Abstract
Introduction:
EGFR-mutant lung cancers are clinically and genomically heterogeneous with concurrent RB1/TP53 alterations identifying a subset at increased risk for small cell transformation. The genomic alterations that induce lineage plasticity are unknown.
Methods:
Patients with EGFR/RB1/TP53-mutant lung cancers, identified by NGS from 2014–2018, were compared to patients with untreated, metastatic EGFR-mutant lung cancers without both RB1- and TP53-alterations. Time to EGFR-tyrosine kinase inhibitor (EGFR-TKI) discontinuation (TTD), overall survival, SCLC transformation rate, and genomic alterations were evaluated.
Results:
Patients with EGFR/RB1/TP53-mutant lung cancers represented 5% (43/863) of EGFR-mutant lung cancers but were uniquely at risk for transformation (18%, 7/39), with no transformations in EGFR-mutant lung cancers without baseline TP53 and RB1 alterations. Irrespective of transformation, patients with EGFR/TP53/RB1-mutant lung cancers had a shorter TTD than EGFR/TP53 and EGFR-mutant only cancers (9.5 vs 12.3 vs 36.6 months respectively, p = 2×10−9). The triple-mutant population had a higher incidence of whole genome doubling (WGD) compared to NSCLC and SCLC at large (80% vs 34%, p < 5×10−9; vs 51%, p < 0.002 respectively) and further enrichment in triple-mutant cancers with eventual small cell histology (7/7 pre-transformed plus 4/4 baseline SCLC vs 23/32 never transformed respectively, p = 0.05). AID/APOBEC mutation signature was also enriched in triple-mutant lung cancers that transformed (FDR = 0.03).
Conclusions:
EGFR/TP53/RB1-mutant lung cancers are at unique risk of histologic transformation, with 25% presenting with de novo SCLC or eventual small cell transformation. Triple-mutant lung cancers are enriched in WGD and AID/APOBEC hypermutation which may represent early genomic determinants of lineage plasticity.
Keywords: EGFR-mutation, TP53, RB1, Whole genome doubling, Small cell histologic transformation
Introduction
Twenty percent of lung adenocarcinomas harbor a sensitizing epidermal growth factor receptor (EGFR) mutation.1 Patients with EGFR-mutant lung cancers have robust responses to EGFR tyrosine kinase inhibitors (EGFR-TKIs) but inevitably their tumors acquire resistance.2 Multiple on-target and off-target mechanisms of acquired resistance to EGFR-TKIs have been described, including secondary mutations in EGFR and activation of other mitogenic signaling pathways.3–6 One particularly aggressive off-target resistance mechanism is transformation of EGFR-mutant lung adenocarcinoma to a small cell lung cancer.4, 7, 8 Small cell histologic transformation occurs in 3–14% of patients with sensitizing EGFR-mutant lung cancers after EGFR-TKI therapy.3, 4 The molecular determinants of lineage plasticity that underlie this histologic transformation are largely unknown.
Small cell lung cancers universally have bi-allelic loss of TP53 and RB1, among a complex genomic landscape that also includes alterations in SOX, NOTCH, PTEN, MYC, and PIK3CA9; alterations in receptor tyrosine kinases such as EGFR are exceedingly rare. Post-transformation, EGFR-mutant small cell lung cancers (SCLCs) continue to harbor the original EGFR-mutation indicating direct evolution from the original non-small cell lung cancer (NSCLC).4, 10, 11 After small cell transformation, the clinical outcomes mimic primary SCLC, with a rapid disease course and a transient response to SCLC-directed chemotherapies. Upon transformation and despite retention of the EGFR-mutation, EGFR protein expression decreases12 and patients have limited benefit from EGFR-TKIs.13 A parallel event occurs in prostate adenocarcinomas receiving androgen receptor-targeted therapy with functional loss of RB1 and TP53 facilitating small cell transformation and reducing sensitivity to anti-androgen therapy.14
Concurrent alterations within EGFR-mutant lung cancers may contribute to heterogeneous outcomes seen and influence the mechanism of resistance that emerges to EGFR-TKI treatment.15, 16 Two thirds of EGFR-mutant lung cancers have concurrent TP53-mutations which are associated with shorter time on EGFR-TKI and shorter overall survival (OS).3, 15, 16 RB1 alterations in EGFR-mutant lung cancers almost always occur concurrently with TP53. EGFR-mutant lung cancers with transformation mimic classical SCLC with RB1 and TP53 biallelic loss. It has not been fully determined whether RB1 and TP53 loss are early events within EGFR-mutant lung cancers, or alternatively, are acquired late in the process of histologic shift. RB1 and TP53 loss appear necessary, but not sufficient, to induce lineage plasticity.10
Single-gene alterations and mutation patterns have been reported in the context of lineage plasticity in other tumor types with alterations in PIK3CA, MYC, MDM, AURKA, FGFR, NOTCH, and TERT implicated in bladder and prostate cancers.14, 17 Beyond single genetic lesions, higher order patterns of mutations may drive or be associated with histological transformation. AID/APOBEC hypermutation signature, previously observed in lung adenocarcinoma,18 was noted to be further enriched in a cohort of lung cancer patients following SCLC transformation.10 Based on methods leveraging Memorial Sloan Kettering (MSK) Integrated Mutation Profiling of Actionable Cancer Targets (IMPACT) targeted next-generation sequencing, whole genome doubling (WGD), seen in nearly 30% of metastatic solid tumors including NSCLC (34%) and SCLC (51%), was found to be even more highly recurrent (72%) in small cell cancers of the bladder that have presumably transformed from urothelial carcinomas.17, 19
Lineage plasticity is an off-target adaptive mechanism to decrease dependence on EGFR signaling. We hypothesize that RB1 and TP53 alterations represent early events in oncogenesis, and that EGFR-mutant lung cancers with RB1/TP53 alterations are at particularly high risk for SCLC transformation. Patients with EGFR/RB1/TP53 mutant lung cancers are an ideal population in which to identify early genomic determinants of small cell histologic transformation.
Materials and Methods
We identified patients with somatic sensitizing EGFR-mutations with concurrent TP53 and RB1 alterations using MSK-IMPACT20 from January 2014 – August 2018. All cases with concomitant alterations in RB1 and TP53 were analyzed given the functional significance of some aberrations remain unknown. This study was undertaken at MSK with the approval of the Institutional Review Board. All patients provided written informed consent.
Time to treatment discontinuation (TTD) was used as a surrogate of progression-free survival and was defined as the time from start of EGFR-TKI to last administered dose before a treatment change. OS was defined as date of diagnosis of metastatic disease to date of death or last follow-up as of August 2018; we utilized the left truncation method to adjust for survival bias.21 For comparison, the clinical data were reviewed for all patients identified over the same time-period with EGFR-mutant metastatic lung cancers without concurrent RB1 and TP53 alterations that were EGFR-TKI naïve at the time of NGS. Differences in outcomes were analyzed using the Mann-Whitney test and the Fisher’s exact test was used to compare the proportions. Tumor mutation burden (TMB) was defined as the total number of missense mutations and indels divided by the total coding region captured and was reported as mutations/megabase (mutations/Mb). For survival and TTD analyses, Kaplan-Meier curves were compared using the Mantel-Cox log-rank test with hazard ratios calculated using a Mantel-Haenszel method. Relative risk was calculated using the Koopman asymptotic score. All patients with archival tissue underwent immunohistochemistry (IHC) for RB1 and TP53.
Single nucleotide variants (SNVs) and copy number variants (CNVs) identified by MSK IMPACT were schematized for analysis on the cBioPortal22. Analysis of gene set enrichment was performed using the DAVID pathway23 database with Fisher’s exact p-values adjusted for multiplicity by the Benjamini-Hochberg method. We used the FACETS algorithm (version 0.5.6)24 to perform allelic copy number analysis, including detection of loss of heterozygosity (LOH). Following previous methodology, we used allelic copy number estimates to define WGD as any instance where > 50% of the autosome contains major copy number (MCN) ⩾ 2 and compared the distribution of WGD to corresponding published distributions of NSCLC and SCLC samples.19 Mutation signature analysis was performed using deconstructSigs, with enrichment of mutation signature assessed by permuting sample labels (see Supplemental Methods).25
Results
Patient characteristics
We evaluated 4,112 patients with lung cancer, identifying 21% (n = 863) with EGFR-mutant lung cancer of whom 43 had concurrent sensitizing EGFR mutations along with both RB1 and TP53-alterations (Supplementary Fig. S1). All patients with the EGFR/RB1/TP53-mutant genotype had metastatic disease. In patients with EGFR-mutant lung cancers with concurrent RB1-alterations, only 11 (11/54) had a wild-type TP53 (Fisher’s exact p < 0.0001). None of these 11 patients had small cell transformation or PTEN loss. Clinical and molecular characteristics of the 43 patients with EGFR/RB1/TP53-mutant lung cancers are shown in Table 1 and Supplementary Table S1. Nine percent (4/43) had small cell histology at initial diagnosis; all were never-smokers (100%) and were comparatively younger than other patient’s whose tumors harbored an EGFR/RB1/TP53 alterations (55 vs 68 years old; Mann Whitney p = 0.009).
Table 1.
Demographics of patients with EGFR/RB1/TP53-mutant lung cancers by small cell lung cancer status during the course of their disease.
| Total | Never Transformed | Transformed SCLC | SCLC at Diagnosis | |
|---|---|---|---|---|
| (N=43) | (N=32) | (N=7) | (N=4) | |
| Median age (range) | 67 (25–86) | 68 (25–86) | 65 (58–73) | 55 (27–60) |
| Sex | ||||
| Female | 26 (60%) | 21 (66%) | 3 (43%) | 3 (75%) |
| Male | 17 | 11 | 4 | 1 |
| Smoking Status | ||||
| Never | 26 (60%) | 18 (56%) | 4 (57%) | 4 (100%) |
| Ever (median; range) | 17 (7; 0–30) | 14 (8; 0–30) | 3 (3; 1–9) | 0 (0) |
| Histology at diagnosis | ||||
| Adenocarcinoma | 39 (91%) | 32 (100%) | 7 (100%) | - |
| Small Cell | 4 | - | - | 4 |
| EGFR-Mutation | ||||
| L858R | 17 (40%) | 13 (41%) | 2 (29%) | 2 (50%) |
| Exon 19 Deletion | 22 (51%) | 16 (50%) | 5 (71%) | 1 (25%) |
| S768I/G719C | 2 | 1 | 0 | 1 |
| L861Q | 2 | 2 | 0 | 0 |
| Never EGFR-TKI | 4 (9%) | 1** (3%) | 0 | 3 (75%) |
| Initial EGFR-TKI used | - | - | - | - |
| Erlotinib | 28 (65%) | 22 (69%) | 6 (86%) | 0 |
| Gefitinib | 1 | 1 | 0 | 0 |
| Afatinib | 5 | 4 | 1 | 0 |
| Osimertinib | 5 | 4 | 0 | 1 |
| NGS showing | ||||
| EGFR/RB1/TP53 | 25 (58%) | 19 (59%) | 2 (29%) | 4 (100%) |
| Pre-EGFR-TKI | 18 | 13 | 5 | 0 |
| Progression on EGFR-TKI | ||||
| Median TMB* (range) | 6.4 (2.6–42.1) | 5.6 (2.6–42.1) | 7.9 (2.6– 16.7) | 6.6 (5.3– 7.9) |
SCLC: small cell lung cancer; TKI: tyrosine kinase inhibitor; NGS: next generation sequencing; TMB: tumor mutation burden.
mutations/megabase
1 never-transformed patient was lost to follow up
Of the patients with lung adenocarcinomas, 18% (7/39) had SCLC transformation during their disease course, and 82% have not had transformation (median follow up 3.2 years). The median time to transformation from start of initial EGFR-TKI was 1.1 years (interquartile range of 9 months to 3.6 years). There was no difference in smoking history in patients with or without histologic transformation to SCLC. Nineteen patients with EGFR/RB1/TP53-mutant lung cancers had available tissue for IHC analysis (7 transformed SCLC and 12 never transformed cases) demonstrating immunoprobing consistent with adenocarcinoma prior to transformation and consistent with SCLC post-transformation (Supplementary Fig. S2). Sixty five percent (28/43) had brain metastasis during their disease course: 18 at diagnosis and 10 developed on treatment. The median time to the development of brain metastases was 2.0 years (range 4.6 months - 6.4 years) from initial metastatic diagnosis.
For comparison, we identified 142 consecutively identified patients with EGFR-mutant metastatic lung cancer during the same period without concurrent RB1 and TP53 alterations (Supplementary Table S2). There were no differences in baseline clinical features between the EGFR/RB1/TP53-mutant group and control groups. There was enrichment for SCLC transformation in patients whose tumors harbored an EGFR/RB1/TP53-mutant genotype compared to EGFR/TP53-mutant RB1-wiltype (relative risk 3.5, 95% CI 2.1–7.7) and EGFR-mutant RB1/TP53-wildtype (relative risk 2.9, 95% CI 1.8–3.8); p = 0.001 (Supplementary Table S3).
Time to treatment discontinuation and overall survival
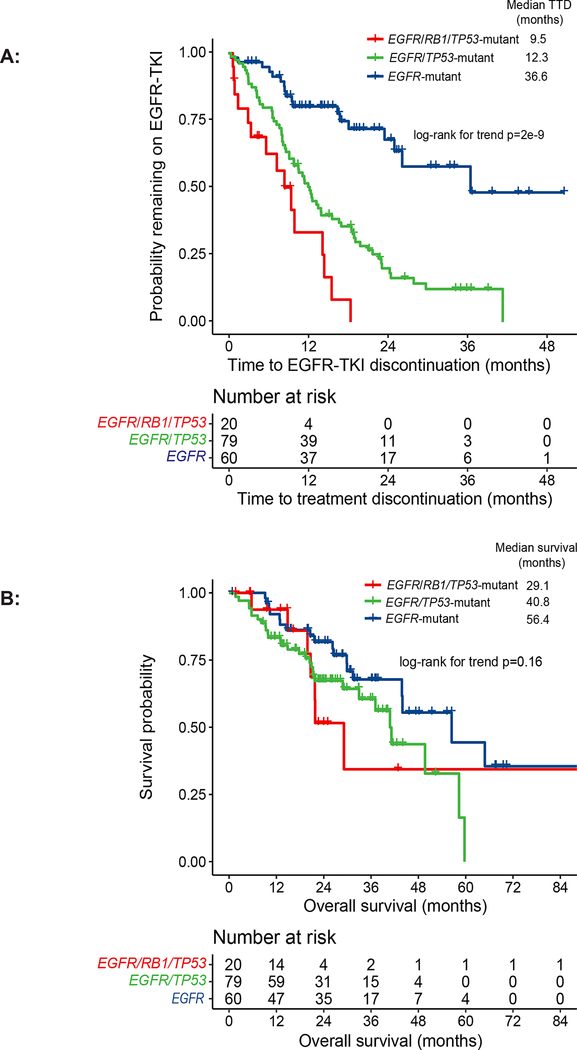
We identified the subset of patients that were EGFR-TKI naïve at the time of molecular testing and used time to EGFR-TKI discontinuation as an indicator of clinical benefit. Time to initial EGFR-TKI discontinuation, which includes treatment beyond radiographic progression, was 9.5 months in the EGFR/RB1/TP53-mutant cohort (n = 20), 12.3 months in the EGFR/TP53-mutant cohort (n = 79), and 36.6 months in the EGFR-mutant only cohort (n = 60; log-rank for trend p = 2e−9) (Fig. 1A). The median overall survival was 29.1 months for the EGFR/RB1/TP53-mutant cohort, 40.8 months for the EGFR/TP53 cohort, and 56.4 months for the EGFR-mutant RB1/TP53 wildtype cohort; log-rank for trend p = 0.16; Fig. 1B).
Figure 1.
Time to treatment discontinuation (TTD) and overall survival (OS) of patients with EGFR/RB1/TP53-mutant lung cancers: patients with EGFR/RB1/TP53-mutant lung cancer without baseline small cell lung cancer (SCLC) who were EGFR-TKI naïve at the time of next-generation sequencing (NGS) (n = 20) versus patients with EGFR/TP53-mutant RB1-wildtype (n = 79) and EGFR-mutant RB1/TP53-wild type lung cancer who were EGFR-TKI naïve at the time of NGS (A) The median TTD for patients with EGFR/RB1/TP53-mutant lung cancer was 9.5 months versus 12.3 months for EGFR/TP53-mutant RB1-wildtype (HR 2.0, 95% CI 1.1 – 3.6) versus 36.6 months in EGFR-mutant RB1/TP53-wiltype groups (HR 7.7, 95% CI 3.6 – 14.2; log-rank for trend p = 2e−9). (B) The median OS of patients with EGFR/RB1/TP53-altered lung cancer was 29.1 months as compared to 40.8 months in EGFR/TP53-mutant RB1-wildtype and 56.4 months in patients with EGFR-mutant RB1/TP53-wildtype (HR 1.0 95% CI 0.4 – 2.4, HR 1.8 95% CI 0.7 – 4.3, respectively; log-rank for trend p = 0.16).
Molecular analyses
Defining the genomic landscape of EGFR/RB1/TP53-mutant lung cancers
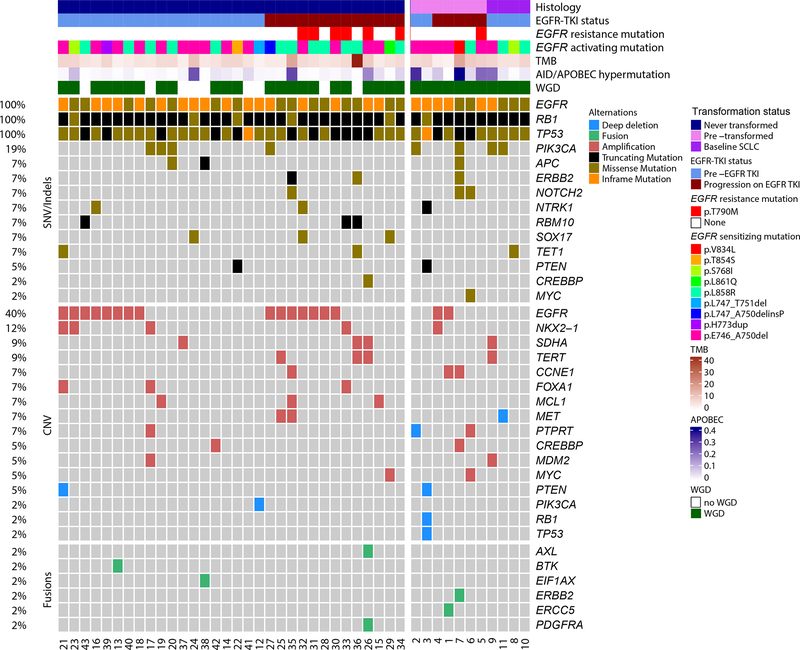
We performed integrated analysis of single nucleotide variants (SNVs), focal copy number variations (CNVs), and fusions in 43 patients with concomitant EGFR, RB1, and TP53 alterations. All had EGFR-sensitizing mutations, including L858R, L861Q, G719C, and in-frame exon 19 deletions (Fig. 2). To determine whether the wild-type allele of TP53 and RB1 was present, we estimated allelic copy number variants24 on samples in which the NGS data allowed (TP53 evaluable in 32, RB1 in 37 samples). The wild-type allele of TP53 was lost in 29 of 32 cases (6 by heterozygous loss and 23 by LOH). Similarly, the majority of RB1 point mutations occurred in conjunction with loss of wild-type allele (31 of 36; 6 by heterozygous loss and 25 by LOH) (Supplementary Fig. S3). Among the 29 samples that had both TP53 and RB1 allelic information available, all 5 samples that retained either TP53 or RB1 wild-type allele remained adenocarcinoma, compared to small cell histology in 5 of the 24 samples with biallelic functional loss of both genes. There was no significant difference in the frequency of biallelic wild-type TP53 or RB1 loss between samples before and after EGFR-TKI therapy. These data confirm functional inactivation of TP53 and RB1 in nearly all tumors. Pooling somatic mutations and copy number changes across all samples with concurrent EGFR/TP53/RB1 alterations, the most frequently co-altered genes were PIK3CA (20%), NTRK1 (11%), MCL1 (11%), NKX2-1 (11%), ERBB2 (9%), FOXA1 (9%), PLCG2 (9%), PTEN (9%), RBM10 (9%), SDHA (9%), SOX17 (9%), and TERT (9%)(Fig. 2).
Figure 2.
Genomic landscape of lung cancer with concurrent EGFR/RB1/TP53 mutations. The type of genetic alteration (missense, in-frame, truncating, amplification, deep (homozygous) deletion, fusion/intragenic alteration) is described in the legend. The frequency of mutations is noted on the right. Mutations present in at least 5% of cases were included in the figure, as well as PIK3CA, MYC, and CREBBP mutations given their known relevance in small cell lung cancer.
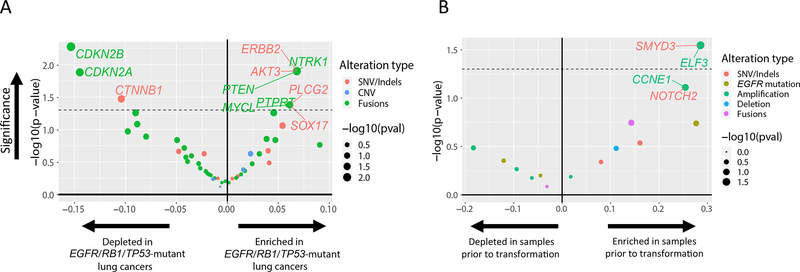
To identify the genomic features enriched in EGFR/RB1/TP53-mutant lung cancers, we compared them to our cohort of 142 EGFR-mutant lung cancers without concurrent RB1/TP53-mutations (Fig. 3A). The most enriched concurrent alterations in the triple mutant cohort included point mutations in ERBB2 (7% versus 0%), AKT3 (7% versus 0%), PLCG2 (7% versus 1%), and SOX17 (7% versus 1%), as well as copy number changes in PTEN loss (7% versus 0%), NTRK1 amplification (7% versus 0%), MYCL amplification (7% versus 1%), and PTPRT amplification (7% versus 1%); no comparisons were significant after adjusting for multiplicity. On the other hand, CTNNB1 mutations (2% versus 13%), CDKN2A (5% versus 19%) and CDKN2B (2% versus 18%) homozygous deletions, were underrepresented in the EGFR/RB1/TP53-mutant cohort compared to EGFR-mutant lung cancer without RB1 and TP53 mutations.
Figure 3.
Enrichment analysis of genomic alterations. (A) Enrichment of mutations in EGFR-mutant lung cancer with concurrent TP53/RB1 mutations versus without concurrent TP53/RB1 mutations. (B) Within EGFR/RB1/TP53-mutant lung cancer, enrichment of mutations in cases with eventual SCLC transformation. Level of enrichment is represented as a volcano plot with the log ratio in frequency between the two states (x-axis) and its significance -log(p-value) (y-axis). The type of alteration is represented by color. The dashed line represents p-value = 0.05.
Early genomic determinants of SCLC transformation in EGFR/RB1/TP53-mutant lung cancer
To assess early genomic determinants of SCLC transformation, we compared 7 samples prior to SCLC transformation to 32 samples that never underwent transformation (Fig. 3B). Candidate genes potentially enriched in cancers that eventually transformed compared to never-transformed cancers included SNVs in SMYD3 (29% versus 0%) and NOTCH2 (29% versus 3%), as well as amplifications in ELF3 (29% versus 0%) and CCNE1 (29% versus 3%) (Fig. 3B). Other recurrent alterations included PIK3CA (29% versus 13%), MYC (14% versus 0%), CREBBP (14% versus 3%), PTEN (14% versus 3%). None of these comparisons were significant after adjusting for multiplicity.
Beyond single genetic lesions, we assessed whether there were any biologically relevant gene sets that were enriched. We found that the 50 most enriched co-mutations in pre-transformed samples (Fisher’s p-value < 0.2) were overrepresented in a number of pathways, including MAPK signaling cascade (ARAF, ERBB2, FGFR1, FGFR3, FGFR4, GRIN2A, MAPK3, MYC) (FDR = 0.002); Jak-STAT signaling (CRLF2, IFNGR1, IL7R, SOCS1, MYC) (FDR = 0.04); ERBB signaling (ARAF, SRC, ERBB2, MAPK3, MYC) (FDR = 0.008); FGFR signaling (FGFR1, FGFR3, FGFR4, MAPK) (FDR = 0.04); MTOR signaling (BRAF, RRAGC, MAPK3, PIK3CG) (FDR = 0.01); and PI3K-Akt signaling (CCNE1, FGFR1/1/3, FLT4, IL7R, MAPK3, RAC1, MYC) (FDR = 0.003).
AID/APOBEC hypermutation in EGFR/RB1/TP53-mutant lung cancer as an early predictor of small cell transformation
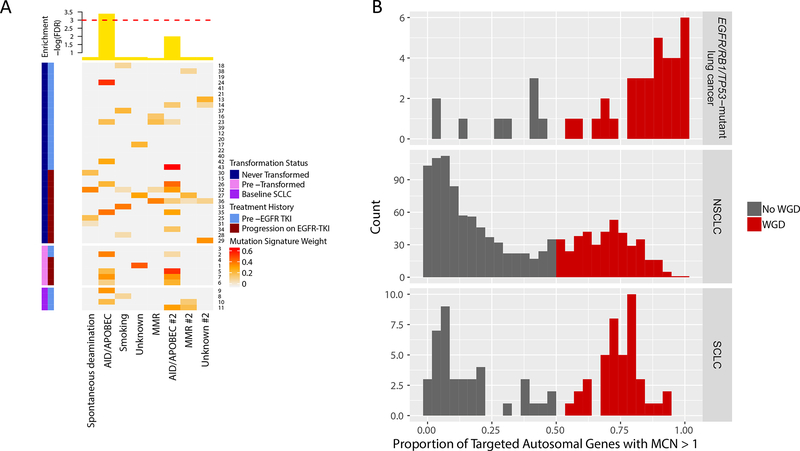
Considering EGFR/RB1/TP53-mutant tumors as a group, the distribution of substitutions identified through NGS showed a strong preference for C>T (24%) and G>A (18%) transitions consistent with cytidine deamination. (Supplementary Fig. S4). We investigated whether our cohort matched any of the 7 canonical mutation signatures known to be associated with lung cancers (signatures 1, 2, 4, 5, 6, 13, 15, and 17 corresponding to spontaneous deamination, AID/APOBEC hypermutation, smoking, unknown, mismatch repair, AID/APOBEC, mismatch repair, and unknown respectively). Prior to the transformation event, EGFR/RB1/TP53-mutant lung cancers destined for transformation were significantly enriched for AID/APOBEC signature compared to those that never transformed (Fig. 4A, FDR = 0.03).
Figure 4.
AID/APOBEC mutation signature and whole genome doubling (WGD) in EGFR/RB1/TP53-mutant lung cancers. (A) Mutation signature analysis identified significant enrichment of AID/APOBEC in pre-transformed SCLC compared to never transformed. Heatmap is calculated based on weights [0,1] measuring how strongly a mutation signature is represented in a given sample. Top annotation plots significance as -log(FDR) for enrichment of a mutation signature in pre-transformed SCLC. The dashed red line corresponds to an FDR of 0.05. (B) The frequency of WGD in lung cancer with concurrent EGFR/RB1/TP53 mutations is higher compared to all NSCLC and SCLC. Plots show the distribution of the proportion of autosomal genes with major copy number of at least 2 in lung cancers with concurrent EGFR/RB1/TP53 mutations, NSCLC, and SCLC respectively. Using a cutoff of 50%, the proportion of samples with WGD is colored in red, corresponding to a WGD frequency of 80% in lung cancer with concurrent EGFR/RB1/TP53 vs 34% in NSCLC (p < 5×10−9) vs 51% in SCLC (p < 0.002).
Whole genome doubling in EGFR/RB1/TP53-mutant lung cancer as an early predictor of small cell transformation
Beyond mutational and sub-chromosomal structural variants, we sought to analyze potential drivers of histologic transformation at the genome level. WGD is a frequent event in oncogenesis, associated with dysregulation of G2/M cell cycle checkpoints. By calculating the proportion of the autosome with major copy number of at least 2, we assessed for WGD in our triple-mutant cohort as well as all NSCLC and SCLC from Bielski, et al. for comparison.19 In contrast to the bimodal distribution of WGD in all lung cancers (Hartigan’s p < 2.2 × 10−16 rejecting unimodality), the EGFR/RB1/TP53-mutant cohort had a distinctly skewed unimodal distribution (Hartigan’s p = 0.94 accepting unimodality). Using 50% as a cutoff, the rate of WGD was elevated at 80% in our cohort compared to 34% of NSCLC (Fisher’s p-value < 5 × 10−9) and 51% of SCLC samples (Fisher’s p-value < 0.002) (Fig. 4B).
Within our cohort of patients with triple-mutant lung cancers, there was further enrichment of WGD in baseline SCLC or pre-transformation samples compared to never-transformed cancers: 4 out of 4 (100%) of baseline SCLC and 7 out of 7 (100%) pre-transformation cancers had WGD compared to 23 of 32 (72%) patients with never-transformed cancers (Fisher’s p-value = 0.05). Using clonality estimates inferred from FACETS analysis, the timing of TP53 and RB1 mutations was assessed in relation to WGD. Of 24 samples that underwent WGD and had sufficient data for unambiguous timing, TP53 mutations preceded WGD in 96% (23/24), and RB1 mutations preceded WGD in 71% of cases (17/24). Only in one case did both TP53 and RB1 mutations follow WGD. Using clonality estimates for allelic copy number variants, we found that 65% of all heterozygous deletions followed a WGD event in our cohort, similar to previously reported rates.19
Discussion
Small cell histologic transformation occurs in the subset of patients with EGFR-mutant lung cancers with concurrent alterations in TP53 and RB1 and is associated with large-scale genomic alterations including both WGD and the APOBEC mutation signature. Patients with co-occurring EGFR/RB1/TP53 alterations (5% of all EGFR-mutant lung cancers) are uniquely at risk for SCLC transformation during their disease course. While not all EGFR/RB1/TP53-mutant lung cancer patients will transform to SCLC during their disease, our observation agrees with previously published findings that RB1/TP53-mutations are necessary, but not sufficient, for lineage plasticity.10 All cases of SCLC transformation occurred among patients with pre-existing mutations in TP53 and RB1 and the frequency of histologic transformation is 6-fold higher in this cohort compared to the EGFR-mutant lung cancer population at large (18% vs 3%).15 Patients with co-occurring EGFR/RB1/TP53-altered NSCLC also have shorter time on EGFR-TKI therapy (p = 0.0007) which was similar to published data by Marcoux et al8 for patients with known histologic transformation. The 18% rate of SCLC transformation (7/39 adenocarcinoma at baseline patients) and shorter time on EGFR-TKI highlight the poorer outcomes seen in this genomic subset of EGFR-mutant lung cancers.
Given the unique clinical features of EGFR/RB1/TP53-mutant lung cancers, we sought to characterize the genomic features that define this cohort. Fewer EGFR T790M mutations were identified among the samples that ultimately transformed (1/7 pre-transformed versus 6/34 never-transformed versus 32/68 relapsed EGFR-mutant samples without concomitant RB1 and TP53 mutations). Concurrent EGFR amplification was also less frequently seen in the pre-transformation and baseline SCLC samples (2/7 pre-transformed versus 15/34 never-transformed versus 42/68 relapsed EGFR-mutant samples without concomitant RB1 and TP53 mutations). Relative absence of EGFR T790M and EGFR amplification is consistent with the loss of EGFR dependence in transformed SCLC.12 That we observe this pattern in pre-transformed samples suggests that loss of EGFR dependence may begin prior to overt histologic transformation.
A number of mutations commonly seen in SCLC are enriched in our cohort compared to EGFR-mutant lung cancer without TP53 and RB1 loss (PIK3CA, PTEN, MYCL), as well as in EGFR/RB1/TP53-mutant samples with eventual transformation compared to those that never transformed (PIK3CA, MYC, CREBBP, PTEN, NOTCH2).8, 10 While SOX17 frequently found in our EGFR/RB1/TP53-mutant cohort has not been directly implicated in neuroendocrine transformation, other SOX family members, including SOX2, play a role in small cell transformation seen in RB1/TP53-mutant prostate cancer.26 In addition, a number of these highlighted alterations are overrepresented in key canonical pathways activated in classical SCLC, including the PI3K-PTEN-AKT pathway.27 These genetic alterations present prior to SCLC transformation represent potential early biomarkers of lineage plasticity.
We also assessed the mutational signatures of these triple mutant lung cancers and found an enrichment of the AID/APOBEC hypermutation signature in EGFR/RB1/TP53-mutant tumors destined for SCLC transformation. This corroborates the increased APOBEC hypermutation seen in post-transformation SCLC cases by Lee et al.10 The enrichment in the adenocarcinomas that eventually transform suggest this hypermutation occurs even prior to transformation and may facilitate the transformation process. Considering its prevalence in SCLC and NSCLC, we assessed for WGD within our cohort, and found a markedly higher prevalence of WGD when compared to previously published WGD rates of lung cancers.19,28 We note that other publications use alternative WGD calling methods,28, 29 but for consistency, we use here the method from Bielski, et al. based specifically on the MSK IMPACT targeted next-generation sequencing platform.
Moreover, there was further enrichment of WGD in samples with histologic transformation, seen prior to the transformation event. Given the role of TP53 and RB1 loss in genomic instability, we assessed the timing of these mutations relative to WGD and found that in all but one case, TP53 mutations and RB1 alterations preceded WGD, suggesting that these lesions may predispose to WGD. While it remains unclear whether WGD itself facilitates transformation, there is a precedent for WGD providing a vehicle for evolutionary change as well as conferring tolerance to further chromosomal instability in other cancers, as evidenced by the higher number of heterozygous deletions following WGD in our cohort.19 To our knowledge, this is the first report suggesting a role for WGD in histologic transformation.
Our study has several limitations. The relatively small number of patients identified with concurrent EGFR/RB1/TP53 alterations limits comparative analyses. Most patients in our cohort received first-line erlotinib or afatinib. Now that osimertinib is the standard initial treatment, we need to assess whether the EGFR-TKI used affects the frequency of transformation. As we treat our patients with more potent and selective EGFR inhibitors, we suspect the frequency of off-target resistance mechanisms such as small cell transformation will increase. Due to the exploratory nature of our analyses, co-mutation analysis was not limited by FDR correction and findings will ultimately need to be validated in larger datasets. Although analysis of pre-transformation samples is valuable to identify early-determinants of lineage plasticity, analysis of paired pre- and post-transformation samples is critical to understand what factors may induce transformation in each specific case. All available tissue samples from the EGFR/RB1/TP53-mutant cohort were evaluated for RB1 and TP53 loss by IHC, however, given limited tissue availability a comprehensive analysis was not feasible.
After initial EGFR-TKI response, a persister cell population remains that serves as the reservoir for eventual clinical progression. Prior trials have evaluated the role of combinatorial first- and second-generation EGFR-TKIs with cytotoxic chemotherapy (pemetrexed or taxol based) in EGFR-mutant lung cancer patients regardless of co-mutation status, showing a potential benefit with the use of the combination.30 Based on those prior studies, we hypothesize that in patients with EGFR/RB1/TP53-mutant lung cancers, the persister cell population might include the subclone that is predisposed to small cell histologic transformation and may benefit from the combination of a potent EGFR-TKI and a neuroendocrine-based chemotherapy regimen. To eradicate this subclone, we have developed a trial of upfront osimertinib and small cell directed chemotherapy (platinum/etoposide) in patients with triple mutant lung cancers ().
In conclusion, patients with EGFR/TP53/RB1-mutant lung cancers demonstrate inferior clinical outcomes and define the population at risk for SCLC transformation. We have identified both small- and large-scale genomic alterations potentially associated with SCLC transformation. This work highlights the importance of understanding genomic heterogeneity to identify high-risk patients that may benefit from treatment intensification and to predict and subsequently prevent mechanisms of resistance to EGFR targeted therapy likely to emerge.
Supplementary Material
Supplemental Methods.docx
Supplemental Tables and Figures.docx
Acknowledgements:
This work was supported by the National Cancer Institute of the National Institutes of Health (P01 CA129243, T32 CA009207, P30 CA008748). The funding source played no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the article for publication. We acknowledge Craig Bielski for providing whole genome doubling calls in unselected lung cancer samples for comparison.
Michael Offin received consulting fees from PharmaMar.
Gregory J. Riely received research funding from Novartis, Roche, Genentech, Millenium, GlaxoSmithKline, Pfizer, Infinity Pharmaceuticals and ARIAD. He has received travel expenses from Merck Sharp & Dohme. He is listed as inventor on a patent application submitted for pulsatile use of erlotinib to treat or prevent brain metastases.
Matthew Hellman receives research funding from Bristol-Myers Squibb; is a paid consultant to Merck, Bristol-Myers Squibb, AstraZeneca, Genentech/Roche, Janssen, Nektar, Syndax, Mirati, and Shattuck Labs; receives travel support/honoraria from AztraZeneca and BMS; and a patent has been filed by MSK related to the use of tumor mutation burden to predict response to immunotherapy (PCT/US2015/062208), which has received licensing fees from PGDx.
Maria Arcila received speaker’s fees from Raindance Technologies.
Dr. Ladanyi reports grants from National Institutes of Health/National Cancer Institute, during the conduct of the study; grants from LOXO Pharmaceuticals, personal fees from Astra-Zeneca, personal fees from Bristol-Myers Squibb, personal fees from Takeda, personal fees from Bayer, personal fees from Merck, and grants from Helsinn Therapeutics, outside the submitted work.
Charles Rudin is a consultant for Abbvie, Amgen, Ascentage, AstraZeneca, Bicycle, Celgene, Chugai, Daiichi Sankyo, Genentech/Roche, GI Therapeutics, Loxo, Novartis, Pharmamar, and Seattle Genetics, and serves on the Scientific Advisory Boards of Elucida and Harpoon.
Mark G. Kris has received consulting fees from AstraZeneca, Pfizer, and Regeneron. He has received honoraria for participation in educational programs from WebMD, OncLive, Physicians Education Resources, AstraZeneca, and Research to Practice. Dr. Kris is an employee of Memorial Sloan Kettering. Memorial Sloan Kettering has received research funding from Genentech Roche and PUMA Biotechnology for trials conducted by Dr. Kris. Memorial Sloan Kettering has a collaboration for the development of Watson for Oncology with IBM and receives royalties from IBM for this activity.
Helena Yu is a consultant for AstraZeneca and has received travel support from Lilly. Her institution, Memorial Sloan Kettering has received research funding from Astellas Pharma, AstraZeneca, Daiichi, Lilly, Novartis and Pfizer for clinical trials she is involved in. She is listed as inventor on a patent application submitted for pulsatile use of erlotinib to treat or prevent brain metastases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Prior Publications:
This manuscript, and the research represented within, has not been published, posted, nor submitted in any other venue.
Conflict of Interest Statement:
All other authors have declared no relevant conflicts of interest.
References
- 1.Jordan EJ, Kim HR, Arcila ME, et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov 2017;7:596–609. 10.1158/2159-8290.cd-16-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113–125. 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- 3.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240–2247. 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piotrowska Z, Isozaki H, Lennerz JK, et al. Landscape of Acquired Resistance to Osimertinib in EGFR-Mutant NSCLC and Clinical Validation of Combined EGFR and RET Inhibition with Osimertinib and BLU-667 for Acquired RET Fusion. Cancer Discov 2018;8:1529–1539. 10.1158/2159-8290.CD-18-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le X, Puri S, Negrao MV, et al. Landscape of EGFR-Dependent and -Independent Resistance Mechanisms to Osimertinib and Continuation Therapy Beyond Progression in EGFR-Mutant NSCLC. Clin Cancer Res 2018;24:6195–6203. 10.1158/1078-0432.ccr-18-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zakowski MF, Ladanyi M, Kris MG. EGFR mutations in small-cell lung cancers in patients who have never smoked. N Engl J Med 2006;355:213–215. 10.1056/NEJMc053610. [DOI] [PubMed] [Google Scholar]
- 8.Marcoux N, Gettinger SN, O’Kane G, et al. EGFR-Mutant Adenocarcinomas That Transform to Small-Cell Lung Cancer and Other Neuroendocrine Carcinomas: Clinical Outcomes. J Clin Oncol 2019;37:278–285. 10.1200/jco.18.01585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47–53. 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JK, Lee J, Kim S, et al. Clonal History and Genetic Predictors of Transformation Into Small-Cell Carcinomas From Lung Adenocarcinomas. J Clin Oncol 2017;35:3065–3074. 10.1200/jco.2016.71.9096. [DOI] [PubMed] [Google Scholar]
- 11.Hanna N, Bunn PA Jr, Langer C, et al. Randomized phase III trial comparing irinotecan/cisplatin with etoposide/cisplatin in patients with previously untreated extensive-stage disease small-cell lung cancer. J Clin Oncol 2006;24:2038–2043. 10.1200/JCO.2005.04.8595. [DOI] [PubMed] [Google Scholar]
- 12.Niederst MJ, Sequist LV, Poirier JT, et al. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun 2015;6:6377 10.1038/ncomms7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oxnard GR, Hu Y, Mileham KF, et al. Assessment of Resistance Mechanisms and Clinical Implications in Patients With EGFR T790M-Positive Lung Cancer and Acquired Resistance to Osimertinib. JAMA Oncol 2018;4:1527–1534. 10.1001/jamaoncol.2018.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mu P, Zhang Z, Benelli M, et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 2017;355:84–88. 10.1126/science.aah4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu HA, Suzawa K, Jordan E, et al. Concurrent Alterations in EGFR-Mutant Lung Cancers Associated with Resistance to EGFR Kinase Inhibitors and Characterization of MTOR as a Mediator of Resistance. Clin Cancer Res 2018;24:3108–3118. 10.1158/1078-0432.ccr-17-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aggarwal C, Davis CW, Mick R, et al. Influence of TP53 Mutation on Survival in Patients With Advanced EGFR-Mutant Non–Small-Cell Lung Cancer. JCO Precis Oncol 2018;2018:1–29. 10.1200/po.18.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang MT, Penson A, Desai NB, et al. Small-Cell Carcinomas of the Bladder and Lung Are Characterized by a Convergent but Distinct Pathogenesis. Clin Cancer Res 2018;24:1965–1973. 10.1158/1078-0432.ccr-17-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGranahan N, Favero F, de Bruin EC, et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med 2015;7:283ra254 10.1126/scitranslmed.aaa1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bielski CM, Zehir A, Penson AV, et al. Genome doubling shapes the evolution and prognosis of advanced cancers. Nat Genet 2018;50:1189–1195. 10.1038/s41588-018-0165-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 2015;17:251–264. 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalbfleisch J, Prentice R. The statistical analysis of failure time data. New York: J. Wiley; 2002. [Google Scholar]
- 22.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 24.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 2016;44:e131 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenthal R, McGranahan N, Herrero J, et al. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 2016;17:31 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ku SY, Rosario S, Wang Y, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 2017;355:78–83. 10.1126/science.aah4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Umemura S, Mimaki S, Makinoshima H, et al. Therapeutic priority of the PI3K/AKT/mTOR pathway in small cell lung cancers as revealed by a comprehensive genomic analysis. J Thorac Oncol 2014;9:1324–1331. 10.1097/jto.0000000000000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez S, Lim E, Huebner A, et al. Whole Genome Doubling mitigates Muller’s Ratchet in Cancer Evolution. 2019:513457 10.1101/513457. [DOI] [Google Scholar]
- 29.Dewhurst SM, McGranahan N, Burrell RA, et al. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov 2014;4:175–185. 10.1158/2159-8290.Cd-13-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seike M, Inoue A, Sugawara S, et al. 1382PDPhase III study of gefitinib (G) versus gefitinib+carboplatin+pemetrexed (GCP) as first-line treatment for patients (pts) with advanced non-small cell lung cancer (NSCLC) with EGFR mutations (NEJ009). Annals of Oncology 2018;29 10.1093/annonc/mdy292.005. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Methods.docx
Supplemental Tables and Figures.docx