Abstract
Background
Expression of neuron-glial antigen 2 (NG2) identifies an aggressive malignant phenotype in glioblastoma (GBM). Mouse models have implicated NG2 in the genesis, evolution, and maintenance of glial cancers and have highlighted potential interactions between NG2 and epidermal growth factor receptor (EGFR). However, it is unknown whether the lineage relationship of NG2+ and NG2− cells follows a hierarchical or stochastic mode of growth. Furthermore, the interaction between NG2 and EGFR signaling in human GBM is also unclear.
Methods
Single GBM NG2+ and NG2− cells were studied longitudinally to assess lineage relationships. Short hairpin RNA knockdown of NG2 was used to assess the mechanistic role of NG2 in human GBM cells. NG2+ and NG2− cells and NG2 knockdown (NG2-KD) and wild type (NG2-WT) cells were analyzed for differential effects on EGFR signaling.
Results
Expression of NG2 endows an aggressive phenotype both at single cell and population levels. Progeny derived from single GBM NG2− or GBM NG2+ cells consistently establish phenotypic equilibrium, indicating the absence of a cellular hierarchy. NG2 knockdown reduces proliferation, and mice grafted with NG2-KD survive longer than controls. Finally, NG2 promotes EGFR signaling and is associated with EGFR expression.
Conclusions
These data support a dynamic evolution in which a bidirectional relationship exists between GBM NG2+ and GBM NG2− cells. Such findings have implications for understanding phenotypic heterogeneity, the emergence of resistant disease, and developing novel therapeutics.
Keywords: glioblastoma, NG2, EGFR signaling
Key Points.
1. Reducing NG2 expression in glioblastoma reduces the proliferative ability of glioblastoma cells.
2. NG2 potentiates EGFR signalling in human glioblastoma.
Importance of the Study
This study demonstrates for the first time that NG2, a developmentally important marker, identifies highly proliferative glioblastoma cells both at single cell and population levels. The expression of NG2 is dynamic and + and − cells establish a phenotypic equilibrium suggesting the absence of a lineage hierarchy. Lack of NG2 expression results in increased latency of tumor formation. Finally, we provide mechanistic insights into NG2 function by showing that NG2 potentiates the EGFR-Akt signaling pathway with implications for targeted therapies.
Glioblastoma (GBM) is characterized by histopathological diversity and poor clinical outcomes, with rapid development of resistance to radiotherapy and chemotherapy. To improve clinical outcomes, novel therapeutic approaches need to be developed based on a better understanding of how malignant cell populations are organized and maintained.
Neuron-glial antigen 2 (NG2; chondroitin sulphate proteoglycan 4 [CSPG4] in humans) is a developmentally important transmembrane proteoglycan found on the surface of NG2+ progenitors, which are the largest population of dividing cells in the adult human brain.1,2 NG2+ progenitors display one of the best-characterized developmental hierarchies, whereby NG2+ progenitors proliferate, downregulate NG2, and differentiate into NG2− progeny expressing mature oligodendrocyte markers.3 These are the cells of origin of low-grade oligodendroglioma4; however, it is unknown whether this well-characterized hierarchy is retained in GBM.
Previous work has demonstrated that NG2+ cells are more proliferative than the corresponding NG2− population,5,6 and targeting NG2 reduces tumor growth.7 Mouse models also demonstrate the contribution of NG2+ progenitors to gliomagenesis: When concurrent p53/neurofibromatosis 1 mutations were induced in neural stem cells, tumor formation occurred only upon differentiation into NG2+ progenitors.8
Work in the normal brain has highlighted the importance of epidermal growth factor receptor (EGFR) signaling in NG2+ progenitors,9–11 and studies using a mouse model of glioma demonstrated that asymmetric segregation and phosphorylation of EGFR in NG2+ progenitors is NG2 dependent.12 EGFR is the most commonly upregulated gene in GBM13 and is essential for tumorigenesis.14 However, the functional importance of interactions between NG2 and EGFR in human GBM is unknown.
Here, we use patient-derived GBM cells to investigate the relationship between GBM NG2+ and NG2− cells, and the role of NG2 in EGFR signaling. Single cell analyses reveal a complex organization in which GBM NG2+ and NG2− cells do not adhere to a cellular hierarchy, but rather have a bidirectional relationship. Within this relationship, GBM NG2+ and NG2− cells coexist in a phenotypic equilibrium that, if perturbed, will result in gradual phenotypic change, ultimately reestablishing the equilibrium. Further analysis reveals that NG2 expression potentiates EGFR signaling, providing mechanistic insights into the role of NG2 in enhancing proliferation in human GBM.
Methods and Materials
Derivation of Cell Lines
Cell lines were derived from surgically resected tumors and cultured as adherent monolayers as described previously15 (see Supplementary Material). Informed consent for tissue collection was obtained from each patient.16 Fetal neural cortical tissue was provided by Professor Roger Barker. All human tissues were obtained according to local ethical guidelines (LREC 04/Q0108/60) in compliance with the Human Tissue Act.
Phoenix (Φnx) cells, used for retroviral transfection, were provided by Dr Masashi Narita (CRUK Cancer Research Institute, Cambridge, UK) and were cultured in 10% v/v fetal calf serum in Dulbecco’s modified Eagle’s medium (Gibco).
All experiments were performed across 3 patient-derived cell lines (details in Supplementary Table 5) with triplicate repeats for each cell line. Statistical analysis was performed on pooled data from triplicate repeats from the 3 cell lines.
MTS and BrdU Assays
Three thousand cells/well in 96-well plates were cultured for 3 days for assay by bromodeoxyuridine (BrdU) and MTS [(3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium)]. MTS assay was performed as described previously.6 The BrdU Cell Proliferation Assay (Millipore) was performed according to the manufacturer’s instructions.
Limiting Dilution Assay
The limiting dilution assay was performed as described previously,15 using 500, 250, 100, 50, 25, and 10 cells/well in non-adherent culture (96-well plates, 200 μL medium), with 20 wells/density. The number of colony forming wells (containing free-floating spherical clusters of cells) was recorded at 1 week. The proportion of colony forming cells was calculated using the online calculator at http://bioinf.wehi.edu.au/software/elda/ with significance assessed by χ2 test.
Short Hairpin RNA Mediated Knockdown of NG2
Synthesis of short hairpin (sh)RNA constructs was performed using a miR30 design by Lixiang Xue (Masashi Narita group). Target sequences for shRNAs were designed using online software (http://cancan.cshl.edu/RNAi_central/step3.cgi). MiR30 oligos, containing the target sequences, were generated for NG2 (sequences in Supplementary Material) and incorporated into MCSV vectors.
As controls, cells were transfected with MCSV vectors containing the sequence for green fluorescent protein (GFP). Multiple NG2 shRNA sequences were used as an internal control. All vectors had puromycin resistance genes to allow for antibiotic based selection.
For transfection, Φnx cells were incubated with calcium phosphate and solution based on HEPES (4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid) (Sigma-Aldrich), with 25 μM chloroquine (Sigma-Aldrich), at 37°C for 10 hours, at which point the medium was replaced. The cells were then incubated at 37°C for a further 36 hours.
For transfection, medium was removed from the Φnx cells, passed through a 0.45 μm filter, added to GBM cells with 5 μg/mL of polybrene, and incubated at 37°C for 4 hours. This process was repeated 3 times, followed by selection with 3 μg/mL puromycin (Sigma-Aldrich).
In Vivo Experimental Work
Orthotopic implantation of patient-derived xenografts was performed as described previously,15 in accordance with the UK Animal (Scientific Procedures) Act 1986 and the Cambridge University Commission for Animal Health. Mice were sacrificed upon development of symptoms by Schedule One procedures.
Antibodies
Antibodies are included in the Supplementary Material.
Single Cell Studies
Using fluorescence activated cell sorting (FACS), single cells from the positive or negative populations were seeded into 96-well plates (1 cell/well [wells were checked by microscopy and excluded if >1 cell was present]) and cultured for 8 weeks. Any colonies (>1 cell/well) that formed were transferred into flasks and cultured for 4 weeks, then collected and analyzed by flow cytometry.
Immunocytochemistry
Cells were plated on sterile coverslips in 24-well plates (10 000 cells/well) and cultured for 3 days. Fixation and staining conditions are included in the Supplementary Material.
Immunofluorescence was imaged using a Leica DM6000B microscope with Leica Application Suite Advanced Fluorescence software. Images were taken at 40x magnification.
Intensity of staining was analyzed by tracing the outline of cells and using the Fiji software intensity calculator within the selected fields. The average intensity for each image was calculated and used for statistical analysis.
Western Blot Analysis
Protein extraction methods and specific conditions for running and imaging western blots are included in the Supplementary Material.
Protein Extraction and PathScan
Protein extraction was performed as described for western blot (see Supplementary Material), and EGFR PathScan (Cell Signaling) was performed according to manufacturer’s instructions.
RNA Purification
RNA was extracted using the Allprep DNA/RNA kit (Qiagen) according to manufacturer’s instructions. Purified RNA was sent for further analysis by microarray platforms.
Microarray Platforms and Analysis
Expression array of NG2+ and NG2− populations was performed using Illumina Platform HumanWG6-V3 (Cancer Research Institute, University of Cambridge, UK). For data analysis, values were filtered according to their log2 fold change of <−0.5 or >0.5 with adjusted false discovery rate P-value of <0.05. Heat map, clustering, and statistical analysis were produced by Roslin Russel (Cancer Research Institute). Gene Ontology analysis was performed using GeneTrail tools.
Data Collection from Oncomine
The Oncomine database (https://www.oncomine.org/resource/login.html) was searched using EGFR and CSPG4 as search terms. Data were filtered for brain and CNS tumor sets.
Statistical Analysis
Statistical comparisons were made using t-tests, 1-way analysis of variance (ANOVA), or 2-way ANOVA as appropriate. If normality testing (Kolmogorov–Smirnov method) failed, the Mann–Whitney rank sum test (U-test) or Kruskal–Wallis 1-way ANOVA on ranks was performed. The degree of correlation between 2 variables was assessed by Pearson’s correlation coefficient or Spearman’s rank correlation coefficient. Statistical analysis of survival following xenografting used the Kaplan–Meier estimate, with time until death as the outcome measure. All error bars generated and depicted in the figures are obtained by pooling data from triplicates. They represent standard error of means. Statistical significance was set at P < 0.05. Statistical analyses were performed using Microsoft Excel, SigmaStat/SigmaPlot (Systel), and MedCalc. Significance in statistical analyses is represented by *<0.05, **<0.01, and ***<0.001.
Results
Differential Expression of NG2 Identifies Distinct Functional Subpopulations of Tumor Cells that Coexist in a Phenotypic Equilibrium
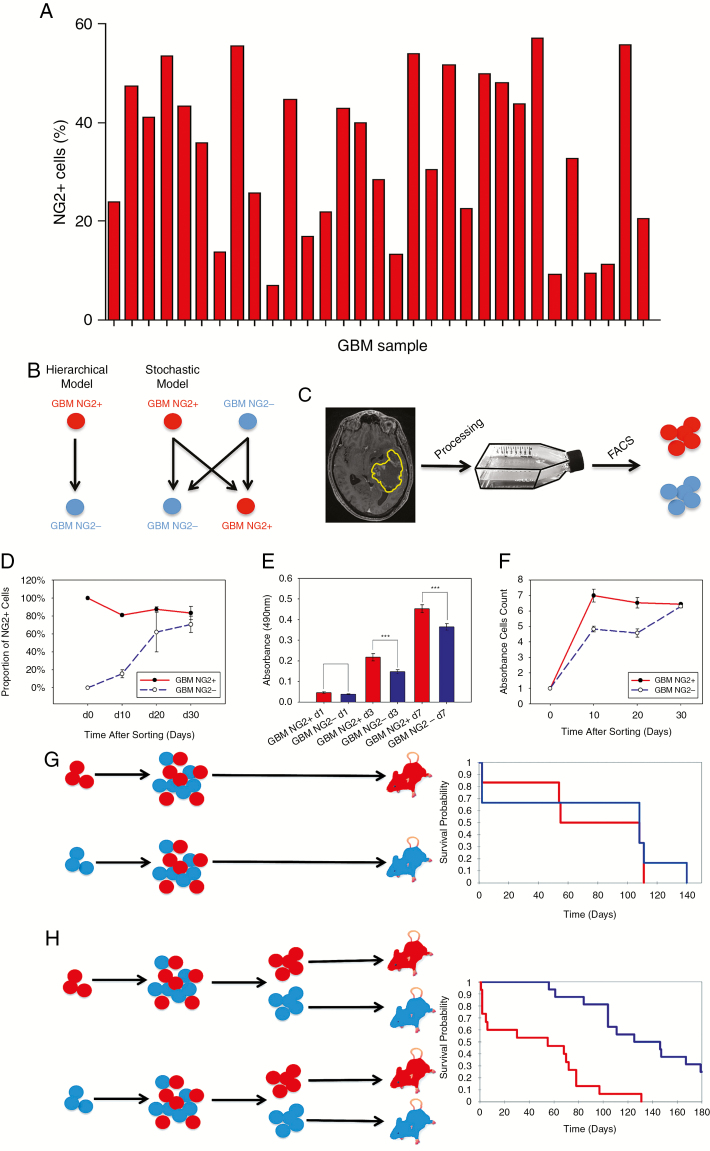
To establish the clinical relevance of NG2 in vitro studies, 31 freshly excised primary GBM tumors were analyzed for NG2 expression among viable cells by flow cytometry (clinical characteristics included in Supplementary Table 1). NG2 expression was found in all samples, ranging from 7% to 57.0% NG2+ cells (median 35.9%; Fig. 1A).
Fig. 1.
Expression of NG2 identifies populations that establish a phenotypic equilibrium. (A) Proportion of NG2+ cells (viable cell population) in 31 freshly derived tumor samples (flow cytometry). (B) Possible lineage relationships between GBM NG2+ and NG2− cells; a hierarchical model (left) where NG2− cells cannot give rise to NG2+ cells, or a non-hierarchical model (right). (C) We used patient-derived GBM cells, FAC-sorted into NG2+ and NG2− fractions. (D) Changes in the proportion of NG2+ cells over time in sorted GBM NG2+ and NG2− populations. (E) MTS assay of GBM NG2+ and NG2− cells at days 1, 3, and 7 post-sorting. (F) Diagram showing the growth of the GBM NG2+ and GBM NG2− cells at days 10, 20, and 30 after sorting. (G, H) Sorted GBM NG2+ and NG2− populations were cultured in vitro, before grafting to NOD-SCID mice. Kaplan–Meier analysis demonstrates no difference in survival (G). However, when these cells were re-sorted into NG2+ and NG2− populations prior to grafting, there is reduced survival in mice grafted with NG2+ cells (P < 0.001, Kaplan–Meier, H).
We hypothesized that NG2+ GBM cells follow a similar hierarchy to the oligodendrocyte lineage in the developing brain, whereby NG2+ cells differentiate into NG2− progeny. However, with the genetic instability and hostile microenvironment present in tumors, this strict hierarchy may be less rigid than in the highly regulated developing brain (Fig. 1B). To investigate the nature of the relationship between NG2+ and NG2− tumor cells, we applied a FACS-based approach to freshly derived human GBM cells (Fig. 1C).
Cell populations were sorted based on NG2 expression (Supplementary Figure 1). After 3 days, 100% of the GBM NG2+ population remained NG2+, while a small proportion (approximately 1%) of the GBM NG2− cells had begun to express NG2 (Supplementary Figure 2). At 10 and 20 days after sorting, the GBM NG2− population contained an increasing proportion of NG2+ cells (Fig. 1D, Supplementary Table 2) with a parallel reduction in the proportion of NG2+ cells in the NG2+ population (Fig. 1D, Supplementary Table 2). By day 30, both sorted populations contained equal proportions of NG2+ cells (Fig. 1D). These observations suggested that both NG2+ and NG2− sorted GBM cells could establish a population containing a balanced equilibrium between NG2+ and NG2− cells.
Based on previous data demonstrating the association between expression of NG2 and a more proliferative cellular phenotype,6 we hypothesized that the acquisition of NG2 expression would lead to an increase in cell proliferation.
To test our hypothesis, GBM cells were FAC-sorted into NG2+ and NG2− fractions, and proliferation was analyzed by MTS assay at different timepoints. GBM NG2+ cells were more proliferative in the initial stages of expansion (Fig. 1E, Supplementary Figure 3) and up to day 20 after initial sorting (Fig. 1F). However, this advantage was lost by day 30 after sorting when the 2 populations had equilibrated in terms of NG2 expression (Fig. 1F). These results confirm that NG2− expression is associated with enhanced proliferation within the context of a phenotypic equilibrium between GBM NG2+ and GBM NG2− cells. To interrogate the phenotype of NG2− cells, transcriptional profiling and Gene Ontology of NG2+ and NG2− cells was performed. Interestingly, the highest upregulated gene in NG2− fractions was Matrix Gla protein (MGP), a vitamin K–dependent, extracellular matrix protein that promotes migration in GBM17 (Supplementary Table 3).
Next, we hypothesized that the enhanced proliferation associated with expression of NG2 in vitro would result in more rapid tumor growth in vivo. Orthotopic grafting of cells derived from FAC-sorted GBM NG2+ and NG2− populations that had equilibrated over 30 days in vitro revealed no difference in time to onset of symptomatic disease (73±18 vs 78±24 days, P = 0.6; Fig. 1G). In contrast, re-sorting these progeny into positive and negative fractions immediately prior to implantation confirmed that NG2 expression was associated with a more aggressive disease phenotype (46±10 vs 139±14 days, P < 0.0001; Fig. 1H).
These observations argue against a unidirectional hierarchy in which GBM NG2+ cells give rise to GBM NG2− cells, and in which the NG2− cells cannot generate GBM NG2+ cells. Rather, our data show that isolated GBM NG2+ and GBM NG2− populations can form mixed progeny which establish a phenotypic equilibrium between NG2+ and NG2− cells similar to the parent tumor. However, within these equilibrated populations, NG2 expression is always associated with a more proliferative phenotype.
Phenotypic Equilibrium Is Established at Single Cell Resolution
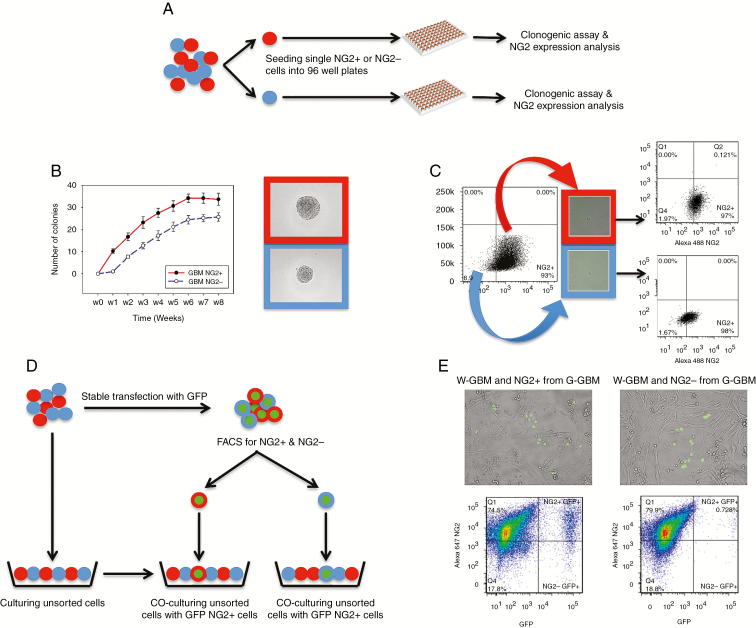
It is possible that the population data could be confounded by contamination with a small number of wrongly sorted cells. To investigate this possibility, we performed single cell analyses in which single GBM NG2+ or GBM NG2− cells were sorted and assayed for their ability to form colonies and generate mixed populations (Fig. 2A).
Fig. 2.
Single GBM NG2+ and GBM NG2− cells generate progeny that establish phenotypic equilibrium. (A) Experimental design for (B) and (C). Single NG2+ (red) or NG2− cells (blue) are seeded in 96-well plates, then tested for clonogenic capacity and NG2 expression by their progeny. (B) NG2+ cells demonstrate increased clonogenicity at all timepoints. Micrographs show examples of colonies formed from single NG2+ or NG2− cells. (C) Progeny of single GBM NG2+ and GBM NG2− cells generate mixed populations of NG2+ and NG2− cells. Illustrative example from G25 cell line. (D) Design of the experiment in (E). Unsorted GBM cells (W-GBM, red and blue) are stably transfected with GFP to form G-GBM population (green center). Single NG2+ (red) and NG2− (blue) cells from the G-GBM population are sorted into plates containing W-GBM cells. (E) Micrographs and flow cytometry analyses of the progeny of: (left) G-GBM NG2+ mixed with W-GBM cells and (right) G-GBM NG2− cells mixed with W-GBM cells. In both cases, the sorted G-GBM cells produce NG2+ and NG2− cells.
Single cells were cultured for 12 weeks, with the number of cells forming colonies quantified weekly. The number of colonies formed by single GBM NG2+ cells was consistently higher than the number of colonies formed by single GBM NG2− cells (P < 0.001; Fig. 2B). These colonies were analyzed by flow cytometry, which confirmed that single GBM NG2+ and single GBM NG2− cells can form mixed populations, each with the same proportion of NG2+ and NG2− cells, similar to the proportion of NG2+ cells in unsorted populations (Fig. 2C, Supplementary Table 4). These data support the results obtained using bulk populations of GBM cells, and show that single GBM NG2+ and NG2− cells can form mixed populations of cells, arguing against a unidirectional hierarchical model. This functional diversity was not replicated by non-malignant human fetal cortex derived NG2+ progenitors (Supplementary Figure 4).
Our data were obtained from a single cell–based model where cells were deprived of cell-to-cell interaction, which may influence tumor growth. Therefore, we also explored whether the in vitro environment and the pressure of single cell cultures influence the single cell results.
To investigate this, GFP-labeled single GBM NG2+ or GBM NG2− cells (G-GBM) were seeded into unsorted, unlabeled GBM cell populations (Fig. 2D). These mixed GFP and unlabeled populations were cultured and analyzed by flow cytometry. We observed that 78.9%±10.1% of the progeny from single G-GBM NG2+ cells expressed NG2, compared with 58%±1% of the progeny from single G-GBM NG2− cells (Fig. 2E).
These data confirm that single NG2+ and NG2− GBM cells have an intrinsic ability to generate mixed cell populations.
NG2 Plays a Functional Role in Enhancing Proliferation of GBM Cells
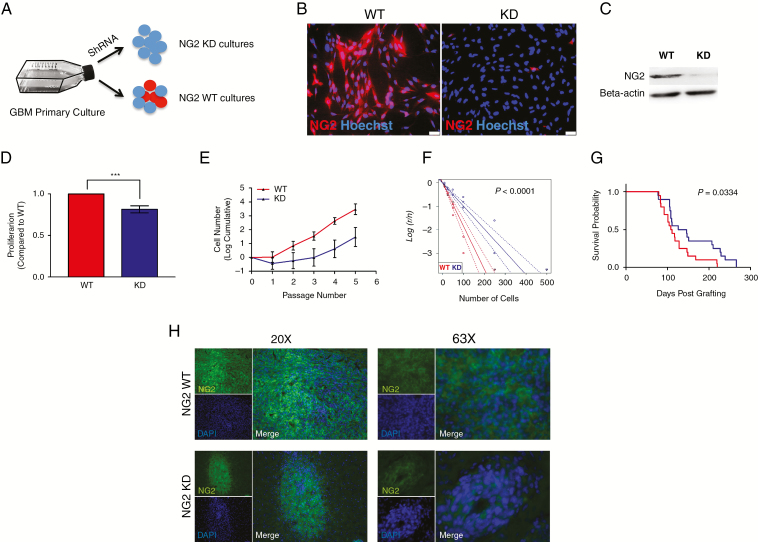
To understand the functional role of NG2 in GBM cells we manipulated the expression of NG2 using shRNA (Fig. 3A). NG2 knockdown was achieved in 3 separate patient-derived cell lines (NG2-KD), with GFP transfection of cells from the same patient used as a wild-type control (NG2-WT). Knockdown at the protein level was confirmed by immunocytochemistry and western blot (Fig. 3B, C). Knockdown did not affect cell viability (Supplementary Figure 5).
Fig. 3.
NG2 knockdown demonstrates a mechanistic role of NG2 in enhancing GBM cell proliferation. (A) Primary cell cultures were derived from clinical samples before stable NG2 shRNA knockdown. (B) Immunofluorescence staining of NG2 in wild-type (WT) culture and its corresponding NG2 knockdown (KD) culture. (C) Western blot of NG2 from WT and KD cells. (D) BrdU assay conducted on NG2-WT and NG2-KD cells, showing the proportion of proliferating cells. (E) Growth curves over several passages of NG2-WT and NG2-KD cells. (F) Limiting dilution assay (LDA) conducted on NG2-WT and NG2-KD cells. (G) Kaplan–Meier curve showing the survival of experimental animals grafted with NG2-WT or NG2-KD cells. (H) Immunofluorescence staining of NG2 in tumors derived from the injection of WT (upper panels) or KD cells (lower panels). The images show magnification at 20x (left) and 63x (right).
Consistent with our observations of NG2− cells, NG2-KD cells were less proliferative (BrdU assay; Fig. 3D) and had attenuated long-term population expansion compared with NG2-WT controls (Fig. 3E). NG2-KD cells were also less clonogenic compared with WT cells, with almost a 50% reduction in the proportion of colony forming cells, as assessed by limiting dilution assay (Fig. 3F). These effects were shown to be specific to NG2 knockdown, by use of a second shRNA construct (Supplementary Figure 5).
Orthotopic xenograft studies were performed to assess whether NG2 knockdown impacted in vivo growth. NOD-SCID mice were grafted with NG2-WT or NG2-KD cells and sacrificed when clinical symptoms appeared. Most mice developed tumors (NG2-WT cells 19/21, NG2-KD cells 20/21). However, analysis of survival confirmed a less aggressive disease phenotype in animals implanted with NG2-KD cells compared with NG2-WT cells (Fig. 3G). Analysis of NG2 expression in a subset of these animals (n = 4) revealed that approximately 45% of the cells in the NG-KD derived tumors were positive for NG2 (Fig. 3H), suggesting that the cell fraction retaining NG2 expression contributed to tumor growth. These results also confirm that NG2 expression is associated with aggressive phenotype, in agreement with our previous findings.6
These results show that NG2 plays a functional role in enhancing proliferation in a patient-derived xenograft model of GBM. Although knockdown of NG2 did not abolish the tumorigenic potential of GBM cells, lack of NG2 expression slowed down disease progression, emphasizing that the absence of NG2 leads to a less aggressive phenotype, but also that NG2− cells retain tumorigenic potential.
NG2 Promotes Cell Proliferation by Potentiating the EGFR‒Phosphatidylinositol-3 Kinase‒Akt Signaling Pathway
Recent work in animal models of glioma has shown that NG2 potentiates the activation of the tyrosine kinase domains of EGFR.12
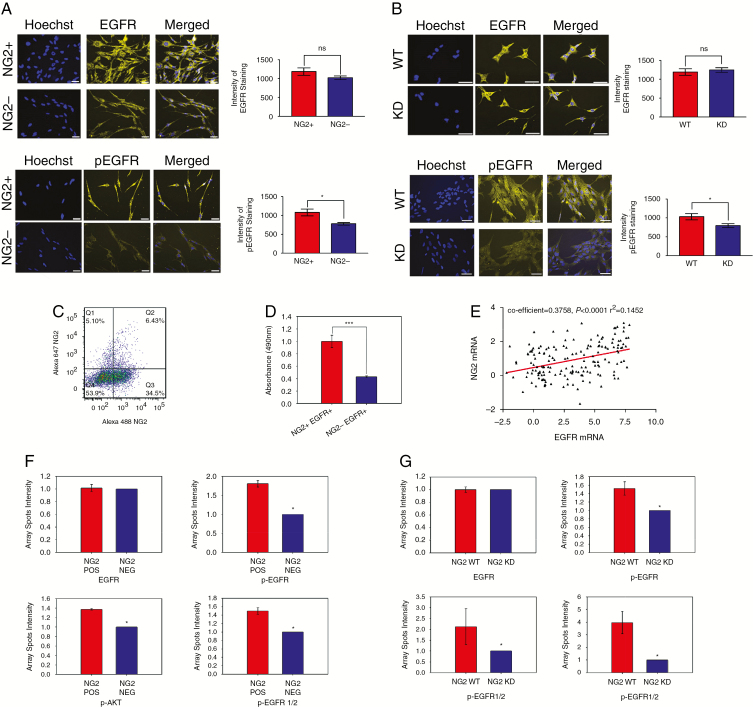
To understand the role of NG2/EGFR interactions in human disease, FAC-sorted NG2+ and NG2− populations were analyzed by immunocytochemistry for the expression of EGFR and phosphorylated EGFR (pEGFR, Tyr1068), one of the key tyrosine residues in EGFR activation.18 We found that the expression of total EGFR was not significantly different between NG2+ and NG2− populations (Fig. 4A). However, phosphorylation of EGFR was increased in NG2+ cells compared with NG2− cells (P = 0.01; Fig. 4A), suggesting that the presence of NG2 was sufficient to enhance activation of EGFR. We also analyzed expression of EGFR and pEGFR in NG2-WT and NG2-KD cells. Similarly to what we observed in NG2+ and NG2− populations, we found a significant reduction in the level of pEGFR in NG2-KD cells compared with NG2-WT cells (P = 0.02; Fig. 4B), despite there being no difference in the expression of total EGFR (Fig. 4B).
Fig. 4.
NG2 promotes activation of EGFR and its intracellular pathway. (A) The total EGFR and pEGFR expression in FAC-sorted GBM NG2+ and GBM NG2− populations. (B) The total EGFR and pEGFR expression in GBM NG2-WT and GBM NG2-KD populations. (C) Flow cytometry analysis of GBM cells for the expression of NG2 and EGFR. (D) MTS assay of FAC-sorted NG2+/EGFR+ and NG2−/EGFR+ subpopulations. (E) Scatterplot showing the correlation of expression of NG2 and EGFR in clinical datasets. (F) PathScan quantification of the expression levels of different components of the EGFR pathway in FAC-sorted GBM NG2+ and GBM NG2− populations. (G) PathScan quantification of the expression levels of different components of the EGFR pathway in NG2-WT and NG2-KD populations. Scale bars, 10 μm. *P < 0.05, ***P < 0.001.
Analysis of the expression of EGFR in 3 patient-derived GBM cell lines revealed EGFR expression of 10.3% ± 1.7% (Fig. 4C, Supplementary Figure 6). Further dissection of the GBM EGFR+ population identified 2 subpopulations: EGFR+/NG2− and EGFR+/NG2+ (Fig. 4C, D). We found that the EGFR+/NG2+ cells were significantly more proliferative compared with EGFR+/NG2− cells (Fig. 4D), suggesting a role for NG2 in potentiating EGFR signaling.
To confirm the functional role of NG2 in enhancing EGFR activation, we used PathScan for analysis of EGFR signaling. Our results revealed significantly higher phosphorylation of several downstream components of EGFR, including p-Akt and phospho–extracellular signal-regulated kinase 1 and 2, in NG2+ cells compared with NG2− cells (Fig. 4F), as well as in NG2-WT cells compared with NG2-KD cells (Fig. 4G). These data support a functional role for NG2 in potentiating EGFR signaling in glioblastoma.
To investigate clinically relevant associations between EGFR and NG2, we looked at the coexpression of the 2 proteins from clinical samples in published datasets. Pooled data from 5 datasets from the Oncomine database with mRNA expression data for EGFR and NG2 were analyzed for correlation between the 2 genes (193 tumors; references in Supplementary Material). These data demonstrated that there was a significant positive correlation between expression of the 2 genes (Fig. 4E). This is consistent with previous work, which has found significant upregulation of NG2 in tumors with EGFR amplification enriched in the classical subtype of GBM.19,20
Our data reveal a previously unrecognized role for the NG2 proteoglycan in human GBM with potentiation of EGFR signaling, leading to enhanced proliferation of EGFR+/NG2+ cells. The association between expression of these markers in patient-derived samples may be of clinical relevance, providing new insights into EGFR signaling that could help to improve the efficacy of anti-EGFR therapies.
Discussion
One of the pathognomonic features of GBM is the significant inter- and intratumoral heterogeneity that exists at both the genetic and histopathological levels.21–23 This drives a dynamic evolutionary process that results in the cellular phenotypic diversity present in tumors by the time of clinical manifestation and which is unlikely to be static.24–26
NG2 cells compose a progenitor population that could act as a cellular substrate for malignant transformation. In the adult brain, NG2+ cells provide the majority of actively proliferating cells.1 An association between NG2 expression and cell proliferation has been found in human GBM,6,27 and NG2 expression identifies an aggressive tumor phenotype in other cancers,28–30 suggesting a conserved role of NG2 in malignant progression in various tissues.
Here we report a dynamic relationship between NG2+ and NG2− cells in GBM. Longitudinal FACS analysis revealed a complex clonal environment in patients with clinically symptomatic GBM. GBM NG2− cells were able to express NG2 and gradually form phenotypically heterogeneous populations similar to the parental unsorted population. GBM NG2+ cells also formed mixed populations, confirming that over time the populations derived from either GBM NG2− or GBM NG2+ cells could become phenotypically similar, consistent with mathematical models of breast cancer development showing that all cells were capable of forming mixed populations.31 This is in striking contrast to the tightly controlled hierarchical organization in normal development. Therefore, the complex relationship between NG2+ and NG2− populations is unlikely to be explained by a hierarchical model. Instead, it suggests that from single cells populations will emerge which contain proliferative NG2+ populations and less proliferative NG2− populations. The consistent association between NG2 expression and a proliferative phenotype argues against the expression of NG2 being purely stochastic. Instead, we propose a hybrid model where NG2 expression identifies a “proliferative cell state” rather than a lineage hierarchy. According to this model, GBM NG2+ and NG2− cells establish a dynamic phenotypic equilibrium (Fig. 5). The point of equilibrium between NG2+ and NG2− cells varies between different patients and may also vary both temporally and spatially. In support of this, studies have shown enrichment of the NG2− population at the tumor–brain interface, with the NG2+ population representing the proliferative core.32 In this respect it is interesting that the NG2− population was enriched for MGP, a protein implicated in tumor invasion,17 suggesting that expression of NG2 represents an important phenotypic switch between proliferative cells (NG2+) and potentially invasive cells (NG2−) (Fig. 5). In the context of the phenotypic equilibrium seen in our cultures, there could be a dynamic bidirectional relationship between proliferative and potentially invasive cells, and could explain the persistence of the NG2− cells despite their more limited proliferative potential.
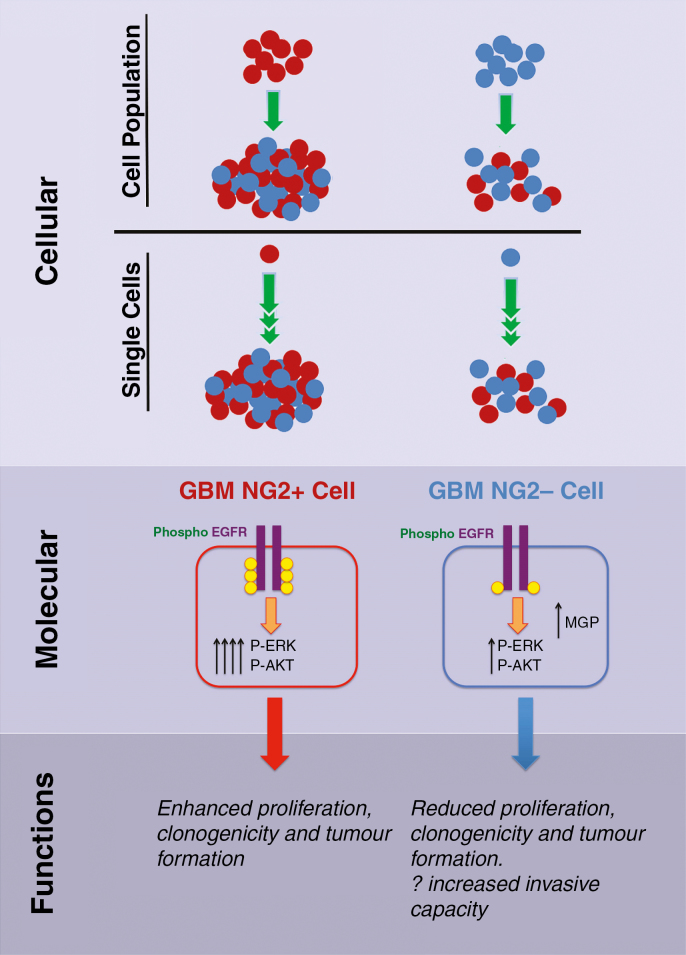
Fig. 5.
Cartoon summarizing the proposed dynamic evolutionary model of NG2 lineage relationships in GBM at cellular, molecular, and functional levels. At a cellular level, both NG2+ (red) and NG2− (blue) cell populations and single cells can form populations of mixed NG2+ and NG2− cells. NG2+ cells remain more proliferative and generate larger populations, supporting a role of NG2+ cells as the main driver of tumor bulk growth. NG2 promotes proliferation by enhanced EGFR signaling, increased EGFR phosphorylation, and increased signaling through extracellular signal-regulated kinase and Akt. This leads to a functional phenotype whereby NG2+ cells are more proliferative, and more aggressive when implanted into mice. Increased MGP expression in NG2− cells may represent an increased invasive phenotype of these cells.
Future work should define the functional properties of the NG2+ and NG2− cells in the context of GBM models where slow-cycling, therapy-resistant stemlike cells are at the apex of the hierarchy and generate rapidly proliferating cells33,34 to determine the role of the proliferative compartment in tumor growth and treatment resistance.
We have shown that NG2 contributes to the phenotype of NG2+ cells by enhancement of EGFR signaling. We found similar expression of EGFR in NG2+ cells and NG2− cells, but significantly higher phosphorylation of EGFR in NG2+ cells. A similar pattern occurred when we compared wild-type GBM cells with NG2 knockdown cells. Analysis of intracellular signaling pathways downstream of EGFR showed increased activation of downstream targets such as Akt and extracellular signal-regulated kinase. Analysis of published datasets confirmed a positive correlation between expression of EGFR and NG2, providing clinical corroboration of our data and supporting a model in which NG2 expression is increased in tandem with EGFR, as an important potentiating co-factor (Fig. 5).
Therefore, NG2 may promote tumor growth by enhancing activity of a known oncogene, rather than as an oncogene itself. NG2 functions in a similar physiological manner as a co-receptor with fibroblast growth factor receptor and platelet derived growth factor receptor A to activate focal adhesion kinase and mitogen-activated protein kinase pathways,35 suggesting a generic role for NG2 in facilitating receptor tyrosine kinase signaling.
Previous work has shown that high expression of NG2 is a poor prognostic factor.36 Analysis of published datasets shows minimal association between NG2 expression and survival (Supplementary Figure 7). However, these analyses use gene expression data and copy number analysis, and in most tumors NG2 copy number and gene expression are similar to the normal brain. Our data suggest that NG2 expression varies in a dynamic manner, and so gene expression at a single timepoint may not be illustrative of its effect on prognosis.
Our data suggest that NG2 may identify GBM cells responsible for accelerating tumor growth that could be better characterized and targeted therapeutically. A first step toward this direction has been made recently in an attempt to develop chimeric antigen receptor redirected T-cell therapy for the treatment of glioblastoma using NG2 as antigen.37 Due to repopulation from NG2− cells, repeated targeting of the aggressive NG2+ population could slow the rate of disease progression.33 Understanding the impact of mechanistic treatment on the evolutionary dynamics and growth patterns of malignant cell populations will yield vital information necessary to manage treatment-resistant disease, the ultimate arbiter of patient survival.
Funding
This work was supported by Addenbrooke’s Charitable Trust (to T.A.M., C.W.), Brain Tumour Charity (C.W.), National Institute for Health Research Cambridge Biomedical Research Centre (C.W., S.G.M.P.), Higher Education Funding Council for England (C.W.), Royal College of Surgeons of Edinburgh (C.W.), and European Commission–Seventh Framework Programme (Marie Curie Intra-European Fellowship to S.G.M.P.).
Supplementary Material
Acknowledgment
We thank Andreas Schwarzer and Simon McCallum for assistance with FACS.
Conflict of interest statement. The authors disclose no potential conflicts of interest.
References
- 1. Geha S, Pallud J, Junier MP, et al. . NG2+/Olig2+ cells are the major cycle-related cell population of the adult human normal brain. Brain Pathol. 2010;20(2):399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. García-Marqués J, Núñez-Llaves R, López-Mascaraque L. NG2-glia from pallial progenitors produce the largest clonal clusters of the brain: time frame of clonal generation in cortex and olfactory bulb. J Neurosci. 2014;34(6):2305–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Trotter J, Karram K, Nishiyama A. NG2 cells: properties, progeny and origin. Brain Res Rev. 2010;63(1-2):72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Persson AI, Petritsch C, Swartling FJ, et al. . Non-stem cell origin for oligodendroglioma. Cancer Cell. 2010;18(6):669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chekenya M, Rooprai HK, Davies D, Levine JM, Butt AM, Pilkington GJ. The NG2 chondroitin sulfate proteoglycan: role in malignant progression of human brain tumours. Int J Dev Neurosci. 1999;17(5-6):421–435. [DOI] [PubMed] [Google Scholar]
- 6. Al-Mayhani MT, Grenfell R, Narita M, et al. . NG2 expression in glioblastoma identifies an actively proliferating population with an aggressive molecular signature. Neuro Oncol. 2011;13(8):830–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang J, Svendsen A, Kmiecik J, et al. . Targeting the NG2/CSPG4 proteoglycan retards tumour growth and angiogenesis in preclinical models of GBM and melanoma. PLoS One. 2011;6(7):e23062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu C, Sage JC, Miller MR, et al. . Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146(2):209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aguirre A, Dupree JL, Mangin JM, Gallo V. A functional role for EGFR signaling in myelination and remyelination. Nat Neurosci. 2007;10(8):990–1002. [DOI] [PubMed] [Google Scholar]
- 10. Galvez-Contreras AY, Quiñones-Hinojosa A, Gonzalez-Perez O. The role of EGFR and ErbB family related proteins in the oligodendrocyte specification in germinal niches of the adult mammalian brain. Front Cell Neurosci. 2013;7:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mich JK, Signer RA, Nakada D, et al. . Prospective identification of functionally distinct stem cells and neurosphere-initiating cells in adult mouse forebrain. Elife. 2014;3:e02669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sugiarto S, Persson AI, Munoz EG, et al. . Asymmetry-defective oligodendrocyte progenitors are glioma precursors. Cancer Cell. 2011;20(3):328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mazzoleni S, Politi LS, Pala M, et al. . Epidermal growth factor receptor expression identifies functionally and molecularly distinct tumor-initiating cells in human glioblastoma multiforme and is required for gliomagenesis. Cancer Res. 2010;70(19):7500–7513. [DOI] [PubMed] [Google Scholar]
- 15. Fael Al-Mayhani TM, Ball SL, Zhao JW, et al. . An efficient method for derivation and propagation of glioblastoma cell lines that conserves the molecular profile of their original tumours. J Neurosci Methods. 2009;176(2):192–199. [DOI] [PubMed] [Google Scholar]
- 16. Guilfoyle MR, Weerakkody RA, Oswal A, et al. . Implementation of neuro-oncology service reconfiguration in accordance with NICE guidance provides enhanced clinical care for patients with glioblastoma multiforme. Br J Cancer. 2011;104(12):1810–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mertsch S, Schurgers LJ, Weber K, Paulus W, Senner V. Matrix gla protein (MGP): an overexpressed and migration-promoting mesenchymal component in glioblastoma. BMC Cancer. 2009;9:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Downward J, Parker P, Waterfield MD. Autophosphorylation sites on the epidermal growth factor receptor. Nature. 1984;311(5985):483–485. [DOI] [PubMed] [Google Scholar]
- 19. Verhaak RG, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wade A, Robinson AE, Engler JR, Petritsch C, James CD, Phillips JJ. Proteoglycans and their roles in brain cancer. FEBS J. 2013;280(10):2399–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bonavia R, Inda MM, Cavenee WK, Furnari FB. Heterogeneity maintenance in glioblastoma: a social network. Cancer Res. 2011;71(12):4055–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sottoriva A, Spiteri I, Piccirillo SG, et al. . Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110(10):4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Patel AP, Tirosh I, Trombetta JJ, et al. . Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324(5935):1670–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Piccirillo SG, Colman S, Potter NE, et al. . Genetic and functional diversity of propagating cells in glioblastoma. Stem Cell Reports. 2015;4(1):7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Piccirillo SG, Spiteri I, Sottoriva A, et al. . Contributions to drug resistance in glioblastoma derived from malignant cells in the sub-ependymal zone. Cancer Res. 2015;75(1):194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chekenya M, Pilkington GJ. NG2 precursor cells in neoplasia: functional, histogenesis and therapeutic implications for malignant brain tumours. J Neurocytol. 2002;31(6-7):507–521. [DOI] [PubMed] [Google Scholar]
- 28. Benassi MS, Pazzaglia L, Chiechi A, et al. . NG2 expression predicts the metastasis formation in soft-tissue sarcoma patients. J Orthop Res. 2009;27(1):135–140. [DOI] [PubMed] [Google Scholar]
- 29. Wang X, Osada T, Wang Y, et al. . CSPG4 protein as a new target for the antibody-based immunotherapy of triple-negative breast cancer. J Natl Cancer Inst. 2010;102(19):1496–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rivera Z, Ferrone S, Wang X, et al. . CSPG4 as a target of antibody-based immunotherapy for malignant mesothelioma. Clin Cancer Res. 2012;18(19):5352–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gupta PB, Fillmore CM, Jiang G, et al. . Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146(4):633–644. [DOI] [PubMed] [Google Scholar]
- 32. Wiranowska M, Ladd S, Smith SR, Gottschall PE. CD44 adhesion molecule and neuro-glial proteoglycan NG2 as invasive markers of glioma. Brain Cell Biol. 2006;35(2-3):159–172. [DOI] [PubMed] [Google Scholar]
- 33. Chen J, Li Y, Yu TS, et al. . A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lan X, Jörg DJ, Cavalli FMG, et al. . Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature. 2017;549(7671):227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goretzki L, Burg MA, Grako KA, Stallcup WB. High-affinity binding of basic fibroblast growth factor and platelet-derived growth factor-AA to the core protein of the NG2 proteoglycan. J Biol Chem. 1999;274(24):16831–16837. [DOI] [PubMed] [Google Scholar]
- 36. Svendsen A, Verhoeff JJ, Immervoll H, et al. . Expression of the progenitor marker NG2/CSPG4 predicts poor survival and resistance to ionising radiation in glioblastoma. Acta Neuropathol. 2011;122(4):495–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pellegatta S, Savoldo B, Di Ianni N, et al. . Constitutive and TNFα-inducible expression of chondroitin sulfate proteoglycan 4 in glioblastoma and neurospheres: implications for CAR-T cell therapy. Sci Trans Meth. 2018. 10(430):eaao2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.