Abstract
Accidental introduction through ballast water and biofouling are currently the main factors responsible for spreading non-indigenous species in the marine realm. In the Southwestern Atlantic, two scleractinian corals, Tubastraea coccinea and T. tagusensis, have been introduced by opportunistic colonization in 1980 and are now widespread along more than 3,500 km of coastline. To better understand the invasion process and the role of vectors in spreading these species, we sampled 306 and 173 colonies of T. coccinea and T. tagusensis from invaded sites, possible vectors and one native population. Analyses revealed a higher diversity of multi-locus genotypes (MLGs) on vectors, suggesting that they were contaminated prior to their arrival in the Southwestern Atlantic, and a high proportion of clones at invaded sites, with few genotypes spread over ~2,000 km. This broad distribution is most likely a result of secondary introductions through the transport of contaminated vectors. Results also suggest the occurrence of multiple invasions, mainly in the northernmost sites. In summary, clonality, secondary introductions, and multiple invasions are the main reasons for the broad spread and invasive success of Tubastraea spp. in the Southwestern Atlantic. Consequently, the correct control of vectors is the most effective approach for management and prevention of new invasions.
Subject terms: Invasive species, Molecular ecology, Population genetics
Introduction
Marine bioinvasion is reshaping the distribution and biogeographic patterns of species worldwide and is reaching unprecedented levels with hundreds of species being transported to new environments every year1–4. Accidental introductions can occur through a number of ways, such as aquaculture, trade in ornamental species, canals linking previously unconnected waters, ballast water and biofouling, the last two being the main factors responsible for spreading non-indigenous species in the marine realm2,5–7. As a consequence of the increasing marine traffic, vessels (e.g. cargo ships, oil platforms, floating docks, buoys; herein called vectors) transport a large number of species (either by ballast water or biofouling), some of which will be able to establish and disperse, becoming invasive in the new environment6,8.
Recently, a review listed 15 non-indigenous species causing major negative impacts in the South Atlantic, including two azooxanthellate corals (Tubastraea coccinea and T. tagusensis)9. T. coccinea was first reported in the Atlantic during the 1940’s in Curaçao and Puerto Rico, and was probably introduced as biofouling on ship hulls traveling from Indo-Pacific waters10–12. Further records of the genus were reported in the Northwestern Atlantic in 2004 (T. coccinea)13, Southwestern Atlantic in the 1980’s (T. coccinea and T. tagusensis)14,15 and Gulf of Mexico in 2010 (T. coccinea and T. micranthus)16. Given the pattern of sea surface currents and previous examples from other invasive species, the Gulf of Mexico and Florida could have been naturally invaded by Caribbean populations12. However, the occurrence of T. micranthus, a species not yet found in the Caribbean, on oil platforms at Gulf of Mexico indicates that human vectors have also been responsible for the introduction into this region17. In the Southwestern Atlantic, Tubastraea spp. were firstly reported as fouling on offshore oil platforms in Rio de Janeiro State12,14. The genus is now widespread on rocky shores and artificial substrates (oil platforms, buoys, wrecks, piers, and drillships) along more than 3,500 km, from Ceará (02°29′S, 39°51′W)18 to Santa Catarina (27°17′S, 48°22′W)19, outcompeting native and endemic species20–24. Although there is no doubt that the introduction in the Southwestern Atlantic was through biofouling12: (1) the invasion history remains unclear; (2) there are no studies elucidating why these two species have been so successful in invading the Brazilian coast; and (3) the role of vectors in spreading them along the coast is unknown.
Successful invasive species frequently share a set of life history and ecological traits that facilitate their establishment, such as rapid growth rate, large number of offspring (r-selected species), sexual and asexual reproduction, early maturity and phenotypic plasticity25. When combined with high propagule pressure, a measure of the number of individuals released and the number of release events, the chances of survival in a new environment are considerably enhanced26,27. Tubastraea spp. possesses all such traits28–33, which may have facilitated their successful establishment and dispersal in the Southwestern Atlantic. Furthermore, considering their reproductive biology and rapid expansion along the Brazilian coastline, it is highly likely that vectors have been playing a key role in Tubastraea spp. dispersion along the Southwestern Atlantic34. Indeed, Tubastraea spp. have been recorded on at least 23 vectors, some of which have been towed along the Brazilian coast without biofouling control12.
The occurrence of multiple introductions has also been correlated with invasion success35–40. When founded by a small number of individuals, recently established populations can suffer a drastic reduction in genetic diversity as a consequence of genetic drift. Ultimately, genetic drift can have negative consequences such as the fixation of deleterious alleles and decrease of the species resilience25,41–45. On the other hand, multiple introduction events of non-indigenous species from more than one native population can lead to an increase in genetic diversity by mixing previously separated populations and increasing the propagule pressure, consequently reducing negative genetic outcomes of the invasion process and enhancing the possibility of a successful invasion7,26,27,36,40,46,47.
A better understanding of the invasion process and the ways by which invasive species are spreading into new environments is essential for improving management effectiveness and control plans that ultimately reduce or prevent future invasions6,25,48. Here we use a set of microsatellite markers34 to: (1) investigate genetic diversity and clonality at invaded sites and on vectors along the Brazilian coast; (2) provide insights into the role of vectors at spreading these invasive corals in the Southwestern Atlantic; and (3) evaluate the population structure at invaded sites with regard to the possibility of multiple introduction events.
Results
Clonality
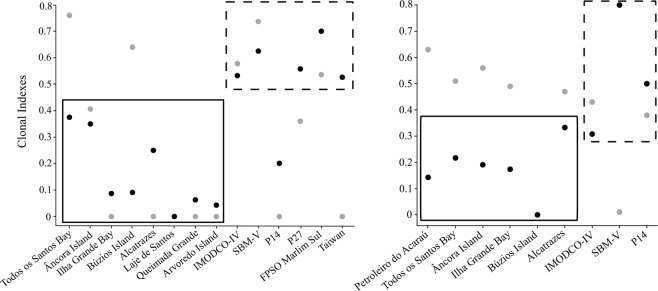
Analyses revealed a high proportion of clones for both T. coccinea and T. tagusensis with only 84 (28%; N = 298) and 30 (18%; N = 166) unique MLGs within all sampled sites. Similar proportion of clones were found when all indivuduals with missing data were removed (T. coccinea: N = 250, MLG = 60 or 20%; T. tagusensis: N = 156, MLG = 22 or 13%). For T. coccinea, invaded sites had, in general, less MLGs when compared to vectors and native population, with five sites holding three or less MLGs (Table 1, Fig. 1). The same was not observed for T. tagusensis, for which the highest number of MLGs observed was at an invaded site (Alcatrazes: MLG = 7, Table 1, Fig. 2). The existence of two MLGs for T. coccinea at the invaded sites Queimada Grande and Santa Catarina is probably a consequence of missing data in one locus and these two populations are likely dominated by only one MLG. For T. coccinea, clonality was higher at invaded sites contrasting to vectors and native population (Fig. 3a). The genotypic evenness did not show a clear pattern, with five invaded sites, one vector and the native site being dominated by one genotype. However, the remaining sites (two invaded sites and three vectors) had more equitable distribution of ramets among the observed MLGs (genotypic evenness (V) close to 1, Fig. 3a). Interestingly, the same pattern for MLGs and clonal richness was not as clear for T. tagusensis. Invaded sites and vectors had similar numbers of MLGs (except for Búzios Island, with one MLG) and clonal richness was slightly lower on vectors, reaching 0.8 in the vector SBM-V (Fig. 3b). The genotypic evenness did not change between invaded sites and vectors for this species (Fig. 3b).
Table 1.
Summary of statistics per samples site for the species Tubastraea coccinea (N = 298) and T. tagusensis (N = 166).
| Status | Site | N | MLG | Psex | A | Ar | Ae | Ho | He | F IS |
|---|---|---|---|---|---|---|---|---|---|---|
| Tubastraea coccinea | ||||||||||
| Invaded | Todos os Santos Bay (TSB) | 25 | 10 | 0 | 22 | 2.2 | 3 | 0.58 | 0.55 | −0.01 |
| Âncora Island (AI) | 21 | 8 | 0 | 20 | 2.2 | 2 | 0.50 | 0.54 | 0.13 | |
| Ilha Grande Bay (IGB) | 24 | 3 | 1 | 13 | 1.8 | 0 | 0.50 | 0.39 | −0.16 | |
| Búzios Island (BI) | 12 | 2 | 1 | 14 | 2.2 | 0 | 0.54 | 0.42 | −0.15 | |
| Alcatrazes (Alc) | 21 | 6 | 0 | 20 | 2.4 | 1 | 0.57 | 0.52 | 0.02 | |
| Laje de Santos (LS) | 24 | 1 | 2 | 11 | 1.8 | 0 | 0.83 | 0.42 | −1 | |
| Queimada Grande (QG) | 17 | 2 | 1 | 11 | 1.8 | 0 | 0.83 | 0.42 | −1 | |
| Arvoredo Island (ArI) | 24 | 2 | 2 | 11 | 1.8 | 0 | 0.83 | 0.42 | −1 | |
| Vectors | IMODCO-IV | 39 | 21 | 0 | 41 | 2.8 | 8 | 0.54 | 0.66 | 0.21* |
| SBM-V | 25 | 16 | 0 | 23 | 2.2 | 0 | 0.41 | 0.48 | 0.18 | |
| P14 | 11 | 3 | 1 | 11 | 1.7 | 0 | 0.42 | 0.32 | −0.18 | |
| P27 | 14 | 8 | 0 | 23 | 2.5 | 0 | 0.54 | 0.57 | 0.12 | |
| FPSO Marlim Sul | 21 | 15 | 0 | 27 | 2.3 | 3 | 0.42 | 0.61 | 0.34* | |
| Native | Taiwan | 20 | 11 | 0 | 22 | 2.2 | 4 | 0.39 | 0.49 | 0.25 |
| Tubastraea tagusensis | ||||||||||
| Invaded | Petroleiro do Acaraú (PA) | 22 | 4 | 2 | 14 | 1.8 | 11 | 0.67 | 0.35 | −0.91 |
| Todos os Santos Bay (TSB) | 24 | 6 | 2 | 17 | 1.9 | 0 | 0.53 | 0.35 | −0.49 | |
| Âncora Island (AI) | 22 | 5 | 0 | 19 | 2.0 | 4 | 0.50 | 0.42 | −0.07 | |
| Ilha Grande Bay (IGB) | 24 | 5 | 3 | 14 | 1.7 | 0 | 0.58 | 0.32 | −0.81 | |
| Búzios Island (BI) | 24 | 1 | 2 | 13 | 1.6 | 0 | 0.62 | 0.31 | −1 | |
| Alcatrazes (Alc) | 19 | 7 | 2 | 16 | 1.9 | 2 | 0.62 | 0.36 | −0.69 | |
| Vectors | IMODCO-IV | 14 | 5 | 1 | 18 | 1.9 | 2 | 0.65 | 0.41 | −0.51 |
| SBM-V | 6 | 5 | 0 | 20 | 2.2 | 5 | 0.46 | 0.45 | 0.09 | |
| P14 | 11 | 6 | 0 | 20 | 2.1 | 4 | 0.57 | 0.41 | −0.30 | |
N = number of sampled individuals, MLG = number of unique multilocus genotypes per site, Psex = number of individuals with Psex ≥ 0.01, A = number of alleles, Ar = allelic richness, Ae = number of exclusive alleles, Ho = observed heterozygosity, He = expected heterozygosity. *Indicates significant deviations from Hardy-Weinberg equilibrium.
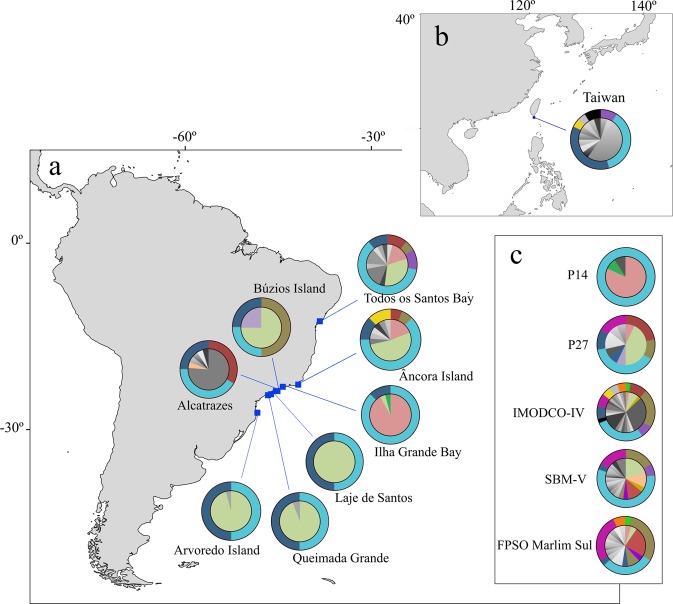
Figure 1.
Sampling sites for T. coccinea (a) along the invasive range in Southwestern Atlantic, (b) at a native population in Taiwan and (c) at five vectors located on the Brazilian coast. Pie diagrams show the number of multilocus genotypes (MLGs) (inner circle) and the allele frequency of the locus Tco 29 (with a total of 13 alleles, outer circle) per population. Colors indicate MLGs or alleles that are shared among two or more sites and gray scale indicate MLGs and alleles that are exclusive for the correspondent site (not observed on any other analyzed site). The locus Tco 29 is the second most diverse for T. coccinea and was chosen to exemplify allele sharing among invaded sites, vectors and Taiwan.
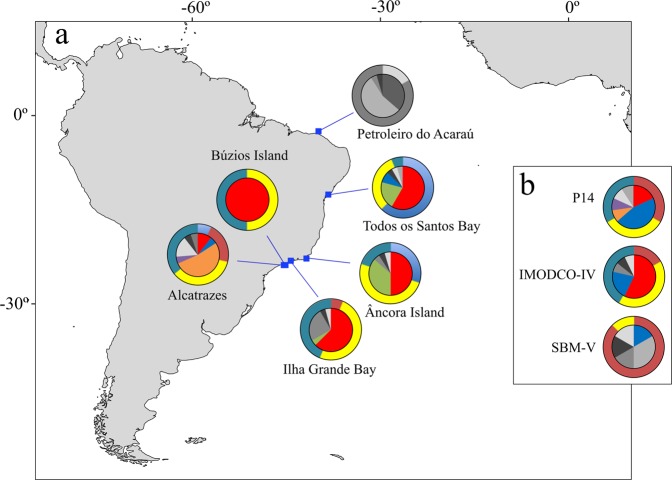
Figure 2.
Sampling sites for T. tagusensis (a) along the invasive range in Southwestern Atlantic and (b) at three vectors located on the Brazilian coast. Pie diagrams show the number of multilocus genotypes (MLGs) (inner circle) and the allele frequency of the locus Tco 34 (with a total of six alleles, outer circle) per population. Colors indicate MLGs or alleles that are shared among two or more sites and gray scale indicate MLGs and alleles that are exclusive for the correspondent site (not observed on any other analyzed site). The locus Tco 34 is the second most diverse for T. tagusensis and was chosen to exemplify allele sharing among invaded sites and vectors.
Figure 3.
Clonal indicess for (a) T. coccinea and (b) T. tagusensis. Black dots indicate the clonal richness (R), ranging from 0 to 1, when all samples analyzed correspond to a different MLG and gray dots indicate the genotypic evenness (V), ranging from 0 to 1, when genets each have the same number of ramets. Continuous and dotted rectangles indicate invaded sites and vectors (plus native Taiwanese population), respectively.
Of the observed MLGs, ten (12%) from T. coccinea and five (17%) from T. tagusensis are shared with one or more sampled sites (Figs 1 and 2). T. coccinea had five of its MLGs shared among invaded sites and vectors, while the remaining five were shared exclusively among vectors. Vectors also showed a higher number of exclusive MLGs, not shared with any other sampled site (Table 1; Fig. 1). Neither the invaded sites nor the vectors shared any MLGs with the native population from Taiwan. One MLG was found on three vectors and all invaded sites (except for Alcatrazes), indicating the occurrence of clones separated by over 1,900 km along the Brazilian coast (Fig. 1). Although for T. coccinea no MLG was shared between Alcatrazes and other invaded sites, the predominant MLG found at this site differed by only one allele from the predominant MLG found at other invaded sites (green MLG showed at the inner circle in Fig. 1). Similarly, for T. tagusensis, the predominant MLG found at Alcatrazes also differed by only one allele from the predominant MLG found at other invaded sites (red MLG showed at the inner circle in Fig. 2). Of the five MLGs observed for T. tagusensis, four were shared between at least one vector and one invaded site and one was found exclusively at invaded sites. The northernmost sampled site, Petroleiro do Acaraú, did not share MLGs with any other invaded site or vectors.
Genetic diversity
For the genetic diversity analyses, all clones found within populations were removed and only one representative of each MLG was included, additionally to those individuals with Psex value higher than 0.01 (Table 1). Within the analysed data set, a total of eight individuals of T. coccinea (3%) and 12 (7%) T. tagusensis had Psex value higher than 0.001, thus being considered products of distinct sexual reproduction events. Two loci from T. coccinea (Tco 1 and Tco 30) and two from T. tagusensis (Tco 4 and Tco 37) showed evidence of linkage disequilibrium with at least two other loci and were excluded from further genetic diversity analyses. There was no evidence of null alleles for T. coccinea, while for T. tagusensis three loci showed evidence of null alleles for one population each (Supplementary Table S1).
For T. coccinea, the number of alleles and allelic richness ranged from 11 to 41 and 1.7 to 2.8, respectively. For this species, exclusive alleles were found at three invaded sites (Todos os Santos Bay, Âncora Island and Alcatrazes), two vectors (SBM-V Araça and FPSO Marlim Sul) and in the native population (Taiwan) (Table 1). The frequency distribution of alleles for one locus (Tco 29, with 13 alleles – Fig. 1, outer circle) shows that invaded sites, vector and the native population share four alleles and that the number of exclusive alleles is higher on vectors and in the native population. All but Tco 5 has at least one allele shared among all sites, and Taiwan shares at least one allele per locus with one or more invaded or vector sites (Supplementary Table S2). Observed (Ho) and expected (He) heterozygosity ranged from 0.39 to 0.83 and 0.32 to 0.66, respectively (Table 1). Only the vectors IMODCO-IV and FPSO Marlim Sul had significant deficits of heterozygosity (Table 1). Although not significant, many sites (e.g., LS, QG and ArI; Table 1) had Ho values twice as high as He, indicating heterozygote excesses. However, it is important to note that these values need to be interpreted with caution, since all sites that displayed Ho excess had low number of individuals analysed. The inbreeding coefficient (Fis) was negative for most invaded sites (except for AI and Alc) and P14, indicating an excess of heterozygotes (Table 1). The remaining vectors and the native population had inbreeding coefficients ranging from 0.12 (i.e. P27) to 0.34 (i.e. FPSO Marlim Sul).
For T. tagusensis, the number of alleles and allelic richness were similar among sampled sites, ranging from 13 to 20 and 1.6 to 2.2, respectively. Exclusive alleles were found at all but three invaded sites (TSB, IGB and BI), with the highest number observed at the northernmost site, Petroleiro do Acaraú (Ae = 11; Table 1). Figure 2 shows the frequency distribution of alleles for the locus Tco 34 (outer circle), with all sites but Petroleiro do Acaraú sharing at least one allele. Five out of eight loci had shared alleles among Petroleiro do Acaraú and at least one other site and only three loci had shared alleles among all samples sites (Supplementary Table S2). Observed heterozygosity (Ho) (ranging from 0.46 to 0.67) was higher than the expected (He) (ranging from 0.31 to 0.45) in all sites (Table 1), with no significant deficits of heterozygosity. The inbreeding coefficient (Fis) was negative for all sites but SBM-V, indicating an excess of heterozygotes (Table 1).
Population structure
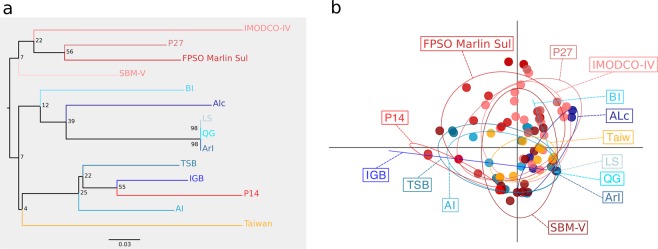
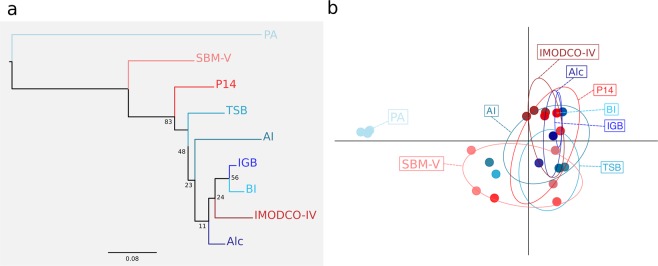
For T. coccinea, the NJ had low support values and PCA analysis showed that 95% confidence ellipses of each site were clearly overlapping, revealing no difference in overall gentic variance amond sites (Fig. 4b). Bayesian clustering analysis also did not recover any clear genetic cluster for any invaded site, vector or the native population of T. coccinea (Supplementary Fig. S1). The two methodologies used to estimate K gave similar results of five, eight or nine possible genetic clusters (Supplementary Fig. S1).
Figure 4.
(a) Neighbor-joining (NJ) tree based on Cavalli-Sforza’s and Edwards chord distance and (b) Principal Components Analysis (PCA) for T. coccinea.
For T. tagusensis, NJ analysis and PCA recovered Petroleiro do Acaraú as the most divergent site (first axis of PCA explaining 53% of the variance), while all remaining sites overlapped, indicating no clear difference in overall gentic variance amond them (Fig. 5b). Bayesian analysis recovered three genetic clusters (Supplementary Fig. S3), suggesting that the Southwestern Atlantic was colonized by more than one native population, as observed for T. coccinea. Except for Petroleiro do Acaraú, the observed genetic clusters were not a function of population structure between localities (Supplementary Fig. S3). No substructure was observed when analyzing higher K. Evidence of interbreeding between two clusters could be observed at Todos os Santos Bay, Alcatrazes and IMODCO-IV (Supplementary Fig. S3).
Figure 5.
(a) Neighbor-joining (NJ) tree based on Cavalli-Sforza’s and Edwards chord distance and (b) Principal Components Analysis (PCA) for T. tagusensis.
Discussion
Forty years after its first record, the genus Tubastraea has spread its range over 3,500 km along the Brazilian coast with increasing densities21,49. Here we show evidence that T. coccinea and T. tagusensis were introduced in the Southwestern Atlantic Ocean in more than one event. Furthermore, our results point to a critical role of the past transport of oil platforms from the Indo-Pacific in introducing these species in the Southwestern Atlantic Ocean, and that buoys and Floating Production Storage and Offloading Vessels (FSPOs) that travel from offshore oil platforms to onshore localities are acting as vectors in spreading these species along the Brazilian coast.
For both T. coccinea and T. tagusensis the Southwestern Atlantic invaded sites have a higher proportion of clones when compared to vectors, with sites dominated by a single genotype (Figs 1 and 2, see also34). Thus, their early reproductive age33 and ability to reproduce asexually are probably the main factors responsible for their success as invaders. When environmental conditions are suitable, such abilities enable large densities from a small starting number of individuals. Interestingly, although T. coccinea from Taiwan had fewer clones when compared to the majority of the Brazilian invaded sites, half of the examined samples were composed of clones of the same genet. This demonstrates that asexual reproduction also plays an important role in its native distribution range. As discussed by previous studies, asexual reproduction in Tubastraea occurs through the asexual production of larvae29,34, a strategy broadly used by anthozoans50–52, but to date only observed in three scleractinian species, T. coccinea, T. diaphana and Pocillopora damicornis29,53,54.
Along the Southwestern Atlantic, T. coccinea and T. tagusensis had one dominant genotype each (shared by 36% and 46% of all analyzed individuals, respectively), showing an over-representation of few genotypes. This may be a consequence of the invasion process where only one or a few genotypes better fitted to the recently invaded environment were able to establish and disperse55,56. A similar pattern was previously observed for Pocillopora damicornis in Hawaii55 and Reunion Islands57, and also for Acropora palmata in the French Antilles58. Nevertheless, those studies analyzed samples collected over small geographic scales (less than 20 km), while the dominant genotype of T. coccinea and T. tagusensis in invaded sites were spread over 1,500 and 2,000 km, respectively (Figs 1 and 2). Three non-exclusive hypotheses could explain such broad clonal distribution: (1) long-distance dispersal of asexual larvae; (2) multiple events of introduction from the same native population; or (3) secondary introduction from one invaded site to another.
Although possible for nearby localities, larval dispersal itself does not explain the broad distribution of clones at sites separated by more than 1,500 km. Tubastraea has a gregarious settlement behavior and most larvae settle within 1 to 3 days30, even though experiments in aquaria have shown that larvae can be competent for about 18 days30,33 and another observation mention competent T. coccinea larvae after 100 days (Richmond, pers. comm.59). Nevertheless, due to the Brazilian surface currents regime, it would be oceanographically not probable that competent larvae would travel long distances. Furthermore, the gregarious settlement with high local clonality observed for Tubastraea follows the “strawberry-coral” model60, when organisms use sexual reproduction to disperse genotypically diverse individuals, while asexual reproduction helps to spread locally adapted genotypes. Interestingly, an opposite hypothesis of dispersal capability was proposed for P. damicornis, where the asexual larvae would be responsible for long-distance dispersal61. Indeed, studies showing evidence of local recruitment from sexual larvae with low clonality corroborate the hypothesis that asexually produced larvae of P. damicornis may travel further62,63. Future studies comparing the size and duration of sexual and asexual larvae of Tubastraea spp. in situ are recommended to test if they follow such pattern.
The second and third hypotheses are the most likely explanations for the observed clonal distribution. The first record of Tubastraea spp. in the Southwestern Atlantic was on offshore oil platforms from the Campos Basin, north of Rio de Janeiro State, in the late 1980s. In such context, oil platforms built by foreign companies abroad that were slow towed to the Southwestern Atlantic may have been colonized with local fauna that were further transported as biofouling. Thus, such structures are the most probable means of introduction of these species along the Brazilian waters12. Once the platforms started to operate, their associated structures (such as buoys) and the year around movement of associated platforms and offshore support fleet shorewards are the most probable vectors for spreading Tubastraea spp. along the Brazilian coast. Indeed, the distributional range of Tubastraea spp. in the Southwestern Atlantic appears to be directly associated to sites with intense ship traffic and waterway terminals. de Paula and Creed64 analyzed the distribution and expansion of Tubastraea spp. at Ilha Grande Bay and found that the Petrobras oil terminal or Verolme shipyard were likely points of introduction of these species at that locality. In addition, Ferreira at el.65 found 22 non-indigenous species when analyzing drill-ships, platforms and cargo ships in Arraial do Cabo, including T. coccinea, and other studies had demonstrated that artificial substrates facilitate invasion66–68. Tubastraea spp. seem to be opportunistic and have been reported on artificial substrates at both invaded12–14,65 and native localities69. To confirm the hypothesis of multiple introduction events from the same native population it is essential to know the origin of the vessels and when they first arrived in the Southwestern Atlantic. However, this information is not easily traceable and could not be verified. Of the five vectors included in the present study, such information was traceable for two: P27 was brought from Singapore to Arraial do Cabo, Brazil, in 1998; and P14 was built in France in 1983, with no information of where or when it has first arrived in Brazil. Singapore is within the natural distributional range of Tubastraea spp.70 and it is highly possible that the platform P27 was infested before its arrival in the Atlantic. Indirect evidence supporting this assumption is the large size of colonies sampled on the platform and the first records of Tubastraea spp. in Arraial do Cabo only one-year after the arrival of P27 in the region12. Regarding the platform P14, it probably arrived free from Tubastraea, as there are no records of this genus in Europe yet. However, it was probably contaminated in Brazilian or Caribbean waters and then became a vector. Although we cannot confirm the origin of all vectors, the (i) occurrence of unique MLGs, (ii) exclusive alleles not found within invaded sites (Figs 1 and 2), and (iii) the higher number of alleles and clonal richness observed at all analyzed vectors (except for P14) compared to invaded sites, suggest that those vectors were already contaminated with Tubastraea spp. prior to their arrival in the Southwestern Atlantic.
The occurrence of secondary introduction from one invaded site to another is supported by information of vessels being transported along the Brazilian coast (Table 2)12 and the occurrence of clones on both invaded sites and vectors. The platforms P14 and P27 and the monobuoy IMODCO-IV were transported at least once along the Brazilian coast after being contaminated (Table 2)12. Furthermore, the traffic of FSPOs from contaminated platforms to onshore terminals might have acted as key vectors as well. Although Tubastraea spp. have been previously recorded at all cited sites prior to the arrival of the analyzed vectors, the transport of contaminated vectors further supports the spread of genotypes that may have not been at an invaded location. Tubastraea spp. were recorded on at least 23 vessels related to oil production (e.g. platforms, drillships, monobuoys)12 of which we have analyzed samples from only five. It is highly possible that vectors not included in this study were the primary responsible for introductions along several localities in the Southwestern Atlantic. Recently, Petrobras (2016) reported that 78% of the 32 structures they operate in the Sergipe region of northeast Brazil were also contaminated with Tubastraea spp.
Table 2.
Locations where the five analyzed vectors were recorded in the Southwestern Atlantic. Tc = T. coccinea; Tt = T. tagusensis.
| Vector | Location (coordinates) | Year | Species | Source |
|---|---|---|---|---|
| P14 | Caravelas field, Itajaí (26°46′2″S, 46°47′2.15″W) | 2000 | Tc | Identified by J. C. Creed from photographic register of Barreiros et al. (2000). |
| Angra dos Reis, Ilha Grande Bay (23°00′53″S, 44°18′59″W) | 2007 | Tc | In port, J. C. Creed (pers. obs.) | |
| Canteiro de São Roque, Todos os Santos Bay (12°51′16″S, 38°50′17″W) | 2014 | Tc/Tt | In port, J. C. Creed (pers. obs.) | |
| P27 | Voador field, Campos Basin (22°22′S, 40°24′W) | 2013 | Tc | Identified by J. C. Creed from photographic register communicated by Ricardo Guedes dos Santos (pers. comm.). |
| Canteiro de São Roque, Todos os Santos Bay (12°51′16″S, 38°50′17″W) | 2014 | Tc | In port, J. C. Creed (pers. obs.) | |
| IMODCO IV | Arraial do Cabo (22°58′21″S, 42°0′49″W) | 2007 | Tc/Tt | Mizrahi (2008) |
| São Sebastião (23°48′48″S, 45°24′11″W) | 2014 | Tc/Tt | In port, J. C. Creed (pers. obs.) | |
| SBM-5 Araça | São Sebastião (23°48′48″S, 45°24′11″W) | 2012 | Tc/Tt | In port, J. C. Creed (pers. obs.) |
| FPSO Marlim Sul | Bacia de Campos (22°32′38″S, 40°01′15″W) | 2016 | Tc | Identified by C. Zilberberg from samples provided by the company SBM-Off-shore |
Our results corroborate the hypothesis of multiple introduction events34, with at least five genetic clusters with no geographic pattern for T. coccinea. The lack of geographic pattern is likely a result of the transport of infested vectors previously discussed. However, as the software Structure is not appropriate for organisms that reproduce mainly asexually71, results should be interpreted with caution. Sammarco et al.72 found similar results when analyzing invasive populations of T. micranthus at two oil platforms in the Gulf of Mexico, with four distinct genetic clusters observed on one single platform, likely resulting from multiple introductions from distinct source populations. In addition to increasing the propagule pressure, multiple introductions can lead to an increase in genetic diversity via the isolate breaking effect by creating new genotypes, potentially benefiting invasive populations and enhancing their chance of survival35–40. Patterns of high genetic diversity of invaded populations associated with multiple introductions have been observed for other marine organisms, such as the green crab Carcinus maenas73, the nassariid gastropod Cyclope neritea74, the caprellid Caprella scaura at the Iberian Peninsula74,75, and others76,77.
NJ analyses had low support values for T. coccinea, showing that there is no specific structure in their distribution. Nevertheless, based on the distribution of MLG it is possible that the northern sites were invaded more than once, as they have higher genotypic diversity when compared to the southern sites. The later was invaded less than ten years ago mostly by the same genotypes, suggesting a single invasion event. For T. tagusensis, both Structure and NJ suggest that the Brazilian northernmost invaded site (Petroleiro do Acaraú) was colonized by a different population, not present at any other invaded site or vector analyzed herein, and is most probably the result from a single introduction event. This site is a shipwreck 1,500 km distant from Todos os Santos Bay, the closest recorded invaded site on the Brazilian coast, and the direction of sea surface currents are westward, which possibly prevents any gene flow between Petroleiro do Acaraú and the remaining Southwestern Atlantic invaded sites. Nevertheless, Mucuripe waterway terminal is ~200 km east from the shipwreck and it is possible that this invasion was also through biofouling on small or large vessels that used this terminal. The remaining invaded sites and vectors are likely derived from two different native populations. The native distribution of T. tagusensis is currently unknown and until an extensive revision of the genus is undertaken (Capel et al. in prep.) further assumptions of the origins of the Southwestern Atlantic populations are challenging.
Supporting preliminary analyses showed by Capel et al.34, we observed an excess of heterozygotes for T. coccinea and a higher genetic diversity on vectors when compared to invaded sites. Lineages where asexual reproduction predominates tend to have high levels of heterozygosity and negative FIS as a consequence of an independent evolution of loci (e.g. “Meselson effect”), accumulating divergence within alleles71,78–80 and a higher genetic diversity on vectors in comparison to invaded sites would be expected when only a few individuals are successufully established on invaded sites. In general, the observed genetic diversity for Tubastraea spp. in the Southwestern Atlantic may result from a combination of factors, such as reproductive strategy, high growth rate, high propagule pressure, occurrence of multiple invasions, and through their dissemination by oil platforms and other shipping.
Understanding the invasion processes and the identification of the vectors are primordial steps for improving management and control of this ever increasing problem. Our results show that clonality and dissemination through vectors are the main reasons for the fast spread and invasive success of Tubastraea spp. in the Southwestern Atlantic. High clonality capability is a common feature among successful invasive species, enabling invaders with low number of individuals/low genetic diversity to reach high densities and successfully dominate the invaded region42,56. We also suggest that the Atlantic population was invaded more than once by different populations from the native region and that the Indo-Pacific is a possible source of the Southwestern Atlantic populations of T. coccinea, although a more extensive sampling of native populations and other invaded sites, such as the Caribbean, are recommended to track the exact origin of the Southwestern Atlantic invaders. We observed that vectors still hold most of the genetic and genotypic diversity and new invasions can worsen the situation by enhancing the diversity and, consequently, increase the resilience of the populations along the coast. Local strategies have been taken to control the expansion of Tubastraea spp. in the Southwestern Atlantic coast81; however, controlling the vectors responsible for introduction and dispersion is the key procedure to turn management more effective and to prevent further invasions/population expansions82.
Methods
Sampling
A total of 306 and 172 colonies of T. coccinea and T. tagusensis, from 14 and nine sites respectively, were sampled by SCUBA diving between 2012 and 2017 (Figs 1 and 2, Supplementary Fig. S3). Samples of T. coccinea and T. tagusensis were taken from eight and six invaded sites along the Southwestern Atlantic, covering the entire range of distribution of each species in the Brazilian coast, and from five and three possible vectors, respectively. The vectors include two monobuoys (IMODCO-IV, SBM-V), two oil platforms (P14 and P27) and one Floating Production Storage and Offloading Vessel (FPSO Marlim Sul). Additionally, a native population of T. coccinea (Taiwan) was sampled for comparison (Fig. 1). At each site, 11–27 colonies of T. coccinea and 6–24 of T. tagusensis were sampled and preserved in 96% ethanol or CHAOS buffer83 prior to extraction.
DNA extraction and microsatellite amplification
Total DNA was extracted using the Phenol:Chloroform method described by Fukami et al.83. Eight and ten microsatellite markers developed by Capel et al.34 were amplified by Polymerase Chain Reactions (PCRs) for all individuals of T. coccinea and T. tagusensis, respectively. PCRs were performed in 10 μl reactions including 0.2 μM of forward primer with M13 tail at their 5′ end (TGT AAA ACG ACG GCC AGT), 0.4 μM of labeled primer (M13 with VIC, NED, PET, or 6-FAM fluorescent dyes)84, 0.8 μM of reverse primer, 1U GoTaq (Promega), 1X PCR Buffer (Promega), 0.20 mM dNTPs (Invitrogen), between 1.5 and 2.5 mM MgCl2 (following Capel et al. 2017), 10 μg BSA (Invitrogen), and 5–10 ng of DNA. Cycling conditions were: 95 °C for 3 min followed by 5 cycles at 95 °C, 30 s; 52–62 °C, 30 s; 72 °C, 45 s; and 30 cycles at 92 °C, 30 s; 52–62 °C, 30 s; 72 °C, 55 s; with a final extension at 72 °C for 30 min85. Final concentration of MgCl2 and annealing temperature followed Capel et al.34. Amplification was verified in 2% agarose gel. PCR products were pooled with GS600-LIZ size standard (Applied Biosystems) and genotyped in the ABI 3500 genetic Analyzer (Applied Biosystems). Genotypes were determined using the program Geneious 7.1.986.
Clonal analyses
The package ‘RClone’87 on R 3.2.388 was used to assess the clonal structure of each species on all sites. A total of eight samples from T. coccinea (3%) and six samples from T. tagusensis (4%) failed to amplify for more than one locus and were excluded from the analyses. Of the remaining samples (298 T. coccinea and 166 T. tagusensis), 48 (16%) and 10 (6%) individuals have missing data at one locus. All individuals with identical alleles at all loci (ramets) were assigned to the same multilocus genotype (MLG, or genets). To check if individuals with the same MLG are truly clones, the probability of finding identical MLGs resulting from distinct sexual reproductive events (Psex) was calculated for each population89. When Psex > 0.001, samples were considered product of distinct sexual reproduction events (not truly clones) and included in analyses of genetic diversity. Two indexes were used to describe the clonal diversity in each population, the clonal richness (R), taking into account the number of individuals sampled (R = (MLG − 1)/(N − 1), ranging from 0 to 1, when all samples analyzed correspond to a different MLG); and the genotypic evenness (V), calculated by the Simpson’s complement evenness index to evaluate equitability in the distribution of the MLG (ranging from 0 to 1, when genets each have the same number of ramets)89,90. For genetic diversity analyses, only unique MLGs per population were considered.
Genetic diversity
The FSTAT program91 was used to test linkage disequilibrium among all pairs of loci. Subsequent analyses were done by removing loci in linkage disequilibrium with more than one other locus. The software INEst was used to evaluate the occurrence of null alleles using the individual inbreeding model (IIM) and taking into account intrapopulation inbreeding92. The presence of null alleles can bias several parameters usually measured in population analyses such as the inbreeding coefficient, the observed heterozigosity and fixation indexes92. To assess each population’s genetic diversity, the number of alleles (A), private alleles (Ap), allelic richness (Ar), observed (Ho) and expected heterozygosities (He) and were calculated using the package ‘diveRsity’93 in R 3.2.388. The inbreeding coefficient (FIS) and deviations from Hardy-Weinberg equilibrium were calculated with the software FSTAT91.
Populations structure
To explore the topology of phylogeographic relationships among sampling sites, a neighbor-joining (NJ) tree based on Cavalli-Sforza’s and Edwards chord distance, suitable for microsatellite data94, was constructed using the software Populations 1.2.3295 and the package ‘ape’96 in R 3.2.388. To visualize possible groups of sites, a Principal Components Analysis (PCA), using sampling sites as grouping factor, was performed using the package ‘Adegenet’97 in R 3.2.388.
To estimate the number of genetic clusters in the dataset, repeated MLGs were removed from the data set, leaving 84 and 30 individuals for T. coccinea and T. tagusensis, respectively. A Bayesian analysis was performed in the new data set using the software Structure v. 2.3.498 with the admixture ancestry model, correlated allele frequency and no sampling locations as prior. The analysis was performed with an initial burn-in of 500,000 cycles followed by 500,000 additional cycles and the number of clusters (K) tested varied from 1 to 14 for T. coccinea and 1 to 9 for T. tagusensis with 15 iterations for each K-value. The most likely K-value was estimated by estimating the “log probability of data” for each value of K (mean LnP(K)) and ΔK criterion98 using Structure Harvester99.
Supplementary information
Acknowledgements
We are greatful to M. Mantellato, E. Faria-Junior, B. Masi, A.F.S. de Oliveira, F. Giannini, M.F. Campos, A. Garrido and C. Cordeiro for the help with collecting samples. The first author is also thankful to Coordenação de Aperfeiçoamento de Pessoal de Nível Superior for the PhD scholarship. Financial support was provided by Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (Joel Creed FAPERJ-E-26/010.003031/2014 PensaRio). M.V.K. thanks the support from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 2014/01332-0) and Conselho Nacional de Pesquisa (CNPq 301436/2018-5). This article is No. 34 from the Projeto Coral-Sol.
Author Contributions
This study was part of K.C.C.C. PhD Thesis. K.C.C.C. participated in part of the field data colletction, collected and analysed the genetic data and drafted the manuscript; J.C. and C.A.C. participated in part of the field data colletction and added ideas to the discussion and critically reviewed; M.V.K. participated in the writing of the manuscript; C.Z. coordinated and conceived this study and participated in the writing of the manuscript. All authors approved the final manuscript.
Data Availability
The datasets generated during and analysed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-50442-3.
References
- 1.Ricciardi A. Are modern biological invasions an unprecedented form of global change? Conserv. Biol. 2007;21:329–336. doi: 10.1111/j.1523-1739.2006.00615.x. [DOI] [PubMed] [Google Scholar]
- 2.Molnar JL, Gamboa RL, Revenga C, Spalding MD. Assessing the global threat of invasive species to marine biodiversity. Front. Ecol. Environ. 2008;6:485–492. doi: 10.1890/070064. [DOI] [Google Scholar]
- 3.Wilson JRU, Dormontt EE, Prentis PJ, Lowe AJ, Richardson DM. Something in the way you move: dispersal pathways affect invasion success. Trends Ecol. Evol. 2009;24:136–144. doi: 10.1016/j.tree.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Seebens H, Gastner MT, Blasius B. The risk of marine bioinvasion caused by global shipping. Ecol. Lett. 2013;16:782–790. doi: 10.1111/ele.12111. [DOI] [PubMed] [Google Scholar]
- 5.Minchin, D., Gollasch, S., Cohen, A. N., Hewitt, C. L. & Olenin, S. Characterizing Vectors of Marine Invasion in Biological Invasions in Marine Ecosystems (eds Rilov, G. & Crooks, J. A.) 109–116 (Springer Berlin Heidelberg, 2009).
- 6.Williams SL, et al. Managing multiple vectors for marine invasions in an increasingly connected world. Bioscience. 2013;63:952–966. doi: 10.1525/bio.2013.63.12.8. [DOI] [Google Scholar]
- 7.Carlton JT. Pattern, process, and prediction in marine invasion ecology. Biol. Conserv. 1996;78:97–106. doi: 10.1016/0006-3207(96)00020-1. [DOI] [Google Scholar]
- 8.Ricciardi A. Tracking marine alien species by ship movements. Proc. Natl. Acad. Sci. 2016;113:5470–5471. doi: 10.1073/pnas.1605152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castro MCT, Fileman TW, Hall-Spencer JM. Invasive species in the Northeastern and Southwestern Atlantic Ocean: A review. Mar. Pollut. Bull. 2017;116:41–47. doi: 10.1016/j.marpolbul.2016.12.048. [DOI] [PubMed] [Google Scholar]
- 10.VAUGHAN THOMAS WAYLAND, WELLS JOHN WEST. Geological Society of America Special Papers. 1943. Revision of the Suborders Families, and Genera of the Scleractinia; pp. 1–394. [Google Scholar]
- 11.Cairns SD, Häussermann V, Försterra G. A review of the Scleractinian (Cnidaria: Anthozoa) of Chile, with the description of two new species. Zootaxa. 2005;46:15–46. doi: 10.11646/zootaxa.1018.1.2. [DOI] [Google Scholar]
- 12.Creed JC, et al. The invasion of the azooxanthellate coral Tubastraea (Scleractinia: Dendrophylliidae) throughout the world: history, pathways and vectors. Biol. Invasions. 2017;19:283–305. doi: 10.1007/s10530-016-1279-y. [DOI] [Google Scholar]
- 13.Fenner D, Banks K. Orange cup coral Tubastraea coccinea invades Florida and the Flower Garden Banks, Northwestern Gulf of Mexico. Coral Reefs. 2004;23:505–507. [Google Scholar]
- 14.Castro CB, Pires D. Brazilian coral reefs: what we already know and what is still missing. Bull. Mar. Sci. 2001;69:357–371. [Google Scholar]
- 15.de Paula AF, Creed JC. Two species of the coral Tubastraea (Cnidaria, Scleractinia) in Brazil: a case of accidental introduction. Bull. Mar. Sci. 2004;74:175–183. [Google Scholar]
- 16.Sammarco PW, Porter SA, Cairns SD. A new coral species introduced into the Atlantic Ocean - Tubastraea micranthus (Ehrenberg 1834) (Cnidaria, Anthozoa, Scleractinia): An invasive threat? Aquat. Invasions. 2010;5:131–140. doi: 10.3391/ai.2010.5.2.02. [DOI] [Google Scholar]
- 17.Sammarco PW, Porter Sa, Sinclair J, Genazzio M. Depth distribution of a new invasive coral (Gulf of Mexico)–Tubastraea micranthus, comparisons with T. coccinea, and implications for control. Manag. Biol. Invasions. 2013;4:291–303. doi: 10.3391/mbi.2013.4.4.04. [DOI] [Google Scholar]
- 18.Soares M, de O, Davis M. & de Macêdo Carneiro, P. B. Northward range expansion of the invasive coral (Tubastraea tagusensis) in the southwestern. Atlantic. Mar. Biodivers. 2016;48:1–4. doi: 10.1007/s12526-016-0623-x. [DOI] [Google Scholar]
- 19.Capel, K. C. C. Scleractinia (Cnidaria: Anthozoa) da Reserva Biológica Marinha do Arvoredo (SC), com ênfase na estrutura espaço-temporal da formação mais meridional de corais recifais no Oceano Atlântico. (Universidade Federal de Santa Catarina, 2012).
- 20.Creed JC. Two invasive alien azooxanthellate corals, Tubastraea coccinea and Tubastraea tagusensis, dominate the native zooxanthellate Mussismilia hispida in Brazil. Coral Reefs. 2006;25:350. doi: 10.1007/s00338-006-0105-x. [DOI] [Google Scholar]
- 21.Mantelatto MC, Creed JC, Mourão GG, Migotto AE, Lindner A. Range expansion of the invasive corals Tubastraea coccinea and Tubastraea tagusensis in the Southwest Atlantic. Coral Reefs. 2011;30:397–397. doi: 10.1007/s00338-011-0720-z. [DOI] [Google Scholar]
- 22.Mantelatto MC, Creed JC. Non-indigenous sun corals invade mussel beds in Brazil. Mar. Biodivers. 2015;45:605–606. doi: 10.1007/s12526-014-0282-8. [DOI] [Google Scholar]
- 23.Miranda RJ, Cruz ICS, Barros F. Effects of the alien coral Tubastraea tagusensis on native coral assemblages in a southwestern Atlantic coral reef. Mar. Biol. 2016;163:45. doi: 10.1007/s00227-016-2819-9. [DOI] [Google Scholar]
- 24.Santos LAH, do. dos, Ribeiro FV, Creed JC. Antagonism between invasive pest corals Tubastraea spp. and the native reef-builder Mussismilia hispida in the southwest Atlantic. J. Exp. Mar. Bio. Ecol. 2013;449:69–76. doi: 10.1016/j.jembe.2013.08.017. [DOI] [Google Scholar]
- 25.Sakai AK, et al. The population biology of invasive species. Annu. Rev. Ecol. Syst. 2001;32:305–332. doi: 10.1146/annurev.ecolsys.32.081501.114037. [DOI] [Google Scholar]
- 26.Lockwood JL, Cassey P, Blackburn T. The role of propagule pressure in explaining species invasions. Trends in Ecology and Evolution. 2005;20:223–228. doi: 10.1016/j.tree.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Lockwood JL, Cassey P, Blackburn TM. The more you introduce the more you get: The role of colonization pressure and propagule pressure in invasion ecology. Divers. Distrib. 2009;15:904–910. doi: 10.1111/j.1472-4642.2009.00594.x. [DOI] [Google Scholar]
- 28.Cairns, S. D. A revision of the Ahermatypic Scleractinia of the Galapagos and Cocos Islands. Smithson. Contrib. to Zool. 1–32, 10.5479/si.00810282.504 (1991).
- 29.Ayre DJ, Resing JM. Sexual and asexual production of planulae in reef corals. Mar. Biol. 1986;90:187–190. doi: 10.1007/BF00569126. [DOI] [Google Scholar]
- 30.Glynn PW, et al. Reproductive ecology of the azooxanthellate coral Tubastraea coccinea in the Equatorial Eastern Pacific: Part V. Dendrophylliidae. Mar. Biol. 2008;153:529–544. doi: 10.1007/s00227-007-0827-5. [DOI] [Google Scholar]
- 31.Dubinsky Zvy, Stambler Noga., editors. Coral Reefs: An Ecosystem in Transition. Dordrecht: Springer Netherlands; 2011. [Google Scholar]
- 32.Capel, K., Migotto, A. E., Zilberberg, C. & Kitahara, M. V. Another tool towards invasion? Polyp “bail-out” in Tubastraea coccinea. Coral Reefs33 (2014).
- 33.de Paula AF, de Oliveira Pires D, Creed JC. Reproductive strategies of two invasive sun corals (Tubastraea spp.) in the southwestern. Atlantic. J. Mar. Biol. Assoc. United Kingdom. 2014;94:481–492. doi: 10.1017/S0025315413001446. [DOI] [Google Scholar]
- 34.Capel KCC, et al. Clone wars: asexual reproduction dominates in the invasive range of Tubastraea spp. (Anthozoa: Scleractinia) in the South-Atlantic Ocean. PeerJ. 2017;5:e3873. doi: 10.7717/peerj.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolbe JJ, et al. Genetic variation increases during biological invasion by a Cuban lizard. Nature. 2004;431:177–81. doi: 10.1038/nature02807. [DOI] [PubMed] [Google Scholar]
- 36.Dlugosch KM, Parker IM. Founding events in species invasions: Genetic variation, adaptive evolution, and the role of multiple introductions. Mol. Ecol. 2008;17:431–449. doi: 10.1111/j.1365-294X.2007.03538.x. [DOI] [PubMed] [Google Scholar]
- 37.Keller SR, Taylor DR. Genomic admixture increases fitness during a biological invasion. J. Evol. Biol. 2010;23:1720–1731. doi: 10.1111/j.1420-9101.2010.02037.x. [DOI] [PubMed] [Google Scholar]
- 38.Verhoeven KJF, Macel M, Wolfe LM, Biere A. Population admixture, biological invasions and the balance between local adaptation and inbreeding depression. Proc. R. Soc. B Biol. Sci. 2011;278:2–8. doi: 10.1098/rspb.2010.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rius M, Darling JA. How important is intraspecific genetic admixture to the success of colonising populations? Trends Ecol. Evol. 2014;29:233–242. doi: 10.1016/j.tree.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 40.Bock DG, et al. What we still don’t know about invasion genetics. Mol. Ecol. 2015;24:2277–2297. doi: 10.1111/mec.13032. [DOI] [PubMed] [Google Scholar]
- 41.Ellstrand NC, Elam DR. Population genetic consequences of small population size: implications for plant conservation. Annu. Rev. Ecol. Syst. 1993;24:217–242. doi: 10.1146/annurev.es.24.110193.001245. [DOI] [Google Scholar]
- 42.Roman J, Darling JA. Paradox lost: genetic diversity and the success of aquatic invasions. Trends Ecol. Evol. 2007;22:454–464. doi: 10.1016/j.tree.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 43.Geller J, Sotka EE, Kado R, Palumbi SR, Schwindt E. Sources of invasions of a northeastern Pacific acorn barnacle, Balanus glandula, in Japan and Argentina. Mar. Ecol. Prog. Ser. 2008;358:211–218. doi: 10.3354/meps07466. [DOI] [Google Scholar]
- 44.Johnson CH, Woollacott RM. Analyses with newly developed microsatellite markers elucidate the spread dynamics of Tricellaria inopinata d’Hondt and Occhipinti-Ambrogi, 1985 - a recently established bryozoan along the New England seashore. Aquat. Invasions. 2015;10:135–145. doi: 10.3391/ai.2015.10.2.02. [DOI] [Google Scholar]
- 45.Wrange A-L, et al. The story of a hitchhiker: population genetic patterns in the invasive barnacle Balanus (Amphibalanus) improvisus Darwin 1854. PLoS One. 2016;11:e0147082. doi: 10.1371/journal.pone.0147082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simberloff D. The role of propagule pressure in biological invasions. Annu. Rev. Ecol. Evol. Syst. 2009;40:81–102. doi: 10.1146/annurev.ecolsys.110308.120304. [DOI] [Google Scholar]
- 47.Blackburn TM, Lockwood JL, Cassey P. The influence of numbers on invasion success. Mol. Ecol. 2015;24:1942–1953. doi: 10.1111/mec.13075. [DOI] [PubMed] [Google Scholar]
- 48.Hewitt, C. L., Gollasch, S. & Minchin, D. The vessel as a vector – biofouling, ballast water and sediments in Bioogial. Invasions in Marine Ecosystems (eds Rilov, G. & Crooks, J. A.) 117–131 (Springer, 2009).
- 49.Silva AG, da, Paula AF, de, Fleury BG, Creed JC. Eleven years of range expansion of two invasive corals (Tubastraea coccinea and Tubastraea tagusensis) through the southwest Atlantic (Brazil) Estuar. Coast. Shelf Sci. 2014;141:9–16. doi: 10.1016/j.ecss.2014.01.013. [DOI] [Google Scholar]
- 50.Ottaway J, Kirby G. Genetic relationships between brooding and brooded Actinia tenebrosa. Nature. 1975;255:221–223. doi: 10.1038/255221a0. [DOI] [PubMed] [Google Scholar]
- 51.Black R, Johnson MS. Asexual viviparity and population genetics of Actinia tenebrosa. Mar. Biol. 1979;53:27–31. doi: 10.1007/BF00386526. [DOI] [Google Scholar]
- 52.Brazeau D, Lasker H. The reproductive cycle and spawning in a Caribbean gorgonian. Biol. Bull. 1989;176:1–7. doi: 10.2307/1541882. [DOI] [Google Scholar]
- 53.Stoddart JA. Asexual production of planulae in the coral Pocillopora damicornis. Mar. Biol. 1983;76:279–284. doi: 10.1007/BF00393029. [DOI] [Google Scholar]
- 54.Combosch DJ, Vollmer SV. Mixed asexual and sexual reproduction in the Indo-Pacific reef coral Pocillopora damicornis. Ecol. Evol. 2013;3:3379–3387. doi: 10.1002/ece3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gorospe KD, Karl SA. Genetic relatedness does not retain spatial pattern across multiple spatial scales: Dispersal and colonization in the coral, Pocillopora damicornis. Mol. Ecol. 2013;22:3721–3736. doi: 10.1111/mec.12335. [DOI] [PubMed] [Google Scholar]
- 56.Caron V, Ede FJ, Sunnucks P. Unravelling the paradox of loss of genetic variation during invasion: Superclones may explain the success of a clonal invader. PLoS One. 2014;9:e97744. doi: 10.1371/journal.pone.0097744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gélin P, et al. Superclone expansion, long-distance clonal dispersal and local genetic structuring in the coral Pocillopora damicornis type β in Reunion Island, South Western Indian Ocean. PLoS One. 2017;12:1–19. doi: 10.1371/journal.pone.0169692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Japaud, A., Fauvelot, C. & Bouchon, C. Populations genetic study of the corals Acropora palmata and Acropora cervicornis of Guadeloupe (French West Indies) in view of their preservation. Proceedings of the 66th Gulf and Caribbean Fisheries Institute 476–482 (2014).
- 59.Fenner D. Biogeography of three Caribbean corals (Scleractinia) and the invasion of Tubastraea coccinea into the Gulf of Mexico. Bull. Mar. Sci. 2001;69:1175–1189. [Google Scholar]
- 60.Williams, G. C. Sex and Evolution. 8 (1975). [PubMed]
- 61.Schmidt-Roach S, Miller KJ, Woolsey E, Gerlach G, Baird AH. Broadcast Spawning by Pocillopora Species on the Great Barrier Reef. PLoS One. 2012;7:e50847. doi: 10.1371/journal.pone.0050847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ayre D, Miller K. Where do clonal coral larvae go? Adult genotypic diversity conflicts with reproductive effort in the brooding coral Pocillopora damicornis. Mar. Ecol. Prog. Ser. 2004;277:95–105. doi: 10.3354/meps277095. [DOI] [Google Scholar]
- 63.Sherman CDH, Ayre DJ, Miller KJ. Asexual reproduction does not produce clonal populations of the brooding coral Pocillopora damicornis on the Great Barrier Reef, Australia. Coral Reefs. 2006;25:7–18. doi: 10.1007/s00338-005-0053-x. [DOI] [Google Scholar]
- 64.de Paula AF, Creed JC. Spatial distribution and abundance of nonindigenous coral genus Tubastraea (Cnidaria, Scleractinia) around Ilha Grande, Brazil. Brazilian J. Biol. 2005;65:661–673. doi: 10.1590/S1519-69842005000400014. [DOI] [PubMed] [Google Scholar]
- 65.Ferreira CEL, Gonçalves JEA, Coutinho R. Ship hulls and oil platforms as potential vectors to marine species introduction. J. Coast. Res. 2006;SI 39:1341–1346. [Google Scholar]
- 66.Glasby TM, Connell SD, Holloway MG, Hewitt CL. Nonindigenous biota on artificial structures: Could habitat creation facilitate biological invasions? Mar. Biol. 2007;151:887–895. doi: 10.1007/s00227-006-0552-5. [DOI] [Google Scholar]
- 67.Dafforn KA, Johnston EL, Glasby TM. Shallow moving structures promote marine invader dominance. Biofouling. 2009;25:277–287. doi: 10.1080/08927010802710618. [DOI] [PubMed] [Google Scholar]
- 68.Ruiz Gregory M., Freestone Amy L., Fofonoff Paul W., Simkanin Christina. Ecological Studies. Berlin, Heidelberg: Springer Berlin Heidelberg; 2009. Habitat Distribution and Heterogeneity in Marine Invasion Dynamics: the Importance of Hard Substrate and Artificial Structure; pp. 321–332. [Google Scholar]
- 69.Ho M-J, Hsu C-M, Chen CA. Wall of orange cup coral, Tubastraea coccinea, at the inlet breakwaters of a nuclear power plant, southern Taiwan. Mar. Biodivers. 2016;47:163–164. doi: 10.1007/s12526-016-0469-2. [DOI] [Google Scholar]
- 70.Cairns SD. A generic revision and phylogenetic analysis of the Dendrophylliidae (Cnidaria: Scleractinia) Smithson. Contrib. to Zool. 2001;615:1–75. doi: 10.5479/si.00810282.615. [DOI] [Google Scholar]
- 71.Halkett F, Simon JC, Balloux F. Tackling the population genetics of clonal and partially clonal organisms. Trends Ecol. Evol. 2005;20:194–201. doi: 10.1016/j.tree.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 72.Sammarco PW, Brazeau DA, McKoin M, Strychar KB. Tubastraea micranthus, comments on the population genetics of a new invasive coral in the western Atlantic and a possible secondary invasion. J. Exp. Mar. Bio. Ecol. 2017;490:56–63. doi: 10.1016/j.jembe.2017.02.003. [DOI] [Google Scholar]
- 73.Roman J. Diluting the founder effect: cryptic invasions expand a marine invader’s range. Proc. R. Soc. B Biol. Sci. 2006;273:2453–2459. doi: 10.1098/rspb.2006.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Simon-Bouhet B, Garcia-Meunier P, Viard F. Multiple introductions promote range expansion of the mollusc Cyclope neritea (Nassariidae) in France: Evidence from mitochondrial sequence data. Mol. Ecol. 2006;15:1699–1711. doi: 10.1111/j.1365-294X.2006.02881.x. [DOI] [PubMed] [Google Scholar]
- 75.Cabezas MP, Xavier R, Branco M, Santos AM, Guerra-García JM. Invasion history of Caprella scaura Templeton, 1836 (Amphipoda: Caprellidae) in the Iberian Peninsula: multiple introductions revealed by mitochondrial sequence data. Biol. Invasions. 2014;16:2221–2245. doi: 10.1007/s10530-014-0660-y. [DOI] [Google Scholar]
- 76.Ashton GV, et al. Mitochondrial DNA reveals multiple Northern Hemisphere introductions of Caprella mutica (Crustacea, Amphipoda) Mol. Ecol. 2008;17:1293–1303. doi: 10.1111/j.1365-294X.2007.03668.x. [DOI] [PubMed] [Google Scholar]
- 77.Facon B, Pointier JP, Jarne P, Sarda V, David P. High genetic variance in life-history strategies within invasive populations by way of multiple introductions. Curr. Biol. 2008;18:363–367. doi: 10.1016/j.cub.2008.01.063. [DOI] [PubMed] [Google Scholar]
- 78.Birky CW. Heterozygosity, heteromorphy, and phylogenetic trees in asexual eukaryotes. Genetics. 1996;144:427–437. doi: 10.1093/genetics/144.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Judson OP, Normark BB. Ancient asexual scandals. Trends Ecol. Evol. 1996;11:41–46. doi: 10.1016/0169-5347(96)81040-8. [DOI] [PubMed] [Google Scholar]
- 80.Balloux F, Lehmann L, De Meeûs T. The population genetics of clonal and partially clonal diploids. Genetics. 2003;164:1635–1644. doi: 10.1093/genetics/164.4.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Christopher Creed J, et al. The Sun-Coral Project: the first social-environmental initiative to manage the biological invasion of Tubastraea spp. in Brazil. Manag. Biol. Invasions. 2017;8:181–19519. doi: 10.3391/mbi.2017.8.2.06. [DOI] [Google Scholar]
- 82.Reaser JK, et al. Ecological and socioeconomic impacts of invasive alien species in island ecosystems. Environ. Conserv. 2007;34:98–111. doi: 10.1017/S0376892907003815. [DOI] [Google Scholar]
- 83.Fukami H, et al. Conventional taxonomy obscures deep divergence between Pacific and Atlantic corals. Nature. 2004;427:832–835. doi: 10.1038/nature02339. [DOI] [PubMed] [Google Scholar]
- 84.Schuelke M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000;18:233–234. doi: 10.1038/72708. [DOI] [PubMed] [Google Scholar]
- 85.Toonen, R. J. Microsatellites for ecologists: non-radioactive isolation and amplification protocols for microsatellite markers. Available at, http://biogeek.ucdavis.edu/Mstats/ (1997).
- 86.Kearse M, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bailleul D, Stoeckel S, Arnaud-Haond S. RClone: a package to identify MultiLocus Clonal Lineages and handle clonal data sets in r. Methods Ecol. Evol. 2016;7:966–970. doi: 10.1111/2041-210X.12550. [DOI] [Google Scholar]
- 88.R Development Core Team. R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, Vienna, Austria).
- 89.Arnaud-Haond S, Duarte CM, Alberto F, Serrão EA. Standardizing methods to address clonality in population studies. Mol. Ecol. 2007;16:5115–5139. doi: 10.1111/j.1365-294X.2007.03535.x. [DOI] [PubMed] [Google Scholar]
- 90.Hurlbert SH. The nonconcept of species diversity: A critique and alternative parameters. Ecology. 1971;52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- 91.Goudet J. FSTAT (version 1.2): A computer note computer program to calculate F-statistics. J. Hered. 1995;86:485–486. doi: 10.1093/oxfordjournals.jhered.a111627. [DOI] [Google Scholar]
- 92.Chybicki IJ, Burczyk J. Simultaneous estimation of null alleles and inbreeding coefficients. J. Hered. 2009;100:106–113. doi: 10.1093/jhered/esn088. [DOI] [PubMed] [Google Scholar]
- 93.Keenan K, McGinnity P, Cross TF, Crozier WW, Prodöhl P. A. diveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol. Evol. 2013;4:782–788. doi: 10.1111/2041-210X.12067. [DOI] [Google Scholar]
- 94.Takezaki N, Nei M. Genetic distances and the setting of conservation priorities. Biol. Conserv. 1996;75:311. [Google Scholar]
- 95.Langella, O. Populations 1.2.31 (2002): a population genetic software. 3–7 (2019).
- 96.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 97.Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 98.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Earl DA, vonHoldt BM. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012;4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and analysed during the current study are available from the corresponding author on reasonable request.