Abstract
Pseudouridine (Ψ) is a post-transcriptional RNA modification that alters RNA-RNA and RNA-protein interactions that affect gene expression. mRNA pseudouridylation was recently discovered as a widespread and conserved phenomenon, but the mechanisms responsible for selective, regulated pseudouridylation of specific sequences within mRNAs were unknown. Here, we have revealed new mRNA targets for five pseudouridine synthases and probed the determinants of mRNA target recognition by the predominant mRNA pseudouridylating enzyme, Pus1, by developing high-throughput kinetic analysis of pseudouridylation in vitro. Combining computational prediction and rational mutational analysis revealed an RNA structural motif that is both necessary and sufficient for mRNA pseudouridylation. Applying this structural context information predicted hundreds of additional mRNA targets, that we showed were pseudouridylated in vivo. These results demonstrate a structure-dependent mode of mRNA target recognition by a conserved pseudouridine synthase and implicate modulation of RNA structure as the likely mechanism to regulate mRNA pseudouridylation.
Introduction
Post-transcriptional RNA modifications are ubiquitous, evolutionarily conserved, and chemically diverse, with more than 100 modifications identified to date primarily in abundant non-coding RNAs (ncRNAs) such as tRNA and rRNA1. The development of high-throughput sequencing techniques to map RNA modifications has dramatically expanded the known complement of modified nucleotides found in mRNAs2, which includes widespread pseudouridylation of mRNAs that is regulated in response to changing cellular environments in yeast, mouse, and human cells3–7. However, the molecular mechanisms underlying regulated mRNA pseudouridylation are not yet understood.
Pseudouridine (Ψ) is formed by isomerization of uridine via breakage of the glycosidic bond, 180° base-rotation, and bond reformation8. Two major classes of pseudouridine synthases (PUS) guide pseudouridylation in either an RNA-dependent, or RNA-independent manner. The RNA-dependent PUS, Cbf5 in yeast and dyskerin in humans, associate with Box H/ACA snoRNPs and recognize their targets through base-pairing between the guide snoRNA and substrate RNA, primarily modifying rRNA. The RNA-independent PUS proteins target site-specific modification of tRNA, snRNA, and rRNA by directly recognizing sequence and structural elements within these molecules8. Most of the enzymes responsible for modifying eukaryotic non-coding RNAs have been identified by genetic analysis in budding yeast and extended to human cells based on homology. Similar genetic, bioinformatic, and biochemical analyses have suggested that the majority of mRNA pseudouridylation events in yeast and human cells are dependent on the tRNA modifying PUS proteins3–7, although genetic data are lacking for many mRNA Ψs, especially in mammalian systems.
How are mRNA substrates recognized by these tRNA modifying enzymes? Sequence motifs found in mRNA targets genetically assigned to Pus4 and Pus7 match the motifs known to be important for their modification of tRNAs, suggesting similar modes of recognition for mRNAs and tRNAs3,4. Consistently, mutational analysis demonstrated the importance of a structure resembling the tRNA TΨC-loop for mRNA recognition by the human Pus4 ortholog, TRUB17. In contrast, the basis for mRNA target recognition by most PUS is unknown. Targeting by Pus1 is particularly perplexing: Pus1 modifies structurally diverse positions in tRNAs as well as sites in the U2 and U6 snRNAs9–12, and the mRNA targets of Pus1 share only a weak sequence motif, HRU, which cannot explain the observed specificity3.
Here we sought to understand the specificity determinants governing mRNA target recognition by yeast Pus1, which we identified as the predominant mRNA pseudouridylating enzyme in growing cells3. We developed a rapid, high-throughput in vitro assay to quantitatively assess pseudouridylation of thousands of sequences in parallel, which validated 83% of mRNA Ψs genetically assigned to yeast Pus1 in vivo. Extending this approach to human mRNA Ψ sites without available genetic data identified novel targets of human TRUB1, PUS7, PUS1, RPUSD2, and TRUB2. We then used this high-throughput pseudouridylation assay to exhaustively probe the importance of mRNA sequence and structural features for modification by yeast Pus1. Based on in silico predictions that were supported by transcriptome-wide structure probing data, we identified a structural motif shared by most mRNA targets of yeast Pus1. Quantitative kinetic analysis revealed a subset of mRNA structural features that are necessary and sufficient for pseudouridylation in vitro. Finally, applying structural context information enabled prediction of additional Pus1 mRNA targets that we showed were modified in vivo. These results demonstrate a structure-dependent mode of target recognition and implicate structure modulation as a likely basis for regulating mRNA pseudouridylation. Our approach is broadly applicable to the study of mRNA modifications.
Results
A High-Throughput In vitro Pseudouridylation Assay
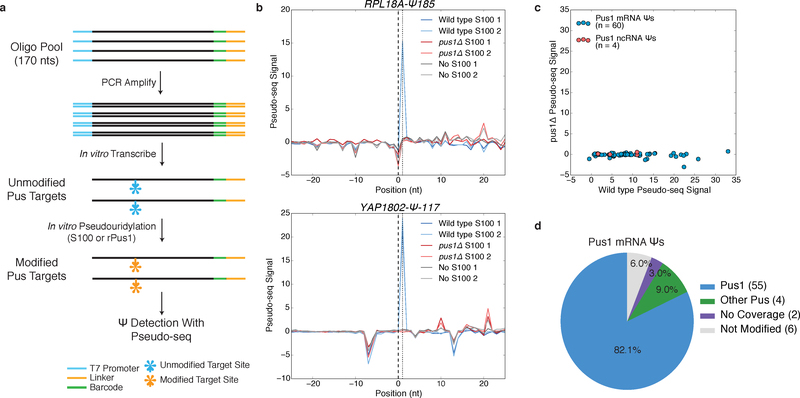
We developed a high-throughput in vitro pseudouridylation and detection assay to validate mRNA Ψs as direct targets of PUS implicated by genetic analysis in vivo and identify the PUS responsible for mRNA Ψs lacking genetic data. Synthetic RNA substrates were prepared by in vitro transcription from DNA oligo pools corresponding to 120 nts flanking mRNA Ψs identified in vivo. These RNAs are then pseudouridylated in vitro using recombinant PUS or cell extract and pseudouridylation is detected using Pseudo-seq (Figure 1a; Online Methods), which takes advantage of the specific labeling of Ψ residues with N-Cyclohexyl-N′-(2-morpholinoethyl)carbodiimide p-toluenesulfonate (CMC) to produce reverse transcriptase stops that can be detected by next-generation sequencing13,14. To validate this in vitro approach, we examined pseudouridylation of known yeast Pus1 ncRNA substrates U2 snRNA (Ψ44) and tRNALysUUU (Ψ27). These control RNAs were efficiently pseudouridylated by wild type, but not pus1Δ extracts and by recombinant Pus1 (Supplementary Figures 1a,b).
Figure 1: A High-Throughput In vitro Pseudouridylation Assay.
a) A schematic of in vitro pseudouridylation of oligo pool-derived RNAs and Ψ detection with Pseudo-seq. b) Pseudo-seq signal for mRNA substrates incubated with wild type S100 (blue), pus1Δ S100 (red), or no extract (gray). RPL18A-Ψ185 (upper), YAP1802-Ψ-117 (lower). c) A scatter plot of Pseudo-seq signal for pools incubated with wild type or pus1Δ S100 extracts. Sequences correspond to Pus1 ncRNA (red, n=4 sequences) and mRNA (blue, n=60 sequences) substrates. Values represent an average of n=2 replicates. d) Summary of in vitro pseudouridylation of PUS1-dependent mRNA Ψs.
We next examined mRNA sites that were detected in wild type but not pus1Δ cells in vivo3 (and Carlile et al. unpublished). Approximately 85% (55/65) of candidate Pus1 targets were pseudouridylated in a Pus1-dependent manner in vitro (Figure 1b–d, Supplementary Dataset 1a). The remaining sites included 4 modified by another PUS and 6 not modified in extracts (Figure 1d). This assay recapitulated the in vivo PUS-dependence for additional enzymes: PUS7-dependent mRNA Ψs were pseudouridylated by wild type and pus1Δ extracts but not by pus7Δ extracts (Supplementary Figure 1c,d, Supplementary Dataset 1b,c), with the exception of PRE6-Ψ392, which was pseudouridylated in vitro in a Pus1-dependent manner. Reexamination of the in vivo Pseudo-seq data for the 5 incorrectly assigned sites showed low coverage and noisy signal. The 6 putative PUS1-dependent sites that were not modified in extracts may represent marginal substrates, false positives, or RNAs that do not fold properly in vitro. Because the median read coverage at mRNA sites in our in vitro Pseudo-seq assay was 18-fold greater than that obtained in transcriptome-wide Pseudo-seq in vivo, both weak positive signal and the absence of signal are more readily interpreted as evidence for or against pseudouridylation. Overall, this assay provides a robust high-throughput method for validating sites of mRNA pseudouridylation by specific PUS enzymes.
Identification of Human PUS mRNA Substrates In vitro
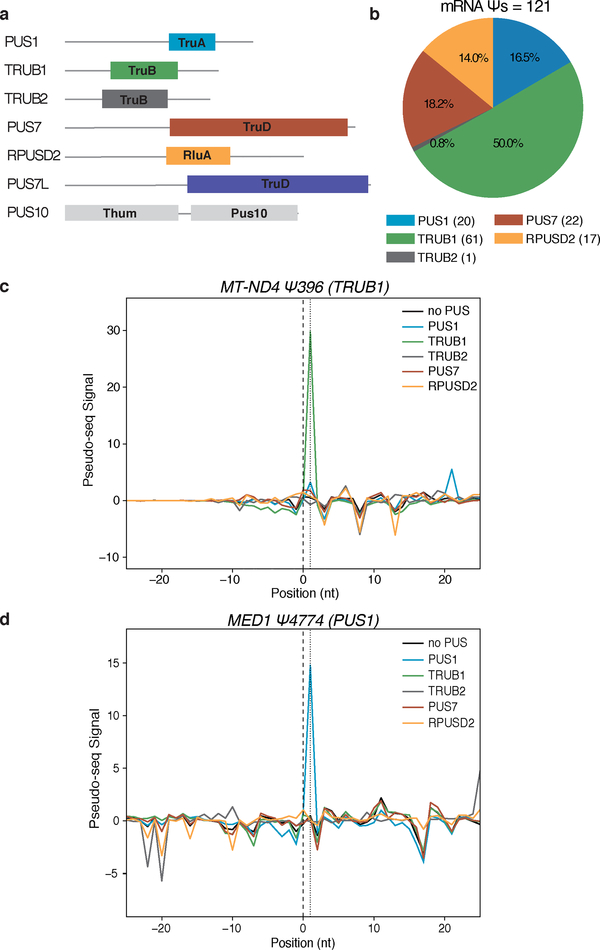
Most human mRNA Ψs identified to date have not been assigned to one of the 13 human PUS. We therefore tested sequences corresponding to 427 novel human mRNA Ψs identified in cells3,4 by in vitro pseudouridylation assays with 7 different human PUS representing 6 PUS families (Figure 2a). In this pool, 121 mRNAs were pseudouridylated by one of the tested PUS (Figure 2b–d, Supplementary Figure 2a–c, Supplementary Dataset 2). TRUB1 modified the largest number of sites (61), followed by PUS7 (22), PUS1 (20), RPUSD2 (17), and TRUB2 (1). PUS7L and PUS10 were active in vitro but did not modify any RNAs in this pool. These in vitro results validated 45 of 53 computationally predicted TRUB1 targets and 5 of 5 predicted PUS7 targets4. The modification of a cytosolic mRNA by TRUB2 was unexpected given the predominantly mitochondrial localization of this enzyme; conceivably another PUS is responsible for this modification in cells. Notably, even closely related PUS such as TRUB1 and TRUB2 did not show cross reactivity towards target mRNAs (Supplementary Dataset 2). The fraction of mRNA sites targeted by each PUS differed between Ψs identified in HEK293T and HeLa cells suggesting cell type specific differences in PUS activity (Supplementary Figure 2d).
Figure 2: Identification of Human PUS mRNA Substrates In vitro.
a) Schematic of the pseudouridine synthase domain structures of 7 human PUS proteins. b-d) RNA pools of sequences of mRNA Ψs from H. sapiens were pseudouridylated with recombinant PUS proteins: PUS1 (blue), TRUB1 (green), TRUB2 (gray), PUS7 (red), RPUSD2 (yellow), no PUS (black). b) A summary of mRNA Ψs assigned to hPUS proteins. c,d) Pseudo-seq signal for (c) a TRUB1 mRNA target: MT-ND4-Ψ396, and (d) a PUS1 mRNA target: MED1-Ψ4774.
The newly assigned TRUB1 targets from mRNA Ψs identified in HeLa cells contain the sequence motif RGUΨCNANYCY, which is found in canonical TRUB1 tRNA substrates (Supplementary Figure 2e)3. Interestingly, 43 of 61 TRUB1 target sites were efficiently modified in vitro by yeast extract (Supplementary Dataset 2), consistent with a conserved mechanism of target recognition by TRUB1 and its orthologue Pus4. The 22 validated PUS7 targets were enriched for a UNΨAR motif, a relaxed version of the yeast Pus7 UGΨAR motif used to predict human PUS7 targets (Supplementary Figure 2f)3,4. In addition, we validated 5 PUS7 and 11 TRUB1 targets that differ from the consensus sequences, and were therefore not predicted previously. The newly identified mRNA targets of PUS1 and RPUSD2 did not occur within consistent sequence contexts and so could not have been predicted computationally (Supplementary Figure 2g,h). Together, these results reveal new mRNA targets for five human PUS proteins that represent four different families of pseudouridine synthases.
We also tested 63 mRNA Ψs genetically assigned to PUS1 in a second study of mRNA pseudouridylation in HEK293T cells6. Unexpectedly, only 4.7% (3/63) were modified by recombinant PUS1 in vitro (Supplementary Dataset 2). By contrast, 82% of genetically assigned yeast Pus1 targets were validated as direct targets (Figure 1d)3. Given that we failed to detect pseudouridylation of some known human PUS1 tRNA targets in vitro (Supplementary Dataset 2), it is possible that some bona fide mRNA targets might also fail to be modified. Alternatively, the criteria used to assign these 63 mRNA Ψs to PUS1 may have been too permissive. Consistent with this possibility, yeast mRNA Ψs with ambiguous PUS1-dependence in vivo validated at a much lower rate than the sites that met our strict criteria for genetic assignment (20% vs. 82%, Supplementary Figures 2i,j). Thus, in addition to assigning novel Ψs to PUS, our approach can validate lower confidence in vivo targets. Taken together, these results establish the utility of high-throughput in vitro pseudouridylation to identify PUS targets, a critical step enabling functional studies of mRNA pseudouridylation in vivo.
A Structural Motif Associated with Yeast Pus1 mRNA Ψs
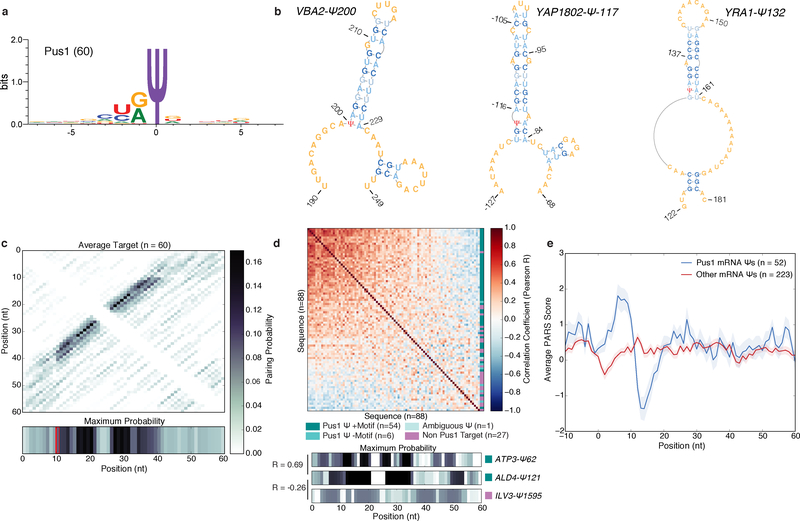
We reasoned that the highly parallel configuration of our in vitro pseudouridylation assay could facilitate mechanistic dissection of PUS target recognition, a task traditionally accomplished by low-throughput testing of mutant RNA substrates. We chose to investigate the mechanism of mRNA target recognition by Pus1 because it is the main yeast mRNA pseudouridine synthase for the conditions that we examined, and its mRNA targets share only a weak sequence motif, HRU, suggesting a primarily structural mode of substrate recognition (Figure 3a)3. To identify structures associated with mRNA pseudouridylation by yeast Pus1, a set of 60 high-confidence Pus1 mRNA targets (Figure 1c, Supplementary Dataset 1a) were folded in silico (Online Methods)15. The pairing probability matrices and predicted minimum free energy (MFE) structures revealed a common structural motif in which the Ψ is located at the 5′ base of a bulged stem loop (Figure 3b, Supplementary Figure 3a). Overall, 54 of 60 high confidence Pus1 mRNA targets were predicted to share this structural motif (Figure 3c), which is similar to the structural context of several known Pus1-dependent ncRNA Ψs including tRNA positions U1, U26–28 (Supplementary Figure 3b), and U34 and U36, which are located near the 5′ bases of the acceptor stem, anticodon stem, and a stem in unspliced tRNA respectively, and U44 in the U2 snRNA, which is located 5′ of stem IIa (Supplementary Figure 3c).
Figure 3: A Structural Motif Associated with Pus1 mRNA Targets in Yeast.
a) The sequence motif surrounding n=60 high confidence PUS1-dependent mRNA Ψs, generated with WebLogo 3.5. b) MFE structures for VBA2-Ψ200 (left), YAP1802-Ψ-117 (middle) YRA1-Ψ132 (right). c) A heatmap of the average pairing probability matrix from RNAfold (upper), and the average maximum pairing probability for each base (lower) for n=60 high-confidence Pus1 targets. d) A heatmap of pairwise correlation coefficients (Pearson R) between the arrays of maximum pairing probabilities for each mRNA site (n=88 Ψs with genetic evidence for Pus1-dependence in vivo). In vitro modified mRNA Ψs with a stem-loop motif (dark teal, n=54 seqeunces), in vitro modified without a stem-loop motif (medium teal, n=6 sequences), with ambiguous in vitro data (light teal, n=1 sequence), and not modified in vitro (purple, n=27 sequences). Rows and columns are ordered by the sum of R values across the row/column. Indicated on the right is the classification of each sequence (upper). The maximum pairing probability and correlation values for 3 sequences (lower). e) Average PARS score ±SEM for our high confidence Pus1 mRNA substrates (blue, n=52 targets), and all other mRNA Ψs (red, n=223 targets) identified in log phase and high density for which there is PARS data.
To rigorously assess the similarity of the structures predicted for Pus1 mRNA targets, we calculated the pairwise Pearson correlation coefficients between all predicted structures, for either the maximum predicted pairing probability at each position (Figure 3d), or the sum of pairing probabilities for each position (Supplementary Figure 3d). Pairing probabilities were generally positively correlated between Pus1 mRNA substrates, and negatively correlated between substrates and non-substrates. This shared structural motif was specific to Pus1 mRNA substrates, as PUS7-dependent mRNA targets lack a similar motif (Supplementary Figure 3e).
Next, we analyzed genome-wide structure-probing data to determine whether the predicted structures for Pus1 mRNA substrates are consistent with experimental observations. Parallel analysis of RNA structure (PARS) measures the intrinsic folding capacity of RNA sequences in vitro after extraction from cells by quantifying cleavage with endonucleases specific for double-stranded or single-stranded RNA16. A higher PARS score indicates increased base pairing. PARS data for high confidence Pus1 mRNA Ψs supported the predicted motif, showing an increase in structure immediately downstream of the Ψs followed by a decrease in structure at positions corresponding to the predicted loops (Figure 3e, Supplementary Figure 3f). This signature was not present in all our other yeast mRNA Ψs nor was it in Pus7 mRNA Ψs (Supplementary Figure 3g), or with candidate Ψs that failed to validate as Pus1 dependent in vitro (Supplementary Figure 3h). Taken together, these data identify a structural motif shared by most Pus1 mRNA targets, and suggest that this motif could be important for Pus1 mRNA substrate modification.
Pus1 Modifies mRNA Targets with Variable Kinetics
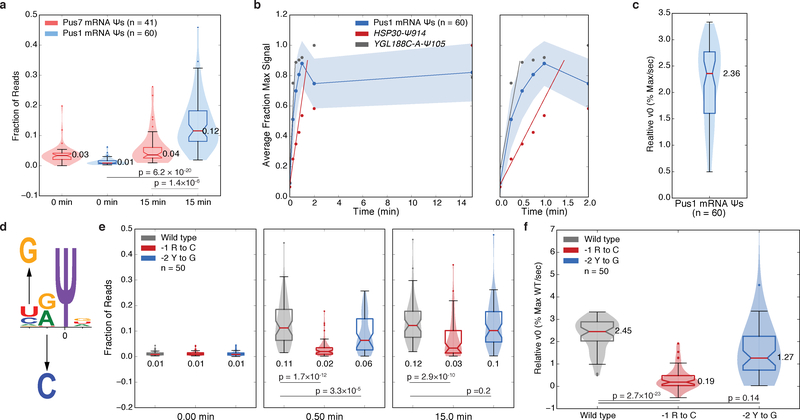
We wished to directly compare pseudouridylation between different substrates and quantify the contributions of various RNA features to pseudouridylation by Pus1. Because endpoint Pseudo-seq signal is affected by the capture biases inherent in library preparation, which is evident from differences in signal at rRNA Ψs that are fully pseudouridylated in vivo3,17, we undertook a kinetic approach to determine the relative initial velocity (v0,rel) for each sequence (Online Methods). RNA pools were incubated with excess recombinant yeast Pus1 and samples were taken across a 15 min time course. As expected, recombinant yeast Pus1 specifically modified the PUS1-dependent mRNA substrates validated in vitro (Figure 4a). Since the fraction of reads mapping to the Ψ-dependent RT stop position correlated well with Pseudo-seq signal (R2=0.88) (Supplementary Figures 4a,b), −CMC libraries were omitted from subsequent experiments. Calculating v0,rel for wild type sequences revealed >6-fold differences between Pus1 mRNA substrates (Figure 4b,c, Supplementary Dataset 4). Because Pus1 was in excess, relative modification rates among wild-type targets may be even more substantial in vivo. Interestingly, the 6 structurally atypical Pus1 targets showed v0,rel values similar to average typical targets suggesting that multiple modes of mRNA target recognition may lead to efficient pseudouridylation by Pus1 (Supplementary Figures 4c,d).
Figure 4: Kinetic Analysis Reveals Sequence Features Important for mRNA Pseudouridylation by Pus1.
a) Violin plots (center lines, medians; notches, 95% confidence intervals; boxes, 25th to 75th percentiles; whiskers, 1.5X inter-quartile range; dots, values outside of the 1.5X IQR) of the distributions of the fraction of reads mapping to the expected RT stop positions for Pus7 mRNA targets (pink, n=41 sequences) and Pus1 mRNA targets (blue, n=61 sequences). Medians, and p-values (unpaired t-test, two-tailed) are indicated. b) The average fraction of maximum signal for n=60 high confidence Pus1 mRNA targets is shown ± standard deviation (blue), for HSP30-Ψ914 (red), YGL188C-A-Ψ105 (gray) on a 0–15 min timescale (left) or a 0–2 min timescale (right). Lines indicating the slope (v0,rel) of the fit are indicated. c) A violin plot (elements as above) of the v0,rel values for n=60 high confidence Pus1 mRNA targets. Median indicated. d) A schematic of sequence motif mutations. e-f) Violin plots (elements as above) of the kinetics of pseudouridylation as indicated by (e) the fraction of reads at expected RT stop positions at indicated timepoints, or (f) v0,rel values for wild type (gray, n=50 sequences), −1 R to C mutants (red, n=50 sequences), and −2 H to G mutants (blue, n=50 sequences). Medians, and p-values (paired t-test, two-tailed) are indicated.
mRNA Sequence and Structural Requirements for Pus1
We used this kinetic framework to systematically probe yeast Pus1 mRNA target recognition by mutational analysis targeting a variety of RNA sequence and structural features. To test the importance of the weak HRU sequence motif, we mutated the −1 position to a C, and the −2 position to a G (Figure 4d). The relative extent of pseudouridylation for the −1 mutants was reduced at both early (30 sec) and late (15 min) time points (Figure 4e) leading to a highly significant reduction in v0,rel (paired, two-tailed t-test, p =2.7×10−23, Figure 4f). The absence of a G at the −2 position modestly slowed pseudouridylation (Figure 4e,f), although these changes failed to meet significance thresholds. Thus, the HR sequence at the −1 and −2 positions is important for efficient pseudouridylation of mRNAs by Pus1.
We next considered the importance of structural features for mRNA pseudouridylation by yeast Pus1. In the majority of Pus1 mRNA substrates, the target uridine is predicted to be base-paired, in either a U-A pair (32/54), or a U-G wobble pair (10/54). We systematically mutated the bases predicted to pair with the target uridine and observed no significant differences in either v0,rel (Supplementary Figures 4e,f), or in the final extent of pseudouridylation (data not shown). This result is consistent with the fact that Pus1 tRNA targets include paired and unpaired uridines, that the catalytic mechanism is thought to involve base flipping8, and that U-A base pairs at the ends of helices are known to fray18.
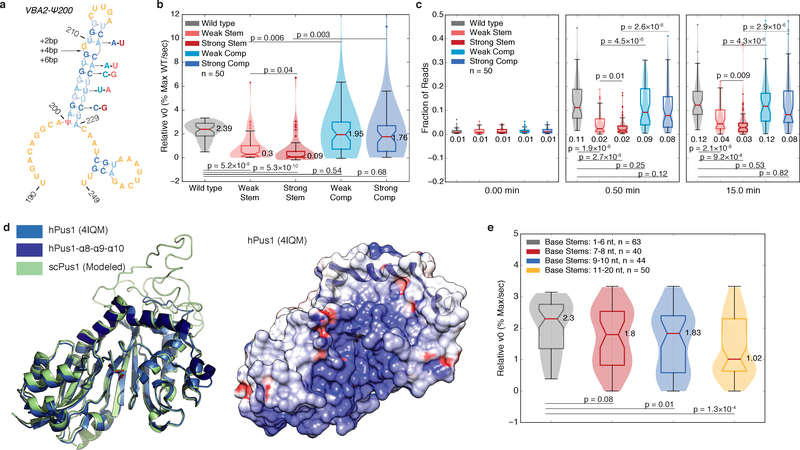
The most striking similarity among Pus1 mRNA targets was the bulged stem loop structure 3′ of the target uridine (Figure 3). To test the importance of this structure we made a series of mutations that disrupt base pairing in these stems to different extents, and compensatory mutations to restore pairing (Figure 5a). The v0,rel values were significantly reduced for both weak and strong stem disrupting mutations with a greater reduction in v0,rel for the stronger mutations. Importantly, both compensatory mutations rescued v0,rel to near wild type levels (Figure 5b). Likewise, the extent of pseudouridylation was significantly reduced by stem-disrupting mutations and was rescued by compensatory mutations (Figure 5c). These data demonstrate that a stem loop structure is critical for pseudouridylation of most Pus1 mRNA substrates.
Figure 5: The Rate of mRNA Pseudouridylation Depends on Stem Length and Stability.
a) A schematic of stem disrupting and compensatory mutations for VBA2-Ψ200. See b-c) for description of color scheme. b-c) Violin plots (center lines, medians; notches, 95% confidence intervals; boxes, 25th to 75th percentiles; whiskers, 1.5X inter-quartile range; dots, values outside of the 1.5X IQR) of (b) v0,rel values, or (c) the fraction of reads at the expected RT stop positions at indicated timepoints for wild type (gray, n=50 sequences), weak stem disrupting mutations (pink, n=50 sequences), strong stem disrupting mutations (red, n=50 sequences), weak compensatory mutations (light blue, n=50 sequences) and strong compensatory mutations (dark blue, n=50 sequences). Medians, and p-values (paired t-test, two-tailed) are indicated. d) The structure of hPus1 (light blue, 4IQM, Czudnochowski et al. 2013) with the three-helical RNA binding channel cap (dark blue) with modelled yeast Pus1 (left). An electrostatic surface map of hPus1, with the helical cap in ribbon form (right). e) Violin plots (elements as in above) of v0,rel for wild type, and stem extension mutants binned by base stem length. 1–6 nt (gray, n=63 targets), 7–8 nt (red, n=40 targets), 9–10 nt (blue, n=44 targets), and 11–20 nt (yellow, n=50 targets) bins are shown. Medians, and p-values (unpaired t-test, two-tailed) are indicated.
Stem Length and Stability Affect mRNA Modification Rate
Despite overall structural similarity, Pus1 mRNA substrates vary in stem length, shape, stability, and composition (Figure 3b, Supplementary Figure 3a). Given this heterogeneity, we designed additional structure-perturbing mutants to determine which stem loop features distinguish good from poor substrates. First, we tested the importance of loop length, which ranges from 3 to 11 nts with 3–4 nt loops the most common (Supplementary Figure 5a). Overall, the effects of mutations perturbing loop length were mild (Supplementary Figures 5b–e). Slight decreases in v0,rel and the relative extent of pseudouridylation at early time points were observed when 4nt loops were extended to 8 nt (paired, two-tailed t-test, p=0.002 and p=0.007 respectively; Supplementary Figures 5c,e). This may reflect a preference of Pus1 for stems with shorter loops. However, it is possible that these loop extension mutants may adopt alternate folds in vitro.
Stems in Pus1 mRNA substrates are long compared to tRNA substrates, ranging in length from 6 to 18 bp (median 11 bp, Supplementary Figure 5f). However, these longer stems tend to have multiple segments interrupted by bulges, with shorter stem segments proximal to the first bulge (Supplementary Figure 5f–h). We hypothesized that bulges may provide conformational flexibility needed to accommodate these longer stems into the RNA binding channel, which is capped by a three α-helical bundle in human Pus119 that is predicted to be conserved in yeast (Figure 5d). To test this hypothesis, we extended the length of the base stems by inserting 2, 4, or 6 base pairs just before the first bulged nucleotides (Figure 5a). These extensions significantly reduced pseudouridylation at early time points (paired t-test, +2 p=0.02, +4 and +6 p<10−5), and a trend of decreasing v0,rel was observed with increasing length of the stem extension (Supplementary Figures 5i–j), although the changes in v0,rel did not meet significance thresholds.
We leveraged the stem extension mutants to further characterize and clarify the effects of base stem length. We recalculated v0,rel values for all extension mutants in the pool, normalizing to the maximum extent of pseudouridylation for each sequence (Supplementary Dataset 5). Binning sequences by base stem length revealed a clear pattern in which sequences with shorter base stems showed significantly higher v0,rel values (Figure 5e). Consistent with this trend, base stem length was globally anti-correlated with v0,rel (R=−0.261, p=0.0002, Supplementary Figure 5k). Taken together, these data suggest that a shorter base stem kinetically favors pseudouridylation, but that longer stems can still be accommodated. Interestingly, deletion of bulged nucleotides to lengthen the base stem had negligible effects on the v0,rel (Supplementary Figure 5l), suggesting a potentially more complex interaction between Pus1 and certain longer stems.
The inhibitory effect of longer stems (Figure 5e) suggested that Pus1 must bend or partially melt the substrate RNA duplex to accommodate the target uridine in the active site. We therefore examined the stability and composition of the stems and their relation to v0,rel. The G-C content of base pairs in the stems ranges from 8 to 75% (median 40%, Supplementary Figure 5m, and the stem motifs alone have predicted stabilities at 30°C of −2.54 to −20.25 kcal/mol (median −10.68 kcal/mol, Supplementary Figure 5n). Relative v0 was negatively correlated with the fraction of G-C base pairs in the distal portion of the stem (R=−0.304, p=0.03), consistent with the model that accommodation of these longer stems requires disruption, or partial disruption of the distal stems. Together, kinetic analysis of 60 wild type and 734 mutant substrates demonstrates a critical role for mRNA folding for pseudouridylation by Pus1.
The Pus1 Structural Motif is Sufficient for Modification
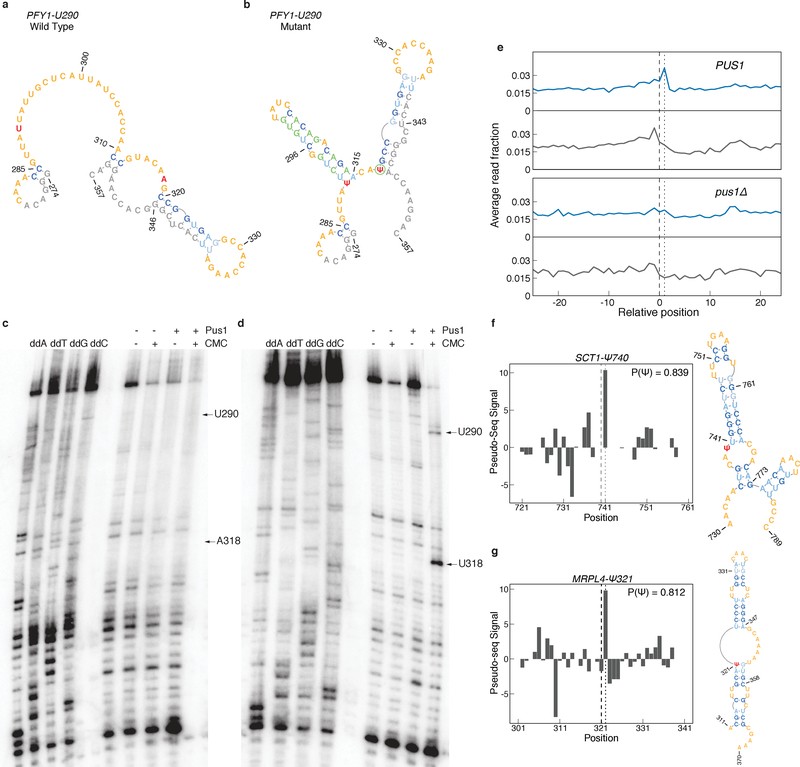
Our computational and biochemical analysis identified mRNA structural features associated with efficient pseudouridylation by yeast Pus1. To determine whether this structural motif is sufficient for Pus1 pseudouridylation, we attempted to rationally design a Pus1 substrate. Endogenous HRU motifs were identified in highly expressed transcripts, and were filtered for sequences in which the nucleotides downstream of and including the U were unpaired in the predicted MFE structures (Online Methods). We chose to manipulate the structural context of PFY1-U290 since PFY1-U86, is a Pus1 target in vivo3; thus the lack of pseudouridylation at U290 in vivo likely reflects an inability to modify rather than failure to access the mRNA (Supplementary Figures 6a–d).
The wild type PFY1-U290 sequence with adapter is predicted to be unpaired for 20 bases downstream of U290 (Figure 6a). We mutated several bases, so that U290 is predicted to be located at the 5′ base of a stem with the median characteristics of a Pus1 mRNA target. In the process of substrate design A318 was mutated to U to disfavor a competing structure. Fortuitously, these mutations positioned U318 in a context resembling typical Pus1 mRNA substrates (Figure 6b). As expected, there is no Pseudo-seq signal at A318 in vivo (Figure S6b,d)3. The wild type and engineered RNA substrates were incubated with recombinant Pus1 and analyzed by primer extension to detect pseudouridylation (Online Methods). Clear Pus1- and CMC-dependent RT stops were observed at both U290 and U318 in the mutant, but not wild type sequences (Figure 6c,d), demonstrating the sufficiency of the structural motif to direct mRNA pseudouridylation by yeast Pus1.
Figure 6: The Pus1 Structural Motif is Sufficient for Pseudouridylation.
a-b) MFE structures for a region surrounding (a) wild type, or (b) mutant PFY1-U290. Target Ψ nucleotides (red), adapter, padding, and T7 sequences (gray), and mutated nucleotides (green) are indicated. c-d) Primer extension gels of (c) PFY1-U290 wild type, and (d) PFY1-U290 mutant sequences. Positions of U290 and A/U318 are indicated. Gels are representative of n=4 replicates. Uncropped gel images can be found in Supplementary Figure 7a,b. e) Metaplot of RT stops for sites predicted to be Pus1 targets with P(Ψ) > 0.8, in +CMC libraries (blue) and −CMC libraries (grey) from high OD in vivo Pseudo-seq data. PUS1 panels show the aggregated reads from knockout libraries for pus2Δ,3Δ,4Δ,5Δ,6Δ,7Δ, and 9Δ. f-g) Pseudo-seq signal from the pooled PUS1 reads, and predicted secondary structure for a putative Pus1 target at (f) position SCT1-Ψ740 and (g) MRPL4-Ψ321.
Structural Context Prediction of New Pus1 In Vivo Targets
Due to the stringent read coverage and reproducibility thresholds we applied for pseudouridine detection in cells3, it is likely that many sites in lowly expressed genes were undetected. We therefore sought to use predicted RNA secondary structures to identify additional Pus1 targets from sub-threshold mRNAs in published in vivo Pseudo-seq data3. We first trained a random forest classifier over a high-confidence set of validated Pus1 target and non-target sites using a feature set informed by the in vitro pseudouridylation experiments and analysis (Online Methods). Features were parsed from the predicted MFE structure for each site (Supplementary Figures 6e–g). The classifier preferentially assigned a high probability of being a Pus1 target (P(Ψ)) to the validated Pus1 sites in the training set: 8 true positive sites had a P(Ψ)≥0.75, and 19 had P(Ψ)>0.50, compared to only 3 negative sites (Supplementary Figure 6e). We then applied the classifier to all ~1.1 million HRU motifs in the transcriptome and identified 603 putative Pus1 targets with a P(Ψ)>0.80 (Supplementary Dataset 6).
We examined in vivo Pseudo-seq signal across the 603 likely Pus1 targets to identify previously undetected Ψs. When examined in aggregate, we observed an increase in CMC-dependent signal in PUS1 libraries compared to pus1Δ libraries, suggesting the presence of sites modified by Pus1 (Figure 6e, Supplementary Figure 6h). Examining individual high P(Ψ) sites revealed 27 sites with peak heights ≥ 5.0, and 9 with peaks ≥ 10.0 (Supplementary Dataset 6). These include SCT1-Ψ740 and MRPL4-Ψ321, which occur in the expected structural context (Figure 6f,g). Most of the computationally predicted target HRU sites did not have sufficient read coverage to determine pseudouridylation status. Intriguingly, some highly expressed RNAs with P(Ψ)>0.80 did not appear to be pseudouridylated in vivo, suggesting that additional factors affect the structure or accessibility of these sites in cells.
Discussion
The mechanisms responsible for selective, regulated pseudouridylation of specific mRNAs are unknown for most pseudouridine synthases. Here we present a high-throughput in vitro approach to quantitatively assess pseudouridylation of thousands of sequences in parallel. Our oligo pool-based in vitro assay is a flexible technique that can be easily adapted to answer a variety of important questions about mRNA modifications. Here we have demonstrated three broadly applicable uses of the approach: identifying the enzymes responsible for mRNA modifications, validating (or invalidating) individual putative mRNA modifications, and dissecting the molecular basis for modification of specific sequences. This assay recapitulated mRNA Ψs genetically assigned to yeast Pus1 in vivo, and identified TRUB1, PUS7, PUS1, RPUSD2, and TRUB2 as the human PUS responsible for modifying mRNA sites without genetic data. Combining in silico RNA folding predictions, systematic substrate mutagenesis, and quantitative kinetic analysis of in vitro pseudouridylation revealed structural features critical for mRNA pseudouridylation by yeast Pus1. These results allowed rational engineering of a Pus1 substrate de novo, demonstrating the sufficiency of a defined structural motif to direct mRNA modification by Pus1. Finally, applying this structural context information allowed us to predict previously unidentified Pus1 mRNA targets that we showed were modified in vivo. Together, our results reveal a structure-dependent mode of target recognition by a conserved pseudouridine synthase, suggesting that mRNA pseudouridylation is likely to be regulated via modulation of RNA substrate structure.
Pus1 mRNA targets identified in vivo share only a short, degenerate sequence motif, HRU, suggesting a structural mode of substrate recognition3. In silico RNA folding revealed a structural motif shared by 90% of our validated Pus1 mRNA targets in which the Ψ is located at the 5′ base of a stem loop, which is supported by genome-wide RNA structure probing data16. The motif has discriminative value, similar structures are not present in sites not verified in vitro, or in mRNA Ψs genetically dependent on another PUS. We further showed that this motif has predictive power to identify new Pus1 targets from in vivo Pseudo-seq data, based on a classifier trained using in vitro-validated Pus1 targets.
We leveraged the throughput of our in vitro assay to systematically probe the importance of sequence and structural motifs associated with Pus1 mRNA targets and identified multiple features that contribute to pseudouridylation. We showed that the −1 purine is essential for pseudouridylation for most sequences, whereas the −2 pyrimidine kinetically promotes pseudouridylation, which could be important in cells when modification must compete with other processes. The presence of a stem loop 3′ of the target uridine is critical for mRNA pseudouridylation by Pus1, as demonstrated by disruptive and compensatory mutagenesis of the substrate stems. Although these stems are somewhat heterogeneous, most contain one or more internal bulged nucleotides. We speculate that these bulges are important for deformation of longer stems to allow positioning of the target uridine in the active site, as most mRNA substrate stems are longer than Pus1 tRNA substrate stems, and the RNA binding channel in hPus1 is capped by a three α-helical bundle that is conserved in yeast Pus119. Consistent with this hypothesis, increasing length of the stem prior to the first bulge and increasing stability of the distal region of the stem were negatively correlated with pseudouridylation. Finally, these results allowed engineering of new Pus1 substrates demonstrating that an HRU sequence motif located at the base of a bulged stem loop is sufficient for pseudouridylation by Pus1.
The critical requirement for mRNA structure for pseudouridylation by Pus1 in vitro strongly argues that its substrates must be structured at the time of modification in vivo. The RNA structure probing data analyzed here measures the intrinsic folding capacity of RNA sequences in vitro after extraction, but does not necessarily reflect mRNA structure in the cell at the time of lysis16. Indeed, mRNA structures are dynamic in vivo through the action of RNA binding proteins, RNA helicases, and cellular processes (e.g. translation). Consistent with the potential for many factors to affect mRNA target accessibility or structure in cells, v0,rel values were not predictive of Pseudo-seq signal observed in vivo. Given the extensive regulation of PUS1-dependent mRNA pseudouridylation, which is not explained by changes in PUS1 expression3, it will be interesting to correlate changes in modification with structural changes in the RNA substrate pool. A number of high-throughput RNA structure-probing assays have been successfully applied to living cells21. However, because the majority of mRNA molecules are cytoplasmic while Pus1 is predominantly nuclear22, it will be necessary to modify these methods to specifically interrogate the structures of nuclear mRNA. The results presented here, together with evidence that additional mRNA modifying PUS require structured RNA features to recognize their targets7,23, suggest that modulation of RNA structure may be a broadly significant mechanism for regulating mRNA pseudouridylation in cells.
Online Methods
Yeast Strains and Plasmids
All S. cerevisiae strains are BY4741 derivatives (BY4741: wild type (YWG506), pus1Δ (YWG1209). The pus1Δ strain was obtained from the Yeast Deletion Collection24. The pET8c-PUS1 construct was a gift of Yuri Motorin. N-terminally 6x-His-tagged full-length (29–427) and truncated (79–427) hPus1p expression plasmids were a gift of Robert Stroud. Human TRUB1, PUS7, PUS7L, TRUB2, RPUSD2 and PUS10 were cloned from human cDNA with Gibson assembly into the BamH1 site of pET15b expression vector.
S100 Extracts
S100 Extracts were prepared as previously described25. Briefly, yeast were grown to mid-log phase, and were lysed by beating with glass beads in Lysis Buffer (50 mM Tris pH 7.5, 100 mM KCl, 0.1 mM EDTA, 10 mM MgCl2, 10% glycerol, 10 mM β-mercaptoethanol, 1 mM PMSF, Mini Complete Protease Inhibitors (Roche)). Lysates were clarified by centrifugation, followed by a 20 min spin at 12,000 × g, and a 60 min spin at 100,000 × g.
scPus1 Purification
pET8c-PUS1-6HIS9 was transformed into E. coli BL21 (DE3) Gold cells [Agilent]. Cells were grown to OD600 0.6 in LB, and scPus1 was induced by addition of 0.5 mM IPTG. Cultures were supplemented with 20 μM zinc acetate at the time of induction. Cells were harvested by centrifugation, and lysed by sonication in Lysis Buffer (50 mM potassium phosphate pH 8.0, 200 mM NaCl, 30 mM Imidazole, 0.1% Triton X-100, 1 mM DTT, 0.5 mg/mL Chicken Egg Lysozyme, 1 mM PMSF, Mini Complete Protease Inhibitors (Roche). Recombinant protein was affinity purified by incubating clarified lysates with Ni-NTA Agarose (QIagen) at 4°C for 30 min with gentle stirring. The resin was washed extensively with Low Salt Buffer (50 mM potassium phosphate pH 8.0, 200 mM NaCl, 30 mM Imidazole, 1 mM DTT), followed by High Salt Buffer (50 mM potassium phosphate pH 8.0, 500 mM NaCl, 30 mM Imidazole, 1 mM DTT). Protein was eluted from the resin with Elution Buffer (50 mM potassium phosphate pH 8.0, 500 mM NaCl, 300 mM Imidazole, 1 mM DTT). The eluate from the Ni-NTA resin was desalted on a HiPrep 26/10 column (GE) into Anion Exchange Start Buffer (20 mM Tris pH 8.5, 100 mM NaCl, 1 mM MgCl2, 1 mM DTT, 10% Glycerol). Fractions containing Pus1 were pooled, loaded on a MonoQ 10/100 GL column (GE), and eluted on a linear gradient of 100 mM to 1 M NaCl. Fractions containing homogenous Pus1 (~300 mM NaCl) were pooled, concentrated, and loaded onto a HiLoad 16/60 Superdex 200 prep grade column (GE Healthcare), and were eluted with Anion Exchange Start Buffer. The S200 fractions containing homogenous recombinant Pus1 were pooled, concentrated, and assayed for activity using primer extension on a YAP1802-Ψ-117 substrate as described.
hPUS Purification
Recombinant hPus1 was purified as described19. Expression was induced in BL21 (DE3) Gold cells [Agilent] with 0.1 mM IPTG at OD600 0.6–0.8. Cells were grown overnight at 16°C, then harvested by centrifugation and resuspended in lysis buffer (50 mM HEPES-KOH pH 7.0, 500 mM NaCl, 5 mM β-mercaptoethanol, 1x Protease Inhibitor Cocktail (Roche)) and lysed by sonication. The lysate was centrifuged at 12,000 rpm for 30 min, bound on a HisTrap column (GE Healthcare) and eluted off the column with 250 mM imidazole. The protein was then dialyzed overnight at 4°C into storage buffer (50 mM HEPES-KOH pH 7.0, 100 mM NaCl, 1 mM β-mercaptoethanol) and further purified by gel filtration over a Superdex-200 column (GE Healthcare). The protein product was concentrated with a centrifugal filter unit (MD Millipore) and concentration determined by Bradford staining against a BSA standard. Human TRUB1, PUS7, PUS7L, TRUB2, RPUSD2 and PUS10 were purified as described below. Rosetta 2 BL21 (DE3) pLysS cells were transformed with the expression vector and an individual colony was grown at 37°C in LB to OD600 0.8. Induction of expression was overnight at 18°C with 1mM IPTG. Protein was affinity purified using the HisTrap HP 5mL column (GE) on an FPLC. Bound protein was washed with wash buffer (50mM potassium phosphate buffer pH 8, 0.5M NaCl, 30mM Imidazole) and then eluted with elution buffer (50mM potassium phosphate buffer pH 8, 0.5M NaCl, 300mM Imidazole). Protein was concentrated with Amicon Ultra Centrifugal Filter Units and stored in storage buffer containing 20mM HEPES (pH 7.5), 200mM NaCl, 10% glycerol, and 1mM DTT. Concentration was determined by Bradford with BSA standards.
Ψ-Site Structure Prediction and Visualization
The structures associated with 60 nts of sequence surrounding Pus1-dependent Ψs (10 nt downstream, Ψ at position 11, and 49 nt upstream) were predicted using the Perl interface for the Vienna RNAfold algorithm15. The pairing probability matrices were obtained using the RNA::pf_fold(), and RNA::get_pr() commands. Bracket notations were obtained using RNAfold with the parameters “-T 30 --noPS”. RNA folds based on bracket notations were visualized using PseudoViewer326, and heat maps of pairing probability matrices were visualized using custom python scripts.
Oligo Pool Design
For yeast pool 1 containing yeast mRNA Ψ’s, 65 nt upstream, and 64 nt downstream of an mRNA Ψ were used. For yeast pool 2 containing wild type yeast Pus1-dependent Ψ’s, sequences, 55 nt upstream, and 64 nt downstream of an mRNA Ψ site were used. Structure perturbing mutants were designed based on the minimum-free energy structure obtained above. Custom python scripts that use the forgi package 27 were used to parse the bracket notations, and to automate the design of mutations. Loop length extensions were made by repeating x bases of the loop sequence, where x is the length of the loop extension. Loop truncations were made by deleting nts from the 3′ end of the loop. Weak stem disrupting and compensatory mutants were made by randomly selecting 25% of base pairs (rounding up), and the strong mutations were made by randomly selecting an additional 25% of base pairs. The base pair +1 to the Ψ was excluded from mutation to minimize effects of library capture bias. Stem disrupting mutants were made by changing bases in the 3′ edge of the stem, and compensatory mutations were made by changing the bases in the 5′ edge of the stem, such that the base pair in the wild type sequence is flipped. Stem extension mutants were made by adding base pairs to the sequence immediately before the first bulged nucleotides in the stem. Base pair composition was matched to that of the base stem. Bulge deletions were made by deleting bulged nucleotides. Sequences containing 65 nt upstream, and 64 nt downstream were used for novel Ψs in human transcripts previously identified3,4, as well as sites genetically assigned to hPus16.
Sequences were prepended with the T7 promoter, and appended with a 10-nucleotide barcode unique to each sequence (yeast pools only) and an adapter sequence (Supplementary Table 1).. All barcodes appended to a given wild type sequence and its mutants had a minimum hamming distance of seven, for barcodes appended to unrelated sequences, the minimum hamming distance was two.
RNA Pool Preparation
Yeast and human oligo pools (Custom Array, Twist Bioscience), were amplified by PCR with primers oBZ131 and oTC_pool_rev or oTC_pool2_rev (Supplementary Table 2) using Phusion Polymerase (NEB). PCR reactions were supplemented with 5M Betaine (Sigma) to a final concentration on 1M. PCR reactions were gel purified. RNA was prepared by in vitro transcription with the MEGAshortscript T7 Transcription Kit (Thermo Fisher), and full length in vitro transcription products were gel purified.
Pus1 Substrate Design
Highly expressed genes were scanned for HRU motifs, and structures associated with these motifs were determined as described above. Sites were then filtered for those with at least 15 predicted unpaired bases downstream of the U. Genes with Pus1-dependent Ψs at other positions were favored, since Pus1 can access these transcripts in vivo. Bases were then mutated to favor the formation of a stem loop. These sequences were prepended with the T7 promoter sequence, and appended with a 3′ adapter sequence (Supplementary Table 2). These sequences were ordered as Ultramers (IDT), and RNA was prepared as described for oligo pools.
In Vitro Pseudouridylation
For all reactions, 15 pmol of in vitro transcribed pool RNA was used as a substrate. Prior to pseudouridylation, RNA was denatured in H2O at 75°C for 2 min, and then cooled on ice for 1 min. RNA was folded at 37°C for 20 min following addition of 5X Pseudouridylation Buffer (500 mM Tris pH 8.0, 500 mM Ammonium Acetate, 25 mM MgCl2, 0.5 mM EDTA) to 1X for the final reaction volume. Pseudouridylation reactions were carried out at 30°C after addition of DTT to 2 mM, and a Pus activity source. Reactions were stopped by snap freezing. For S100 extracts, 2.5 μL of extract were used for a 50 μL reaction, followed by incubation at 30°C for 1 hr. Pools were pseudouridylated with recombinant yeast Pus1 at a final concentration of 600 nM in 500 μL reaction volumes. 150 ng of designed Pus1 substrates were incubated with 300 nM Pus1 for 30 min in 50 μL reaction volumes. Human pool RNAs were in vitro transcribed and 30 pmol of RNA was pseudouridylated as described above with recombinant human PUS1, TRUB1, TRUB2, RPUSD2, PUS7, PUS7L or PUS10 at a final concentration of 600nM by incubating for 45 minutes at 30°C.
Pseudo-seq Library Preparation
Libraries were prepared essentially as described in with the indicated changes28. Briefly, pool RNA was isolated from pseudouridylation reactions by extraction with acid phenol, followed by isopropanol precipitation. CMC modification (0.4 M final) and reversal were carried out as described. Mock (−CMC) reactions were carried out for samples pseudouridylated with S100 extracts, but were omitted for samples pseudouridylated with recombinant scPus1. Reverse transcription was carried out as described with the primer oTC_RT-L2_3′10N (yeast pools) or oTC_RT-L2 or ONM_RT-L2 (human pools). Truncated cDNAs from 120–190 nts (yeast pools) and 140–170 nts (human pools) were gel purified. cDNAs were circularized for 6 hours as described. Libraries were PCR amplified with primers RP1 and a unique barcode primer (BC) for each PUS incubation, gel purified, and sequenced in paired-end mode on an Illumina HiSeq or Illumina MiSeq. All p rimer sequences are contained in Supplementary Table 2.
Sequencing Data Analysis
Sequencing data was analyzed with in house Bash and Python scripts. To allow mapping of reads with short inserts, primer sequences associated with 5′ end of the amplicon were trimmed from the reverse read using cutadapt (-G parameter)29. Trimmed paired-end reads were then merged with PEAR (default settings)30. PCR duplicates were then collapsed using fastx_collapser (http://hannonlab.cshl.edu/fastx_toolkit/), and collapsed reads were trimmed of 10 nts on the 3′ end, and of 3′ adapter sequence using cutadapt (yeast pools only). Processed reads were mapped to a bowtie index of pool sequences using tophat231. Multiply mapping reads were excluded from analysis using SAMtools, and the resulting mapped reads were processed with in house Python scripts.
Pseudo-seq signal was as follows. For each position in a 51 nt window centered at a given Ψ, the fraction of reads in the window whose 5′ ends map to said position was calculated. Pseudo-seq signal is the difference in fractional reads between the +CMC and −CMC libraries, scaled by the window size. According to the equations below, where NR+ and NR− refer to the normalized reads at the RT stop +CMC and −CMC respectively.
For a Ψ, the reported pseudo-seq signal corresponds to the expected RT stop position 1 nt 3′ of the Ψ.
For libraries prepared from pools pseudouridylated with recombinant yeast Pus1, signal was calculated as the fraction of reads whose 5′ ends map to the expected stop position in the 1–91 nt region of each sequence.
Identification of High Confidence PUS Targets
The set of high confidence yeast Pus1 mRNA targets was defined as those sites genetically dependent upon PUS1 in vivo, and which were pseudouridylated in Pus1-dependent manner by S100 extract, or by recombinant yeast Pus1. For these sites, we first required that the average pseudo-seq signal from wild type extracts be ≥ 2.0, and ≤ 0.5 for pus1Δ extracts (51 sites). For sites with a wild type peak value in the range 1.0–2.0, we required a fold change in peak values of ≥ 5.0 between wild type and pus1Δ extracts (3 sites). We manually examined the remainder of sites for Pus1-dependent modification in S100 extract (2 sites), and for modification by recombinant S. cerevisiae Pus1 (3 sites).
Assignment of human PUS to pseudouridylated targets was carried out by using the Grubbs outlier test with significance level alpha set to 0.05 to identify sites that had peak height values that deviated from the normal distribution of peak heights for all the other conditions (all other PUS and no PUS). A target site was assigned to a PUS if it was called as an outlier exclusively in the corresponding PUS sample and had a peak height greater than > 1.0.
Primer Extension
RNA was isolated from pseudouridylation reactions as described above for pool RNA, and CMC treatment and reversal was carried out as described, except samples were both treated with CMC and mock treated. oTC_pool_rev was radiolabeled with γ-ATP (Perkin-Elmer) by treatment T4 PNK (NEB). Primers extension was carried out with AMV RT (Promega). Briefly, primers were annealed in 1X AMV RT buffer by incubation for 5 min 65°C, followed by 5 min on ice, and 5 min at room temperature. Reverse transcription was carried out at 42°C for 30 min. Reactions were quenched with 2X Stop Solution (0.5X TBE, 90% Formamide, 0.05% w/v Bromophenol Blue, 0.05% w/v Xylene Cyanol). Reactions were then run on 10% TBE-Urea sequencing gel.
Pus1 Target Prediction
A random forest classifier was trained over a high-confidence set of Pus1 targets and non-target sites. The true positives in the training set were the yeast Pus1 sites validated in vitro in this paper. The set of true negative sites consisted of yeast transcriptome sites that met the three following conditions: (1) the sites contained the HRU sequence motif, (2) were in a 50-nucleotide window with at least 120 read 5′ ends mapped in 14/16 libraries from in vivo Pseudo-seq profiling3, and (3) consistently had a Pseudo-seq peak height < 0.6 in all 16 libraries. The finalized training set consisted of 49 true positive sites and 404 true negative sites. Using the training set, the random forest approach generates multiple decision trees, each of which uses a random subset of the features to classify each site as a target or non-target. The classifier combines the outputs of all the decision trees to assign each site a probability, P(Ψ), that it is or is not a Pus1 target. The classifier was applied to all yeast mRNA sites that contained the HRU sequence motif.
For all sites, we obtained the sequence starting 10-nt upstream of the position of interest, and ending 50-nt downstream. Secondary structure predictions were obtained using RNAfold, with -T 30. All features were parsed from the RNAfold output, and included the following: the AU content of the stem, the GC content of the stem, the relative position of the 5′ end of the base stem, the length of the loop, the ΔG of the ensemble, the ΔG of the minimum free energy structure, the length and position of the 3′ bulge, the length and position of the 5′ bulge, the length of the base stem, and the relative position of the 3′ end of the base stem.
Training and applying the random forests was carried out using functions from the randomForest R package (https://www.stat.berkeley.edu/~breiman/RandomForests/) We chose our model parameters by comparing different parameter combinations. Specifically, we tested mtry values between 2 and 10 (number of features to use in each tree) and maxnodes between 5 and 30 (maximum number of nodes in each tree), to find a combination that minimized the out-of-bag error rate. Ultimately, we chose to train 10 random forests, using parameters mtry=4 and maxnodes=17. Each of these forests was used to predict P(Ψ) for each HRU site, and we use the median P(Ψ) for all subsequent analyses. The median, as well as the 10th and 90thpercentiles, are reported (Supplementary Dataset 6).
To examine modification of putative Pus1 targets, Pseudo-seq libraries were merged based on growth states, CMC-treatment, and whether they were prepared from PUS1 or pus1Δ cells. From these merged libraries metaplots of normalized, aggregated RT stops were made, and the fraction of reads at the expected RT stop positions 1 nt 3′ of putative Ψ sites in PUS1 merged libraries and pus1Δ merged libraries were compared.
Other Analyses
Motifs were generated using WebLogo 3.5 using default settings, and the modified position was changed to a Ψ after logo generation32. PARS data was downloaded, the PARS score for each position was calculated as described, and the average PARS score was calculated for the indicated sets of mRNA Ψs16. For pairwise correlations between Pus1 structures, Pearson R values were calculated for either the maximum pairing probability for each position in a sequence, or the sum of all pairing probabilities for each position in a sequence. Rows and columns were ordered by the sum of R values across the row/column. Yeast Pus1 was modelled onto hPus1 (4IQM) using SWISS-MODEL19,33.
Relative v0 (v0,rel) values were calculated as follows. First, the background fraction of reads for each sequence, calculated as the average of two 0 min replicates, was subtracted from all time points, and resulting negative values were set to 0.0. Then, values were normalized to the highest signal obtained across the time course for the wild type sequence. Linear regression was used to obtain the initial velocity (slope) for both 0–30 sec, and 0–45 sec time points. The slope for the fit with the better R value was used. For the analysis shown in Figure 6a, v0 values were calculated as above, except normalization to the highest values obtained for each sequence, wild type or mutant.
Data Availability
Yeast strains and plasmids are available upon request. All sequencing data and oligo pool sequences have been deposited in GEO, accession GSE99487.
Code Availability
Custom bash and python code used for analysis is available on request.
Supplementary Material
Acknowledgments
Acknowledgements
We thank Yuri Motorin (Université de Lorraine) for the yeast Pus1 expression plasmid, and Robert Stroud (University of California, San Francisco) for human Pus1 expression plasmids. We also thank Gina Mawla and Caitlyn Mason for technical assistance and members of the Gilbert lab for critical reading of the manuscript. This work was supported by NIH (GM101316) (W.V.G.), American Cancer Society (RSG-13-396-01-RMC) (W.V.G.), American Cancer Society Post-doctoral Fellowship (T.M.C.), Jane Coffin Childs Post-doctoral Fellowship (N.M.M.), NSF Graduate Research Fellowship (C.S.), and NIH Pre-Doctoral Training grant (T32GM007287) (T.A.B.).
Footnotes
The authors declare no competing financial interests.
References
- 1.Boccaletto P et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. (2018). doi: 10.1093/nar/gkx1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert WV, Bell TA & Schaening C Messenger RNA modifications: Form, distribution, and function. Science (80-. ). 352, 1408–1412 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlile TM et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwartz S et al. Transcriptome-wide Mapping Reveals Widespread Dynamic-Regulated Pseudouridylation of ncRNA and mRNA. Cell 159, 148–62 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lovejoy AF, Riordan DP & Brown PO Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One 9, e110799 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol 11, 592–7 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Safra M, Nir R, Farouq D, Slutzkin IV & Schwartz S TRUB1 is the predominant pseudouridine synthase acting on mammalian mRNA via a predictable and conserved code. Genome Res. (2017). doi: 10.1101/gr.207613.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamma T & Ferre-D’Amare AR Pseudouridine synthases. Chem Biol 13, 1125–1135 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Motorin Y et al. The yeast tRNA:pseudouridine synthase Pus1p displays a multisite substrate specificity. RNA 4, 856–69 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Behm-Ansmant I et al. A previously unidentified activity of yeast and mouse RNA:pseudouridine synthases 1 (Pus1p) on tRNAs. RNA (2006). doi: 10.1261/rna.100806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massenet S et al. Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase Pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol. Cell. Biol 19, 2142–2154 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basak A & Query CC A pseudouridine residue in the spliceosome core is part of the filamentous growth program in yeast. Cell Rep. (2014). doi: 10.1016/j.celrep.2014.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bakin A & Ofengand J Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry 32, 9754–62 (1993). [DOI] [PubMed] [Google Scholar]
- 14.Carlile TM, Rojas-Duran MF & Gilbert WV Pseudo-Seq: Genome-Wide Detection of Pseudouridine Modifications in RNA. Methods Enzymol. 560, 219–245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorenz R et al. ViennaRNA Package 2.0. Algorithms Mol. Biol (2011). doi: 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kertesz M et al. Genome-wide measurement of RNA secondary structure in yeast. Nature (2010). doi: 10.1038/nature09322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taoka M et al. The complete chemical structure of Saccharomyces cerevisiae rRNA: Partial pseudouridylation of U2345 in 25S rRNA by snoRNA snR9. Nucleic Acids Res. (2016). doi: 10.1093/nar/gkw564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia T et al. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry 37, 14719–35 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Czudnochowski N, Wang AL, Finer-Moore J & Stroud RM In human pseudouridine synthase 1 (hPus1), a C-terminal helical insert blocks tRNA from binding in the same orientation as in the Pus1 bacterial homologue TruA, consistent with their different target selectivities. J. Mol. Biol (2013). doi: 10.1016/j.jmb.2013.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz S et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155, 1409–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwok CK, Tang Y, Assmann SM & Bevilacqua PC The RNA structurome: Transcriptome-wide structure probing with next-generation sequencing. Trends in Biochemical Sciences 40, 221–232 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Breker M, Gymrek M, Moldavski O & Schuldiner M LoQAtE - Localization and Quantitation ATlas of the yeast proteomE. A new tool for multiparametric dissection of single-protein behavior in response to biological perturbations in yeast. Nucleic Acids Res. (2014). doi: 10.1093/nar/gkt933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Urban A, Behm-Ansmant I, Branlant C, Motorin Y & Motorlin Y RNA sequence and two-dimensional structure features required for efficient substrate modification by the Saccharomyces cerevisiae RNA:Ψ-synthase Pus7p. J. Biol. Chem 284, 5845–58 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Winzeler EA et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science (80-. ). 285, 901–6 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Jiang HQ, Motorin Y, Jin YX & Grosjean H Pleiotropic effects of intron removal on base modification pattern of yeast tRNA(Phe): An in vitro study. Nucleic Acids Res. (1997). doi: 10.1093/nar/25.14.2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Byun Y & Han K PseudoViewer3: Generating planar drawings of large-scale RNA structures with pseudoknots. Bioinformatics (2009). doi: 10.1093/bioinformatics/btp252 [DOI] [PubMed] [Google Scholar]
- 27.Kerpedjiev P, Höner Zu Siederdissen C & Hofacker IL Predicting RNA 3D structure using a coarse-grain helix-centered model. RNA (2015). doi: 10.1261/rna.047522.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlile TM, Rojas-Duran MF & Gilbert WV Transcriptome-Wide Identification of Pseudouridine Modifications Using Pseudo-seq. Curr. Protoc. Mol. Biol 112, 4.25.1–4.25.24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal (2011). doi: 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 30.Zhang J, Kobert K, Flouri T & Stamatakis A PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics (2014). doi: 10.1093/bioinformatics/btt593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim D et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. (2013). doi: 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crooks GE, Hon G, Chandonia JM & Brenner SE WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waterhouse A et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. (2018). doi: 10.1093/nar/gky427 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Yeast strains and plasmids are available upon request. All sequencing data and oligo pool sequences have been deposited in GEO, accession GSE99487.