Abstract
Genomic DNA is constantly assaulted by both endogenous and exogenous damaging agents. The resulting DNA damage, if left unrepaired, can interfere with DNA replication and be converted into mutations. Genomic DNA is packaged into a highly compact yet dynamic chromatin structure, in order to fit into the limited space available in the nucleus of eukaryotic cells. This hierarchical chromatin organization serves as both the target of DNA damaging agents and the context for DNA repair enzymes. Biochemical studies have suggested that both the formation and repair of DNA damage are significantly modulated by chromatin. Our understanding of the impact of chromatin on damage and repair has been significantly enhanced by recent studies. We focus on the nucleosome, the primary building block of chromatin, and discuss how the intrinsic structural properties of nucleosomes, and their associated epigenetic modifications, affect damage formation and DNA repair, as well as subsequent mutagenesis in cancer.
Keywords: UV damage, skin cancer, nucleotide excision repair, base excision repair, mutagenesis, nucleosome
1. Introduction
DNA damage is a major threat to genomic integrity in living cells [1]. Different types of DNA damage, including small base modifications, bulky DNA lesions, and strand breaks, can interfere with DNA replication and gene transcription, causing cytotoxicity or mutagenesis. Small base damage such as oxidation, alkylation, deamination, and uracil misincorporation arise frequently in the cell from endogenous sources, and are therefore largely unavoidable [2]. Base excision repair (BER) is the primary mechanism that removes base lesions. Several enzymes are required for BER, including a DNA glycosylase, apurinic/apyrimidinic (AP) endonuclease, gap-filling DNA polymerase, and DNA ligase [3]. These enzymes are coordinated in a sequential manner to ensure that the repair product from the upstream enzyme serves as the substrate for the subsequent enzyme. This coordination has been shown to be important for efficient repair and preventing accumulation of more deleterious BER intermediates such as AP sites or strand breaks [4].
Additionally, environmental factors such as UV radiation and chemotherapeutic drugs (e.g., cisplatin) can induce bulky lesions, which cause significant distortion to the structure of the DNA double helix. Bulky lesions are generally more harmful to the cell than base damage, because they can strongly block DNA replication and transcription [1], The major repair pathway for bulky lesions is nucleotide excision repair (NER), which is a multi-step process comprising ~30 repair proteins [1]. Mutations of many NER genes are associated with severe human diseases such as Xeroderma pigmentosum (XP) and Cockayne syndrome (CS) [5], XP patients have extremely high risk for developing skin cancers, while CS patients are characterized by neurological degeneration and premature aging. Collectively, excision repair (ER), including BER and NER, acts as an essential ‘first-line’ defense mechanism in the cell to maintain genome stability by removing a wide spectrum of common DNA lesions. On the other hand, ER is a double-edged sword for chemotherapy. DNA adducts induced by chemotherapeutic agents may be removed by ER factors in cancer cells. The removal of these lethal DNA adducts in tumors can compromise chemotherapy and cause drug resistance [6].
Genomic DNA in eukaryotes is organized into chromatin. The basic repeating unit of chromatin is nucleosome, which is a complex consisting of ~147bp DNA wrapped in ~1.7 left-handed superhelical turns around a core histone octamer [7]. Neighboring nucleosomes are connected by linker DNA and packaged into higher-order chromatin structures. Chromatin compaction strongly affects DNA accessibility and represents a fundamental regulatory mechanism for DNA replication, transcription, and repair. For transcription, actively transcribed chromatin is ‘preset’ in an open state and enriched with modified nucleosomes that allow access of proteins to DNA [8], However, ER proteins have to recognize and remove damage in all structural domains of chromatin and at different levels of chromatin compaction [9], Therefore, a central challenge of DNA repair studies in eukaryotes is to understand how repair factors cope with the wide diversity of chromatin landscapes to efficiently repair DNA damage and avoid mutations. Biochemical data indicates that even the first level of chromatin compaction – the nucleosome – can significantly inhibit BER and NER [10,11]. These studies suggest that DNA damage in nucleosome-occupied DNA is more likely to persist and potentially cause mutations. In addition to affecting repair, the nucleosome structure has also been shown to modulate the formation of certain types of DNA lesions (e.g., UV photoproducts) [9].
Our understanding of DNA damage formation and ER in the context of chromatin has been significantly advanced by new technologies for mapping nucleosomes and measuring DNA damage and repair at high-resolution and across the genome [12-15]. The development of next generation sequencing (NGS)-based methods for mapping DNA damage provides an unprecedented opportunity to measure the formation and repair of DNA lesions in different chromatin contexts. In this review, we will summarize recent progress in our understanding of how chromatin organization impacts DNA damage formation and repair, and how these processes shape the genomic distribution of somatic mutations in human cancers.
2. DNA damage formation in chromatin and its impact on cancer mutations
2.1. DNA damage formation is modulated by nucleosomes
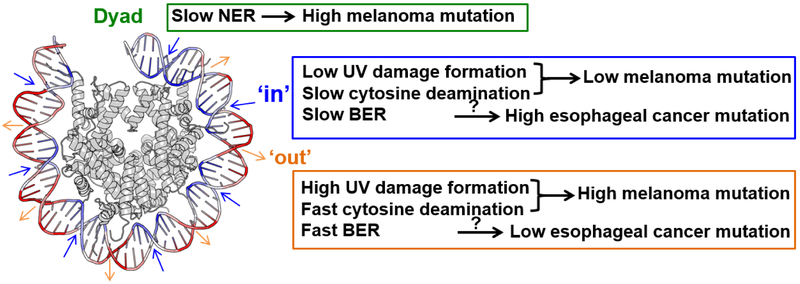
Solar UV is the major environmental carcinogen for melanoma and nonmelanoma skin cancers. Major photolesions induced by UV include cyclobutane pyrimidine dimers (CPDs) and (6-4) photoproducts (6,4-PPs), both of which occur at dipyrimidine DNA sequences (i.e., TT, TC, CT, and CC) [1]. Although it has been assumed that UV damage formation is largely uniform in chromatin (e.g., [16]), previous studies have shown that nucleosomes significantly modulate CPD formation in UV-irradiated chromatin, generating an ~10 bp periodicity of CPD lesions along the nucleosomal DNA [17]. This periodic CPD pattern is consistent with the nucleosome rotational settings (i.e., DNA backbone orientation relative to histone proteins), with high CPD formation at outward rotational (‘out’) and low CPD formation at inward rotational settings (‘in’) [17,18], The elevated CPD formation at ‘out’ positions is likely due to increased DNA mobility, which can facilitate the [2+2] cycloaddition reaction upon UV irradiation [18].
This unique oscillatory CPD pattern in nucleosomal DNA has been recapitulated in UV-irradiated cells using NGS-based damage mapping methods. Using a genome-wide method named CPD-seq (cyclobutane pyrimidine dimer sequencing), the periodic CPD pattern is apparent in nucleosomes in both yeast and human cells [18-20]. In CPD-seq, genomic DNA fragments are ligated to an adaptor DNA and all the free 3’ ends are blocked with dideoxyATP. CPD damage is then processed by the DNA repair enzymes T4 endonuclease V (T4 endo) and AP endonuclease (APE1) to generate a new ligatable 3’-OH, which is then ligated to a second adaptor DNA for sequencing [19]. Using this method, the locations of CPD lesions can be mapped at single nucleotide resolution. By overlaying CPD-seq data with genomic nucleosome maps (e.g., [12,13]), it was revealed that strongly positioned nucleosomes exhibit a striking 10 bp periodicity of CPD formation, particularly for the more mutagenic CPDs (mCPDs) associated with TC, CT, and CC dipyrimidines [20]. Consistent with previous data [17], the CPD-seq data revealed that CPD (or mCPD) peaks and valleys are associated with ‘out’ and ‘in’ rotational settings, respectively [19,20]. Furthermore, this periodicity is not caused by the underlying DNA sequence context. UV-irradiation of naked (i.e., non-nucleosomal) genomic DNA has the opposite pattern of CPD formation, with elevated CPD levels at ‘in’ positions, likely due to an increased frequency of lesion-forming TT dinucleotides at inward rotational settings [19]. Normalization of cellular CPD-seq by naked DNA CPD-seq reveals even stronger CPD enrichment at outward rotational settings [18-20], suggesting that the nucleosomal rotational settings can override intrinsic DNA sequence preferences (i.e., higher TT dinucleotides at 'in' positions) to promote UV damage formation at 'out' positions. Furthermore, the CPD periodicity is not observed in weakly positioned nucleosomes that have potentially reduced occupancy and poorly established rotational settings [19].
The distribution of UV damage across the human genome has also been mapped by a method named HS-Damage-seq (high sensitivity damage sequencing) [21]. HS-Damage-seq exploits the observation that bulky DNA damage, such as 6-4PPs and CPDs, block DNA replication, in order to map the location of these DNA lesions. This method uses a specific antibody to enrich for DNA containing bulky lesions, such as CPDs. A new strand is subsequently synthesized using the damaged DNA strand as a template, so that the nascent DNA strand terminates at the site of DNA damage. By mapping the location of the 3' end of the nascent DNA strand (i.e., the site of the replication block), HS-Damage-seq is able to identify the DNA damage site. Analysis of HS-Damage-seq data from UV-irradiated human fibroblasts also revealed a periodic damage pattern, with ~10 bp periodicity in human nucleosomes [22].
Surprisingly, this analysis revealed that CPDs are elevated at ‘in’ rotational settings relative to ‘out’ positions in the HS-Damage-seq data [22], which is at odds with past studies [17]. Normalization of cellular HS-Damage-seq data by the 'expected' CPD levels, based on the pentanucleotide DNA sequence context, revealed more CPD enrichment at ‘out’ rotational settings [22], as expected. This suggests that the increased CPD formation at ‘in’ positions in the un-normalized HS-Damage-seq data is probably caused by DNA sequence biases. It has been previously shown that nucleosomal DNA has an intrinsic sequence preference to form CPDs at 'in' positions in the absence of bound histones [19]. For example, TT dinucleotide sequences are enriched at ‘in’ rotational settings to accommodate the sharp DNA bending in the nucleosome structure [13,19]. One potential explanation for increased effects of DNA sequence context on the HS-Damage-seq data is that this method employs a CPD-specific antibody to enrich for damaged DNA strands [21]. Some CPD antibodies have been reported to have some degree of sequence specificity (i.e., bias toward TT and CT lesions) [23], which could potentially explain why the TT (and CT) dinucleotides are overrepresented, while TC and CC lesions are underrepresented in the HS-Damage-seq data [21]. Furthermore, HS-Damage-seq reads associated with CPDs at TC and CC dinucleotides, which are likely the major contributors to melanoma mutations [24], were excluded from the publically accessible data (GEO accession number: GSE98025), so that only TT and CT lesions were analyzed in nucleosomes [21,22]. The high enrichment of TT dinucleotides, and resultant TT lesions, at ‘in’ rotational settings can explain why CPD levels were elevated at ‘in’ positions in the unnormalized HS-Damage-seq data [22].
Oxidatively induced DNA damage (i.e., 8-oxoguanine [8-oxoG]) also appears to be modulated by nucleosomes. A recent study mapped the endogenous 8-oxoG lesions at single nucleotide resolution across the yeast genome [25]. It was shown that 8-oxoG lesions are more abundant in nucleosome-occupied DNA. In contrast, nucleosome-depleted regions such as transcription start sites (TSS), transcription termination sites (TTS), and autonomously replicating sequences (ARS), had low 8-oxoG lesions [25]. However, the endogenous 8-oxoG lesion distribution is affected by the equilibrium between DNA susceptibility to oxidation damage and ongoing BER. Because BER is inhibited by nucleosomes in vitro and in vivo (see Section 3.3), the increased 8-oxoG levels in nucleosomes may be caused by lower BER activity in nucleosomes, not by elevated damage formation [25]. Further studies, for example, mapping the genome-wide distribution of 8-oxoG in a BER-deficient strain (i.e., OGG1 mutant), may provide insights into how nucleosome structure affects the formation of oxidation damage.
2.2. DNA damage formation in nucleosomes affects the distribution of cancer mutations
The vast majority of somatic mutations in melanoma are UV signature mutations, characterized by C>T transitions at dipyrimidine sites [24]. Comparison of genome-wide maps of UV-induced lesions and melanoma mutations suggests that increased susceptibility to UV damage at 'out' positions in nucleosomal DNA may promote mutagenesis in melanoma. Indeed, analyses of somatic melanoma mutations show a striking ~10 bp periodicity in strongly positioned nucleosomes [20,22]. Similar to the mutagenic CPD lesion pattern, melanoma mutations are elevated at outward but repressed at inward rotational settings [20,22]. In addition to lesion formation, mutations at CPD lesions are also stimulated by cytosine (or methylated cytosine [mC]) deamination in CPDs, which leads to cytosine-to-uracil or mC-to-thymine transitions [26]. Importantly, in vitro data indicate that mC deamination in CPDs is significantly accelerated if the lesion is located at outward rotational settings, but deamination rates are inhibited at inward settings in nucleosomes [27,28]. The synergistic modulation of both CPD formation and deamination may contribute to the strong rotational periodicity of somatic mutations in melanomas [28] (Figure 1). However, to test this model, it will be important to map the cytosine deamination rates in CPD lesions throughout the genome.
Figure 1: Nucleosome translational and rotational settings regulate UV damage formation, DNA repair, and cancer mutagenesis.
NER activity is inhibited at the nucleosome dyad center, which is associated with increased mutation density in melanoma. At inward-facing rotational settings (‘in’), where the DNA minor groove faces toward histones, both UV damage formation and deamination of methylated cytosine in CPDs are reduced, which is associated with decreased mutations at ‘in’ rotational positions in melanoma. BER activity is decreased at ‘in’ rotational settings, which may contribute to the high mutation frequency at ‘in’ positions in esophageal and gastric cancer. However, further studies are needed to establish the direct correlation between BER and cancer mutation variations in the nucleosome. In contrast to the ‘in’ positions, UV damage formation, deamination of methylated cytosine in CPDs, and BER activity are elevated at ‘out’ rotational settings.
A survey of somatic mutations from different types of cancer indicates that the 10 bp rotational periodicity is a widespread phenomenon in nucleosome-occupied DNA [22]. Similar to melanomas, lung cancer tumors also exhibit strong 10 bp periodicity, with high mutation density at ‘out’ and low mutation at ‘in’ rotational positions [22]. The tobacco carcinogen benzo[a]pyrene (BaP) is a major cause for lung cancer [29]. BaP is converted into the mutagen BaP diol epoxide (BPDE) by enzymatic metabolism in the cell, and BPDE can covalently attach to guanines and form bulky DNA adducts such as BPDE-deoxyguanosines (BPDE-dGs) [30,31]. The similar rotational mutation pattern between melanoma and lung cancer suggests that BPDE-induced bulky lesions may also preferentially occur at outward rotational settings. The genome-wide repair map for BPDE-dG has been reported recently [30], but the damage distribution map is not yet available. A high-resolution map of BPDE-dG adducts may help us understand how nucleosomes affect formation of lung cancer-associated DNA damage.
Intriguingly, other cancers (e.g., esophageal and gastric cancer) have significantly elevated mutation density at inward rotational settings [22], which has the opposite phase of somatic mutations derived from melanoma or lung cancers. It has been suggested that inhibited BER activity at ‘in’ positions in nucleosomes (see Section 3.3) could explain this periodicity [22], since DNA base damage may be more persistent at 'in' locations in nucleosomes. Esophageal and gastric cancer genomes have complicated mutation signatures [32,33], and it has been suggested that oxidative damage to the nucleotide pool substantially contributes to their mutagenesis [34]. The elevated mutation density at inward rotational settings in esophageal and gastric cancer genomes may be caused by the reduced BER activity for oxidative damage at ‘in’ positions in nucleosomes.
2.3. Transcription factor binding sites (TFBS) modulate UV damage formation and melanoma mutation distribution
In addition to nucleosomes, studies have shown the important role of transcription factor (TF) binding in affecting CPD formation. TF binding can both stimulate and suppress CPD formation in gene promoters in human cells, in a TF-dependent manner [35]. High-resolution mapping of CPD lesions across the yeast genome using CPD-seq revealed that DNA binding by Abf1 and Reb1 [19], two well-studied yeast transcription factors [36], generally suppressed CPD formation. However, it is not known if DNA binding by other yeast TFs can promote CPD formation.
If TF binding generally inhibits UV damage formation, reduced mutation rates would be expected at TFBS in skin cancers. Interestingly, analyses of melanoma mutations reveal the opposite: highly elevated mutation frequency was found at TFBS and transcription initiation sites [37,38]. Elevated mutation rates at TFBS have been attributed to decreased NER activity, likely due to impaired accessibility to the DNA lesion by the binding of TFs [37,38]. However, it was not clear to what extent variations in UV damage formation also contribute to high mutation rates at TFBS.
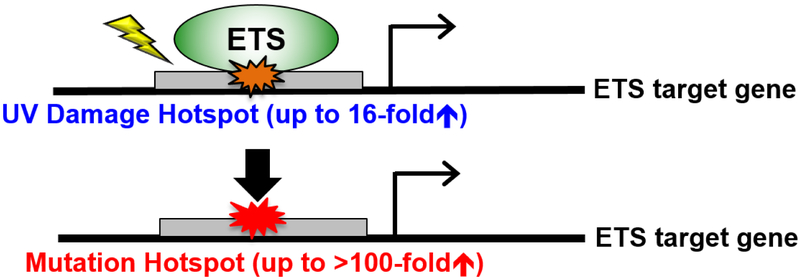
Two independent groups recently showed that specific classes of TFs can promote CPD formation in human cells upon UV treatment. Using CPD-seq for genome-wide damage mapping in human fibroblast cells, we found that CPD formation was elevated at active TFBS [39]. Among 82 different transcription factors analyzed, two classes of TFs showed a striking induction of CPD lesions at their binding sites: the ETS (E26 Transformation-specific) TF family and Nuclear Transcription Factor Y (NFYA/B) family. NFYA/B primarily induced CPD lesions at a TT dinucleotide in its binding motif, which is typically not a mutagenic lesion in human cells. This may explain why somatic mutations in melanoma are not significantly elevated at NFYA/B sites, despite elevated CPD levels. In contrast, analysis at ETS binding sites reveals unique damage and mutation hotspots, with up to 16-fold increase in CPD formation and over 100-fold increase in melanoma mutation density [39]. Additionally, ETS protein binding can directly stimulate CPD formation upon UV irradiation in vitro, likely by altering the torsion angle between two adjacent pyrimidines to facilitate the [2+2] cycloaddition reaction [39]. Elliott et al. also found highly increased CPD formation at ETS binding sites in human melanoma cells after UV treatment [40]. In fact, certain ETS binding sites (e.g., the RPL13 gene promoter) are so sensitive to UV irradiation that a single low dose of UVB treatment (20 J/m2) can induce mutations in the RPL13A ETS motif in cultured cells [40]. The occurrence of ETS mutation hotspots in cultured cells was independent of either GG-NER or TC-NER, pinpointing elevated CPD formation as the major mechanism for high mutations at ETS binding sites [40] (Figure 2).
Figure 2: ETS binding promotes UV damage formation and melanoma mutations.
DNA binding by ETS transcription factors significantly induces UV damage formation. The UV damage hotspots at ETS binding sites are associated with highly recurrent non-coding mutations in melanoma, which may alter transcription of ETS target genes.
While mutation hotspots at ETS binding sites arise from high susceptibility to UV damage, the increased mutation frequency associated with other TFs and at transcription initiation sites appears to be caused by inhibited repair [37-39]. Additionally, formation of 6,4-PPs can be significantly elevated at the binding sites of specific TFs in human gene promoters [35], and unrepaired 6,4-PPs may also cause mutations [41].
3. Excision repair in chromatin and its impact on cancer mutations
3.1. Chromatin state strongly modulates NER and affects melanoma mutations
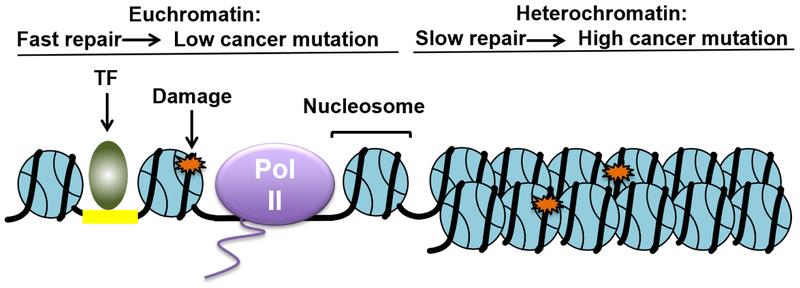
Genome-wide surveys of NER activity using the excision repair-sequencing (XR-seq) method in human fibroblast cells have revealed that UV damage repair is strongly modulated by the chromatin state [16,21]. Specifically, fast repair of both CPDs and 6,4-PPs preferentially occurs in open chromatin regions characterized with DNase I hypersensitivity and active post-translational histone modifications (e.g., H3K4me3 and H3K27ac). In contrast, slow repair is observed in closed chromatin, particularly for CPDs [16]. Repair of 6,4-PPs in general is fast and most lesions are repaired within 4 hours post-UV treatment, although 6,4-PPs are still preferentially repaired in open chromatin at earlier time points (as early as 5 minutes) [16]. TC-NER, which is a subpathway of NER and only repairs the transcribed strand of actively transcribed genes [42], is also more efficient in open chromatin regions.
The impact of chromatin states on NER is correlated with the mutation density in the melanoma genome. Closed chromatin regions, which usually are repaired less efficiently by NER, are associated with high somatic mutations in melanomas [16]. Consistent with this finding, a significant correlation is observed between melanoma mutation density and chromatin accessibility in melanocyte cells [43]. High mutation density is frequently found in poorly accessible chromatin regions [43], where low NER activity is observed [16]. These studies indicate that variable NER activity dictated by the chromatin state plays an important role in shaping global mutation heterogeneity in the melanoma genome (Figure 3).
Figure 3: Open and closed chromatin states significantly affect excision repair and cancer mutation distribution.
DNA damage (e.g., UV, cisplatin, or benzo[a]pyrene adducts) in open chromatin regions is repaired more efficiently, while damage in closed chromatin such as heterochromatin is repaired less efficiently. Consequently, high mutation densities in cancer are frequently found in closed chromatin, while low mutation densities are associated with an open chromatin state.
Chromatin state may also affect NER of other types of DNA damage, including DNA adducts induced by anti-cancer drug cisplatin (e.g., Pt-d(GpG) intrastrand crosslinks) and tobacco carcinogen benzo[a]pyrene (e.g., BPDE-dG) [44]. Using XR-seq, the genome-wide NER kinetics for Pt-d(GpG) crosslinks and BPDE-dG adducts has been characterized [30,45]. Similar to the repair of CPDs, repair of both Pt-d(GpG) and BPDE-dG is regulated by the chromatin state. High NER activity is associated with open chromatin states, such as gene promoters, enhancers, and transcribed gene bodies, while low repair efficiency is observed in closed chromatin [30,45], indicating that NER activity is generally modulated by the chromatin state, independent of the damage type.
3.2. Translational setting of the DNA lesion in a nucleosome affects NER and melanoma mutations
In vitro repair data and cellular repair analysis at individual gene loci in yeast indicate that NER is inhibited by nucleosomes (e.g., [11,46]). Analysis of genome-wide CPD-seq data in yeast indicates that the nucleosome translational setting (i.e., linear distance from the nucleosome dyad) plays a key role in affecting NER efficiency [19]. Specifically, CPDs located near the nucleosomal dyad axis are repaired less efficiently relative to damage near the nucleosome edges. This translational setting-dependent repair pattern is consistent with the observation that nucleosome dynamics are low at the central nucleosome dyad, but increase progressively toward the nucleosomal DNA ends [18,47].
In human cells, analysis of XR-seq data [21] generated in human fibroblast cells (after normalizing to CPD damage) shows that NER is generally slower near the nucleosome dyad in strongly positioned nucleosomes (over 1 million human nucleosomes in total) [20]. Importantly, the slower repair at the nucleosome center is correlated with elevated mutation density near the dyad axis in cutaneous melanomas [20]. The genome-wide damage and repair data indicates that both the rotational and translational settings of the lesion within the nucleosome play important roles in modulating mutation distribution in the melanoma genome, but through different mechanisms. The rotational setting affects UV damage formation (and potentially mC deamination rates) [20,22,27,28], but the translational setting appears to primarily affect repair efficiency [19,20] (Figure 1). It is likely that repair of other helix-distorting damage (e.g., cisplatin and benzo[a]pyrene damage) is similarly regulated by the nucleosomal translational setting (e.g., [45]).
3.3. BER is regulated by nucleosomes, which may promote mutagenesis in cancer
General inhibition of BER by nucleosomes has been observed in vitro using purified BER enzymes and reconstituted nucleosomes (reviewed in [48]). By changing the damage location to different rotational positions (e.g., ‘out’ and ‘in’), it has been shown that BER of uracil damage is significantly modulated by the nucleosomal rotational settings [49,50], presumably because uracil lesions at ‘out’ rotational settings are more easily recognized by DNA glycosylases, such as uracil-DNA glycosylase (UDG).
Genome-wide mapping of base damage has provided new insights into BER kinetics in chromatin. Using a method named NMP-seq (N-methylpurine sequencing), the precise location and relative abundance of alkylation lesions (i.e., N7-methylguarnine [7meG] and N3-methyladenine [3meA]) was mapped in the yeast genome upon treatment with the alkylating agent methyl methanesulfonate (MMS) [51]. By mapping remaining lesions after repair and normalizing to the initial damage distribution, high-resolution BER profiles at different repair time points were generated [51]. The BER maps indicate that repair of the most abundant alkylation damage, 7meG, is strongly modulated by nucleosome organization in gene coding regions. When ~5,000 yeast genes were aligned at their transcription start sites (TSS) [51], there was a striking repair periodicity that correlated with the stereotypic nucleosome positioning (e.g., +1, +2, and so on nucleosomes) [52]. BER is elevated in nucleosome-depleted regions (NDR) in gene promoters and also in nucleosomal linker DNA, but repressed within nucleosomes, particularly near the dyad axis, indicating that the translational setting of a 7meG affects its repair efficiency in vivo [51]. As a control, analysis of NMP-seq data in a BER-deficient mutant (i.e., mag1Δ) revealed no difference in 7meG removal between the dyad axis and the nucleosomal DNA ends [51]. Consistent with these findings, it was also discovered that the mutation density in MMS-treated yeast cells is elevated near the nucleosome center [51], suggesting that the reduced repair of alkylation damage near the central dyad promotes mutagenesis.
It was also found that epigenetic marks, such as histone post-translational modifications (PTMs), regulate BER efficiency in yeast chromatin [53]. Using genome-wide NMP-seq data, it was shown that histone PTMs enriched toward the 5’ end of active genes (e.g., H3K14ac, H3K4me3) [14] appear to promote BER near the nucleosome ends, but paradoxically repress repair at the dyad, while PTMs that are enriched toward the 3’ end of genes (e.g., H3K36me3 and H3K79me3) display the opposite trend [51]. Previous studies have suggested that H3K14ac can promote repair in part by enhancing binding of the chromatin remodeler RSC (Remodels Structure of Chromatin) to lesion-containing nucleosomes [54]. RSC-remodeled nucleosomes may have increased DNA accessibility near the nucleosome edges, which could explain the faster repair of 7meG toward the DNA ends of H3K14ac nucleosomes [51]. On the other hand, H3K14ac has been shown to strongly inhibit the gap-filling activity of DNA polymerase β in BER near the dyad center [55], which may contribute to the reduced repair of 7meG at the dyad center in H3K14ac-enriched nucleosomes in vivo [51]. Although these studies provide a potential mechanism for how histone PTMs affect BER, clearly further studies are needed.
NMP-seq analysis indicates that the nucleosome rotational settings also affect BER of 7meG, with more efficient repair at ‘out’ rotational positions than ‘in’ positions [51]. This finding is consistent with in vitro studies (e.g., [49]) and indicates that the high DNA accessibility at outward rotational positions facilitates repair of 7meG by the yeast glycosylase Magi This periodic BER pattern in nucleosomes may in some cases promote a similar mutation periodicity in certain types of human cancer, such as esophageal cancer and gastric cancer [22]. Low BER activity at ‘in’ rotational positions could promote the persistence of unrepaired base damage at inward facing positions, thereby promoting mutagenesis at these positions in cancers. In contrast, the high BER activity at ‘out’ positions may reduce mutation accumulation at outward rotational settings in nucleosomes (Figure 1).
Intriguingly, genome-wide analysis of BER using NMP-seq reveals asymmetric BER in yeast nucleosomes. Plotting of 7meG repair in the nucleosomal DNA shows more robust BER in the 5’ half of each DNA strand, particularly at the ‘out’ rotational settings [51]. This repair polarity may be caused by differential DNA accessibility in the 5’ and 3’ halves of each strand, because DNase-seq data shows that the ‘out’ positions 5’ of the dyad axis are more accessible to DNase I cleavage [12,51]. It will be interesting to investigate if other repair pathways such as NER are also affected by this asymmetric property of nucleosomal DNA.
4. Conclusions
Development of genome-wide methods for mapping DNA damage and repair has provided an important opportunity to investigate how chromatin organization regulates both lesion distribution and repair kinetics in eukaryotes. These studies shed new light on the impact of different levels of chromatin compaction, from the primary nucleosome structure to higher-order chromatin domains, on excision repair. High-resolution analysis in precisely mapped nucleosomes have further uncovered that intrinsic structural properties of the nucleosome, such as rotational and translational settings, and DNA strand polarity associated with its left-handed wrapping around histones, significantly modulate DNA damage formation and excision repair. Importantly, genome-wide damage and repair profiles provide mechanistic insights into mutation patterns in human cancers, highlighting that variations in both damage formation and excision repair, largely dictated by the underlying chromatin structure, profoundly affect the mutation landscape in cancer genomes.
Acknowledgements
We thank Drs. Michael Smerdon and Steven Roberts for critical reading of the manuscript. Research related to this work is supported by grants from NIEHS (R03ES027945 to P.M., R21ES029302 to P.M. and J.J.W., R01ES028698, R21ES029655, and R21ES027937 to J.J.W).
Abbreviations
- NER
nucleotide excision repair
- BER
base excision repair
- ER
excision repair
- XP
xeroderma pigmentosum
- CS
Cockayne syndrome
- NGS
nextgeneration sequencing
- CPD
cyclobutane pyrimidine dimer
- 6,4-PPs
(6-4) photoproducts
- mCPD
mutagenic cyclobutane pyrimidine dimer
- 8-oxoG
8-oxoguanine
- BaP
benzo[a]pyrene
- BPDE
BaP diol epoxide
- BPDE-dGs
BPDE-deoxyguanosines
- CPD-seq
cyclobutane pyrimidine dimer sequencing
- XR-seq
excision repair sequencing
- HS-Damage-seq
high sensitivity damage sequencing
- NMP-seq
N-methylpurine sequencing
- TFBS
transcription factor binding site
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no conflicts of interest.
References
- [1].Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenburger T, DNA repair and mutagenesis, in: DNA Repair and Mutagenesis., ASM Press, 2006. [Google Scholar]
- [2].De Bont R, van Larebeke N, Endogenous DNA damage in humans: a review of quantitative data, Mutagenesis. 19 (2004) 169–185. [DOI] [PubMed] [Google Scholar]
- [3].Krokan HE, Bjørås M, Base excision repair, Cold Spring Harb Perspect Biol. 5 (2013) a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Prasad R, Beard WA, Batra VK, Liu Y, Shock DD, Wilson SH, A Review of Recent Experiments on Step-to-Step “Hand-off” of the DNA Intermediates in Mammalian Base Excision Repair Pathways, Mol Biol (Mosk). 45 (2011) 586–600. [PMC free article] [PubMed] [Google Scholar]
- [5].Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JHJ, Understanding nucleotide excision repair and its roles in cancer and ageing, Nature Reviews Molecular Cell Biology. 15 (2014) 465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- [6].Martin LP, Hamilton TC, Schilder RJ, Platinum Resistance: The Role of DNA Repair Pathways, Clin Cancer Res. 14 (2008) 1291–1295. doi: 10.1158/1078-0432.CCR-07-2238. [DOI] [PubMed] [Google Scholar]
- [7].Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ, Crystal structure of the nucleosome core particle at 2.8 A resolution, Nature. 389 (1997) 251. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- [8].Li B, Carey M, Workman JL, The Role of Chromatin during Transcription, Cell. 128 (2007) 707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- [9].Smerdon MJ, Conconi A, Modulation of DNA Damage and DNA Repair in Chromatin, in: Moldave K (Ed.), Progress in Nucleic Acid Research and Molecular Biology, Academic Press, 1998: pp. 227–255. doi: 10.1016/S0079-6603(08)60509-7. [DOI] [PubMed] [Google Scholar]
- [10].Rodriguez Y, Smerdon MJ, The Structural Location of DNA Lesions in Nucleosome Core Particles Determines Accessibility by Base Excision Repair Enzymes, Journal of Biological Chemistry. 288 (2013) 13863–13875. doi: 10.1074/jbc.M112.441444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hara R, Mo J, Sancar A, DNA Damage in the Nucleosome Core Is Refractory to Repair by Human Excision Nuclease, Mol Cell Biol. 20 (2000) 9173–9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhong J, Luo K, Winter PS, Crawford GE, Iversen ES, Hartemink AJ, Mapping nucleosome positions using DNase-seq, Genome Research. 26 (2016) 351–364. doi: 10.1101/gr.195602.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Brogaard K, Xi L, Wang J-P, Widom J, A map of nucleosome positions in yeast at base-pair resolution, Nature. 486 (2012) 496. doi: 10.1038/nature11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Weiner A, Hsieh T-HS, Appleboim A, Chen HV, Rahat A, Amit I, Rando OJ, Friedman N, High-Resolution Chromatin Dynamics during a Yeast Stress Response, Molecular Cell. 58 (2015) 371–386. doi: 10.1016/j.molcel.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wyrick JJ, Roberts SA, Genomic approaches to DNA repair and mutagenesis, DNA Repair. 36 (2015) 146–155. doi: 10.1016/j.dnarep.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Adar S, Hu J, Lieb JD, Sancar A, Genome-wide kinetics of DNA excision repair in relation to chromatin state and mutagenesis, PNAS. (2016) 201603388. doi: 10.1073/pnas.1603388113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gale JM, Nissen KA, Smerdon MJ, UV-induced formation of pyrimidine dimers in nucleosome core DNA is strongly modulated with a period of 10.3 bases., Proceedings of the National Academy of Sciences. 84 (1987) 6644–6648. doi: 10.1073/pnas.84.19.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mao P, Wyrick JJ, Roberts SA, Smerdon MJ, UV-Induced DNA Damage and Mutagenesis in Chromatin, Photochemistry and Photobiology. 93 (2017) 216–228. doi: 10.1111/php.12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mao P, Smerdon MJ, Roberts SA, Wyrick JJ, Chromosomal landscape of UV damage formation and repair at single-nucleotide resolution, PNAS. 113 (2016) 9057–9062. doi: 10.1073/pnas.1606667113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Brown AJ, Mao P, Smerdon MJ, Wyrick JJ, Roberts SA, Nucleosome positions establish an extended mutation signature in melanoma, PLOS Genetics. 14 (2018) e1007823. doi: 10.1371/journal.pgen.1007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hu J, Adebali O, Adar S, Sancar A, Dynamic maps of UV damage formation and repair for the human genome, PNAS. 114 (2017) 6758–6763. doi: 10.1073/pnas.1706522114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pich O, Muiños F, Sabarinathan R, Reyes-Salazar I, Gonzalez-Perez A, Lopez-Bigas N, Somatic and Germline Mutation Periodicity Follow the Orientation of the DNA Minor Groove around Nucleosomes, Cell. 175 (2018) 1074–1087.e18. doi: 10.1016/j.cell.2018.10.004. [DOI] [PubMed] [Google Scholar]
- [23].Mori T, Nakane M, Hattori T, Matsunaga T, Ihara M, Nikaido O, Simultaneous Establishment of Monoclonal Antibodies Specific for Either Cyclobutane Pyrimidine Dimer or (6-4)photoproduct from the Same Mouse Immunized with Ultraviolet-Irradiated Dna, Photochemistry and Photobiology. 54 (1991) 225–232. doi: 10.1111/j.1751-1097.1991.tb02010.x. [DOI] [PubMed] [Google Scholar]
- [24].Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, Patch A-M, Kakavand H, Alexandrov LB, Burke H, Jakrot V, Kazakoff S, Holmes O, Leonard C, Sabarinathan R, Mularoni L, Wood S, Xu Q, Waddell N, Tembe V, Pupo GM, De Paoli-lseppi R, Vilain RE, Shang P, Lau LMS, Dagg RA, Schramm S-J, Pritchard A, Dutton-Regester K, Newell F, Fitzgerald A, Shang CA, Grimmond SM, Pickett HA, Yang JY, Stretch JR, Behren A, Kefford RF, Hersey P, Long GV, Cebon J, Shackleton M, Spillane AJ, Saw RPM, López-Bigas N, Pearson JV, Thompson JF, Scolyer RA, Mann GJ, Whole-genome landscapes of major melanoma subtypes, Nature. 545 (2017) 175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- [25].Wu J, McKeague M, Sturla SJ, Nucleotide-Resolution Genome-Wide Mapping of Oxidative DNA Damage by Click-Code-Seq, Journal of the American Chemical Society. 140 (2018) 9783–9787. doi: 10.1021/jacs.8b03715. [DOI] [PubMed] [Google Scholar]
- [26].Ikehata H, Ono T, The Mechanisms of UV Mutagenesis, Journal of Radiation Research. 52 (2011) 115–125. doi: 10.1269/jrr.10175. [DOI] [PubMed] [Google Scholar]
- [27].Song Q, Cannistraro VJ, Taylor J-S, Rotational Position of a 5-Methylcytosine-containing Cyclobutane Pyrimidine Dimer in a Nucleosome Greatly Affects Its Deamination Rate, Journal of Biological Chemistry. 286 (2011) 6329–6335. doi: 10.1074/jbc.M110.183178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Song Q, Cannistraro VJ, Taylor J-S, Synergistic modulation of cyclobutane pyrimidine dimer photoproduct formation and deamination at a TmCG site over a full helical DNA turn in a nucleosome core particle, Nucleic Acids Res. 42 (2014) 13122–13133. doi: 10.1093/nar/gku1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cruz CSD, Tanoue LT, Matthay RA, Lung Cancer: Epidemiology, Etiology, and Prevention, Clinics in Chest Medicine. 32 (2011) 605–644. doi: 10.1016/j.ccm.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Li W, Hu J, Adebali O, Adar S, Yang Y, Chiou Y-Y, Sancar A, Human genome-wide repair map of DNA damage caused by the cigarette smoke carcinogen benzo[a]pyrene, PNAS. (2017) 201706021. doi: 10.1073/pnas.1706021114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Denissenko MF, Pao A, Tang M, Pfeifer GP, Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53, Science. 274 (1996) 430–432. [DOI] [PubMed] [Google Scholar]
- [32].the Oesophageal Cancer Clinical and Molecular Stratification (OCCAMS) Consortium, Secrier M, Li X, de Silva N, Eldridge MD, Contino G, Bornschein J, MacRae S, Grehan N, O’Donovan M, Miremadi A, Yang T-P, Bower L, Chettouh H, Crawte J, Galeano-Dalmau N, Grabowska A, Saunders J, Underwood T, Waddell N, Barbour AP, Nutzinger B, Achilleos A, Edwards PAW, Lynch AG, Tavare S, Fitzgerald RC, Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance, Nature Genetics. 48 (2016) 1131–1141. doi: 10.1038/ng.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Li X, Wu WKK, Xing R, Wong SH, Liu Y, Fang X, Zhang Y, Wang M, Wang J, Li L, Zhou Y, Tang S, Peng S, Qiu K, Chen L, Chen K, Yang H, Zhang W, Chan MTV, Lu Y, Sung JJY, Yu J, Distinct Subtypes of Gastric Cancer Defined by Molecular Characterization Include Novel Mutational Signatures with Prognostic Capability, Cancer Res. 76 (2016) 1724–1732. doi: 10.1158/0008-5472.CAN-15-2443. [DOI] [PubMed] [Google Scholar]
- [34].Tomkova M, Tomek J, Kriaucionis S, Schuster-Böckler B, Mutational signature distribution varies with DNA replication timing and strand asymmetry, Genome Biology. 19 (2018). doi: 10.1186/s13059-018-1509-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tornaletti S, Pfeifer GP, UV Light as a Footprinting Agent: Modulation of UV-induced DNA Damage by Transcription Factors Bound at the Promoters of Three Human Genes, Journal of Molecular Biology. 249 (1995) 714–728. doi: 10.1006/jmbi.1995.0331. [DOI] [PubMed] [Google Scholar]
- [36].Kasinathan S, Orsi GA, Zentner GE, Ahmad K, Henikoff S, High-resolution mapping of transcription factor binding sites on native chromatin, Nat. Methods. 11 (2014) 203–209. doi: 10.1038/nmeth.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sabarinathan R, Mularoni L, Deu-Pons J, Gonzalez-Perez A, López-Bigas N, Nucleotide excision repair is impaired by binding of transcription factors to DNA, Nature. 532 (2016) 264–267. doi: 10.1038/nature17661. [DOI] [PubMed] [Google Scholar]
- [38].Perera D, Poulos RC, Shah A, Beck D, Pimanda JE, Wong JWH, Differential DNA repair underlies mutation hotspots at active promoters in cancer genomes, Nature. 532 (2016) 259–263. doi: 10.1038/nature17437. [DOI] [PubMed] [Google Scholar]
- [39].Mao P, Brown AJ, Esaki S, Lockwood S, Poon GMK, Smerdon MJ, Roberts SA, Wyrick JJ, ETS transcription factors induce a unique UV damage signature that drives recurrent mutagenesis in melanoma, Nature Communications. 9 (2018). doi: 10.1038/s41467-018-05064-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Elliott K, Böstrom M, Filges S, Lindberg M, den Eynden JV, Ståhlberg A, Clausen AR, Larsson E, Elevated pyrimidine dimer formation at distinct genomic bases underlies promoter mutation hotspots in UV-exposed cancers, PLOS Genetics. 14 (2018) e1007849. doi: 10.1371/journal.pgen.1007849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Otoshi E, Yagi T, Mori T, Matsunaga T, Nikaido O, Kim ST, Hitomi K, Ikenaga M, Todo T, Respective roles of cyclobutane pyrimidine dimers, (6-4)photoproducts, and minor photoproducts in ultraviolet mutagenesis of repair-deficient xeroderma pigmentosum A cells, Cancer Res. 60 (2000) 1729–1735. [PubMed] [Google Scholar]
- [42].Hanawalt PC, Spivak G, Transcription-coupled DNA repair: two decades of progress and surprises, Nat. Rev. Mol. Cell Biol. 9 (2008) 958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- [43].Polak P, Karlić R, Koren A, Thurman R, Sandstrom R, Lawrence M, Reynolds A, Rynes E, Vlahoviček K, Stamatoyannopoulos JA, Sunyaev SR, Cell-of-origin chromatin organization shapes the mutational landscape of cancer, Nature. 518 (2015) 360–364. doi: 10.1038/nature14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nouspikel T, DNA Repair in Mammalian Cells: Nucleotide excision repair: variations on versatility, Cellular and Molecular Life Sciences. 66 (2009) 994–1009. doi: 10.1007/s00018-009-8737-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hu J, Lieb JD, Sancar A, Adar S, Cisplatin DNA damage and repair maps of the human genome at single-nucleotide resolution, Proc. Natl. Acad. Sci. U.S.A. 113 (2016) 11507–11512. doi: 10.1073/pnas.1614430113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Li S, Ding B, LeJeune D, Ruggiero C, Chen X, Smerdon MJ, The roles of Rad16 and Rad26 in repairing repressed and actively transcribed genes in yeast, DNA Repair (Amst). 6 (2007) 1596–1606. doi: 10.1016/j.dnarep.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Anderson JD, Widom J, Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites, J. Mol. Biol. 296 (2000) 979–987. doi: 10.1006/jmbi.2000.3531. [DOI] [PubMed] [Google Scholar]
- [48].Rodriguez Y, Hinz JM, Smerdon MJ, Accessing DNA damage in chromatin: Preparing the chromatin landscape for base excision repair, DNA Repair. 32 (2015) 113–119. doi: 10.1016/j.dnarep.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Olmon ED, Delaney S, Differential Ability of Five DNA Glycosylases to Recognize and Repair Damage on Nucleosomal DNA, ACS Chemical Biology. 12(2017) 692–701. doi: 10.1021/acschembio.6b00921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hinz JM, Rodriguez Y, Smerdon MJ, Rotational dynamics of DNA on the nucleosome surface markedly impact accessibility to a DNA repair enzyme, PNAS. 107 (2010) 4646–4651. doi: 10.1073/pnas.0914443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Mao P, Brown AJ, Male EP, Mieczkowski PA, Smerdon MJ, Roberts SA, Wyrick JJ, Genome-wide maps of alkylation damage, repair, and mutagenesis in yeast reveal mechanisms of mutational heterogeneity, Genome Res. (2017). doi: 10.1101/gr.225771.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Jiang C, Pugh BF, Nucleosome positioning and gene regulation: advances through genomics, Nat Rev Genet. 10 (2009) 161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Mao P, Wyrick JJ, Emerging roles for histone modifications in DNA excision repair, FEMS Yeast Res. 16 (2016). doi: 10.1093/femsyr/fow090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Duan M-R, Smerdon MJ, Histone H3 lysine 14 (H3K14) acetylation facilitates DNA repair in a positioned nucleosome by stabilizing the binding of the chromatin Remodeler RSC (Remodels Structure of Chromatin), J. Biol. Chem. 289 (2014) 8353–8363. doi: 10.1074/jbc.M113.540732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Rodriguez Y, Hinz JM, Laughery MF, Wyrick JJ, Smerdon MJ, Site-specific Acetylation of Histone H3 Decreases Polymerase β Activity on Nucleosome Core Particles in Vitro, Journal of Biological Chemistry. 291 (2016) 11434–11445. doi: 10.1074/jbc.M116.725788. [DOI] [PMC free article] [PubMed] [Google Scholar]