Abstract
The RecQ family of helicases is highly conserved both structurally and functionally from bacteria to humans. Defects in human RecQ helicases are associated with genetic diseases that are characterized by cancer predisposition and/or premature aging. RecQ proteins exhibit 3′-5′ helicase activity and play critical roles in genome maintenance. Recent advances in single-molecule techniques have revealed the reiterative unwinding behavior of RecQ helicases. However, the molecular mechanisms involved in this process remain unclear, with contradicting reports. Here, we characterized the unwinding dynamics of the Caenorhabditis elegans RecQ helicase HIM-6 using single-molecule fluorescence resonance energy transfer measurements. We found that HIM-6 exhibits reiterative DNA unwinding and the length of DNA unwound by the helicase is sharply defined at 25–31 bp. Experiments using various DNA substrates revealed that HIM-6 utilizes the mode of ‘sliding back’ on the translocated strand, without strand-switching for rewinding. Furthermore, we found that Caenorhabditis elegans replication protein A, a single-stranded DNA binding protein, suppresses the reiterative behavior of HIM-6 and induces unidirectional, processive unwinding, possibly through a direct interaction between the proteins. Our findings shed new light on the mechanism of DNA unwinding by RecQ family helicases and their co-operation with RPA in processing DNA.
INTRODUCTION
Helicases are molecular motors that couple the hydrolysis of triphosphates (typically ATP) to the unwinding of poly-nucleic acid structures and have multifaceted roles in virtually all aspects of nucleic acid metabolism, including DNA replication, repair, recombination, transcription, chromosome segregation and telomere maintenance (1). The significance of helicases to cellular homeostasis is highlighted by the fact that defects in human RecQ helicases are linked to distinct heritable diseases. RecQ helicases belong to the SF2 family of helicases and are conserved from bacteria to humans. RecQ is a DNA structure-specific helicase that unwinds 3′-tailed duplexes, bubble structures, DNA displacement loops (D-loops), Holliday junctions and G-quadruplexes, thereby maintaining genomic stability (2,3). In humans, there are five RecQ helicases and mutations in three of the corresponding genes can lead to severe genetic disorders, namely, Werner syndrome (mutation of the WRN gene), Bloom syndrome (mutation of the BLM gene) and Rothmund-Thomson syndrome (mutation of the RECQL4 gene), which are associated with predisposition to cancer and/or premature aging.
A number of studies show that BLM functions in homologous recombination, stabilization and repair of stalled forks, and sister chromatid segregation, which are associated with its unwinding activity and protein interactions (4). Recently, single-molecule studies showed that BLM unwinds forked DNAs, DNA hairpins, and G-quadruplexes in a highly repetitive fashion by alternating between unwinding and rewinding modes, a process referred to as ‘reiterative unwinding’ (5–7). Other RecQ helicases also show such repetitive dynamics (8), and reiterative unwinding behavior seems to be a general feature of RecQ helicases. This behavior is thought to be important for in vivo DNA metabolism, for instance, processing stalled replication forks, eliminating potentially deleterious recombination intermediates, or stripping off other DNA binding proteins, as observed for single-stranded DNA (ssDNA) translocases.
Several basic aspects of reiterative unwinding remain elusive. It is unclear how many base pairs (bp) are unwound before RecQ helicase changes direction, and which strand the helicase moves along during rewinding. In addition, the mechanism by which RecQ makes a return is unknown, and it is unclear how the reiterative dynamics can be modulated by partner proteins. Several single-molecule studies suggest that the number of base pairs that are unwound before rewinding varies depending on the specific DNA substrate and assay used. Single-molecule fluorescence resonance energy transfer (smFRET) measurements of forked DNA substrates estimated that human BLM unwinds fewer than 34 bp before rewinding (5). By contrast, measurements on DNA hairpins using magnetic tweezers suggested that human BLM unwinds ∼15 bp before rewinding, a value that was estimated from the unwinding time (6). Human WRN and its Arabidopsis thaliana homolog RECQ2 (AtRECQ2) also reiteratively unwind DNA hairpins (9,10), but the unwinding lengths have not been determined for these proteins. A few different models of DNA rewinding by RecQ helicases have been proposed. In one model, human BLM and AtRECQ2 switch strands for rewinding (5,10), whereas in another, human and chicken WRN slide back along the translocated strand (9,11), leaving the exact molecular mechanism of DNA rewinding to be elucidated.
RecQ helicases interact with several proteins, and their helicase activity is affected accordingly (3). Human replication protein A (RPA), a ssDNA binding protein, changes the reiterative unwinding mediated by human WRN into unidirectional unwinding (9), but does not inhibit the reiterative unwinding activity of human BLM (5). Such selective effects suggest that RPA may interact directly with RecQ helicases to modulate their unwinding activities. However, the exact mechanism by which RPA modulates the unwinding activity of RecQ helicases remains elusive. RPA may protect the unwound DNA to force unidirectional unwinding, or form a complex with the helicase, thereby modulating its conformation in favor of processive translocation.
Multicellular organisms possess several RecQ homologs. There are four members of the RecQ family in Caenorhabditis elegans, namely, recq-1, wrn-1, him-6, and rcq-5 (12–15), and him-6 is an ortholog of human BLM (14). The HIM-6 protein utilizes DNA-dependent adenosine triphosphatase (ATPase) and 3′-5′ DNA helicase activities to unwind forked DNAs, D-loops, and Holliday junctions (16). In the current study, we used smFRET techniques to characterize the DNA unwinding activity of HIM-6 and reveal the molecular mechanism of its reiterative unwinding mode and transition to the processive mode. We found that HIM-6 consistently unwinds 25–31 nt from the ss/dsDNA junction of forked DNA and then moves backwards along the translocated strand without strand-switching. In addition, we found that the reiterative unwinding activity of HIM-6 changes to unidirectional unwinding activity in the presence of C. elegans RPA (CeRPA). Our findings provide profound insights into how reiterative unwinding of DNA and its modulation control the functions of RecQ helicases in DNA metabolism.
MATERIALS AND METHODS
Preparation of DNA substrates
The DNA oligomers used in this study were synthesized to include biotin and amine modifications for dye labeling (Integrated DNA Technologies, USA). The Cy3 and Cy5 NHS esters were purchased from GE Healthcare (USA) and conjugated to the internal dT of DNA oligos or terminal positions via a C6 amino linker (17). For each substrate, biotin-conjugated DNA strands (1 μM final concentration) and complementary strands (1.2 μM) were annealed by heating to 90°C for 5 min and then slowly cooling to 4°C at a rate of −1°C/min. The sequences and structures of the DNA oligomers are listed in Supplementary Table S1.
Protein expression and purification
The C. elegans HIM-6/BLM protein was purified as described previously (16). The HIM-6 cDNA was cloned into expression vector pDEST17 for six-His-tagged protein (Gateway Cloning System, Invitrogen, Carlsbad, CA, USA). The recombinant His6-tagged HIM-6 proteins were expressed in Escherichia coli BL21AI and purified with a Ni+-NTA agarose (Invitrogen, Carlsbad, CA, USA). The C. elegans RPA protein was purified as reported previously (18,19). CeRPA proteins were expressed in E. coli BL21(DE3) and purified using Ni+-NTA agarose and Hitrap-Q-sepharose. The purity of the proteins was checked on an sodium dodecyl sulphate-polyacrylamide gelelectrophoresis gel with coomassie blue staining. Purified proteins were verified using a MALDI TOF-TOF 4700 proteomics analyzer (POSTECH Biotech Center, Korea). Enzymatic activity of HIM-6 proteins was confirmed using DNA unwinding assay with radiolabeled DNA.
Single-molecule FRET assay
Pre-cleaned quartz slides and glass coverslips were coated with a mixture of polyethylene glycol (m-PEG-SVA-5000; Laysan Bio, USA) and biotinylated-PEG (biotin-PEG-SC-5000; Laysan Bio) at a ratio of 40:1. Surface immobilization of the biotinylated DNA was mediated by NeutrAvidin (Thermo Scientific, USA). The standard reaction/imaging buffer contained 50 mM HEPES (pH 7.5), 20 mM KCl, 2 mM Dithiothreitol (DTT), 0.1 mg/ml bovine serum albumin (New England Biolabs, USA), and saturated Trolox (∼3 mM), with an oxygen scavenging system (0.1 mg/ml glucose oxidase, 0.6% (w/v) glucose and 0.06 mg/ml catalase) added prior to each measurement. The buffer contained equal concentrations of ATP and Mg2+. ATP was used at a concentration of 100 μM in all experiments unless otherwise specified. A custom-built prism-type total internal reflection fluorescence microscope was used to perform the measurements. Imaging was performed at room temperature (22 ± 1°C).
Analysis of single-molecule FRET data
The FRET efficiency was calculated from the fluorescence intensities of the donor (ID) and acceptor (IA) using the formula 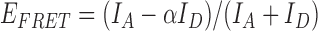 where α = 0.06 represents donor leakage correction. The smFRET histograms were generated from the traces containing a single pair of donor and acceptor, taking the frames before the photobleaching of dyes. smFRET histograms were fitted by single or double Gaussian peak curves. Data analysis was carried out by scripts custom written in MATLAB, and data fitting to Michaelis–Menten and gamma distribution curves was performed using the Origin package (Origin Lab Corporation, USA). Post-synchronized FRET density plots were constructed from individual unwinding cycles. The traces were synchronized at the moment that the FRET efficiency reached the bottom and increased back. The fragmented unwinding cycles were overlaid and shown as density maps using the contour plot as described earlier (20). Kinetic rate constants were obtained by taking the inverse of the averaged dwell time in each stage. The fraction of Cy3 spots remaining in the imaging area was obtained by taking the average number of Cy3 spots from five images taken at different locations per each minute, in order not to be affected by the photobleaching of the dyes. This was repeated three times to obtain the averaged curves shown with standard deviation.
where α = 0.06 represents donor leakage correction. The smFRET histograms were generated from the traces containing a single pair of donor and acceptor, taking the frames before the photobleaching of dyes. smFRET histograms were fitted by single or double Gaussian peak curves. Data analysis was carried out by scripts custom written in MATLAB, and data fitting to Michaelis–Menten and gamma distribution curves was performed using the Origin package (Origin Lab Corporation, USA). Post-synchronized FRET density plots were constructed from individual unwinding cycles. The traces were synchronized at the moment that the FRET efficiency reached the bottom and increased back. The fragmented unwinding cycles were overlaid and shown as density maps using the contour plot as described earlier (20). Kinetic rate constants were obtained by taking the inverse of the averaged dwell time in each stage. The fraction of Cy3 spots remaining in the imaging area was obtained by taking the average number of Cy3 spots from five images taken at different locations per each minute, in order not to be affected by the photobleaching of the dyes. This was repeated three times to obtain the averaged curves shown with standard deviation.
RESULTS
HIM-6 reiteratively unwinds forked DNA
HIM-6 unwinds dsDNA in the 3′-5′ direction (14,16,21). To determine the molecular characteristics of the DNA unwinding process, we performed smFRET measurements using a total internal reflection fluorescence microscope. We designed a forked duplex DNA substrate, named FK-50, containing a 50 bp duplex region, a 3′ single-stranded tail of (dT)30 labeled with Cy3 (donor) fluorophore at the ss/dsDNA junction and a 5′ single-stranded tail of (dT)17 labeled with Cy5 (acceptor) fluorophore located 7 nt away from the junction and biotinylated at the end for surface immobilization (Figure 1A). The structures and sequences of the DNA substrates are included in Supplementary Table S1. The substrate was immobilized on a polyethylene glycol-coated quartz surface via biotin-neutravidin interaction, and pre-incubated with HIM-6 (30 nM) for 5 min. The unwinding reaction was then initiated by introducing 100 μM ATP after washing out unbound proteins.
Figure 1.
HIM-6 reiteratively unwinds forked DNA substrate. (A) Schematic representation of the forked DNA substrate, FK-50, where Cy3 was located at the ss/dsDNA junction and Cy5 was located 7 nt away from the junction to the ssDNA side. The procedure used to initiate the unwinding reaction is shown. (B) A representative smFRET time trace of reiterative DNA unwinding by HIM-6, measured at 100 ms time resolution, showing the Cy3 intensity (green), Cy5 intensity (magenta) and calculated FRET efficiency (blue). (C) smFRET histograms of FK-50 DNA only (left, n = 216 molecules), and FK-50 with 30 nM HIM-6 and 100 μM ATP (right, n = 184). The histograms were normalized for the relative populations. (D) Time course of the density of Cy3 spots remaining for FK-50 only (blue) and FK-50 with HIM-6 and 100 μM ATP (red). Data represent the mean ± SD of three independent measurements.
As shown by the representative FRET trace in Figure 1B, there was a gradual decrease of the initially high FRET level, followed by a fast increase and then a pause until the next repetitive cycle. A large number of traces (39%) showed this repetitive pattern of FRET changes, with FRET efficiencies (EFRET) fluctuating between 0.25 and 0.85 until the fluorophores were photobleached. The FRET dynamics were ATP hydrolysis-dependent because such fluctuations were not observed in the presence of ATPγS, a non-hydrolyzable analog of ATP or with a helicase-defective HIM-6 mutant in which lysine 275 was mutated to alanine (K275A; Supplementary Figure S1). The high EFRET of 0.85 corresponded to the intact forked DNA, in which the fluorophores were located close together, as demonstrated by the fact that the same FRET level was observed for the bare FK-50 substrate prior to the addition of proteins (Figure 1C). The subsequent decrease in EFRET indicated unwinding of the forked DNA by the helicase activity of HIM-6. If HIM-6 had fully separated the two strands of FK-50, it would have resulted in complete disappearance of the fluorescence signal due to dissociation of the Cy3-labeled translocated strand. However, the FRET level paused at 0.25 during each repetitive cycle, as indicated by the corresponding peak in the FRET population histogram (Figure 1B and C). This peak at an EFRET of 0.25 was likely associated with the cessation of DNA unwinding activity. The subsequent rapid increase in EFRET back to its initial level was interpreted as the rewinding of duplex DNA, and the whole process started again after a pause. The repetitive cycle must have occurred via reinitiation of unwinding by the same helicase protein, because the unbound proteins in solution were washed away prior to the addition of ATP. Full unwinding of FK-50 occurred only rarely, as shown by a minor decrease in the fraction of Cy3 spots in the presence of active HIM-6 (Figure 1D). These observations indicate that HIM-6 is capable of repetitively unwinding DNA but rarely performs full unwinding of its substrate.
DNA unwinding and waiting times depend on the ATP concentration
Each cycle of the observed FRET changes could be dissected into three stages: the unwinding stage (U), during which the FRET level gradually decreased to 0.25; the rewinding/reannealing stage (R), during which the FRET level rapidly returned to 0.85; and the waiting stage (W), during which the FRET signal remained at 0.85 until the next unwinding cycle (Figure 2A). To visualize the variations in FRET dynamics between different unwinding cycles or different HIM-6 molecules, we separated the individual cycles and overlaid the traces after synchronizing them at the boundary between the unwinding and rewinding stages. This produced density plots showing the distribution of the FRET level changing over time (Figure 2B). A comparison of the density plots generated in reactions using 100 μM or 1 mM ATP clearly showed a dependence of the unwinding speed on ATP concentration, as indicated by a temporally narrower pattern at 1 mM ATP than at 100 μM ATP. Remarkably, the density plots also revealed a highly homogeneous pattern of FRET changes between different cycles or different HIM-6 molecules. In addition, rewinding consistently brought the FRET level back to the initial level seen for the intact duplex DNA. These results suggest that HIM-6 performs the unwinding cycle in a highly regular manner, rather than stochastically switching between the cycle stages.
Figure 2.
The unwinding and waiting stages are ATP-dependent. (A) Dissection of a representative smFRET time trace into the unwinding (U), rewinding (R), and waiting (W) stages. (B) FRET population maps synchronized at the transition from unwinding and rewinding stages at 100 μM ATP (n = 84) or 1 mM ATP (n = 76). (C–E) Kinetic rates of each stage, measured using the FK-50 substrate and varying ATP concentrations. Data represent the mean ± SEM of three independent measurements. The ATP-dependent rates of completing the unwinding and waiting stages were fitted to the Michaelis–Menten curve to find Vmax and Km values. (F–H) Dwell time distributions for the unwinding (F), rewinding (G) and waiting (H) stages measured at 100 μM ATP. Each histogram was fitted to a gamma distribution,  , where α, β and Г(α) are shape parameter, rate parameter and gamma function (red curve).
, where α, β and Г(α) are shape parameter, rate parameter and gamma function (red curve).
Next, we measured the duration of each stage of the unwinding process with varying ATP concentrations (50 μM to 3 mM) and calculated kinetic rates for transitions between the stages, which were obtained by taking the inverse of the average dwell time in each stage. The speed of the unwinding stage exhibited clear ATP-dependence (Figure 2C). The ATP-dependent unwinding rate was fit to the Michaelis–Menten equation, 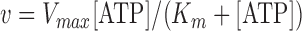 , and yielded a maximum rate (Vmax) of 1.29 ± 0.07 s−1 and a Michaelis–Menten constant (Km) of 0.31 ± 0.05 mM. On the other hand, the rewinding rate was not altered at varying ATP concentrations, giving consistent values of ∼1.4 s−1 (Figure 2D). This finding implies that the helicase is likely to passively slide back along one of the separated DNA strands without consuming ATP. Rapid rezipping of the duplex DNA is possibly the driving force to push the helicase back along the strand during this stage. Like the unwinding stage, the waiting stage was also ATP concentration-dependent, fitting to the Michaelis–Menten curve with a Vmax of 0.87 ± 0.08 s−1 and a Km of 0.14 ± 0.04 mM (Figure 2E). This finding indicates that the helicase is not merely at rest after completing rewinding, but actively re-engages in the next unwinding cycle, possibly by translocating along the ssDNA tail(s) to return to the fork junction. The distributions of the dwell times at each stage of the cyclic unwinding-rewinding process were fit to gamma distributions with shape parameters of 7.17 ± 0.63, 7.50 ± 2.03 and 4.12 ± 0.59 for the unwinding, rewinding, and waiting stages, respectively (Figure 2F–H), suggesting that each stage consists of multiple sub-steps rather than a single dominating step. However, the possible sub-steps were not directly resolved from our measurements. Also, the existence of ATP-driven motions in the rewinding stage cannot be completely ruled out because helicase activity was not clearly observable at ATP concentration below 50 μM.
, and yielded a maximum rate (Vmax) of 1.29 ± 0.07 s−1 and a Michaelis–Menten constant (Km) of 0.31 ± 0.05 mM. On the other hand, the rewinding rate was not altered at varying ATP concentrations, giving consistent values of ∼1.4 s−1 (Figure 2D). This finding implies that the helicase is likely to passively slide back along one of the separated DNA strands without consuming ATP. Rapid rezipping of the duplex DNA is possibly the driving force to push the helicase back along the strand during this stage. Like the unwinding stage, the waiting stage was also ATP concentration-dependent, fitting to the Michaelis–Menten curve with a Vmax of 0.87 ± 0.08 s−1 and a Km of 0.14 ± 0.04 mM (Figure 2E). This finding indicates that the helicase is not merely at rest after completing rewinding, but actively re-engages in the next unwinding cycle, possibly by translocating along the ssDNA tail(s) to return to the fork junction. The distributions of the dwell times at each stage of the cyclic unwinding-rewinding process were fit to gamma distributions with shape parameters of 7.17 ± 0.63, 7.50 ± 2.03 and 4.12 ± 0.59 for the unwinding, rewinding, and waiting stages, respectively (Figure 2F–H), suggesting that each stage consists of multiple sub-steps rather than a single dominating step. However, the possible sub-steps were not directly resolved from our measurements. Also, the existence of ATP-driven motions in the rewinding stage cannot be completely ruled out because helicase activity was not clearly observable at ATP concentration below 50 μM.
The return point of HIM-6
The persistent reiteration of unwinding without fully separating the dsDNA substrate suggests that HIM-6 stops after unwinding a certain length of DNA and before reaching the end of the duplex substrate, and then moves back to the original fork junction. To identify the position at which HIM-6 returns, we generated multiple DNA substrates in which Cy3/Cy5 pairs were placed at varying distances from the junction (Figure 3A and Supplementary Table S1). The FK-50(D0) substrate was identical to the FK-50 substrate used in Figure 1. In the FK-50(D18), FK-50(D25) and FK-50(D31) substrates, the Cy3 dye was located at the 18th, 25th and 31st nt from the ss/dsDNA junction on the 3′ strand, respectively, and the Cy5 dye was located at the 15th, 22nd and 28th nt from the junction on the 5′ strand, respectively. Although the reiterative unwinding dynamics of FK-50(D18) and FK-50(D25) were similar to that of FK-50(D0) (Figure 3B), the FRET level of the low FRET population increased with increasing distance between the labels and the junction (Figure 3C). This finding indicates that the 18th and 25th nucleotides were closer to the limiting position of unwinding than the first nucleotide. Even though the FRET levels were different, the frequencies of the unwinding events were comparable between the D0, D18 and D25 substrates (Figure 3D), indicating that HIM-6 unwinds DNA consistently up to 25 bp without returning to the fork junction. By contrast, the D31 substrate did not exhibit any FRET dynamics, but showed a stable high FRET population (Figure 3B and C). The sharp contrast in the dynamics between FK-50(D25) and FK-50(D31) suggests that the returning position of HIM-6 is narrowly defined between the 25th and 31st nt.
Figure 3.
HIM-6 unwinds a narrowly defined length of DNA before returning to the fork junction. (A) Schematic representation of the forked DNA substrates with fluorophores positioned at different locations. (B) Representative smFRET time traces of the Cy3 (green) and Cy5 (magenta) signals for each DNA substrate in the presence of HIM-6 and 100 μM ATP. Time traces were measured at 100 ms resolution. (C) FRET histograms of FK-50(D0) (n = 184), FK-50(D18) (n = 42), FK-50(D25) (n = 29) and FK-50(D31) (n = 35). The histograms were normalized for the relative populations. (D) Frequencies of the reiterative unwinding events for each substrate. Data represent the mean ± SEM of three independent measurements. (E) Schematic representation of the forked DNA substrate, FK-18, containing a shorter dsDNA region of 18 bp. (F) Two representative time traces of FK-18 unwinding in the presence of HIM-6 and 100 μM ATP. The dotted line indicates the point at which ATP was added. (G) Time course of the density of Cy3 spots remaining for FK-18 only (blue) and FK-18 with HIM-6 and 100 μM ATP (red). Data represent the mean ± SD of three independent measurements.
To determine if the returning point is determined by either the length of the duplex DNA remaining ahead of the helicase or the length of the unwound DNA, we designed FK-18 substrate, which contained a shorter duplex region of 18 bp and the same ssDNA tails as FK-50 (Figure 3E). In experiments using FK-18, the FRET level dropped upon the addition of HIM-6 and ATP, immediately followed by disappearance of the Cy3 signal, indicating full separation of the duplex FK-18 substrate (Figure 3F). Accordingly, the number of Cy3 spots per imaging area decreased rapidly upon loading 30 nM helicase and 1 mM ATP (Figure 3G).
We tested another set of DNA substrates which contains a 60-bp duplex region, labeled at D0, D25 or D31 positions, named FK-60(D0), FK-60(D25) and FK-60(D31), respectively (Supplementary Figure S2A, sequence information in Supplementary Table S1). FK-60(D0) and FK-60(D25) displayed similar unwinding signals as FK-50 substrates upon the addition of HIM-6 and ATP while FK-60(D31) did not show unwinding signals (Supplementary Figure S2B–G), confirming that HIM-6 does not go beyond the 31st nt regardless of the substrate length. These results unambiguously show that the returning point is determined by the number of unwound base pairs rather than the remaining DNA duplex length.
HIM-6 actively translocates along the 3′ ssDNA tail to reinitiate unwinding
Since the waiting time of the cyclical HIM-6-mediated unwinding process was ATP-dependent (Figure 2E), we conjectured that the helicase must be actively translocating along one or both of the DNA tails during the waiting stage, in preparation to re-engage in the next unwinding event. If HIM-6 stays on the translocated strand (the strand with the 3′ ssDNA tail) throughout the rewinding and waiting stages, the waiting time would only depend on the length of the 3′ ssDNA tail. By comparison, if HIM-6 switches to the displaced strand (the strand with the 5′ ssDNA tail), more steps will be required to place the helicase back at the junction; specifically, the helicase would have to switch to the displaced strand, return along the displaced strand, switch back to the translocated strand and then translocate to the junction. In this case, the waiting time is likely to depend on the lengths of both the 5′ and 3′ ssDNA tails. To identify which DNA strand HIM-6 uses during the waiting stage, we measured the waiting times on four forked DNA substrates with varying lengths of ssDNA tails (Figure 4A and Supplementary Table S1). For example, the 5′T03′T10 substrate contained no 5′ ssDNA tail and a 3′ ssDNA tail of (dT)10, and the 5′T103′T30 substrate contained a 5′ ssDNA tail of (dT)10 and a 3′ ssDNA tail of (dT)30. The smFRET time traces for 5′T03′T10, 5′T103′T10, 5′T03′T30, 5′T103′’T30, 5′T303′T30 and 5′T103′T50 substrates indicated similar reiterative unwinding patterns but the waiting time varied between the substrates with different tail lengths. The waiting time turned out to be nearly proportional to the length of the 3′’ ssDNA tail, resulting in a more frequent onset of the unwinding cycle for the substrates with shorter 3′ tail (Figure 4A and B). On the other hand, it did not vary upon changing the length of the 5′ ssDNA tail and 5′T03′T30, 5′T103′T30 and 5′T303′T30 showed a similar waiting time. These results suggest that the 5′ ssDNA tail is not translocated by HIM-6 in the waiting stage, and that HIM-6 uses the 3′ ssDNA tail (translocated strand) exclusively. It is still possible that HIM-6 switches to the displaced strand at certain point and then returns to the translocated strand before reaching the ss/dsDNA junction. Substrates containing either a short 3′ tail (5′T03′T10 and 5′T103′T10) or no 5′ tail (5′T03′T10 and 5′T03′T30) showed higher FRET levels in the unwinding stage than the other substrates (Figure 4A). Thus, HIM-6 may interact with both strands during unwinding and the initial contact of HIM-6 with DNA strands at the ss/dsDNA junction may determine the extent of strand separation during unwinding.
Figure 4.
HIM-6 translocates along the 3′ overhang during the waiting stage. (A) Schematic representation of the structures of the DNA substrates with varying overhang lengths, and corresponding representative smFRET time traces (Cy3 in green, Cy5 in magenta and FRET efficiency in blue). (B) Waiting times for the 5′T03′T10 (n = 122), 5′T103′T10 (n = 87), 5′T03′T30 (n = 153), 5′T103′T30 (n = 98), 5′T303′T30 (n = 115) and 5′T103′T50 (n = 59) substrates. Data represent the mean ± SD of three independent measurements. (C and D) Structures of the flapped substrates generated by hybridizing complementary oligomers to either the 3′ overhang (C; 5′ FLAP) or the 5′ overhang (D; 3′ FLAP) of FK-50. Representative smFRET time traces are shown on the right. (E) Waiting times for the 5′ FLAP (n = 64) and 3′ FLAP (n = 68) substrates compared with that of the forked substrate FK-50 (n = 72). Data represent the mean ± SD of three independent measurements.
Next, we tried to block HIM-6 translocation along each ssDNA tail using flapped substrates formed by annealing another DNA oligomer to the 5′ or 3′ ssDNA tail (named 3′ FLAP and 5′ FLAP, respectively; Figure 4C and D). Similar to what was observed for the substrate with the shorter 3′ tail, 5′ FLAP substrate showed a reduced waiting time, resulting in a more frequent onset of the unwinding cycle (Figure 4D and E). By contrast, 3′ FLAP substrate showed an even longer waiting time than the non-flapped substrate. The duplex tail of 3′ FLAP substrate might modulate the relocation of HIM-6 to the junction. For example, a domain of HIM-6, e.g. the helicase-and-RNase-D-C-terminal (HRDC) domain, may interact with the duplex tail, affecting the relocation of HIM-6 to the junction. Based on these results, we propose that HIM-6 actively translocates along the 3′ ssDNA tail of the translocated strand during the waiting stage.
HIM-6 slides back along the translocated strand during rewinding
The fact that the rapid FRET increase during the rewinding stage was ATP-independent suggests that HIM-6 slides back along the DNA strand in a passive manner during the rewinding stage, possibly caused by the rapid rezipping of DNA. To determine which strand HIM-6 moves along during the rewinding stage, we designed DNA substrates with no 5′ ssDNA tail of the displaced strand and placed Cy5 at this end of the strand (Figure 5A) and speculated the following scenarios. If HIM-6 moves along the displaced strand during rewinding, the junction of the DNA substrate would remain partially separated until the helicase switches back to the translocated strand, because it must be holding several terminal nucleotides of the displaced strand. Thus, at the end of rewinding, the FRET level would not immediately return to that of the intact duplex DNA, but rather stay at a lower level until the helicase switches back (Figure 5A). We tested two such DNA substrate designs without 5′ ssDNA tails and with a Cy3 label on the translocated strand either upstream or downstream of the ss/dsDNA junction (Figure 5B and F). On both substrates, the FRET level immediately returned to the original level of the bare DNA at the end of rewinding (Figure 5C–E and G–I). These results suggest that the rewinding process occurs not by switching to and sliding along the displaced strand, but by sliding back along the translocated strand (Figure 5A). It still cannot be ruled out that HIM-6 switched to the displaced strand at the end of unwinding and returned to the translocated strand before reaching the original junction, or it returned at the end of rewinding quickly beyond the imaging time resolution (100 ms).
Figure 5.
Reiterative unwinding dynamics on a 3′-overhang DNA. (A) Schematic representation of two possible rewinding modes on a 3′-tailed partial duplex substrate. In the strand-switching mode, once HIM-6 has translocated along the displaced strand or slid back to the end of the 5′ tail, the FRET level would remain at a lower level until HIM-6 switches back to the 3′ tail. In the sliding-back mode without strand-switching, as HIM-6 slides back along the translocated strand and passes by the dye position, the FRET level would immediately go back to its original high level. (B) Schematic representation of the Cy3 upstream substrate comprising a 3′-overhang DNA molecule containing a 50 bp duplex region, a 3′ ssDNA tail of (dT)30 labeled with Cy3 (green), located 7 nt from the junction and Cy5 (magenta) labeled at the 5′ end of the opposite strand at the junction. (C andD) Representative smFRET time traces of the Cy3 upstream DNA substrate only (C) and the same substrate with HIM-6 and 100 μM ATP (D). (E) FRET histograms corresponding to the conditions in (C) and (D), taken from more than 100 molecules each. (F) Schematic representation of the Cy3 downstream substrate. The substrate is similar to the one shown in (B), but the duplex region was extended to cover Cy3 which was positioned 7 nt into the duplex region. (G–I) The same analyses as (C–E) for the Cy3 downstream substrate
Based on the results described above, we propose the following model for the reiterative unwinding of DNA by HIM-6: (i) HIM-6 actively unwinds 25–31 bp of DNA by translocating in the 3′-5′ direction (unwinding stage); (ii) subsequently, HIM-6 shifts to a returning mode, possibly by losing its grip on DNA strands through a conformational change and slides back along the same strand in a passive manner beyond the original forked junction, allowing the DNA to rapidly rezip (rewinding stage); (iii) HIM-6 then actively translocates along the 3′ ssDNA tail of the same strand to get back to the junction and reinitiate unwinding (waiting stage).
CeRPA switches the reiterative unwinding of HIM-6 to unidirectional unwinding
A previous study shows that human BLM can unwind duplex DNA molecules longer than 200 bp in the presence of human RPA (hRPA) in bulk unwinding reactions (22). However, in single-molecule assays, BLM shows repetitive unwinding of short duplex DNA substrates (5). To resolve this discrepancy, we investigated the modulation of the reiterative unwinding of HIM-6 by CeRPA. A 73 kDa subunit (RPA-1) and a 32 kDa subunit (RPA-2) form an active, stable CeRPA complex that is sufficient for RPA function (18,19). First, we examined the ssDNA binding activity of CeRPA by smFRET measurements. A partial duplex DNA labeled with Cy3 at the end of the 3′ ssDNA tail and Cy5 at the ss/dsDNA junction showed a high FRET efficiency of 0.8, which went down to 0.4 within seconds after the addition of 20 nM CeRPA (Supplementary Figure S3A). The FRET decrease indicated rapid binding of CeRPA to the ssDNA tail, causing tail extension. We also confirmed that the E. coli ssDNA binding protein, SSB, assembled rapidly on the ssDNA tail (Supplementary Figure S3B).
The FK-50(D31) substrate was incubated with HIM-6 (30 nM), unbound HIM-6 was washed out and then 100 μM ATP was added (Figure 6A). No FRET change was observed at this point as the labels were positioned past the unwinding limit of HIM-6 (Figure 3B and C). However, adding 20 nM CeRPA while maintaining ATP level reduced the FRET level to ∼0.2 (Figure 6B). There were no reiterative FRET changes and the Cy3 signal subsequently disappeared, representing dissociation of the translocated strand. Accordingly, the number of Cy3 spots in the imaging area decreased rapidly (Figure 6F, green circle). These results show that HIM-6 progressively and fully unwound the FK-50(D31) substrate in the presence of CeRPA. As a control, the addition of CeRPA alone, without HIM-6, to the FK-50(D31) substrate did not cause any FRET changes and resulted in minimal decrease in the number of Cy3 spots (Supplementary Figure S3C and D). Analyses of the dynamics of HIM-6/CeRPA-mediated unwinding of the FK-50(D0) substrate revealed that several reiterative unwinding cycles occurred before the full separation of DNA (Figure 6C), representing the time required for CeRPA proteins to cover the unwound strands. These results suggest that CeRPA proteins assemble on the unwound ssDNA region as HIM-6 unwinds the DNA and moves forward, preventing the DNA from reannealing and/or preventing HIM-6 from sliding back, resulting in processive, full unwinding of the duplex. Interestingly, the inducing effect on processive unwinding was specific for CeRPA because E. coli SSB did not induce the full separation of FK-50(D31) and FK-50(D0), but only interfered with the reiterative unwinding behavior (Figure 6F, purple circle; Supplementary Figure S3E and F).
Figure 6.
CeRPA switches the HIM-6-mediated reiterative unwinding to processive unwinding. (A) Schematic diagram showing the reaction steps of adding HIM-6, ATP and CeRPA to the FK-50 substrates. In the first experiment, 30 nM HIM-6 was added, unbound proteins were flushed out, 100 μM ATP was added and 20 nM CeRPA was added while maintaining ATP levels. In the second experiment, 20 nM CeRPA was added, unbound proteins were flushed out, 30 nM HIM-6 was added, unbound proteins were flushed out while adding 100 μM ATP. (B) A representative smFRET time trace from the first experiment, showing the full unwinding of the FK-50(D31) substrate. The dotted line indicates the point at which CeRPA was added. (C) Same as (B) for the FK-50(D0) substrate. (D) A representative smFRET time trace from the second experiment, again showing the full unwinding of the FK-50(D31) substrate. (E) Same as (D) for the FK-50(D0) substrate. (F) Time courses of the density of Cy3 spots remaining for the FK-50(D31) substrate only (blue), supplied with HIM-6/ATP (red), supplied with HIM-6/ATP and then 20 nM Escherichia coli SSB /ATP (purple), supplied with CeRPA/ATP and then HIM-6/ATP (orange), and supplied with HIM-6/ATP and then CeRPA/ATP (green). Data represent the mean ± SD of three independent measurements.
Next, the order in which the proteins were added to the reaction was changed. The FK-50(D31) substrate was first incubated with 20 nM CeRPA and then, after washing away unbound CeRPA, 30 nM HIM-6 and 100 μM ATP were added. Notably, similar to what was observed in the above experiment, the FRET efficiency dropped rapidly to ∼0.4, followed by dissociation of the translocated strand (Figure 6D). The number of Cy3 spots in the imaging area also decreased, but at a slower rate than when CeRPA was present in solution (Figure 6F, orange circle). When CeRPA was preloaded to FK-50(D0) and flushed out before unwinding was initiated with HIM-6, HIM-6 fully unwound the substrate in one cycle (Figure 6E). These results are intriguing because there was no CeRPA available to protect the newly exposed unwound ssDNA region, but HIM-6 was still able to fully unwind the DNA. It is possible that there is direct interaction between HIM-6 and CeRPA, which was previously observed in a human system (22). HIM-6 loaded at the forked junction may bind preloaded CeRPA, and these proteins may translocate together while unwinding the DNA. These results suggest that CeRPA plays a crucial role in switching HIM-6 between the reiterative and processive unwinding modes, thereby regulating the processing of long DNAs.
DISCUSSION
In this study, we addressed the key molecular characteristics of reiterative DNA unwinding by the C. elegans BLM helicase HIM-6. We found that HIM-6 unwinds 25–31 bp by translocating in the 3′-5′ direction from the ss/dsDNA junction of forked DNA substrates, and then returns along the same translocated strand via rapid backsliding. This cycle of unwinding and rewinding occurs in a highly repetitive fashion, but long duplex DNAs are not fully unwound. The reiterative unwinding process transitions into unidirectional unwinding in the presence of CeRPA, thereby allowing full unwinding of long duplex DNA substrates.
Several RecQ family helicases are able to reiteratively unwind DNA (5,6,9–11). However, it has remained unclear how far the helicases translocate along the DNA before turning back. A previous smFRET study by Yodh et al. suggested that human BLM unwinds fewer than 34 bp of forked DNA substrates (5). In that study, the unwound DNA length was estimated by comparing the unwinding times and chances of full unzipping for DNA substrates of varying lengths. Another study of BLM by Wang and colleagues observed unwinding-rewinding cycles on DNA hairpins using magnetic tweezers and estimated the unwound DNA length to be ∼15 bp (6). Other studies with magnetic tweezers have shown that human WRN and AtRecQ2 reiteratively unwind DNA hairpins containing a stem region of ∼40–50 bp (9,10). However, these studies have not directly shown how many base pairs separate, making it difficult to determine how the returning position is distributed. Our smFRET measurements of DNA substrates with internal labels at varying positions revealed that HIM-6 returns at a narrowly distributed position between the 25th and 31st nt. To our knowledge, the current study is the first direct identification of the returning position of a RecQ family helicase showing reiterative unwinding. Our findings raise the question of how RecQ helicases precisely sense the unwound DNA length and change their mode of dynamics accordingly.
It has long been of interest to identify which strand RecQ helicase uses for moving backward. Three possible modes have been proposed: (i) strand-switching and active translocation along the displaced strand, (ii) strand-switching and sliding back along the displaced strand and (iii) sliding back along the translocated strand without strand-switching. Different mechanisms have been proposed for different RecQ helicases. The ‘strand-switching and translocation’ mode, in which the helicase switches strands after unwinding the DNA duplex and translocates along the displaced strand in an ATP-dependent manner, has been suggested for human BLM (5). However, a later study of human BLM using DNA hairpins suggested the ‘strand-switching and sliding back’ mode, in which the helicase returns by passively sliding along the displaced strand (6). A study of AtRECQ2, a WRN homolog, also proposed the ‘strand-switching and sliding back’ mode (10). More recently, human WRN and Gallus gallus (chicken) WRN were shown to follow the third mode of ‘sliding back along the translocated strand’ (11). Notably, a recent study of E. coli RecQ proposed that either the ‘strand-switching’ or ‘sliding back’ mode is possible, depending on the involvement of the HRDC domain (23). If HIM-6 follows the first two models, the cycle of repetitive unwinding would depend on the length of the 5′ ssDNA tail of the forked substrates. However, our results revealed that the waiting time depends on the length of the 3′ ssDNA tail, but not the 5′ ssDNA tail (Figure 4), providing no support for the first two models.
Our experiments using partial duplex DNA substrates without a 5′ tail further support the third mode of ‘sliding back along the translocated strand’. If HIM-6 were to switch to the displaced strand to translocate or slide back, and then switch back to the translocated strand, it would spend a certain amount of time at the end of the displaced strand for strand-switching, during which the FRET level would not revert to its original FRET level (Figure 5A). However, our results showed that the FRET level immediately returned to its original high level at the end of the rewinding stage, which does not support the existence of such strand-switching steps. If HIM-6 were to slide back on the translocated strand without strand-switching, these behaviors can be coherently explained. The rewinding mode appears not to be influenced by the structure of DNA substrates because our data revealed consistent, reiterative unwinding behavior on several kinds of DNA substrates containing overhang, flap, or fork structure, and the rewinding time was measured to be in the same range. Transition to the rewinding mode could be explained by either the interaction of RecQ helicase with DNA or a conformational change in the protein. Most RecQ helicases have three conserved domains: the helicase domain, RecQ C-terminal (RQC) domain and HRDC domain. A previous study of human BLM using DNA hairpins proposed that the RQC domain interacts with the DNA substrate (6). In this model, BLM uses its helicase domain to bind to the translocated strand, and the RQC domain to bind to both the dsDNA and the displaced strand during unwinding. When the helicase domain is released from the translocated strand, the enzyme switches strands and slides back quickly along the displaced strand. These interactions of RecQ domains with DNA have been demonstrated for BLM and may explain the ‘strand-switching’ model (24). On the other hand, as a possible explanation of the ‘sliding back on the translocated strand’ model, a study of E. coli RecQ proposed that the HRDC domain plays a role in DNA interaction (23). If the HRDC domain binds to the displaced strand, the tethered helicase may slide back along the translocated strand after unwinding a certain length of the duplex. In this case, the extent of HRDC-tethered backsliding is probably limited by the length and flexibility of the linker region connecting the RQC and HRDC domains. If the HRDC domain binds to the helicase core (consisting of the helicase and RQC domains), the interaction between the helicase and the translocated strand will allow RecQ to slide toward the 3′ end of the translocated strand, resulting in rapid reannealing. An interaction of the HRDC domain with the helicase core was recently demonstrated for human BLM (25).
A similar mechanism of controlling the DNA unwinding dynamics by conformational changes has been described for other kinds of helicases. Rep, PcrA and UvrD helicases spontaneously transition between a ‘closed’ conformation, which actively unwinds DNA, and an ‘open’ conformation, which reverses to rezip the DNA, and binding partner proteins can selectively stabilize the open conformation to promote processive unwinding (26,27). Since RecQ members belonging to the same family use distinct mechanisms, it remains to be elucidated how the domains of RecQ helicases interplay to control the translocating and unwinding dynamics of the proteins. Although structural information about HIM-6 is currently unavailable, the detailed mechanism of unwinding used by HIM-6 could be elucidated by experiments using modified HIM-6 to identify the domains involved in each stage of reiterative unwinding motion.
Because the reiterative unwinding behavior of RecQ helicases has been observed using purified recombinant proteins, it is still unclear whether such behavior actually occurs in the cellular environment. Helicases interact with many different kinds of proteins; thus it is of interest to determine if partner proteins can change the reiterative unwinding into processive, unidirectional unwinding. Our results showed that CeRPA changes the behavior of HIM-6 from reiterative unwinding into processive unwinding, allowing it to completely unwind a 50 bp duplex DNA (Figure 6). Also, a longer substrate, FK-60, was completely unwound by HIM-6 in the presence of CeRPA (Supplementary Figure S2H–J). This finding is in contrast to what has been observed for human BLM, which persistently showed reiterative unwinding in the presence of hRPA (5). However, a recent study of human WRN found that the interaction of multiple hRPAs enables WRN to unwind dsDNA unidirectionally for longer than 1 kbp (9). Although a direct, physical interaction of HIM-6 with CeRPA has not been demonstrated, we speculate that the processive unwinding observed here must be due to their direct interaction. Our observation that preloaded CeRPA induces the full separation of the DNA substrate by HIM-6 in the absence of free CeRPA proteins to cover the unwound DNA strands strongly supports that their interaction persists while HIM-6 unwinds the DNA. Combined with earlier observations, processive unwinding of HIM-6 stimulated by CeRPA implies a universal role of RPA in generating a long ssDNA region, which is possibly involved in DNA repair processes. Together with the ability of RecQ helicases to sense the length of unwound DNA, these results provide clues as to how RecQ helicases decide and control the mode of translocation and length of DNA to unwind.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea [NRF-2018R1D1A1B07048882 to B.A.; NRF-2017R1D1A1B03036239, 2017M3A9E2062181 to H.K.]; Institute for Basic Science [IBS-R022-D1 to H.K.]. Funding for open access charge: NRF-2018R1D1A1B07048882 and 2017M3A9E2062181.
Conflict of interest statement. None declared.
REFERENCES
- 1. Singleton M.R., Dillingham M.S., Wigley D.B.. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007; 76:23–50. [DOI] [PubMed] [Google Scholar]
- 2. Singh D.K., Ghosh A.K., Croteau D.L., Bohr V.A.. RecQ helicases in DNA double strand break repair and telomere maintenance. Mutat. Res. 2012; 736:15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Croteau D.L., Popuri V., Opresko P.L., Bohr V.A.. Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 2014; 83:519–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Renty C., Ellis N.A.. Bloom's syndrome: Why not premature aging?: A comparison of the BLM and WRN helicases. Ageing Res. Rev. 2017; 33:36–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yodh J.G., Stevens B.C., Kanagaraj R., Janscak P., Ha T.. BLM helicase measures DNA unwound before switching strands and hRPA promotes unwinding reinitiation. EMBO J. 2009; 28:405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang S., Qin W., Li J.H., Lu Y., Lu K.Y., Nong D.G., Dou S.X., Xu C.H., Xi X.G., Li M.. Unwinding forward and sliding back: an intermittent unwinding mode of the BLM helicase. Nucleic Acids Res. 2015; 43:3736–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chatterjee S., Zagelbaum J., Savitsky P., Sturzenegger A., Huttner D., Janscak P., Hickson I.D., Gileadi O., Rothenberg E.. Mechanistic insight into the interaction of BLM helicase with intra-strand G-quadruplex structures. Nat. Commun. 2014; 5:5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Budhathoki J.B., Maleki P., Roy W.A., Janscak P., Yodh J.G., Balci H.. A comparative study of G-Quadruplex unfolding and DNA reeling activities of human RECQ5 helicase. Biophys. J. 2016; 110:2585–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee M., Shin S., Uhm H., Hong H., Kirk J., Hyun K., Kulikowicz T., Kim J., Ahn B., Bohr V.A. et al.. Multiple RPAs make WRN syndrome protein a superhelicase. Nucleic Acids Res. 2018; 46:4689–4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Klaue D., Kobbe D., Kemmerich F., Kozikowska A., Puchta H., Seidel R.. Fork sensing and strand switching control antagonistic activities of RecQ helicases. Nat. Commun. 2013; 4:2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu W.Q., Hou X.M., Zhang B., Fossé P., René B., Mauffret O., Li M., Dou S.X., Xi X.G.. Single-molecule studies reveal reciprocating of WRN helicase core along ssDNA during DNA unwinding. Sci. Rep. 2017; 7:43954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kusano K., Berres M.E., Engels W.R.. Evolution of the RECQ family of helicases: a drosophila homolog, Dmblm, is similar to the human bloom syndrome gene. Genetics. 1999; 151:1027–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee S.J., Yook J.S., Han S.M., Koo H.S.. A Werner syndrome protein homolog affects C. elegans development, growth rate, life span and sensitivity to DNA damage by acting at a DNA damage checkpoint. Development. 2004; 131:2565–2575. [DOI] [PubMed] [Google Scholar]
- 14. Wicky C., Alpi A., Passannante M., Rose A., Gartner A., Müller F.. Multiple genetic pathways involving the Caenorhabditis elegans Bloom's syndrome genes him-6, rad-51, and top-3 are needed to maintain genome stability in the germ line. Mol. Cell Biol. 2004; 24:5016–5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sekelsky J.J., Brodsky M.H., Rubin G.M., Hawley R.S.. Drosophila and human RecQ5 exist in different isoforms generated by alternative splicing. Nucleic Acids Res. 1999; 27:3762–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jung H., Lee J.A., Choi S., Lee H., Ahn B.. Characterization of the caenorhabditis elegans HIM-6/BLM Helicase: unwinding recombination intermediates. PLoS One. 2014; 9:e102402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roy R., Hohng S., Ha T.. A practical guide to single-molecule FRET. Nat. Methods. 2008; 5:507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim D.H., Lee K.H., Kim J.H., Ryu G.H., Bae S.H., Lee B.C., Moon K.Y., Byun S.M., Koo H.S., Seo Y.S.. Enzymatic properties of the Caenorhabditis elegans Dna2 endonuclease/helicase and a species-specific interaction between RPA and Dna2. Nucleic Acids Res. 2005; 33:1372–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hyun M., Park S., Kim E., Kim D.H., Lee S.J., Koo H.S., Seo Y.S., Ahn B.. Physical and functional interactions of Caenorhabditis elegans WRN-1 helicase with RPA-1. Biochemistry. 2012; 51:1336–1345. [DOI] [PubMed] [Google Scholar]
- 20. Kim H., Abeysirigunawarden S.C., Chen K., Mayerle M., Ragunathan K., Luthey-Schulten Z., Ha T., Woodson S.A.. Protein-guided RNA dynamics during early ribosome assembly. Nature. 2014; 506:334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grabowski M.M., Svrzikapa N., Tissenbaum H.A.. Bloom syndrome ortholog HIM-6 maintains genomic stability in C. elegans. Mech. Ageing Dev. 2005; 126:1314–1321. [DOI] [PubMed] [Google Scholar]
- 22. Brosh R.M. Jr, Li J.L., Kenny M.K., Karow J.K., Cooper M.P., Kureekattil R.P., Hickson I.D., Bohr V.A.. Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity. J. Biol. Chem. 2000; 275:23500–23508. [DOI] [PubMed] [Google Scholar]
- 23. Harami G.M., Seol Y., In J., Ferencziová V., Martina M., Gyimesi M., Sarlós K., Kovács Z.J., Nagy N.T., Sun Y. et al.. Shuttling along DNA and directed processing of D-loops by RecQ helicase support quality control of homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E466–E475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Swan M.K., Legris V., Tanner A., Reaper P.M., Vial S., Bordas R., Pollard J.R., Charlton P.A., Golec J.M., Bertrand J.A.. Structure of human Bloom's syndrome helicase in complex with ADP and duplex DNA. Acta Crystallogr. D. Biol. Crystallogr. 2014; 70:1465–1475. [DOI] [PubMed] [Google Scholar]
- 25. Newman J.A., Savitsky P., Allerston C.K., Bizard A.H., Özer Ö., Sarlós K., Liu Y., Pardon E., Steyaert J., Hickson I.D. et al.. Crystal structure of the Bloom's syndrome helicase indicates a role for the HRDC domain in conformational changes. Nucleic Acids Res. 2015; 43:5221–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Comstock M.J., Whitley K.D., Jia H., Sokoloski J., Lohman T.M., Ha T., Chemla Y.R.. Direct observation of structure-function relationship in a nucleic acid–processing enzyme. Science. 2015; 348:352–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arslan S., Khafizov R., Thomas C.D., Chemla Y.R., Ha T.. Engineering of a superhelicase through conformational control. Science. 2015; 348:344–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.








