Abstract
The human opportunistic pathogen Staphylococcus aureus produces numerous small regulatory RNAs (sRNAs) for which functions are still poorly understood. Here, we focused on an atypical and large sRNA called RsaC. Its length varies between different isolates due to the presence of repeated sequences at the 5′ end while its 3′ part is structurally independent and highly conserved. Using MS2-affinity purification coupled with RNA sequencing (MAPS) and quantitative differential proteomics, sodA mRNA was identified as a primary target of RsaC sRNA. SodA is a Mn-dependent superoxide dismutase involved in oxidative stress response. Remarkably, rsaC gene is co-transcribed with the major manganese ABC transporter MntABC and, consequently, RsaC is mainly produced in response to Mn starvation. This 3′UTR-derived sRNA is released from mntABC-RsaC precursor after cleavage by RNase III. The mature and stable form of RsaC inhibits the synthesis of the Mn-containing enzyme SodA synthesis and favors the oxidative stress response mediated by SodM, an alternative SOD enzyme using either Mn or Fe as co-factor. In addition, other putative targets of RsaC are involved in oxidative stress (ROS and NOS) and metal homeostasis (Fe and Zn). Consequently, RsaC may balance two interconnected defensive responses, i.e. oxidative stress and metal-dependent nutritional immunity.
INTRODUCTION
Staphylococcus aureus is a major human opportunistic pathogen responsible for a wide variety of infections, from superficial to deep-seated infections with high mortality (e.g. infective endocarditis, osteomyelitis and necrotizing pneumonia). Professional phagocytes (i.e. neutrophils and macrophages) represent the first line of host defence and are important components of innate immunity against S. aureus (1,2). After migration to the infection site, immune effectors deploy an arsenal of offensive and defensive strategies including phagocytosis, prevention of S. aureus dissemination by extracellular traps (ETs) (3), and synthesis of a battery of antimicrobial effectors such as reactive oxygen species (ROS) (1). ROS include superoxide anion (O2.−), hydrogen peroxide (H2O2) and hydroxyl radical (.OH), which are also natural products of metabolism formed when oxygen becomes partially reduced during aerobic respiration (4). They have the potential to damage nucleic acids, proteins and lipids. Neutrophils also locally secrete the chelating agent calprotectin to sequester critical ions for bacterial growth such as manganese (Mn) and zinc (Zn) (5).
To evade immune defences and colonize numerous organs, S. aureus produces a wide range of virulence factors and stress-response proteins. They notably impede phagocytosis, escape ETs, extract nutrients from host cells and detoxify ROS (2,3). For example, superoxide dismutases (SOD) and catalases are produced to convert O2.− to H2O2, and then to water and oxygen (6). SODs are metalloenzymes classified according to their loaded cofactor: copper/zinc (Cu/Zn-SOD), manganese (Mn-SOD), nickel (Ni-SOD) or iron (Fe-SOD) (7). In S. aureus, two SODs are involved in oxidative response (8,9). The major SOD enzyme SodA is strictly Mn-dependent while the cambialistic enzyme SodM is active when loaded with Mn or Fe and is therefore important in manganese limiting conditions (10). Remarkably, SodM is specific to S. aureus as coagulase-negative staphylococci lack sodM gene (9,10). There are conflicting reports regarding the role of SODs in virulence. An early study suggested no correlation between SOD activity and lethality in a mouse infection model (11). However, SOD activity was found higher in S. aureus strains isolated from infected patients (12). Deletion of sodAM reduced virulence in abscess or retro-orbital infection models (5,13). More recently, the contribution of each Sod enzyme was deciphered during mice infection (10). SodA plays a crucial role when Mn is abundant while SodM becomes the main enzyme assuring the resistance to oxidative stress when Mn is scarce. Nevertheless, the regulation of their synthesis is still poorly understood.
The production of S. aureus virulence factors is regulated by a combination of transcriptional regulators and regulatory RNAs (sRNAs) (14). Many sRNAs form base-pairings with their mRNA targets to control their expression and play key functions in adaptive processes and virulence (15). The most studied sRNA in S. aureus is RNAIII, one of the main intracellular effectors of the quorum sensing agr system. RNAIII represses the expression of virulence factors such as protein A, immunoglobulin-binding protein Sbi, coagulase, and of the repressor of toxins Rot while it activates the synthesis of hemolysin α (16–21). In this study, we have analyzed the function of another very peculiar and large sRNA, named RsaC, which was found highly expressed in acute osteomyelitis, a bone infection usually caused by S. aureus (22). Here, we identified sodA mRNA as a primary target of the 3′-UTR derived sRNA RsaC, which is mainly produced upon Mn starvation. By repressing the translation of the Mn-containing enzyme SodA, RsaC avoids the synthesis of a non-functional enzyme. Concurrently, RsaC indirectly enhances the oxidative stress response mediated by the cambialistic enzyme SodM, which in absence of manganese, uses Fe as a cofactor to restore the ROS detoxification pathway. We also demonstrated that RsaC might have a broader role in oxidative stress response (ROS and NOS) and in metal homeostasis (Mn, Fe and Zn). The consequences of this regulation on S. aureus pathogenicity will be discussed.
MATERIALS AND METHODS
Strains, plasmids and growth conditions
Staphylococcus aureus strains and plasmids used in this study are listed in Supplementary Table S1. Escherichia coli strain DH5α (Promega) was used as host strain for plasmid constructions. Staphylococcus aureus strain RN4220 (23) or E. strain DC10B (24) were used as host strains for plasmid amplification before electroporation in S. aureus HG001. Transformation of E. coli DH5α or DC10B was performed by heat shock and S. aureus strains were transformed by electroporation (Bio-Rad gene Pulser).
Escherichia coli strains were cultivated in lysogeny broth (LB). Staphylococcus aureus strains were isolated on glucose peptone (GP), tryptic soy (TS) or blood agar plates. Overnight cultures in brain-heart infusion (BHI) or in tryptic soy broth (TSB) ± erythromycin (5 μg/ml) were diluted 50-fold in fresh medium and grown at 37°C (180 rpm, 5:1 flask-to-medium ratio). To induce Mn starvation, cells were grown in BHI-chelex medium as followed. We added 5% Chelex-100 resin (Sigma) to BHI medium and mix 6h at 20°C. The medium was sterile filtered and, according to Kehl-Fie et al. (25), we complemented it with metal ions at the following concentrations: 100 μM CaCl2, 25 μM ZnCl2, 1 mM MgCl2, 1 μM FeSO4. As a control, BHI-chelex medium was also supplemented with 25 μM MnCl2.
Deletion of rsaC1116 gene (ΔrsaC) in HG001 strain was performed using pMAD (26). Chromosomal regions upstream and downstream of the rsaC sequence were amplified by PCR (RsaC-PCR1-Rev/RsaC-PCR1-For and RsaC-PCR2-Rev/RsaC-PCR2-For, respectively) (Supplementary Table S2) and then cloned into pMAD vector. The resulting plasmid (pLUG1110) was electroporated into RN4220 recipient strain and then transferred to HG001. Growth at non-permissive temperature (44°C) was followed by several subcultures at 30°C and 37°C to favor double crossing over as previously described (26). As control, we verified that deletion of rsaC does not alter the expression of surrounding genes. Indeed, rsaC gene is located between mntC and nhaK_1 genes, which encodes a subunit of a manganese ABC transporter and a proton pump, respectively (Supplementary Figure S1A). Using qRT-PCR experiments, we have verified that no difference existed in the expression level of downstream and upstream genes between wild type and mutant strains (Supplementary Figure S1B). To complement ΔrsaC strain (rsaC+), rsaC1116 gene was amplified using PstI-RsaC1116-For/BamHI-RsaC1116-Rev oligonucleotides (Supplementary Table S2). The amplicon RsaC1116 was digested by PstI and BamHI restriction enzymes and inserted into a PstI/BamHI digested pCN51-P3 plasmid. The quorum sensing-dependent P3 promoter allows an overexpression of the downstream gene in stationary phase of growth. To generate the RsaC1116mut construct (deletion of nucleotides +1000 to +1006), two independent PCR reactions were performed with the following oligonucleotides: PstI-RsaC1116-For/RsaCmut-Rv and RsaCmut-For/BamHI-RsaC-Rev. The two PCR products were then mixed to serve as template for a third PCR (PstI-RsaC1116-For/BamHI-RsaC1116-Rev). The amplicon RsaC1116mut was digested by PstI and BamHI restriction enzymes and inserted into a PstI/BamHI digested pCN51-P3 plasmid. To obtain a pJET-T7-sodA, a PCR fragment was amplified using oligos T7-sodA-For/T7-sodA-Rev (Supplementary Table S2) and then cloned into pJET1.2/blunt vector (ThermoFisher). For MAPS experiment, MS2 aptamer was fused to the 5′ end of RsaC544 using oligonucleotides PstI-MS2-RsaC-For and BamHI-RsaC-Rev (Supplementary Table S2). Obtained fragment was cloned into PstI/BamHI-digested pCN51-P3. Total DNA and plasmid DNA were prepared using DNAeasy tissue Kit and Qiaprep Miniprep respectively (Qiagen).
Northern blot analysis
Total RNAs were prepared from different volumes of S. aureus cultures taken at 2 h (OD600nm∼0.4; 15 ml), 4 h (OD600 nm ∼2.5; 10 ml) and 6 h (OD600 nm ∼5; 5 ml). After centrifugation, bacterial pellets were resuspended in RNA Pro Solution (MP Biomedicals). Lysis was performed with FastPrep apparatus (MP Biomedicals). RNA purification followed strictly the procedure described for the FastRNA Pro Blue Kit (MP Biomedicals).
Electrophoresis of total RNA (10–20 μg) was performed on 1% agarose gel containing 25 mM guanidium thiocyanate. After migration, RNAs were vacuum transferred on Hybond N+ nitrocellulose membrane (GE Healthcare Life Sciences). Hybridization with specific digoxygenin (DIG)-labelled probe complementary to RsaC or 5S sequence followed by luminescent detection were carried out as described previously (16). For mntA, we used a radiolabelled DNA probe (Supplementary Table S2). The labeling of the 5′ end of the oligonucleotide was performed with T4 polynucleotide kinase (Fermentas) and [γ32P] ATP. For determination of molecular weights, we used the Dynamarker® Prestain Marker for RNA high (CliniSciences).
MS2-affinity purification coupled with RNA sequencing (MAPS)
The MS2 aptamer, which is recognized by the MS2 coat protein, was fused to the 5′ end of RsaC544. Here, we used a shorter form of RsaC due to nonspecific interaction of RsaC1116 with the affinity column. RsaC544 construct carries the whole 3′ domain of RsaC (447 nt) and 97 nucleotides from the 5′ part, which enables to preserve the secondary structure of the 3′ domain of RsaC (Figure 1B). This construct was expressed in vivo under the control of the quorum sensing-dependent promoter P3 and confirmed as functional when compared to the full-length RsaC1116 and MS2-RsaC1116 (Supplementary Figure S2). Indeed, the overexpression of the three RNAs induces similar ROS accumulation, revealing that the 3′ domain of RsaC is sufficient to modulate the oxidative stress response.
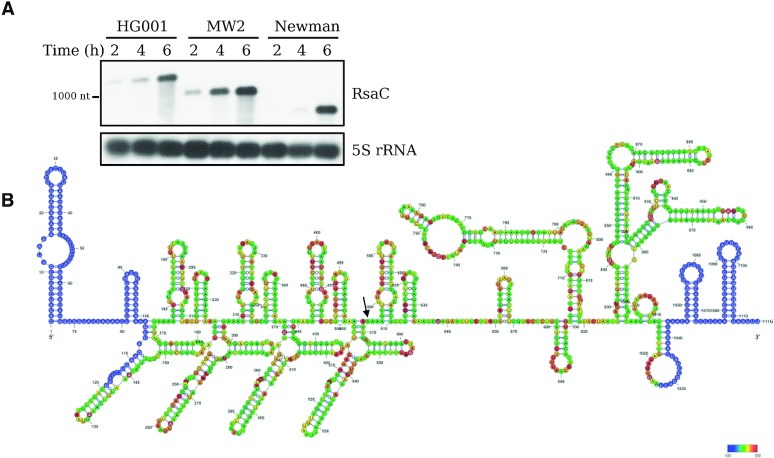
Figure 1.
RsaC is a large and highly structured sRNA. (A) Northern blot analysis of RsaC sRNA in three different S. aureus strains (HG001, MW2 and Newman). Total RNA was extracted after 2, 4 and 6 h of growth in BHI at 37°C. 5S rRNA was used as loading control. Data are representative of three independent experiments. (B) Secondary structure model of RsaC. The structure of RsaC (from HG001 strain) was mapped using selective 2′-hydroxyl acylation by benzol cyanide and the reactivity was analyzed by primer extension (SHAPE). Data were analyzed, and each modification was quantified using QuSHAPE software. The secondary structure was drawn using RNA structure and VARNA considering the reactivity of each ribose. The regions, which were not mapped, are shown in blue. The color code for each reactivity is given on the right side of the secondary structure model, red is for the highest reactivity while green is for the lowest reactivity. A black arrow indicates the +1 of RsaC544.
Crude extracts from ΔrsaC strain expressing MS2-RsaC544 or from WT strain expressing MS2 tag alone (control) were harvested after 6 h of growth at 37°C in BHI medium. MS2-affinity purifications were performed as previously described (27,28). All steps were performed at 4°C. Poly-Prep chromatography column (Biorad) containing amylose resin (300 μl; NEB) were washed three times with 10 ml of Buffer A (20 mM Tris–HCl pH 8, 150 mM KCl, 1 mM MgCl2, 1 mM DTT), before addition of 3600 pmol of MS2-MBP. Crude bacterial extracts (4 ml) were directly loaded, followed by three washes with 10 ml of Buffer A. Finally, RNAs were eluted from the column with 1 ml of Buffer E (20 mM Tris–HCl pH 8, 150 mM KCl, 1 mM MgCl2, 1 mM DTT, 0,1% Triton X-100, 12 mM maltose). Eluted RNAs were extracted with phenol:chloroform:isoamylalcohol (25:24:1 (v/v), Roth) and precipitated with 3 volumes of cold absolute ethanol in the presence of 0.3 M sodium acetate. RNA samples were then treated with DNase I prior to RNA-seq analysis. MS2-affinity purifications were performed in duplicates.
RNA quality and quantity assessments were performed on Agilent Nano Chip on Bioanalyzer 2100. RNA samples were ribo-depleted (Ribo-Zero rRNA Removal Kit (Bacteria) Illumina) and cDNA libraries were prepared using the Random Hexamer approach and sequenced by Fasteris (Switzerland). Libraries were sequenced using Hi-Seq 2500 system (Illumina). RNA-seq analysis was performed according to Lalaouna et al. (2018) (27). Reads were processed and aligned on HG001 genome (29) using the Galaxy platform (30). We used DEseq2 to estimate enrichment values for MAPS (n = 2) (P-value < 0.05; Fold change (FC) > 2) (Table 1). The whole list of RNA enriched with MS2-RsaC544 is available in Supplementary Table S3. MAPS data that support our findings are available in the GEO database with the accession code GSE128741.
Table 1.
List of RNAs significantly co-purified with MS2-RsaC544
| ID | Gene name | Function | Fold change MS2-RsaC/Ctrl | P-value |
|---|---|---|---|---|
| HG001_00569 | - | RsaC sRNA | 9.981 | 3.33E-23 |
| HG001_01477 | sodA | Superoxide dismutase SodA | 8.088 | 2.44E-18 |
| HG001_01627 | gloB_1 | Hydroxyacylglutathione hydrolase | 5.406 | 4.20E-13 |
| HG001_02152 | fba | Fructose-bisphosphate aldolase | 4.082 | 4.84E-10 |
| HG001_00556 | sarA | Transcriptional regulator SarA | 3.156 | 1.88E-06 |
| HG001_00934 | ptsI | Phosphoenolpyruvate-protein phosphotransferase | 3.122 | 5.80E-07 |
| HG001_00252 | - | FtsX-like permease family protein | 3.093 | 5.72E-07 |
| HG001_02633 | envR | DNA-binding transcriptional regulator EnvR | 2.877 | 7.08E-06 |
| HG001_00671 | fecD_1 | Fe(3+) dicitrate transport system permease protein FecD | 2.721 | 4.27E-05 |
| HG001_00933 | ptsH | Phosphocarrier protein HPr | 2.692 | 1.30E-05 |
| HG001_01479 | znuB | High-affinity zinc uptake system membrane protein ZnuB | 2.642 | 4.16E-05 |
| HG001_00967 | - | Cysteine-rich secretory protein family protein | 2.613 | 2.67E-05 |
| HG001_01416 | srrB | Sensor protein SrrB | 2.576 | 2.71E-05 |
| HG001_02376 | - | Hypothetical protein | 2.491 | 8.80E-05 |
| HG001_01478 | zur | Zinc-specific metallo-regulatory protein Zur | 2.446 | 0.000206 |
| HG001_01220 | msrR | Regulatory protein MsrR | 2.443 | 9.65E-05 |
| HG001_01721 | menE | 2-succinylbenzoate–CoA ligase | 2.351 | 0.000382 |
| HG001_02519 | - | Hypothetical protein | 2.222 | 0.000486 |
| HG001_00968 | - | Hypothetical protein | 2.21 | 0.000569 |
| HG001_01480 | znuC | High-affinity zinc uptake system ATP-binding protein ZnuC | 2.176 | 0.001228 |
| HG001_02411 | - | hypothetical protein | 2.146 | 0.001107 |
| HG001_01710 | yvgN | Glyoxal reductase | 2.124 | 0.001455 |
| HG001_01657 | ccpA | Catabolite control protein A | 2.101 | 0.001521 |
| HG001_00540 | ywqN_2 | Putative NAD(P)H-dependent FMN-containing oxidoreductase YwqN | 2.065 | 0.001444 |
| HG001_01626 | nhaX | Stress response protein NhaX | 2.056 | 0.001481 |
| HG001_01845 | lytN_3 | putative cell wall hydrolase LytN precursor | 2.056 | 0.001999 |
| HG001_01925 | cobQ | Cobyric acid synthase | 2.035 | 0.002683 |
| HG001_00932 | - | Hypothetical protein | 2.029 | 0.001941 |
| HG001_02069 | rex | Redox-sensing transcriptional repressor Rex | 2.023 | 0.001773 |
| HG001_00568 | nhaK_1 | Sodium, potassium, lithium and rubidium/H(+) antiporter | 2.005 | 0.002142 |
| HG001_02589 | - | Virus attachment protein p12 family protein | 2.002 | 0.004538 |
Differential proteomic analysis on cytoplasmic proteins
Label free spectral count analysis was performed in triplicate on cytoplasmic proteins extracts prepared from tested strains using nanoLC–MS/MS. Cells were harvested after 6 h of growth in BHI (Supplementary Table S4) or in BHI-chelex supplemented or not with 25 μM of MnCl2 (Supplementary Table S5). Proteins were extracted from cytoplasm of tested strains and were digested with sequencing-grade trypsin (Promega, Fitchburg, MA, USA) as previously described (28). The samples were analyzed by nanoLC-MS/MS either on a NanoLC-2DPlus system (with nanoFlex ChiP module; Eksigent, Sciex, Concord) coupled to a TripleTOF 5600 mass spectrometer (AB Sciex) or on a QExactive+ mass spectrometer coupled to an EASY-nanoLC-1000 (Thermo-Fisher Scientific, USA). Data were searched against S. aureus HG001 database using Mascot algorithm (version 2.5, Matrix Science, London, UK) through ProlineStudio 1.4 package (http://proline.profiproteomic.fr/). Peptides were validated on Mascot pretty rank equal to 1, an ion score cut-off equal to 25, and 1% FDR on both peptide spectrum matches (PSM) and protein sets (false discovery rate-based ion score). The total number of spectra was recorded for all the proteins, allowing to quantify these proteins across all the samples (quantification by spectral count). Data were submitted to a negative-binomial test using an edgeR GLM regression through R (R v3.5.0), normalized according to a median-to-ratio method to calculate fold changes (FC) and P-values. The mass spectrometric data that support our findings are available via ProteomeXchange with identifier PXD013225.
Preparation of RNAs for in vitro experiments
PCR fragments containing T7-RsaC544, T7-RsaC544mut, T7-ldh1 (from –167 to +418), T7-rex (from –103 to +205), T7-sarA (full-length; from –147 to +375), T7-sodM (full-length; from –28 to +651), T7-sufC (full-length; from –100 to +762), T7-sufD (from –97 to +626), T7-znuB (full-length; from –60 to +786), T7-znuC (from –22 to +646) and T7-zur (full-length; from –212 to 412) were used for in vitro transcription with T7 RNA polymerase (Supplementary Table S2). We used HG001 genomic DNA as DNA template for most constructs. For T7-RsaC544 and T7-RsaC544mut, we used pCN51-P3-MS2-RsaC544 and pCN51-P3-RsaC1116mut, respectively. The transcription of full-length mRNA sodA was performed from a DraI/XhoI-digested pJET-T7-sodA plasmid (from –28 to +600). RNAs were then purified using a 6% polyacrylamide–8 M urea gel electrophoresis. After overnight elution in Elution Buffer (0.5 M ammonium acetate pH 6.5, 1 mM EDTA and 0.1% SDS), RNAs were precipitated in cold absolute ethanol and washed with 80% ethanol. RNAs were dephosphorylated using Fast AP (ThermoFisher), according to the manufacturer protocol. The labeling of the 5′ end of dephosphorylated RNA was performed with T4 polynucleotide kinase (Fermentas) and [γ32P] ATP as previously described (16).
Gel retardation assays
5′-radiolabeled RsaC544 (15000 cpm/sample, concentration <1 pM) and cold mRNAs were renaturated separately by incubation at 90°C for 1 min in Buffer GR- (20 mM Tris–HCl pH 7.5, 60 mM KCl, 40 mM NH4Cl, 3 mM DTT), cooled 1 min on ice, and incubated 15 min at RT in presence of 10 mM MgCl2. For each experiment, increasing concentrations of cold mRNA were added to the 5′ end labelled RsaC544 in a total volume of 10 μl containing Buffer GR+ (20 mM Tris–HCl pH 7.5, 60 mM KCl, 40 mM NH4Cl, 3 mM DTT, 10 mM MgCl2). Complex formation was performed at 37°C during 15 min. After incubation, 10 μl of glycerol blue was added and samples were loaded on a 6% polyacrylamide gel containing 10 mM MgCl2 under non-denaturing conditions (8 h, 300 V, 4°C). Quantification of data corresponding to RsaC544 and to RsaC544/mRNA complex was done with ImageQuant TL software (GE Healthcare Life Sciences). Under these conditions where the concentration of the labeled RNA is negligible, the Kd dissociation constant can be evaluated as the concentration of the cold RNA that showed 50% of binding. The same protocol was used with 5′-radiolabeled RsaC544mut.
Toe-printing assays
The preparation of S. aureus 30S ribosomal subunits, the formation of a simplified translational initiation complex (constituted by S. aureus 30S ribosomal subunit, initiator tRNAfMet and sodA mRNA), and the extension inhibition conditions were performed as described previously (31). Increasing concentrations of RsaC544 (25–400 nM) were used to monitor their effects on the formation of the initiation complex (250 nM of 30S S. aureus; 4 μM of tRNAfMet) with sodA mRNA (50 nM).
Primer extension assays
Primer extension assays were performed using 30 μg of total RNA, incubated with 5′-radiolabeled oligonucleotide (PE-RsaC-Rev, Supplementary Table S2), 2.5 mM dNTPs and AMV reverse transcriptase (4 units, NEB). RNA template was removed by addition of 3 µl of 3 M KOH and 20 µl of Destroy Buffer (50 mM Tris–HCl pH 8.0, 0.5% SDS, 7.5 mM EDTA) for 3 min at 90°C, followed by 1 h at 37°C. Samples were finally precipitated and migrated on a denaturing 10% polyacrylamide gel, next to sequencing ladder. The sequencing ladder was obtained with a DNA template (PCR with oligonucleotides PE-RsaC-For and PE-RsaC-Rev, from genomic DNA). Here, ddNTP were added to stop the reaction performed by the Vent (exo-) DNA polymerase (2 units, NEB). As an example, the A reaction is composed of 0.25 mM ddATP, 0.025 mM dATP, 0.05 mM dGTP, 0.05 mM dCTP and 0.05 mM dTTP. 0.1% Triton was also added to the reaction. PCR was performed as follows: 1 min at 95°C, 1 min at 52°C and 1 min at 72°C (25 cycles). The primer PE-RsaC-Rev is located at nucleotides +42 to +59 (from the +1 of RsaC).
Footprinting assays
RsaC544-sodA mRNA complex formation was carried out at 37°C for 15 min in Native Buffer (20 mM Tris–HCl pH 7.5, 10 mM MgCl2 and 150 mM KCl). Enzymatic hydrolysis was performed on the unlabelled and renatured RsaC sRNA (50 nM) in absence or in presence of sodA mRNA (75, 150 or 300 nM). The reaction was performed in Native Buffer with RNase V1 (0.5 U) or with RNase T2 (0.05 U) at 20°C for 5 min in presence of 1 μg of total tRNA. The reactions were stopped by phenol extraction followed by RNA precipitation. The enzymatic cleavages in RsaC were detected by primer extension with AMV reverse transcriptase according to Fechter et al. (32).
Determination of RNA structure with SHAPE
To elucidate the secondary structure of RsaC sRNA, SHAPE reactivity assays were performed as previously described by Rice et al. (2014) (33). RNA folding was probed with benzoyl cyanide (BzCN), which modified 2′-hydroxyl of riboses. 1 pmol of RNA was denatured and renatured in SHAPE-BzCN buffer with 2 μg of yeast tRNA. Then, BzCN was added with different final concentrations (10-50-150 mM). After 1 min at 20°C, RNA was precipitated. Thereafter, reverse transcription was performed with VIC-labelled oligonucleotides (Supplementary Table S2). In parallel, ddGTP sequencing of 1 pmole RNA was made with the same oligonucleotide labelled with NED fluorophore. cDNA samples were analysed by capillary electrophoresis and data processed by QuSHAPE software (34). SHAPE data (Supplementary Table S6) were used to constrain structure prediction by RNAstructure (35). The secondary structure of RsaC was finally visualized using VARNA (36) (Figure 1B).
RT-PCR
RT-PCR was performed using OneStep RT-PCR Kit (Qiagen) according to the manufacturer's instructions. Total RNA extracted after 4 or 6 h of growth in BHI medium was used as template (after DNase I treatment). Primers are described in Supplementary Table S2. The same experiment was performed without RT enzyme as a control.
Determination of the 5′ end status of RsaC
To determine if the 5′ end of RsaC is mono- or tri-phosphorylated, we used Terminator™ 5′-Phosphate-Dependent Exonuclease Terminator (Epicentre) according to manufacturer's protocol. Total RNA (10 μg) was incubated with Terminator Exonuclease (1U) and Terminator 1X Reaction Buffer A at 30°C for 1 h. Subsequently, we performed phenol extraction and ethanol precipitation. Northern blot analysis was performed on 1% agarose gel containing 25 mM guanidium thiocyanate.
Superoxide stress assays
Sensitivity to methyl viologen (MV) was assessed by viability test (13). Fresh BHI broth was inoculated with an overnight culture to an initial OD600nm of 0.05 and cultivated at 37°C with shaking (200 rpm). Cells were challenged with MV (10 mM) in early-exponential phase (2 h). Samples were collected at appropriate intervals post-challenge. Viability of bacteria was determined by plating onto GP agar supplemented with 5 μg/ml erythromycin. Experiments were performed in triplicate.
SOD activity assays
SOD activity assays were performed as previously described (37). Basically, when exposed to light, riboflavin produces O2.−, which reduces tetrazolium to the dark blue formazan dye. The presence of SOD enzymes hinders this reaction and can be visualized by an achromatic area (negative staining). Samples were collected after 8 h of growth in absence or presence of methyl viologen (10 mM; BHI) or after 6h in BHI-chelex ±25 μM MnCl2. Cells were then washed in PBS and lysed (8). Total protein concentration was determined by Bradford assay (Bio-Rad). SodA and SodM activities were determined by negative staining on native polyacrylamide gel (9). Equal amounts of protein (20 μg) were resolved on 12% polyacrylamide gels by electrophoresis in glycine buffer without sodium dodecyl sulfate. After the run, the gel was soaked in 1.225 mM of nitro blue tetrazolium (NBT) solution for 45 min and then washed with sterile distilled water. Subsequently, the gel was soaked in a solution containing 0.028 mM riboflavin and 28 mM TEMED (tetramethylethylene diamine) for another 45 min. The gel was exposed to light to initiate the photochemical reaction. The SOD activity was monitored as a clear zone surrounded by a dark blue background.
Endogenous ROS production
Cells were harvested (1 ml at OD600nm = 1) after 6h of growth and washed twice with PBS. The probe 2′,7′-dichlorofluorescein diacetate (Sigma) was then added to cell suspensions to a final concentration of 10 μM. Fluorescence intensity (FI) was acquired during 5 h using 96-well spectrofluorimeter (Ex/Em = 485/538 nm). The FI value was normalized to the OD600 nm of each culture. The experiment was performed in triplicate.
RESULTS
RsaC is a long and structured RNA whose length varies between isolates
We first analyzed by Northern blot the expression of RsaC in three different strains (HG001, MW2 and Newman) during growth in BHI at 37°C (Figure 1A). RsaC is mainly produced in stationary phase and its length varies depending on tested strains. To understand this variation, the 5′ end of the longest form of RsaC sRNA (in HG001) was mapped using primer extension. The experiment shows a RT stop corresponding to a fragment with a length of 1116 nt (Supplementary Figure S3).
By performing a comparative multiple sequence alignment with MW2 and Newman strains, we observed that the 3′ domain of RsaC (comprising 447 nt) is highly conserved (>95%) (Supplementary Figure S4A), while its 5′ domain differs in strains due to variation in the number of repeats (from 2 to 4; 133 nt) (Supplementary Figure S4B). A truncated repeat of 58 nt marks the end of the 5′ part of RsaC. Therefore, RsaC is 850 nt-long (2 repeats) in Newman, 984 nt-long (3 repeats) in MW2 and 1116 nt-long (longest form; four repeats) in HG001. We expanded our analysis to all bacterial genomes using a sequence similarity search program (BLASTn; Supplementary Table S7) and observed that RsaC is restricted to S. aureus and the closely related S. argenteus species. The length of RsaC sRNA is ranging from 584 nt (a single truncated repeat) to 1116 nt (four repeats and a truncated repeat). This highlights the wide variability in length and composition of the 5′ part of RsaC.
We probed the secondary structure of RsaC (from HG001 strain) using selective 2′-hydroxyl acylation of riboses with benzoyl cyanide (BzCN) (33). Resulting 2′-O-adducts were detected by primer extension and data were analysed and quantified using QuSHAPE software (34). The BzCN reactivity (Supplementary Table S6) provides information on the local nucleotide flexibility and the highest reactivities of riboses are indeed mainly located in unpaired regions. The proposed secondary structure model, which takes into account the ribose reactivities, shows that RsaC is a highly structured sRNA characterized by two large and independent domains (Figure 1B). The 5′ part is constituted of four repeat motifs, each of them carrying four distinct well-characterized stem-loop structures. The region from nucleotides +920 to +1070 was also probed with various ribonucleases specific either for unpaired nucleotides (RNase T2) or for double-stranded regions (RNase V1). These data support the proposed secondary structure model since the RNase T2 cleavages were preferentially located in the apical loops while major RNase V1 cuts occurred in paired regions (Figure 2D). The 3′ part of RsaC is characterized by two large hairpin structures interrupted by internal loops and by several interhelical unpaired regions (Figure 1B).
Figure 2.
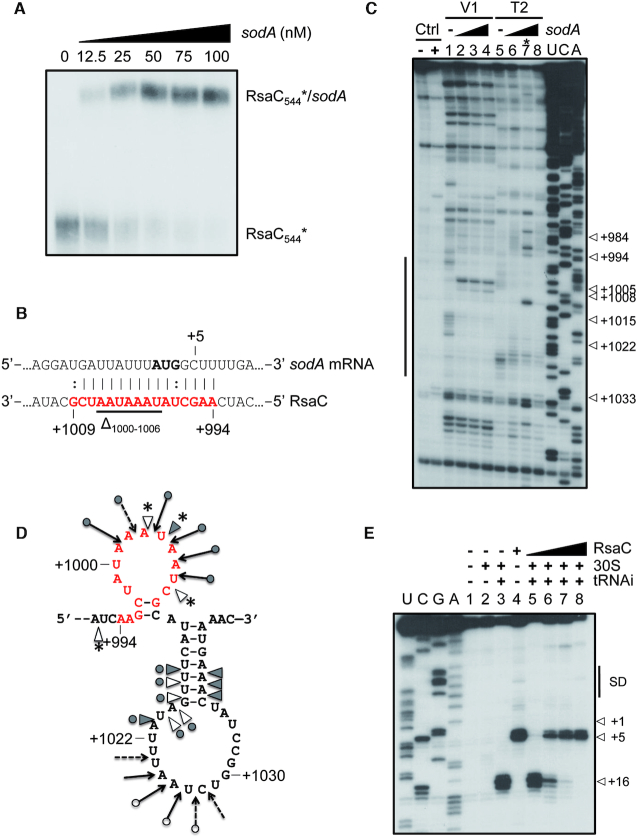
RsaC binds to sodA mRNA and blocks its translation. (A) Gel retardation assays using RsaC544 sRNA and full-length sodA mRNA. 5′ end-radiolabeled RsaC544 (*) was incubated with increasing concentrations of sodA mRNA (from 0 to 100 nM). (B) The RsaC:sodA pairing site predicted using IntaRNA software is indicated in red. The AUG start codon is in bold. Deleted nucleotides (Δ1000–1006) in RsaCmut sequence are underlined. (C) Footprint of sodA mRNA on RsaC. Incubation controls (Ctrl) were performed in absence of RNases on RsaC either free (–) or bound to sodA (50 nM, +). Reactions with either RNase V1 (V1) or RNase T2 (T2) were performed with RsaC (50 nM) in absence (lanes 1 and 5) or in presence of increasing concentrations of sodA mRNA (lanes 2–4 and 6–8; 75, 150 or 300 nM, respectively). U, C, A: sequencing ladders. *Lane 7 should not be considered due to a technical glitch. On the left side, a bar denotes changes in cleavages as the result of sodA binding. (D) Schematic representation of sodA pairing site on RsaC sRNA revealed by footprinting assays. RNase V1 cleavages are denoted by grey triangles for strong cut and empty triangles for moderate cut. RNase T2 cleavages are shown by filled arrows for strong cut and dashed arrows for weak cut. New RNase V1 cleavages are labelled by stars. The predicted pairing site by IntaRNA software is indicated in red. (E) Toe-printing assay monitoring the effect of RsaC on the formation of the ternary ribosomal initiation complex comprising S. aureus 30S subunit, initiator tRNAfMet and sodA mRNA (50 nM). Lanes 1–4: incubation controls (lane 1: sodA mRNA alone; lane 2: sodA mRNA with 30S subunit; lane 3: sodA mRNA with 30S subunit and initiator tRNAfMet (tRNAi); lane 4: sodA mRNA with RsaC); lanes 5–8: formation of the initiation complex in presence of increasing concentrations of RsaC (25, 50, 100, 400 nM). U, C, G, A: sequencing ladders; SD: Shine-Dalgarno sequence; +1: start codon; +5: RT stop induced by RsaC binding; +16: toe-printing signal. Data are representative of two independent experiments.
Characterization of RsaC targetome
To identify RsaC targets, we performed MS2-affinity purification coupled with RNA sequencing (MAPS) in ΔrsaC background complemented with MS2-RsaC544. Computer prediction suggested that this 3′ part of RsaC forms an independent and conserved domain, which would adopt the same structure as in full-length RsaC. The MS2-RsaC544 construct was expressed in vivo under the control of the quorum sensing-dependent promoter P3 and was confirmed as functional (Supplementary Figure S2). Crude extracts from ΔrsaC strain expressing MS2-RsaC544 or from WT strain expressing MS2 alone were harvested after 6h of growth in BHI medium. After MS2-affinity purification and RNAseq analysis, we identified 31 RNAs significantly enriched with MS2-RsaC544 (Fold change > 2, P-value < 0.05; Table 1). The best enrichment was for sodA mRNA (reads obtained with MS2-RsaC544 versus MS2 control), which encodes a superoxide dismutase. Moreover, other candidates code for proteins involved in oxidative stress response (e.g. sarA and rex), metal homeostasis (e.g. fecD_1, znuB, zur and znuC) and carbon metabolism (e.g. fba, ptsI, ptsH and ccpA).
To unravel RsaC effect on identified candidates, differential proteomic analysis was performed on cytoplasmic proteins extracts prepared from WT and ΔrsaC strains using LC–MS/MS coupled to spectral count analysis. Using stringent criteria (Supplementary Table S4), we did not observe major changes (Fold change <0.5 or >2). However, the level of SodA was found slightly but reproducibly decreased in WT strain as compared to ΔrsaC strain. These minor effects suggested that the expression of RsaC under tested conditions (i.e. rich medium conditions) is not optimal to reveal the full spectra of its regulatory activities.
The combination of data suggests that sodA mRNA is one of the major targets of RsaC and that its regulation would occur at the translational level. Indeed, using qRT-PCR (Supplementary Figure S5), we demonstrated that the steady-state level of sodA mRNA remains equal in WT and mutant strains as well as in ΔrsaC strain complemented with a plasmid overexpressing RsaC (∼10-fold). In the following, we focused our study on sodA mRNA target.
RsaC blocks translation initiation of sodA mRNA
Using gel retardation assays, we first demonstrated that the 5′ end radiolabeled RsaC544 forms a stable complex with sodA mRNA in vitro (Kd ≈24 nM) (Figure 2A). This result correlates well with the fact that the seed region of RsaC is located in its 3′ part (Figure 2B) and is accessible for efficient binding (Figures 1B and 2D). Indeed, the in silico prediction of the pairing site (Figure 2B) identified base pairing complementarities involving nucleotides +994 to +1009 of RsaC and –9 to +7 of sodA mRNA.
In agreement with the in silico prediction, enzymatic footprinting experiments (Figure 2C) demonstrated that binding of sodA mRNA induces changes in the unpaired region of RsaC encompassing the predicted pairing site (from A+994 to G+1009, Figure 2C and D). Our data support the existence of a double-stranded region with sodA as revealed by the appearance of new RNase V1 cleavages at positions A+1003, U+1004 and U+1007 (Figure 2D). Concomitantly, nucleotides A+1001 to A+1006 are found protected against RNase T2 hydrolysis. Remarkably, nucleotides A+999 to A+1006 are highly modified by BzCN (Figure 1B), indicating that this region is accessible for efficient pairing with sodA mRNA. Additional changes in the enzymatic cleavage patterns were also observed in distant regions of RsaC including two surrounding and accessible apical loops, encompassing nucleotides +950 to +975, and +1015 to +1030 (Figure 2D and Supplementary Figure S6A). Whether these effects might result from conformational changes of RsaC or from the formation of additional base pairing interactions with sodA mRNA, remained to be demonstrated. Notably, base pairing complementarities are predicted between nucleotides -26 to -23 at the 5′ end of sodA with nucleotides +1027 to +1030, and between nucleotides +362 to +367 in the coding sequence of sodA with nucleotides +957 to +962 of RsaC (Supplementary Figure S6B).
To further probe the importance of the pairing site of RsaC, we deleted the nucleotides +1000 to +1006 in RsaC (RsaCmut; as indicated in Figure 2B) and analyzed the ability of RsaCmut to form a complex with sodA (Supplementary Figure S7). Data showed that RsaC mutation completely abolished RsaC:sodA complex formation. This demonstrates that the sequestration of the initiation codon of sodA is the critical site, which confers high stability to the complex.
Because RsaC sequesters the AUG initiation codon of sodA mRNA, we further addressed the consequences of RsaC binding on sodA translation initiation. Toe-printing assays were used to monitor the formation of the ternary ribosomal initiation complex (Figure 2E). We observed that RsaC efficiently hinders translation initiation to the SD sequence of sodA mRNA. Indeed, the toe-print signal (+16) fades when RsaC and sodA are equimolar and completely disappears when RsaC is supernumerary (Figure 2E, lanes 4–8). Simultaneously, we noticed a RT stop at nucleotide +5 in presence of RsaC, hinting that RsaC pairs near the AUG start codon
All in all, our data showed that RsaC binds to the translation initiation site of sodA mRNA and, consequently, prevents the recruitment of ribosomes to inhibit translation initiation.
RsaC modulates oxidative stress resistance
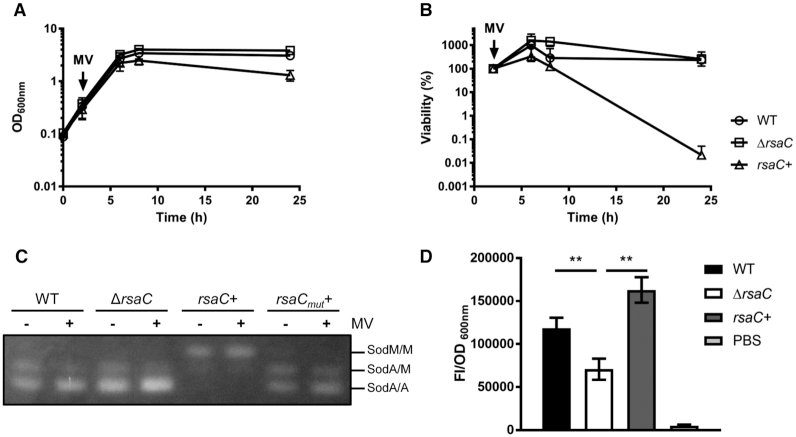
As SodA is involved in oxidative stress resistance, we analyzed if the expression of RsaC could influence it. Hence, WT/pCN51-P3 (WT), ΔrsaC/pCN51-P3 (ΔrsaC) and ΔrsaC/pCN51-P3-RsaC1116 (rsaC+) strains were cultured in BHI (2h) and exposed to the ROS-generating reagent methyl viologen (MV) (Figure 3), which is commonly used to mimic the oxidative stress encountered in vivo. Growth (OD600nm; Figure 3A) and cell viability (Figure 3B) were measured for 24 h. While the deletion of rsaC had no impact on bacterial growth, the overexpression of RsaC induced a significant growth defect and rendered cells highly sensitive to oxidative stress. Hence, in presence of high level of RsaC, bacteria hardly survived to the internal oxidative stress induced by MV.
Figure 3.
RsaC is involved in oxidative stress response via the repression of SOD activity. (A) Growth monitoring of WT/pCN51-P3 (○), ΔrsaC/pCN51-P3 (□) and ΔrsaC/pCN51-P3-RsaC1116 (rsaC+) (Δ) upon internal superoxide stress (10 mM methyl viologen (MV)). MV was added after 2h of growth. (B) Viability assays of WT/pCN51-P3 (○), ΔrsaC/pCN51-P3 (□) and ΔrsaC/pCN51-P3-RsaC1116 (rsaC+) (Δ) grown in BHI and challenged with 10 mM MV at early-exponential phase (2 h). Samples were taken thereafter, and viability was determined by performing viable counts on GP plates containing 5 μg/ml erythromycin. All data shown represent mean ± SD of three independent experiments. (C) Monitoring of SOD activity in WT/pCN51-P3 (WT), ΔrsaC/pCN51-P3 (ΔrsaC), ΔrsaC/pCN51-P3-RsaC1116 (rsaC+) and ΔrsaC/pCN51-P3-RsaC1116mut (rsaCmut+) after 8h of growth (in presence or absence of MV). SOD activity was determined by negative staining on native polyacrylamide gel (NBT/riboflavin method). Results are representative of two independent experiments. (D) Measurement of reactive oxygen species (ROS) accumulation using fluorescence assays (after 6 h of growth in TSB) in WT/pCN51-P3 (black), ΔrsaC/pCN51-P3 (white) and ΔrsaC/pCN51-P3-RsaC1116 (rsaC+) (gray). PBS buffer (light grey) was used as a control. Data represent mean ± SD of three independent experiments. Statistical analysis with ANOVA, ** P<0.005. Strains and plasmid constructs are given in Supplementary Table S1.
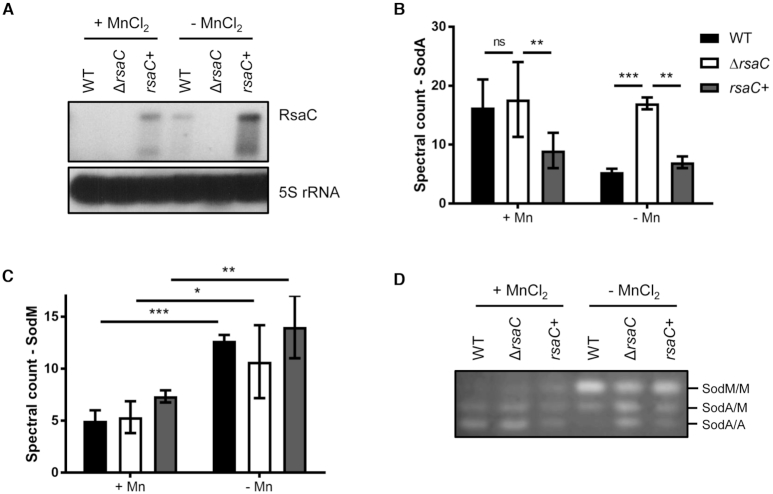
To understand the relationship between oxidative stress resistance and the cellular level of RsaC sRNA, we performed SOD activity assays in presence and absence of MV in the same strains (Figure 3C). As it was previously published (38), three bands corresponding to SodM homodimers (top), SodA/SodM heterodimers (middle) and SodA homodimers (bottom) were detected. In tested conditions, the first band was hardly visible in WT background suggesting that SodM homodimer contribution is marginal compared to SodA homodimer. While no clear difference was observed between WT and ΔrsaC mutant, SodA activity was nearly abolished in presence of overexpressed RsaC while SodM activity was enhanced (Figure 3C). Interestingly, the RsaC-dependent regulation of SOD activity was hindered in a strain overexpressing RsaC1116mut, which carries the deletion of sodA mRNA pairing site (from nucleotides +1000 to +1006; Figure 2B). This experiment showed that the repression of SodA synthesis is dependent on RsaC:sodA mRNA interaction in vivo.
As SOD activity assays are semi-quantitative, we quantified internal ROS level using fluorescence assays. We noticed the presence of higher ROS levels in WT strain than in ΔrsaC mutant (Figure 3D) suggesting that SOD activity is enhanced in ΔrsaC mutant. In presence of overexpressed full-length RsaC (Figure 3D) or its 3′ domain (Supplementary Figure S2), the accumulation of ROS was even higher than in WT strain. These results demonstrated that ROS level was correlated to enhance RsaC synthesis.
Because oxidative stress is a defence mechanism used by host cells to kill pathogens, we tested the ability to persist in macrophages (Supplementary Figure S8A) or to resist to opsonophagocytosis mediated by neutrophils (Supplementary Figure S8B). We observed that ΔrsaC mutant strain tends to be more persistent in macrophages and more resistant to opsonophagocytosis than WT strain. Conversely, the rsaC overexpressing strain was less persistent in macrophages and less resistant to opsonophagocytosis. Even if these observations are indirect, they are consistent with a higher SodA activity in absence or limited concentration of RsaC.
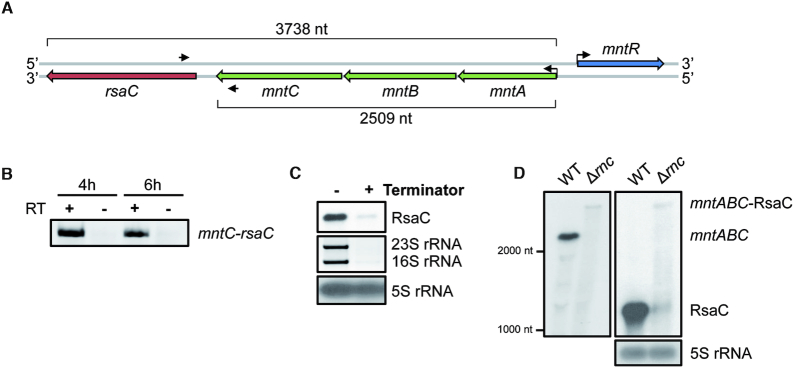
RsaC is released from the 3′UTR of mntABC operon
Our data suggest that RsaC is a noxious sRNA within cells, as high RsaC level renders them sensitive to oxidative stress. Hence, RsaC must be only produced under specific conditions and its endogenous expression should be firmly controlled. rsaC gene is located downstream the mntABC operon coding for the major manganese ABC transporter (Figure 4A). MntA is the ATP-binding protein, MntB the permease, and MntC the metal binding protein. The synteny is conserved in S. aureus and in closely related S. argenteus genomes (Supplementary Table S7) and only 113 nt separate mntC and rsaC genes, hinting that RsaC could be part of mntABC operon. By performing RT-PCR with primers in both RsaC and mntC (Figure 4A, B), we found that RsaC is co-transcribed with mntABC operon. We then checked if RsaC is issued from the maturation of the mntABC-RsaC precursor or from an internal promoter. The 5′-phosphate-dependent exonuclease Terminator fully digested RsaC as the large processed rRNAs (Figure 4C). Because this enzyme degrades only processed transcripts, we concluded that the 5′ end of RsaC is monophosphorylated. Previous works revealed that RsaC might be a target of the double-stranded endoribonuclease III (RNase III) (39). Northern blot analysis on RsaC performed in WT and Δrnc (RNase III mutant) strains (Figure 4D, right panel) revealed that in absence of RNase III, RsaC signal drastically drops while a far larger band appears (>3 kb). Using a probe complementary to mntA (Figure 4D, left panel), we confirmed that this very large RNA corresponds to the unprocessed mntABC-RsaC transcript.
Figure 4.
rsaC is part of the polycistronic operon mntABC and is released after RNase III cleavage. (A) Genomic context of mntABC operon. Oligonucleotides used for RT-PCR are indicated with black arrows. (B) RT-PCR using oligonucleotides in both mntC and rsaC genes and total RNA extracted after 4 or 6 h of growth in BHI medium. Same experiment was performed without RT enzyme (–) as a control. (C) Determination of the 5′ end status of RsaC sRNA. 10 μg of total RNA extracted after 6 h of growth in BHI medium were treated with the Terminator™ 5′-Phosphate-Dependent Exonuclease. 16S and 23 rRNAs were used as a positive control and 5S rRNA as a negative control. (D) Northern blot analysis of mntABC-RsaC transcript in wild type strain (WT, HG001) and the isogenic RNase III mutant (Δrnc) strain. Total RNA was extracted after 6 h of growth in BHI at 37°C. We used a probe targeting mntA (left panel) or RsaC (right panel). 5S rRNA was used as loading control. Data are representative of three independent experiments.
Because transcription of mntABC is repressed by the cognate repressor MntR in presence of Mn (40), we verified that RsaC level varies in function of Mn bioavailability (Figure 5A). Tested strains were grown in BHI-chelex medium supplemented or not with 25 μM MnCl2 for 6 h at 37°C. In absence of Mn, we observed a strong increase of endogenous RsaC level, which is not visible in presence of Mn in WT strain. Only a weak signal was detected after overexposition of the autoradiography (Supplementary Figure S9A). Conversely, when BHI medium was supplemented with 25 or 100 μM MnCl2 (Supplementary Figure S9B), RsaC synthesis was drastically reduced.
Figure 5.
RsaC is involved in Mn homeostasis. (A) Northern blot analysis of RsaC level in WT/pCN51-P3 (WT), ΔrsaC/pCN51-P3 (ΔrsaC) and ΔrsaC/pCN51-P3-RsaC1116 (rsaC+) strains. Total RNA was extracted after 6 h of growth in BHI-chelex supplemented or not with 25 μM MnCl2 at 37°C. 5S rRNA was used as loading control. When RsaC is highly produced, a shorter form is observed. Data are representative of three independent experiments. Differential proteomic analysis of SodA (B) and SodM (C) in presence or absence of 25 μM MnCl2 (BHI-chelex). Data represent spectral count mean ± SD of three independent experiments. Statistical analysis is based on a negative-binomial test using an edgeR GLM regression through R, *P< 0.05; **P< 0.005; ***P< 0.0005; ns: not significant. (D) Monitoring of SOD activity in WT/pCN51-P3 (WT), ΔrsaC/pCN51-P3 (ΔrsaC) and ΔrsaC/pCN51-P3-RsaC1116 (rsaC+) after 6h of growth (in BHI-chelex ± 25 μM MnCl2). SOD activity was determined by negative staining on native polyacrylamide gel (NBT/riboflavin method). Results are representative of three independent experiments. Strains and plasmid constructs are given in Supplementary Table S1.
RsaC modulates oxidative stress response upon Mn starvation
SodA is the major SOD enzyme in rich medium but is supplanted by SodM notably when cells are Mn-starved (10). In Figure 3C, we noticed that the overexpression of RsaC completely abolished SodA activity, but at the opposite increased SodM activity. While sodA mRNA was validated as a main target of RsaC, sodM mRNA was not enriched with RsaC in MAPS (Table 1). Moreover, RsaC interacts with sodM mRNA albeit with a very poor affinity in vitro (>500 nM) compared to sodA mRNA (≈24 nM) due to a partial conservation of the pairing region (Supplementary Figure S10). Hence, RsaC-dependent regulation of SodM activity must be indirect.
Beside Northern blot analysis performed in BHI-chelex ± 25 μM MnCl2 (Figure 5A), the cytoplasmic proteins content was monitored using mass spectrometry (Supplementary Table S5). We observed that MntC is highly produced in absence of Mn (Supplementary Figure S11), confirming that cells are Mn starved. Focusing on SOD enzymes, no variation in SodA level was found in WT and ΔrsaC background in presence of 25 μM MnCl2 when RsaC is barely detectable (Figure 5A, B). Only the overexpression of RsaC from a Mn-independent promoter had a negative impact on SodA synthesis (Figure 5B). In absence of Mn, SodA synthesis was reduced (∼3-fold) in strains where RsaC was either endogenously produced or overproduced (Figure 5B). RsaC had no significant effect on SodM synthesis although a tendency to increased SodM yield is observed when RsaC is overexpressed (Figure 5C). Remarkably, a clear increase of SodM level was noticed in absence of Mn (∼2.5-fold) (Figure 5C).
We then verified if SOD activities were correlated to proteins level (Figure 5D). Proteins were extracted in the same conditions as above. As expected, SodA activity was maximal when Mn was present, but was strongly impaired by high levels of RsaC. Activities of SodA homodimers and SodA/SodM heterodimers remained very low when RsaC was produced from a plasmid or from the chromosome in response to Mn starvation (Figure 5D). Interestingly, SodM homodimer activity strongly increased in absence of Mn and was significantly enhanced in WT and complemented (rsaC+) strains.
Our data showed that Mn starvation strongly induces RsaC expression, leading to a specific repression of SodA synthesis and to an enhanced activity of SodM, which is most likely occurring in an indirect manner.
RsaC has a broader role in oxidative stress response and metal homeostasis
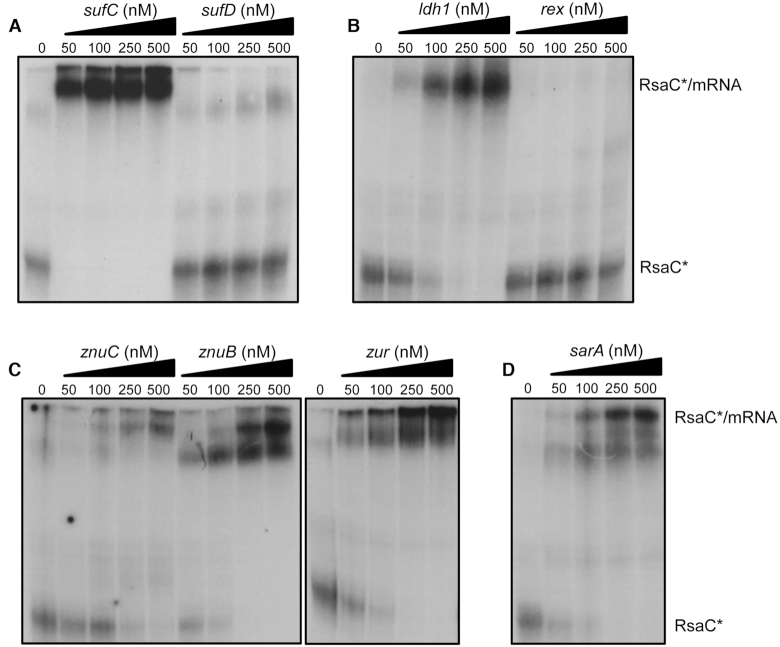
In the differential proteomic analysis (Supplementary Table S5), we identified additional proteins, such as SufC, SufD and Ldh1, whose synthesis follows a regulatory pattern similar to SodA protein (Supplementary Figure S12). Indeed, their synthesis was significantly repressed in conditions where RsaC level is high (i.e. WT/ΔrsaC (-Mn), rsaC+/ΔrsaC (–Mn), rsaC+/ΔrsaC (+Mn) and WT (–Mn)/WT (+Mn)) and remained equal when RsaC level is low (WT/ΔrsaC (+Mn)). These proteins code for components of the Suf iron-sulfur cluster biosynthetic system and the lactate dehydrogenase 1, respectively (Supplementary Figure S12). None of them were co-purified with MS2-RsaC544 (Table 1). However, MAPS experiments were performed after 6h of growth in BHI, in which Mn concentration was not controlled, hinting that some targets could have been missed. Therefore, we performed gel retardation assays to analyze the binding of RsaC to several other mRNAs, an indication that RsaC might regulate the synthesis of corresponding proteins (Figure 6). Our data show that RsaC efficiently binds to sufC transcript (Figure 6A) and ldh1 mRNA (Figure 6B). We suggest that RsaC pairing with sufC might be sufficient to regulate the whole sufCDSUB operon.
Figure 6.
RsaC potentially regulates additional mRNA targets involved in oxidative stress response and metal homeostasis. Gel retardation assays using RsaC544 sRNA and several mRNA targets identified by MAPS and/or proteomic analysis. 5′ end-radiolabeled RsaC544 (*) was incubated with increasing concentrations of (A) sufC and sufD, (B) ldh1 and rex, (C) znuC, znuB and zur, and (D) sarA mRNA (from 0 to 500 nM). Data are representative of two independent experiments. All transcripts have been described in Material and Methods.
Most of the targets enriched by MAPS were not or only poorly detected in our proteomic analysis (Supplementary Table S5). Nevertheless, we also tested the interaction of RsaC with several enriched mRNA candidates identified by MAPS that are involved in related functions. These mRNAs encode the zinc uptake system and the transcriptional regulators SarA, Zur and Rex, all related to metal homeostasis and oxidative stress (Table 1). Using gel retardation assays, we observed that RsaC binds to every member of znuB-znuC-zur operon and sarA mRNAs, but not with rex mRNA (Figure 6).
Using IntaRNA software (41), pairing sites were predicted between RsaC and each targeted mRNAs validated by gel retardation assays (Supplementary Figure S13). RsaC potentially binds to the ribosome binding site of sufC, ldh1, znuC, znuB, zur and sarA mRNAs, using several exposed C/U-rich sequences located in its 3′ part. Consequently, RsaC could repress the translation of additional targets, mainly involved in oxidative stress response and metal homeostasis.
DISCUSSION
RsaC, a connection node between oxidative stress and manganese bioavailability
In this study, we have shown that RsaC is a long atypical sRNA conserved in all S. aureus strains (Supplementary Table S7), characterized by two structured and independent domains (Figure 1). We showed that its 3′ domain was sufficient to reduce the translation of its major target SodA, identified by a combination of approaches including MAPS. Found in all staphylococci, SodA is the major SOD enzyme which is crucial to protect cells against damage caused by ROS produced upon reduction of O2 or by immune cells (oxidative burst). We demonstrated that RsaC specifically pairs to the ribosome binding site of sodA mRNA to block its translation (Figure 2). RsaC does not use a classical C-rich sequence to base-pair with sodA mRNA as it was shown for many S. aureus sRNAs (42). Indeed, RsaC binding site overlaps the AUG start codon of sodA mRNA, not its Shine-Dalgarno sequence. This RsaC-dependent repression of sodA might explain why cells expressing a high level of RsaC are very sensitive to internally induced ROS production (Figure 3) and more sensitive to host immune attacks (Supplementary Figure S8). However, these harmful effects were mostly due to artificial overexpression of RsaC. Indeed, we hardly noticed an effect of rsaC mutation on S. aureus oxidative stress resistance when compared to WT. Only opsonophagocytosis assays (Supplementary Figure S8B) showed that endogenously synthesized RsaC reduced cells survival but this cannot be confidently attributed to a greater sensitivity to oxidative stress generated by immune cells.
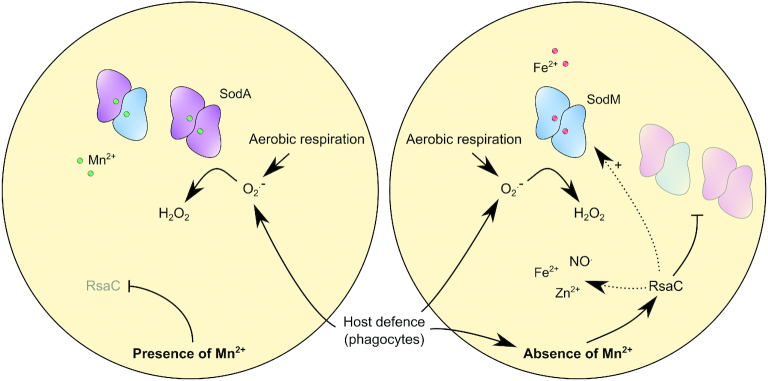
We showed that RsaC level increased considerably in manganese-starved cells along with a drastic reduction of SodA activity indicating that RsaC exerts its regulatory activities under these specific physiological conditions (Figure 5). In S. aureus, SodM is an alternative and cambialistic SOD enzyme, which can utilize either Mn or Fe as cofactor (10). SodM compensates for the lack of functional SodA in manganese-starved cells and promotes resistance to oxidative stress (10,13). We confirmed that SodM becomes the major SOD enzyme in response to Mn limitation but also to RsaC level (Figures 3C and 5D). Unlike SodA, SodM is restricted to S. aureus and S. argenteus strains and potentially confers them a selective advantage over other staphylococci in human tissue environment. Remarkably, rsaC gene was also acquired exclusively by S. aureus and S. argenteus species. The proposed model is that, upon Mn starvation, RsaC negatively regulates the Mn-containing SodA enzyme to spare Mn and avoid translation of a useless enzyme. The absence of SodA also indirectly favors the formation of SodM homodimers, key players of the alternative ROS detoxification pathway, which is activated by both Mn starvation and RsaC (Figure 7). This situation is reminiscent of iron homeostasis in E. coli (43,44) since RyhB sRNA re-establishes iron homeostasis when iron is limiting. In E. coli, SodA and SodB synthesis are utterly opposed as RyhB negatively regulates the non-essential iron-containing SodB but not the Mn-dependent SodA enzyme (45,46). Consequently, SodA replaces SodB in iron-starved cells. However, the role of RyhB in oxidative stress response is still poorly explored.
Figure 7.
RsaC sRNA modulates oxidative stress response during Mn starvation. SodA is a Mn-dependent superoxide dismutase which is crucial to response to oxidative stress generated by aerobic respiration or by host defence cells (e.g. macrophages, neutrophils). In absence of Mn, RsaC is highly produced and negatively regulates sodA mRNA translation. It enables to spare Mn for essential Mn-containing proteins and avoids the synthesis of a non-functional enzyme. SodM, an alternative SOD enzyme using Fe as co-factor, replaces SodA to re-establish the ROS detoxification pathway. RsaC may indirectly activate SodM synthesis via the repression of the transcriptional regulator SarA. In addition, RsaC could have a broader role in oxidative stress response, notably through the regulation of Fe and Zn homeostasis (transport and storage) and NO. resistance.
We propose that RsaC directly links oxidative stress response and nutritional immunity, both encounter at infection sites. Nutritional immunity refers to the sequestration of metals (i.e iron, manganese or zinc) by host organism to limit the growth of pathogens, which acquire these metals directly from host organism during infection (47–50). These metals are essential for proteins folding and for numerous enzymatic functions as cofactors (e.g. DNA synthesis, respiration or response to reactive oxygen and nitrogen species). The major innate immune effector involved in Mn depletion is the calprotectin (CP) protein, which sequesters Mn2+ with a high affinity (Kd < 1.3 nM) (5). Neutrophils release a large amount of CP (50 μM) to completely restrict Mn bioavailability at infection sites. For example, staphylococcal abscesses are completely Mn-depleted (51). To counteract Mn limitation, S. aureus has two Mn uptake systems, the ABC transporter MntABC and the proton-dependent Nramp homolog MntH (25,52). Both are under the control of the Mn2+-sensing transcriptional regulator MntR (52). The MntR-mediated repression of mntABC is released in Mn-starved cells while mntH expression seems to be constitutive and activated by MntR-Mn2+. The major manganese transporter MntABC not only promotes resistance to Mn starvation but also contributes to oxidative stress response and virulence (25,53). Indeed, Mn depletion by CP severely impacts SOD activity (5), lowering the resistance of S. aureus to oxidative burst generated by immune cells.
Remarkably, RsaC is co-transcribed with MntABC and, consequently, is repressed by MntR in presence of Mn. We showed that RsaC is released and stabilized from mntABC-RsaC operon after cleavage by RNase III. RNase III is involved in numerous functions including 5′UTR overlapping mRNAs processing, mRNA processing and turnover, and sRNA-mediated regulation (54). Here, we identified a new function of RNase III, which is responsible for sRNA release from its large precursor. The precise mechanism requires further analysis but the large 5′ hairpin structure encompassing the +1 of RsaC might be part of the binding site of RNase III (Figure 1B). Alternatively, two antisense RNAs have been previously identified (39,55) in this region, which can serve as guides for the RNase III-dependent cleavage. Indeed, we previously co-purified a fragment of 246 nt encompassing the +1 of RsaC (from +176 to –71 nt) and an antisense RNA (as_RsaC) using RNase III as bait in S. aureus RN6390 (39). Furthermore, another antisense RNA has been identified in this region in S. aureus SH1000, SbrC (55). This σB-dependent ∼310 nt-long RNA also overlaps the +1 of RsaC but predicted positions differ (from +12 to –297 nt). Attempts to validate its extremities were unsuccessful, presumably due to the too low level of SbrC. RsaC is the first example of 3′’UTR-derived sRNA in S. aureus, but in other bacteria several sRNAs were found released from the 3′UTR of precursor mRNAs by the single-strand specific endoribonuclease RNase E (56–58). Notably, SorX sRNA is co-transcribed with a gene encoding an OmpR-like transcriptional regulator in the α-Proteobacteria Rhodobacter sphaeroides (58). SorX counteracts oxidative stress (singlet oxygen and organic hydroperoxides) presumably by regulating a polyamine transporter.
Other physiological consequences of RsaC-dependent regulation
Using MAPS technology, 31 mRNAs were significantly enriched with RsaC (Table 1), suggesting that they could interact with RsaC. Here, not only SodA is involved in oxidative stress response, but also the staphylococcal accessory regulator SarA (59,60) and the redox-sensing transcriptional repressor Rex (61). SarA represses trxB, coding for the thioredoxin reductase, which is involved in ROS scavenging (60). SarA is also a transcriptional repressor of sodM (59). It is tempting to speculate that via sarA translational repression, RsaC could increase SodM level in response to Mn starvation, as suggested by the activation of SodM activity in presence of RsaC (Figures 3C and 5D). Remarkably, we demonstrated that RsaC binds to sarA mRNA in vitro and, according to the in silico prediction, could negatively regulate its translation by base pairing with its SD sequence (Figure 6D and Supplementary Figure S13). Therefore, via the repression of SarA, RsaC might indirectly activate SodM synthesis and, by extent, its cellular activity.
In addition to ROS, phagocytes produce nitrogen-based radicals such as nitric oxide (NO.) (1). Both SodA and MntABC somehow contribute to NO. resistance so that S. aureus can adapt its metabolism to limit deleterious effects (62). At high level, NO. disrupts respiration and, consequently, perturbs the metabolic state, notably NAD+/NADH homeostasis. In response, Rex is inactivated, and cells switch to several metabolic pathways allowing NAD+ regeneration and ATP synthesis (61,63). For example, the lactate dehydrogenase Ldh1, which belongs to Rex regulon, enables the shift to heterolactic fermentation (61). The synthesis of Ldh1 protein appears to be repressed by RsaC (Supplementary Figure S12C). Moreover, we demonstrated that RsaC binds to ldh1 mRNA and potentially block its translation (Figure 6B and Supplementary Figure S13). The level of other proteins involved in NO. stress resistance and belonging to Rex regulon (Supplementary Figure S14) is slightly decreased in response to Mn depletion in WT strain but does not respond to RsaC (Supplementary Table S5). Although, rex mRNA was copurified with MS2-RsaC544, RsaC does not bind to rex mRNA in vitro (Figure 6B). Further studies will be required to better address the involvement of RsaC in redox homeostasis.
MAPS also revealed putative interactions with mRNAs encoding Fe-related proteins (Table 1). Metal ions are essential for all forms of life, but their over-accumulation is toxic to cells. For example, excess of Fe leads to ROS production via Fenton reaction (64). Hence, it is not surprising that the Suf-dependent Fe-S cluster synthesis is negatively regulated by RsaC (SufC and SufD; Supplementary Figure S12). We also validated a pairing of RsaC with sufCDSUB operon (Figure 6A), where RsaC seems to interact with the SD sequence of sufC mRNA (Supplementary Figure S13). Moreover, RsaC potentially controlled a Fe ABC transporter (fecD_1; Table 1). Although we cannot distinguish between positively and negatively regulated targets using MAPS, we can assume that RsaC might suspend iron import and promote iron storage in order to reduce oxidative stress generated by free intracellular iron. Upon manganese starvation, the iron-binding ferritin-like Dps is actively synthetized and YfmC_2, which is part of a Fe3+-citrate ABC transporter, is negatively regulated (Supplementary Table S5), supporting our assumption. Here, S. aureus likely uses both RsaC-dependent and -independent regulatory mechanisms to regulate Fe homeostasis. Putative interactions revealed by MAPS also highlighted a potential role of RsaC in the regulation of zinc uptake, i.e. znuC-znuB-zur operon. According to the Irving-Williams series (65), the stability of complexes formed by Mn2+ is very poor compared to Zn2+. Therefore, an overabundance of Zn is toxic due to competition for binding to Mn-dependent enzymes. Both MntC and MntR are particularly vulnerable to an irreversible Zn2+ binding, affecting Mn homeostasis (66,67). Therefore, S. aureus must limit the presence of intracellular Zn2+. Although we could not detect the protein involved in Zn uptake (Supplementary Table S5), certainly due to the presence of 25 μM ZnCl2 in the medium, interactions with znuC, znuB and zur were validated in vitro (Figure 6C). Therefore, RsaC could hinder zinc import by blocking the translation of Znu transporter (Supplementary Figure S13). Besides, the RsaC-dependent control of Zur cellular level could lead to the derepression of its whole regulon. Further investigations will be necessary to understand the role of RsaC in metal homeostasis.
Links between oxidative stress and metal ion homeostasis are tightly regulated by metal-dependent transcriptional regulators such as PerR (68), Fur (69), Zur (70) and MntR (52). Notably, PerR controls oxidative stress resistance and iron storage in response to Mn2+ and Fe2+ (68,71). Again, this demonstrates the interdependency between oxidative stress response and metal homeostasis, but also the need to balance relative intracellular concentrations of Mn, Fe and Zn.
In this manuscript, we characterized a singular sRNA as a connection node between Mn bioavailability and ROS detoxification. Nevertheless, we expect that RsaC could have a wider role in oxidative stress response, via the regulation of metals homeostasis and NO. resistance. It also remains to be addressed whether the unusual repeat sequences in the 5′ domain of RsaC might play additional functional roles.
DATA AVAILABILITY
MAPS data have been deposited to GEO under accession GSE128741.
The mass spectrometric data were deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (72) with the dataset identifier PXD013225.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Thomas E. Kehl-Fie (University of Illinois, USA) for fruitful discussions. We thank Anne-Catherine Helfer for her technical support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
labEx NetRNA [ANR-10-LABX-0036 to P.R., ANR-17-EURE-0023 to P.R.] as part of the investments for the future program from the state managed by the French National Research Agency (ANR); ANR [ANR-16-CE11-0007-01 to P.R. and F.V.]; European Union's Horizon 2020 research and innovation programme [753137 to D.L.]; Mass spectrometry instrumentation was funded by University of Strasbourg, IdEx Equipement mi-lourd 2015 and labEx NetRNA. Funding for open access charge: RIBOSTAPH [ANR-16-CE11-0007-01].
Conflict of interest statement. None declared.
REFERENCES
- 1. Flannagan R.S., Heit B., Heinrichs D.E.. Antimicrobial mechanisms of macrophages and the immune evasion strategies of Staphylococcus aureus. Pathogens. 2015; 4:826–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boe D.M., Curtis B.J., Chen M.M., Ippolito J.A., Kovacs E.J.. Extracellular traps and macrophages: new roles for the versatile phagocyte. J. Leukocyte Biol. 2015; 97:1023–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brinkmann V., Zychlinsky A.. Neutrophil extracellular traps: is immunity the second function of chromatin. J. Cell biol. 2012; 198:773–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Imlay J.A. Pathways of oxidative damage. Annu. Rev. microbiol. 2003; 57:395–418. [DOI] [PubMed] [Google Scholar]
- 5. Kehl-Fie T.E., Chitayat S., Hood M.I., Damo S., Restrepo N., Garcia C., Munro K.A., Chazin W.J., Skaar E.P.. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe. 2011; 10:158–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Imlay J.A. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol. 2013; 11:443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Abreu I.A., Cabelli D.E.. Superoxide dismutases-a review of the metal-associated mechanistic variations. Biochim. Biophys. Acta. 2010; 1804:263–274. [DOI] [PubMed] [Google Scholar]
- 8. Clements M.O., Watson S.P., Foster S.J.. Characterization of the major superoxide dismutase of Staphylococcus aureus and its role in starvation survival, stress resistance, and pathogenicity. J. Bacteriol. 1999; 181:3898–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Valderas M.W., Gatson J.W., Wreyford N., Hart M.E.. The superoxide dismutase gene sodM is unique to Staphylococcus aureus: absence of sodM in coagulase-negative staphylococci. J. Bacteriol. 2002; 184:2465–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garcia Y.M., Barwinska-Sendra A., Tarrant E., Skaar E.P., Waldron K.J., Kehl-Fie T.E.. A Superoxide dismutase capable of functioning with iron or manganese promotes the resistance of staphylococcus aureus to calprotectin and nutritional Immunity. PLoS Pathog. 2017; 13:e1006125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mandell G.L. Catalase, superoxide dismutase, and virulence of Staphylococcus aureus. In vitro and in vivo studies with emphasis on staphylococcal–leukocyte interaction. J Clin Invest. 1975; 55:561–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kanafani H., Martin S.E.. Catalase and superoxide dismutase activities in virulent and nonvirulent Staphylococcus aureus isolates. J. Clin. Microbiol. 1985; 21:607–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karavolos M.H., Horsburgh M.J., Ingham E., Foster S.J.. Role and regulation of the superoxide dismutases of Staphylococcus aureus. Microbiology. 2003; 149:2749–2758. [DOI] [PubMed] [Google Scholar]
- 14. Somerville G.A. Staphylococcus: Genetics and Physiology. 2016; [Google Scholar]
- 15. Wagner E.G., Romby P.. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv.Genet. 2015; 90:133–208. [DOI] [PubMed] [Google Scholar]
- 16. Boisset S., Geissmann T., Huntzinger E., Fechter P., Bendridi N., Possedko M., Chevalier C., Helfer A.C., Benito Y., Jacquier A. et al.. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007; 21:1353–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huntzinger E., Boisset S., Saveanu C., Benito Y., Geissmann T., Namane A., Lina G., Etienne J., Ehresmann B., Ehresmann C. et al.. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005; 24:824–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bronesky D., Wu Z., Marzi S., Walter P., Geissmann T., Moreau K., Vandenesch F., Caldelari I., Romby P.. Staphylococcus aureus RNAIII and its regulon link quorum sensing, stress responses, metabolic adaptation, and regulation of virulence gene expression. Annu. Rev. Microbiol. 2016; 70:299–316. [DOI] [PubMed] [Google Scholar]
- 19. Chevalier C., Boisset S., Romilly C., Masquida B., Fechter P., Geissmann T., Vandenesch F., Romby P.. Staphylococcus aureus RNAIII binds to two distant regions of coa mRNA to arrest translation and promote mRNA degradation. PLoS Pathog. 2010; 6:e1000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chabelskaya S., Bordeau V., Felden B.. Dual RNA regulatory control of a Staphylococcus aureus virulence factor. Nucleic Acids Res. 2014; 42:4847–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gupta R.K., Luong T.T., Lee C.Y.. RNAIII of the Staphylococcus aureus agr system activates global regulator MgrA by stabilizing mRNA. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:14036–14041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Szafranska A.K., Oxley A.P., Chaves-Moreno D., Horst S.A., Rosslenbroich S., Peters G., Goldmann O., Rohde M., Sinha B., Pieper D.H. et al.. High-resolution transcriptomic analysis of the adaptive response of Staphylococcus aureus during acute and chronic phases of osteomyelitis. MBio. 2014; 5:e01775-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kreiswirth B.N., Lofdahl S., Betley M.J., O’Reilly M., Schlievert P.M., Bergdoll M.S., Novick R.P.. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983; 305:709–712. [DOI] [PubMed] [Google Scholar]
- 24. Monk I.R., Shah I.M., Xu M., Tan M.W., Foster T.J.. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. MBio. 2012; 3:e00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kehl-Fie T.E., Zhang Y., Moore J.L., Farrand A.J., Hood M.I., Rathi S., Chazin W.J., Caprioli R.M., Skaar E.P.. MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect. Immun. 2013; 81:3395–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Arnaud M., Chastanet A., Debarbouille M.. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 2004; 70:6887–6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lalaouna D., Desgranges E., Caldelari I., Marzi S.. Chapter Sixteen - MS2-Affinity Purification Coupled With RNA Sequencing Approach in the Human Pathogen Staphylococcus aureus. Methods Enzymol. 2018; 612:393–411. [DOI] [PubMed] [Google Scholar]
- 28. Tomasini A., Moreau K., Chicher J., Geissmann T., Vandenesch F., Romby P., Marzi S., Caldelari I.. The RNA targetome of Staphylococcus aureus non-coding RNA RsaA: impact on cell surface properties and defense mechanisms. Nucleic Acids Res. 2017; 45:6746–6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Caldelari I., Chane-Woon-Ming B., Noirot C., Moreau K., Romby P., Gaspin C., Marzi S.. Complete genome sequence and annotation of the Staphylococcus aureus strain HG001. Genome Announcements. 2017; 5:e00783-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Afgan E., Baker D., van den Beek M., Blankenberg D., Bouvier D., Cech M., Chilton J., Clements D., Coraor N., Eberhard C. et al.. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016; 44:W3–w10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fechter P., Chevalier C., Yusupova G., Yusupov M., Romby P., Marzi S.. Ribosomal initiation complexes probed by toeprinting and effect of trans-acting translational regulators in bacteria. Methods Mol. Biol. 2009; 540:247–263. [DOI] [PubMed] [Google Scholar]
- 32. Fechter P., Parmentier D., Wu Z., Fuchsbauer O., Romby P., Marzi S.. Traditional chemical mapping of RNA structure in vitro and in vivo. Methods Mol. Biol. 2016; 1490:83–103. [DOI] [PubMed] [Google Scholar]
- 33. Rice G.M., Leonard C.W., Weeks K.M.. RNA secondary structure modeling at consistent high accuracy using differential SHAPE. RNA. 2014; 20:846–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karabiber F., McGinnis J.L., Favorov O.V., Weeks K.M.. QuShape: rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA. 2013; 19:63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reuter J.S., Mathews D.H.. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010; 11:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Darty K., Denise A., Ponty Y.. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009; 25:1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Beauchamp C., Fridovich I.. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 1971; 44:276–287. [DOI] [PubMed] [Google Scholar]
- 38. Valderas M.W., Hart M.E.. Identification and characterization of a second superoxide dismutase gene (sodM) from Staphylococcus aureus. J. Bacteriol. 2001; 183:3399–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lioliou E., Sharma C.M., Caldelari I., Helfer A.C., Fechter P., Vandenesch F., Vogel J., Romby P.. Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLoS Genet. 2012; 8:e1002782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Handke L.D., Hawkins J.C., Miller A.A., Jansen K.U., Anderson A.S.. Regulation of Staphylococcus aureus MntC expression and its role in response to oxidative stress. PLoS One. 2013; 8:e77874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wright P.R., Georg J., Mann M., Sorescu D.A., Richter A.S., Lott S., Kleinkauf R., Hess W.R., Backofen R.. CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res. 2014; 42:W119–W123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Geissmann T., Chevalier C., Cros M.J., Boisset S., Fechter P., Noirot C., Schrenzel J., Francois P., Vandenesch F., Gaspin C. et al.. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res. 2009; 37:7239–7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Salvail H., Masse E.. Regulating iron storage and metabolism with RNA: an overview of posttranscriptional controls of intracellular iron homeostasis. Wiley Interdiscipl. Rev. RNA. 2012; 3:26–36. [DOI] [PubMed] [Google Scholar]
- 44. Chareyre S., Mandin P.. Bacterial iron homeostasis regulation by sRNAs. Microbiol. Spectrum. 2018; 6:doi:10.1128/microbiolspec.RWR-0010-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Prevost K., Desnoyers G., Jacques J.F., Lavoie F., Masse E.. Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev. 2011; 25:385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Niederhoffer E.C., Naranjo C.M., Bradley K.L., Fee J.A.. Control of Escherichia coli superoxide dismutase (sodA and sodB) genes by the ferric uptake regulation (fur) locus. J. Bacteriol. 1990; 172:1930–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kehl-Fie T.E., Skaar E.P.. Nutritional immunity beyond iron: a role for manganese and zinc. Curr. Opin. Chem. Biol. 2010; 14:218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Grim K.P., San Francisco B., Radin J.N., Brazel E.B., Kelliher J.L., Parraga Solorzano P.K., Kim P.C., McDevitt C.A., Kehl-Fie T.E.. The metallophore staphylopine enables Staphylococcus aureus to compete with the host for zinc and overcome nutritional immunity. MBio. 2017; 8:e01281-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kelliher J.L., Kehl-Fie T.E.. Competition for manganese at the host-pathogen interface. Prog. Mol. Biol. Translat. Sci. 2016; 142:1–25. [DOI] [PubMed] [Google Scholar]
- 50. Nairz M., Dichtl S., Schroll A., Haschka D., Tymoszuk P., Theurl I., Weiss G.. Iron and innate antimicrobial immunity-Depriving the pathogen, defending the host. J. Trace Elem. Med. Biol. 2018; 48:118–133. [DOI] [PubMed] [Google Scholar]
- 51. Corbin B.D., Seeley E.H., Raab A., Feldmann J., Miller M.R., Torres V.J., Anderson K.L., Dattilo B.M., Dunman P.M., Gerads R. et al.. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008; 319:962–965. [DOI] [PubMed] [Google Scholar]
- 52. Horsburgh M.J., Wharton S.J., Cox A.G., Ingham E., Peacock S., Foster S.J.. MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol. Microbiol. 2002; 44:1269–1286. [DOI] [PubMed] [Google Scholar]
- 53. Handke L.D., Gribenko A.V., Timofeyeva Y., Scully I.L., Anderson A.S.. MntC-dependent manganese transport is essential for Staphylococcus aureus oxidative stress resistance and virulence. mSphere. 2018; 3:e00336-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lioliou E., Sharma C.M., Altuvia Y., Caldelari I., Romilly C., Helfer A.C., Margalit H., Romby P.. In vivo mapping of RNA-RNA interactions in Staphylococcus aureus using the endoribonuclease III. Methods. 2013; 63:135–143. [DOI] [PubMed] [Google Scholar]
- 55. Nielsen J.S., Christiansen M.H., Bonde M., Gottschalk S., Frees D., Thomsen L.E., Kallipolitis B.H.. Searching for small sigmaB-regulated genes in Staphylococcus aureus. Arch Microbiol. 2011; 193:23–34. [DOI] [PubMed] [Google Scholar]
- 56. Chao Y., Vogel J.. A 3′ UTR-derived small RNA provides the regulatory noncoding arm of the inner membrane stress response. Mol. Cell. 2016; 61:352–363. [DOI] [PubMed] [Google Scholar]
- 57. Miyakoshi M., Chao Y., Vogel J.. Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA. EMBO J. 2015; 34:1478–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Peng T., Berghoff B.A., Oh J.I., Weber L., Schirmer J., Schwarz J., Glaeser J., Klug G.. Regulation of a polyamine transporter by the conserved 3' UTR-derived sRNA SorX confers resistance to singlet oxygen and organic hydroperoxides in Rhodobacter sphaeroides. RNA Biol. 2016; 13:988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ballal A., Manna A.C.. Regulation of superoxide dismutase (sod) genes by SarA in Staphylococcus aureus. J. Bacteriol. 2009; 191:3301–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ballal A., Manna A.C.. Control of thioredoxin reductase gene (trxB) transcription by SarA in Staphylococcus aureus. J. Bacteriol. 2010; 192:336–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pagels M., Fuchs S., Pane-Farre J., Kohler C., Menschner L., Hecker M., McNamarra P.J., Bauer M.C., von Wachenfeldt C., Liebeke M. et al.. Redox sensing by a Rex-family repressor is involved in the regulation of anaerobic gene expression in Staphylococcus aureus. Mol. Microbiol. 2010; 76:1142–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Grosser M.R., Paluscio E., Thurlow L.R., Dillon M.M., Cooper V.S., Kawula T.H., Richardson A.R.. Genetic requirements for Staphylococcus aureus nitric oxide resistance and virulence. PLoS Pathog. 2018; 14:e1006907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ravcheev D.A., Li X., Latif H., Zengler K., Leyn S.A., Korostelev Y.D., Kazakov A.E., Novichkov P.S., Osterman A.L., Rodionov D.A.. Transcriptional regulation of central carbon and energy metabolism in bacteria by redox-responsive repressor Rex. J. Bacteriol. 2012; 194:1145–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Winterbourn C.C. Toxicity of iron and hydrogen peroxide: the Fenton reaction. Toxicol. Lett. 1995; 82–83:969–974. [DOI] [PubMed] [Google Scholar]
- 65. Irving H., Williams R.J.P.. 637. The stability of transition-metal complexes. J. Chem. Soc. (Resumed). 1953; 3192–3210. [Google Scholar]
- 66. Gribenko A., Mosyak L., Ghosh S., Parris K., Svenson K., Moran J., Chu L., Li S., Liu T., Woods V.L. Jr et al.. Three-dimensional structure and biophysical characterization of Staphylococcus aureus cell surface antigen-manganese transporter MntC. J. Mol. Biol. 2013; 425:3429–3445. [DOI] [PubMed] [Google Scholar]
- 67. Lieser S.A., Davis T.C., Helmann J.D., Cohen S.M.. DNA-binding and oligomerization studies of the manganese(II) metalloregulatory protein MntR from Bacillus subtilis. Biochemistry. 2003; 42:12634–12642. [DOI] [PubMed] [Google Scholar]
- 68. Horsburgh M.J., Clements M.O., Crossley H., Ingham E., Foster S.J.. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. immun. 2001; 69:3744–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Horsburgh M.J., Ingham E., Foster S.J.. In Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and Is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 2001; 183:468–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lindsay J.A., Foster S.J.. zur: a Zn(2+)-responsive regulatory element of Staphylococcus aureus. Microbiology. 2001; 147:1259–1266. [DOI] [PubMed] [Google Scholar]
- 71. Morrissey J.A., Cockayne A., Brummell K., Williams P.. The staphylococcal ferritins are differentially regulated in response to iron and manganese and via PerR and Fur. Infect. Immun. 2004; 72:972–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Martens L., Hermjakob H., Jones P., Adamski M., Taylor C., States D., Gevaert K., Vandekerckhove J., Apweiler R.. PRIDE: the proteomics identifications database. Proteomics. 2005; 5:3537–3545. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MAPS data have been deposited to GEO under accession GSE128741.
The mass spectrometric data were deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (72) with the dataset identifier PXD013225.