Abstract
To adequately connect zebrafish medical models to human biology, it is essential that gene nomenclature reflects gene orthology. Analysis of gene phylogenies and conserved syntenies shows that the zebrafish gene currently called wnt11 (ENSDARG00000004256, ZFIN ID: ZDB-GENE-990603-12) is not the ortholog of the human gene called WNT11 (ENSG00000085741); instead, the gene currently called wnt11r (ENSDARG00000014796, ZFIN ID: ZDB-GENE-980526-249) is the zebrafish ortholog of human WNT11. Genomic analysis of Wnt11-family genes suggests a model for the birth of Wnt11-family gene ohnologs in genome duplication events, provides a mechanism for the death of a Wnt11-family ohnolog in mammals after they diverged from birds, and suggests revised nomenclature to better connect teleost disease models to human biology.
Keywords: Wnt signaling, Wnt11, Wnt11r, conserved synteny, ohnolog, whole-genome duplication
Introduction
Wnt signaling plays diverse roles during development, including the regulation of gene activities by short-range cell signaling and the orchestration of cell behaviors that shape morphologies.1–3 Most mammalian genomes possess 19 Wnt ligands that can be divided into 12 subclasses.4 This large number of Wnt ligands provides a model system to understand the evolution and consequence of gene duplication and gene loss in a phylogenetic context, and reflects the complexity of establishing relationships among gene family members in different species by evolutionary analyses.5–8
The Wnt5a group of Wnt ligands includes Wnt4, Wnt5a, and Wnt11; often, but not exclusively, Wnt5a-group ligands utilize the “noncanonical” pathway of Wnt signaling, which does not involve cytoplasmic stabilization and nuclear migration of beta-catenin to activate target genes that control cell proliferation and differentiation, but instead alters intracellular calcium ion and JNK signaling to modulate cell polarity and motility.2,9,10 The cell behavior functions of Wnt11 are important in gastrulation and in the development of the skeleton, kidneys, habenula, and heart, in the response of fish embryos to toxic chemicals, in gonad development, and in the metastatic invasion of cancers, including squamous cell carcinomas of the head and neck, an enormous problem for young adults with Fanconi Anemia.11–19
Zebrafish has two Wnt11-family members; one, currently called “wnt11” (ZFIN ID: ZDB-GENE-990603-12, ENSDARG00000004256, on chromosome Dre5), was first identified by its mutant phenotype (silberblick), a defect in convergent-extension cell behavior in gastrulation.20 This gene was named before the full content of the zebrafish genome had become available due to phylogenetic analyses that showed that its closest human family member was Wnt11. The other Wnt11-family member, which is currently called “wnt11r” (“wnt11-related,” ZFIN ID: ZDB-GENE-980526-249, ENSDARG00000014796, chromosome Dre10), is expressed in mesoderm and promotes the in-pouching of the endodermal epithelium in craniofacial development,21,22 among other functions.
The complexity of the vertebrate Wnt11 gene family has led to several explanations of gene origins and confusing gene nomenclature. The goal of the work presented here is to enhance connectivity among vertebrate genomes by clarifying the historical origins, orthology relationships, and nomenclature of vertebrate Wnt11-family genes.
The human genome has a single copy of the WNT11 gene family (WNT11, ENSG00000085741), but nonmammalian vertebrates, including chicken, xenopus, zebrafish, and other ray-finned fish, have two genes related to Wnt11.5–8 This situation raises the question: What evolutionary mechanisms resulted in different copies of Wnt11-family genes in different vertebrates? According to one hypothesis, ancestral vertebrates had two Wnt11-family genes and one copy was lost during mammalian evolution. Alternatively, vertebrate ancestors might have had a single Wnt11 gene, and gene duplications, from either tandem duplications or genome duplications, either of which could be shared or lineage specific, resulted in two copies of Wnt11-family genes in various extant species.
To test these hypotheses, we conducted analyses of phylogenies and conserved syntenies. Results showed that the human gene called WNT11 grouped with chicken “Wnt11,” xenopus “Wnt11-R,” zebrafish “wnt11r,” and mouse Wnt11. In contrast, Wnt11-family genes that do not exist in mammals were originally called “Wnt11b” in chicken, “Wnt11” in xenopus, and “wnt11” in zebrafish.23 In zebrafish, the Wnt11-family gene that is missing from mammals is still called “wnt11” (alias silberblick, the original mutant name20), even though substantial work shows that it is not the ortholog of the mammalian WNT11 gene.5–8 This problem arose because phylogenetic trees can mislead conclusions when gene content in a species is incompletely known.
Confusing nomenclature of the different Wnt11 orthologs has made comparison of their functions challenging and sometimes misleading. Based on improvements in genome annotation and conserved synteny analyses, work presented here provides an updated model for evolutionary relationships among Wnt11-family members, suggests evolutionary mechanisms for gene birth and gene death by genome duplication and chromosome rearrangements, and proposes a change in the nomenclature of zebrafish Wnt11-family genes to more accurately reflect historical relationships.
Materials and Methods
Phylogenetic analysis used the ComparaTree method as implemented at Ensembl,24 which harnesses phylogenetic distance to resolve gene duplications. Conserved synteny analyses used the Synteny Database.25
Results
The problem
In humans, WNT11 (ENSG00000085741) is on the long arm of chromosome 11 (Hsa11). Zebrafish (Danio rerio) has two genes related to WNT11: one currently called “wnt11” (ENSDARG00000004256) located on Dre5 (Danio rerio chromosome 5), and the other called “wnt11r” (wnt11-related, ENSDARG00000014796) on Dre10. What is the evolutionary relationship and origin of the human gene and the two zebrafish genes? Which, if either, zebrafish gene is the ortholog of the human WNT11 gene? Under one hypothesis, the two zebrafish genes originated from the teleost genome duplication (TGD)26–30 and each is co-orthologous to human WNT11. Under an alternative hypothesis, “wnt11r” and “wnt11” were already present as duplicates from the vertebrate genome duplication events (VGDs31) in the last common ancestor of zebrafish and humans, but one was lost at some time in the human lineage after it diverged from ray-finned fish.
Phylogenetic analysis
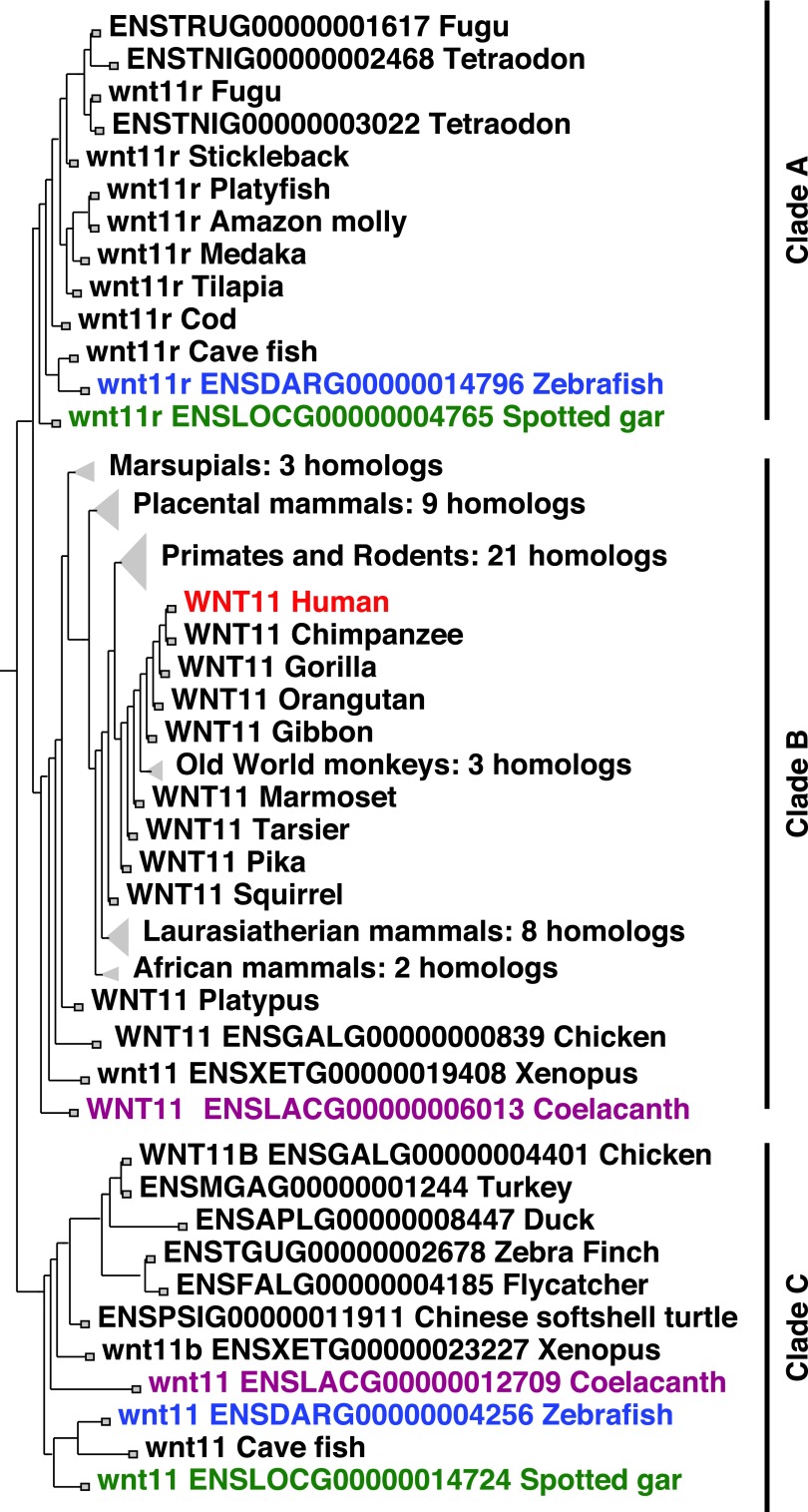
Phylogenetic analysis shows that the human WNT11 gene has orthologs in other mammals and in birds (e.g., chicken ENSGALG00000000839), turtles (e.g., ENSGAGG00000013775),32 and amphibians (e.g., ENSXETG00000019408); further, this clade is rooted on the “WNT11” gene (ENSLACG00000006013) in coelacanth,33 a basally diverging lobe-finned vertebrate (Fig. 1). These results are as expected if this gene existed in the last common ancestor of all lobe-finned fish (sarcopterygians).
FIG. 1.
A phylogeny for the Wnt11 family. Clade A, a gene clade in ray-finned fish. Clade B, a gene clade of lobe-finned fish that is the sister of Clade A. Clade C, a gene clade found in many ray-finned vertebrates, but not percomorph fish, and in many lobe-finned fish, including many tetrapods, but not mammals.
A clade of genes in ray-finned (actinopterygian) fish (Clade A in Fig. 1), including “wnt11r” in zebrafish (ENSDARG00000014796), branches as sister to the lobe-finned WNT11 gene clade (Clade B in Fig. 1) that contains human WNT11 (ENSG00000085741) (Fig. 1). This ray-finned gene clade (Clade A) is rooted on the “wnt11r” gene (ENSLOCG00000004765) in spotted gar, a basally diverging ray-finned vertebrate,34 and has representatives in ostariophysan teleosts (including zebrafish) “wnt11r” (ENSDARG00000014796) and Mexican tetra cavefish (ENSAMXG00000040853), as well as in percomorph fish, including stickleback (ENSGACG00000020196), and even in elephantfish (ENSPKIG00000013749), a member of the bony tongues, a basally diverging teleost group.
This ray-finned clade (Clade A) and its sister clade in lobe-finned fish (Clade B), including amphibians, turtles, lizards, and birds, support the contention that the last common ancestor of all extant bony vertebrates had a gene that became the WNT11 gene in human and the “wnt11r” gene in zebrafish.
The second WNT11-family gene in zebrafish is currently called “wnt11” (ENSDARG00000004256) and occupies Clade C in Figure 1. Other ostariophysan teleosts, including Mexican tetra cavefish (ENSAMXG00000007607), piranha (ENSPNAG00000023848), and channel catfish (ENSIPUG00000016221), also have an ortholog of this gene, as does the basally diverging bony tongue teleost elephantfish (ENSPKIG00000013763), while the nonteleost ray-finned fish spotted gar (ENSLOCG00000014724) roots the ray-finned fish portion of Clade C (Fig. 1). In contrast, all sequenced percomorph teleost genomes in Ensembl, including stickleback and medaka, lack this gene (Fig. 1, Clade C). Because zebrafish, cavefish, elephantfish, and spotted gar contain an ortholog of “wnt11” (ENSDARG00000004256), we conclude that this gene was present in the last common ancestor of all ray-finned fish.
The ray-finned WNT11-family Clade C that contains “wnt11” (ENSDARG00000004256) has a sister gene clade in lobe-finned fish, which includes coelacanth (ENSLACG00000012709), a frog (ENSXETG00000023227), a turtle (ENSPSIG00000011911), and several birds (including chicken, ENSGALG00000004401), but mammals appear to lack a representative of this Wnt11-family clade (Fig. 1, Clade C). Because at least some nonmammalian lobe-finned fish and some ray-finned fish both contain representatives of this gene, the last common ancestor of all bony fish had an ortholog of the zebrafish gene currently called “wnt11” (ENSDARG00000004256), but that gene is now missing from the lobe-finned fish we call mammals.
This phylogenetic analysis suggests the hypothesis that the last common ancestor of all bony fish had two WNT11-family genes. The descendants of one of those genes are now called “wnt11r” in zebrafish and “WNT11” in human. The descendants of the other gene are called “wnt11” in zebrafish and, while orthologs of zebrafish “wnt11” are found in some tetrapods, it is missing from humans and other mammals.
Conserved syntenies
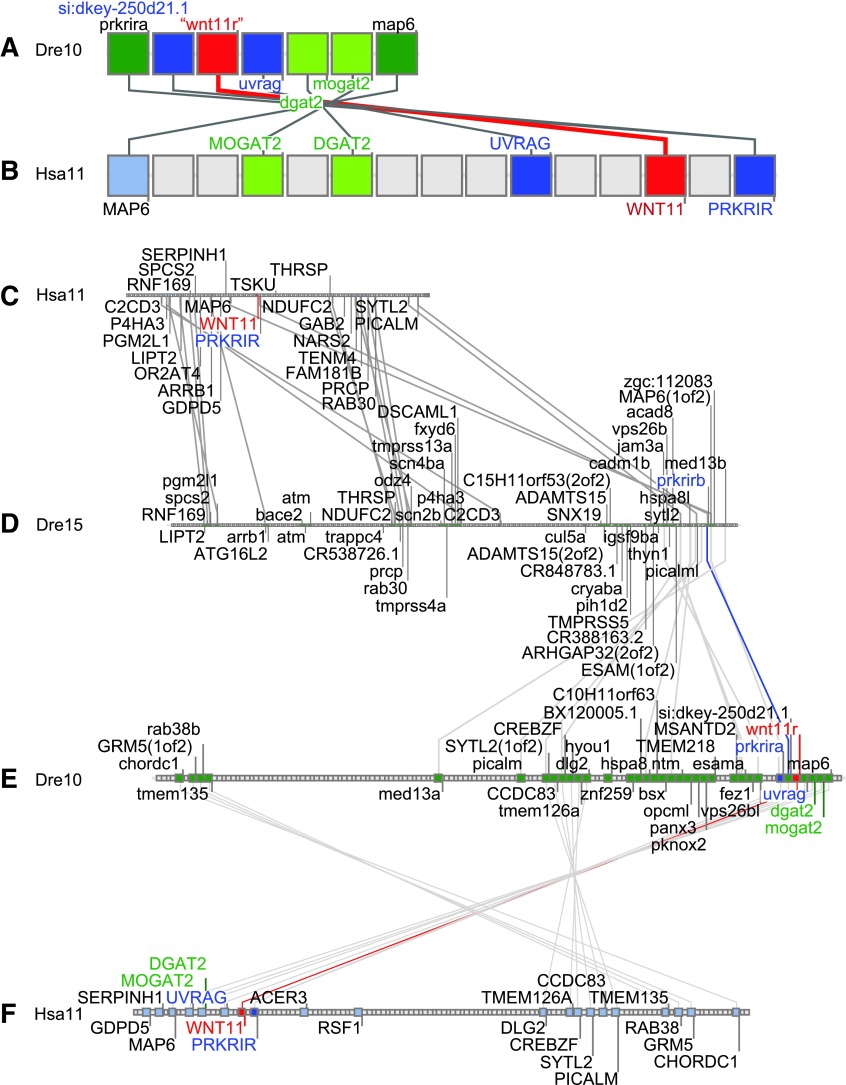
The hypothesis generated from the phylogeny—that “wnt11r” in zebrafish is the ortholog of WNT11 in human—predicts that the chromosomal segment containing “wnt11r” in zebrafish should have strong conserved syntenies with the chromosome segment encompassing WNT11 in humans. Conserved synteny analysis showed that a group of seven contiguous genes on zebrafish chromosome Dre10 that includes “wnt11r” shares conserved syntenies with human orthologs occupying a small portion of human chromosome 11 surrounding WNT11 (Fig. 2A, B). This evidence supports the hypothesis suggested by the phylogenetic analysis that “wnt11r” and WNT11 are orthologs.
FIG. 2.
Conserved synteny analysis for Wnt11-family genes. (A) A portion of zebrafish chromosome 10 (Dre10) containing “wnt11r.” (B) A portion of human chromosome 11 (Hsa11) containing WNT11. (C) Genes surrounding WNT11 on human chromosome 11. (D) Genes on zebrafish chromosome 15 (Dre15) showing conserved syntenies with the human chromosome 11 WNT11 containing region, but without the expected wnt11-family gene. (E) Genes on zebrafish chromosome 10 (Dre10) showing the TGD duplicated region corresponding to zebrafish chromosome 15 and the expected “wnt11r” gene. (F) Genes surrounding WNT11 on human chromosome 11 showing conserved syntenies with “wnt11r.”
The portion of Dre10 that contains “wnt11r” has a duplicate region in Dre15 that arose in the TGD,26–30,35 with duplicated copies of PRKRIR(THAP12), one of which is adjacent to “wnt11r” on Dre10 (compare Fig. 2D, E). This portion of Dre15 has no Wnt11-family gene, and, while gene orders of this region in Dre10 and Hsa11 are locally conserved (Fig. 2A, B), gene orders in the corresponding portion of Dre15 show chromosome rearrangements both with respect to the zebrafish TGD ohnologous region on Dre10 and with respect to the region in the human genome that contains WNT11 (Fig. 2C–E). We conclude that before the TGD, the region containing “wnt11r” was relatively well conserved in gene order comparing the zebrafish and human lineages, and that the TGD produced two copies of “wnt11r,” one of which was lost; the lost TGD ohnolog is at a position associated with the breakpoints of chromosome rearrangements on Dre15, suggesting gene loss by a chromosome break. Furthermore, this gene loss occurred early in the teleost radiation because elephantfish, representing one of the earliest diverging teleost clades, appears to lack a duplicate of “wnt11r.”
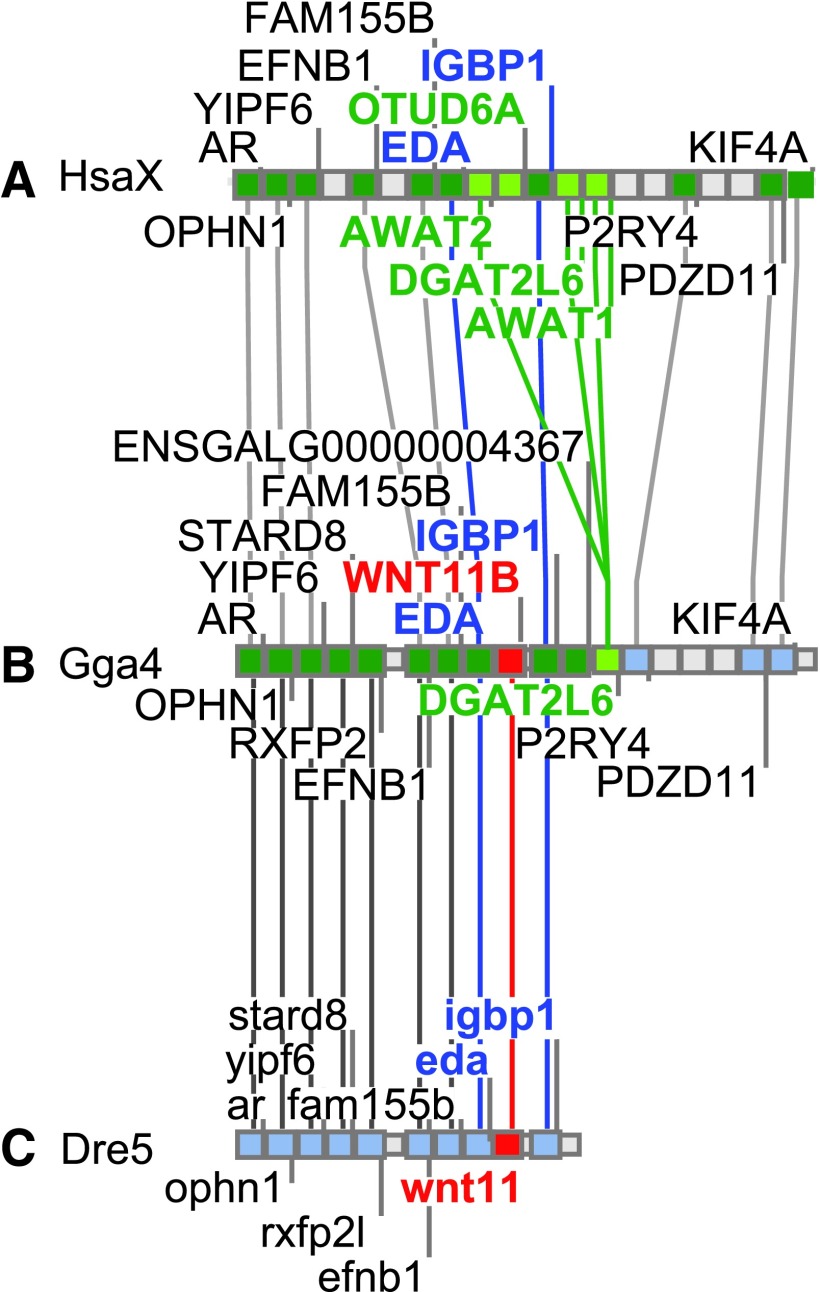
The hypothesis generated from the phylogeny (Fig. 1)—that humans and other mammals have lost their ortholog of the zebrafish “wnt11” gene—predicts first that the segment of Dre5 that contains “wnt11” should be orthologous to the region of chicken (Gallus gallus) chromosome 4 (Gga4) that contains the WNT11-family gene “WNT11B” (ENSGALG00000004401), and second, that this chromosome segment in chicken should have an orthologous segment in the human genome that lacks a WNT11-family gene. Figure 3 shows that “wnt11” in zebrafish and “WNT11B” (ENSGALG00000004401) in chicken are flanked by eda(EDA) and igbp1(IGBP1), and that the human orthologs of these two genes are within a few genes of each other but do not have a WNT11-family gene between them. This arrangement is what would be predicted if the human ortholog of chicken “WNT11B” (ENSGALG00000004401) were lost in the mammalian lineage after it diverged from the “reptile” + bird lineage.
FIG. 3.
Conserved syntenies suggest a mechanism for the loss of a wnt11-family gene from the mammalian genome. (A) A portion of human chromosome X (HsaX) showing the expected location of the ortholog of the gene called zebrafish “wnt11.” (B) A portion of chicken chromosome 4 (Gga4) showing the location of “WNT11B” with conserved syntenies to a portion of the human X chromosome (HsaX). (C) A portion of zebrafish chromosome 5 (Dre5) containing zebrafish “wnt11” showing conserved syntenies with chicken.
In the position predicted to contain the human ortholog of zebrafish “wnt11” and chicken Wnt11B genes, the human genome has instead two genes, AWAT2 and OTUD6A, that are inserted between EDA and IGBP1 (Fig. 3A). A phylogenetic analysis of acyltransferases shows that AWAT2, AWAT1, and DGAT2L6 (acyl-CoA wax alcohol acyltransferase-2, acyl-CoA wax alcohol acyltransferase-1, and diacylglycerol O-acyltransferase-2-like-6, which are co-orthologs of the yeast gene DGA1, diacylglycerol acyltransferase), branch as nested sister clades and are present only in mammals.36 While in human, AWAT2 occupies the position expected for a WNT11-related gene between EDA and IGBP1, DGAT2L6 is located adjacent to, but on the other side of, IGBP1, while AWAT1 lies beside it. AWAT2 is transcribed in a direction opposite to that of DGAT2L6 and AWAT1. Chicken also has an acyltransferase gene (ENSGALG00000004367) at the orthologous location to the right of the EDA—”WNT11B”—IGBP1 trio. Thus, the phylogenetic relationship and adjacent or nearby location of these acyltransferase genes would be as expected by mammalian-specific tandem gene duplication events that placed one of the acyltransferase copies in the location expected for a WNT11-family gene. In addition, two acyltransferase genes dgat2 and mogat2 (diacylglycerol O-acyltransferase-2 and monoacylglycerol O-acyltransferase-2) are adjacent to the nearest neighbor (UVRAG) on the 3′ side of WNT11 in human and “wnt11r” in zebrafish. The location of tandemly duplicated acyltransferase genes only one gene away from WNT11 in human and “wnt11r” zebrafish and the location of the sister clade of these acyltransferase genes within one gene of “wnt11” in zebrafish are as expected if the ancestral condition before the divergence of zebrafish and human lineages was a WNT11-family gene adjacent to an acyltransferase gene, and the mammalian WNT11-family gene in this location was lost in a chromosome rearrangement event associated with the duplication of a neighboring acyltransferase gene.
Origin of WNT11-family genes by chromosome duplication
Phylogenetic analyses and conserved synteny studies both support the hypothesis that the last common ancestor of extant bony fish had two WNT11-family genes, but that one of these genes was lost in the mammalian lineage associated with the tandem duplication of an adjacent gene. What was the origin of these two WNT-family genes? Under one hypothesis, they emerged from two rounds of vertebrate genome duplication (VGD1 and VGD2) thought to precede the diversification of vertebrates31 (but see37). Under another hypothesis, they derived from tandem duplication events followed by chromosome translocations. A third hypothesis involves retrotransposition. Each of these hypotheses makes specific predictions regarding gene structure and conserved syntenies.
Genes that originate by retrotransposition generally do not have introns, but both “wnt11” and “wnt11r” have several introns mainly in positions orthologous to each other and to their human paralog, arguing against retrotransposition as a mechanism for the origin of either of the extant zebrafish WNT11-family genes.
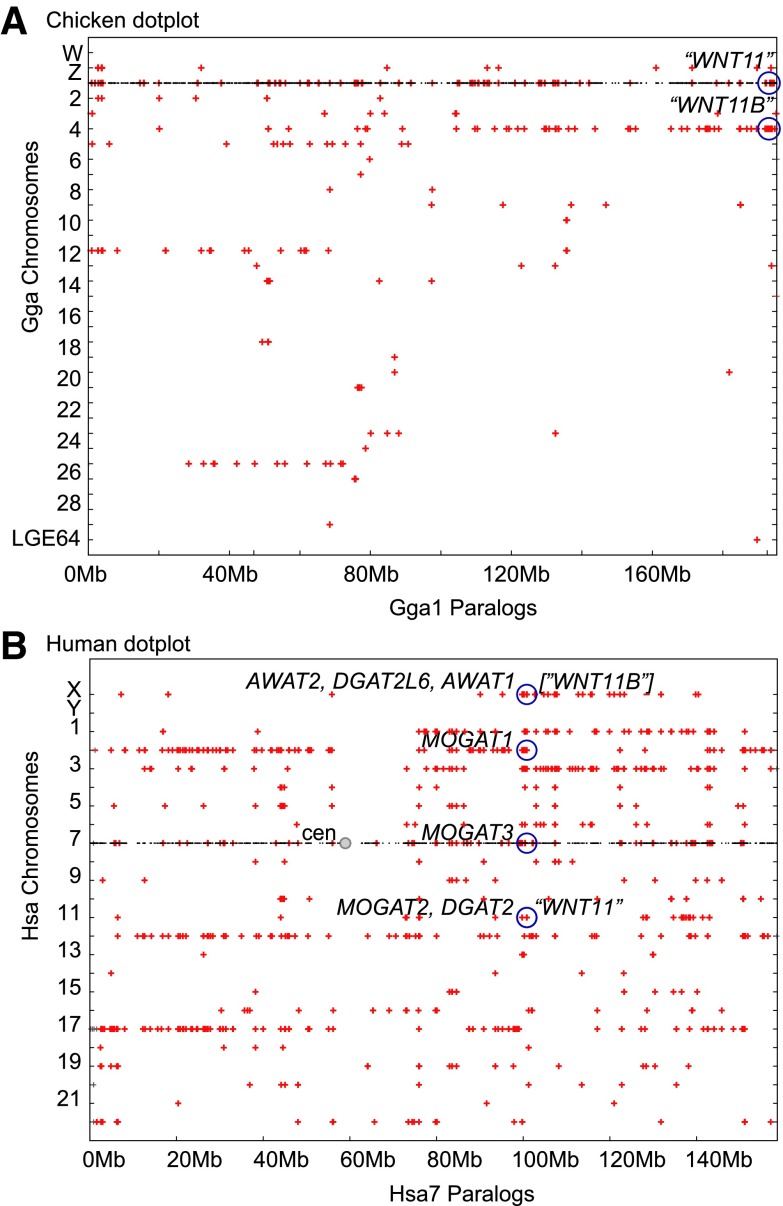
A dot plot analysis can provide evidence regarding gene origins by duplication events. Figure 4A shows a dot plot that displays the location of genes on chicken chromosome Gga1 (black dots) along the horizontal axis and directly beneath each Gga1 gene, the plot shows paralogs (red pluses) in the vertical axis.25 The circle at the right end of Gga1 represents the location of the chicken ortholog of the human WNT11 gene on that chromosome and directly below it lies the paralogous chicken WNT11-family gene on Gga4. The plot (Fig. 4A) shows that the right half of Gga1, the longest chicken chromosome, is paralogous to much of chromosome Gga4, including the region that contains chicken “WNT11” (which is orthologous to zebrafish “wnt11r”) and chicken WNT11B (ENSGALG00000004401) (which does not have a human ortholog).
FIG. 4.
Dot plot analysis consistent with genome duplication. (A) Genes on chicken (Gallus gallus) chromosome 1 (Gga1) with orthologs and paralogs plotted directly above or below the position of each gene on Gga1. (B) Dot plot of genes on human chromosome 7 (Hsa7) with orthologs and paralogs shown directly above or below each Hsa7 gene demonstrating paralogons due to large-scale, presumably genome, duplications.
This result is as expected if a substantial portion of Gga1 and Gga4 arose from large-scale duplication events. While these paralogous chromosome regions are to be expected from the VGD events, results show only two paralogons rather than the four expected. The left portion of Gga1 up to ∼90Mb, in contrast, shows all four paralogous segments: Gga1, Gga5, Gga12, and Gga25 (Fig. 4A), as expected from two rounds of whole-genome duplication.
A dot plot analysis of the human genome (Fig. 4B) shows that the eight closely related acyltransferase genes in the human genome occupy four chromosomal regions, including HsaX with three genes adjacent to EDA and the predicted ancestral site of the human ortholog of the zebrafish “wnt11” gene, and paralogs on Hsa2, Hsa7, and notably on Hsa11 at a location adjacent to “WNT11” (Fig. 4B). Two rounds of genome duplication would be predicted to make four paralogous regions, which are clearly evident for the short (left) arm of Hsa7 (“cen,” centromere) on Hsa2, 12, and 17, but are more chaotic for the long (right) arm. For the region of Hsa7 from ∼70 Mb to 110 Mb, paralogous regions are evident on HsaX, Hsa1, Hsa2, and Hsa11, but other chromosomes also appear to be paralogous. We conclude that the evidence from dot plots does not rule out the hypothesis of the origin of the “WNT11 (Hsa11)” and “wnt11 (HsaX)” regions in the vertebrate whole-genome duplication events, but other hypotheses for the origin of these paralogs are also not strongly ruled out.
Discussion
A model for the origin of WNT11-family genes
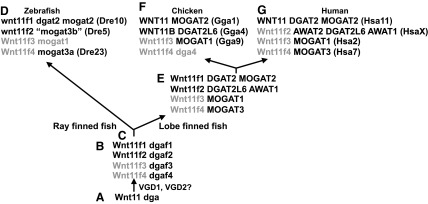
These data suggest the following model for the origin of vertebrate WNT11-family genes. Evidence supports the model that the founding member of the WNT11-family in a nonvertebrate chordate was adjacent to a diacylglycerol acyltransferase gene (Fig. 5A). After the whole-genome duplication events thought to precede the vertebrate radiation (VGDs 1 and 2, VGD1 and VGD2), this model assumes that the founding wnt11-family gene and its adjacent dga (diacylglycerol acyltransferase) gene each became four ohnologs, which could be called wnt11f1 to wnt11f4 for wnt11-family member-1 to wnt11-family member-4. According to the model, two of the wnt11-family genes were lost, and the other two became what are now called “wnt11” and “wnt11r” in zebrafish. The model suggests that the dga ohnologs diversified in function and all four were maintained (Fig. 5B).
FIG. 5.
A model for Wnt11-family evolution. (A) Wnt11 and dga genes lying adjacent to a prevertebrate chordate. (B) Two rounds of the vertebrate genome duplication would give four copies of this region, but two duplicates of Wnt11-family genes would soon disappear. (C) The divergence of ray-finned fish from lobe-finned fish. (D–G) The current situation in zebrafish, chicken, and human genomes. Chromosome locations in parentheses, Dre: (Danio rerio, zebrafish), Gga (Gallus gallus, chicken), and Hsa (Homo sapiens, human).
After the divergence of ray-finned and lobe-finned fish lineages (Fig. 5C), the line leading to zebrafish lost the mogat1 ohnolog but maintained the other dga paralogs (although the gene currently called “mogat3b” appears to be an ortholog of DGAT2L6, its enzymatic function is unknown) (Fig. 5D).
In the lobe-finned lineage, several tandem duplications may have occurred (Fig. 5E). Coelacanth, frog, turtle, and several birds maintain an ortholog of both zebrafish wnt11-family genes today (Fig. 5F). In the human lineage, an AWAT gene duplicated and one copy ended up between the EDA and IGBP1 genes at the location expected for the ortholog of the zebrafish “wnt11” gene, which disappeared (Fig. 5G). It is parsimonious to think that the insertion of AWAT2 into this location may have disrupted the human ortholog of the zebrafish “wnt11” gene, destroyed its function, and the gene sequence gradually decayed. The opossum, a marsupial mammal, has AWAT2 adjacent to EDA and no WNT11-family gene at this location, so the deletion event likely occurred before the origin of therian mammals.
Solving a connectivity problem
The nomenclature of Wnt11-family genes presents a substantial problem because the zebrafish ortholog of the human WNT11 gene is now called “wnt11r” and the zebrafish gene now called “wnt11” has no human ortholog. This nomenclature situation is not optimal because it (1) implies incorrect orthologies, which obfuscates connecting experimental results from zebrafish functional genetic studies to human biology; and (2) contradicts zebrafish nomenclature conventions, which state, “Genes should be named after the mammalian orthologue whenever possible” (https://wiki.zfin.org/display/general/ZFIN+Zebrafish+Nomenclature+Guidelines#ZFINZebrafishNomenclatureGuidelines-1.1).
To solve this problem, we suggest to change zebrafish “wnt11r” (ENSDARG00000014796, ZFIN ID: ZDB-GENE-980526-249) (shown to be the ortholog of human WNT11) to wnt11 (alias wnt11f1, wnt11-family member-1) and zebrafish “wnt11” (ENSDARG00000004256, ZFIN ID: ZDB-GENE-990603-12) to wnt11f2 (wnt11-family member-2). This change has the unfortunate drawback of introducing confusion with regard to previously published papers, but moving forward, has the advantage of emphasizing the relatedness of the two genes while, importantly, eliminating the error implied by the previous nomenclature that zebrafish “wnt11” is the ortholog of human WNT11. This type of revision is somewhat common, for example, the gene currently called wnt5b (ENSDARG00000102464, ZFIN ID: ZDB-GENE-980526-87) was formerly called wnt5a, contrary to human orthology.
In conclusion, analysis of phylogenies and conserved syntenies from sequenced genomes clarifies the evolutionary history and nomenclature of the Wnt11 gene family to better connect teleost models of human disease to human biology.
Acknowledgments
We acknowledge National Institutes of Health grants 5R01OD011116 and R01GM085318 (J.H.P.), 1R01DC015488-01A1 (T.P.), and institutional support from the Stowers Institute for Medical Research (T.P.), the Ditch Bros. for their grand support, and Amy Singer and Ken Frazer of ZFIN for important conversations.
Disclosure Statement
No competing financial interests exist.
References
- 1. Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 2004;20:781–810 [DOI] [PubMed] [Google Scholar]
- 2. van Amerongen R, Nusse R. Towards an integrated view of Wnt signaling in development. Development 2009;136:3205–3214 [DOI] [PubMed] [Google Scholar]
- 3. Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 2004;20:781–810 [DOI] [PubMed] [Google Scholar]
- 4. Prud'homme B, Lartillot N, Balavoine G, Adoutte A, Michel Vervoort M. Phylogenetic Analysis of the Wnt Gene Family: insights from Lophotrochozoan Members. Curr Biol 2002;12:1395–1400 [DOI] [PubMed] [Google Scholar]
- 5. Garriock RJ, D'Agostino SL, Pilcher KC, Krieg PA. Wnt11-R, a protein closely related to mammalian Wnt11, is required for heart morphogenesis in Xenopus. Dev Biol 2005;279:179–192 [DOI] [PubMed] [Google Scholar]
- 6. Garriock RJ, Warkman AS, Meadows SM, D'Agostino S, Krieg PA. Census of vertebrate Wnt genes: isolation and developmental expression of Xenopus Wnt2, Wnt3, Wnt9a, Wnt9b, Wnt10a, and Wnt16. Dev Dyn 2007;236:1249–1258 [DOI] [PubMed] [Google Scholar]
- 7. Hardy KM, Garriock RJ, Yatskievych TA, D'Agostino SL, Antin PB, Krieg PA.: Non-canonical Wnt signaling through Wnt5a/b and a novel Wnt11 gene, Wnt11b, regulates cell migration during avian gastrulation. Dev Biol 2008;320:391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kuraku S, Kuratani S. Genome-wide detection of gene extinction in early mammalian evolution. Genome Biol Evol 2011;3:1449–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tian S, Hu J, Tao K, Wang J, Chu Y, Li J, et al. Secreted AGR2 promotes invasion of colorectal cancer cells via Wnt11-mediated non-canonical Wnt signaling. Exp Cell Res 2018;364:198–207 [DOI] [PubMed] [Google Scholar]
- 10. Tao Q, Yokota C, Puck H, Kofron M, Birsoy B, Yan D, et al. Maternal wnt11 activates the canonical wnt signaling pathway required for axis formation in Xenopus embryos. Cell 2005;120:857–871 [DOI] [PubMed] [Google Scholar]
- 11. Rosenberg PS, Greene MH, Alter BP. Cancer incidence in persons with Fanconi anemia. Blood 2003;101:822–826 [DOI] [PubMed] [Google Scholar]
- 12. Velleuer E, Dietrich R. Fanconi anemia: young patients at high risk for squamous cell carcinoma. Mol Cell Pediatr 2014;1:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Vliet PP, Lin L, Boogerd C, Martin J, Andelfinger G, Grossfeld P, et al. Tissue specific requirements for WNT11 in developing outflow tract and dorsal mesenchymal protrusion. Dev Biol 2017;429:249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nagy I, Xu Q, Naillat F, Ali N, Miinalainen I, Samoylenko A, et al. Impairment of Wnt11 function leads to kidney tubular abnormalities and secondary glomerular cystogenesis. BMC Dev Biol 2016;16:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lako M ST, Bullen P, Wilson D, Robson S, Lindsay S. Isolation, characterisation and embryonic expression of WNT11, a gene which maps to 11q13. 5 and has possible roles in the development of skeleton, kidney and lung. Gene 1998;219:101–110 [DOI] [PubMed] [Google Scholar]
- 16. Vincent-Chong VK, Ismail SM, Rahman ZA, Sharifah NA, Anwar A, Pradeep PJ, et al. Genome-wide analysis of oral squamous cell carcinomas revealed over expression of ISG15, Nestin and WNT11. Oral Dis 2012;18:469–476 [DOI] [PubMed] [Google Scholar]
- 17. Pandey S, Shekhar K, Regev A, Schier AF. Comprehensive identification and spatial mapping of habenular neuronal types using single-cell RNA-Seq. Curr Biol 2018;28:1052–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schuttler A, Reiche K, Altenburger R, Wibke Busch W. The transcriptome of the zebrafish embryo after chemical exposure: a meta-analysis. Toxicol Sci 2017;157:291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duncan RN, Panahi S, Piotrowski T, Dorsky RI. Identification of Wnt genes expressed in neural progenitor zones during zebrafish brain development. PLoS One 2015;10:e0145810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heisenberg C-P, Tada M, Rauch G-J, Saude L, Concha ML, Geisler R, et al. Silberblick/Wnt11 mediates convergent extension movements during zebrafish gastrulation. Nature 2000;405:76–81 [DOI] [PubMed] [Google Scholar]
- 21. Choe CP, Collazo A, Trinh LA, Pan L, Moens CB, Crump JG. Wnt-dependent epithelial transitions drive pharyngeal pouch formation. Dev Cell 2013;24:296–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Choe GP, Crump JG. Tbx1 controls the morphogenesis of pharyngeal pouch epithelia through mesodermal Wnt11r and Fgf8a. Development 2014;141:3583–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hardy KM, Garriock RJ, Yatskievych TA, D'Agostino SL, Antin PB, Krieg PA. Non-canonical Wnt signaling through Wnt5a/b and a novel Wnt11 gene, Wnt11b, regulates cell migration during avian gastrulation. Dev Biol 2008;320:391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Flicek P, et al. Ensembl's 10th year. Nucleic Acids Res 2010;38:D557–D562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Catchen JM, Conery JS, Postlethwait JH. Automated identification of conserved synteny after whole-genome duplication. Genome Res 2009;19:497–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Amores AFA, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, et al. Zebrafish hox clusters and vertebrate genome evolution. Science 1998;282:1711–1714 [DOI] [PubMed] [Google Scholar]
- 27. Postlethwait J, Amores A, Force A, Yan Y. The zebrafish genome. Methods Cell Biol 1999;60:149–163 [PubMed] [Google Scholar]
- 28. Postlethwait JHYY, Gates MA, Horne S, Amores A, Brownlie A, Donovan A, et al. Vertebrate genome evolution and the zebrafish gene map. Nat Genet 1998;18:345–349 [DOI] [PubMed] [Google Scholar]
- 29. Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res 2003;13:382–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate protokaryotype. Nature 2004;431:946–957 [DOI] [PubMed] [Google Scholar]
- 31. Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol 2005;3:e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Z, Anaya JP, Zadissa A, Li W, Niimura Y, Huang Z, et al. The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nat Genet 2013;45:701–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Amemiya CT, Lee AP, Fan S, Philippe H, Maccallum I, Braasch I, et al. The African coelacanth genome provides insights into tetrapod evolution. Nature 2013;496:311–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Braasch I, Gehrke A, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, et al. The spotted gar genome illuminates vertebrate evolution and facilitates human-to-teleost comparisons. Nat Genet 2016;48:427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Naruse K, Tanaka M, Mita K, Shima A, Postlethwait J, Mitani H. A medaka gene map: the trace of ancestral vertebrate proto-chromosomes revealed by comparative gene mapping. Genome Res 2004;14:820–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Holmes RS. Comparative genomics and proteomics of vertebrate diacylglycerol acyltransferase (DGAT), acyl CoA wax alcohol acyltransferase (AWAT) and monoacylglycerol acyltransferase (MGAT). Comp Biochem Physiol Part D 2010;5:45–54 [DOI] [PubMed] [Google Scholar]
- 37. Smith JJ, Keinath MC. The sea lamprey meiotic map improves resolution of ancient vertebrate genome duplications. Genome Res 2015;25:1081–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]