Abstract
Common fragile sites (CFSs) are genomic regions prone to breakage under replication stress conditions recurrently rearranged in cancer. Many CFSs are enriched with AT-dinucleotide rich sequences (AT-DRSs) which have the potential to form stable secondary structures upon unwinding the double helix during DNA replication. These stable structures can potentially perturb DNA replication progression, leading to genomic instability. Using site-specific targeting system, we show that targeted integration of a 3.4 kb AT-DRS derived from the human CFS FRA16C into a chromosomally stable region within the human genome is able to drive fragile site formation under conditions of replication stress. Analysis of >1300 X chromosomes integrated with the 3.4 kb AT-DRS revealed recurrent gaps and breaks at the integration site. DNA sequences derived from the integrated AT-DRS showed in vitro a significantly increased tendency to fold into branched secondary structures, supporting the predicted mechanism of instability. Our findings clearly indicate that intrinsic DNA features, such as complexed repeated sequence motifs, predispose the human genome to chromosomal instability.
INTRODUCTION
Common fragile sites (CFSs) are specific genomic regions within the normal chromosomal structure, which appear as gaps and breaks in metaphase chromosomes of cells grown under mild replication stress conditions impeding DNA synthesis. CFSs are recurrently rearranged in tumor cells and are preferentially unstable already in early stages of cancer development (1–4).
The landscape of CFS expression is cell type specific (5,6) and more recently was shown to be oncogene-specific (7), indicating plasticity in the molecular basis underlying the specific fragile site sensitivity to breakage. No single mechanism can account for the sensitivity of CFSs to replication stress, and their preferred instability is governed by both genetic and epigenetic factors. The key features suggested to underlie CFS fragility include late replication timing, delayed replication completion, failure to activate dormant origins, origin paucity along large genomic regions, collision between replication and transcription complexes along large genes and the presence of AT-dinucleotide rich sequences (AT-DRSs) [reviewed in (8,9)].
AT-DRSs tend to form unusual DNA structures upon the unwinding of the double helix (10), which can potentially impair replication fork progression and may cause fork collapse and increased chromosomal breakage [reviewed in (11)]. The resolution of these abnormal structures and stabilization of stalled forks by the Werner and Bloom helicases, the Fanconi anemia proteins and CtIP have been implicated in the stability of fragile sites harboring AT-DRSs (12–16). AT-DRSs were shown to be difficult-to-replicate sequences in various in vitro systems and have been identified as particular unstable sequences driving cancer-related chromosomal instability [reviewed in (11)]. Many CFSs harbor AT-DRSs, which have been suggested to play a major role in the preferred instability of these regions (10,17–22).
In the present work, we investigated the direct effect of AT-DRS elements on genomic stability by exploring the ability of AT-DRSs per se to drive fragility. For this, we targeted a long 3.4 kb AT-DRS derived from the endogenous CFS on human chromosome 16, FRA16C, into a chromosomally non-fragile ectopic site. We have previously shown that this AT-DRS impedes the progression of replication forks by causing replication blocks under normal and replication stress conditions (20). Here we show that integration of this long AT-DRS into a non-fragile region drives fragile site expression, providing a clear evidence for the direct role of AT-DRSs in chromosomal fragility.
MATERIALS AND METHODS
Experimental system
The hypoxanthine phosphoribosyl transferase (HPRT) gene targeting vector (pHPRThyg) (23) served as a site-specific carrier for targeting an exogenous AT-DRS derived from the endogenous FRA16C. The integration was targeted via homologous recombination (HR) to the HPRT locus in the human diploid male fibrosarcoma cell line HT1080. As these cells contain a single copy of the X-linked HPRT gene, integrating into these cells ensures that all X chromosomes in the positive clones harbor the integrated AT-DRS. The use of the pHPRThyg vector as a tool for gene targeting has several advantages: (i) the targeted region is not known to carry recurrent breakage (see Supplementary Table S1); (ii) it enables introduction of one copy of the DNA construct, avoiding fragility due to integration of multiple copies of AT-DRSs; (iii) the homology arms of the vector, composed of DNA sequences of the HPRT gene, are HT1080 isogenic, thus improving the targeting frequency in the HT1080 cells (24); (iv) cloning of the AT-DRS into the pHPRThyg vector preserves the homology arms almost intact, hence preserving the original HR efficiency.
Vector construction
The original targeting vector pHPRThyg (14.2 kb) (Figure 1A) was cloned by the Gene Targeting Group, Imperial College School of Medicine, Hammersmith Hospital, London, UK (23), and was kindly given to us by Dr Andy Porter (Imperial College Faculty of Medicine, London UK). The pHPRThyg vector includes an 8.5 kb DNA fragment containing HPRT intron I, exon II, intron II, exon III, intron III and a hygromycin B phosphotransferase expression cassette, inserted within HPRT exon II.
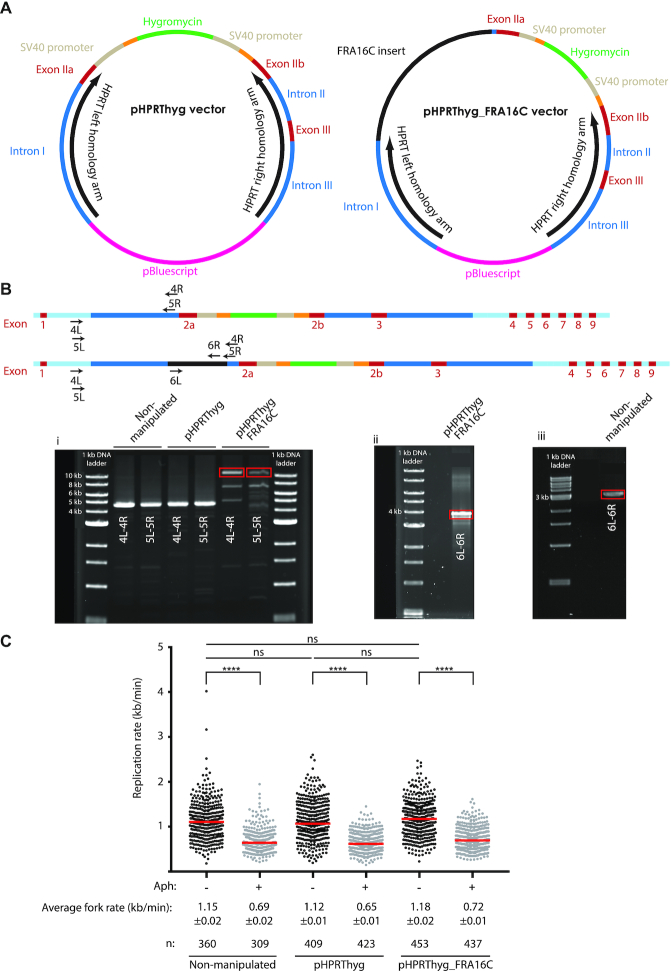
Figure 1.
Integration of FRA16C-derived AT-DRS using an HPRT gene targeting system. (A) The original pHPRThyg vector (left) and the newly cloned pHPRThyg_FRA16C vector (right) were designed to allow HR and construct integration within the HPRT locus. (B) A scheme of the HPRT locus after the integration of the pHPRThyg (upper panel) or pHPRThyg_FRA16C (lower panel) vector. The positions of primer sets 4 (4L-4R), 5 (5L-5R) and 6 (6L-6R) are illustrated. Light blue represents genomic regions that are not present in the vector. (i) Genomic DNA, isolated from HT1080 cells integrated with the pHPRThyg_FRA16C vector, was subjected to a long PCR across the AT-DRS at its ectopic integration site (HPRT locus), using two sets of chromosome X specific primers (4L-4R and 5L-5R). The same two sets of primers were also used to amplify genomic DNA isolated from HT1080 non-manipulated cells and cells integrated with the pHPRThyg vector. The red boxes mark the expected 10.6 kb PCR products harboring the 3.4 kb AT-DRS. (ii) The 10.6 kb DNA fragment, generated in the PCR of the pHPRThyg_FRA16C integrated cells, was subjected to nested PCR, using chromosome 16 specific primers (6L-6R), flanking the 3.4 kb AT-DRS. The red box marks the expected 3.8 kb PCR product harboring the 3.4 kb AT-DRS. (iii) The length of the AT-DRS at its endogenous site within FRA16C on chromosome 16 was determined by PCR on genomic DNA isolated from non-manipulated HT1080, using chromosome 16 specific primers (6L-6R). The red box marks a 3.2 kb PCR product suggesting that the AT-DRS in the endogenous FRA16C region is ∼2.8 kb. (C) Whole genome DNA replication rate analysis was performed by DNA combing, under normal or replication stress (0.1 μM aphidicolin) conditions. The distribution of replication fork rates in the non-manipulated and integrated HT1080 cells (pHPRThyg and pHPRThyg_FRA16C) is shown. The results are from three independent experiments. The median of each group is shown in red. The average fork rate of each group ±SE and the number of measurements (n) are indicated. Under normal growth conditions (-aphidicolin) the replication rate in the non-manipulated cells was not significantly different from the rate in the cells integrated with the pHPRThyg (P = 0.89) or with pHPRT_FRA16C (P = 0.72). There is also no significant difference between the replication rates in the pHPRThyg-integrated cells and the pHPRT_FRA16C-integrated cells (P = 0.07). Aphidicolin treatment led to a significant reduction in the mean rate in all clones (****P < 0.0001).
For the cloning of FRA16C-derived AT-DRS into the original pHPRThyg, we used the bacterial artificial chromosome (BAC) clone AC123909.1 (hg16_dna range=chr16:65107783-65268248, hg38_dna range=chr16:65298743-65458026). According to the NCBI34/hg16 genome reference, this BAC clone contains a 3.4 kb AT-DRS. This AT-dinucleotide rich element is composed of several types of repeated sequences, generating a complex polymorphism in the population due to variation in the length of this AT-DRS among individuals (25). It should be noted that the AT-DRS length according to genome reference GRCh38/hg38 is 2.2 kb, probably reflecting the length polymorphism among individuals.
For the cloning we isolated from BAC clone AC123909.1 a 6 kb sequence which harbors a 3.4 kb AT-DRS element and flanking sequences in two steps (see Supplementary Figure S1 for the sequence of the 6 kb cloned insert). First, a 7.1 kb fragment was isolated by BamHI/EcoRI digestion of the BAC clone and cloned into a pcDNA3 vector (Invitrogen) (Supplementary Figure S2, lane 2). The BamHI/EcoRI digested 7.1 kb fragment is expected from a sequence containing 3.4 kb of AT-DRS. Second, the 7.1 kb fragment was isolated and digested using the restriction enzymes HincII/XmnI, creating the 6 kb insert (Supplementary Figure S2, lane 3). For the cloning of the 6 kb insert, the pHPRThyg vector was restricted with the Pm1l enzyme (one digestion site) for linearization, treated with phosphatase (Antarctic phosphatase, New England Biolabs) and ligated with the 6 kb insert (T4 DNA ligase, Fermentas) by blunt end ligation (O/N, 16°C). The molar ratio between the vector and the insert was 1:9 (30:112.5 ng). Pm1l digestion reduced the length of the left homology arm from 4.6 to 4.4 kb. The resulting vector, termed pHPRThyg_FRA16C (20.0 kb) (Figure 1A), was transformed into Stable3 competent cells by heat shock transformation. Following transformation, plasmids were isolated from 8/58 colonies. The plasmids were subjected to HincII/XmnI enzymatic restriction, for validating correct ligation and preservation of the AT-DRS during the cloning. Gel electrophoresis of the reaction products showed in 2/8 purified plasmids the presence of the various expected digestion products, among them an ∼6.6 kb long fragment, indicating correct ligation of the vector and preservation of the AT-DRS length (Supplementary Figure S2, lane 4). In order to determine the insert ligation orientation, the pHPRThyg_FRA16C vector was sequenced using a primer (#7, see Supplementary Table S2 for primer sequence) that anneals 88 bp upstream the ligation position in HPRT intron 1. The sequencing results further confirmed the correct ligation of the 6 kb AT-DRS into the vector generating pHPRThyg_FRA16C. All the restriction enzymes used were manufactured by New England Biolabs. The work presented here was carried out on one of these clones.
Cell culture and vector transfection
Human fibrosarcoma HT1080 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Hyclone). For vector transfection, 10 × 106 cells were electroporated (BTX, Harvard Apparatus) with 15 μg of linearized (SalI restriction) pHPRThyg or pHPRThyg_FRA16C. Forty eight hours after transfection, selection with hygromycin (90 μg/ml) was started followed by double selection with 6-Thioguanine (6-TG) (15 μg/ml) after 96 h. Resistant colonies (HR positive colonies) were counted after 7 days of both hygromycin and 6-TG selection.
In order to verify that the HPRT remains inactive during culturing, following thawing of the cells, they were exposed to hygromycin and 6-TG selection. However, cell cultures designated for all experiments were grown without hygromycin and 6-TG selection during the last 48 h prior to the experiment as well as during the experiment.
Verification that the ectopically integrated sequences are preserved
Following HR of the pHPRThyg vector, a 2.7 kb sequence containing a hygromycin B expression cassette was inserted within HPRT exon II (Figure 1B, upper scheme). Following HR of the pHPRThyg_FRA16C vector, an 8.9 kb sequence, containing the 6 kb AT-rich insert and the hygromycin B expression cassette, was inserted between HPRT intron I and exon IIb (Figure 1B, lower scheme). In order to validate that the ectopically integrated sequences are preserved, genomic DNA was isolated from the non-manipulated HT1080 cells, HT1080 cells integrated with the pHPRThyg vector and HT1080 cells integrated with the pHPRThyg_FRA16C vector and various polymerase chain reaction (PCR) reactions were performed. In order to avoid amplification of vector sequences that were randomly integrated into the genome, one primer in each pair was designed to anneal to genomic sequences on chromosome X, which are not present in the vector. PCR using primers amplifying sequences of the hygromycin resistance cassette (1L-1R) was performed under the following conditions: 94°C for 2 min, 25 cycles of 94°C for 30 s, 53°C for 30 s, 68°C for 7 min. PCR using two sets of primers amplifying sequences flanking the AT-DRS (2L-2R and 3L-3R), was performed under the following conditions: set 2–94°C for 2 min, 25 cycles of 94°C for 30 s, 56°C for 30 s, 68°C for 7 min; set 3–94°C for 2 min, 25 cycles of 94°C for 30 s, 55°C for 30 s, 68°C for 9 min. These PCRs were performed using the Platinum Taq DNA polymerase (Invitrogen), using 50–100 ng genomic DNA.
For validating that the length of the integrated AT-DRS has not been changed during the integration process, additional PCRs were performed using two set of primers (4L-4R and 5L-5R) under the following conditions: 94°C for 3 min, 35 cycles of 94°C for 20 s, 60°C for 30 s, 60°C for 15 min and at last 60°C for 6 min. The resulted 10.6 kb PCR product was isolated and served as a template for a nested PCR, using primers specific to chromosome 16 that flank the AT-DRS (6L-6R). The nested PCR to amplify the integrated AT-DRS was performed under the same PCR conditions as for primers 4L-4R and 5L-5R. Following verification of its length by gel electrophoresis, this PCR product was isolated and sequenced (Sanger Sequencing, GenScript USA Inc.). These PCRs were performed using the Kapa long range hot start PCR kit (Kapa Biosystems) with 10 ng genomic DNA. The sequences of all primer sets are listed in Supplementary Table S2.
Fluorescent in situ hybridization (FISH) analysis
For the induction of CFS expression, cells were treated with 0.1–0.3 μM aphidicolin and 0.7 mM caffeine. Following 24 h, the cells were treated with 100 ng/ml colcemid (Biological Industries, Israel) for 40 min in a 5% CO2 incubator. Then, the cells were collected by trypsinization, treated with hypotonic solution at 37°C for 30 min and fixed with multiple changes of methanol:acetic acid 3:1. Fixed cells were kept at minus 20°C until analysis. For fluorescent in situ hybridization (FISH) analysis, we used specific probes that map to different regions on the X chromosome: BAC clones RP11-1151L12 (hg38_chrX:134323531-134470399) and RP11-671P4 (hg38_chrX:134376988-134571881) were used for HPRT locus (hg38_chrX:134460165-134500668) detection, BAC clones RP11-279C14 (hg38_chrX:102562640-102727121) and RP11-15E22 (hg38_chrX:101416342-101594002) were used for FRAXC (Xq22.1) detection and the plasmid PDMX1 was used for centromere detection. For the evaluation of endogenous FRA16C expression we used the BAC clone RP11-27L11 (GenBank: AC123909.1) which contains the 3.4 kb AT-DRS. Our previous FISH analysis using this BAC clone showed that it spans the gaps and constrictions of FRA16C (10). It is worth noting that the chromosome 16 RP11-27L11 BAC clone could not be used for the detection of the integrated FRA16C-derived 6 kb AT-DRS in chromosome X, as the short hybridization length is below the possible FISH detections (Supplementary Figure S3).
The probes were labeled by nick translation using DY-505-aadUTP (Spectral Imaging). The slides with metaphase spreads were treated with RNase and then fixated with 1% formaldehyde/PBS-MgCl2 at room temperature (RT) for 10 min followed by dehydration with serial ethanol washing steps. Probes were suspended in denaturation/hybridization solution (50% deionized formamide/50% hybridization buffer), followed by 60 min vortex at RT. The slides were denatured with deionized formamide/20×SSC for 2 min at 67°C and then dehydrated with serial ethanol washing steps. The probes were applied to the slides and incubated overnight at 37°C in a humidified chamber. The slides were then washed with 2×SSC for 1 min at RT followed by two washes for 5 min each with 0.5×SSC at 72°C. An additional wash with 4 × SSC/0.01% Tween for 2 min at RT was followed by a 1-min wash in DDW. Slides were air dried and mounted with Vectashield mounting medium with propidium iodide (Vector Laboratories). FISH images were acquired with an inverted fluorescence microscope (Olympus IX81, lens × 60 oil). Fragmented metaphases were not included in the analysis. Only sites with a breakage frequency above 1% from the analyzed chromosome (X or 16) were considered as fragile sites.
Molecular combing
The molecular combing approach allows high-resolution analysis of replication dynamics of single genomic DNA molecules. The principle of the combing method is that individual DNA molecules, labeled with thymidine analogs, are combed on special surfaces, with a constant stretching factor enabling accurate quantification of replication fork speed and inter-origin distance (26). For labeling nascent DNA in the different HT1080 cultures, non-treated or aphidicolin-treated unsynchronized cells were pulse-labeled for 30 min with 100 μM of the thymidine analog 5-iodo-2′-deoxyuridine (IdU, Sigma). At the end of the first labeling period, the cells were washed three times with warm medium and pulse labeled for 30 min with 100 μM of a second thymidine analog, 5-chloro-2′-deoxyuridine (CldU, Sigma). At the end of the second labeling period, the cells were washed three times with phosphate-buffered saline (PBS) and then harvested. Aphidicolin treatment (0.1 μM) was initiated 1 h before the analog pulses and persisted through the labeling period. For preparation of agarose plugs, the cells were re-suspended in equal volumes of PBS and molten low melting agarose 1% (NuSieve GTG agarose) to a final concentration of 0.5% agarose and mixed gently. The mixture was immediately transferred into a plug mold and plugs were incubated at 4°C until solidified. Plugs were incubated overnight at 50°C with freshly prepared Proteinase K digestion solution (2 mg/ml proteinase K, 1% sodium dodecyl sulphate and 100 mM ethylenediaminetetraacetic acid (EDTA)). The following day, the proteinase K digestion solution was removed and replaced with TE wash buffer (10 mM Tris pH 7.5/1 mM EDTA). The plugs were washed for 6 h on a test tube rotator; the TE was replaced every hour. Each agarose plug was transferred into a round bottom microtube containing 0.5M MES buffer pH5.5 and melted at 68°C for 20 min followed by incubation at 42°C for 10 min. The extracted genomic DNA was combed on silanized coverslips and analyzed as previously described (26,27). The primary antibody for fluorescence detection of IdU was mouse anti-BrdU (Becton Dickinson) and the secondary antibody was goat anti-mouse Alexa Fluor 488 (Invitrogen). The primary antibody for fluorescence detection of CldU was rat anti-BldU (Abcam). The secondary antibody was goat anti-rat Alexa Fluor 594 (Invitrogen). The length of the replication signals was measured in micrometers and converted to kilo bases according to a constant and sequence-independent stretching factor (1 μm = 2 kb), as previously reported (27). DNA Replication rates (kb/min) were calculated by dividing the measured length of the replication signal (kb) in 30 min.
Computational predictions of DNA secondary-structure formation
To evaluate DNA conformational flexibility, we used a measure of the potential local variations in the DNA structure, expressed as fluctuations in the twist angle (28). This measure provides average fluctuations in the twist angle for each of the possible dinucleotides and thus enables the evaluation of the flexibility of a DNA sequence by summation of these values. This analysis enables the identification of AT-DRSs, which have the highest flexibility among all dinucleotides (28). To carry out the analysis, we used the TwistFlex computer program (http://margalit.huji.ac.il/TwistFlex/index.html), which calculates flexibility measures of DNA sequences (10). The analysis was performed in overlapping windows of 100 bp. Dinucleotide values were summed along the window and averaged by the window length. Windows with values of >13.7° were considered as flexibility peaks (10).
To evaluate the potential of single stranded DNA (ssDNA) to form stable secondary structures, we performed Mfold analysis of secondary structure-forming potential, using a chromosomal fragility prediction threshold (29). For this, the HPRT locus was divided into segments of 300 nt, with a 150 nt shift window, and each segment was analyzed by the Mfold program (30). A fragility threshold of at least seven consecutive segments with a free energy value <−40 kcal/mol, characterizing aphidicolin-induced CFSs was used.
Reduplexing assay
DNA fragments, derived from the integrated 3.4 kb AT-DRS (AT1 and AT2) or control sequences from the vector pHPRThyg (Ctrl1 and Ctrl2), were dephosphorylated at the 5′ end with calf intestinal alkaline phosphatase (New England Biolabs) and end-labeled with [γ-32P] ATP (PerkinElmer) using T4 polynucleotide kinase (New England Biolabs). End-labeled DNA (1 ng) was added to solutions of 500 μl containing 0.1 M NaCl in TE buffer and incubated at 95°C for 5 min. The samples were slowly cooled to room temperature, and the DNA was ethanol-precipitated in the presence of glycogen (Roche). The DNA pellets were then air-dried and resuspended in TE buffer. DNA samples were electrophoresed in a 4% native polyacrylamide gel cast in TBE at 150 V for 3 h at room temperature. The gel was dried and visualized by phosphorimaging (GE HealthCare).
Statistical analysis
For statistical comparison of the fork progression rate an ANOVA test was used. For comparison of the breakage extent at the HPRT locus versus the endogenous site FRA16C, Fisher's exact test was used. For statistical comparison of the extent of slow migrating reduplexed DNA, Student's t-test was used. For statistical analysis of the distribution of AT-DRS >3000 bp among chromosomal bands harboring fragile sites, a chi square test was used. In all analysis, statistical significance difference was considered as P-value of ≤0.05.
RESULTS
The ectopic site for targeting FRA16C-derived AT-DRS is chromosomally stable and is devoid of CFS features
As the ectopic site for integration, we chose the HPRT locus on chromosome X (hg38_chrX:134460165-134500668, q26.2-26.3), for several reasons. First, this locus is not harbored within the described CFSs in the human genome (Supplementary Table S1), and particularly not in the described repertoire of CFSs in fibroblasts (7,31,32). Importantly, the HPRT locus is non-fragile in the HT1080 cells studied here as described below. Yet, a very low-expressed aphidicolin-induced CFS (<0.3%) was reported in Xq26 in lymphocytes (33). Second, the AT/TA dinucleotide content in the HPRT locus is <16% of all possible dinucleotides, in comparison to the very high level (66%) of the AT/TA dinucleotide in the 3.4 kb fragment. In addition, the HPRT locus is devoid of AT-DRSs (>250 bp in length), as shown by DNA flexibility calculations performed on 500 kb encompassing the HPRT region (hg38_dna range=chrX:134230000-134730000). Moreover, Mfold analysis of secondary structure-forming potential, using a chromosomal fragility prediction threshold (29) showed that the HPRT gene region does not contain any consecutive sequence segments predicted to fold into stable secondary structures, a feature characterizing fragile regions (see ‘Materials and Methods’ section for details).
We further examined the replication timing and large gene content in ∼500 kb flanking the HPRT locus, as it has been proposed that these features contribute to the sensitivity of CFSs to replication stress and breakage (7,11,34). The analysis revealed that the region is relatively early replicating in most studied cell types (http://www.replicationdomain.com) (35) and is lacking large genes (≥300 kb) (https://genome.ucsc.edu). Moreover, the HPRT gene enables targeting of ectopic sequences using a unique gene targeting system in the human diploid male HT1080 fibrosarcoma cell line containing only one copy of the X chromosome (23). Thus, the HPRT locus does not have FS characteristics, making it to an excellent ectopic site for integration.
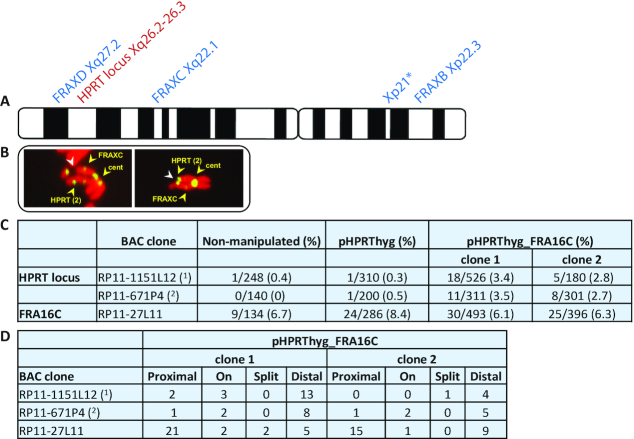
Since the repertoire of CFSs is cell type specific (5,6) and oncogene dependent (7), we analyzed the chromosomal stability of the HPRT region in the analyzed HT1080 fibrosaroma cells. For this, we grew the cells 24 h under conditions of mild replication stress using low concentrations of the DNA polymerase inhibitor aphidicolin and the ataxia-telangiectasia mutated (ATM)/Rad3-related (ATR) inhibitor caffeine, which abrogates the G2/M checkpoint enabling fragile site detection (36,37). As expected, caffeine treatment alone resulted in no recurrent fragility in over 90 analyzed metaphases. FISH analysis using fluorescently labeled BAC clones from the HPRT region was performed on metaphase chromosomes. Analysis of 388 metaphase X chromosomes (Figure 2C) revealed no recurrent gaps and breaks, supporting previous FS mapping studies in fibroblasts (7,31,32). We also evaluated the breakage frequency of the low-expressed CFS FRAXD (Xq27.2), identified in lymphocytes and mapped ∼6.7 Mb distal to the HPRT locus (33). For the evaluation of FRAXD instability, we searched for gaps and breaks distal to the HPRT locus, marked by fluorescently labeled BAC clones from the HPRT region, which are located ∼14% of the Xq arm from the telomere. In the 388 analyzed X chromosomes, no fragility was detected at the FRAXD region under the replication stress conditions, supporting previous studies which profiled CFS expression in fibroblasts and did not find FRAXD expression (7,31,32). Altogether, the various analyses indicate that the HPRT locus is chromosomally stable and is devoid of CFS features.
Figure 2.
The integrated AT-DRS creates a novel fragile site. (A) A schematic illustration of the known CFSs on chromosome X expressed in different cell types and the HPRT locus. *Represents a CFS described in fibroblasts and epithelial cells (5,31). (B) Examples of metaphase X chromosomes from HT1080 cells integrated with the pHPRThyg_FRA16C vector grown under mild replication stress. White arrowheads mark breaks at the HPRT region. Yellow arrowheads mark FISH probes for the HPRT locus [RP11-1151L12 (1) or RP11-671P4 (2)], FRAXC region and the centromere X. (C) Breakage frequency of the novel FS at the HPRT locus and the frequency of the endogenous FRA16C site in the non-manipulated HT1080 cells, pHPRThyg integrated cells and two independent clones integrated with the pHPRThyg_FRA16C vector. The breakage frequency is calculated as the number of breaks at chromosome X or chromosome 16 relative to the number of the specific analyzed chromosomes. (D) The number of chromosomes with FISH signals proximal, on, or distal to the novel fragile sites.
The ectopically integrated FRA16C-derived AT-DRS is preserved
In order to target the FRA16C-derived AT-DRS, we cloned a 6 kb fragment from BAC clone AC123909.1 which includes the 3.4 kb AT-DRS and flanking sequences into the HPRT gene targeting system (pHPRThyg) (23) in two steps that led to creation of the pHPRThyg_FRA16C vector (Figure 1A, see the detailed description of these steps in the ‘Materials and Methods’ section). This AT-DRS is composed of several types of AT-dinucleotide rich repeats, among which are minisatellites with 32, 33 and 37 bp repeat motifs. The 33 bp repeat is expanded in the rare fragile site FRA16B (25).
The vector was transfected into HT1080 cells by electroporation. Successful insertion of the targeted sequences into the HPRT locus by HR led to HPRT inactivation and resistance of the targeted HT1080 cells to both hygromycin and 6-TG (see also ‘Materials and Methods’ section). Validation of correct integration of the vector sequences was performed by several PCR reactions amplifying the vector sequences (primer set 1) or sequences flanking the AT-DRS (primer sets 2 and 3) (‘Materials and Methods’ section, Supplementary Table S2 and Figure S4). In order to validate that the integrated AT-DRS has not been amplified, deleted or rearranged during the integration process, we performed a long PCR across the AT-DRS at its ectopic integration site. For this we used two sets of chromosome X specific primers (primer set 4, 4L-4R and primer set 5, 5L-5R) which are expected to amplify a 10.6 kb fragment if the integrated 3.4 kb AT-DRS is preserved (‘Materials and Methods’ section, Supplementary Table S2). As can be seen in Figure 1B (i), gel electrophoresis of the PCR products revealed a 10.6 kb DNA fragment, indicating the presence of the integrated AT-DRS. Using the same two sets of PCR primers on genomic DNA of the HT1080 non-manipulated cells and cells integrated with the pHPRThyg vector generated an expected ∼4.5 kb product corresponding to the normal sequence of this HPRT genomic region. We next determined the length of the integrated AT-DRS by subjecting the purified 10.6 kb PCR product to nested PCR, using chromosome 16 specific primers (primer set 6, 6L-6R), flanking the 3.4 kb AT-DRS, predicted to amplify a 3.8 kb fragment. As can be seen in Figure 1B (ii), a 3.8 kb fragment was generated, indicating the presence of a preserved AT-DRS. At last, the sequencing of this 3.8 kb fragment, using primers 6L and 6R, revealed a perfect match with the AT-rich sequence (Supplementary Figure S5). Altogether, this data demonstrates that the ectopically integrated FRA16C-derived AT-DRS is preserved at the HPRT locus on X chromosomes integrated with the pHPRThyg_FRA16C vector.
The integrated clones exhibit normal whole genome replication dynamics
Normal DNA replication requires a balanced and sufficient cellular nucleotide pool (1). In eukaryotes, nucleotide levels are maintained by de novo synthesis as well as the salvage pathway, in which the HPRT is a central enzyme. Although HPRT deficiency is accompanied by activation of purine synthesis via the de novo pathway (38), we investigated the effect of HPRT deficiency in the integrated clones on whole genome DNA replication dynamics. This analysis aimed to ensure that the perturbed salvage by itself did not lead to perturbed replication, which could interfere with the evaluation of the genomic stability following AT-DRS integration. Analysis of whole genome DNA replication rates using DNA combing revealed no significant differences in the replication rate among the various clones under normal growth conditions (Figure 1C). In addition, treatment of all clones with aphidicolin led to a significant reduction in the mean fork rate, which was similar in the different clones (39–42% of the normal rate) (****P < 0.0001). These results indicate that HPRT deficiency due to the integrated sequences had no effect on whole genome DNA replication dynamics and that the integrated HT1080 clones are a suitable cellular system for investigating the effect of AT-DRS on genomic instability.
The integrated AT-DRS creates a novel fragile site
To investigate the effect of the integrated AT-DRS on the chromosomal stability at the integration site under replication stress conditions, we treated cells from two different clones integrated with the pHPRThyg_FRA16C with low concentrations of aphidicolin and caffeine and performed FISH analysis using two BAC clones encompassing the HPRT region (RP11-1151L12 and RP11-671P4). The integration of the original pHPRThyg vector, which does not harbor the AT-DRS, served as a control. Screening of X chromosomes targeted with the control vector revealed no recurrent chromosomal instability at the HPRT locus (Figure 2C). Strikingly, 42/1318 (3.2%) of the X chromosomes targeted with the AT-DRS showed recurrent gaps and breaks at the HPRT locus, indicating that the AT-DRS is sufficient to generate a novel FS (Figure 2C). The observed FISH signals of clones RP11-1151L12 and RP11-671P4 were proximal, overlapping and distal to the breaks in different chromosomes, implying that these clones span the unstable HPRT region (Figure 2D).
We also analyzed the breakage frequency of the endogenous FRA16C in the HT1080 cells integrated with the pHPRThyg_FRA16C. The analysis revealed a significantly higher breakage frequency of the endogenous FRA16C compared to the novel ectopic fragile site in each of the HT1080 clones integrated with the pHPRThyg_FRA16C vector. In clone 1, 30/493 (6.1%) of chromosome 16 showed instability at the endogenous FRA16C while at the novel ectopic fragile sites 29/837 (3.5%) X chromosomes showed instability (*P < 0.05) (Figure 2C). In clone 2, 25/396 (6.3%) of chromosome 16 showed instability at the endogenous site while at the novel fragile sites 13/481 (2.7%) X chromosomes showed instability (*P < 0.05) (Figure 2C). We next determined the length of the endogenous AT-DRS in FRA16C in the non-manipulated HT1080 cells. PCR using primers 6L-6R flanking this AT-DRS generated an ∼3.2 kb fragment, suggesting that its length is ∼2.8 kb (Figure 1B (iii)). This indicated that the higher fragility frequency of the endogenous site is affected by additional factors. Indeed, the endogenous FRA16C site harbors additional AT-DRSs (>400 bp in length) that were previously found to perturb the replication progression under mild replication stress conditions (20). Altogether, these results indicate that the integrated AT-DRS is able in itself to generate fragility under replication stress and is a major cause predisposing the FRA16C region to instability. Yet, an effect of epigenetic changes, due to the integration of the AT-DRS, on the stability of the region cannot be excluded.
Secondary structure formation by AT-DRSs
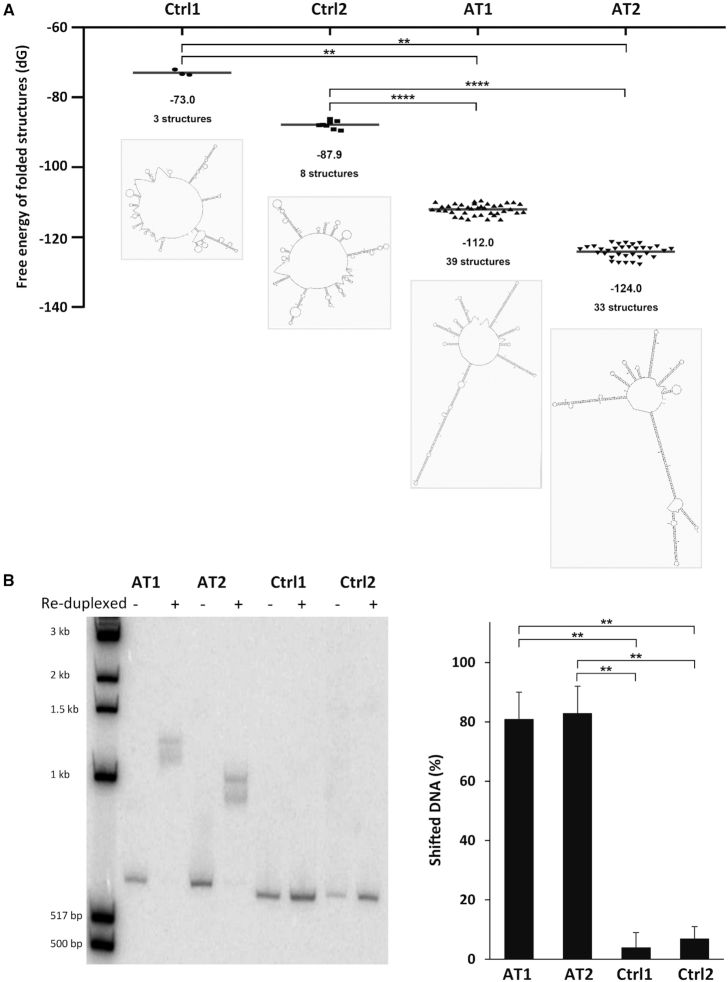
To experimentally explore whether the genomic instability of the integrated AT-DRS sequence could be due to the formation of secondary structures, we analyzed the DNA folding potential by the reduplexing assay aimed to detect slow migrating branched molecules. The assay has a fragment size limitation, thus we analyzed AT-DRS and non AT-DRS (control) fragments of ∼ 550 bp, derived from the integrated 3.4 kb AT-DRS and the pHPRThyg control vector (see Supplementary Figure S6 for fragments sequences). Each of the analyzed fragments contains AT-dinucleotide level similar to the level of the entire sequence from which it was derived: the AT/TA dinucleotide content of the entire 3.4 kb AT-DRS is very high (66%), and the two derived fragments AT1 and AT2 are also highly enriched with AT/TA dinculeotides, 71 and 61%, respectively. The AT/TA content of the 2.7 kb control sequence is 11% and the two derived fragments Ctrl1 and Ctrl2 contain 11 and 9%, respectively. First, in silico predictions of the folding ability of these DNA fragments were performed by the Mfold algorithm (30), which predicts nucleic acid folding and hybridization. For each DNA fragment, the free energy values of predicted folded structures, the average free energy and the number of predicted secondary structures were analyzed. As can be seen in Figure 3A, the potential to form multiple stable secondary structures was significantly higher for each of the AT-dinucleotide rich fragments relative to each of the control sequences (**P < 0.01, ****P < 0.0001).
Figure 3.
Secondary structure formation by FRA16C AT-DRSs. (A) In silico predictions of the folding ability of ∼550 bp long DNA fragments derived from the integrated 3.4 kb AT-DRS (AT1 and AT2) or control sequences from the vector pHPRThyg (Ctrl1 and Ctrl2) were performed by the Mfold algorithm (30). The fragments sequences are shown in Supplementary Figure S6. Dots represent the free energy values of predicted folded structures. The average free energy and the number of predicted secondary structures are shown below the dots. An illustration of the most stable secondary structure for each fragment is also shown. **P < 0.01, ****P < 0.0001. (B) The same DNA fragments analyzed by Mfold, represented in (A) and Supplementary Figure S6 (AT1, AT2, Ctrl1 and Ctrl2), were subjected to denaturation and reduplexing reaction and separated by gel electrophoresis. **P < 0.002.
Next we experimentally analyzed the folding potential of AT and control DNA fragments (AT1, AT2, Ctrl1 and Ctrl2). For this the fragments were synthesized (GeneArt) and subjected to reduplexing assay to observe the formation of DNA secondary structures. Low concentrations of DNA fragments were first denaturated, and the single-stranded DNAs were allowed to re-anneal in the presence of 100 nM NaCl. Reduplexing was performed 8 times for Ctrl1, Ctrl2 and AT1 and 4 times for AT2. Slower migrating products, quantified as shifted DNA, represent the formation of stable secondary structures. Separation by polyacrylamide gel electrophoresis showed that the reduplexed AT-DRS fragments give rise to significantly more slow-migrating products compared to the control sequences (**P < 0.002), suggesting an increased tendency to form stable secondary structures under the single-stranded state (Figure 3B).
Long AT-DRSs (≥3000 bp) are involved in chromosomal instability
The discovery that a long AT-DRS can itself drive genomic instability led us to explore whether similar long AT-DRSs predispose other genomic regions to fragility. To do so, we identified the genomic location of all AT-DRSs ≥3000 bp, along the human genome and analyzed whether they reside within known FSs. Using the TwistFlex program (10), 31 AT-DRSs were identified (Supplementary Table S3). Strikingly, 14 of these (45.2%) reside in chromosomal bands harboring recurrently expressed CFSs (Supplementary Tables S1 and S3), implying a non-random distribution of these AT-DRSs (P = 0.0003). The fact that not all long AT-DRSs reside in the currently known CFS regions further highlights the complex basis of chromosomal instability that involves an interplay between various factors, together governing the preferred sensitivity of specific genomic loci to replication stress (11).
DISCUSSION
In the present study, we have demonstrated that integration of a long AT-DRS into a non-fragile region can drive fragile site expression. This provides a clear evidence for the direct role of AT-DRSs in driving chromosomal fragility. We further found that DNA fragments derived from the integrated AT-DRS have an increased tendency to form stable secondary structures under single stranded state which explains their ability to perturb DNA replication leading to genomic instability (20). The observation that a major proportion (∼45%) of long AT-DRSs along the human genome reside within cytogenetically defined unstable chromosomal regions further highlighting that AT-dinucleotide repeats are a key factor predisposing chromosomal regions to fragility.
The breakage frequency of the newly generated FS was significantly lower than the frequency at the endogenous FRA16C, from which the AT-DRS was derived. This indicated that other factors are contributing to the FRA16C fragility. Interestingly, in FRA16C, in addition to the long AT-DRS studied here, there are three additional long (>400 bp) AT-DRSs, which were shown to stall the replication fork progression under replication stress conditions (20) and are probably contributing to the higher fragility in the endogenous site. Moreover, replication timing data (http://www.replicationdomain.com) reveals that the endogenous FRA16C region is replicating later in S-phase relative to the HPRT locus, thus may further increase the difficulty to complete the replication along this region before entry into mitosis. It will be interesting to further characterize the novel generated fragile site, by studying the effect of the AT-DRS on the ability of the region to complete the replication during S-phase. Recent finding showed that fragile regions fail to complete their replication during S-phase and the under-replicated sequences are subjected to repair synthesis during mitosis termed MiDAS (34).
The question of whether integration of CFS sequences are sufficient to recapitulate FS-like instability was previously addressed using whole BACs containing FRA3B sequences (39). Integration of two adjacent FRA3B BACs, 150 kb each, into a non-fragile ectopic site in human cells was able to confer FS-like instability, implying an inherent instability of these sequences. However, a specific sequence motif responsible for the observed fragility in that study could not be identified. In the present work, however, we showed that a long AT-DRS is able and sufficient to drive the formation of an FS in a non-fragile region in the studied cellular system.
A number of fragile sites are enriched with AT-DRSs that have the potential to form alternative non-B DNA secondary structures (10,21,29,40). AT-DRSs >200 bp in length are predicted to readily form secondary structures following unwinding of the DNA double helix during replication (10). Under aphidicolin treatment, which leads to uncoupling between the DNA polymerase and the helicase (41) longer stretches of ssDNA are exposed, which may enhance the formation of stable secondary structures of the AT-DRSs. Three in vitro studies tested the replication dynamics within plasmids containing different AT-DRSs derived from FRA16D (21,22) and an AT-DRS derived from FRA16B (40). In one study (21), an ∼500 bp sequence which spans a FRA16D region highly enriched with perfect AT repeats, was shown to stall replication fork progression in yeast in a manner depending on the repeat length. Mfold predictions and gel electrophoresis migration analysis of these fragments suggested that the observed replication perturbation involves secondary structure formation (21). This suggestion was recently reinforced by showing that this AT-DRS is targeted by the MUS81 structure-specific endonuclease which cleaves the replication forks stalled at DNA secondary structures along this region (42). In another study, cell-free assays showed alleviated polymerase stalling within FRA16D-derived AT-DRSs by the addition of the Werner helicase, implicated in processing of secondary structures arising during replication (22). In the third study, a FRA16B-derived AT-rich fragment, cloned into a SV40 replication plasmid, was able to fold into branched secondary structures, promoting polymerase stalling (40). The probability to fold into secondary structures was increased when the structure-prone strand served as the lagging strand template (40). Recently, using HR reporters it was shown that the Bloom helicase activity and the FANCM fork reversal activity are required for preventing double strand breaks (DSBs) formation along AT-DRSs derived from FRA3B, FRA16C and FRA16D (14,15). In an additional recent study, genome-wide analysis in human cells identified structure forming repeats, including quasi palindromic AT-rich repeats, as principal sites of fork collapse upon ATR inhibition (43). All these studies demonstrate that AT-DRSs can perturb the replication progression and generate DSBs. In the current study however, we have directly demonstrated that an AT-DRS that was shown to stall the replication fork progression (20) and form highly stable secondary structures (Figure 3B) has the ability to create recurrent chromosomal fragility in a non-fragile region. This provides for the first time a direct evidence for the role of the difficult to replicate AT-DRSs in the mechanism underlying fragile site formation.
DNA DSBs induced by hydroxyurea treatment were recently detected within poly(dA:dT) tracts in the CFSs FRA3B and FRA16D, suggesting that in addition to AT-DRSs, other sequences with potential to form non-B DNA sequences may also contribute to the fragility of CFSs under various stress conditions (44).
The landscape of CFS expression differs across cell types. This has been attributed to differences in the availability of replication origins and differences in replication timing programs among cell types (6). However, for example, FRA16D and FRA3B that are highly expressed in lymphocytes in which they exhibit a paucity of replication initiation events (45), are also expressed in fibroblasts (although to a lesser extent) in which replication initiation events are distributed along the fragility core (32,45,46). This intrinsic instability suggests that features as AT-DRSs which reside along FRA16D and FRA3B (18,47) predispose these regions to breakage, independently of the tissue specific replication program and together with other key features underlying the fragility along CFSs.
CFSs are preferentially unstable in precancerous lesions and during cancer development [reviewed in (9,48)]. A comprehensive study of the gene pairs involved in all recurrent chromosomal translocations in tumor cells found that over half of breakpoints are mapped to human fragile sites (49). Sequences within and flanking three randomly selected pairs of translocations prone genes showed enrichment of AT-DRSs, that were shown to form highly stable secondary structures (49). Another study analyzed ∼20 000 translocation breakpoints in cancer genomes and found significant association with potential non-B DNA forming sequences, including AT-DRSs (50). AT-DRSs were also found as hot spots for translocations causing genetic syndromes, as in the case of the translocation between chromosomes 11 and 22, t(11;22), which underlies the Emanuel syndrome or the supernumerary-der(22)t(11;22) syndrome. In most patients the translocation breakpoint is found at the center of the AT-DRS (51,52). All these studies highlight the role of AT-DRSs in genomic instability driving cancer and genetic diseases. In summary, the results presented in the current work highlight the deleterious effect of intrinsic DNA features such as the AT-DRSs in driving recurrent genome instability.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Christine Tran and Naomi Atkin for technical assistance, Dr Danny Kitsberg for helpful advice, Tamar Golan-Lev for manuscript graphics and all members of the Kerem lab for thoughtful discussions and advice.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Israel Science Foundation [176/11 to B.K.]; The Chief Scientist Office of the Israel Ministry of Health [3-00000-6014 to B.K.]; The Israel Science Foundation and the National Natural Science Foundation of China (NSFC-ISF) Joint Research Program [2535/16 to B.K.]; Israeli Centers for Research Excellence (I-CORE), Gene Regulation in Complex Human Disease [41/11 to B.K.]; NIH [RO1GM101192 to Y.H.W.]. Funding for open access charge: ISF.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bester A.C., Roniger M., Oren Y.S., Im M.M., Sarni D., Chaoat M., Bensimon A., Zamir G., Shewach D.S., Kerem B.. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011; 145:435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Di Micco R., Fumagalli M., Cicalese A., Piccinin S., Gasparini P., Schurra C., Garre M., Nuciforo P.G., Bensimon A. et al.. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006; 444:638–642. [DOI] [PubMed] [Google Scholar]
- 3. Bartkova J., Horejsi Z., Koed K., Kramer A., Tort F., Zieger K., Guldberg P., Sehested M., Nesland J.M., Lukas C. et al.. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005; 434:864–870. [DOI] [PubMed] [Google Scholar]
- 4. Gorgoulis V.G., Vassiliou L.-V.F., Karakaidos P., Zacharatos P., Kotsinas A., Liloglou T., Venere M., Ditullio R.A., Kastrinakis N.G., Levy B. et al.. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005; 434:907–913. [DOI] [PubMed] [Google Scholar]
- 5. Hosseini S.A., Horton S., Saldivar J.C., Miuma S., Stampfer M.R., Heerema N.A., Huebner K.. Common chromosome fragile sites in human and murine epithelial cells and FHIT/FRA3B loss-induced global genome instability. Genes. Chromosomes Cancer. 2013; 52:1017–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Debatisse M., Le Tallec B., Letessier A., Dutrillaux B., Brison O.. Common fragile sites: mechanisms of instability revisited. Trends Genet. 2012; 28:22–32. [DOI] [PubMed] [Google Scholar]
- 7. Miron K., Golan-Lev T., Dvir R., Ben-David E., Kerem B.. Oncogenes create a unique landscape of fragile sites. Nat. Commun. 2015; 6:7094. [DOI] [PubMed] [Google Scholar]
- 8. Ozeri-Galai E., Tur-Sinai M., Bester A.C., Kerem B.. Interplay between genetic and epigenetic factors governs common fragile site instability in cancer. Cell Mol. Life Sci. 2014; 71:4495–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sarni D., Kerem B.. The complex nature of fragile site plasticity and its importance in cancer. Curr. Opin. Cell Biol. 2016; 40:131–136. [DOI] [PubMed] [Google Scholar]
- 10. Zlotorynski E., Rahat A., Skaug J., Ben-Porat N., Ozeri E., Hershberg R., Levi A., Scherer S.W., Margalit H., Kerem B.. Molecular basis for expression of common and rare fragile sites. Mol. Cell Biol. 2003; 23:7143–7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ozeri-Galai E., Bester A.C., Kerem B.. The complex basis underlying common fragile site instability in cancer. Trends Genet. 2012; 28:295–302. [DOI] [PubMed] [Google Scholar]
- 12. Pirzio L.M., Pichierri P., Bignami M., Franchitto A.. Werner syndrome helicase activity is essential in maintaining fragile site stability. J. Cell Biol. 2008; 180:305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Madireddy A., Kosiyatrakul S.T., Boisvert R.A., Herrera-Moyano E., García-Rubio M.L., Gerhardt J., Vuono E.A., Owen N., Yan Z., Olson S. et al.. FANCD2 facilitates replication through common fragile sites. Mol. Cell. 2016; 64:388–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang H., Li S., Zhang H., Wang Y., Hao S., Wu X.. BLM prevents instability of structure-forming DNA sequences at common fragile sites. PLoS Genet. 2018; 14:e1007816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang H., Li S., Oaks J., Ren J., Li L., Wu X.. The concerted roles of FANCM and Rad52 in the protection of common fragile sites. Nat. Commun. 2018; 9:2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang H., Li Y., Truong L.N., Shi L.Z., Hwang P.Y.-H., He J., Do J., Cho M.J., Li H., Negrete A. et al.. CtIP maintains stability at common fragile sites and inverted repeats by end resection-independent endonuclease activity. Mol. Cell. 2014; 54:1012–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Palumbo E., Matricardi L., Tosoni E., Bensimon A., Russo A.. Replication dynamics at common fragile site FRA6E. Chromosoma. 2010; 119:575–587. [DOI] [PubMed] [Google Scholar]
- 18. Mishmar D., Rahat A., Scherer S.W., Nyakatura G., Hinzmann B., Kohwi Y., Mandel-Gutfroind Y., Lee J.R., Drescher B., Sas D.E. et al.. Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:8141–8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mishmar D., Mandel-Gutfreund Y., Margalit H., Kerem B.. Common fragile sites: G-Band characteristics within an R-band. Am. J. Hum. Genet. 1999; 64:908–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ozeri-Galai E., Lebofsky R., Rahat A., Bester A.C., Bensimon A., Kerem B.. Failure of origin activation in response to fork stalling leads to chromosomal instability at fragile sites. Mol. Cell. 2011; 43:122–131. [DOI] [PubMed] [Google Scholar]
- 21. Zhang H., Freudenreich C.H.. An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in S. cerevisiae. Mol. Cell. 2007; 27:367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shah S.N., Opresko P.L., Meng X., Lee M.Y.W.T., Eckert K.A.. DNA structure and the Werner protein modulate human DNA polymerase delta-dependent replication dynamics within the common fragile site FRA16D. Nucleic Acids Res. 2010; 38:1149–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yáñez R.J., Porter A.C.. Gene targeting is enhanced in human cells overexpressing hRAD51. Gene Ther. 1999; 6:1282–1290. [DOI] [PubMed] [Google Scholar]
- 24. Deng C., Capecchi M.R.. Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol. Cell Biol. 1992; 12:3365–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu S., Mangelsdorf M., Hewett D., Hobson L., Baker E., Eyre H.J., Lapsys N., Le Paslier D., Doggett N.A., Sutherland G.R. et al.. Human chromosomal fragile site FRA16B is an amplified AT-rich minisatellite repeat. Cell. 1997; 88:367–374. [DOI] [PubMed] [Google Scholar]
- 26. Conti C., Sacca B., Herrick J., Lalou C., Pommier Y., Bensimon A.. Replication fork velocities at adjacent replication origins are coordinately modified during DNA replication in human cells. Mol. Biol. Cell. 2007; 18:3059–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Michalet X., Ekong R., Fougerousse F., Rousseaux S., Schurra C., Hornigold N., van Slegtenhorst M., Wolfe J., Povey S., Beckmann J.S. et al.. Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science. 1997; 277:1518–1523. [DOI] [PubMed] [Google Scholar]
- 28. Sarai A., Mazur J., Nussinov R., Jernigan R.L.. Sequence dependence of DNA conformational flexibility. Biochemistry. 1989; 28:7842–7849. [DOI] [PubMed] [Google Scholar]
- 29. Dillon L.W., Pierce L.C.T., Ng M.C.Y., Wang Y.-H.. Role of DNA secondary structures in fragile site breakage along human chromosome 10. Hum. Mol. Genet. 2013; 22:1443–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Le Tallec B., Dutrillaux B., Lachages A.-M., Millot G.A., Brison O., Debatisse M.. Molecular profiling of common fragile sites in human fibroblasts. Nat. Struct. Mol. Biol. 2011; 18:1421–1423. [DOI] [PubMed] [Google Scholar]
- 32. Murano I., Kuwano A., kajii t.. Fibroblast-specific common fragile sitesinduced by aphidicolin. Hum. Genet. 1989; 83:45–48. [DOI] [PubMed] [Google Scholar]
- 33. Mrasek K., Schoder C., Teichmann A., Behr K., Franze B., Wilhelm K., Blaurock N., Claussen U., Liehr T., Weise A.. Global screening and extended nomenclature for 230 aphidicolin-inducible fragile sites, including 61 yet unreported ones. Int. J. Oncol. 2010; 36:929–940. [DOI] [PubMed] [Google Scholar]
- 34. Minocherhomji S., Ying S., Bjerregaard V.A., Bursomanno S., Aleliunaite A., Wu W., Mankouri H.W., Shen H., Liu Y., Hickson I.D.. Replication stress activates DNA repair synthesis in mitosis. Nature. 2015; 528:286–290. [DOI] [PubMed] [Google Scholar]
- 35. Helmrich A., Ballarino M., Tora L.. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell. 2011; 44:966–977. [DOI] [PubMed] [Google Scholar]
- 36. Cheng C.H., Kuchta R.D.. DNA polymerase epsilon: aphidicolin inhibition and the relationship between polymerase and exonuclease activity. Biochemistry. 1993; 32:8568–8574. [DOI] [PubMed] [Google Scholar]
- 37. Yunis J.J.J., Soreng A.L.. Constitutive fragile sites and cancer. Science. 1984; 226:1199–1204. [DOI] [PubMed] [Google Scholar]
- 38. Fasullo M., Endres L.. Nucleotide salvage deficiencies, DNA damage and neurodegeneration. Int. J. Mol. Sci. 2015; 16:9431–9449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ragland R.L., Glynn M.W., Arlt M.F., Glover T.W.. Stably transfected common fragile site sequences exhibit instability at ectopic sites. Genes Chromosom. Cancer. 2008; 47:860–872. [DOI] [PubMed] [Google Scholar]
- 40. Burrow A.A., Marullo A., Holder L.R., Wang Y.H.. Secondary structure formation and DNA instability at fragile site FRA16B. Nucleic Acids Res. 2010; 38:2865–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walter J., Newport J.. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol. Cell. 2000; 5:617–627. [DOI] [PubMed] [Google Scholar]
- 42. Kaushal S., Wollmuth C.E., Das K., Hile S.E., Regan S.B., Barnes R.P., Haouzi A., Lee S.M., House N.C.M., Guyumdzhyan M. et al.. Sequence and nuclease requirements for breakage and healing of a Structure-Forming (AT)n sequence within fragile site FRA16D. Cell Rep. 2019; 27:1151–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shastri N., Tsai Y.-C., Hile S., Jordan D., Powell B., Chen J., Maloney D., Dose M., Lo Y., Anastassiadis T. et al.. Genome-wide identification of Structure-Forming repeats as principal sites of fork collapse upon ATR inhibition. Mol. Cell. 2018; 72:222–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tubbs A., Sridharan S., van Wietmarschen N., Maman Y., Callen E., Stanlie A., Wu W., Wu X., Day A., Wong N. et al.. Dual roles of Poly(dA:dT) tracts in replication initiation and fork collapse. Cell. 2018; 174:1127–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Letessier A., Millot G.A., Koundrioukoff S., Lachages A.M., Vogt N., Hansen R.S., Malfoy B., Brison O., Debatisse M., Lachagès A.-M.. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011; 470:120–123. [DOI] [PubMed] [Google Scholar]
- 46. Le Tallec B., Millot G.A., Blin M.E., Brison O., Dutrillaux B., Debatisse M.. Common fragile site profiling in epithelial and erythroid cells reveals that most recurrent cancer deletions lie in fragile sites hosting large genes. Cell Rep. 2013; 4:420–428. [DOI] [PubMed] [Google Scholar]
- 47. Ried K., Finnis M., Hobson L., Mangelsdorf M., Dayan S., Nancarrow J.K., Woollatt E., Kremmidiotis G., Gardner A., Venter D. et al.. Common chromosomal fragile site FRA16D sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum. Mol. Genet. 2000; 9:1651–1663. [DOI] [PubMed] [Google Scholar]
- 48. Roukos V., Burman B., Misteli T.. The cellular etiology of chromosome translocations. Curr. Opin. Cell Biol. 2013; 25:357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Burrow A.A., Williams L.E., Pierce L.C.T., Wang Y.-H.. Over half of breakpoints in gene pairs involved in cancer-specific recurrent translocations are mapped to human chromosomal fragile sites. BMC Genomics. 2009; 10:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bacolla A., Tainer J.A., Vasquez K.M., Cooper D.N.. Translocation and deletion breakpoints in cancer genomes are associated with potential non-B DNA-forming sequences. Nucleic Acids Res. 2016; 44:5673–5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kato T., Kurahashi H., Emanuel B.S.. Chromosomal translocations and palindromic AT-rich repeats. Curr. Opin. Genet. Dev. 2012; 22:221–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kurahashi H., Emanuel B.S.. Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum. Mol. Genet. 2001; 10:2605–2617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.