Abstract
Although long overlooked, it is now well understood that DNA does not systematically assemble into a canonical double helix, known as B-DNA, throughout the entire genome but can also accommodate other structures including DNA hairpins, G-quadruplexes and RNA:DNA hybrids. Notably, these non-canonical DNA structures form preferentially at transcriptionally active loci. Acting as replication roadblocks and being targeted by multiple machineries, these structures weaken the genome and render it prone to damage, including DNA double-strand breaks (DSB). In addition, secondary structures also further accumulate upon DSB formation. Here we discuss the potential functions of pre-existing or de novo formed nucleic acid structures, as bona fide repair intermediates or repair roadblocks, especially during Transcription-Coupled DNA Double-Strand Break repair (TC-DSBR), and provide an update on the specialized protein complexes displaying the ability to remove these structures to safeguard genome integrity.
Keywords: DNA Double-strand break Repair, RNA:DNA hybrid, R-loop, G-quadruplex, Transcription, Chromatin
Non-canonical DNA structures mainly accumulate in the genome at transcribed loci
The DNA double helix is naturally structured as a right handed helix known as B-DNA [1]. Yet, in specific contexts, non-canonical DNA secondary structures can also form, including R-loops (composed of a RNA:DNA hybrid and the displaced ssDNA) (reviewed in [2]) and intra- or inter-molecular G-quadruplexes (G4), which can adopt various topologies (reviewed in [3]). For instance, negative supercoils behind the transcription machinery melt the DNA double helix, generating ssDNA further available for G4 or/and RNA:DNA hybrids assembly (for review [4]). In that respect, transcribed GC-skewed sequences, by both providing a substrate for intramolecular G4 formation and a C-rich DNA strand template that can stably hybridize with the complementary G-rich RNA transcript, are particularly prone to R-loop formation (reviewed in [5]).
While DNA secondary structures have been experimentally reported in vitro, and predicted to form across the genomes for decades, it is only recently that their existence in vivo is no longer debated, owing to the explosion of available probes (ligands and antibodies) [3,6-11], In particular, the use of antibodies directed against G4 (BG4) and RNA:DNA hybrids (S9.6), or a mutant form of RNase H, have recently allowed genome-wide maps of the distribution of G4 and R-loops across the genome [12-17], Of interest, these studies revealed that secondary DNA structures distribution is strongly biased toward transcriptionally active loci. On mammalian genomes, R-loops form co-transcriptionally, mainly accumulating at 5’ and 3’ ends of GC-skewed, active, hypomethylated genes [12,13,16,18], In budding yeast genomes, R-loops were also found to accumulate on dA:dT tracks [15], although it is yet unclear whether these loci are also prone to R-loops in higher eukaryotes. On the other hand, G4s tend to form on hypomethylated promoter regions with a bias for genes exhibiting RNA Polymerase II (Pol II) pausing [14,19].
The strong conservation of these secondary structures across evolution [20,21] suggest that they fulfill important genome regulatory functions. These structures not only impose physical constraints on the DNA double helix, thereby directly promoting or counteracting the activity and processivity of machineries on DNA, but can also act as recruitment platforms for proteins involved in DNA, RNA and chromatin metabolism. Mounting evidence has established their control of essential genetic processes including transcription, replication, telomere maintenance and Class Switch Recombination. Given that this has been extensively reviewed elsewhere (see [5,22-25]), we will provide a few recent examples below.
R-loops and G4s both tightly control transcription. Indeed, R-loops can promote surrounding chromatin changes, for example H3K9 dimethylation over the terminator 3′ end regions of human genes allowing efficient termination [26], or PcG protein recruitment on a subset of regulated genes to ensure transcriptional repression [27]. G4 structures can also act as docking sites for transcription factors, as on the promoters of c-myc, hTERT or p21 where G4 motifs are required for regulating transcriptional activity [28-30], and contribute in regulating the local epigenomic landscape by sequestrating and inhibiting DNA methylation [19].
Beyond regulating transcription, multiple pieces of evidence also place G4s and R-loops as direct players in priming replication (for review [23,31]). This role was actually the first biological function proposed for bacterial R-loops [32] and more recently, in eukaryotes, persistent RNA:DNA hybrids were found to initiate DNA synthesis in ribosomal DNA in a replication origin-independent manner [33]. Genome-wide mapping of human replication origins also showed that the majority of constitutive origins are associated with CpG island promoters, and more precisely with G4 motifs [34,35]. Further evidence for a role of G4 in priming replication in mammals came from the report of the preferential binding of origin recognition complex (ORC) to these structures [36] and with the observation that some G4 motifs are required for efficient origin activation [37].
Finally, DNA secondary structures also initiate Class Switch Recombination (CSR) in antigen-activated B cells. CSR occurs at the Immunoglobulin heavy chain (IgH) locus and results in a deletion event after recombination between two Switch sequences, the whole process being initiated by the single-strand DNA-specific cytidine deaminase AID (for review, [38]). Mammalian Switch regions were among the first described G-rich R-loop forming sequences [39] and were shown by Electron Microscopy to produce so-called “G-loops”, which contain both a G4 DNA on one strand and a stable RNA:DNA hybrid that forms co-transcriptionally on the other strand [40]. It has long been known that these secondary structures were necessary to initiate CSR but it is only recently that G4 structures were identified as the preferred recognition substrates for AID binding [41].
While R-loops and G4 structures accumulate over transcribed loci and play important biological functions during several essential genetic processes, the assembly of these structures provide unique challenges for genome integrity.
Hyper-sensitivity of non-canonical DNA structures to breakage
R-loops, G4 and other DNA hairpins are very potent inducers of endogenous DNA damage, especially DSBs, a notion supported by an incredibly extensive amount of evidence both in yeast and mammals (reviewed in [42-44]). Yet, once again it is only recently that DSB distribution across the genomes has started to be characterized at the molecular level thanks to genome-wide methodologies. These include ChIP-seq against DSB repair proteins (such as [45]), methods measuring the outputs of DNA transactions following break production (HTGTS, Strand-seq, or GUIDE-seq [46-49]) or techniques allowing to directly infer the position of DNA ends themselves at a nucleotide resolution such as BLESS, BLISS, DIG-seq, End-seq, DSB-capture and more recently BrlTL [46,50-55]. These approaches (reviewed in [56]) have recently provided valuable insights regarding the genome wide distribution of DSBs, in various organisms and experimental conditions (see Table 1). Altogether, these studies pointed toward DNA secondary structures as prominent (although not systematic) hallmarks of DSB production. Indeed, endogenous DSBs accumulate in transcribed loci and active Transcriptional Start Sites (TSS) [48,57-64], with a bias toward those prone to form G4s [49,50,65]. Additionally, DSBs levels further increase at these loci in conditions where secondary structures accumulate, for instance upon depletion of the BLM helicase or treatment with pyridostatin both triggering G4 stabilization [11,49], as well as in yeast strains unable to unwind RNA:DNA hybrids, where DSBs precisely occur at the 3’end of a subset of R-loops [66]). Finally, upon replication stress, DSBs also accumulate on loci prone to form secondary structures such as active gene-enriched loci, repeated sequences and dA:dT long stretches [45,52,67]. Hence, altogether these new insights regarding DSB distribution across the genome revealed a compelling connection between DNA secondary structures and DNA breakage (Table 1).
Table 1.
Genome-wide DSB mapping: DSBs are essentially distributed over DNA secondary structure-forming sequences.
| DSBs distribution | Organism | Experimental condition1 | Method | Study2 |
|---|---|---|---|---|
| Active TSSs | mouse | WT and AID−/− B cells | HTGTS | [48] |
| Active genes, TSSs | mouse | WT and AID−/− B cells | TC-seq | [60] |
| Predicted G4-containing gene bodies | human | pyridostatin | γH2AX ChIP-seq | [11] |
| Highly transcribed genes, repeated sequences and GC-rich regions | human | none and hydroxyurea (replication stress), WT, XRCC2−/−53BP1−/− cells | RPA, γH2AX, BRCA1, SMC5 ChIP-seq | [45] |
| Alpha-type satellite repeats prone to hairpins, CFSs and long active genes | human | none and aphidicolin (replication stress) | BLESS | [52] |
| Promoters of high and medium expressed genes | human | etoposide | DSB-seq | [64] |
| Preferentially occur at genes transcriptionally induced by HU | yeast | hydroxyurea, WT and Mec1 mutant cells | Break-seq | [159] |
| Promoter/TSS of active genes | mouse | none, etoposide, doxorubicin and aclarubicin | BLESS | [61] |
| Breakpoint hotspots near centromeric or telomeric regions | human | none | GUIDE-seq | [46] |
| H3K4me3 enriched loci (specific mark of active gene promoters) | human | none | RAFT | [62] |
| Topo IIβ-induced DSBs at promoter regions of neuronal early-response genes | mouse | etoposide, Top2b shRNAs | γH2AX ChIP-seq | [160] |
| Active TSSs | mouse | Neural Stem Cell Progenitors, WT, Xrcc4−/− p53−/− and ATM −/− | HTGTS | [59] |
| Within long, actively transcribed and late replicating neural genes | mouse | Neural Stem Cell Progenitors, WT, Xrcc4−/− p53−/−, aphidicolin (replication stress) | LAM-HTGTS | [58] |
| G4-forming sequences, CTCF binding sites, highly transcribed loci, 5’UTRs, promoters and TSSs | human | none | DSB-capture | [50] |
| RSS heptamer cleavage sites at Ig and TCR loci and RAG off-targets at cryptic RSSs | mouse | ATM −/−, Lig4−/− | END-seq | [51] |
| TSSs and along the gene body of highly expressed genes | human, mouse |
etoposide | BLISS | [53] |
| Loop anchors (CTCF and Top2 binding sites) | mouse | none and etoposide | END-seq | [161] |
| CFSs (very long active genes) and ERFSs | chicken | aphidicolin (replication stress) | FANCD2 ChIP-seq | [57] |
| G4 motifs on active genes (higher on transcribed DNA strands) and at promoters | mouse, human |
BLM−/− cells | Strand-seq (SCE mapping) | [49] |
| rDNA and 3' end of R-loops | yeast | Sen1 and Rnases-H Δ triple strain | Rad52 ChIP-seq | [66] |
| Preferentially within introns and repeated sequences | mouse | differentiating spermatids | Breakome | [162] |
| Intergenic, replication origins, single-strand and unprotected poly(dA:dT) tracts within ERFSs, CFSs and rDNA, prone to form non-B DNA structures | mouse, human |
none and hydroxyurea (replication stress) | END-seq | [67] |
| G4-forming sequences | yeast | none and Pif1 mutant cells | i-BLESS | [65] |
| CTCF binding sites, repetitive DNA that form stable intrastrand secondary structures (microsatellites, inverted retroelement repeats, and quasi-palindromic AT-rich repeats) | mouse, human |
ATR inhibitor, aphidicolin, TIMELESS shRNAs | RPA ChIP-seq and BrITL | [55] |
(genetic background or drug treatment)
(references in chronological order)
Several mechanisms account for DSBs production in non-canonical DNA structures. First, all these structures are strong roadblocks for DNA replication. Replication fork collisions with these structures and/or with the RNA polymerase machinery itself stalled at these structures can further trigger fork collapse, leading to DSBs (reviewed in [68-70]). Second, these structures are preferential targets for nucleases and other enzymes inducing DNA modifications that can further be converted into DSBs. One of the best illustration is found on the IgH switch region, where both G4 and R-loop accumulate (see above, [40,71,72]) and directly contribute to recruit AID, triggering deamination of the ssDNA strand cytidines and further DSB production to ensure CSR [41]. In addition, R-loops are also direct targets for the endonucleases XPG and XPF [73] or CtIP [74], which allow the resolution of these structures thanks to the production of ssDNA nicks, that can further be converted into a DSB [73-76]. In that respect, DSBs precisely map at the 3’end of a subset of R-loops in yeast strains lacking Sen1 and RNAse H1/2 [66], further illustrating nicely the tight relationship between R-loops and DSB production.
No matter the mechanism(s) involved, it is hence clear that the DNA secondary structures R-loops and G4s coincide on the genome with DSBs, especially at transcribed loci. One of the questions that arise therefore is how do repair mechanisms accommodate these potentially dangerous structures to ensure faithful genomic information recovery. Recently, a part of this answer was provided by the identification of specialized repair mechanisms that operate in actively transcribed loci, which we describe in the next section.
Repair of broken transcriptionally active loci: a complex process
Several pathways ensure repair of DSBs in higher eukaryotes [77]. On one hand, classical Non-Homologous End-Joining (NHEJ) displays the ability to rapidly reseal both DNA ends in a manner that depends on the Ku heterodimer-binding to DNA ends and sealing of the ends by XRCC4/Ligase 4 complex. While generally faithful, this pathway can trigger localized (few bp) mutations. NHEJ is also the main pathway involved in generating translocations or distant End joining (such as during CSR). On the other extreme hand lies the Homologous Recombination (HR) repair pathway, which uses an intact copy of the broken locus as a template, providing highly accurate repair when operating on unique sequences in S and G2 cell cycle phases when a sister chromatid template is available. Yet, HR can also trigger dramatic genomic events when improperly controlled, for example when operating on repeated sequences or homologous chromosomes. Consequently, this pathway is severely downregulated during the G1 cell cycle phase when no sister chromatid is available [78]. In addition to these two main DSB pathways, other mechanisms can ensure the restoration of the linearity of DNA, including alternative NHEJ pathway(s) and Single-Strand Annealing. Resection at DNA ends (i.e the generation of ssDNA through the action of endo and exonucleases) is a critical step which lies at the heart of choosing which DSB repair pathway to engage and which strongly influences the accuracy of the final repair product [79]. Of interest, specialized mechanisms ensure DSB repair of damaged active genes: first the damaged genes (and potentially genes within close proximity to the break) undergo a transcriptional arrest, which is further required to ensure proper repair by a yet unclear repair pathway.
Tuning down surrounding transcriptional activity
It has been observed that DNA breaks located in active genes result in repression of transcription locally and transiently surrounding the break. These activities following DNA damage may function to avert potential conflicts that may exist between two DNA-templated processes, transcription and DNA repair. In eukaryotes, the transcription cycle is a highly regulated process from initiation, to elongation, termination and Pol II recycling [80]. Repression of transcription following DNA damage relies heavily on direct alterations of chromatin and Pol II, triggered by the DNA Damage Response (DDR) kinases ATM [81] and DNA-PK [82], as well as the DDR Poly(ADP-ribose) polymerase (PARP1) enzyme [83] (Fig. 1).
Figure 1. Repression of transcription at damage-associated loci.
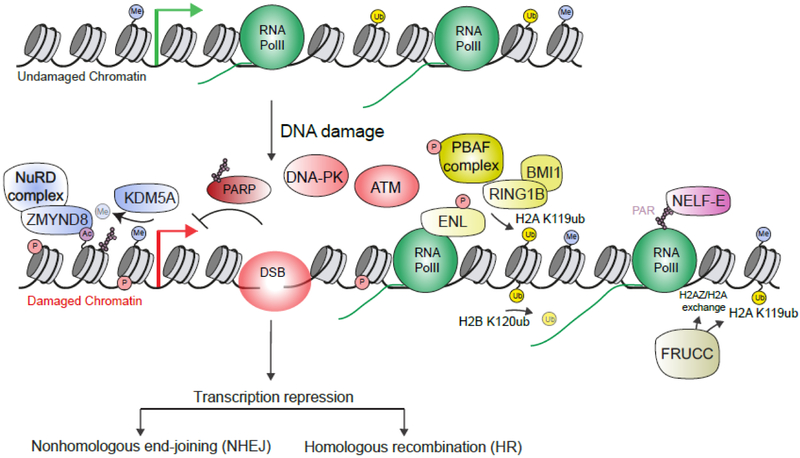
Undamaged chromatin is decorated by histone modifications that promote transcription. Upon DSB formation, chromatin is highly modified and participates along with pre-existing marks to facilitate repressive chromatin and altered RNA Pol II, which collectively tune down transcription to coordinate repair activities within these damage gene loci. DSB – DNA double-strand break.
On one hand, evidence suggests that the Pol II cycle is directly affected by a nearby DSB. Indeed, using a powerful reporter assay to monitor transcription in single cells following DNA damage, it was observed that the Ser2 of the Pol II CTD (associated with productive elongation) is dephosphorylated in an ATM dependent manner [81]. Interestingly, DSB-induced ATM activation also triggers the phosphorylation of the transcription elongation factor ENL, a component of the super elongation complex, which regulates Pol II elongation. ENL phosphorylation is further required for silencing transcription [84]. In addition, it was also reported that DNA-PK promotes the removal of Pol II from damage sites in a proteasome-dependent manner [82]. Finally, the pausing, negative elongation factor NELF complex, which contains NELF-E, is recruited to DNA damage sites and represses transcription in a PARP1-dependent manner [85]. NELF-E binds PAR directly through a domain in its N-terminus, explaining the potential role of PARP in regulating NELF complex. Interestingly, NELF-E also requires Pol II to associate with DNA damage, suggesting that ADP-ribosylation of Pol II may act as the signal to recruit NELF to damage sites. NELF-E binding to nascent transcripts is also required as mutation of the RNA binding domain in NELF-E was not able to support its repressive activity at damage sites.
On another hand, transcriptional repression is also achieved via multiple changes of the chromatin structure assembled on the damaged gene. It was observed that H2AK119ub, a histone mark associated with repressive chromatin [86], is induced after DSB formation in an ATM-dependent manner [81]. Several studies have identified likely factors that perform this function. H2AK119ub is regulated by the polycomb repressive complexes 1 (PRC1), which contain the E3 Ub ligases RING1B and BMI1 [86]. The PRC1 complex ubiquitylates H2AK119 and functions along with the PRC2 complex, which methylates H3K27me3, collectively resulting in repressive chromatin that silences transcription. Members of both the PRC1 and PRC2 complexes, including RING1B, BMI1 and EZH2, localize to DNA damage sites [84,87-90]. Both the PRC1 and PRC2 complexes are required for transcriptional repression after DNA damage, including by regulating H2AK119ub at break sites [84,87,91]. Several additional complexes have been identified that regulate H2AK119ub. For example, the PBAF chromatin remodeling complex is required for H2AK119ub at DNA break sites, in a manner that depends on the ATM- mediated phosphorylation of BAF180, a component of the PBAF complex [91]. Additionally, deficiency in ENL results in reduced H2AK119ub and phosphorylated ENL (see above) was found to associate with the PRC1 E3 ligase BMI1 [84]. FBXL10-RNF68-RNF2, members of a large complex called FRRUC which display ubiquitin ligase activity, are also recruited to DNA damage sites in a PARP/TIMELESS-dependent manner where they act to ubiquitylate H2AK119 [92] and are required to suppress transcription at DNA damage sites.
Beyond H2AK119 ubiquitylation, other chromatin changes participate in DSB-induced transcriptional repression. FRRUC-dependent H2AZ loading at DSBs was shown to be required for transcriptional repression at DSBs [92]. The role of H2AZ in this process is unclear as effector H2AZ proteins have not been identified and these effects could be through structural changes in chromatin that occur when H2AZ is loaded versus H2A. H2AZ is ubiquitylated by RNF168 [93] and other ubiquitylations on this histone variant could also facilitate transcription effects at DNA lesions, similarly to H2AK119. Additionally, the cohesin complex is also required for transcription inhibition following DSB [94].
Furthermore, the histone demethylase KDM5A demethylates the transcription active mark, H3K4me3, which is reduced after DNA damage [95,96]. The demethylation of H3K4me3 may participate in transcriptional repression by reducing this active mark but also by allowing the stable association of the NuRD chromatin remodeling complex with damaged chromatin in actively transcribed genes. The NuRD complex also requires the ZMYND8 bromodomain-containing protein to facilitate its association with break sites [97-99], ZMYND8 binds to acetylated lysines and the HAT TIP60 regulates its association with DSBs, which are required to support the recruitment of NuRD to damage sites. Collectively, these observations suggest that there is a demethylation-acetylation pathway involving KDM5A-ZMYND8-NuRD axis that functions to remodel chromatin to repress transcription and promote DSB repair. Finally, H2BK120 deubiquitylation was recently reported to take place at site-specific DSBs in human cancer cells [100]. Of interest, H2B ubiquitination have been proposed to facilitate Pol II release from pausing and to contribute to productive elongation [101,102]. H2B deubiquitylation may therefore participate in dephosphorylating Pol II Ser2 in response to DSB, hence contributing to alter Pol II cycle and turn off transcription.
Collectively, these studies uncovered a complex picture, involving multiple pathways, to down-regulate transcription of DSB nearby genes (Fig. 1). Additional experiments are warranted to determine the interplay between these multiple pathways that orchestrate chromatin and transcription changes surrounding DNA breaks [103], and to establish whether they act in concert or independently at damage sites.
Transcription-coupled DSB Repair (TC-DSBR) pathway
Beyond the signals that end up turning off transcription of damage-associated genes, evidence is starting to accumulate for a specific pathway that repairs DSBs occurring in transcriptionally active loci, that we refer to here as Transcription-Coupled DSB Repair (TC-DSBR) (see [104] and Fig. 2). Unbiased, genome-wide ChIP-seq analysis of repair protein recruitment at DSBs generated by a restriction enzyme revealed that in human asynchronous transformed cells, active genes (and active promoters) display the ability to robustly recruit the Rad51 recombinase, involved in HR, which is not the case for other DSBs induced -with an equal efficiency- elsewhere in euchromatin (silent genes or intergenic sequence) [100,105]. Notably, turning off gene transcription using transcription inhibitors decreases Rad51 recruitment [105-108], while conversely turning on transcription of the targeted locus, prior to break induction, switches the repair pathway choice toward HR [105,108,109], indicating that the transcriptional status of a broken locus is a decisive determinant for DNA repair pathway choice. Consistently, DSBs induced by AID off-target activity-which mostly reside in transcriptionally active and potentially G4-rich genes [41,110]- exhibit a strong requirement for HR [111].
Figure 2. Transcription-coupled DSB Repair pathway (TC-DSBR) repairs DSBs occurring in transcriptionally active loci.
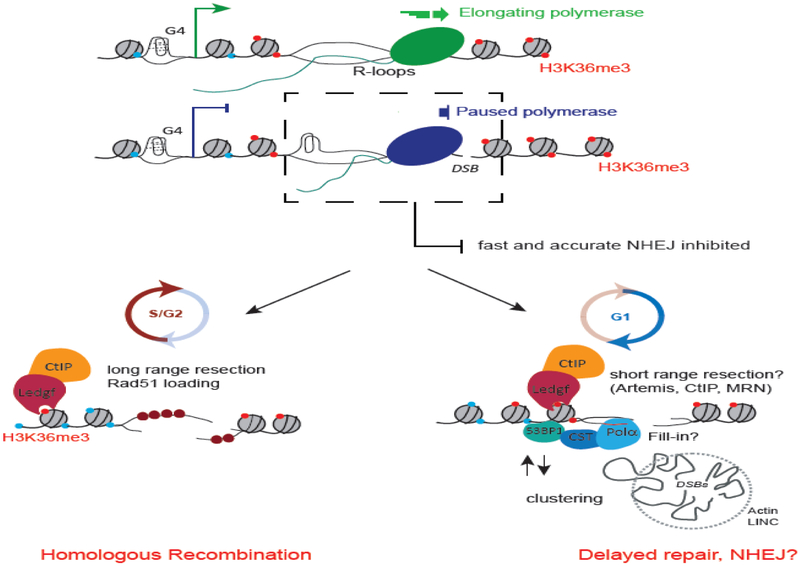
R-loops and G4 structures accumulate at transcribed genes, mainly over TSS and promoter regions, where they can induce DSBs, resulting in a rapid transcriptional arrest of the local elongating RNA polymerases (RNA Pol II) (see Fig. 1). We propose that secondary DNA structures may inhibit fast and accurate NHEJ pathway, consequently triggering DNA ends processing, further stimulated by CtIP recruitment through LEDGF/H3K36me3 interaction. In G1 cells, this short range resection may delay repair and promote DSB clustering [117]. These processed DNA ends may require DNA Polα dependent fill-in to complete repair by NHEJ. In S/G2 cells, long range resection of the 5’ strand allows formation of a 3’ nucleoprotein filament further available for HR repair using the available sister chromatid as a template.
Notably, the chromatin status, via the SETD2-dependent elongation-associated H3K36me3 histone mark, which preexists on active genes prior DSB, contributes to channeling broken transcribed loci to HR repair [105,112,113]. In addition, other active genes associated histones modifications also promote HR usage (such as H4K16ac [114,115], reviewed in [104,116]), suggesting that the chromatin structure assembled at active genes may create a platform suitable for HR protein recruitment via various “reader” protein/histone mark-mediated interactions.
Of importance, this preference of transcribed loci for HR repair is restricted to S/G2 phases [105,107] and the mechanism(s) that ensure repair of these loci in G1 are still unclear (Fig. 2). In G1, DSBs induced in PolII-bound loci display delayed repair compared to DSBs induced in intergenic or silent genes [117], similarly to AID-induced DSBs at off-target genomic sites [118], Interestingly this delayed repair coincides with the ability of broken genes to “cluster” together (i.e. get spatially closer within the nuclear space) [117]. We proposed that secondary DNA structures, for example in cis embedded RNAs or Pol II machinery occupancy, may counteract fast classical NHEJ repair, at transcriptionally active loci, leaving a time window for resection to occur (Fig. 2). At these processed breaks, further repair “pausing” (potentially ensured by clustering or roadblocks by in cis factors?) may be required for faithful recovery of the genetic information [104,117], In this respect, evidence indicates that resection can occur in G1 [119], in a manner that depends on ATM, Artemis, CtIP, and Mre11 exonuclease activity [119,120]. This G1-specific resection process was proposed to occur at heterochromatic breaks but also potentially at transcriptionally active broken genes (discussed in [104,121]). Last year, a major breakthrough into 53BP1 function in counteracting resection was uncovered with the discovery that it brings CST-DNA Pol α at sites of resected DSB for fill-in [122]. Hence a tempting hypothesis is that at broken active genes, CtIP, Mre11, Artemis- dependent resection that occurs in G1 is counterbalanced by the accumulation of 53BPl-CST-Polα that allows to reconstitute compatible DNA ends for accurate repair by NHEJ. Future work is necessary to further validate this hypothesis and to determine whether such fill-in reactions can occur in G1 or necessitate entry into S phase. In this respect, the fact that G1-formed, AID-induced DSBs (both on the IgH locus as well as off-target loci, which display active transcription and R-loops/G4 structures) are resolved in early S phase [118,123] and require DNA synthesis [124], further supports the idea that an S phase environment may be critical to complete repair at active loci when they experience breakage in G1.
The data presented above delineate a specific pathway dedicated to signal and repair DSBs in active genes, which entails both transcription extinction and specialized repair events. Of importance, the proneness to form secondary structures and the presence of RNAs and Pol II complexes at active genes and promoters, confer to these damaged loci the ability to further accumulate secondary structures including RNA:DNA hybrids and G4 post-damage (described below). Whether these structures represent bona fide repair intermediates or roadblocks requiring dissolution for proper repair is discussed.
Transcription-Coupled DSB repair entails RNA:DNA hybrid accumulation
There is now a large body of evidence showing that RNA:DNA hybrids specifically accumulate at sites of DNA damage. Indeed, accumulation of mutant RNase HI and GFP-tagged hybrid-binding domain of RNase H1 (GFP-HB), as well as structures detected by the S9.6 antibody, are evident at sites of laser micro-irradiation or ROS-induced damage [107,109,125]. Further experiments using a duplex specific nuclease (DSN) and RNase H identified RNA:DNA hybrids at sites of DSB induced by I-SceI [126]. Finally, the use of S9.6 to immunoprecipitate RNA:DNA hybrid loci (by the so called DRIP technique) followed by qPCR [106,127,128] or high throughput sequencing [129,130] further established that hybrids accumulate in cis to DSBs induced by endonucleases. Strikingly, genome-wide analysis of hybrid distribution around multiple DSBs indicated that hybrids mostly accumulate at loci exhibiting transcriptional activity and/or high Pol II binding [129,130], although few exceptions with very low levels of hybrid formation were also reported at intergenic, Pol II-unbound, loci [129]. Consistently, ROS-induced damage display RNA:DNA hybrids accumulation only if the targeted locus is transcriptionally active [109].
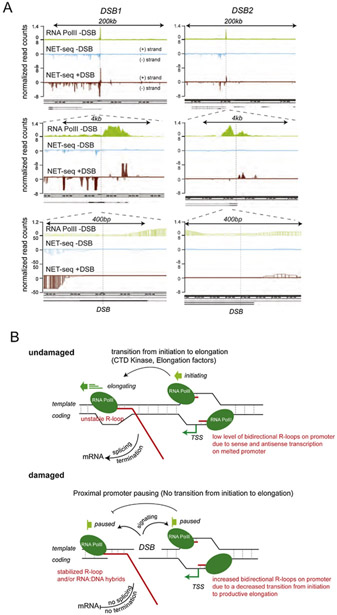
The exact mechanisms that underlie accumulation of R-loops is still under investigation (Fig. 3). It has been proposed that they could be (i) either a consequence of DSB-induced transcriptional pausing [129], a process known to favor R-loop formation [16,131,132], (ii) a consequence of in cis co-transcriptional pre-mRNA annealing with the resected strand [126] or (iii) due to de novo recruitment of Pol II and transcription at DNA ends, which would produce a long non coding RNA (lncRNA) complementary to the resected strand [106,127,133,134] (Fig. 3). Notably, RNA:DNA hybrids specifically accumulate in S-G2 [106] and inhibition of resection, by depleting CtIP, ExoI or MRN, decreases hybrid formation [106,107] suggesting that indeed these hybrids form, or are stabilized via hybridization with available resected ssDNA ends. Yet, genome-wide ChIP-seq experiments against RNA Pol II failed to demonstrate de novo recruitment at intergenic DSBs ([135], and our unpublished observations). Along the same line, genome-wide analyses of Pol II-embedded RNAs by strand-specific NET-seq also confirmed that RNAs only accumulate at DSBs induced in genic sequences [127]. This, together with the strong bias for DSB-induced RNA:DNA hybrids to form at active loci compared to inactive loci [109,129], suggest that proximity of pre-bound RNA Pol II fosters (if not accounts for) the accumulation of RNAs at DSBs. Whether at these loci, proximal Pol II can be de novo loaded onto ssDNA to ensure RNA production from the ends (Fig. 3 iii) is a matter of debate, which needs additional experimental clarification. Indeed, this hypothesis was recently proposed from RT-qPCR analyses [106,134] as well as NET-seq experiments ([127] and see Fig. 4A top panel at 200-kb scale). However, zooming in on this same data to a 4 kb and 400-bp scale failed to detect Pol II-embedded transcripts starting at the DSB ends (see Fig. 4A lower panel, data from [127]), which may suggest other alternative hypotheses. Collectively, experimental data may support the idea that R-loops accumulate at loci where Pol II pauses following DSB. Transcripts embedded in paused Pol II would further hybridize with either the double-stranded DNA and/or the ssDNA after resection takes place, accounting for increased RNA:DNA hybrids, and potentially other processed RNAs, around DSBs (Fig. 4B). Further characterization of these hybrids using for example DRIPc-seq, which allows both strands specificity and exact length of hybrids to be determined, may help resolve this issue and provide a better understanding of the nature of the DSB-induced RNA:DNA hybrids.
Figure 3. Hypotheses for RNA:DNA hybrids and/or R-loops formation at transcriptionally active broken loci.
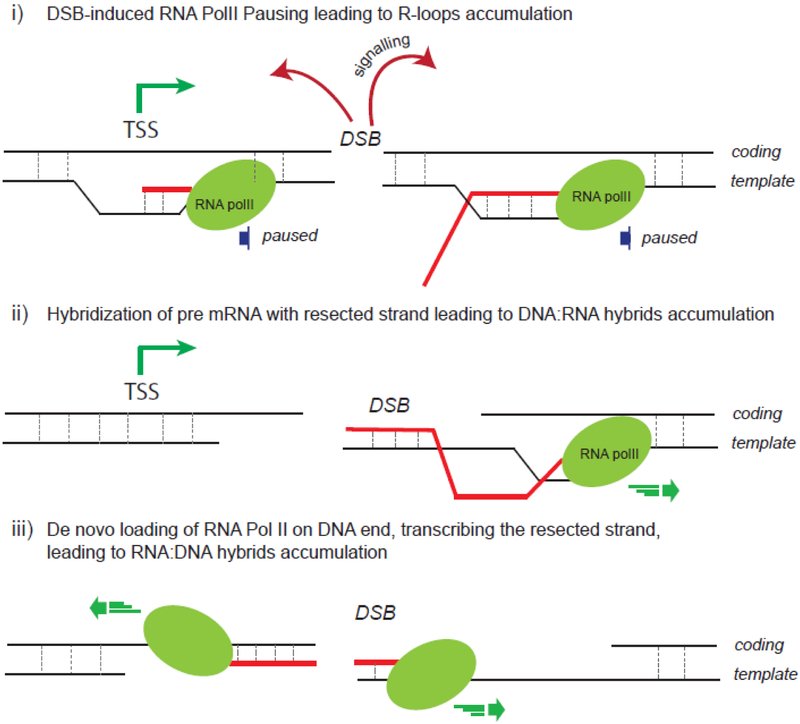
(i) The signaling of a DSB occurring in a Pol II-bound locus induces transcriptional pausing known to trigger R-loops formation. (ii) Nearby pre-mRNA hybridizes with the single-strand DNA following resection. (iii) Pol II is de novo recruited and uses the single-strand DNA available following resection as a template to generate a RNA:DNA hybrid.
Figure 4. A proposed model for RNA production around DSB.
A. Genome browser screenshots showing Pol II occupancy (ChIP-seq data in undamaged DIvA cells [129]) and NET-seq data from [127] in undamaged U2OS cells (NET-seq -DSB) and in damaged DIvA cells (NET−seq +DSB), at different scales, around two AsiSI-induced DSBs. The DSB position is indicated with a vertical dotted line. Note that Pol II-embedded transcripts are not exactly starting at the DSB position even following damage. B. A proposed model of RNA:DNA hybrids and R-loops accumulation after DSB occurring in transcriptionally active genes. Top panel: In normal conditions (no DSB), sense and antisense transcription can produce R-loops on melted promoters [17] (see for instance Fig. 4A, 4kb scale, NET-seq data in absence of damage, in blue, where low level of antisense transcription is detected upstream TSS). Downstream of the TSS, elongating Pol II can also produce transient RNA:DNA hybrids during the process of transcription. Lower panel: As a consequence of DSB induction (damaged locus), the transition from initiation to elongation is inhibited. Non elongating Pol II accumulate at melted promoters of damaged genes, increasing sense and antisense transcription (see for example Fig. 4A, at 4kb and 400bp scale, the NET-seq data in presence of DSB, in red), further enhancing R-loops. Moreover, downstream TSS, DSB-induced Pol II pausing, or simply a decrease in Pol II elongation rate, triggers RNA:DNA hybrids accumulation, which may also be further stabilized as the resection machinery uncovers single-strand DNA.
Regardless of the exact mechanism for generating these structures, DSB-induced RNA:DNA hybrids may fulfill important functions during DSB repair (Fig. 5). RNA:DNA hybrids may contribute to repair factor recruitment, as was proposed for CSB, Rad51, Rad52 and BRCA1 loading [106,107,109,130,136], as well as 53BP1 [127,130] andMDCl [127]. Of interest, BRCA1, Rad52 and CSB were shown to directly interact in vitro with RNA:DNA hybrids [106,109,136]. RNA:DNA hybrids may further contribute to the production of DDRNAs, a class of double-stranded small RNAs required for full DDR foci assembly (reviewed in [137]). DDRNAs are reported as a general feature of DSBs, yet some evidence also suggests that their accumulation may not be universal [130,138-140] and may be related to the expression level of the host locus, as well as the presence of introns and splicing machinery [138,139]. Hence, future studies are required to understand the frequency and contribution of DDRNAs during DSR repair in general and TC-DSBR more specifically (Fig. 5). RNAs within RNA:DNA hybrids could serve as a template for repair [141], by a yet undeciphered mechanism (reviewed in [137,142]) which requires at least CSB and Rad52 [108,136](Fig. 5). In this respect, the unique strand exchange activity of RAD52 with homologous ssRNA in vitro [143] further points toward a potential role of RNA:DNA hybrids (independently shown to recruit Rad52, see above) in RNA-templated repair. Finally, RNA:DNA hybrids have important roles in controlling resection and/or ssDNA binding factor assembly (Fig. 5). On one hand, some data indicate that they could promote resection: indeed Drosha depletion, necessary for hybrid accumulation, decreases resection [130] and transcription inhibition using DRB (expected to decrease hybrids) impairs RPA recruitment [107], In contrast, in S.pombe, overexpression of RNAse H (which also decreases hybrids) enhanced the extent of resection as detected by the spreading of RPA binding and single-strand annealing (SSA) frequency [133], suggesting a potential anti-resection role for RNA:DNA hybrids. Lastly, other studies have suggested that RNA:DNA hybrids act downstream of resection and rather regulate the recruitment of ssDNA binding factors. For example, transcription inhibition by α amanitin-treatment was reported to have no effect on resection (as detected by BrdU staining) and to rather increase RPA binding [106]. Secondly, depletion of Dead box 1 (DDX1), a RNA:DNA hybrid helicase, does not impact CtIP recruitment (resection initiation) but does decrease RPA foci assembly [126]. Finally, the depletion of senataxin (SETX), another RNA:DNA helicase specifically recruited at damaged active genes and not at other DSB induced elsewhere on the genome, does not affect resection but reduces Rad51 binding [129]. Collectively, all these data suggest that RNA:DNA hybrids form at DSBs-induced in active genes. More work is required to know whether these are just a side effect of transcription extinction (Fig. 4B) or result from an active process dedicated to produce such intermediates de novo (Fig. 3 iii), and to fully determine whether they display important functions during repair or are rather obstacles that need to be displaced. Of importance, these structures possess the capability of regulating ssDNA accessibility, which has profound impacts on the repair output (see next section).
Figure 5. Possible functions of RNA:DNA hybrids during DSB repair.
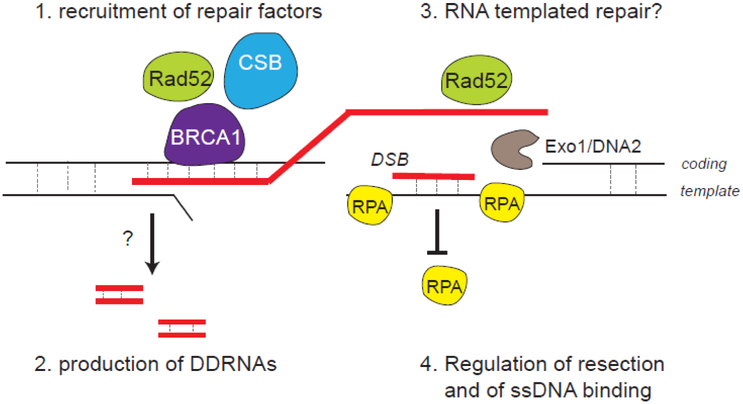
RNA:DNA hybrids formed around the break could serve: 1. as a platform to recruit DSB repair proteins; 2. to produce DNA damage response RNAs (DDRNAs); 3. as a template for a RNA-mediated repair mechanism 4. to control resection and single-strand binding proteins assembly, thereby regulating downstream events such as Rad51 nucleofilament assembly and homologous recombination.
Removal of secondary structures is necessary for repair completion
The RNA:DNA hybrids referenced in the above section, by assembling on the resected strand or/and by controlling the resection rate, strongly regulate ssDNA availability for further steps in homologous recombination (RPA assembly, homology search, strand invasion). In agreement, depletion of SETX or DDX1, both shown to be recruited at DSB forming RNA:DNA hybrids (see above), impairs Rad51 loading and HR [126,129]. Notably, XPG, a R-loop processing factor [73] is also recruited at DSBs and necessary for TC-DSBR completion by HR, along with RAD52 [107], as is RNase H2 [106] (Fig. 6, right side of the break). Recently, RPA has been described as able to assemble on the displaced ssDNA of R-loop structures and, more importantly, to trigger RNase H targeting at sites of R-loops on the genome [144]. This raises the interesting possibility that this could also occur on resected-RPA-coated DSB ends, thereby creating a feedforward enhancing loop: as soon as a small patch of ssDNA would be made available thanks to RNA:DNA hybrid unwinding activities, RPA binding would further recruit RNase H, fostering hybrid removal and further RPA binding/spreading to allow HR repair.
Figure 6: Pre-existing and DSB-induced G4 and RNA:DNA hybrids shall be dissolved for repair completion.
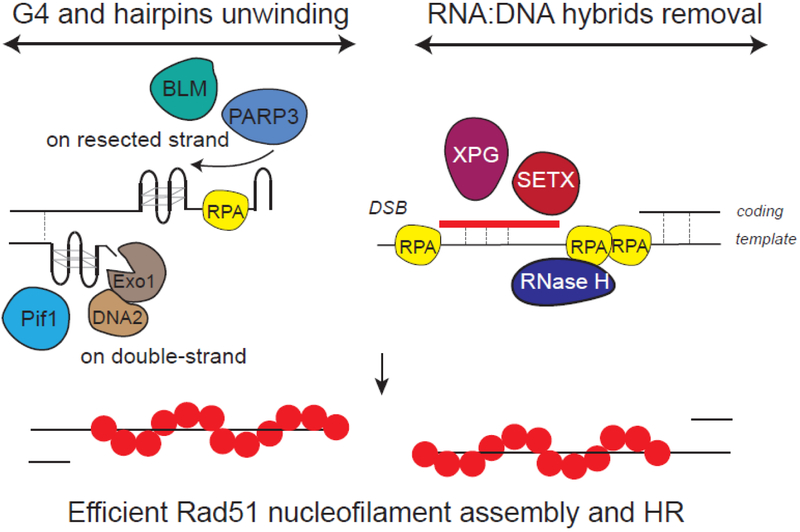
Right side of the DSB: RNA:DNA hybrids that pre-exist and accumulate following DSB shall be removed to ensure RPA loading. SETX, XPG and RNAse H participate in hybrids dissolution. In this respect, RNAse H recruitment by RPA may allow processive hybrids unwinding and RPA spreading. Left side of the DSB: G4 shall also be disassembled for repair. Pif1 is necessary for DNA double-strand G4 unwinding, allowing resection. On another hand, G4 were also proposed to form on the resected strand which may require BLM and PARP3 for dissolution and RPA binding. Together these processes allows Rad51 nucleofilament assembly and HR.
Defects in hybrid removal during DSB repair have profound impact on repair outcome and cell survival. In human cells, depletion of SETX triggers a strong increase in translocation frequency and DSB-induced lethality [129]. Similarly, rnaseh1/rnaseh2 mutant yeast strain display persistent Rad52 foci, increased usage of Break Induced Replication and increased lethality, phenotypes suppressed by BIR inactivation [145]. Altogether these data suggest that upon inactivation of RNA:DNA hybrid removal mechanisms, HR is compromised and alternative repair pathways take over, eventually leading to impaired repair capacities and loss of viability.
Beyond R-loops and/or RNA:DNA hybrids repair intermediates removal, evidence is accumulating that G4s can also act as roadblocks for DSB repair (Fig. 6 left side of the break). Notably, various proteins that display G4 binding or melting properties (reviewed in [146]) are recruited at DSBs and/or promote repair. This includes BLM and WRN, two G4 unwinding helicases with 3’to 5’ directionality [146,147], Pif1, FANCJ, DNA2 and RTEL1, all 5’ to 3’ G4 unwinding helicases [148-151], CSB [152], as well as Rif1, a G4-interacting 53BP1 effector [153,154], Even though their G4 helicase function may not be the sole role of these proteins in DSB responses, it is interesting to note that most of them are involved in hereditary syndromes with genetic instability and cancer susceptibility.
G4 structures that pre-exist before break induction may block or reduce resection processivity as suggested by the recent finding that the helicase activity of Pif1 is required for resection [149]. Consistently, Pif1 depletion strongly affected HR [149]. Notably, artificial introduction of TG repeats on one side of a DSB dramatically impairs resection on the TG-rich side of the break and triggers translocation in yeast [155]. While in this study the authors did not investigate whether such behavior is due to secondary structure formation, it raises the important possibility that resection processivity and symmetry at natural loci contribute to the efficiency of HR and to the translocation outcome in human cells.
In addition, G4 and hairpins can also accumulate in cis to DSB on the resected strand (Fig. 6). In this respect, RPA was shown to be necessary for elimination of hairpins [156,157]. Moreover, break induction by CRISPR/Cas9 was recently reported to increase G4 structures in the vicinity of DSB [158]. The authors proposed that free ssDNA may fold to form G4 structures which then require BLM and PARP3 for unwinding and proper HR [158]. At last, it is likely that RNA:DNA hybrids and G4/hairpins crosstalk during DSB repair: on double-stranded DNA, G-quadruplexes formation stabilize R-loops, and hence combined R-loop/G4 structures may act as strong roadblocks for resection and repair. In contrast, on ssDNA, it can be envisaged that RNA hybridization could rather be antagonistic to hairpin and G4 assembly.
Concluding remarks
Altogether, recent studies have then determined that i) transcribed loci are prone to DNA secondary structures such as G4 and R-loops, ii) these secondary structures are potent inducers of DSBs, and iii) a specialized pathway acts to repair these DSBs induced in Pol II-bound, transcribed, secondary structure-forming, loci. Moreover, DSB production also further trigger accumulation of these non-canonical DNA/RNA structures, due to the regulation of Pol II and chromatin at damaged genes and to the generation of single-strand DNA by resection. Hence, repairing these loci represent a tremendous challenge that nevertheless shall be accomplished with minimal genetic change, given the function of transcribed loci as essential bricks for cell function and fate. Yet the exact nature of RNA:DNA hybrids and DNA secondary structures at DSBs is still unclear, and the processes that are responsible for their accumulation and dissolution, as well as how these factors are switched between damaged and undamaged states within chromatin, are only starting to be deciphered. Future work will be necessary to fully apprehend the complexity and metabolism of non-canonical DNA/RNA structures as well as their consequences on genome integrity.
Acknowledgements
We apologize to our colleagues whose works could not be included in this review owing to space limitations. The K.M.M. laboratory is supported by the NIH National Cancer Institute (R01CA198279 and RO1CA201268) and the American Cancer Society (RSG-16-042-01-DMC). The G.L. laboratory is funded by grants from the European Research Council (ERC-2014-CoG 647344), Agence Nationale pour la Recherche (ANR-14-CE10-0002-01 and ANR-13-BSV8-0013), the Institut National contre le Cancer (INCA), and the Ligue Nationale contre le Cancer (LNCC). NP is an INSERM researcher.
Abbreviations:
- AID
activation-induced cytidine deaminase
- CSR
class switch recombination
- CTD
carboxy-terminal domain
- DDR
DNA damage response
- DDRNAs
DNA damage response RNAs
- DRIP
DNA-RNA immunoprecipitation
- DSB
double-strand break
- G4
G-quadruplex
- HR
homologous recombination
- IgH
immunoglobulin heavy chain
- lncRNA
long non coding RNA
- NHEJ
non-homologous end joining
- Pol II
RNA polymerase II
- ssDNA
single-strand DNA
- TC-DSBR
transcription-coupled DNA double-strand break repair
- TSS
transcriptional start site
Footnotes
Competing interests
The authors have no competing interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- [1].Watson JD, Crick FH, Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid., Nature. 171 (1953)737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- [2].Chédin F, Nascent Connections: R-Loops and Chromatin Patterning., Trends Genet. 32 (2016) 828–838. doi: 10.1016/j.tig.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hänsel-Hertsch R, Di Antonio M, Balasubramanian S, DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential., Nat. Rev. Mol. Cell Biol. 18 (2017)279–284. doi: 10.1038/nrm.2017.3. [DOI] [PubMed] [Google Scholar]
- [4].Corless S, Gilbert N, Effects of DNA supercoiling on chromatin architecture., Biophys. Rev. 8 (2016) 51–64. doi: 10.1007/s12551-016-0242-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Belotserkovskii BP, Tornaletti S, D’Souza AD, Hanawalt PC, R-loop generation during transcription: Formation, processing and cellular outcomes., DNA Repair (Amst.). 71 (2018) 69–81. doi: 10.1016/j.dnarep.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kwok CK, Merrick CJ, G-Quadruplexes: Prediction, Characterization, and Biological Application., Trends Biotechnol. 35 (2017) 997–1013. doi: 10.1016/j.tibtech.2017.06.012. [DOI] [PubMed] [Google Scholar]
- [7].Zeraati M, Langley DB, Schofield P, Moye AL, Rouet R, Hughes WE, et al. , I-motif DNA structures are formed in the nuclei of human cells., Nat. Chem. 10 (2018) 631–637. doi: 10.1038/s41557-018-0046-3. [DOI] [PubMed] [Google Scholar]
- [8].Biffi G, Tannahill D, McCafferty J, Balasubramanian S, Quantitative visualization of DNA G-quadruplex structures in human cells., Nat. Chem. 5 (2013) 182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Vanoosthuyse V, Strengths and Weaknesses of the Current Strategies to Map and Characterize R-Loops., Noncoding RNA. 4 (2018). doi: 10.3390/ncrna4020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].García-Rubio M, Barroso SI, Aguilera A, Detection of DNA-RNA Hybrids In Vivo., Methods Mol. Biol. 1672 (2018) 347–361. doi: 10.1007/978-1-4939-7306-4_24. [DOI] [PubMed] [Google Scholar]
- [11].Rodriguez R, Miller KM, Forment JV, Bradshaw CR, Nikan M, Britton S, et al. , Small-molecule-induced DNA damage identifies alternative DNA structures in human genes., Nat. Chem. Biol. 8 (2012) 301–310. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sanz LA, Hartono SR, Lim YW, Steyaert S, Rajpurkar A, Ginno PA, et al. , Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals., Mol. Cell. 63 (2016) 167–178. doi: 10.1016/j.molcel.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ginno PA, Lott PL, Christensen HC, Korf I, Chédin F, R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters., Mol. Cell. 45 (2012) 814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hänsel-Hertsch R, Beraldi D, Lensing SV, Marsico G, Zyner K, Parry A, et al. , G-quadruplex structures mark human regulatory chromatin., Nat. Genet. 48 (2016) 1267–1272. doi: 10.1038/ng.3662. [DOI] [PubMed] [Google Scholar]
- [15].Wahba L, Costantino L, Tan FJ, Zimmer A, Koshland D, S1-DRIP-seq identifies high expression and poly A tracts as major contributors to R-loop formation., Genes Dev. 30 (2016) 1327–1338. doi: 10.1101/gad.280834.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chen L, Chen J-Y, Zhang X, Gu Y, Xiao R, Shao C, et al. , R-ChIP Using Inactive RNase H Reveals Dynamic Coupling of R-loops with Transcriptional Pausing at Gene Promoters., Mol. Cell. 68 (2017) 745–757.e5. doi: 10.1016/j.molcel.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dumelie JG, Jaffrey SR, Defining the location of promoter-associated R-loops at near-nucleotide resolution using bisDRIP-seq., Elife. 6 (2017). doi: 10.7554/eLife.28306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Manzo SG, Hartono SR, Sanz LA, Marinello J, De Biasi S, Cossarizza A, et al. , DNA Topoisomerase I differentially modulates R-loops across the human genome., Genome Biol. 19 (2018) 100. doi: 10.1186/s13059-018-1478-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mao S-Q, Ghanbarian AT, Spiegel J, Martínez Cuesta S, Beraldi D, Di Antonio M, et al. , DNA G-quadruplex structures mold the DNA methylome., Nat. Struct. Mol. Biol. 25 (2018) 951–957. doi: 10.1038/s41594-018-0131-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hartono SR, Korf IF, Chédin F, GC skew is a conserved property of unmethylated CpG island promoters across vertebrates., Nucleic Acids Res. 43 (2015) 9729–9741. doi: 10.1093/nar/gkv811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Saranathan N, Vivekanandan P, G-Quadruplexes: More Than Just a Kink in Microbial Genomes., Trends Microbiol. 27 (2019) 148–163. doi: 10.1016/j.tim.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kim N, The interplay between G-quadruplex and Transcription., Curr. Med. Chem. (2017). doi: 10.2174/0929867325666171229132619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Valton A-L, Prioleau M-N, G-Quadruplexes in DNA Replication: A Problem or a Necessity?, Trends Genet. 32 (2016) 697–706. doi: 10.1016/j.tig.2016.09.004. [DOI] [PubMed] [Google Scholar]
- [24].Toubiana S, Selig S, DNA:RNA hybrids at telomeres - when it is better to be out of the (R) loop., FEBS J. 285 (2018) 2552–2566. doi: 10.1111/febs.14464. [DOI] [PubMed] [Google Scholar]
- [25].Pavri R, R loops in the regulation of antibody gene diversification., Genes (Basel). 8 (2017). doi: 10.3390/genes8060154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ, R-loops induce repressive chromatin marks over mammalian gene terminators., Nature. 516 (2014) 436–439. doi: 10.1038/nature13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Skourti-Stathaki K, Torlai Triglia E, Warburton M, Voigt P, Bird A, Pombo A, R-Loops Enhance Polycomb Repression at a Subset of Developmental Regulator Genes., Mol. Cell. 73 (2019) 930–945.e4. doi: 10.1016/j.molcel.2018.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hussain T, Saha D, Purohit G, Kar A, Kishore Mukherjee A, Sharma S, et al. , Transcription regulation of CDKN1A (p21/CIP1/WAF1) by TRF2 is epigenetically controlled through the REST repressor complex., Sci. Rep. 7 (2017) 11541. doi: 10.1038/s41598-017-11177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Saha D, Singh A, Hussain T, Srivastava V, Sengupta S, Kar A, et al. , Epigenetic suppression of human telomerase (hTERT) is mediated by the metastasis suppressor NME2 in a G-quadruplex-dependent fashion., J. Biol. Chem. 292 (2017) 15205–15215. doi: 10.1074/jbc.M117.792077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Thakur RK, Kumar P, Haider K, Verma A, Kar A, Parent J-L, et al. , Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression., Nucleic Acids Res. 37 (2009) 172–183. doi: 10.1093/nar/gkn919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lombraña R, Almeida R, Álvarez A, Gómez M, R-loops and initiation of DNA replication in human cells: a missing link?, Front. Genet. 6 (2015) 158. doi: 10.3389/fgene.2015.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Itoh T, Tomizawa J, Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H., Proc. Natl. Acad. Sci. USA. 77 (1980) 2450–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Stuckey R, García-Rodríguez N, Aguilera A, Wellinger RE, Role for RNA:DNA hybrids in origin-independent replication priming in a eukaryotic system., Proc. Natl. Acad. Sci. USA. 112 (2015) 5779–5784. doi: 10.1073/pnas.1501769112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Besnard E, Babied A, Lapasset L, Milhavet O, Parrinello H, Dantec C, et al. , Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs., Nat. Struct. Mol. Biol. 19 (2012) 837–844. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- [35].Picard F, Cadoret J-C, Audit B, Arneodo A, Alberti A, Battail C, et al. , The spatiotemporal program of DNA replication is associated with specific combinations of chromatin marks in human cells., PLoS Genet. 10 (2014) e1004282. doi: 10.1371/journal.pgen.1004282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hoshina S, Yura K, Teranishi H, Kiyasu N, Tominaga A, Kadoma H, et al. , Human origin recognition complex binds preferentially to G-quadruplex-preferable RNA and single-stranded DNA., J. Biol. Chem. 288 (2013) 30161–30171. doi: 10.1074/jbc.M113.492504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Valton A-L, Hassan-Zadeh V, Lema I, Boggetto N, Alberti P, Saintomé C, et al. , G4 motifs affect origin positioning and efficiency in two vertebrate replicators., EMBO J. 33 (2014) 732–746. doi: 10.1002/embj.201387506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Matthews AJ, Zheng S, DiMenna LJ, Chaudhuri J, Regulation of immunoglobulin class-switch recombination: choreography of noncoding transcription, targeted DNA deamination, and long-range DNA repair., Adv Immunol. 122 (2014) 1–57. doi: 10.1016/B978-0-12-800267-4.00001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Reaban ME, Griffin JA, Induction of RNA-stabilized DNA conformers by transcription of an immunoglobulin switch region., Nature. 348 (1990) 342–344. doi: 10.1038/348342a0. [DOI] [PubMed] [Google Scholar]
- [40].Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N, Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA., Genes Dev. 18 (2004) 1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Qiao Q, Wang L, Meng F-L, Hwang JK, Alt FW, Wu H, AID recognizes structured DNA for class switch recombination., Mol. Cell. 67 (2017) 361–373.e4. doi: 10.1016/j.molcel.2017.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Freudenreich CH, R-loops: targets for nuclease cleavage and repeat instability., Curr. Genet. 64 (2018) 789–794. doi: 10.1007/s00294-018-0806-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wang G, Vasquez KM, Effects of Replication and Transcription on DNA Structure-Related Genetic Instability., Genes (Basel). 8 (2017). doi: 10.3390/genes8010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sollier J, Cimprich KA, Breaking bad: R-loops and genome integrity., Trends Cell Biol. 25 (2015) 514–522. doi: 10.1016/j.tcb.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Barlow JH, Faryabi RB, Callén E, Wong N, Malhowski A, Chen HT, et al. , Identification of early replicating fragile sites that contribute to genome instability., Cell. 152 (2013) 620–632. doi: 10.1016/j.cell.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, et al. , GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases., Nat. Biotechnol. 33 (2015) 187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang X, Wang Y, Wu X, Wang J, Wang Y, Qiu Z, et al. , Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors., Nat. Biotechnol. 33 (2015) 175–178. doi: 10.1038/nbt.3127. [DOI] [PubMed] [Google Scholar]
- [48].Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho Y-J, et al. , Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells., Cell. 147 (2011) 107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].van Wietmarschen N, Merzouk S, Halsema N, Spielings DCJ, Guryev V, Lansdorp PM, BLM helicase suppresses recombination at G-quadruplex motifs in transcribed genes., Nat. Commun. 9 (2018) 271. doi: 10.1038/s41467-017-02760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lensing SV, Marsico G, Hänsel-Hertsch R, Lam EY, Tannahill D, Balasubramanian S, DSBCapture: in situ capture and sequencing of DNA breaks., Nat. Methods. 13 (2016) 855–857. doi: 10.1038/nmeth.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Canela A, Sridharan S, Sciascia N, Tubbs A, Meltzer P, Sleckman BP, et al. , DNA Breaks and End Resection Measured Genome-wide by End Sequencing., Mol. Cell. 63 (2016) 898–911. doi: 10.1016/j.molcel.2016.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Crosetto N, Mitra A, Silva MJ, Bienko M, Dojer N, Wang Q, et al. , Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing., Nat. Methods. 10 (2013) 361–365. doi: 10.1038/nmeth.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Yan WX, Mirzazadeh R, Garnerone S, Scott D, Schneider MW, Kallas T, et al. , BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks., Nat. Commun. 8 (2017) 15058. doi: 10.1038/ncomms15058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kim D, Kim J-S, DIG-seq: a genome-wide CRISPR off-target profiling method using chromatin DNA., Genome Res. 28 (2018) 1894–1900. doi: 10.1101/gr.236620.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Shastri N, Tsai Y-C, Hile S, Jordan D, Powell B, Chen J, et al. , Genome-wide Identification of Structure-Forming Repeats as Principal Sites of Fork Collapse upon ATR Inhibition., Mol. Cell. 72 (2018) 222–238.e11. doi: 10.1016/j.molcel.2018.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Bouwman BAM, Crosetto N, Endogenous DNA Double-Strand Breaks during DNA Transactions: Emerging Insights and Methods for Genome-Wide Profiling., Genes (Basel). 9 (2018). doi: 10.3390/genes9120632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Pentzold C, Shah SA, Hansen NR, Le Tallec B, Seguin-Orlando A, Debatisse M, et al. , FANCD2 binding identifies conserved fragile sites at large transcribed genes in avian cells., Nucleic Acids Res. 46 (2018) 1280–1294. doi: 10.1093/nar/gkx1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Wei P-C, Chang AN, Kao J, Du Z, Meyers RM, Alt FW, et al. , Long neural genes harbor recurrent DNA break clusters in neural stem/progenitor cells., Cell. 164 (2016) 644–655. doi: 10.1016/j.cell.2015.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Schwer B, Wei P-C, Chang AN, Kao J, Du Z, Meyers RM, et al. , Transcription-associated processes cause DNA double-strand breaks and translocations in neural stem/progenitor cells., Proc. Natl. Acad. Sci. USA. 113 (2016) 2258–2263. doi: 10.1073/pnas.1525564113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Klein IA, Resch W, Jankovic M, Oliveira T, Yamane A, Nakahashi H, et al. , Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements inB lymphocytes., Cell. 147 (2011) 95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Yang F, Kemp CJ, Henikoff S, Anthracyclines induce double-strand DNA breaks at active gene'' promoters., Mutat. Res. 773 (2015) 9–15. doi: 10.1016/j.mrfmmm.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Tchurikov NA, Kretova OV, Fedoseeva DM, Chechetkin VR, Gorbacheva MA, Snezhkina AV, et al. , Genome-wide mapping of hot spots of DNA double-strand breaks in human cells as a tool for epigenetic studies and cancer genomics., Genom. Data. 5 (2015) 89–93. doi: 10.1016/j.gdata.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kravatsky YV, Chechetkin VR, Tchurikov NA, Kravatskaya GI, Genome-wide study of correlations between genomic features and their relationship with the regulation of gene expression., DNA Res. 22 (2015) 109–119. doi: 10.1093/dnares/dsu044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Baranello L, Kouzine F, Wojtowicz D, Cui K, Przytycka TM, Zhao K, et al. , DNA break mapping reveals topoisomerase II activity genome-wide., Int. J. Mol. Sci. 15 (2014) 13111–13122. doi: 10.3390/ijms150713111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Biernacka A, Zhu Y, Skrzypczak M, Forey R, Pardo B, Grzelak M, et al. , i-BLESS is an ultra-sensitive method for detection of DNA double-strand breaks., Commun. Biol. 1 (2018) 181. doi: 10.1038/s42003-018-0165-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Costantino L, Koshland D, Genome-wide Map of R-Loop-Induced Damage Reveals How a Subset of R-Loops Contributes to Genomic Instability., Mol. Cell. 71 (2018) 487–497.e3. doi: 10.1016/j.molcel.2018.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Tubbs A, Sridharan S, van Wietmarschen N, Maman Y, Callen E, Stanlie A, et al. , Dual roles of poly(da:dt) tracts in replication initiation and fork collapse., Cell. 174 (2018) 1127–1142.e19. doi: 10.1016/j.cell.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Gadaleta MC, Noguchi E, Regulation of DNA Replication through Natural Impediments in the Eukaryotic Geno me., Genes (Basel). 8 (2017). doi: 10.3390/genes8030098. [DOI] [Google Scholar]
- [69].García-Muse T, Aguilera A, Transcription-replication conflicts: how they occur and how they are resolved., Nat. Rev. Mol. Cell Biol. 17 (2016) 553–563. doi: 10.1038/nrm.2016.88. [DOI] [PubMed] [Google Scholar]
- [70].Gaillard H, Aguilera A, Transcription as a threat to genome integrity., Annu. Rev. Biochem. 85 (2016) 291–317. doi: 10.1146/annurev-biochem-060815-014908. [DOI] [PubMed] [Google Scholar]
- [71].Yu K, Chedin F, Hsieh C-L, Wilson TE, Lieber MR, R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells., Nat. Immunol. 4 (2003)442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- [72].Ribeiro de Almeida C, Dhir S, Dhir A, Moghaddam AE, Sattentau Q, Meinhart A, et al. , RNA Helicase DDX1 Converts RNA G-Quadruplex Structures into R-Loops to Promote IgH Class Switch Recombination., Mol. Cell. 70 (2018) 650–662.e8. doi: 10.1016/j.molcel.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Sollier J, Stork CT, García-Rubio ML, Paulsen RD, Aguilera A, Cimprich KA, Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability., Mol. Cell. 56 (2014) 777–785. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Makharashvili N, Arora S, Yin Y, Fu Q, Wen X, Lee J-H, et al. , Sae2/CtIP prevents R-loop accumulation in eukaryotic cells., Elife. 7 (2018). doi: 10.7554/eLife.42733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].García-Pichardo D, Cañas JC, García-Rubio ML, Gómez-Gonzïlez B, Rondón AG, Aguilera A, Histone Mutants Separate R Loop Formation from Genome Instability Induction., Mol. Cell. 66 (2017) 597–609.e5. doi: 10.1016/j.molcel.2017.05.014. [DOI] [PubMed] [Google Scholar]
- [76].Stork CT, Bocek M, Crossley MP, Sollier J, Sanz LA, Chédin F, et al. , Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage., Elife. 5 (2016). doi: 10.7554/eLife.17548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Mladenov E, Magin S, Soni A, Iliakis G, DNA double-strand-break repair in higher eukaryotes and its role in genomic instability and cancer: Cell cycle and proliferation-dependent regulation., Semin. Cancer Biol. 37-38 (2016) 51–64. doi: 10.1016/j.semcancer.2016.03.003. [DOI] [PubMed] [Google Scholar]
- [78].Hustedt N, Durocher D, The control of DNA repair by the cell cycle., Nat. Cell Biol. 19 (2016) 1–9. doi: 10.1038/ncb3452. [DOI] [PubMed] [Google Scholar]
- [79].Jimeno S, Mejías-Navarro F, Prados-Carvajal R, Huertas P, Controlling the balance between chromosome break repair pathways., Adv. Protein Chem. Struct. Biol. 115 (2019) 95–134. doi: 10.1016/bs.apcsb.2018.10.004. [DOI] [PubMed] [Google Scholar]
- [80].Harlen KM, Churchman LS, The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain., Nat. Rev. Mol. Cell Biol. 18 (2017) 263–273. doi: 10.1038/nrm.2017.10. [DOI] [PubMed] [Google Scholar]
- [81].Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA, ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks., Cell. 141 (2010) 970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Pankotai T, Bonhomme C, Chen D, Soutoglou E, DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks., Nat. Struct. Mol. Biol. 19(2012)276–282. doi: 10.1038/nsmb.2224. [DOI] [PubMed] [Google Scholar]
- [83].Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, et al. , A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage., Proc. Natl. Acad. Sci. USA. 107 (2010) 18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Ui A, Nagaura Y, Yasui A, Transcriptional elongation factor ENL phosphorylated by ATM recruits polycomb and switches off transcription for DSB repair., Mol. Cell. 58 (2015) 468–482. doi: 10.1016/j.molcel.2015.03.023. [DOI] [PubMed] [Google Scholar]
- [85].Awwad SW, Abu-Zhayia ER, Guttmann-Raviv N, Ayoub N, NELF-E is recruited to DNA double-strand break sites to promote transcriptional repression and repair., EMBO Rep. 18 (2017) 745–764. doi: 10.15252/embr.201643191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. , Role of histone H2A ubiquitination in Polycomb silencing., Nature. 431 (2004) 873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- [87].Sanchez A, De Vivo A, Uprety N, Kim J, Stevens SM, Kee Y, BMI1-UBR5 axis regulates transcriptional repression at damaged chromatin., Proc. Natl. Acad. Sci. USA. 113 (2016) 11243–11248. doi: 10.1073/pnas.1610735113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Ismail IH, Andrin C, McDonald D, Hendzel MJ, BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair., J. Cell Biol. 191 (2010) 45–60. doi: 10.1083/jcb.201003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Ginjala V, Nacerddine K, Kulkami A, Oza J, Hill SJ, Yao M, et al. , BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair., Mol. Cell. Biol. 31 (2011) 1972–1982. doi: 10.1128/MCB.00981-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Campbell S, Ismail IH, Young LC, Poirier GG, Hendzel MJ, Polycomb repressive complex 2 contributes to DNA double-strand break repair., Cell Cycle. 12 (2013) 2675–2683. doi: 10.4161/cc.25795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Kakarougkas A, Ismail A, Chambers AL, Riballo E, Herbert AD, Künzel J, et al. , Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin., Mol. Cell. 55 (2014) 723–732. doi: 10.1016/j.molcel.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Rona G, Roberti D, Yin Y, Pagan JK, Homer H, Sassani E, et al. , PARP1-dependent recruitment of the FBXL10-RNF68-RNF2 ubiquitin ligase to sites of DNA damage controls H2A.Z loading., Elife. 7 (2018). doi: 10.7554/eLife.38771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].O’Connor HF, Lyon N, Leung JW, Agarwal P, Swaim CD, Miller KM, et al. , Ubiquitin-Activated Interaction Traps (UBAITs) identify E3 ligase binding partners., EMBO Rep. 16(2015) 1699–1712. doi: 10.15252/embr.201540620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Meisenberg C, Pinder SI, Hopkins SR, Wooller SK, Benstead-Hume G, Pearl FMG, et al. , Repression of Transcription at DNA Breaks Requires Cohesin throughout Interphase and Prevents Genome Instability., Mol. Cell. 73 (2019) 212–223.e7. doi: 10.1016/j.molcel.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Gong F, Clouaire T, Aguirrebengoa M, Legube G, Miller KM, Histone demethylase KDM5A regulates the ZMYND8-NuRD chromatin remodeler to promote DNA repair., J. Cell Biol. 216 (2017) 1959–1974. doi: 10.1083/jcb.201611135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Gong F, Miller KM, Double duty: ZMYND8 in the DNA damage response and cancer., Cell Cycle. 17 (2018) 414–420. doi: 10.1080/15384101.2017.1376150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Savitsky P, Krojer T, Fujisawa T, Lambert J-P, Picaud S, Wang C-Y, et al. , Multivalent Histone and DNA Engagement by a PHD/BRD/PWWP Triple Reader Cassette Recruits ZMYND8 to K14ac-Rich Chromatin., Cell Rep. 17 (2016) 2724–2737. doi: 10.1016/j.celrep.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Spruijt CG, Luijsterburg MS, Menafra R, Lindeboom RGH, Jansen PWTC, Edupuganti RR, et al. , ZMYND8 Co-localizes with NuRD on Target Genes and Regulates Poly(ADP-Ribose)-Dependent Recruitment of GATAD2A/NuRD to Sites of DNA Damage., Cell Rep. 17 (2016) 783–798. doi: 10.1016/j.celrep.2016.09.037. [DOI] [PubMed] [Google Scholar]
- [99].Gong F, Chiu L-Y, Cox B, Aymard F, Clouaire T, Leung JW, et al. , Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination., Genes Dev. 29 (2015) 197–211. doi: 10.1101/gad.252189.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Clouaire T, Rocher V, Lashgari A, Amould C, Aguirrebengoa M, Biemacka A, et al. , Comprehensive Mapping of Histone Modifications at DNA Double-Strand Breaks Deciphers Repair Pathway Chromatin Signatures., Mol. Cell. 72 (2018) 250–262.e6. doi: 10.1016/j.molcel.2018.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Xiao T, Kao C-F, Krogan NJ, Sun Z-W, Greenblatt JF, Osley MA, et al. , Histone H2B ubiquitylation is associated with elongating RNA polymerase II., Mol. Cell. Biol. 25 (2005) 637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Wu L, Li L, Zhou B, Qin Z, Dou Y, H2B ubiquitylation promotes RNA Pol II processivity via PAF1 and pTEFb., Mol. Cell. 54 (2014) 920–931. doi: 10.1016/j.molcel.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Clouaire T, Legube G, A Snapshot on the Cis Chromatin Response to DNA Double-Strand Breaks., Trends Genet. (2019). doi: 10.1016/j.tig.2019.02.003. [DOI] [PubMed] [Google Scholar]
- [104].Marnef A, Cohen S, Legube G, Transcription-Coupled DNA Double-Strand Break Repair: Active Genes Need Special Care., J. Mol. Biol. 429 (2017) 1277–1288. doi: 10.1016/j.jmb.2017.03.024. [DOI] [PubMed] [Google Scholar]
- [105].Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, Briois S, et al. , Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks., Nat. Struct. Mol. Biol. 21 (2014) 366–374. doi: 10.1038/nsmb.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].D’Alessandro G, Whelan DR, Howard SM, Vitelli V, Renaudin X, Adamowicz M, et al. , BRCA2 controls DNA:RNA hybrid level at DSBs by mediating RNase H2 recruitment., Nat. Commun. 9 (2018) 5376. doi: 10.1038/s41467-018-07799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Yasuhara T, Kato R, Hagiwara Y, Shiotani B, Yamauchi M, Nakada S, et al. , Human Rad52 Promotes XPG-Mediated R-loop Processing to Initiate Transcription-Associated Homologous Recombination Repair., Cell. 175 (2018) 558–570.e11. doi: 10.1016/j.cell.2018.08.056. [DOI] [PubMed] [Google Scholar]
- [108].Wei L, Nakajima S, Böhm S, Bernstein KA, Shen Z, Tsang M, et al. , DNA damage during the G0/G1 phase triggers RNA-templated, Cockayne syndrome B-dependent homologous recombination., Proc. Natl. Acad. Sci. USA. 112 (2015) E3495–504. doi: 10.1073/pnas.1507105112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Teng Y, Yadav T, Duan M, Tan J, Xiang Y, Gao B, et al. , ROS-induced R loops trigger a transcription-coupled but BRCA1/2-independent homologous recombination pathway through CSB., Nat. Commun. 9 (2018) 4115. doi: 10.1038/s41467-018-06586-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Wang Q, Oliveira T, Jankovic M, Silva IT, Hakim O, Yao K, et al. , Epigenetic targeting of activation-induced cytidine deaminase., Proc. Natl. Acad. Sci. USA. Ill (2014) 18667–18672. doi: 10.1073/pnas.1420575111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Hasham MG, Donghia NM, Coffey E, Maynard J, Snow KJ, Ames J, et al. , Widespread genomic breaks generated by activation-induced cytidine deaminase are prevented by homologous recombination., Nat. Immunol. 11 (2010) 820–826. doi: 10.1038/ni.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Pfister SX, Ahrabi S, Zalmas L-P, Sarkar S, Aymard F, Bachrati CZ, et al. , SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability., Cell Rep. 7 (2014) 2006–2018. doi: 10.1016/j.celrep.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Daugaard M, Baude A, Fugger K, Povlsen LK, Beck H, Sørensen CS, et al. , LEDGF (p75) promotes DNA-end resection and homologous recombination., Nat. Struct. Mol. Biol. 19 (2012) 803–810. doi: 10.1038/nsmb.2314. [DOI] [PubMed] [Google Scholar]
- [114].Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, et al. , Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination., Nat. Struct. Mol. Biol. 20 (2013) 317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Jacquet K, Fradet-Turcotte A, Avvakumov N, Lambert J-P, Roques C, Pandita RK, et al. , The TIP60 Complex Regulates Bivalent Chromatin Recognition by 53BP1 through Direct H4K20me Binding and H2AK15 Acetylation., Mol. Cell. 62 (2016) 409–421. doi: 10.1016/j.molcel.2016.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Clouaire T, Legube G, DNA double strand break repair pathway choice: a chromatin based decision?, Nucleus. 6 (2015) 107–113. doi: 10.1080/19491034.2015.1010946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Aymard F, Aguirrebengoa M, Guillou E, Javierre BM, Bugler B, Arnould C, et al. , Genome-wide mapping of long-range contacts unveils clustering of DNA double-strand breaks at damaged active genes., Nat. Struct. Mol. Biol. 24 (2017) 353–361. doi: 10.1038/nsmb.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Hasham MG, Snow KJ, Donghia NM, Branca JA, Lessard MD, Stavnezer J, et al. , Activation-induced cytidine deaminase-initiated off-target DNA breaks are detected and resolved during S phase., J. Immunol. 189 (2012) 2374–2382. doi: 10.4049/jimmunol.1200414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Yamane A, Robbiani DF, Resch W, Bothmer A, Nakahashi H, Oliveira T, et al. , RPA accumulation during class switch recombination represents 5’-3' DNA-end resection during the S-G2/M phase off the cell cycle., Cell Rep. 3 (2013) 138–147. doi: 10.1016/j.celrep.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Biehs R, Steinlage M, Barton O, Juhász S, Künzel J, Spies J, et al. , DNA Double-Strand Break Resection Occurs during Non-homologous End Joining in G1 but Is Distinct from Resection during Homologous Recombination., Mol. Cell. 65 (2017) 671–684.e5. doi: 10.1016/j.molcel.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Löbrich M, Jeggo P, A Process of Resection-Dependent Nonhomologous End Joining Involving the Goddess Artemis., Trends Biochem. Sci. 42 (2017) 690–701. doi: 10.1016/j.tibs.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Mirman Z, Lottersberger F, Takai H, Kibe T, Gong Y, Takai K, et al. , 53BP1-RIF1-shieldin counteracts DSB resection through CST- and Polα-dependent fill-in., Nature. 560 (2018) 112–116. doi: 10.1038/s41586-018-0324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Wiedemann E-M, Peycheva M, Pavri R, DNA Replication Origins in Immunoglobulin Switch Regions Regulate Class Switch Recombination in an R-Loop-Dependent Manner., Cell Rep. 17 (2016) 2927–2942. doi: 10.1016/j.celrep.2016.11.041. [DOI] [PubMed] [Google Scholar]
- [124].He M, Cortizas EM, Verdun RE, Severinson E, Cyclin-dependent kinases regulate Ig class switching by controlling access of AID to the switch region., J. Immunol. 194 (2015) 4231–4239. doi: 10.4049/jimmunol.1402146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Britton S, Dernoncourt E, Delteil C, Froment C, Schiltz O, Salles B, et al. , DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal., Nucleic Acids Res. 42 (2014) 9047–9062. doi: 10.1093/nar/gku601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Li L, Germain DR, Poon H-Y, Hildebrandt MR, Monckton EA, McDonald D, et al. , DEAD Box 1 Facilitates Removal of RNA and Homologous Recombination at DNA Double-Strand Breaks., Mol. Cell. Biol. 36 (2016) 2794–2810. doi: 10.1128/MCB.00415-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Burger K, Schlackow M, Gullerova M, Tyrosine kinase c-Abl couples RNA polymerase II transcription to DNA double-strand breaks., Nucleic Acids Res. (2019). doi: 10.1093/nar/gkz024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Zhao H, Zhu M, Limbo O, Russell P, RNase H eliminates R-loops that disrupt DNA replication but is nonessential for efficient DSB repair., EMBO Rep. 19 (2018). doi: 10.15252/embr.201745335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Cohen S, Puget N, Lin Y-L, Clouaire T, Aguirrebengoa M, Rocher V, et al. , Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations., Nat. Commun. 9 (2018) 533. doi: 10.1038/s41467-018-02894-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Lu W-T, Hawley BR, Skalka GL, Baldock RA, Smith EM, Bader AS, et al. , Drosha drives the formation of DNA:RNA hybrids around DNA break sites to facilitate DNA repair., Nat. Commun. 9 (2018) 532. doi: 10.1038/s41467-018-02893-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Zhang X, Chiang H-C, Wang Y, Zhang C, Smith S, Zhao X, et al. , Attenuation of RNA polymerase II pausing mitigates BRCA1-associated R-loop accumulation and tumorigenesis., Nat. Commun. 8 (2017) 15908. doi: 10.1038/ncomms15908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Shivji MKK, Renaudin X, Williams ÇH, Venkitaraman AR, BRCA2 Regulates Transcription Elongation by RNA Polymerase II to Prevent R-Loop Accumulation., Cell Rep. 22 (2018) 1031–1039. doi: 10.1016/j.celrep.2017.12.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Ohle C, Tesorero R, Schermann G, Dobrev N, Sinning I, Fischer T, Transient RNA-DNA Hybrids Are Required for Efficient Double-Strand Break Repair., Cell. 167 (2016) 1001–1013.e7. doi: 10.1016/j.cell.2016.10.001. [DOI] [PubMed] [Google Scholar]
- [134].Michelini F, Pitchiaya S, Vitelli V, Sharma S, Gioia U, Pessina F, et al. , Damage-induced IncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks., Nat. Cell Biol. 19 (2017) 1400–1411. doi: 10.1038/ncb3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Iannelli F, Galbiati A, Capozzo I, Nguyen Q, Magnuson B, Michelini F, et al. , A damaged genome’s transcriptional landscape through multilayered expression profiling around in situ-mapped DNA double-strand breaks., Nat. Commun. 8 (2017) 15656. doi: 10.1038/ncomms15656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Welty S, Teng Y, Liang Z, Zhao W, Sanders LH, Greenamyre JT, et al. , RAD52 is required for RNA-templated recombination repair in post-mitotic neurons., J. Biol. Chem. 293 (2018) 1353–1362. doi: 10.1074/jbc.M117.808402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Michelini F, Jalihal AP, Francia S, Meers C, Neeb ZT, Rossiello F, et al. , From “cellular” RNA to “smart” RNA: multiple roles of RNA in genome stability and beyond., Chem. Rev. 118 (2018) 4365–4403. doi: 10.1021/acs.chemrev.7b00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Merk K, Breinig M, Böttcher R, Krebs S, Blum H, Boutros M, et al. , Splicing stimulates siRNA formation at Drosophila DNA double-strand breaks., PLoS Genet. 13 (2017) e1006861. doi: 10.1371/journal.pgen.1006861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Vítor AC, Sridhara SC, Sabino JC, Afonso AI, Grosso AR, Martin RM, et al. , Single-molecule imaging of transcription at damaged chromatin., Sci. Adv. 5 (2019) eaau1249. doi: 10.1126/sciadv.aau1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Bonath F, Domingo-Prim J, Tarbier M, Friedländer MR, Visa N, Next-generation sequencing reveals two populations of damage-induced small RNAs at endogenous DNA double-strand breaks., Nucleic Acids Res. 46 (2018) 11869–11882. doi: 10.1093/nar/gky1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Keskin H, Shen Y, Huang F, Patel M, Yang T, Ashley K, et al. , Transcript-RNA-templated DNA recombination and repair., Nature. 515 (2014) 436–439. doi: 10.1038/nature13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Meers C, Keskin H, Storici F, DNA repair by RNA: Templated, or not templated, that is the question., DNA Repair (Amst.). 44 (2016) 17–21. doi: 10.1016/j.dnarep.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Mazina OM, Keskin H, Hanamshet K, Storici F, Mazin AV, Rad52 Inverse Strand Exchange Drives RNA-Templated DNA Double-Strand Break Repair., Mol. Cell. 67 (2017) 19–29.e3. doi: 10.1016/j.molcel.2017.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Nguyen HD, Yadav T, Giri S, Saez B, Graubert TA, Zou L, Functions of replication protein A as a sensor of R loops and a regulator of rnaseh1., Mol. Cell. 65 (2017) 832–847.e4. doi: 10.1016/j.molcel.2017.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Amon JD, Koshland D, RNase H enables efficient repair of R-loop induced DNA damage., Elife. 5 (2016). doi: 10.7554/eLife.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Mendoza O, Bourdoncle A, Boulé J-B, Brosh RM, Mergny J-L, G-quadruplexes and helicases., Nucleic Acids Res. 44 (2016) 1989–2006. doi: 10.1093/nar/gkw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Croteau DL, Popuri V, Opresko PL, Bohr VA, Human RecQ helicases in DNA repair, recombination, and replication., Annu. Rev. Biochem. 83 (2014) 519–552. doi: 10.1146/annurev-biochem-060713-035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Dhar S, Brosh RM, BLM’s balancing act and the involvement of FANCJ in DNA repair., Cell Cycle. 17 (2018) 2207–2220. doi: 10.1080/15384101.2018.1520567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Jimeno S, Camarillo R, Mejías-Navarro F, Fernández-Ávila MJ, Soria-Bretones I, Prados-Carvajal R, et al. , The Helicase PIF1 Facilitates Resection over Sequences Prone to Forming G4 Structures., Cell Rep. 25 (2018) 3543. doi: 10.1016/j.celrep.2018.12.029. [DOI] [PubMed] [Google Scholar]
- [150].León-Ortiz AM, Panier S, Sarek G, Vannier J-B, Patel H, Campbell PJ, et al. , A distinct class of genome rearrangements driven by heterologous recombination., Mol. Cell. 69 (2018) 292–305.e6. doi: 10.1016/j.molcel.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Masuda-Sasa T, Polaczek P, Peng XP, Chen L, Campbell JL, Processing of G4 DNA by Dna2 helicase/nuclease and replication protein A (RPA) provides insights into the mechanism of Dna2/RPA substrate recognition., J. Biol. Chem. 283 (2008) 24359–24373. doi: 10.1074/jbc.M802244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Scheibye-Knudsen M, Tseng A, Borch Jensen M, Scheibye-Alsing K, Fang EF, Iyama T, et al. , Cockayne syndrome group A and B proteins converge on transcription-linked resolution of non-B DNA., Proc. Natl. Acad. Sci. USA. 113 (2016) 12502–12507. doi: 10.1073/pnas.1610198113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Kobayashi S, Fukatsu R, Kanoh Y, Kakusho N, Matsumoto S, Chaen S, et al. , Both a Unique Motif at the C Terminus and an N-Terminal HEAT Repeat Contribute to G-Quadruplex Binding and Origin Regulation by the Rif1 Protein., Mol. Cell. Biol. 39 (2019). doi: 10.1128/MCB.00364-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Kanoh Y, Matsumoto S, Fukatsu R, Kakusho N, Kono N, Renard-Guillet C, et al. , Rif1 binds to G quadruplexes and suppresses replication over long distances., Nat. Struct. Mol. Biol. 22 (2015) 889–897. doi: 10.1038/nsmb.3102. [DOI] [PubMed] [Google Scholar]
- [155].Marcomini I, Shimada K, Delgoshaie N, Yamamoto I, Seeber A, Cheblal A, et al. , Asymmetric Processing of DNA Ends at a Double-Strand Break Leads to Unconstrained Dynamics and Ectopic Translocation., Cell Rep. 24 (2018) 2614–2628.e4. doi: 10.1016/j.celrep.2018.07.102. [DOI] [PubMed] [Google Scholar]
- [156].Chen H, Lisby M, Symington LS, RPA coordinates DNA end resection and prevents formation of DNA hairpins., Mol. Cell. 50 (2013) 589–600. doi: 10.1016/j.molcel.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Sugiyama T, Zaitseva EM, Kowalczykowski SC, A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein., J. Biol. Chem. 272 (1997) 7940–7945. [DOI] [PubMed] [Google Scholar]
- [158].Day TA, Layer JV, Cleary JP, Guha S, Stevenson KE, Tivey T, et al. , PARP3 is a promoter of chromosomal rearrangements and limits G4 DNA., Nat. Commun. 8 (2017) 15110. doi: 10.1038/ncomms15110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Hoffman EA, McCulley A, Haarer B, Arnak R, Feng W, Break-seq reveals hydroxyurea-induced chromosome fragility as a result of unscheduled conflict between DNA replication and transcription., Genome Res. 25 (2015) 402–412. doi: 10.1101/gr.180497.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Madabhushi R, Gao F, Pfenning AR, Pan L, Yamakawa S, Seo J, et al. , Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes., Cell. 161 (2015) 1592–1605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Canela A, Maman Y, Jung S, Wong N, Callen E, Day A, et al. , Genome organization drives chromosome fragility., Cell. 170 (2017) 507–521.el8. doi: 10.1016/j.cell.2017.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Grégoire M-C, Leduc F, Morin MH, Cavé T, Arguin M, Richter M, et al. , The DNA double-strand “breakome” of mouse spermatids., Cell Mol. Life Sci. 75 (2018) 2859–2872. doi: 10.1007/s00018-018-2769-0. [DOI] [PMC free article] [PubMed] [Google Scholar]