bs),17, 18 respectively. The pivotal mechanism of this sensing approach in fetal bovine serum (FBS) is the transduction of protein capture and release events into a high-fidelity electrical readout via a movable polypeptide adaptor (O), which was fused to the untethered end of the bait. Therefore, our single-polypeptide chain protein sensor is OBn(GGS)2t-FhuA, a 583-residue pore-based nanostructure.
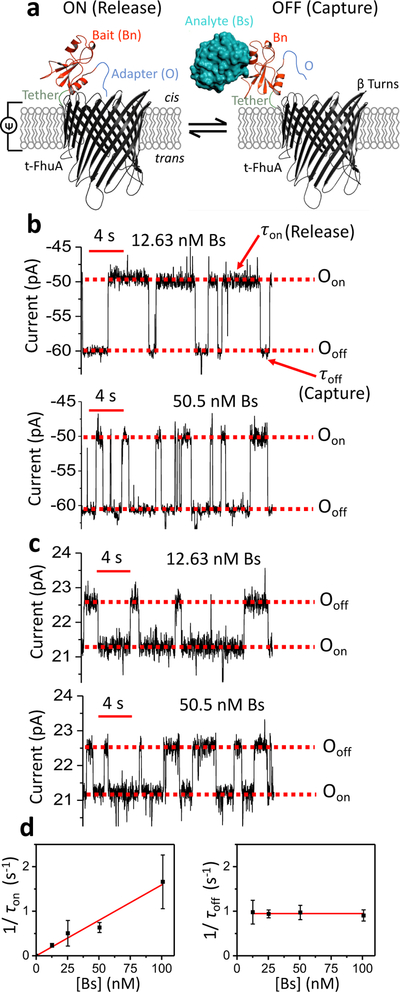
Figure 1: Single-molecule protein detection at single-tethered receptor resolution.
(a) Stochastic protein sensing of barstar (Bs) protein using OBn(GGS)2t-FhuA. This protein sensor encompasses a truncated t-FhuA protein pore, a short (GGS)2 hexapeptide tether, a barnase (Bn) protein receptor, and a dodecapeptide adapter (O). This model was generated in Pymol using the pdb files 1BY3 and 1BRS for (FhuA)11 and (Bn-Bs),18 respectively. (b) Representative single-channel electrical traces of OBn(GGS)2t-FhuA in the presence of 12.63 nM Bs (top) and 50.5 nM Bs (bottom) at an applied transmembrane potential of −40 mV. The control experiment in the absence of Bs is shown in Supporting Information, Fig. S1. Single-channel electrical traces were further processed using a 20 Hz low-pass 8-pole Bessel filter. Oon indicates the Bs-released open substate; Ooff shows the Bs-captured open substate. (c) Representative single-channel electrical traces of OBn(GGS)2t-FhuA in the presence of 12.63 nM Bs (top) and 50.5 nM Bs (bottom) at an applied transmembrane potential of +15 mV. The control experiment in the absence of Bs is shown in Supporting Information, Fig. S6. Single-channel electrical traces were further processed using a 20 Hz low-pass 8-pole Bessel filter. Oon and Ooff have the same meanings as those stated in (b). (d) Both diagrams show dependence of 1/τon (left) and 1/τoff (right) on Bs concentration, [Bs]. Here, these kinetic rate constants in the form of mean ± s.e.m. are kon = (1.59 ± 0.09) × 107 M−1s−1 and koff = 0.95 ± 0.02 s−1. Data points in both panels represent mean ± s.d. obtained from n distinct experiments. In this case, n was 5, 3, 3, and 4, for a [Bs] of 12.63, 25.25, 50.5, and 100.01 nM, respectively. Experimental conditions in (d) were the same as those stated in (c).
Our selective nanopore sensor permits single-channel electrical recordings12, 19 over long periods under these challenging conditions. In this work, specific protein captures, which occur outside the nanopore, lead to uniform current openings. In contrast, the FBS constituents, which partition into the t-FhuA pore lumen, bring about irregular current blockades. This readout feature makes the protein captures unambiguously different from those events resulting from nonspecific pore penetrations of nontarget FBS constituents. This strategy for distinguishing between protein captures and releases from other nonspecific residual events is a fundamental distinction of this work from prior detection studies using conventional nanopores in homogeneous solutions.1, 20–37 Here, we hypothesized that the SNR value might be further amplified under optimized recording conditions. Moreover, we show that time-resolved protein capture and release events can be recorded in a heterogeneous solution at an applied potential as low as 15 mV, because of a satisfactory SNR value. Yet, in these challenging circumstances the bait-analyte complex formation followed a simple bimolecular association process, whereas the analyte release underwent a unimolecular dissociation process. Such a membrane protein design, which makes this method highly specific and quantitative, enables accurate determinations of single-molecule protein detection in FBS. Finally, the kinetics of binding and unbinding in a heterogeneous solution was identical to those determined in a homogeneous solution, confirming that specific and nontarget single-molecule events are independent of each other.
RESULTS
Maximization of the SNR value in FBS.
Our aim was to record single protein captures at a maximized SNR and at a minimized interference of FBS-induced current transitions. OBn(GGS)2t-FhuA spontaneously inserted itself from the cis side into the membrane with a preferred orientation (Fig. 1a). At a transmembrane potential of −40 mV with respect to the cis side (grounded) and in 300 mM KCl, 10 mM Tris-HCl, pH 8.0, OBn(GGS)2t-FhuA showed a uniform single-channel current with a mean value of −46.9 ± 2.5 pA (n = 4 experiments) (Supporting Information, Fig. S1). When Bs was added to the cis side at a low-nanomolar Bs concentration, [Bs], transient current transitions were readily observed between Oon and Ooff (Fig. 1b). The current amplitudes of these open substates were −47.5 ± 2.1 pA (n = 4) and −58.2 ± 3.1 pA (n = 4), respectively. We have recently shown that these current transitions represent reversible release (Oon) and capture (Ooff) events of Bs by Bn.10 We used a single-site mutant of Bn (H102A), a catalytically-inactive protein bait. In this way, we examined more weakly-binding protein captures (e.g., Kd of ~10−9 M)10, 38 than strongly-binding protein captures (e.g., wild-type Bn, Kd of ~10−14 M).38 This tactic permitted the recording of single-molecule Bs capture events at equilibrium. The Ooff and Oon current amplitudes enabled a clear signal separation of ~10 pA for the capture and release events, respectively. Enhancing the Bs concentration increased the event frequency (Fig. 1b). Standard histograms of the release (τon) and capture (τoff) durations are presented in Supporting Information (Fig. S2). The statistically significant fit model was determined by using a standard logarithm likelihood ratio (LLR) test.39 At a confidence level C = 0.95, the best model was one-exponential fit, unless otherwise stated (see below terms resulting from serum constituents). This outcome suggests single-barrier transitions of the free-energy landscape of these single-molecule protein capture and release events. However, the incubation of the cis side of the chamber in FBS within the range of 1–5% (v/v) FBS resulted in appearance of large-current amplitude and long-lived blockades at a transmembrane potential of −40 mV (Supporting Information, Fig. S3). For example, at an FBS concentration of 5% (v/v), the duration of some of these long-lived events was even longer than 10 s.
Therefore, these long-lived FBS-induced events interfered with our ability to detect single-molecule protein captures. t-FhuA is an acidic β-barrel (PK/PCl is ~5.5).13 Thus, the pore lumen acts as an energetic barrier for the partitioning of negatively-charged FBS constituents. In contrast, positively-charged FBS impurities are electrostatically attracted into the pore interior. A negative transmembrane potential would amplify this process, because FBS was added to the cis side of the chamber. On the other hand, a positive transmembrane potential would counteract on positive FBS constituents, whereas the negatively-charged FBS impurities would face an energetic barrier due to the cation selectivity of t-FhuA. These outcomes led us to systematically examine the voltage dependence of the differential current, ΔIoff-on (I (Ooff) – I (Oon)), between the capture and release substates. For example, at a negative voltage bias, ΔIoff-on was ~17.6 pA and ~2.5 pA at −50 mV and −15 mV, respectively (Supporting Information, Fig. S4). This inspection revealed greater ΔIoff-on values for negative potentials than those values recorded at positive potentials. For example, ΔIoff-on spanned a range of 1.2–1.8 pA between +15 and +40 mV, respectively. This finding motivated us to conduct our single-molecule protein capture measurements at a higher positive potential. Surprisingly, we noted that increase in the positive voltage bias at a value of +40 mV deteriorated the SNR and induced transient current fluctuations in the stability of the substates Oon and Ooff (Supporting Information, Fig. S5).
Therefore, we then tested single-molecule Bs captures at a potential of +15 mV. In these conditions, OBn(GGS)2t-FhuA exhibited a uniform unitary current of 19.9 ± 1.5 pA (n = 5) (Supporting Information, Fig. S6). When Bs was added to the cis side at a low-nanomolar concentration, reversible protein captures in the form of transient current transitions were noted between Oon and Ooff. The mean currents of these open substates were 20.1 ± 1.6 pA (n = 5) and 21.3 ± 1.5 pA (n = 5), respectively. This results in a ΔIoff-on of ~1.2 ± 0.1 pA (n=5). Representative single-channel traces of these protein captures, when either 12.63 nM Bs (top trace) or 50.5 nM Bs (bottom trace) was added to the cis side, are illustrated in Fig. 1c. Standard histograms of the release (τon) and capture (τoff) durations are presented in Supporting Information (Fig. S7).
Time analyses of these events were used to determine the association (kon) and dissociation (koff) rate constants of the Bn-Bs interactions, where kon = 1/([Bs]τon) and koff = 1/τoff. Here, τon and τoff denote the release and capture durations of the free Bs by the tethered Bn, respectively. The frequency of the single Bs captures in the form of 1/τon was linearly dependent on [Bs], suggesting a bimolecular association process (Fig. 1d). The slope of the linear fit of event frequency was kon, with a mean value of (1.59 ± 0.09) × 107 M−1s−1. The reciprocal of the τoff duration, koff, with a mean value of 0.95 ± 0.02 s−1, was independent of [Bs], indicating a unimolecular dissociation process (Supporting Information, Table S1). These values correspond to a Kd of 60 ± 4 nM (Supporting Information, Table S2). The kinetic and equilibrium constants determined at +15 mV were in excellent accord with those parameters obtained in an earlier study at a voltage bias of −40 mV (kon = (1.34 ± 0.04) × 107 M−1s−1, koff = 0.86 ± 0.02 s−1, and Kd = 64 ± 02 nM).10 These values are also in agreement with prior bulk-phase kinetic determinations.16, 38 Furthermore, kon and koff were not sensitive to changes in the transmembrane potential when the voltage bias was negative (Supporting Information, Fig. S8).
Single-molecule protein detection in FBS.
Based upon the above findings, we then examined single-molecule Bs captures at a transmembrane potential of +15 mV and in the presence of 5% (v/v) FBS (Fig. 2a). Under these conditions, FBS-induced current blockades were in the range of milliseconds. Two representative single-channel traces are shown when either 12.63 nM Bs (top trace) or 50.5 nM Bs (bottom trace) were added to the cis side (Fig. 2b). FBS-induced large-amplitude blockades featured a current amplitude (ΔIFBS) of 17.2 ± 1.2 pA (n = 3). These current blockades were unambiguously distinguished from specific Bs capture-induced, low-amplitude current transitions between the Oon and Ooff substates (ΔIoff-on was 1.4 ± 0.3 pA (n = 3)). We highlight that single-molecule Bs captures were detected outside the nanopore, whereas the nonspecific FBS-induced events were probed inside the nanopore. Therefore, we hypothesize that these two categories of events are independent of each other. Indeed, we noticed that any reversible, large-amplitude, FBS-induced current transition departing from either Oon or Ooff substate always returned to the same substate. This confirms their independent mechanisms.
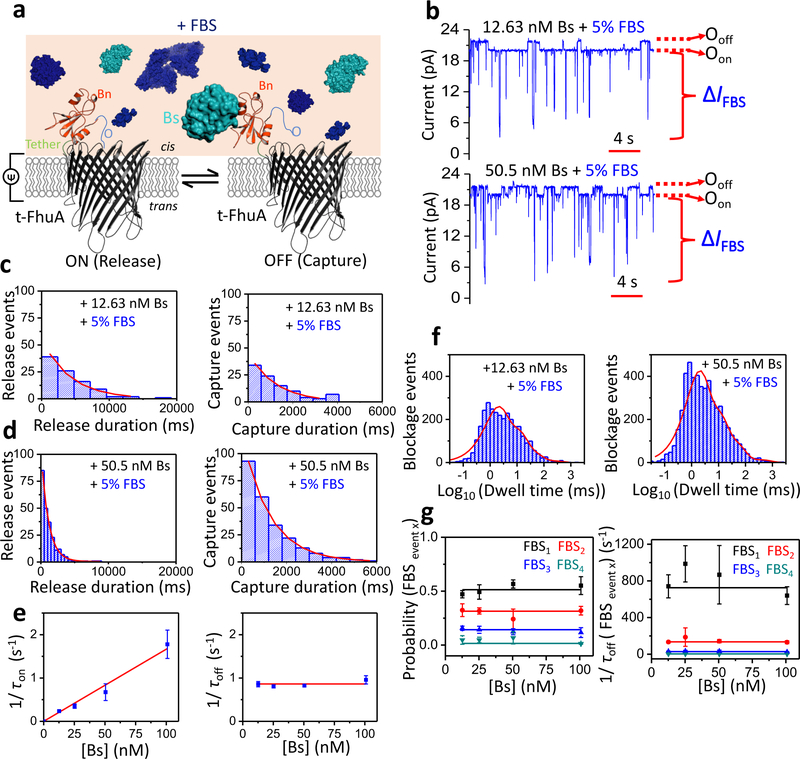
Figure 2: Single-molecule protein detection at single-tethered receptor resolution in unprocessed fetal bovine serum (FBS).
(a) Schematic representation of stochastic protein sensing of Bs using OBn(GGS)2t-FhuA in the presence of FBS. (b) Representative single-channel electrical traces of OBn(GGS)2t-FhuA in 5% (v/v) FBS and in the presence of 12.63 nM Bs (top) and 50.5 nM Bs (bottom). Experiments were conducted at an applied transmembrane potential of +15 mV. Single-channel electrical traces were further processed using a 20 Hz low-pass 8-pole Bessel filter. Bs was added to the cis side of the chamber. Oon and Ooff stand for the same meanings as in Fig. 1a. Single-molecule protein captures are indicated by upwards current transitions (to Ooff) from basal current level (Oon) of OBn(GGS)2t-FhuA. ΔIFBS represents spectrum of FBS-induced current transitions either departed from either Oon or Ooff substate. (c) Representative standard histograms of the release (τon; left) and capture (τoff; right) durations at 12.63 nM Bs. The τon and τoff values obtained from the fits (mean ± s.e.m.) were 4,188 ± 429 ms (n = 96) and 1,220 ± 90 ms (n = 97), respectively. (d) Representative standard histograms of the release (τon; left) and capture (τoff; right) durations at 50.5 nM Bs. The τon and τoff values obtained from the fits were 1,150 ± 57 ms (n = 239) and 1,223 ± 25 ms (n = 243), respectively. Experimental conditions in (c) and (d) were the same as in (b). (e) Dependence of 1/τon (left) and 1/τoff (right) on [Bs] when measurements were conducted in 5% FBS. Here, kon = (1.67 ± 0.09) × 107 M−1s−1 and koff = 0.86 ± 0.03 s−1. Data points in both panels represent mean ± s.d. obtained from n = 3 distinct experiments. (f) Event dwell time histograms of FBS-induced current blockades in 5% (v/v) FBS as well as in the presence of 12.63 nM Bs (left) and 50.5 nM Bs (right). The numbers of FBS-induced events were 3,040 and 5,007, respectively. The best-fit model was a multiexponential function with four terms (Supporting Information, Tables S4–S5). (g) Event probability (left) and dwell time (right) values of FBS-induced current blockades (FBSevent x) are independent of [Bs]. Data points in both panels represent mean ± s.d. obtained from n = 3 distinct experiments. Experimental conditions in panels (c) - (g) were the same as those stated in (b).
Histograms of the release and capture durations in the presence of 5% (v/v) FBS, and at either 12.63 or 50.5 nM Bs, are displayed in Fig. 2c and Fig. 2d, respectively. Remarkably, the presence of FBS did not affect single-exponential distributions of these time constants. Moreover, (1/τon) was linearly dependent on [Bs], whereas (1/τoff) was independent of [Bs], suggesting that these events resulted from a bimolecular association process and a unimolecular dissociation mechanism in the presence of FBS. Again, kon and koff of the single-molecule protein captures were determined in the presence of FBS using linear fits of the dependences of 1/τon ([Bs]) and 1/τoff ([Bs]) (Fig. 2e; Supplementary Information, Table S3). These values were (1.67 ± 0.09) × 107 M−1s−1 and 0.86 ± 0.03 s−1, respectively, which corresponded to a Kd of 52 ± 3 nM (Supplementary Information, Table S2). Remarkably, these kinetic and equilibrium constants determined in FBS are closely similar to those acquired in a homogeneous solution. This outcome is significant, because it also reveals that no FBS impurity interacts with the binding interface of either Bn or Bs. To test whether the presence of Bs affected the partitioning of nontarget FBS constituents into the pore lumen, we conducted the time analysis of the large-amplitude current transitions (IFBS) at [Bs] in the range of 0–100 nM. For the time analysis of the Bn-Bs interactions, we used a 20 Hz low-pass Bessel filter. Yet, in the case of the FBS-induced current blockades, an 0.5 kHz filter frequency was employed (Supporting Information, Fig. S9), which corresponded to an event deadtime, Td, of 0.36 ms.40 An LLR test analysis indicated that the FBS-induced events followed a four-exponential distribution, regardless of [Bs] (Fig. 2f, Fig. 2g; Supporting Information, Tables S4–S9). We conclude that the Bn-Bs interactions are also quantitatively independent of the FBS-induced current blockades.
Single-molecule protein detection in heat-inactivated FBS (HI-FBS).
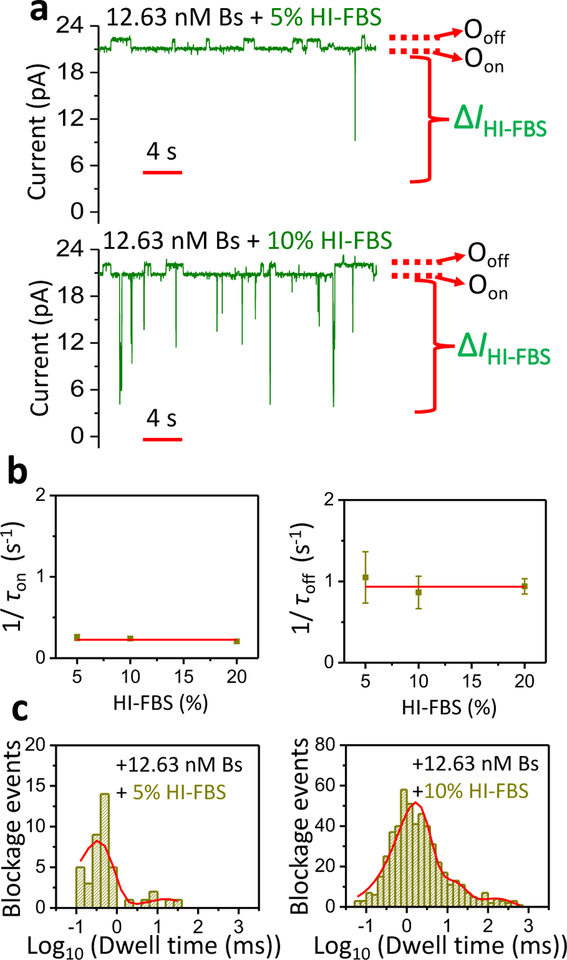
The enhancement of protein-detection sensitivity might also be achieved by reducing the highly frequent and short-lived current blockades produced by the FBS impurities. Studies of protein detection using other bioanalytical methods show that the inhibition of background noise is usually conducted either by gradual dilutions or by heat inactivation (HI) of the serum.41–44 Here, we examined single-molecule Bs captures by the tethered Bn when heat-inactivated FBS (HI-FBS) was added in increased volumetric concentrations. Two single-channel traces are illustrated when either 5% (v/v) HI-FBS (top trace) or 10% (v/v) HI-FBS (bottom trace) were added to the cis side at 12.63 nM Bs (Fig. 3a; Supporting Information, Fig. S10). Again, concurrent Bs capture-induced, low-amplitude current transitions (ΔIoff-on) and HI-FBS-induced, large-amplitude blockades (ΔIHI-FBS) were noted. The total frequency of HI-FBS-induced events at 12.63 nM Bs and 5% (v/v) HI-FBS was 0.14 ± 0.05 s−1 (n=3). Notably, this value was much lower than that determined in the presence of 5% (v/v) FBS (4.48 ± 1.04 s−1, n=3). This finding highlights the significance of removal of FBS-induced residual blockades by HI of the serum. As in the case of FBS-incubated samples, the release and capture durations obeyed single-exponential event distributions (Supporting Information, Fig. S11). Moreover, these time constants were independent of the HI-FBS concentration (Fig. 3b; Supporting Information, Table S10). Therefore, we determined no statistically distinctive rate constants at increased HI-FBS concentrations. Finally, HI-FBS-induced events followed a multi-exponential distribution that included four or less exponential terms (Fig. 3c, Supporting Information, Tables S11–S13).
Figure 3: Single-molecule protein detection in HI-FBS.
(a) Single-channel electrical traces of OBn(GGS)2t-FhuA in either 5% (v/v) (top) or 10% (bottom) HI-FBS and in presence of 12.63 nM Bs. The applied transmembrane potential was +15 mV. Single-channel electrical traces were further processed using a 20 Hz low-pass 8-pole Bessel filter. Bs was added to the cis side of the chamber. Oon and Ooff have the same meanings as in Fig. 1a. ΔIHI-FBS represents spectrum of HI-FBS-induced current transitions. (b) Dependence of 1/τon (left) and 1/τoff (right) on the HI-FBS concentration. Linear fits of both plots provide evidence that the average of 1/τon and 1/τoff of the Bs captures at 12.63 nM Bs are independent of the tested HI-FBS concentration. Here, 1/τon = 0.227 ± 0.014 s−1 and 1/τoff =0.934 ± 0.030 s−1. Data points represent mean ± s.d. obtained from n = 3 distinct experiments. Experimental conditions in panel (b) were the same as those stated in (a). (c) Dwell time histograms of HI-FBS-induced current blockades in either 5% (left) or 10% HI-FBS (right), and in the presence of 12.63 nM Bs. The number of HI-FBS-induced events were 42 and 515, respectively. For the top graph, the best fit model was a double-exponential distribution function with corresponding dwell time and probability values (mean ± s.e.m.), as follows: τoff-1 = 0.33 ± 0.23 ms, P1 = 0.88 ± 0.14, τoff-2 = 16.57 ± 2.09 ms; P2 = 0.12 ± 0.13. For the bottom graph, the best fit model was a four-exponential distribution function with corresponding dwell time and probability values (mean ± s.e.m.), as follows: τoff-1 = 1.06 ± 0.33 ms, P1 = 0.63 ± 0.39, τoff-2 = 2.93 ± 1.74 ms, P2 = 0.19 ± 0.31, τoff-3 = 13.67 ± 1.14 ms, P3 = 0.13 ± 0.13, τoff-4 = 131.28 ± 1.19 ms, P4 = 0.06 ± 0.05.
DISCUSSION
In this article, we show single-molecule discrimination of proteins in FBS using a highly specific nanopore sensor. The detection readout is not acquired at the expense of protein unfolding,29, 30 because single-molecule capture events occur outside the nanopore20, 22, 25 and they are accurately time resolved via discrete current transitions. Measurements using t-FhuA can be conducted at low salt concentrations,13 elevated temperatures,13 and increased osmotic pressures at a semi-dilute regime of crowding agents.45 The polypeptide adaptor facilitates protein sensing outside the nanopore, a difficult process exacerbated by the fact that the dimensions of the folded, interacting proteins (e.g., bait and analyte) are greater than the cross-sectional diameter of the nanopore. Therefore, the adaptor enables the conversion of electrostatically-driven Bs captures and releases into a high-fidelity electrical readout. This mechanism for protein detection is fundamentally distinct from those developed by our predecessors.20, 22, 25, 46 Previously, single-molecule protein detection has been conducted using protein nanopores equipped with small to medium-sized tethered ligands. Protein detection was also examined using solid-state nanopores by chemical attachment of protein recognition partners,21, 47 but the single-molecule precision of the attachment of these large functional protein groups remains difficult. Protein detection in FBS can be further optimized by making the nanopore even more acidic and measuring the protein captures at a lower positive potential (e.g., ~5 mV). The t-FhuA tolerates extensive alterations in the overall internal charge of the pore interior without affecting the stability of the open-state current.48, 49 Therefore, repelling a large amount of negatively-charged FBS impurities by the more-acidic pore interior will likely be instrumental in pushing temporal resolution toward detection of weak-affinity protein analytes in FBS. Another opportunity for a reduction in the FBS-induced noise is the use of the HI-FBS. Indeed, the frequency of FBS-induced irregular current blockades was drastically diminished by HI-FBS, suggesting that enhancement in the temporal resolution can be achieved. Other studies have highlighted the role of HI in enhancing protein detection.43, 44 Of course, this approach might only be used if the targeted protein analyte does not undergo denaturing under HI conditions.
In summary, we show quantitative protein detection in mammalian serum using a protein bait-containing biological nanopore. Measuring ion current modulation in the vicinity of the pore via a polypeptide tail adaptor is the most distinctive feature of this work from prior studies involving conventional nanopores. This process significantly differentiates our method from simple pore-plugging recordings, drastically enhancing its specificity. Because this selective nanopore sensor is engineered in a modular fashion as a single-polypeptide chain protein, there is no requirement for tedious purification steps of the targeted oligomer from other products of the assembly reaction, as many other multimeric biological nanopores necessitate. Moreover, our method circumvents the need for chemical modification of the t-FhuA stem, as well as covalent attachment of fluorophores or other reactive groups. Finally, the ability to discriminate a specific protein from many others present in a heterogeneous solution will have a transformative impact on molecular biomedical diagnostics and protein biotechnology.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Edward Lipson, Walter Freeman, Eleni Degaga, Motahareh Ghahari Larimi, and Lauren Mayse for their comments and stimulating discussions. This work was supported by US National Institutes of Health grants GM088403 (to L.M.) and GM129429 (to L.M.).
Footnotes
SUPPORTING INFORMATION. (i) Cloning and mutagenesis of the nanopore sensor and protein analyte; (ii) Protein expression and purification; (iii) Membrane protein refolding; (iv) Single-molecule electrophysiology using planar lipid bilayers; (v) Control single-channel electrical trace of the OBn(GGS)2t-FhuA protein pore at −40 mV; (vi) Standard histograms of the Bs capture and release events; (vii) FBS constituents produced long-lived current blockades at a high negative potential; (viii) Voltage dependence of the differential current; (ix) The SNR of the Oon and Ooff substates is improved at an applied transmembrane potential of +15 mV; (x) Control single-channel electrical trace of the OBn(GGS)2t-FhuA protein pore at +15 mV; (xi) Standard histograms of the Bs capture and release events at +15 mV; (xii) Analyses of the kinetic rate constants in homogeneous (FBS-free) and heterogeneous (FBS) solutions; (xiii) Other pieces of supporting information; (xiv) Supporting references. These materials are available free of charge via the Internet at http://pubs.acs.org
COMPETING INTERESTS.
A.K.T. and L.M. are named inventors on one non-provisional patent application, US 16/177,554, filed by Syracuse University on this work.
REFERENCES
- 1.Restrepo-Perez L; Joo C; Dekker C, Paving the way to single-molecule protein sequencing. Nat. Nanotechnol 2018, 13, 786–796. [DOI] [PubMed] [Google Scholar]
- 2.Heikenfeld J; Jajack A; Feldman B; Granger SW; Gaitonde S; Begtrup G; Katchman BA, Accessing analytes in biofluids for peripheral biochemical monitoring. Nat. Biotechnol 2019, 37, 407–419. [DOI] [PubMed] [Google Scholar]
- 3.Wienken CJ; Baaske P; Rothbauer U; Braun D; Duhr S, Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun 2010, 1, 100. [DOI] [PubMed] [Google Scholar]
- 4.Wang S; Haque F; Rychahou PG; Evers BM; Guo P, Engineered nanopore of Phi29 DNA-packaging motor for real-time detection of single colon cancer specific antibody in serum. ACS nano 2013, 7, 9814–9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fahie MA; Yang B; Mullis M; Holden MA; Chen M, Selective Detection of Protein Homologues in Serum Using an OmpG Nanopore. Analytical chemistry 2015, 87, 11143–11149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kukwikila M; Howorka S, Nanopore-based electrical and label-free sensing of enzyme activity in blood serum. Analytical chemistry 2015, 87, 9149–9154. [DOI] [PubMed] [Google Scholar]
- 7.Yoo J; Lee TS; Choi B; Shon MJ; Yoon TY, Observing Extremely Weak Protein-Protein Interactions with Conventional Single-Molecule Fluorescence Microscopy. J. Am. Chem. Soc 2016, 138, 14238–14241. [DOI] [PubMed] [Google Scholar]
- 8.Sze JYY; Ivanov AP; Cass AEG; Edel JB, Single molecule multiplexed nanopore protein screening in human serum using aptamer modified DNA carriers. Nat. Commun 2017, 8, 1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thakur AK; Larimi MG; Gooden K; Movileanu L, Aberrantly Large Single-Channel Conductance of Polyhistidine Arm-Containing Protein Nanopores. Biochemistry 2017, 56, 4895–4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thakur AK; Movileanu L, Real-Time Measurement of Protein-Protein Interactions at Single-Molecule Resolution using a Biological Nanopore. Nat. Biotechnol 2019, 37, 96–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Locher KP; Rees B; Koebnik R; Mitschler A; Moulinier L; Rosenbusch JP; Moras D, Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell 1998, 95, 771–778. [DOI] [PubMed] [Google Scholar]
- 12.Mohammad MM; Howard KR; Movileanu L, Redesign of a plugged beta-barrel membrane protein. J. Biol. Chem 2011, 286, 8000–8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohammad MM; Iyer R; Howard KR; McPike MP; Borer PN; Movileanu L, Engineering a Rigid Protein Tunnel for Biomolecular Detection. J. Am. Chem. Soc 2012, 134, 9521–9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z; Ghai I; Winterhalter M; Schwaneberg U, Engineering Enhanced Pore Sizes Using FhuA Delta1–160 from E. coli Outer Membrane as Template. ACS Sens. 2017, 2, 1619–1626. [DOI] [PubMed] [Google Scholar]
- 15.Wolfe AJ; Si W; Zhang Z; Blanden AR; Hsueh YC; Gugel JF; Pham B; Chen M; Loh SN; Rozovsky S; Aksimentiev A; Movileanu L, Quantification of membrane protein-detergent complex interactions. J. Phys. Chem. B 2017, 121, 10228–10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schreiber G; Fersht AR, Interaction of barnase with its polypeptide inhibitor barstar studied by protein engineering. Biochemistry 1993, 32, 5145–5150. [DOI] [PubMed] [Google Scholar]
- 17.Deyev SM; Waibel R; Lebedenko EN; Schubiger AP; Pluckthun A, Design of multivalent complexes using the barnase*barstar module. Nat. Biotechnol 2003, 21, 1486–1492. [DOI] [PubMed] [Google Scholar]
- 18.Buckle AM; Schreiber G; Fersht AR, Protein-protein recognition: crystal structural analysis of a barnase-barstar complex at 2.0-A resolution. Biochemistry 1994, 33, 8878–8889. [DOI] [PubMed] [Google Scholar]
- 19.Larimi MG; Mayse LA; Movileanu L, Interactions of a Polypeptide with a Protein Nanopore Under Crowding Conditions. ACS nano 2019, 13, 4469–4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Movileanu L; Howorka S; Braha O; Bayley H, Detecting protein analytes that modulate transmembrane movement of a polymer chain within a single protein pore. Nat. Biotechnol 2000, 18, 1091–1095. [DOI] [PubMed] [Google Scholar]
- 21.Wei R; Gatterdam V; Wieneke R; Tampe R; Rant U, Stochastic sensing of proteins with receptor-modified solid-state nanopores. Nat. Nanotechnol 2012, 7, 257–263. [DOI] [PubMed] [Google Scholar]
- 22.Rotem D; Jayasinghe L; Salichou M; Bayley H, Protein Detection by Nanopores Equipped with Aptamers. J. Am. Chem. Soc 2012, 134, 2781–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin Q; Fleming AM; Johnson RP; Ding Y; Burrows CJ; White HS, Base-excision repair activity of uracil-DNA glycosylase monitored using the latch zone of alpha-hemolysin. J. Am. Chem. Soc 2013, 135, 19347–13353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ying ,YZ, Xing; Liu Yu; Xue Mengzhu; Li Honglin; Long Yitao, Single Molecule Study of the Weak Biological Interactions Between P53 and DNA. Acta Chimica Sinica 2013, 71, 44–50. [Google Scholar]
- 25.Fahie M; Chisholm C; Chen M, Resolved single-molecule detection of individual species within a mixture of anti-biotin antibodies using an engineered monomeric nanopore. ACS nano 2015, 9, 1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohammad MM; Movileanu L, Excursion of a Single Polypeptide into a Protein Pore: Simple Physics, but Complicated Biology. Eur. Biophys. J 2008, 37, 913–925. [DOI] [PubMed] [Google Scholar]
- 27.Niedzwiecki DJ; Grazul J; Movileanu L, Single-molecule observation of protein adsorption onto an inorganic surface. J. Am. Chem. Soc 2010, 132, 10816–10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bikwemu R; Wolfe AJ; Xing X; Movileanu L, Facilitated Translocation of Polypeptides Through a Single Nanopore. J. Phys.: Condens. Matter 2010, 22, 454117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nivala J; Marks DB; Akeson M, Unfoldase-mediated protein translocation through an alpha-hemolysin nanopore. Nat. Biotechnol 2013, 31, 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez-Larrea D; Bayley H, Multistep protein unfolding during nanopore translocation. Nat. Nanotechnol 2013, 8, 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larkin J; Henley RY; Muthukumar M; Rosenstein JK; Wanunu M, High-bandwidth protein analysis using solid-state nanopores. Biophys. J 2014, 106 (3), 696–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kennedy E; Dong Z; Tennant C; Timp G, Reading the primary structure of a protein with 0.07 nm3 resolution using a subnanometre-diameter pore. Nat. Nanotechnol 2016, 11, 968–976. [DOI] [PubMed] [Google Scholar]
- 33.Yusko EC; Bruhn BR; Eggenberger OM; Houghtaling J; Rollings RC; Walsh NC; Nandivada S; Pindrus M; Hall AR; Sept D; Li J; Kalonia DS; Mayer M, Real-time shape approximation and fingerprinting of single proteins using a nanopore. Nat. Nanotechnol 2017, 12, 360–367. [DOI] [PubMed] [Google Scholar]
- 34.Howorka S, Building membrane nanopores. Nat. Nanotechnol 2017, 12, 619–630. [DOI] [PubMed] [Google Scholar]
- 35.Wloka C; Van Meervelt V; van Gelder D; Danda N; Jager N; Williams CP; Maglia G, Label-Free and Real-Time Detection of Protein Ubiquitination with a Biological Nanopore. ACS nano 2017, 11, 4387–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoogerheide DP; Gurnev PA; Rostovtseva TK; Bezrukov SM, Real-Time Nanopore-Based Recognition of Protein Translocation Success. Biophys. J 2018, 114, 772–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varongchayakul N; Song J; Meller A; Grinstaff MW, Single-Molecule Protein Sensing in a Nanopore: a Tutorial. Chem. Soc. Rev 2018, 47, 8512–8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schreiber G; Fersht AR, Energetics of protein-protein interactions: analysis of the barnase-barstar interface by single mutations and double mutant cycles. J. Mol. Biol 1995, 248, 478–486. [DOI] [PubMed] [Google Scholar]
- 39.Couoh-Cardel S; Hsueh YC; Wilkens S; Movileanu L, Yeast V-ATPase Proteolipid Ring Acts as a Large-conductance Transmembrane Protein Pore. Sci. Rep 2016, 6, 24774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sackmann B; Neher E, Single-Channel Recording. Second Edition ed.; Kluwer Academic/Plenum Publishers: New York, 1995. [Google Scholar]
- 41.Soltis RD; Hasz D; Morris MJ; Wilson ID, The effect of heat inactivation of serum on aggregation of immunoglobulins. Immunology 1979, 36, 37–45. [PMC free article] [PubMed] [Google Scholar]
- 42.Cancado EL; Vilas-Boas LS; Abrantes-Lemos CP; Novo NF; Porta G; Da Silva LC; Laudanna AA, Heat serum inactivation as a mandatory procedure for antiactin antibody detection in cell culture. Hepatology 1996, 23, 1098–104. [DOI] [PubMed] [Google Scholar]
- 43.Namekar M; Kumar M; O’Connell M; Nerurkar VR, Effect of serum heat-inactivation and dilution on detection of anti-WNV antibodies in mice by West Nile virus E-protein microsphere immunoassay. PloS one 2012, 7, e45851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kennedy SN; Wilhite B; Margaret Castellini J; Rea LD; Kuhn TB; Ferrante A; O’Hara TM, Enhanced quantification of serum immunoglobulin G from a non-model wildlife species, the Steller sea lion (Eumetopias jubatus), using a protein A ELISA. J. Immunol. Methods 2018, 462, 42–47. [DOI] [PubMed] [Google Scholar]
- 45.Niedzwiecki DJ; Mohammad MM; Movileanu L, Inspection of the Engineered FhuA deltaC/delta4L Protein Nanopore by Polymer Exclusion. Biophys. J 2012, 103, 2115–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harrington L; Cheley S; Alexander LT; Knapp S; Bayley H, Stochastic detection of Pim protein kinases reveals electrostatically enhanced association of a peptide substrate. Proc. Natl. Acad. Sci. U.S.A 2013, 110, E4417–E4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ying YL; Yu RJ; Hu YX; Gao R; Long YT, Single antibody-antigen interactions monitored via transient ionic current recording using nanopore sensors. Chem. Commun. (Camb) 2017, 53, 8620–8623. [DOI] [PubMed] [Google Scholar]
- 48.Wolfe AJ; Mohammad MM; Thakur AK; Movileanu L, Global redesign of a native beta-barrel scaffold. Biochim. Biophys. Acta 2016, 1858, 19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolfe AJ; Hsueh YC; Blanden AR; Mohammad MM; Pham B; Thakur AK; Loh SN; Chen M; Movileanu L, Interrogating Detergent Desolvation of Nanopore-Forming Proteins by Fluorescence Polarization Spectroscopy. Analytical chemistry 2017, 89, 8013–8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.