Abstract
To develop an antibody (Ab) therapeutic against Staphylococcal Enterotoxin B (SEB), a potential incapacitating bioterrorism agent and a major cause of food poisoning, we developed a “Class T” anti-SEB neutralizing Ab (GC132) targeting an epitope on SEB distinct from that of previously developed “Class M” Abs. A systematic engineering approach was applied to affinity mature Ab GC132 to yield an optimized therapeutic candidate (GC132a) with sub-nanomolar binding affinity. Mapping of the binding interface by hydrogen deuterium exchange coupled to mass spectrometry (HDX-MS) revealed that the Class T epitope on SEB overlapped with the T-cell receptor (TCR) binding site, whereas other evidence suggested that the Class M epitope overlapped with the binding site for the major histocompatibility complex. In the IgG format, GC132a showed ~50-fold more potent toxin-neutralizing efficacy than the best Class M Ab in vitro, and fully protected mice from lethal challenge in a toxic shock post-exposure model. We also engineered bispecific Abs (bsAbs) that bound tetravalently by utilizing two Class M binding sites and two Class T binding sites. The bsAbs displayed enhanced toxin neutralization efficacy compared with the respective monospecific Ab subunits as well as a mixture of the two, indicating that enhanced efficacy was due to heterotypic tetravalent binding to two non-overlapping epitopes on SEB. Together, these results suggest that Class T anti-SEB Ab GC132a is an excellent candidate for clinical development and for bispecific Ab engineering.
Keywords: synthetic antibody, phage display, SEB toxin, protein engineering, hydrogen-deuterium exchange coupled to mass spectrometry
Introduction
Staphylococcus aureus is a pathogenic commensal gram-positive bacterium that has evolved to be resistant to many antibiotics and has become a global public health threat. [1] S. aureus is involved in an array of diverse human pathologies ranging from relatively mild skin infections to severe, sometimes lethal, sepsis, pneumonia, endocarditis, and toxic shock syndrome. [2] Staphylococcal superantigens (SAgs), together with a wide array of cell-surface-associated components, extracellular enzymes and cytolytic toxins, contribute to the pathology of S. aureus disease. [3] SAgs are a large family of secreted exotoxins that are produced primarily by S. aureus and group A streptococci, consisting of staphylococcal toxic shock syndrome toxin 1 (TSST-1), staphylococcal enterotoxins (SEs), and the streptococcal pyogenic exotoxins. SAgs are among the most potent virulence factors that contribute to fatal bacterial infections through immune evasion, and more than 20 have been identified. In addition, SAgs may also play a role in autoimmune disorders as well as some other abnormal immunologic states. [4] SAgs mediate their pathological effects by forming a bridge between certain subsets of T-cell receptor (TCR) β chains on T lymphocytes and class II major histocompatibility complex (MHC-II) molecules of antigen-presenting cells outside of the conventional peptide-binding groove without the requirement of prior antigen processing. This peptide-independent cross-linking leads to indiscriminate stimulation of T-cells and consequent massive production of proinflammatory cytokines, such as interferon-gamma (INFγ) and tumor necrosis factor alpha (TNFα), which in turn leads to toxic shock syndrome (TSS) that is characterized by high fever, rash and hypertension, and can rapidly progress to multiple organ failure and death. [2, 4] Besides TSS and multiorgan failure [5], staphylococcal SAgs have been specifically implicated in the pathogenesis of sepsis [6], infective endocarditis [7–9], acute kidney injury [7], atopic dermatitis [10], and Kawasaki’s disease [11].
Among SAgs, staphylococcal enterotoxin B (SEB) is the prototypical enterotoxin. [12] SEB is the primary cause of food poisoning and the major enterotoxin associated with non-menstrual TSS not caused by TSST-1. [13, 14] There have also been reports of rare incidences of laboratory-acquired SEB intoxications. [15] Moreover, SEB is one of the two most important toxin threats in bioterrorism, and in the 1960s, was extensively investigated as an incapacitating agent in the US biological warfare program. [16] SEB is attractive as a biological weapon because it is highly toxic and stable, easy to produce in large quantities and to disperse in various forms, difficult to diagnose, and has no effective treatment or vaccine. [17] Due to its potential as a bioweapon or bioterrorist agent, SEB is recognized by the National Institute for Allergy and Infectious Diseases as one of the Category B priority pathogens that pose the second highest risk to national security and public health. Therefore, there is an urgent need for countermeasures of SEB intoxication or SEB-induced TSS following accidental or malicious exposure.
However, there are currently no approved preventative measures or therapies for SEB exposure. Attempts to develop therapies for SEB with varying degrees of success have been reported using peptide antagonists [18–20], synthetic chimeric mimics of MHC-II/TCR complexes [21–23] and engineered TCRs [24]. In particular, the most recent development that utilized engineered picomolar-affinity TCRs to block SEB action in vitro showed promising results when tested in vivo in a rabbit model. [24] Although the reported TCR variant had a short half-life of several hours in rabbits, this study demonstrated the value of blocking the TCR-binding site and the necessity for extremely potent neutralizing agents, since SEB is highly toxic even at extremely low concentrations. Some promising results have also been obtained for active immunization to induce anti-SEB antibodies in the human body using synthetic peptides [25], SEB toxoid or recombinant SEB vaccines [26–30]. We recently completed a Phase I clinical trial on a recombinant SEB vaccine (STEBVax), which demonstrated safety and immunogenicity. [31]
Besides preventive measures for SEB exposure, passive immunotherapy with engineered antibodies (Abs) has garnered much interest due to several desirable characteristics of Ab drugs, including long serum half-lives, high potency, and low off-target toxicity. In addition to hyperimmune intravenous immunoglobulin [32, 33], Abs targeting SEB have been developed to combat SEB intoxications using traditional hybridoma methods [34–38] and in vitro display technologies [39, 40]. At the molecular level, these antagonists act by preventing or disrupting the formation of the ternary TCR/SEB/MHC-II complex. The crystal structures of SEB/MHC-II, SEB/TCR and the ternary TCR/SEB/MHC-II complexes have been reported [41–43]. Structural analysis indicates that the MHC-II and TCR binding sites on SEB are spatially distinct, suggesting that effective SEB neutralization could be achieved by targeting either or both of these epitopes.
We previously generated a class of high-affinity human Abs against SEB by using phage display technology. [40] We have shown that these “Class M” Abs are capable of neutralizing SEB in vitro and can completely protect mice from lethal SEB challenge. Here, we report a second class of anti-SEB neutralizing Abs that target an epitope distinct from that of Class M Abs. “Class T” anti-SEB Ab GC132 was derived from the same phage-displayed synthetic human Ab library that was used to produce the Class M Abs. Through systematic engineering, Ab GC132 was further optimized to achieve sub-nanomolar binding affinity for SEB. Optimized Class T Abs neutralized SEB and blocked toxin-mediated activity in vitro, and the best Class T Ab in the IgG format (IgG-GC132a) showed complete protection in a mouse toxin shock post-exposure model. Moreover, considering SEB requires the formation of a ternary complex with MHC-II and TCR to induce host immune response, we hypothesized more effective toxin neutralization may be achieved if interactions of SEB with MHC-II and TCR were blocked simultaneously. Thus, we engineered bispecific Abs (bsAbs) that could interact simultaneously with the two non-overlapping epitopes on SEB by combining the most potent Class M IgG with single-chain variable fragments (scFvs) of Class T Abs in a single tetravalent entity. These bsAbs showed improved in vitro toxin neutralizing efficacy. IgG-GC132a and the engineered bsAbs represent ideal candidates for development of Ab therapeutics for prophylactic and post-exposure treatment of SEB-induced disease and lethality.
Results and Discussion
Isolation and Characterization of Class T anti-SEB Ab GC132
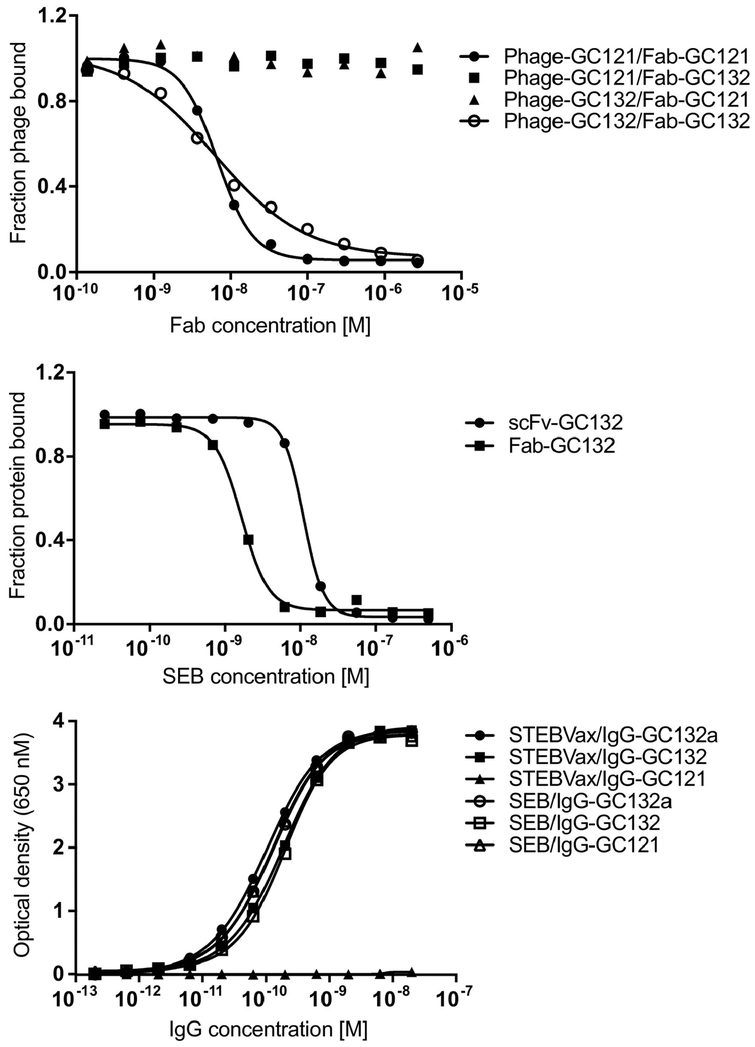
We previously engineered Class M neutralizing Abs for SEB, of which IgG-GC121 was the most potent and showed efficient toxin neutralization and protection in mouse models. [40] IgG-GC121 bound SEB with sub-nanomolar affinity but failed to bind to STEBVax (see below), a recombinant form of SEB containing three substitutions (L45R/Y89A/Y94A) that disrupt the interaction of the toxin with human MHC-II and render the protein non-toxic while retaining immunogenicity [28]. To develop another class of neutralizing Abs for SEB that bind to a different epitope, we performed a selection with a phage-displayed synthetic antigen-binding fragment (Fab) library [44] in the presence of excess Fab-GC121 to block the Class M epitope and bias the selection for binding to other non-overlapping epitopes. After four rounds of binding selections, we identified Class T Ab GC132 (Table 1), which bound to SEB and was not blocked by Ab GC121 (Figure 1a). Purified Fab-GC132 exhibited an IC50 value of 1.6 nM for SEB by competitive enzyme-linked immunosorbent assay (ELISA) (Figure 1b) and neutralized SEB in vitro, but with lower efficacy than Ab GC121 (data not shown). Moreover, IgG-GC132 bound strongly to both SEB and STEBVax by direct ELISA, whereas IgG-GC121 bound to SEB but not to STEBVax (Figure 1c). Taken together, these results show that Class M Ab GC121 binds to an epitope that likely overlaps with the MHC-II binding site and neutralizes SEB toxicity by preventing interaction with the human MHC-II complex, whereas Class T Ab GC132 was able to neutralize SEB toxicity by binding to an epitope that is distinct from that of the Class M Ab and likely does not overlap with the MHC-II binding site.
Table 1.
CDR sequences of Class T anti-SEB Abs
| ID | CDR-L3 | CDR-H1 | CDR-H2 | CDR-H3 | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 107 | 108 | 114 | 115 | 116 | 29 | 30 | 35 | 36 | 37 | 38 | 39 | 55 | 56 | 57 | 58 | 59 | 62 | 63 | 64 | 65 | 66 | 107 | 108 | 109 | 110 | 111 | 111.1 | 112.2 | 112.1 | 112 | 113 | 114 | 115 | |
| GC132 | S | W | W | P | I | N | L | S | Y | S | Y | I | S | I | Y | S | S | S | S | Y | T | S | S | Y | Y | A | G | S | Y | F | H | Y | G | M |
| Libra ry |
X | . | . | . | X | D |
FI L |
AGS T |
AGS T |
YSA D |
AGSD YW |
I M |
. | . | . | . | . | . | . | . | . | . | . | . | . | S | X | X | . | . | . | . | . | X |
| GC132 a |
R | . | . | . | P | D | . | G | A | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | S | L | L | . | . | . | . | . | W |
| GC132 b |
K | . | . | . | L | D | . | G | G | . | . | M | . | . | . | . | . | . | . | . | . | . | . | . | . | S | A | H | . | . | . | . | . | L |
| GC132 c |
K | . | . | . | V | D | F | A | G | . | . | M | . | . | . | . | . | . | . | . | . | . | . | . | . | S | A | A | . | . | . | . | . | F |
| GC132 d |
R | . | . | . | V | D | F | G | G | A | . | M | . | . | . | . | . | . | . | . | . | . | . | . | . | S | F | A | . | . | . | . | . | W |
| GC132 e |
G | . | . | . | C | D | I | T | S | A | . | M | . | . | . | . | . | . | . | . | . | . | . | . | . | S | E | W | . | . | . | . | . | W |
| GC132 f |
D | . | . | . | E | D | . | A | S | . | . | M | . | . | . | . | . | . | . | . | . | . | . | . | . | S | T | G | . | . | . | . | . | W |
CDR sequences are numbered according to IMGT numbering scheme [56]. Residues that are identical to the parental clone are shown in dots. The design of the affinity maturation library is shown below the GC132 sequence, with diversified positions shown in bold italics. At each diversified position, allowed amino acids are denoted by the single-letter code. X denotes a mixture of 20 amino acids encoded by the degenerate codon NNK (N = A/G/C/T, K = G/T). Compared with the GC132, the library design also encoded to fixed substitutions in the heavy chain (N29D/A110S). CDR-L1 and CDR-L2 are not shown, because they were not diversified.
Fig. 1. Phage and protein ELISAs for anti-SEB Abs.
(a) Competitive Fab-phage ELISAs to assess blocking of Fab-phage binding to immobilized SEB by Fab proteins. The binding of the indicated phage-displayed Fab (y-axis) was assessed in the presence of serial dilutions of the indicated solution-phase Fab protein (x-axis). Curves were normalized to binding in the absence of solution-phase Fab. (b) Competitive ELISAs for assessing inhibition ofFab-GC132 (IC50 = 1.6 nM) or scFv-GC132 (IC50 = 11 nM) binding to immobilized SEB (y-axis) by serial dilutions of solution-phase SEB (x-axis). IC50 was defined as the concentration of SEB that blocked 50% of Ab binding to immobilized SEB. Curves were normalized to binding in the absence of solution-phase SEB. (c) ELISAs for assessment of serial dilutions of the indicated IgG protein (x-axis) binding to immobilized SEB or STEBVax (y-axis).
Shotgun homolog scanning of Ab GC132
We displayed GC132 on phage in the scFv format and constructed two homolog scanning libraries that covered 17 or 28 complementarity-determining region (CDR) residues in the light or heavy chain, respectively. In shotgun homolog scanning libraries, each scanned position is represented by a binary codon that encodes the wild-type (wt) and a homologous amino acid [45]. Each library was subjected to a display selection with an anti-FLAG Ab to capture displayed protein by virtue of a FLAG epitope fused to the N-terminus of the scFv, and separately, to an antigen selection to capture functional scFvs capable of binding to SEB. As shown previously, dividing the wt/mutant ratio from the antigen selection by the wt/mutant ratio from the display selection generates a function ratio (Fwt/mut) that provides a quantitative assessment of the effect of each mutation on antigen binding, with ratios greater or less than one indicating mutations that are deleterious or beneficial relative to the wt, respectively (Table 2). Thus, the homolog scan provides a comprehensive map of how CDR side chains contribute to the formation of a functional antigen-binding site. A few residues in each of the three light chain CDRs exhibited Fwt/mut values greater than 5, whereas in contrast, all five residues in the heavy chain with Fwt/mut greater than 5 were located in a continuous stretch of CDR-H3, indicating that these residues are important for antigen recognition.
Table 2.
Shotgun homolog scan of scFv-GC132
| Residuea | wt/mut ratiosb | Fwt/mutc | Residue | wt/mut ratios | Fwt/mut | ||
|---|---|---|---|---|---|---|---|
| Antigen selection |
Display se1ection |
Antigen se1ection |
Disp1ay se1ection |
||||
| 1S2 8 | 1 | 1.5 | 0.67 | hL30* | 2.6 | 1.4 | 1.8 |
| 1V2 9 | 3.4 | 1.8 | 1.9 | hS35* | 0.49 | 0.35 | 1.4 |
| 1S36 | 21 | 1.1 | 19 | hY36* | 3.6 | 5.6 | 0.64 |
| 1S37 | 5.3 | 3.5 | 1.5 | hS37* | 0.42 | 1.5 | 0.27 |
| 1A38 | >44 | 0.54 | >81 | hY3 8* | 0.94 | 2 | 0.47 |
| 1S56 | 43 | 2.1 | 21 | hI39* | 0.39 | 0.69 | 0.57 |
| 1A57 | >44 | 0.67 | >65 | hS55 | >64 | 21 | >3 |
| 1S65 | 1.2 | 1.9 | 0.65 | hI56 | 3.3 | 3.1 | 1.0 |
| 1S66 | 3 | 1.6 | 1.9 | hY57 | 2.8 | 3.7 | 0.74 |
| 1L67 | 2.4 | 6 | 0.40 | hS58 | 3.3 | 2.9 | 1.1 |
| 1Y68 | 2.4 | 2.3 | 1.0 | hS59 | 2.8 | 1.9 | 1.5 |
| 1S69 | 0.91 | 1.6 | 0.58 | hS62 | 2.0 | 2.1 | 0.96 |
| 1S107* | 3 | 0.83 | 3.6 | hS63 | 1.4 | 1.4 | 1.0 |
| 1W108 | >44 | 3.8 | >11 | hY64 | 0.42 | 0.94 | 0.45 |
| 1W114 | >44 | 1.9 | >23 | hT65 | 0.94 | 0.89 | 1.1 |
| 1P115 | >44 | 3.5 | >12 | hS66 | 3.9 | 2 | 2.0 |
| 1I116* | 3.9 | 2.5 | 1.6 | hS107 | 2.6 | 4.1 | 0.63 |
| hY108 | 6.1 | 1.5 | 4.0 | ||||
| hY109 | 2.2 | 1.1 | 1.9 | ||||
| hA110 | 0.33 | 0.89 | 0.38 | ||||
| hG111* | 0.94 | 3.7 | 0.25 | ||||
| hS111.1* | 1.6 | 0.83 | 1.9 | ||||
| hY112.2 | 6.1 | 1.1 | 5.8 | ||||
| hF112.1 | 7 | 1 | 7 | ||||
| hH112 | >64 | 0.40 | >158 | ||||
| hY113 | >64 | 1.1 | >56 | ||||
| hG114 | >64 | 1.3 | >50 | ||||
| hM115* | 0.56 | 1.4 | 0.39 | ||||
A residue is denoted by a letter in lower case indicating the heavy or light chain (h or l, respectively), followed by the single letter amino acid code and the IMGT number of the position in the chain. Asterisks (*) indicated residues that were diversified in the affinity maturation library.
The wt/mut ratio for each residue was determined from the sequences of binding clones isolated after selection for binding to either SEB (antigen selection) or an anti-FLAG antibody (display selection).
The function ratio (Fwt/mut) was determined by dividing the wt/mut ratio from the antigen selection by the wt/mut ratio from the display selection. Residues with Fwt/mut >5 are shown in bold text.
Affinity maturation of Ab GC132
We used the data from the homolog scan to design a library for affinity maturation of Ab GC132. Given that Ab GC132 bound to SEB with reasonably high affinity, we reasoned that it would be most effective to diversify residues close to those identified as important by homolog scanning, with the goal of optimizing the periphery of the functional paratope (Table 1). Thus, we completely diversified two or three residues flanking the important residues in CDR-L3 or CDR-H3, respectively. Also, because CDR-H1 lies adjacent to CDR-H3 in the Ab fold, we diversified six positions in CDR-H1 with degenerate codons that encoded for residues that occur frequently at each position in functional Abs. Finally, in the heavy chain, we substituted Asp for Asn at position 29 to remove a potential glycosylation site and we substituted Ser for Ala at position 110, which was favored in the homolog scan. The resulting library was cycled through five rounds of selection for binding to SEB and subsequent screening of approximately 100 clones by phage ELISA yielded 33 unique sequences. Competitive phage ELISAs were used to rank order these 33 clones based on apparent affinities (data not shown) and the six best clones were purified as scFv proteins. Direct binding ELISAs with the purified scFvs revealed that four exhibited better apparent affinities than scFv-GC132 (Figure S1). Most importantly, these four scFv variants exhibited significantly improved potency compared with scFv-GC132 in SEB neutralization assays conducted with peripheral blood mononuclear cells (PBMCs) from multiple donors using IFNγ release as a measure of activation (Table 3). In particular, scFv-GC132a exhibited a 250-fold improvement in potency (IC50 = 0.04 nM), and this variant was converted into Fab and full-length IgG formats for further studies, which are described below.
Table 3.
IC50 values for toxin neutralization by anti-SEB Abs determined with human PBMCs
| Antibody | IC50 (nM) | ||||
|---|---|---|---|---|---|
| Donor 1 | Donor 2 | Donor 3 | Mean | S.D. | |
| scFv-GC132 | 6.23 | 4.53 | 19.5 | 10 | 8 |
| scFv-GC132a | 0.02 | 0.04 | 0.066 | 0.04 | 0.02 |
| scFv-GC132b | 0.077 | 0.18 | 0.12 | 0.13 | 0.05 |
| scFv-GC132c | 0.152 | 1.03 | 0.125 | 0.4 | 0.5 |
| scFv-GC132d | 0.12 | 1.41 | 0.064 | 0.5 | 0.8 |
| scFv-GC132e | 0.094 | 1.78 | 0.199 | 0.7 | 0.9 |
| scFv-GC132f | 0.62 | 9.08 | 0.23 | 3 | 5 |
| IgG-GC121 | 0.1356 | 0.067 | 0.0699 | 0.09 | 0.04 |
| IgG-GC132 | 0.0591 | 0.141 | 0.182 | 0.13 | 0.06 |
| IgG-GC132a | 0.0025 | 0.0011 | 0.0016 | 0.0017 | 0.0007 |
| bsAb-121/132 | 0.0273 | 0.0425 | 0.0095 | 0.03 | 0.02 |
| IgG-GC121+scFv-GC132 | 0.173 | 0.484 | 0.181 | 0.3 | 0.2 |
| bsAb-121/132a | 0.0064 | 0.0021 | 0.0034 | 0.004 | 0.002 |
| IgG-GC121+scFv-GC132a | 0.0086 | 0.0034 | 0.0049 | 0.006 | 0.003 |
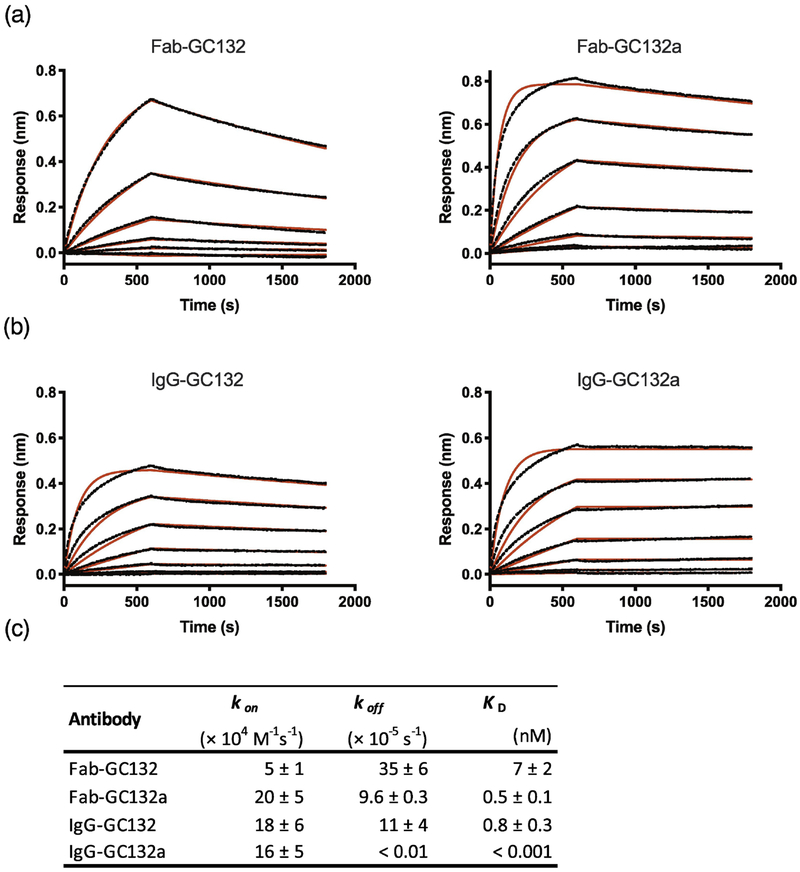
To define binding kinetics, we used bio-layer interferometry (BLI) to compare GC132a to its parent GC132 (Figure 2). Fab-GC132a exhibited sub-nanomolar affinity, which was ~14-fold tighter than Fab-GC132. The higher affinity of Fab-GC132a relative to Fab-GC132 was due to a 4- and ~3-fold improvement in the on and off rates, respectively. In the bivalent IgG format, both Abs exhibited improved affinities, presumably due to avidity effects. In particular, IgG-GC132a exhibited an extremely slow off rate, which resulted in an apparent dissociation constant in the low picomolar range. Taken together, these results show that our best Class T Ab GC132a binds SEB with high affinity in the Fab format, and affinity is further improved in the IgG format, suggesting that IgG-GC132a should be a potent inhibitor of SEB activity in vitro and in vivo. Consistent with affinity measurements in vitro, native mass spectrometry (MS) experiments showed that STEBVax and IgG-GC132a formed a stable 2:1 complex (Figure S2). The complex dissociates into a 1:1 complex with increased collision energy (Figure S2b), which further confirms the identity of the complex.
Fig. 2. Kinetic analysis of Abs binding to SEB.
BLI was used to assess binding of serial dilutions of SEB (0.14–100 nM) to immobilized Abs with a 600 second association step followed by a 1200 second dissociation step. (a) Traces for Fab-GC132 and Fab-GC132a. (b) Traces for IgG-GC132 and IgG-GC132a. Curve fitting is shown in red. (c) kon, koff and KD values for Abs binding to SEB.
Mapping the GC132a paratope and STEBVax epitope
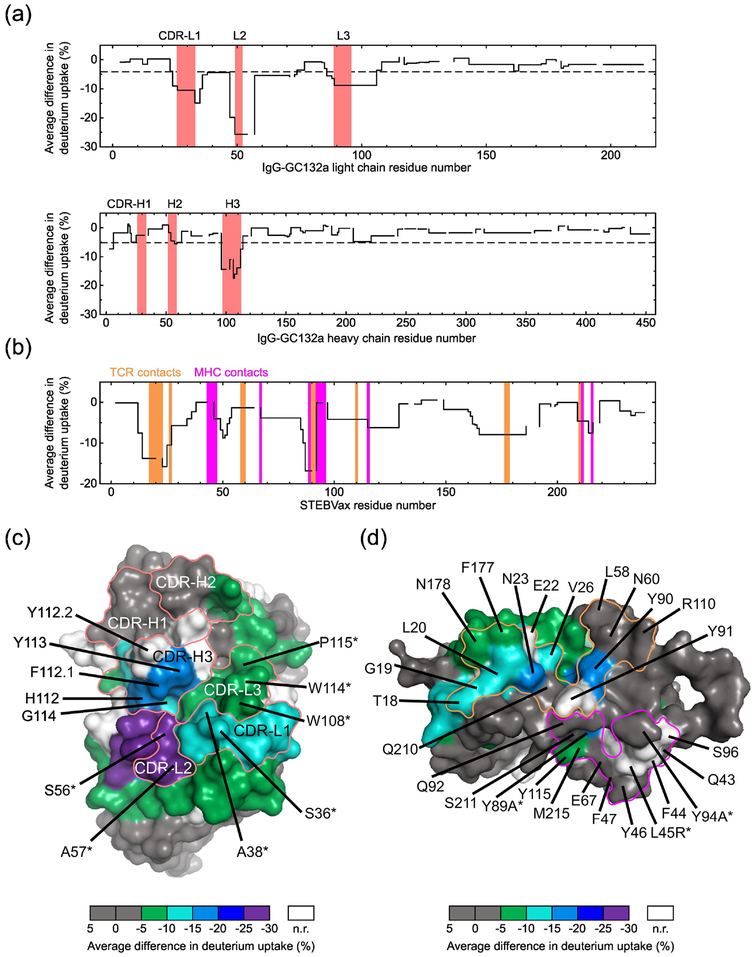
To map the IgG-GC132a paratope and STEBVax epitope, we conducted hydrogen-deuterium exchange coupled to mass spectrometry (HDX-MS), where amide hydrogens on the protein backbone are exchanged to deuterium from a D2O buffer, and MS of peptides derived from the proteins is used to determine levels of deuterium uptake [46]. Peptides in the binding regions are expected to show lower deuterium uptake in the bound state than the unbound state, as these regions become less solvent accessible and/or more hydrogen-bonded in the bound state. On the Ab, in agreement with the shotgun homolog scan data, we observed decreased deuterium uptake in CDRs L1, L2, L3 and H3 (Figure 3a and Figure S3a), but not in CDRs H1 and H2. Moreover, HDX plots of the peptides from the paratope regions showed different kinetic behavior (Figure S2b). Deuterium uptake differences of the three peptides from CDRs L1, L3 and H3 remained after 16 hours, suggesting slow conversion between bound and unbound states. In contrast, the HDX curves for the peptide from CDR-L2 gradually converged, indicating that deuterium uptake differences for this peptide became less apparent as the exchange reaction progressed. Mapping of the average deuterium uptake differences onto the structure of an Ab with the same framework as Ab GC132a revealed a large paratope that was protected from deuterium uptake, and notably, the residues that were shown to be functionally important by homolog scanning were located in a contiguous patch within this paratope (Figure 3c).
Fig. 3. HDX-MS analysis of the IgG-GC132a paratope and STEBVax epitope.
(a) Average deuterium uptake differences (y-axis) between IgG-GC132a only and IgG-GC132a-STEBVax complex plotted against residue numbers (x-axis) of IgG-GC132a light chain (top) and heavy chain (bottom). CDRs defined by the IMGT scheme [56] are shaded red. The dashed lines indicate the maximum average differences between % deuterium uptake for IgG-GC132a only and for IgG-GC132a-STEBVax complex observed in the constant region of the Ab and are used as the thresholds in determining residues responsible for binding. (b) Average deuterium uptake differences (y-axis) between STEBVax only and IgG-GC132a-STEBVax complex plotted against STEBVax residue numbers (x-axis). Regions that make contact with MHC-II or TCR [43] are shaded magenta or orange, respectively. (c) Average deuterium uptake differences on IgG-GC132a, unbound or bound to STEBVax, mapped on the surface of a Fab (PDB entry 1FVC) sharing the same framework. CDRs defined by the IMGT scheme are labeled and outlined by red lines. Residues are colored according to differences in deuterium uptake, as shown in the color bar at the bottom (n.r. denotes no results due to lack of peptide coverage in HDX-MS). Residues determined by shotgun homolog-scanning to be favorable for antigen binding (Fwt/mut >5) are labeled and asterisks (*) indicate light chain residues. (d) Average deuterium uptake differences on STEBVax, unbound or bound to IgG-GC132a, mapped on the surface of SEB (PDB entry 3SEB). Residues are colored as in (c). Residues on SEB that interact with MHC-II or TCR [43] are outlined by magenta or orange lines, respectively; residues are labeled and asterisks (*) indicate the three residues that differ between SEB and STEBVax, as shown by the residue type to the left or right of the number, respectively.
Compared with the unbound state, the HDX-MS analysis also identified several regions with decreased deuterium uptake in STEBVax bound to the Ab (Figure 3b and 3d). Among the 14 residues that comprise the TCR binding interface [43], six residues (Thr18, Gly19, Leu20, Asn23, Val26 and Tyr90) exhibited significant decreases and two others (Phe177, Asn178) exhibited moderate decreases in HDX upon binding. Of the remaining six residues, Glu22 and Tyr91 were not detected in the HDX-MS analysis but are close to this region and may also be involved in interactions with the Ab. In contrast, among the 14 residues that comprise the MHC-II binding site, only three residues (Tyr89, Tyr115, Met215) exhibited decreases in HDX upon binding, and these were all in close proximity to the TCR binding site. The HDX-MS results reinforce the hypothesis, based on competitive ELISA, that the epitope for Ab GC132a overlaps with the TCR binding site on SEB and STEBVax, and consequently, Ab GC132a neutralizes toxin activity by competitively inhibiting TCR binding.
Toxin Neutralization in vitro
Co-administration of two anti-SEB Abs or a combination of an anti-SEB Ab and an intracellular inhibitor of T-cell activation has been shown to result in synergistic neutralization efficacy in vitro and/or better protection against toxin challenge in vivo [36, 38, 47]. To explore whether potency could be enhanced by simultaneously targeting two SEB epitopes with one molecule, we constructed tetravalent bsAbs consisting of IgG-GC121 with the addition of scFv-GC132 (bsAb-121/132) or scFv-GC132a (bsAb-121/132a) fused to the C-terminus of the Fc. The potencies of the bsAbs and the Class M and Class T IgGs for neutralization of SEB activity in vitro were assessed using PBMCs (Table 3). Compared with the Class M IgG-GC121 (IC50 = 0.09 nM), the Class T IgG-GC132 was essentially equipotent (IC50 = 0.13 nM), whereas the optimized IgG-GC132a was ~50-fold more potent (IC50 = 0.0017 nM). The bsAbs 121/132 (IC50 = 0.03 nM) and 121/132a (IC50 = 0.004 nM) were more potent than IgG-GC121 or mixtures of IgG-GC121 and scFv-GC132 (IC50 = 0.3 nM) or scFv-GC132a (IC50 = 0.006 nM), but they did not match the potency of IgG-GC132a. However, it is worth noting that, in the absence of Ab-toxin complex structures and clear epitope information, it is not clear whether the space between the two antigen-binding sites in the same molecule is sufficient for simultaneous toxin binding. Also, the alternative option of using a Class T Ab as the full-length IgG portion of the bsAb molecule is still kept open. Hence, optimization of the linker between the IgG and the scFv and different combinations and arrangements of IgG and scFv portions, may result in bsAb molecules with improved efficacy. By and large, the bsAb format reported here is a simple, yet effective, strategy for bsAb construction that could potentially improve the efficacy of antibody treatment in general. Taken together, our results showed that IgG-GC132a was the best neutralizing agent, and thus, we focused on it for assessment of protection from toxic shock in vivo.
Toxin neutralization in vivo
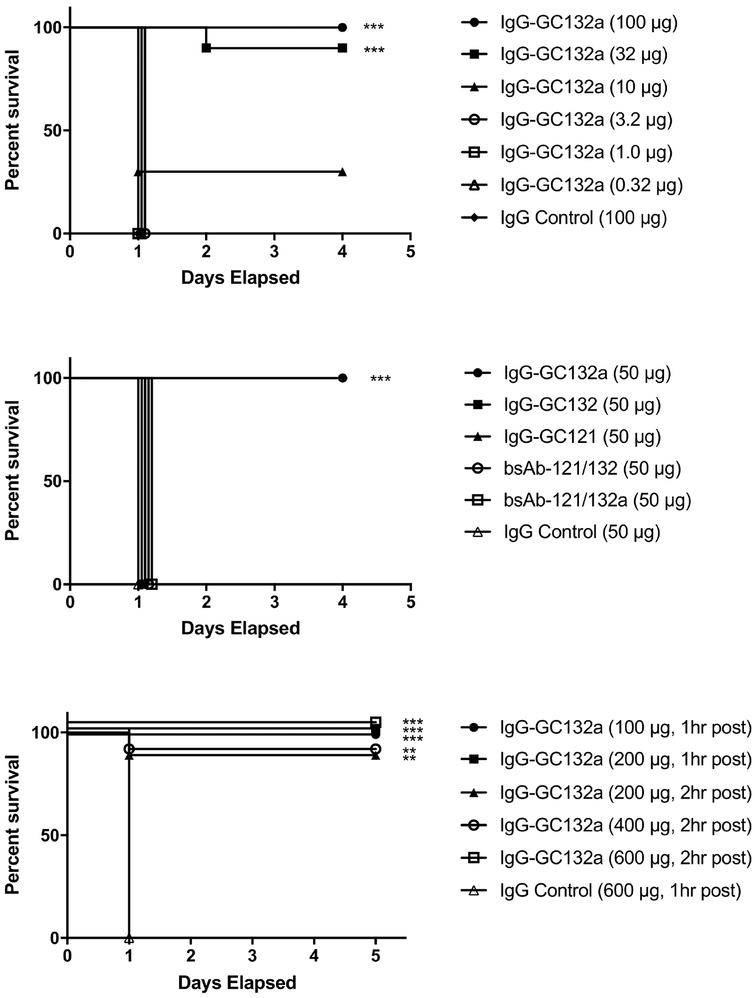
To assess in vivo efficacy, we tested anti-SEB Abs in a mouse model of parenteral challenge toxic shock. To determine appropriate dosing for the lipopolysaccharide (LPS) potentiated challenge model [48], groups of 10 BALB/c mice were treated with various amounts of IgG-GC132a, and after 2 hours, were challenged with SEB followed 4 hours later by LPS. Mice treated with 100, 32 or 10 μg of IgG-GC132a exhibited 100%, 90% or 30% survival, respectively, whereas those treated with lower doses or a control Ab all died within a day after challenge (Figure 4a). In the same assay with 50 μg of Ab, IgG-GC132a provided complete protection, whereas no protection was observed with IgG-GC132, IgG-GC121, bsAb-121/132 or bsAb-121/132a (Figure 4b). Finally, we conducted a highly stringent test of efficacy by assessing survival of mice that were challenged with SEB and LPS prior to treatment with Ab. When treated 1 hour after challenge, all mice treated with 600 μg of control Ab died within one day, whereas all mice similarly treated with 100 or 200 μg of IgG-GC132a survived (Figure 4c). Moreover, even when IgG-GC132a was administered 2 hours after challenge, 100% protection was observed with a dose of 600 μg and 90% protection was observed with a dose of 200 or 400 μg (Figure 4c). Previously reported Abs had displayed partial [37, 39, 49] or complete [40] protection only as far as 0.5–1 hour post challenge. Thus, ours is the first report of any significant protection against SEB with a monoclonal Ab in this model beyond the window of 1 hour post exposure. Taken together, these results show that IgG-GC132a is extremely effective in vivo as a neutralizing agent for the toxic effects of SEB.
Fig. 4. Toxin neutralization in vivo.
(a) Effects of various IgG-GC132a doses on survival. Survival of BALB/c mice (n = 10 per group) treated with the indicated dose of IgG-GC132a (y-axis) was determined over time (x-axis). Mice were injected intraperitoneally with the indicated IgG dose, 2 hours later with 3 μg SEB, and another 4 hours later with 40 μg LPS. (b) Effects of various IgGs and bsAbs on survival. Survival of BALB/c mice (n = 10 per group) treated with the indicated Ab (y-axis) was determined over time (x-axis). Mice were injected intraperitoneally with 50 μg indicated Ab, 2 hours later with 3 μg SEB, and another 4 hours later with 40 μg LPS. (c) Effects on survival of IgG-GC132a treatment after challenge with SEB. Survival of BALB/c mice (n = 10 per group) treated with the indicated dose of IgG-GC132a (y-axis) was determined over time (x-axis). Mice were injected intraperitoneally with 3 μg SEB, 1 or 2 hours later with the indicated IgG dose, and 4 hours later with 40 μg LPS. **, p < 0.005 compared with IgG control; ***,p < 0.001 compared with IgG control.
The superior in vitro toxin neutralizing efficacy and highly effective in vivo protection afforded by Class T Ab IgG-GC132a are most likely due to its extremely tight affinity and consequent effective blocking of the TCR binding site on SEB. With an affinity in the low picomolar range, IgG-GC132a bound SEB much more tightly than did our best Class M Ab IgG-GC121, which exhibited an affinity in the high subnanomolar range [40]. Although differences in effectiveness may also be due to effects inherent to blocking the MHC-II or TCR binding site, which may depend on the region of the Ab and the direction of approach to the toxin, extremely tight binding is likely one of the main reasons why IgG-GC132a is better than IgG-GC121 in both neutralizing and protecting against SEB.
Conclusions
SEB links MHC-II molecules on antigen-presenting cells to TCRs on T-cells to induce massive cytokine production and consequent toxic shock. The toxic effects of SEB could be blocked by Abs that prevent binding of MHC-II, TCR, or both. We previously developed the Class M anti-SEB Ab GC121, which likely blocked MHC-II binding, and here we report development of the Class T Ab GC132a, which likely blocks TCR binding. Both Abs were effective for inhibition of SEB activity in vitro, and in particular, IgG-GC132a was extremely potent with activity in the picomolar range. Most importantly, IgG-GC132a also proved to be extremely potent at neutralizing toxicity in vivo, providing strong protection even in an extremely stringent mouse model in which Ab treatment was administered after SEB challenge. Moreover, initial studies with bsAbs, which contained both Class M and Class T antigen-binding sites, suggest that efficacy may be improved further by increasing valency and simultaneously blocking both MHC-II and TCR engagement. Taken together, our results provide a framework to characterize new antibodies and strong support for IgG-GC132a as an excellent lead for bsAb engineering and clinical development of anti-SEB therapeutics.
Materials and Methods
Chemicals and reagents
Unless otherwise stated, chemical reagents were purchased from BioShop Canada (Burlington, ON, Canada), restriction enzymes were purchased from New England Biolabs (Pickering, ON, Canada), and DNA oligonucleotides were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA, USA).
Bacterial superantigens and endotoxin
Biotinylated SEB used for phage display selections and SEB for in vitro PBMC and in vivo toxin neutralization assays (TNAs) were purchased from Toxin Technology (Sarasota, FL, USA) and reconstituted with deionized water. Toxins were aliquoted and stored at −80 °C until use. Lipopolysaccharide (LPS, Escherichia coli 055:B5) was purchased from List Biological Laboratories, Inc. (Campbell, CA, USA) and reconstituted with phosphate-buffered saline (PBS) buffer (10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4) prior to use.
Selection and characterization of anti-SEB Fabs
Phage display selections, direct phage ELISAs and competitive phage ELISAs were performed as described [50] with minor modifications. Briefly, phage particles from the libraries were cycled through rounds of binding selections with SEB coated directly on 96-well Maxisorp immunoplates (Fisher Scientific, Nepean, ON, Canada) or biotinylated SEB captured by immobilized neutravidin (Thermo Fisher Scientific, Rockford, IL, USA) as the capture target. After four or five rounds of selections, phage particles were produced from individual clones grown in a 96-well format, and the culture supernatants were used in phage ELISAs to detect specific binding clones. Clones that bound specifically to SEB were subjected to DNA sequencing to decode the sequences of the displayed Fabs. To assess competition between Abs for binding to SEB antigen, a competitive phage ELISA was used, whereby binding of Fab-phage to immobilized SEB was assessed in the presence of solution-phase Fab protein, as described [50].
Shotgun homolog-scanning and affinity maturation of scFv-GC132
A phagemid (pscFvHis-GC132) was designed for display of scFv-GC132 on bacteriophage, as described [51]. The scFv-GC132 we engineered is in the VL-linker-VH format and was constructed by linking VL and VH regions of GC132 with a linker of 16 amino acids (GTTAASGSSGGSSSGA) identified from a phage library designed to optimize the linker between VL and VH of an anti-maltose binding protein scFv [51]. For homolog-scanning of the light and heavy chains, respectively, libraries 132HL and 132HH were constructed, as described [45]. Briefly, for each library, we used a “stop template” version of pscFvHis-GC132, which contained TAA stop codons within each of the CDRs to be mutated. Mutagenic oligonucleotides designed to simultaneously repair the stop codons and introduce mutations at the designed sites were used to construct libraries. Each codon within the CDR sequences to be scanned (Table 2) was replaced with a degenerate codon that encoded for equal proportions of wt and a homologous amino acid. Phage pools representing each library were cycled separately through four rounds of binding selections using immobilized anti-FLAG M2 antibody (Sigma-Aldrich, Oakville, ON, Canada) or SEB as capture agents. From each selection, at least 40 unique sequences were obtained by DNA sequencing, and these were used to determine the wt:mutant ratio at each varied position. Based on the results of homolog-scanning, library 132v2 was constructed for affinity maturation of scFv-GC132. A stop template version of pscFvHis-GC132 was used as the template for site-directed mutagenesis with mutagenic oligonucleotides designed to diversify positions in CDR-L1, CDR-L3 and CDR-H3 (Table 1). Phage pools representing library 132v2 were cycled through five rounds of binding selection with immobilized SEB and a single-point competitive phage ELISA was used to estimate the affinities of phage-displayed anti-SEB scFvs for solution-phase SEB, as described [50]. Clones that exhibited better estimated affinities than scFv-GC132 were subjected to DNA sequence analysis to decode the sequences of the displayed scFvs.
Production and characterization of Abs
Fab and scFv proteins were expressed and purified from E. coli 55244 or BL21(DE3)pLysS, respectively, as described [40, 52]. IgG proteins were expressed and purified using the pMAZ mammalian expression vector system [53] in HEK293T cells, as described [40]. The bsAb was constructed by fusing the scFv fragment with the format described above onto the C-terminus of the Fc region of a full-length IgG with the classic spacer linker (Gly4Ser)3. The bsAb proteins were expressed and purified in the same way as IgG proteins. ELISAs for assessment of Fab, scFv, IgG and bsAb proteins binding directly to immobilized SEB or STEBVax were performed as described [40]. ELISAs to assess competition of solution-phase SEB with immobilized SEB for binding to Fab-GC132 or scFv-GC132 were performed as described [50].
BLI assays
BLI was performed on an Octet HTX system (ForteBio, Fremont, CA) in flat, black 96-well plates at 25 °C with shaking at 1,000 rpm. Fab or IgG proteins were immobilized onto FAB2G sensors (Pall ForteBio, Fremont, CA) for 120 s followed by a 60 s baseline step to ensure stable ligand loading before each ligand was dipped into SEB analyte for a 600 s association step in assay buffer (PBS, 1% BSA, 0.05% Tween20) and a buffer blank followed by a 1200 s dissociation step in assay buffer. SEB was used at concentrations ranging from 0.14–100 nM. To control for non-specific binding of SEB to sensors in the absence of ligand, signals from reference sensors for each concentration of SEB were subtracted from total response. Data analyses were performed with PRISM 7 software (GraphPad Software, La Jolla, CA). Specific binding curves were fit globally to a 1:1 Langmuir binding model to obtain kon, koff, and KD values.
Native MS
Native MS was performed as described [54] with minor modifications. Briefly, stock solutions of STEBVax and IgG-GC132a were buffer-exchanged to 150 mM ammonium acetate (pH 6.8) using Vivaspin 500 centrifugal concentrators (Sartorius, Göttingen, Germany). The working solution was prepared by mixing Ab and antigen solutions with a molar ratio of ~2:1 and a final total protein concentration of ~2 μM. The solution was loaded into NanoES spray capillaries (Thermo Fisher Scientific) on a Nanospray Flex ion source (Thermo Fisher Scientific). Mass spectra were collected on an Exactive Plus EMR mass spectrometer (Thermo Fisher Scientific). The in-source collisional ionization dissociation energy was 20 eV, spray voltage was 1.3 kV, capillary temperature was 100 °C and collision energy was 100 or 200 a.u.
HDX-MS
STEBVax and IgG-GC132a stock solutions were prepared by dialyzing into PBS buffer, and 2 μL protein stock solution (20 μM each protein) were added to 18 μL D2O-based PBS buffer to initiate deuterium exchange at 25 °C. At various time points, the reaction was quenched by adding 30 μL quench solution (4 M guanidine hydrochloride, 0.5 M TCEP, pH 2.4) with rapid pipet mixing. The mixture was incubated at 25 °C for 3 min to allow protein denaturation and disulfide bond reduction, and the sample was injected into a LC device with online pepsin digestion, as described [46]. Mass spectra for each sample were collected in duplicate with an LTQ-FT Ultra mass spectrometer operating with an ESI source (Thermo Fisher Scientific). Data were processed with the HDExaminer software (Sierra Analytics, Modesto, CA) to generate deuterium uptake percentage on peptides. HDX heat map data were exported from HDExaminer as plots of the average differences in deuterium uptake across STEBVax and IgG-GC132a. The first two N-terminal residues of each peptide were subject to rapid back exchange during LC, and they were not considered when calculating the heat map displaying deuterium uptake of overlapping peptides by the software.
In vitro toxin neutralization assay
PBMCs were isolated from heparinized blood of healthy human donors by Ficoll gradient centrifugation as described [55]. Normal human blood donors complete a blood donor questionnaire. An IRB approved consent form was obtained from each donor giving permission to collect blood and use for research purposes. PBMCs were frozen in 10% DMSO in heat inactivated fetal bovine serum (HI-FBS) and stored in liquid nitrogen for future use. For the assay, cell pellet was re-suspended in RPMI 1640 with 10% FBS, and cells were washed, enumerated and adjusted to 2 × 106 cells/mL. 100 μL of the cell suspension (2 × 105 cells) with a viability of >95% was added to duplicate wells of 96-well plates containing 50 μL semi-log diluted mAb and 50 μL SEB. Wells containing medium with toxin only were used as controls. The cultures were incubated at 37 °C with 5% CO2 for 48 hours. Cells were pelleted, culture supernatants were harvested, and IFNγ production was assessed by ELISA (R&D Systems, Minneapolis, MN) following the manufacturers’ protocol. Plates were read at 450 nm using a VersaMax plate reader and data were analyzed in Microsoft Office Excel. Cells stimulated with toxin in the absence of a neutralizing agent served as a positive control and this value was considered as 0% IFNγ inhibition. Accordingly, inhibition of IFNγ production in the presence of Ab was calculated as the difference between positive control and sample. The IC50 value for each Ab was determined using a 4-parameter logistic model (equation 205, XLFit v5.2).
Animal studies
10–12 week-old female BALB/c mice were purchased from Charles River (Willmington, MA). Mice were maintained under pathogen-free conditions and fed laboratory chow and water ad libitum. All mouse work was conducted in accordance with protocols approved by institutional animal care and use committees. Challenge studies were performed in BALB/c mice by intraperitoneal (IP) injection of 10 LD50 (3 μg) of SEB followed by potentiation by 40 μg LPS administered intraperitoneally four hours later. At various time points, mice were treated by IP injection with various doses of anti-SEB Abs or a negative control Ab in 200 μL PBS. Mice were monitored for morbidity (weight loss, hunched posture, lethargy, ruffled fur) and mortality for 4 days. The time period was expanded to 5 days in the post-challenge model.
Statistical Analysis
Data were analyzed using PRISM (GraphPad Software, La Jolla, CA) or XLFit (IDBS, Boston, MA). Curve fitting was performed using a standard four-parameter logistic equation.
Supplementary Material
Fig. S1. ELISAs for assessment of serial dilutions of the indicated scFv protein (x-axis) binding to immobilized SEB (y-axis). The plots represent the average of results from two independent experiments.
Fig. S2. Native MS analysis of the STEBVax-IgG-GC132a complex. Native MS was performed with higher-energy collision dissociation (HCD) energy of (a) 100 a.u. and (b) 200 a.u. A STEBVax-IgG-GC132a complex with 2:1 stoichiometry was observed along with excess STEBVax. Triplet peaks were observed for IgG-GC132a, representing 0, 1 or 2 units of glycosylation. Increased HCD energy partially dissociates the 2:1 complex into a 1:1 complex, which verifies the identities of the observed components.
Fig. S3. HDX-MS analysis of the STEBVax-IgG-GC132a complex. (a) HDX plots of representative peptides from 6 CDR and 2 framework (non-CDR) regions on light (top) and heavy (bottom) chains of IgG-GC132a only (orange) or IgG-GC132a-STEBVax complex (black) samples. (b) HDX plots of representative peptides from STEBVax only (green) and STEBVax-IgG-GC132a complex (black). The data shown are the average of two replicates.
Highlights.
SEB is a potent incapacitating agent and a major cause of food poisoning.
SEB neutralizing antibodies were identified from a synthetic antibody phage library.
An antibody with picomolar binding affinity was generated by antibody engineering.
The best antibody demonstrated superior in vitro and in vitro anti-toxin efficacy.
An excellent lead for further development of anti-SEB therapeutics was reported.
Acknowledgments
We thank Itai Benhar and George Georgiou for kindly providing IgG expression vectors. This work was supported, in whole or in part, by National Institutes of Health, NIAID, Grants U01 AI078023–05 and R01AI111205 (to Integrated BioTherapeutics, Inc.). The mass spectrometry was supported by NIGMS of the NIH (Grant P41GM103422).
Abbreviations used:
- Ab
antibody
- BLI
bio-layer interferometry
- bsAb
bispecific antibody
- CDR
complementarity-determining region
- ELISA
enzyme-linked immunosorbent assay
- Fab
antigen-binding fragment
- HDX-MS
hydrogen deuterium exchange coupled to mass spectrometry
- INFγ
interferon-gamma
- LPS
lipopolysaccharide
- MHC-II
class II major histocompatibility complex
- MS
mass spectrometry
- PBMC
peripheral blood mononuclear cell
- PBS
phosphate-buffered saline
- SAg
staphylococcal superantigen
- scFv
single-chain variable fragment
- SEB
staphylococcal enterotoxin B
- SEs
staphylococcal enterotoxins
- TCR
T-cell receptor
- TNA
toxin neutralization assay
- TNFα
tumor necrosis factor alpha
- TSS
toxic shock syndrome
- TSST-1
toxic shock syndrome toxin 1
- wt
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none.
References
- [1].Chambers HF, DeLeo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nature Reviews Microbiology. 2009;7:629–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Dinges MM, Orwin PM, Schlievert PM. Exotoxins of Staphylococcus aureus. Clinical Microbiology Reviews. 2000;13:16–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Foster TJ. Immune evasion by Staphylococci. Nature Reviews Microbiology. 2005;3:948–58. [DOI] [PubMed] [Google Scholar]
- [4].McCormick JK, Yarwood JM, Schlievert PM. Toxic shock syndrome and bacterial superantigens: An update. Annual Review of Microbiology. 2001;55:77–104. [DOI] [PubMed] [Google Scholar]
- [5].Bohach GA, Fast DJ, Nelson RD, Schlievert PM. STAPHYLOCOCCAL AND STREPTOCOCCAL PYROGENIC TOXINS INVOLVED IN TOXIC SHOCK SYNDROME AND RELATED ILLNESSES. Critical Reviews in Microbiology. 1990;17:251–72. [DOI] [PubMed] [Google Scholar]
- [6].Humphreys H, Keane CT, Hone R, Pomeroy H, Russell RJ, Arbuthnott JP, et al. ENTEROTOXIN PRODUCTION BY STAPHYLOCOCCUS-AUREUS ISOLATES FROM CASES OF SEPTICEMIA AND FROM HEALTHY CARRIERS. Journal of Medical Microbiology. 1989;28:163–72. [DOI] [PubMed] [Google Scholar]
- [7].Salgado-Pabon W, Breshears L, Spaulding AR, Merriman JA, Stach CS, Horswill AR, et al. Superantigens Are Critical for Staphylococcus aureus Infective Endocarditis, Sepsis, and Acute Kidney Injury. Mbio. 2013;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Spaulding AR, Lin YC, Merriman JA, Brosnahan AJ, Peterson ML, Schlievert PM. Immunity to Staphylococcus aureus secreted proteins protects rabbits from serious illnesses. Vaccine. 2012;30:5099–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Nienaber JJC, Kuinkel BKS, Clarke-Pearson M, Lamlertthon S, Park L, Rude TH, et al. Methicillin-Susceptible Staphylococcus aureus Endocarditis Isolates Are Associated With Clonal Complex 30 Genotype and a Distinct Repertoire of Enterotoxins and Adhesins. Journal of Infectious Diseases. 2011;204:704–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Geoghegan JA, Irvine AD, Foster TJ. Staphylococcus aureus and Atopic Dermatitis: A Complex and Evolving Relationship. Trends in Microbiology. 2018;26:484–97. [DOI] [PubMed] [Google Scholar]
- [11].Matsubara K, Fukaya T. The role of superantigens of group A Streptococcus and Staphylococcus aureus in Kawasaki disease. Current Opinion in Infectious Diseases. 2007;20:298–303. [DOI] [PubMed] [Google Scholar]
- [12].Ulrich RG, Sidell S, Taylor TJ, Wilhelmsen CL, Franz DR. Staphylococcal enterotoxin B and related pyogenic toxins In: Zajtchuk R, editor. Textbook of Military Medicine: Medical Aspects of Chemical and Biological Warfare Washington, DC:: US Dept of the Army, The Surgeon General, and the Borden Institute; 1997. p. 621–30. [Google Scholar]
- [13].Schlievert PM. Staphylococcal Enterotoxin B And Toxic Shock Syndrome Toxin-1 Are Significantly Associated With Non-Menstrual TSS. Lancet. 1986;1:1149–50. [DOI] [PubMed] [Google Scholar]
- [14].Schlievert PM. Role Of Superantigens In Human Disease. Journal of Infectious Diseases. 1993;167:997–1002. [DOI] [PubMed] [Google Scholar]
- [15].Rusnak JM, Kortepeter M, Ulrich R, Poli M, Boudreau E. Laboratory exposures to Staphylococcal enterotoxin B. Emerging Infectious Diseases. 2004;10:1544–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Madsen JM. Toxins as weapons of mass destruction. A comparison and contrast with biological-warfare and chemical-warfare agents. Clinics in Laboratory Medicine. 2001;21:593–605. [PubMed] [Google Scholar]
- [17].Ahanotu E, Alvelo-Ceron D, Ravita T, Gaunt E. Staphylococcal Enterotoxin B as a Biological Weapon: Recognition, Management, and Surveillance of Staphylococcal Enterotoxin. Applied Biosafety. 2006; 11:120–6. [Google Scholar]
- [18].Arad G, Levy R, Hillman D, Kaempfer R. Superantigen antagonist protects against lethal shock and defines a new domain for T-cell activation. Nature Medicine. 2000;6:414–21. [DOI] [PubMed] [Google Scholar]
- [19].Arad G, Hillman D, Levy R, Kaempfer R. Superantigen antagonist blocks Th1 cytokine gene induction and lethal shock. Journal of Leukocyte Biology. 2001;69:921–7. [PubMed] [Google Scholar]
- [20].Arad G, Hillman D, Levy R, Kaempfer R. Broad-spectrum immunity against superantigens is elicited in mice protected from lethal shock by a superantigen antagonist peptide. Immunology Letters. 2004;91:141–5. [DOI] [PubMed] [Google Scholar]
- [21].Lehnert NM, Allen DL, Allen BL, Catasti P, Shiflett PR, Chen M, et al. Structure-based design of a bispecific receptor mimic that inhibits T cell responses to a superantigen. Biochemistry. 2001;40:4222–8. [DOI] [PubMed] [Google Scholar]
- [22].Hong-Geller E, Mollhoff M, Shiflett PR, Gupta G. Design of chimeric receptor mimics with different TcRV beta isoforms - Type-specific inhibition of superantigen pathogenesis. Journal of Biological Chemistry. 2004;279:5676–84. [DOI] [PubMed] [Google Scholar]
- [23].Mollhoff M, Vander Zanden HB, Shiflett PR, Gupta G. Modeling of receptor mimics that inhibit superantigen pathogenesis. Journal of Molecular Recognition. 2005;18:73–83. [DOI] [PubMed] [Google Scholar]
- [24].Buonpane RA, Churchill HRO, Moza B, Sundberg EJ, Peterson ML, Schlievert PM, et al. Neutralization of staphylococcal enterotoxin B by soluble, high-affinity receptor antagonists. Nature Medicine. 2007;13:725–9. [DOI] [PubMed] [Google Scholar]
- [25].Visvanathan K, Charles A, Bannan J, Pugach P, Kashfi K, Zabriskie JB. Inhibition of bacterial superantigens by peptides and antibodies. Infect Immun. 2001;69:875–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lowell GH, Colleton C, Frost D, Kaminski RW, Hughes M, Hatch J, et al. Immunogenicity and efficacy against lethal aerosol staphylococcal enterotoxin B challenge in monkeys by intramuscular and respiratory delivery of proteosome-toxoid vaccines. Infect Immun. 1996;64:4686–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lowell GH, Kaminski RW, Grate S, Hunt RE, Charney C, Zimmer S, et al. Intranasal and intramuscular proteosome-staphylococcal enterotoxin B (SEB) toxoid vaccines: Immunogenicity and efficacy against lethal SEB intoxication in mice. Infect Immun. 1996;64:1706–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ulrich RG, Olson MA, Bavari S. Development of engineered vaccines effective against structurally related bacterial superantigens. Vaccine. 1998;16:1857–64. [DOI] [PubMed] [Google Scholar]
- [29].Stiles BG, Garza AR, Ulrich RG, Boles JW. Mucosal vaccination with recombinantly attenuated staphylococcal enterotoxin B and protection in a murine model. Infect Immun. 2001;69:2031–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Boles JW, Pitt MLM, LeClaire RD, Gibbs PH, Torres E, Dyas B, et al. Generation of protective immunity by inactivated recombinant staphylococcal enterotoxin B vaccine in nonhuman primates and identification of correlates of immunity. Clinical Immunology. 2003;108:51–9. [DOI] [PubMed] [Google Scholar]
- [31].Chen WH, Pasetti MF, Adhikari RP, Baughman H, Douglas R, El-Khorazaty J, et al. Safety and Immunogenicity of a Parenterally Administered, Structure-Based Rationally Modified Recombinant Staphylococcal Enterotoxin B Protein Vaccine, STEBVax. Clinical and Vaccine Immunology. 2016;23:918–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Darenberg J, Ihendyane N, Sjolin J, Aufwerber E, Haidl S, Follin P, et al. Intravenous immunoglobulin G therapy in streptococcal toxic shock syndrome: A European randomized, double-blind, placebo-controlled trial. Clinical Infectious Diseases. 2003;37:333–40. [DOI] [PubMed] [Google Scholar]
- [33].Darenberg J, Soderquist B, Normark BH, Norrby-Teglund A. Differences in potency of intravenous polyspecific immunoglobulin G against streptococcal and staphylococcal superantigens: Implications for therapy of toxic shock syndrome. Clinical Infectious Diseases. 2004;38:836–42. [DOI] [PubMed] [Google Scholar]
- [34].Hamad ARA, Herman A, Marrack P, Kappler JW. Monoclonal Antibodies Defining Functional Sites on the Toxin Superantigen Staphylococcal Enterotoxin B Journal of Experimental Medicine. 1994;180:615–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pang LTY, Ku WWS, Chow AW. Inhibition of staphylococcal enterotoxin B-induced lymphocyte proliferation and tumor necrosis factor alpha secretion by MAb5, an anti-toxic shock syndrome toxin 1 monoclonal antibody. Infect Immun. 2000;68:3261–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tilahun ME, Rajagopalan G, Shah-Mahoney N, Lawlor RG, Tilahun AY, Xie C, et al. Potent Neutralization of Staphylococcal Enterotoxin B by Synergistic Action of Chimeric Antibodies. Infect Immun. 2010;78:2801–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Drozdowski B, Zhou Y, Kline B, Spidel J, Chan YY, Albone E, et al. Generation and characterization of high affinity human monoclonal antibodies that neutralize staphylococcal enterotoxin B. Journal of immune based therapies and vaccines. 2010;8:9-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Varshney AK, Wang X, Cook E, Dutta K, Scharff MD, Goger MJ, et al. Generation, Characterization, and Epitope Mapping of Neutralizing and Protective Monoclonal Antibodies against Staphylococcal Enterotoxin B-induced Lethal Shock. Journal of Biological Chemistry. 2011;286:9737–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Larkin EA, Stiles BG, Ulrich RG. Inhibition of Toxic Shock by Human Monoclonal Antibodies against Staphylococcal Enterotoxin B. Plos One. 2010;5:e13253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Karauzum H, Chen G, Abaandou L, Mahmoudieh M, Boroun AR, Shulenin S, et al. Synthetic human monoclonal antibodies toward staphylococcal enterotoxin B (SEB) protective against toxic shock syndrome. Journal of Biological Chemistry. 2012;287:25203–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Jardetzky TS, Brown JH, Gorga JC, Stern LJ, Urban RG, Chi YI, et al. 3-DIMENSIONAL STRUCTURE OF A HUMAN CLASS-II HISTOCOMPATIBILITY MOLECULE COMPLEXED WITH SUPERANTIGEN. Nature. 1994;368:711–8. [DOI] [PubMed] [Google Scholar]
- [42].Li HM, Llera A, Tsuchiya D, Leder L, Ysern X, Schlievert PM, et al. Three-dimensional structure of the complex between a T cell receptor beta chain and the superantigen staphylococcal enterotoxin B. Immunity. 1998;9:807–16. [DOI] [PubMed] [Google Scholar]
- [43].Rodstrom KEJ, Elbing K, Lindkvist-Petersson K. Structure of the Superantigen Staphylococcal Enterotoxin B in Complex with TCR and Peptide-MHC Demonstrates Absence of TCR-Peptide Contacts. J Immunol. 2014;193:1998–2004. [DOI] [PubMed] [Google Scholar]
- [44].Persson H, Ye W, Wernimont A, Adams JJ, Koide A, Koide S, et al. CDR-H3 diversity is not required for antigen recognition by synthetic antibodies. Journal of Molecular Biology. 2013;425:803–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Vajdos FF, Adams CW, Breece TN, Presta LG, De Vos AM, Sidhu SS. Comprehensive functional maps of the antigen-binding site of an anti-ErbB2 antibody obtained with shotgun scanning mutagenesis. Journal of Molecular Biology. 2002;320:415–28. [DOI] [PubMed] [Google Scholar]
- [46].Johnson B, McConnell P, Kozlov AG, Mekel M, Lohman TM, Gross ML, et al. Allosteric Coupling of CARMIL and V-1 Binding to Capping Protein Revealed by Hydrogen-Deuterium Exchange. Cell Reports. 2018;23:2795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Tilahun ME, Kwan A, Natarajan K, Quinn M, Tilahun AY, Xie C, et al. Chimeric Anti-Staphylococcal Enterotoxin B Antibodies and Lovastatin Act Synergistically to Provide In Vivo Protection against Lethal Doses of SEB. Plos One. 2011;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Stiles BG, Bavari S, Krakauer T, Ulrich RG. TOXICITY OF STAPHYLOCOCCAL ENTEROTOXINS POTENTIATED BY LIPOPOLYSACCHARIDE - MAJOR HISTOCOMPATIBILITY COMPLEX CLASS-II MOLECULE DEPENDENCY AND CYTOKINE RELEASE. Infect Immun. 1993;61:5333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Varshney AK, Wang XB, MacIntyre J, Zollner RS, Kelleher K, Kovalenko OV, et al. Humanized Staphylococcal Enterotoxin B (SEB)-Specific Monoclonal Antibodies Protect From SEB Intoxication and Staphylococcus aureus Infections Alone or as Adjunctive Therapy With Vancomycin. Journal of Infectious Diseases. 2014;210:973–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Fellouse FA, Sidhu SS. Making antibodies in bacteria In: Howard GC, Kaser MR, editors. Making and Using Antibodies: A Practical Handbook. Boca Raton, FL: CRC Press; 2006. p. 157–80. [Google Scholar]
- [51].Van Deventer JA, Kelly RL, Rajan S, Wittrup KD, Sidhu SS. A switchable yeast display/secretion system. Protein Engineering Design & Selection. 2015;28:317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Luz D, Chen G, Maranhao AQ, Rocha LB, Sidhu S, Piazza RMF. Development and Characterization of Recombinant Antibody Fragments That Recognize and Neutralize In Vitro Stx2 Toxin from Shiga Toxin-Producing Escherichia coli. Plos One. 2015;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Mazor Y, Barnea I, Keydar I, Benhar I. Antibody internalization studied using a novel IgG binding toxin fusion. Journal of Immunological Methods. 2007;321:41–59. [DOI] [PubMed] [Google Scholar]
- [54].Zhang Y, Cui WD, Wecksler AT, Zhang H, Molina P, Deperalta G, et al. Native MS and ECD Characterization of a Fab-Antigen Complex May Facilitate Crystallization for X-ray Diffraction. Journal of the American Society for Mass Spectrometry. 2016;27:1139–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Berthold F ISOLATION OF HUMAN-MONOCYTES BY FICOLL DENSITY GRADIENT CENTRIFUGATION. Blut. 1981;43:367–71. [DOI] [PubMed] [Google Scholar]
- [56].Lefranc MP, Pommie C, Ruiz M, Giudicelli V, Foulquier E, Truong L, et al. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Developmental and Comparative Immunology. 2003;27:55–77. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. ELISAs for assessment of serial dilutions of the indicated scFv protein (x-axis) binding to immobilized SEB (y-axis). The plots represent the average of results from two independent experiments.
Fig. S2. Native MS analysis of the STEBVax-IgG-GC132a complex. Native MS was performed with higher-energy collision dissociation (HCD) energy of (a) 100 a.u. and (b) 200 a.u. A STEBVax-IgG-GC132a complex with 2:1 stoichiometry was observed along with excess STEBVax. Triplet peaks were observed for IgG-GC132a, representing 0, 1 or 2 units of glycosylation. Increased HCD energy partially dissociates the 2:1 complex into a 1:1 complex, which verifies the identities of the observed components.
Fig. S3. HDX-MS analysis of the STEBVax-IgG-GC132a complex. (a) HDX plots of representative peptides from 6 CDR and 2 framework (non-CDR) regions on light (top) and heavy (bottom) chains of IgG-GC132a only (orange) or IgG-GC132a-STEBVax complex (black) samples. (b) HDX plots of representative peptides from STEBVax only (green) and STEBVax-IgG-GC132a complex (black). The data shown are the average of two replicates.