Abstract
DdrO is an XRE family transcription repressor that, in coordination with the metalloprotease PprI, is critical in the DNA damage response of Deinococcus species. Here, we report the crystal structure of Deinococcus geothermalis DdrO. Biochemical and structural studies revealed the conserved recognizing α-helix and extended dimeric interaction of the DdrO protein, which are essential for promoter DNA binding. Two conserved oppositely charged residues in the HTH motif of XRE family proteins form salt bridge interactions that are essential for promoter DNA binding. Notably, the C-terminal domain is stabilized by hydrophobic interactions of leucine/isoleucine-rich helices, which is critical for DdrO dimerization. Our findings suggest that DdrO is a novel XRE family transcriptional regulator that forms a distinctive dimer. The structure also provides insight into the mechanism of DdrO-PprI-mediated DNA damage response in Deinococcus.
INTRODUCTION
Transcription factors participate in various cellular processes by controlling the timing and level of gene expression. These proteins are able to bind to a specific DNA sequence to promote (as an activator) or block (as a repressor) the transcription of relevant genes (1,2). For example, the activator TFIID from eukaryotes is one of several general transcription factors composed of TATA-binding protein (TBP) and its associated factors, TAFs (3–5). In contrast, LexA protein functions as a repressor of the expressions of recA and other stress-induced genes, which is the key element involved in the SOS response system in most characterized bacterial species (6). It is well understood that after DNA damage, RecA-ssDNA-ATP filaments activate the autocleavage of free LexA proteins, which in turn decrease the cellular pool of LexA proteins (6). Thus, LexA dissociates from the pre-inhibited promoter region, promoting the transcription of SOS genes for DNA repair (7). When DNA repair is complete, the coprotease activity of the RecA filaments is eliminated, and LexA re-accumulates and binds to the targeted promoters, thus repressing the overexpression of the SOS genes (6,7).
Bacteria belonging to the genus Deinococcus are usually highly resistant to environmental stress, including high doses of ionizing radiation, oxidation and long periods of desiccation. Their robustness is mainly contributed by an enhanced antioxidant system and enhanced DNA repair capability (8–11). The antioxidant system in Deinococcus is well documented and consists of catalase, peroxidase, superoxide dismutase, carotenoids and manganese ion antioxidant complex (12,13). However, Deinococcus does not appear to apply active error-prone DNA repair systems, such as translesion synthesis and nonhomologous end joining (13). It is well accepted that the error-free DNA repair pathway, especially homologous recombination, accounts for its efficient DNA repair.
Transcription factors in Deinococcus are important for its growth and robust adaptation to various stresses. Gfh and NusA are involved in the regulation of transcription initiation, pausing and termination in Deinococcus (14–16). DdrI, the cAMP receptor protein, is involved in cell division and mega-plasmid stability and is critical for the adaptation of Deinococcus to environmental stresses such as heat shock treatment (17). OxyR, which is a peroxide sensor, is activated after oxidative treatment (18). DrRRA is involved in the two-component signal transduction system, which contributes to the cellular resistance to environmental stress (19). Notably, two LexA-related proteins are encoded in D. radiodurans but are not involved in RecA induction, indicating the absence of the classic error-prone SOS response system (20).
Among all these transcription factors, DdrO, a xenobiotic-response element (XRE) family protein, appears to be one of the most important proteins involved in gene regulation in response to DNA damage (21). XRE proteins comprises a large family of proteins in bacteria that control gene expression involved in various metabolic functions (e.g. toxin-antitoxin system, nitrogen regulation system) (22–25). It was recently revealed that DdrO, together with PprI (also called IrrE), mediates a novel DNA damage response pathway (26–29). Both DdrO and PprI are highly conserved species-specific proteins that regulate the expression of DNA repair genes, such as recA and pprA (30–32). While DdrO is required for viability (28), cells lacking PprI shows a complete loss of radiation resistance (31). The crystal structure of PprI from Deinococcus deserti has been determined and revealed an N-terminal metalloprotease domain and a C-terminal GAF-like domain interconnected by an HTH motif (33). It was proposed that under normal growth conditions, the constitutively expressed DdrO functions as a repressor binding to the conserved palindromic motif (radiation/desiccation response motif, RDRM) at the promoter regions of a series of DNA repair genes including ddrO itself. Despite that the mechanism of PprI activation remains unclear, DdrO is cleaved by PprI protein after DNA damage, which in turn relieves the repression of the DNA repair genes. When DNA repair is completed, the protease activity of PprI is eliminated, enabling the repression of DdrO to resume (26–29). Moreover, substitution of the native DdrO with the uncleavable protein drastically sensitized the bacteria to DNA damage (27).
To help elucidate the molecular basis of the DdrO-PprI-mediated DNA response, we report the crystal structure of D. geothermalis DdrO (DG-DdrO). This structure, together with mutagenesis and biochemical studies, provides mechanistic insights into DNA binding and derepression by DdrO. Furthermore, comparative analyses of DdrO and LexA revealed a distinctive mechanism for the efficient DNA damage response in Deinococcus.
MATERIALS AND METHODS
Strains and culture
Deinococcus geothermalis strains (DSM 11300) were grown at 45°C in TGY broth (0.5% tryptone, 0.3% yeast extract, 0.1% glucose) or on TGY plates with 1.5% (w/v) agar powder. Escherichia coli strains, including trans5α and BL21 (DE3), were cultivated in LB broth (1% tryptone, 0.5% yeast extract, 1% NaCl) or on LB plates with 1.5% (w/v) agar at 37°C. The antibiotics used to select E. coli were kanamycin (40 μg/ml). All the strains and plasmids are listed in Supplementary Table S1.
Cloning and strain constructions
Entire DG-DdrO truncated after residue 11 (residues 12–140) or before α8 (residues 12–117) were amplified by PCR and cloned into the pET28a expression vector using NdeI and BamHI restriction enzyme sites. And DG-DdrO proteins used for crystallization and biochemical assays contained an N-terminal 6 × His-tag (MGSSHHHHHHSSGLVPRGSH). Amplification and site-directed mutagenesis were carried out using PrimeSTAR HS DNA polymerase (Takara). For the site directed mutagenesis, the amplified product was treated with DpnI and transformed into DH5α following the temperature cycling. All vectors were verified before transformed into the expression cell BL21 (DE3). All the primers and DNA substrates are listed in Supplementary Table S2.
Protein expression and purification
The expression strains of DG-DdrO were grown in LB broth containing kanamycin at 37°C to an optical density at 600 nm of 0.6–0.8. The protein was induced at 16°C for 16 hr by adding isopropyl-β-d-thioga-lactopyranoside (IPTG) at a final concentration of 0.2 mM. After harvesting and washing with PBS (phosphate saline), cell pellets were resuspended in lysis buffer (1 M NaCl, 20 mM Tris–HCl 7.5, 5% (w/v) glycerol, 1 mM PMSF and 20 μg/ml lysozyme) and lysed by sonication and then centrifuged at 14000 rpm for 30 min at 4°C. The cell debris was discarded, and the supernatant was purified by an AKTA Purifier system. The supernatant was loaded onto a HisTrap HP column after equilibration with buffer A (1 M NaCl, 20 mM Tris–HCl 7.5 and 5% (w/v) glycerol). After washing with 50 mM imidazole, the protein was finally eluted with 250 mM imidazole. Further purification was carried out using a Heparin HP column by gradient elution from 250 mM to 1 M NaCl. The protein was finally purified by gel filtration chromatography (Superdex 75 column with 250 mM NaCl, 20 mM Tris–HCl 7.5) and analyzed by Tricine-SDS-PAGE. Purified proteins for subsequent use were concentrated to ∼3 mg/ml and aliquoted in gel filtration buffer respectively, flash frozen and stored at -80°C. DR-DdrO and other DG-DdrO mutants and truncations were expressed and purified by similar methods to those used for wild-type of DG-DdrO. DR-PprI was induced and purified as reported previously (27). DG-PprI was induced and purified similar to the wild-type of DR-PprI.
Crystallization and structure determination
DG-DdrO (residues 12–140) containing an N-terminal 6 × His-tag (MGSSHHHHHHSSGLVPRGSH) was used for crystallization. Native crystals were grown by the drop vapor diffusion method at 289 K over wells containing 2.4 M LiCl and 0.1 M HEPES (pH 7.0) after freshly purified DdrO was concentrated to ∼15 mg/ml. Cryocooling was achieved by stepwise soaking with crystals in reservoir solution containing 10, 20 and 30% (w/v) glycerol for 3 min, followed by flash freezing in liquid nitrogen. The diffraction intensities were recorded on beamline BL17U at Shanghai Synchrotron Radiation Facility (Shanghai, China) and were integrated and scaled with the XDS suite. The structure was determined by molecular replacement using a published HipB structure (PDB ID: 4PU7) as the search model (25). Structures were refined using PHENIX (34) and interspersed with manual model building using COOT (35). Later stages of refinement utilized TLS group anisotropic B-factor refinement. DG-DdrO forms a dimer in the crystallographic asymmetric unit. The refined structure includes 256 aa of DG-DdrO (residues 12–139 from each protomer) and five lithium ions, which showed large positive peaks (Fo – Fc) after refinement. All the residues are in the most favorable (98.4%) and allowed regions (1.6%) of the Ram All the residues are in the most favorable and allowed regions of the Ramachandran plot. All structural figures were rendered in PyMOL (www.pymol.org).
Electrophoretic mobility shift assay (EMSA)
The final reaction mixture consisted of 150 mM NaCl, 20 mM Tris–HCl 8.0, 0.1 mg/ml Albumin from bovine serum (BSA), 100 nM 5′-FAM-labeled RDRM-contained DNA and proteins at 0, 1 and 2 μM. The mixtures were incubated at room temperature for 30 min. Samples were separated on 8% native polyacrylamide gels in 1× TB buffer and the gels were imaged in fluorescence mode (FAM) on Typhoon FLA 9500 (GE).
DdrO cleavage assay
DdrO cleavage assays were carried out in the conditions of 200 mM NaCl, 20 mM Tris–HCl 8.0, 1 mM DTT and 2 mM MnCl2 with a final concentration of 8 μM DG/DR-DdrO and 1 μM DG/DR-PprI at 37°C or 45°C for 30 min. The cleavage results were detected by Tricine-SDS-PAGE.
RESULTS
Protein characterization and crystallization
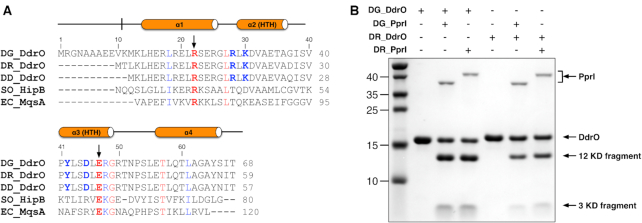
Despite its low sequence identity with other family members, DG-DdrO is an XRE family protein containing an additional C-terminal 70 amino acids outside the core DNA binding domain. Within Deinococcus species, DG-DdrO is highly conserved, sharing 93% and 94% amino acid identity with the D. radiodurans DdrO (DR-DdrO) and D. deserti DdrO (DD-DdrO) (Figure 1A and Supplementary Figure S1). Despite the additional 11 amino acids at the N-terminus of DG-DdrO, the cleavage site region (CSR) of DG-DdrO (residues 116–121, EL↓RGKR) is strictly conserved among different Deinococcus species (Supplementary Figure S1). To test whether PprI cleavage of DdrO is universal among Deinococcus species, either DG-DdrO or DR-DdrO was incubated with DG-PprI or DR-PprI in the presence of divalent cations. Consistent with the cleavage reaction observed previously (26,27,29), both two DdrO proteins were cleaved, which resulted in two product fragments (Figure 1B). These results indicate that the DdrO-PprI system is likely shared among Deinococcus species.
Figure 1.
Protein characterization and sequence alignment. (A) Structural based sequence alignment of DNA binding domains of XRE family proteins. DdrO proteins from D. geothermalis, D. radiodurans and D. deserti are denoted by DG-DdrO, DR-DdrO and DD-DdrO, respectively. HipB from S. oneidensis and MqsA from E. coli are denoted by SO-HipB and EC-MqsA, respectively. Residues conserved in XRE family proteins and DdrO proteins are highlighted in red and blue, respectively. The black arrowheads indicate the signature RE pair of XRE family proteins. (B) SDS-PAGE gel showing the DdrO cleavage by PprI. For the reaction, 8 μM of DG-DdrO or DR-DdrO was incubated with 1 μM of DG-PprI or DR-PprI in the presence of Mn2+ at 37°C for 30 min. DdrO, PprI and two product fragments are indicated by black arrowheads.
Overall structure and the signature RE pair
Based on the sequence alignment, we crystalized the entire DG-DdrO except for a truncation up to residue 11 (residues 12–140). The crystals were grown in the presence of lithium, and the crystal structure was determined at 2.3 Å using the molecular replacement method using a published HipB structure (PDB ID: 4PU7) as the search model (25). DG-DdrO crystallizes in space group C2221 with a twofold symmetric dimer molecule in the crystallographic asymmetric unit (Figure 2a). After refinement, these two protomers are superimposable on each other with a root mean square deviation (rmsd) of 0.533 Å over 122 pairs of Cα atoms. The crystal data, together with the data collection and refinement statistics, are summarized in Table 1.
Figure 2.
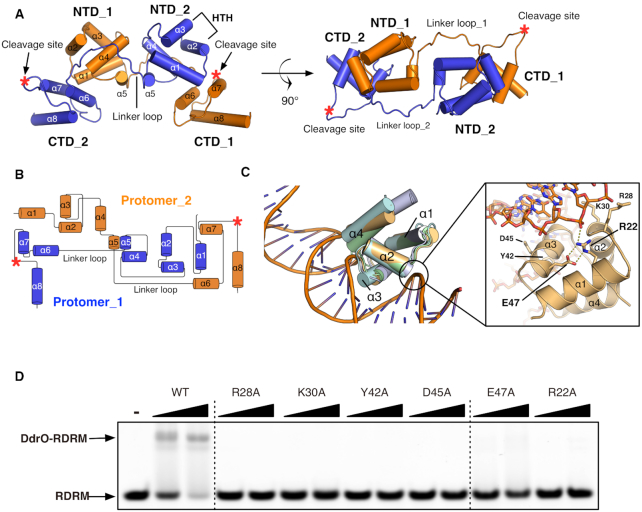
Overall structure of and the signature RE pair of DG-DdrO. (A) DdrO dimer protein is shown as cartoon and two protomers are colored in orange and blue respectively. The α-helices, HTH motifs, N-terminal (NTD), C-terminal (CTD) and the linker region of each protomer are labeled. The cleavage site is located at α7-α8 loop and indicated by red star. (B) Topology diagram of the protein fold of DG-DdrO dimer. (C) Superposition of the HTH-containing N-terminal domains of XRE family proteins. DG-DdrO, SO-HipB (PDB ID: 4Z5C), EC-MqsA-DNA (PDB ID: 3O9X) and GraA form Pseudomonas putida (PDB ID: 6FIX) are colored wheat, green, cyan and light blue, respectively. The signature RE pair and four residues (Arg28, Lys30, Tyr42 and Asp45) possibly involved in the promoter binding are shown as stick and labeled. The salt bridge interactions between arginine and glutamic acid and the interaction between Arg22 and DNA phosphate backbone are indicated by the yellow dashed lines. (D) EMSA showing abolished promoter DNA binding of DG-DdrO mutant proteins. 5′-FAM-labeled DNA containing RDRM sequence (100 nM) was incubated with 1 or 2 μM of DdrO mutant protein.
Table 1.
Data collection, phasing and refinement statistics
| DG-DdrO | |
|---|---|
| Data collection | |
| Space group | C2221 |
| Cell dimensions | |
| a, b, c (Å) | 35.95 |
| 127.90 | |
| 156.41 | |
| Wavelength (Å) | 0.979 |
| Resolution (Å) | 30.0–2.30 |
| R sym (%) | 4.9 (69.2) |
| I/σI | 16.8 (2.0) |
| Completeness (%) | 98.7 (96.9) |
| Redundancy | 4.6 (4.4) |
| Refinement | |
| Resolution (Å) | 30.0–2.30 |
| No. reflections | 16282 |
| R work/ R free | 23.7/26.0 |
| No. atoms | |
| Protein | 2068 |
| Ion | 5 |
| Water | 8 |
| B-factors | |
| Protein | 46.8 |
| Ion | 51.1 |
| Water | 49.8 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 0.720 |
*Highest resolution shell is shown in parenthesis.
DG-DdrO, which is composed of eight α-helices, contains two domains: an HTH-containing N-terminal domain (residues 12–74) and a C-terminal domain (residues 89–140). These two domains are interconnected by a 15 amino acid linker loop (Figure 2A and B). As a member of XRE family protein, the HTH-containing N-terminal domain of DG-DdrO consists of five α-helices and could be aligned well with the DNA binding domain of XRE family proteins, including HipB (PDB ID: 4Z5C, rmsd of 0.689 Å over 33 pairs of Cα atoms) and MqsA (PDB ID: 3O9X, rmsd of 1.413 Å over 40 pairs of Cα atoms) from Escherichia coli (Figures 1A and 2C). Indeed, the first three α-helices of these proteins containing the helix-turn-helix motif (α2-α3) are well aligned (Figure 2C), suggesting the possible interactions between DG-DdrO and the promoter DNA. We performed a search with the DALI server (36) using the C-terminal domain of DG-DdrO as the query. The C-terminal domain of DG-DdrO, consisting of three α-helices, did not show any significant sequence similarity to any proteins with solved structures. Although the search gave several similar structure hits, none of them could be manually aligned with the C-terminal α-helices of DG-DdrO, suggesting a novel fold of the C-terminal domain. The CSR of DG-DdrO, with weak electron density, is located in the loop region between the last two α-helices (α7–α8 loop) (Figure 2A and B). Analyses of the distribution of the electrostatic surface potential showed that this region, in addition to the HTH motif, exhibits a positively charged surface (Supplementary Figure S2).
To further investigate the functional implications of our structure, we mutated conserved solvent-exposed amino acid residues in the HTH motif. We postulated that mutation of these residues would affect the promoter DNA binding of DG-DdrO. Based on the superposition between DdrO and MqsA-DNA complex (PDB ID: 3O9X), four residues, Arg28, Lys30, Tyr42 and Asp45, were chosen and mutated to alanine (22), and electrophoretic gel mobility shift assays (EMSA) with RDRM-containing DNA were performed (Figure 2D). Compared with that of the wild-type protein, the DNA binding of these mutant proteins was almost eliminated, indicating that these residues are required for the specific binding of promoter DNA. Moreover, the DALI server analysis identified a delicate motif (RE pair) shared by most XRE family proteins, located at the α1 and α3 helices (Figure 1A and Supplementary Figure S3a). This pair consists of two oppositely charged residues, arginine (Arg22) and glutamic acid (Glu47), which form a salt bridge interaction in DG-DdrO (Figure 2D). The arginine is solvent-exposed and interacts directly with the phosphate backbone of promoter DNA in other solved XRE-DNA complexes (Figure 2D). Alanine mutations of these two residues drastically impaired the DNA binding of DG-DdrO (Figure 2D), suggesting that the RE pair is critical for the promoter DNA binding of XRE family proteins. Interestingly, while R22A DG-DdrO retained dimer formation in solution, the mutation of Glu47 to alanine shifted the peak toward the monomer, indicating that Glu47 is also critical for the dimer formation of DG-DdrO (Supplementary Figure S3b).
Distinct dimerization of DG-DdrO
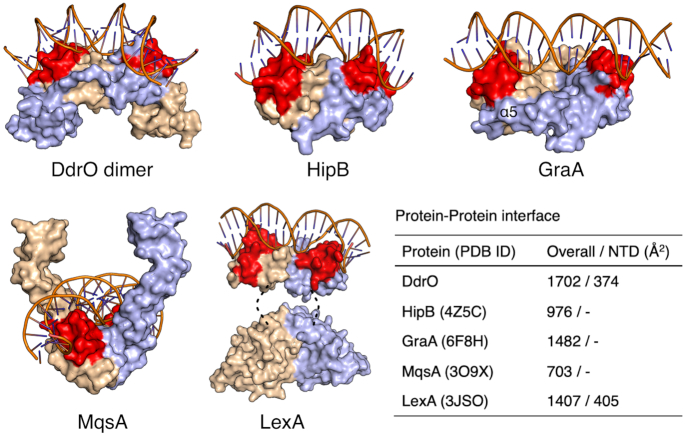
DG-DdrO contains an additional C-terminal domain not present in other XRE family proteins (Figure 2A and B). In contrast to previous solved structures of XRE family proteins, whose dimerization is mediated by the HTH motif, the DG-DdrO dimer involves three protein-protein interfaces: (i) interactions between the α5 helices from two protomers; (ii) interactions between the α4 helix of one protomer and the linker region of the other protomer; (iii) interactions between the HTH motif (α1 and α4 helices) of one protomer and the C-terminal domain (α6 and α7 helices) of the other protomer. Thus, our structure suggests that DG-DdrO forms a tight dimer with an extended interface between the two protomers (Figures 2A and 3). Indeed, predicted by the PISA server (37), 1702 Å2 of total surface area is buried at the dimeric interface of DG-DdrO, which is more extensive than that of other XRE family proteins such as HipB, GraA and MqsA (Figure 3 and Supplementary Figure S4) (22,24,38). It is worth noting that the interactions between the HTH motifs of the two protomers contribute only 22% (374 Å2) of the total dimeric interface, suggesting that the additional 70 C-terminal residues are important for DG-DdrO dimerization. As mentioned above, DG-DdrO and LexA are both cleaved to activate gene expression under environmental stresses. We also compared the dimer formation between these two proteins. Similarly, the LexA protein consists of both the N-terminal domain (HTH motif) and the C-terminal domain, which are interconnected by a short loop (Figure 3). In the presence of DNA, the dimerization of LexA is mediated by both the flexible N-terminal wing regions (405 Å2) and the C-terminal domains (1002 Å2) of two protomers (Figure 3) (39). It has been reported that these two wing regions interact with the minor groove of DNA, thereby determining the binding affinities for promoter DNA (39). However, this wing region is absent in the HTH motif of DG-DdrO, which makes the dimerization of the two HTH motifs more rigid. Indeed, the RDRM motif is invariable, and modifying any one of the consensus sequences or shortening the spacer length severely impaired the promoter binding of DG-DdrO (Supplementary Figure S5).
Figure 3.
Structural comparison of the dimerization between DG-DdrO and XRE family proteins. DG-DdrO, HipB (PDB ID: 4Z5C), MqsA (PDB ID: 3O9X), GraA (PDB ID: 6FIX) and LexA (PDB ID: 3JSO) are shown as surface and two protomers are colored in wheat and light blue, respectively. The HTH motifs are highlighted in red. For DdrO protein, DNA from MqsA-DNA complex was docked onto the DG-DdrO protein by superposition between HTH motifs of DG-DdrO and MqsA. The position of α5 in GraA is labeled. The disordered link regions in LexA are indicated by dashed lines. The dimeric interface between two protomers were calculated by PDBePISA (37) and listed in the table.
Derepression mechanism of DG-DdrO
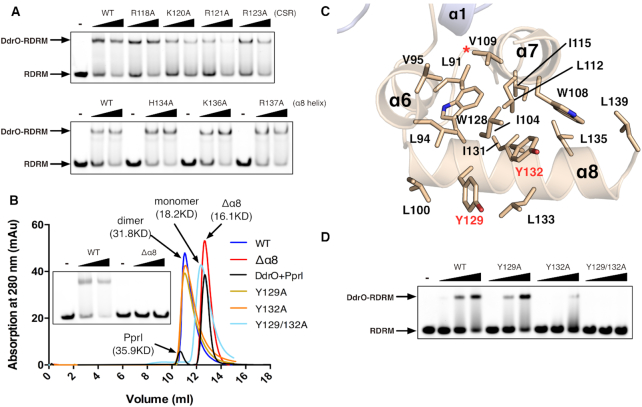
During the DNA damage response, the scissile peptide bond between Leu117 and Arg118 of DG-DdrO is destroyed, resulting in two pieces of fragments including the very C-terminal helix (α8 helix) with molecular weight of ∼3 kD (Figure 1B). To further investigate the derepression mechanism, we tested whether this CSR or the α8 helix of the DG-DdrO is involved in the direct DNA binding. Seven solvent accessible residues, including four positively charged residues from the CSR (Arg118, Lys120, Arg121 and Arg123) and three residues from the α8 helix (His134, Lys136 and Arg137), were mutated to alanine, and the mutants were incubated with RDRM-containing promoter DNA. The EMSA results revealed that none of these substitutions weakened the DG-DdrO binding to the promoter DNA, indicating that the cleavage may not directly affect the protein-DNA interactions (Figure 4A). However, DG-DdrO lacking the α8 helix was unable to bind the promoter DNA and eluted as a monomer during size exclusion chromatography (Figure 4B).
Figure 4.
The derepression mechanism of DG-DdrO. (A) EMSA showing the unaffected promoter DNA binding of DG-DdrO mutant proteins. 5′-FAM-labeled DNA containing RDRM sequence (100 nM) was incubated with 1 or 2 μM of DdrO mutant protein. (B) Size exclusion chromatography of wild type, mutant (Y129A, Y132A and Y129/132A), truncated (Δα8) DG-DdrO proteins and cleaved DG-DdrO protein (DdrO+PprI, cleavage reaction at 45°C for 30 min) on Superdex 75 10/300 GL column. The peaks correspond to monomeric or dimeric DG-DdrO proteins and their calculated molecular weights are labeled and colored differently. (C) A cut away view shows that the C-terminal domain of DG-DdrO forms a stable hydrophobic core. Conserved hydrophobic residues are shown as stick and labeled. The red star indicates the cleavage site. (D) EMSA assays of mutant DG-DdrO proteins using the same reaction conditions as in panel A.
Since the α8 helix is not located at the dimeric interface between two protomers, how does its cleavage disturb the dimerization of DG-DdrO? Sequence alignments revealed the enrichment of hydrophobic residues, predominantly leucine and isoleucine residues, in the C-terminal α-helices of DG-DdrO (Supplementary Figure S1). Notably, these hydrophobic residues are concentrated in the α8 helix and interact with the α6 and α7 helices, which form a stable hydrophobic core of the C-terminal domain of DG-DdrO (Figure 4C). While the single mutation of two conserved tyrosine residues, Tyr129 and Tyr132, retained the DG-DdrO dimer, the double mutant was folded and existed as a monomer in the solution (Figure 4B and Supplementary Figure S6). This finding was consistent with the EMSA results showing the loss of the promoter DNA binding of the double-mutant protein (Figure 4D). These results indicate that PprI cleavage of DG-DdrO destabilizes the hydrophobic core of the C-terminal domain, further eliminating the dimer formation of DG-DdrO.
DISCUSSION
XRE family proteins, widely distributed in bacteria, archaea and eukaryotes, play a broad role in the regulation of cellular metabolism. Members of this family of proteins bind to the promoter region and usually serve as repressors under normal conditions. Despite the low sequence similarities and diverse protein length, the XRE family proteins share an HTH motif and usually exist as dimers. Analyses of the DG-DdrO structure provide insight into the conserved N-terminal HTH motif and the distinctive C-terminal domain (Figure 2A). Similar to the reported XRE protein NHTF (23) and GraA (24,40), DG-DdrO consists entirely of α-helices, and the core structure of the HTH motif (α1–α4) can be well aligned (Figure 2C), suggesting a similar DNA binding mode. However, the key residues involved in the protein-DNA interface are not conserved, which may explain the varied sequence specificity. Notably, the N-terminal and C-terminal domains of DG-DdrO are connected by a long linker loop. To our knowledge, such a protein architecture has not been observed in any previously reported XRE family proteins. A pair of oppositely charged residues (arginine/glutamic acid) presenting in most XRE family proteins were identified and found to form a salt bridge interaction at the predicted protein–DNA interface (Figure 2C). Alanine substitutions of any of these two residues abolished the promoter DNA binding of DG-DdrO (Figure 2D), suggesting the critical role of these two residues. In addition, the E47A mutant protein eluted as a monomer during size exclusion chromatography (Supplementary Figure S3B), indicating that the RE pair might also be important for the dimerization of XRE family proteins.
Residues in the HTH motif outside the core structure contribute greatly to the dimerization interface of XRE family proteins. For example, a long α-helix (α5) next to the α4 helix is present at the dimerization interface of GraA and forms strong helix-helix interactions (Figure 3) (24). However, the DG-DdrO dimer structure revealed a unique upside-down V-shaped dimeric conformation (Figure 2A). Despite the helix-helix interactions between two short α5 helices, the HTH-containing N-terminal domains of two DG-DdrO protomers form limited interactions, less than in other XRE family proteins such as HipB and MqsA (Figure 3). Destabilization of the C-terminal domain disrupted the protein dimer (Figure 4B and D), indicating that the protein-protein interface between the HTH motif of one protomer and the C-terminal domain of the other protomer is important for DG-DdrO dimerization. Notably, the long linker loop of one protomer also extensively interacts with the N-terminal domain of the other protomer. Taken together, our structure revealed the extended dimerization interactions of DG-DdrO, which relied on both the conserved HTH motif and the unconserved C-terminus of the protein.
As mentioned above, the LexA/RecA-mediated SOS response appears to be inactive in Deinococcus, and the DdrO/PprI system was recently identified as an alternative protease-based DNA damage response pathway. Comparisons of the structures of DG-DdrO and LexA yields interesting similarities and differences (Figure 3). Both DG-DdrO and LexA consist of an HTH motif and a C-terminal domain, which is required for protein dimerization. Despite the disordered short linker region, LexA contains a wing region, and the N-terminal domain is highly flexible (39). And the reorientation of the N-terminal DNA binding domains with respect to the C-terminal domain is required for stable operator binding of LexA. However, this wing region is absent in DG-DdrO, and the linker loop is restrained at the dimerization interface. Thus, in contrast to LexA, the protein-DNA interface of DG-DdrO appears to be pre-formed, which is in line with the selective and efficient promoter DNA binding of DG-DdrO (Figure 3). The cleavage site of DG-DdrO is lying in a loop at the C-terminal domain. Similar to LexA autocleavage, cleavage of this loop by PprI protein causes destabilization of the dimerization interface of DG-DdrO, further dissociating the protein dimer. While the CSR in LexA has a strand-loop-strand topology, the CSR of Deinococcus include a solvent-exposed helix-loop-helix motif, which lies outside the C-terminal hydrophobic core (Figure 2A).
Collectively, we reported the crystal structure of DG-DdrO, which is the key component involved in DNA damage response in Deinococcus species. The biochemical and structural studies revealed that DG-DdrO is a distinct XRE family protein dimer with conserved HTH motif and the RE pair. The novel C-terminal domain forms a compact hydrophobic core, which plays a critical role in DG-DdrO dimerization. Additionally, cleavage of DG-DdrO destabilizes the dimerization interface, which discloses the mechanism for the derepression of DdrO proteins.
DATA AVAILABILITY
The coordinates and structure factors have been deposited to Protein Data Bank with accession codes 6JQ1.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the staff at the Shanghai Synchrotron Radiation Facility (SSRF in China) for assistance in data collection.
Author contributions: Y.H. and Y.Z. conceived the project. H.L. designed the experiments and carried out the construction, purification, crystallization, acquisition of data and biochemical experiments. L.W. and S.L. carried out mutagenesis, purification and acquisition of data, C.P., K.C. and Y.L. carried out mutagenesis and purification. H.X. and B.T. performed EMSA assays and acquisition of data. Y.Z. determined and analyzed the structure. Y.H., Y.Z. and H.L. wrote the manuscript. All authors took part in data analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Basic Research Program of China [2015CB910600]; National Key Research and Development Program of China [2017YFA0503900]; National Natural Science Foundation of China [31670065, 31870051, 31500656, 31670819]; Zhejiang Provincial Natural Science Foundation for Outstanding Young Scientists [LR16C050002]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Jin J., He K., Tang X., Li Z., Lv L., Zhao Y., Luo J., Gao G.. An arabidopsis transcriptional regulatory map reveals distinct functional and evolutionary features of novel transcription factors. Mol. Biol. Evol. 2015; 32:1767–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., Chen X., Taipale J., Hughes T.R., Weirauch M.T.. The human transcription factors. Cell. 2018; 172:650–665. [DOI] [PubMed] [Google Scholar]
- 3. Kornberg R.D. The molecular basis of eukaryotic transcription. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:12955–12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Akhtar W., Veenstra G.J.. TBP-related factors: a paradigm of diversity in transcription initiation. Cell Biosci. 2011; 1:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Teves S.S., An L., Bhargava-Shah A., Xie L., Darzacq X., Tjian R.. A stable mode of bookmarking by TBP recruits RNA polymerase II to mitotic chromosomes. eLife. 2018; 7:e35621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Butala M., Klose D., Hodnik V., Rems A., Podlesek Z., Klare J.P., Anderluh G., Busby S.J., Steinhoff H.J., Zgur-Bertok D.. Interconversion between bound and free conformations of LexA orchestrates the bacterial SOS response. Nucleic Acids Res. 2011; 39:6546–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Butala M., Zgur-Bertok D., Busby S.J.. The bacterial LexA transcriptional repressor. Cell Mol. Life Sci.: CMLS. 2009; 66:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blasius M., Sommer S., Hubscher U.. Deinococcus radiodurans: what belongs to the survival kit. Crit. Rev. Biochem. Mol. Biol. 2008; 43:221–238. [DOI] [PubMed] [Google Scholar]
- 9. Cox M.M., Battista J.R.. Deinococcus radiodurans - the consummate survivor. Nat. Rev. Microbiol. 2005; 3:882–892. [DOI] [PubMed] [Google Scholar]
- 10. Makarova K.S., Omelchenko M.V., Gaidamakova E.K., Matrosova V.Y., Vasilenko A., Zhai M., Lapidus A., Copeland A., Kim E., Land M. et al.. Deinococcus geothermalis: the pool of extreme radiation resistance genes shrinks. PLoS One. 2007; 2:e955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lim S., Jung J.H., Blanchard L., de Groot A.. Conservation and diversity of radiation and oxidative stress resistance mechanisms in Deinococcus species. FEMS Microbiol. Rev. 2019; 43:19–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sharma A., Gaidamakova E.K., Grichenko O., Matrosova V.Y., Hoeke V., Klimenkova P., Conze I.H., Volpe R.P., Tkavc R., Gostincar C. et al.. Across the tree of life, radiation resistance is governed by antioxidant Mn(2+), gauged by paramagnetic resonance. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E9253–E9260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Slade D., Radman M.. Oxidative stress resistance in Deinococcus radiodurans. Microbiol. Mol. Biol. Rev: MMBR. 2011; 75:133–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Esyunina D., Agapov A., Kulbachinskiy A.. Regulation of transcriptional pausing through the secondary channel of RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:8699–8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Agapov A., Esyunina D., Pupov D., Kulbachinskiy A.. Regulation of transcription initiation by Gfh factors from Deinococcus radiodurans. Biochem. J. 2016; 473:4493–4505. [DOI] [PubMed] [Google Scholar]
- 16. Agapov A., Olina A., Esyunina D., Kulbachinskiy A.. Gfh factors and NusA cooperate to stimulate transcriptional pausing and termination. FEBS Lett. 2017; 591:946–953. [DOI] [PubMed] [Google Scholar]
- 17. Meyer L., Coste G., Sommer S., Oberto J., Confalonieri F., Servant P., Pasternak C.. DdrI, a cAMP receptor protein family member, acts as a major regulator for adaptation of Deinococcus radiodurans to various stresses. J. Bacteriol. 2018; 200:e00129-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen H., Xu G., Zhao Y., Tian B., Lu H., Yu X., Xu Z., Ying N., Hu S., Hua Y.. A novel OxyR sensor and regulator of hydrogen peroxide stress with one cysteine residue in Deinococcus radiodurans. PLoS One. 2008; 3:e1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang L., Xu G., Chen H., Zhao Y., Xu N., Tian B., Hua Y.. DrRRA: a novel response regulator essential for the extreme radioresistance of Deinococcus radiodurans. Mol. Microbiol. 2008; 67:1211–1222. [DOI] [PubMed] [Google Scholar]
- 20. Jolivet E., Lecointe F., Coste G., Satoh K., Narumi I., Bailone A., Sommer S.. Limited concentration of RecA delays DNA double-strand break repair in Deinococcus radiodurans R1. Mol. Microbiol. 2006; 59:338–349. [DOI] [PubMed] [Google Scholar]
- 21. Liu Y., Zhou J., Omelchenko M.V., Beliaev A.S., Venkateswaran A., Stair J., Wu L., Thompson D.K., Xu D., Rogozin I.B. et al.. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:4191–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brown B.L., Wood T.K., Peti W., Page R.. Structure of the Escherichia coli antitoxin MqsA (YgiT/b3021) bound to its gene promoter reveals extensive domain rearrangements and the specificity of transcriptional regulation. J. Biol. Chem. 2011; 286:2285–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang H.C., Ko T.P., Wu M.L., Ku S.C., Wu H.J., Wang A.H.. Neisseria conserved protein DMP19 is a DNA mimic protein that prevents DNA binding to a hypothetical nitrogen-response transcription factor. Nucleic Acids Res. 2012; 40:5718–5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Talavera A., Tamman H., Ainelo A., Konijnenberg A., Hadzi S., Sobott F., Garcia-Pino A., Horak R., Loris R.. A dual role in regulation and toxicity for the disordered N-terminus of the toxin GraT. Nat. Commun. 2019; 10:972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wen Y., Behiels E., Felix J., Elegheert J., Vergauwen B., Devreese B., Savvides S.N.. The bacterial antitoxin HipB establishes a ternary complex with operator DNA and phosphorylated toxin HipA to regulate bacterial persistence. Nucleic Acids Res. 2014; 42:10134–10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ludanyi M., Blanchard L., Dulermo R., Brandelet G., Bellanger L., Pignol D., Lemaire D., de Groot A.. Radiation response in Deinococcus deserti: IrrE is a metalloprotease that cleaves repressor protein DdrO. Mol. Microbiol. 2014; 94:434–449. [DOI] [PubMed] [Google Scholar]
- 27. Wang Y., Xu Q., Lu H., Lin L., Wang L., Xu H., Cui X., Zhang H., Li T., Hua Y.. Protease activity of PprI facilitates DNA damage response: Mn2+-dependence and substrate sequence-specificity of the proteolytic reaction. PLoS One. 2015; 10:e0122071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Devigne A., Ithurbide S., Bouthier de la Tour C., Passot F., Mathieu M., Sommer S., Servant P.. DdrO is an essential protein that regulates the radiation desiccation response and the apoptotic-like cell death in the radioresistant Deinococcus radiodurans bacterium. Mol. Microbiol. 2015; 96:1069–1084. [DOI] [PubMed] [Google Scholar]
- 29. Blanchard L., Guerin P., Roche D., Cruveiller S., Pignol D., Vallenet D., Armengaud J., de Groot A.. Conservation and diversity of the IrrE/DdrO-controlled radiation response in radiation-resistant Deinococcus bacteria. MicrobiologyOpen. 2017; 6:e477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Earl A.M., Mohundro M.M., Mian I.S., Battista J.R.. The IrrE protein of Deinococcus radiodurans R1 is a novel regulator of recA expression. J. Bacteriol. 2002; 184:6216–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hua Y., Narumi I., Gao G., Tian B., Satoh K., Kitayama S., Shen B.. PprI: a general switch responsible for extreme radioresistance of Deinococcus radiodurans. Biochem. Biophys. Res. Commun. 2003; 306:354–360. [DOI] [PubMed] [Google Scholar]
- 32. Narumi I., Satoh K., Cui S., Funayama T., Kitayama S., Watanabe H.. PprA: a novel protein from Deinococcus radiodurans that stimulates DNA ligation. Mol. Microbiol. 2004; 54:278–285. [DOI] [PubMed] [Google Scholar]
- 33. Vujicic-Zagar A., Dulermo R., Le Gorrec M., Vannier F., Servant P., Sommer S., de Groot A., Serre L.. Crystal structure of the IrrE protein, a central regulator of DNA damage repair in Deinococcaceae. J. Mol. Biol. 2009; 386:704–716. [DOI] [PubMed] [Google Scholar]
- 34. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D, Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D, Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Holm L., Laakso L.M.. Dali server update. Nucleic Acids Res. 2016; 44:W351–W355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 38. Schumacher M.A., Balani P., Min J., Chinnam N.B., Hansen S., Vulic M., Lewis K., Brennan R.G.. HipBA-promoter structures reveal the basis of heritable multidrug tolerance. Nature. 2015; 524:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang A.P., Pigli Y.Z., Rice P.A.. Structure of the LexA-DNA complex and implications for SOS box measurement. Nature. 2010; 466:883–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Talavera A., Tamman H., Ainelo A., Hadaei S., Garcia-Pino A., Horak R., Konijnenberg A., Loris R.. Production, biophysical characterization and crystallization of Pseudomonas putida GraA and its complexes with GraT and the graTA operator. Acta Crystallogr. F. Struct. Biol. Commun. 2017; 73:455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structure factors have been deposited to Protein Data Bank with accession codes 6JQ1.