Abstract
HMGB1 is a ubiquitous non-histone protein, which biological effects depend on its expression and subcellular location. Inside the nucleus, HMGB1 is engaged in many DNA events such as DNA repair, transcription and telomere maintenance. HMGB1 has been reported to bind preferentially to bent DNA as well as to noncanonical DNA structures like 4-way junctions and, more recently, to G-quadruplexes. These are four-stranded conformations of nucleic acids involved in important cellular processes, including telomere maintenance. In this frame, G-quadruplex recognition by specific proteins represents a key event to modulate physiological or pathological pathways. Herein, to get insights into the telomeric G-quadruplex DNA recognition by HMGB1, we performed detailed biophysical studies complemented with biological analyses. The obtained results provided information about the molecular determinants for the interaction and showed that the structural variability of human telomeric G-quadruplex DNA may have significant implications in HMGB1 recognition. The biological data identified HMGB1 as a telomere-associated protein in both telomerase-positive and -negative tumor cells and showed that HMGB1 gene silencing in such cells induces telomere DNA damage foci. Altogether, these findings provide a deeper understanding of telomeric G-quadruplex recognition by HMGB1 and suggest that this protein could actually represent a new target for cancer therapy.
INTRODUCTION
Mechanisms of regulation of telomere maintenance are the subject of extensive investigation, because of their direct relation with genome stability, aging and cancer (1). The telomere is a highly specialized functional structure located at the end of eukaryotic chromosomes whose main role is to maintain genomic stability. In normal cells, telomere is shortened during every DNA replication until its loss eventually triggers apoptosis (2). It consists of tandem repeats of a DNA sequence and a number of associated proteins. In humans, telomeric DNA is a double-stranded array of TTAGGG repeats, which terminates in a 3′ single-stranded G-rich overhang capable of forming noncanonical structures known as G-quadruplexes (G4s) (3,4). Telomeric G4s have been shown to have regulatory roles in telomere extension and maintenance (5). Indeed, G4 formation interferes with the activity of telomerase, a ribonucleoprotein complex overexpressed in ∼85% of cancers that elongates the single-stranded telomeric overhang, thus leading to cell immortality (6).
Besides telomerase, several proteins have been shown to interact with telomeric DNA with different biological functions (7,8). Some of these proteins are able to unfold the G4 structure promoting telomerase activity, while others hinder the interaction between telomeric DNA and telomerase (9,10). Recently, through a chemoproteomic-driven approach, some of us have identified novel binding partners of human telomeric G4 DNA, thus suggesting a previously unknown role for these proteins at telomeric level (11). Among the identified proteins is the nuclear protein High Mobility Group B1 (HMGB1), a highly abundant vertebrate nuclear protein involved in a number of DNA activity-associated events (12,13). Besides G4 DNA, HMGB1 binds with high affinity to other noncanonical DNA structures like 4-way junctions and hemicatenated DNA loops. In addition, it binds to B-form DNA without sequence specificity and causes distortion of the DNA helix, facilitating the interaction of DNA with other nuclear proteins (12). Thus, HMGB1 acts as a DNA chaperone in transcription, replication, recombination, and repair. When released in the extracellular space, HMGB1 accomplishes its function by activating signaling pathways in combination with other cytokines and chemokines. Its high levels are usually associated with tumor development, proliferation, invasion and metastasis, but paradoxically HMGB1 has also been reported to promote tumor suppression (14,15).
In parallel to its identification as telomeric (and later also as non-telomeric) G4-interacting protein (11,16), HMGB1 was found to be involved in the regulation of telomere homeostasis by an independent research group (17). In addition, previous results clearly showed that knockout of HMGB1 gene in mouse embryonic fibroblasts resulted in reduced telomerase activity and telomere dysfunction (18). Moreover, purified HMGB1 was unable to enhance telomerase activity in vitro, indicating that this effect was not a consequence of a direct binding of the protein to active telomerase complex (18).
HMGB1 is structurally composed by two tandem box domains, box-A and box-B, connected by a basic linker region. Solution structures of the isolated domains determined by NMR are reported in the protein data bank (box-A, PDB codes: 2RTU (19) and 2LY4 (20); box-B, PDB code: 1HMF (21)). The two box domains present the same global fold with a L-shaped architecture, containing a short arm comprising helices I and II, and a long arm comprising helix III and the N-terminal extended strand. However, the different relative helices orientation and the low sequence homology (about 29%) account for different ways of binding to DNA of the two domains.
NMR relaxation measurements, performed on the full-length protein, indicated that the two box domains behave as rigid structures connected by a highly flexible linker region and do not interact with each other (22). This was also confirmed by the family of NMR structures of the free HMGB1 full-length protein (PDB code: 2YRQ) that is composed by a bundle of conformations with different relative inter-domain orientations. Unfortunately, the high interdomain flexibility of HMGB1 and the lack of any DNA-sequence specificity preclude the generation of a well-defined and stable complex and prevent the structure determination of a full-length protein/DNA complex both by X-ray crystallography and NMR. Nevertheless, structures of complexes of the isolated box domains or of chimeric proteins with duplex DNA are available in the protein data bank, namely, the X-ray structure of isolated box-A in complex with a duplex DNA (PDB code: 4QR9) (23), and the NMR structure of the complex formed by a hybrid HMGB1 protein with duplex-DNA (PDB code: 2GZK) (24). In the latter, box-A was replaced by the HMG box of the sequence-specific transcription factor SRY. The analysis of these structures revealed that the basis of the B-DNA distortion induced by the binding of HMG boxes is the intercalation of bulky hydrophobic amino acid side-chains between successive base-pairs.
G4 DNA structures differ considerably from the double helical structure of B-DNA in the number and orientation of strands, groove width, and presence of loops (25). Such substantial structural differences obviously result in considerable differences in the shape and electrostatic potential of the DNA molecules that are expected to strongly influence protein recognition.
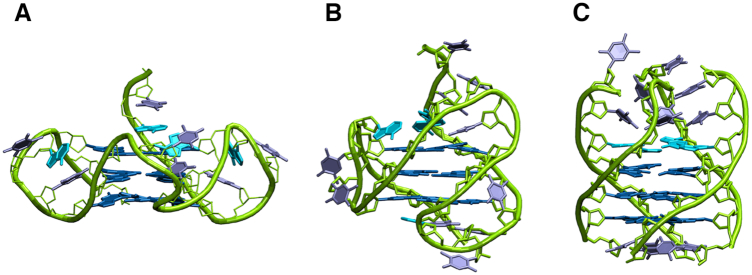
In this context, this work provides a detailed biophysical examination of the interaction between HMGB1 and the G4-forming 26-mer truncation of human telomeric DNA d[(TTAGGG)4TT]. We mainly focused on the unimolecular parallel-stranded G4 structure adopted by this sequence (hereafter referred to as TelG4-up, Figure 1A), which was previously used as a probe to fish out HMGB1 from a nuclear protein matrix (11), and that seems to be the most favored under cell-mimicking conditions (26). Since the human telomeric G4s are structurally heterogeneous and in order to evaluate the possible role of loops in the G4/protein recognition, experiments were also performed on the hybrid [3+1] G4 conformation adopted by such telomeric DNA (TelG4-uh, Figure 1B), and on the tetramolecular parallel G4 structure formed by the d(TTAGGGT) sequence (TelG4-tp, Figure 1C). The results of our biophysical studies provide useful insights into the structural and energetic aspects of the HMGB1/G4 recognition, including the role of the two protein domains in the molecular recognition of these DNA structures and the G4 structural determinants for an optimal interaction. To shed light on the possible interaction mechanism occurring between HMGB1 and TelG4-up, models of the complex have been generated by performing docking calculations driven by experimental data. Biological analyses have been also performed to observe the telomeric localization of HMGB1 as well as the effect of silencing of HMGB1 encoding gene at the telomeric level in both telomerase-positive and -negative tumor cell lines.
Figure 1.
Representation of (A) unimolecular parallel (TelG4-up), (B) unimolecular hybrid (TelG4-uh) and (C) tetramolecular parallel (TelG4-tp) G4 structures formed by truncations of human telomeric DNA sequence.
MATERIALS AND METHODS
Protein expression and purification
The plasmids encoding the three protein constructs were cloned as described in Supplementary Material. GST-fused HMGB11-166 (residues 1–166) and GST-fused box-B (residues 98–166) constructs were both expressed in Escherichia coli C41(DE3) strain cells, whereas GST-fused box-A (residues 1–81) construct was expressed in E. coli BL21(DE3) Codon Plus RIPL (simply RIPL) cells (Supplementary Figures S1–S3, Supplementary Material). The pETG-30A-transformed cells were cultured in 13C,15N-enriched Silantes OD2 medium supplied with 0.1 mg ml−1 ampicillin (and even 34 μg ml−1 chloramphenicol in the case of RIPL cells), grown at 310 K until A600 nm reached 0.6, then induced with 1 mM IPTG. Cells were further let grow at 298 K overnight (for C41(DE3) cells) and at 310 K for 3 h (for RIPL cells), then harvested by centrifugation at 7500 rpm for 15 min at 277 K. The extraction and purification steps were performed with the same protocol for all the three constructs. The bacterial pellet was resuspended in 50 mM Tris–HCl, pH 8.0, 500 mM NaCl buffer (lysis buffer, 45 ml for liter of culture) and incubated at 277 K for 20 min upon stirring. The suspension was sonicated on ice for 12 cycles, alternating 30 s of pulse and 180 s of resting, and then ultracentrifuged at 40 000 rpm at 277 K for 40 min. The supernatant was again incubated on ice and treated with polyethylenimine (PEI): small fractions of ∼5% (w/v) PEI solution (pH 7.9) were gradually added up to reach a PEI concentration of 0.8% w/v in the suspension, which was then centrifuged at 8000 rpm for 20 min. The precipitate of nucleic acids was removed, whereas the supernatant was treated with aliquots of solid (NH4)2SO4 up to 70% (w/v) of saturation, upon stirring at 277 K. Again, the suspension was centrifuged at 10 000 rpm for 30 min. The supernatant was discarded, whereas the precipitate was washed three times with a 70% (w/v) saturated (NH4)2SO4 solution. The final protein pellet was stored at 277 K. The purification steps before and after TEV cleavage were carried out by a Ni-affinity chromatography. The GST-fused protein pellet was resuspended in 50 mM Tris–HCl, pH 8.0, 500 mM NaCl, 5 mM imidazole buffer (binding buffer, 50 ml for liter of culture) and loaded onto a pre-equilibrated 5 ml Ni-column. After washing with the binding buffer, the elution was carried out using 20 mM imidazole. Fractions of pure GST-fused protein were combined and buffer-exchanged to 50 mM Tris–HCl, pH 8.0, 150 mM NaCl. The solution was concentrated down to 2 mg ml−1 of GST-fused protein, then supplied with 0.5 mM EDTA and 1.0 mM DTT. A stock of glycerolized TEV solution was added and the mixture incubated at 298 K overnight. After TEV cleavage, 5 mM imidazole was added to the solution which was then loaded onto a pre-equilibrated 5 ml Ni-column. The protein (without the GST fragment) was eluted with 40 mM imidazole. The solution of protein was then further purified by gel filtration using a Superdex 75 16/60 column. The protein was eluted in 10 mM KH2PO4 buffer, pH 7.0, containing 70 mM KCl, and the fractions containing pure protein were identified by an SDS-PAGE and combined. The solution was concentrated to 4.5 μM, supplied with 250 mM LiCl and 10% D2O, then stored at 277 K.
Preparation of DNA samples
DNA oligonucleotides were chemically synthesized on an ABI 394 DNA/RNA synthesizer (Applied Biosystem) at 5 or 1 μmol scale, using the standard β-cyanoethylphosphoramidite solid phase chemistry as described elsewhere (27). In particular, the following oligonucleotides were synthesized: d[(TTAGGG)4TT] and d(TTAGGGT), corresponding to two different truncations of human telomeric DNA sequence. After synthesis, the oligomers were detached from the support and deprotected by treatment with concentrated aqueous ammonia at 55°C for 17 h. The combined filtrates and washings were concentrated under reduced pressure, dissolved in H2O, and purified by high-performance liquid chromatography (HPLC) employing standard protocols. The isolated oligomers were proved to be >98% pure by NMR. The concentration of oligonucleotides was determined by UV adsorption measurements at 90°C using appropriate molar extinction coefficient values ϵ (λ = 260 nm), calculated by the nearest-neighbor model (28). All G4s were prepared in 10 mM KH2PO4 buffer containing 70 mM KCl, pH 7.0. Samples were then heated at 90°C for 5 min, gradually cooled to room temperature overnight, and finally incubated at 4°C for 24 h, before data acquisition. The G4 parallel arrangement of the 26-mer telomeric sequence (TelG4-up) was prepared and checked as previously described (29,30).
Circular dichroism (CD) experiments
CD experiments were recorded on a Jasco J-815 spectropolarimeter equipped with a PTC-423S/15 Peltier temperature controller. All the experiments were carried out in 10 mM KH2PO4 buffer (pH 7.0) containing 70 mM KCl. CD spectra were recorded at 20°C in a quartz cuvette with 1 mm path length in the 190–340 nm wavelength range and averaged over three scans. A concentration of 1 μM was used for both G4s and proteins. The scan rate was set to 100 nm/min, with 1 s response time and 1 nm bandwidth. Buffer baseline was subtracted from each spectrum. CD melting experiments of G4s in the absence or presence of 1 molar equiv. of HMGB11–166 or box-A were carried out at 1°C/min heating rate by following the CD signal at the wavelength of maximum intensity in the spectral region where only the G4 chromophores absorb (263 nm for TelG4-up and TelG4-tp, and 289 nm for TelG4-uh). Melting temperatures were determined from curve fitting using Origin 7.0 (OriginLab, Northampton, MA, USA).
NMR measurements
NMR titrations of the HMGB11-166 protein and of the isolated box domains (A and B) with G4 DNA were performed at 298 K in buffered solution: 10 mM KH2PO4, 70 mM KCl, 250 mM LiCl, pH 7.0. All the titrations were performed at 25 μM protein concentration. Increasing aliquots of TelG4-up, TelG4-uh and TelG4-tp (to reach concentrations of 0.25, 0.5, 1, 2, 25 μM) have been added to the HMGB11–166 protein solution. Titrations of the isolated box domains were performed only with TelG4-up, by adding increasing amounts of DNA to the box-A (to reach concentrations of 0.25, 0.5, 1, 2, 4, 25 μM) and box-B (to reach concentrations of 0.25, 0.5, 1, 2, 12.5, 25 μM) solutions. At the end of box-B titration, a solution of box-A was added to the final box-B/TelG4-up solution to obtain a box-A/box-B/TelG4-up mixture in the 1:1:1 molar ratio. The effects have been monitored through 2D 1H–15N HSQC NMR (1H spectral window = 15 ppm; 15N spectral window = 35 ppm; relaxation delay = 1.2 s; number of scans = 32; number of points in F1 dimension = 128). The NMR titrations of the HMGB11–166 protein and isolated box-A were performed also in the presence of the well-known G4 ligands Braco-19 (31) or RHPS4 (32). The ligand (2.5 mM) was first added to the protein solutions; then, increasing amounts of TelG4-up (0.25, 0.5, 1, 2, 4, 8 μM) were added. For box-A, the effects of the equimolar concentration of TelG4-up (25 μM) in the presence of Braco-19 were also analyzed. The spectra were collected on Bruker Advance III NMR spectrometers operating at 900 and 950 MHz, 1H Larmor frequencies, processed with the Bruker's TopSpin software and analyzed using the software CARA (33). The protein assignment was based on the data reported in the Biological Magnetic Resonance Data Bank (34) under the accession code 11147.
Fluorescence titrations
Fluorescence titration experiments were performed at 25°C on a FP-8300 spectrofluorometer (Jasco) equipped with a Peltier temperature controller system (Jasco PCT-818). A sealed quartz cuvette with a path length of 1 cm was used. Titrations were carried out by stepwise addition (5 μl) of a G4 DNA solution (150–200 μM) to a cell containing a fixed concentration of HMGB11–166 solution (3–4 μM) in 10 mM KH2PO4 and 70 mM KCl, at pH 7.0. Experiments were performed in the presence of Braco-19 or RHPS4 (1:1 ligand/DNA). Protein was excited at 280 nm, and emission spectra were recorded between 285 and 500 nm. Both excitation and emission slit widths were set at 5 nm. After each G4 addition, the solution was stirred and allowed to equilibrate for 5 min before spectrum acquisition. The fraction of bound protein (α) at each point of the titration was calculated following the changes of fluorescence intensity at 327 nm, using the following relationship:
 |
where I327 is the fluorescence intensity at the various protein/DNA ratios investigated;  and
and  are the fluorescence intensities of the free and fully bound protein, respectively. Titration curves were obtained by plotting α versus the G4 concentration. The equilibrium dissociation constant (Kd) and the stoichiometry of interaction were estimated from this plot by fitting the resulting curve, using nonlinear regression, to an independent and equivalent binding site model as previously described (35). The protein concentration was corrected for dilution effects resulting from the change in volume due to DNA solution addition.
are the fluorescence intensities of the free and fully bound protein, respectively. Titration curves were obtained by plotting α versus the G4 concentration. The equilibrium dissociation constant (Kd) and the stoichiometry of interaction were estimated from this plot by fitting the resulting curve, using nonlinear regression, to an independent and equivalent binding site model as previously described (35). The protein concentration was corrected for dilution effects resulting from the change in volume due to DNA solution addition.
Surface plasmon resonance (SPR) experiments
SPR experiments were performed on a Biacore X100 (GE Healthcare), using a research-grade CM5 sensor chip. HMGB11–166 protein was immobilized using amine-coupling chemistry and HBS-EP as running buffer (HEPES 10 mM, NaCl 150 mM, EDTA 3 mM, 0.005% surfactant P20, pH 7.4). The surfaces of flow cells were activated with a 1:1 mixture of 0.1 M NHS (N-hydroxysuccinimide) and 0.1 M EDC (3-(N,N-dimethylamino)propyl-N-ethylcarbodiimide) at a flow rate of 10 μl/min. The protein at a concentration of 50 μg/ml in 10 mM sodium acetate, pH 4.5, was immobilized on the sample flow cell, leaving the reference cell as blank. Unreacted activated groups were blocked by injection of 1.0 M ethanolamine at 10 μl/min over the chip surface. DNA samples were injected at 25°C at various concentrations (from 2 to 32 μM), using 10 mM KH2PO4 buffer (pH 7.0) containing 70 mM KCl as running solution. Injections were performed at a flow rate of 30 μl/min, with association and dissociation times of 80 and 600 s, respectively. The sensor surface was regenerated by using 10 mM NaOH for 12 s. Data were fit to a simple 1:1 kinetic interaction model, using the global data analysis option available within the Biacore Evaluation software provided with the instrument.
Modeling of the complexes of HMGB1 with TelG4-up
The models of the complex between either the isolated box domains or the HMGB11–166 protein and the human telomeric G4 were obtained by performing docking calculations with the software HADDOCK 2.2 (36) on the WeNMR GRID (http://www.wenmr.eu) (37) using the Guru interface. In all calculations, during the rigid-body docking, 1000 complexes were generated, then 200 structures were selected for the semi-flexible simulated annealing in torsion angle space, and finally refined in Cartesian space with explicit solvent. The first model of HMGB1 NMR structures’ family (PDB code: 2YRQ) provided the input coordinates for both the HMGB11–166 protein and the single domains (box-A residues: K7-P81; box-B residues: N93-G166). As input coordinates for unimolecular parallel telomeric G4 (TelG4-up), the PDB 2LD8 structure, formed by the 23-mer d[TAGGG(TTAGGG)3] sequence (38), was used in the calculations. In the docking calculations of box-A with TelG4-up, the residues identified during the NMR titration of this isolated domain were ambiguously restrained to either the third G-tetrad plane (residues G5, G11, G17, G23) or to the residues of the first two loop regions of the G4 (T6, T7, A8, T12, T13, A14). For box-B, no specific region of interaction could be identified in the NMR titration of the isolated domain; thus, the residues of box-B experiencing the highest decrease of signal intensity in the titration of HMGB11–166 were ambiguously restrained to the third G-tetrad plane (residues G5, G11, G17, G23). In the multi-body docking calculations, box-A and box-B were simultaneously docked on TelG4-up using the interacting residues previously selected for the calculations of the two separated domains; box-A was docked either on the third G-tetrad plane (residues G5, G11, G17, G23) or on the loop region (residues T6, T7, A8, T12, T13, A14), while box-B was docked on the first G-tetrad plane (residues G3, G9, G15, G21). The best water-refined HADDOCK models showing a distance between the two domains compatible with the length of the linker connecting the two domains, were selected as input templates to Modeller (39) to re-build the linker between box-A and box-B. The obtained models were then subjected to a further refinement protocol in HADDOCK and scored according to the HADDOCK-scores.
Immunofluorescence
Cells were fixed in 2% formaldehyde in phosphate buffered saline (PBS) for 10 min at room temperature (RT) and permeabilized in 0.25% Triton X-100 in PBS for 5 min at RT. For immune-labeling, cells were incubated with primary antibody for 2 h at RT, washed twice in PBS and finally incubated with the secondary antibodies for 1 h. The following primary antibodies were used: Rabbit pAb anti-HMGB1 (Abcam Ltd, Cambridge, UK), Mouse mAb anti-γH2AX (Millipore, Billerica, MA, USA) and Rabbit pAb anti-TRF1 N19 (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The following secondary antibodies were used: Anti-Mouse IgG (H+L), F(ab’)2 Fragment (Alexa Fluor 488 Conjugate) (Cell Signaling) and Anti-rabbit IgG (H+L), F(ab’)2 Fragment (Alexa Fluor 555 Conjugate) (Cell Signaling). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma). Fluorescence signals were recorded by using a Leica DMIRE2 microscope equipped with a Leica DFC 350FX camera and elaborated by Leica FW4000 deconvolution software (Leica, Solms, Germany). For quantitative analysis of γH2AX positivity, 300 cells on triplicate slices were scored and for TIF analysis, 30 γH2AX-positive cells were scored. Cells with at least four co-localizations (γH2AX/TRF1) were considered as TIF-positive. Where reported, cells were incubated with the indicated doses of Braco-19 for 24 h.
Western blotting
Western blot analysis was performed as previously reported (40). Expression levels of HMGB1 were evaluated by using the Rabbit pAb anti-HMGB1 (Abcam Ltd). Actin signal was detected by Mouse mAb anti-β-actin (Sigma Aldrich) and used as loading control.
Chromatin immunoprecipitation assay (ChIP)
Formaldehyde-cross-linked chromatin fragments obtained from HeLa cervical cancer cells were immunoprecipitated with Rabbit pAb anti-HMGB1 (Abcam Ltd.) and with Rabbit pAb anti-TRF1 N19 (Santa Cruz Biotechnology) as positive control for telomeric sequences. Chromatin fragments immunoprecipitated without the antibody (No Ab) and with Normal Rabbit immunoglobulins (IgG) (Santa Cruz Biotechnology) were used as negative controls of the ChIP assay. After precipitation, the assay was performed as previously described (41).
RESULTS
Protein purification and assignment of protein residues
HMGB11-166, box-A, and box-B were expressed as fusion proteins containing a N-terminal GST (glutathione-S-transferase) tag that allows an easy purification by using standard TEV protease-based methods (see Materials and Methods). The identity and purity of the samples were then ascertained by SDS-PAGE analysis of the purified proteins (Supplementary Figure S4, Supplementary Material). The 2D 1H–15N HSQC NMR spectrum of HMGB11-166 showed that the purified protein was correctly folded (Supplementary Figure S5, Supplementary Material). Moreover, the cross-peaks of the isolated box-A and box-B overlaid well with the spectrum of the N-terminal and C-terminal regions of HMGB11–166, respectively (Supplementary Figure S6, Supplementary Material), thus indicating that they adopted their native conformation as in the entire protein. The chemical shift assignments obtained from the BMRB entry 11147 were directly mapped to the 2D 1H–15N HSQC spectra of the HMGB11-166 protein and of the isolated domains, allowing us to detect the protein regions involved in the G4 DNA recognition.
CD characterization of the investigated G-quadruplexes
G-rich human telomeric motifs can, in the presence of K+, fold into different G4 topologies depending on the sequence and experimental conditions (42). Since several studies suggest that the parallel G4 fold is prevalent in the overcrowded conditions of cells (26,38), and given that this conformation is the one used as a probe to fish out HMGB1 from a nuclear protein matrix (11), we selected it as the main candidate for studying HMGB1/G4 interaction. Therefore, we prepared a d[(TTAGGG)4TT] sample at high DNA concentration conditions in order to promote the formation of the unimolecular parallel G4 conformation (43), characterized by the presence of three double-chain-reversal loops running along three of the four grooves having the same width (TelG4-up, Figure 1A) (38).
To investigate the HMGB1 binding properties to different telomeric G4s, we also prepared a d[(TTAGGG)4TT] sample at low concentration in which it is expected to form the so-called hybrid [3+1] fold as major conformation, characterized by mixed parallel/antiparallel G-strands, one double-chain-reversal and two lateral loops, and grooves of different width (TelG4-uh, Figure 1B) (44). Finally, we also considered the tetramolecular G4 structure formed by the short d(TTAGGGT) truncation, having all parallel-oriented strands and no loops (TelG4-tp, Figure 1C) (45). In this way, we could investigate the binding to G4 structures differing in the presence/absence or conformation of loops, and in the width or accessibility of grooves.
The folding adopted by each G4 sample was verified by CD spectroscopy, a well-established technique for determining the presence and the overall topology of G4 structures (46). TelG4-up and TelG4-tp showed very similar CD spectra, with a positive band at 264 nm and a negative band at ∼240 nm (Figure 2), characteristic of the parallel G4 conformations. On the other hand, TelG4-uh showed a CD spectrum with two positive bands at 290 and 268 nm, and a weak negative band at ∼240 nm (Figure 2), in agreement with the hybrid G4 folding topology. CD melting experiments were also performed to get information on the stability of the investigated G4s (Supplementary Figure S7, Supplementary Material). The thermal denaturation of G4s was monitored at the wavelength of maximum CD intensity, i.e. 264 nm for TelG4-up and TelG4-tp, and 290 nm for TelG4-uh. The melting experiments showed sigmoidal transition curves still indicative of the formation of the structures, while the melting temperatures (Tm) obtained from the curve fitting were 65.6 (±1.0), 54.8 (±0.5) and 61.8 (±0.5)°C for TelG4-up, TelG4-uh and TelG4-tp, respectively. These data are in agreement with the previously published results for these DNA molecules in the same experimental conditions (11,47).
Figure 2.
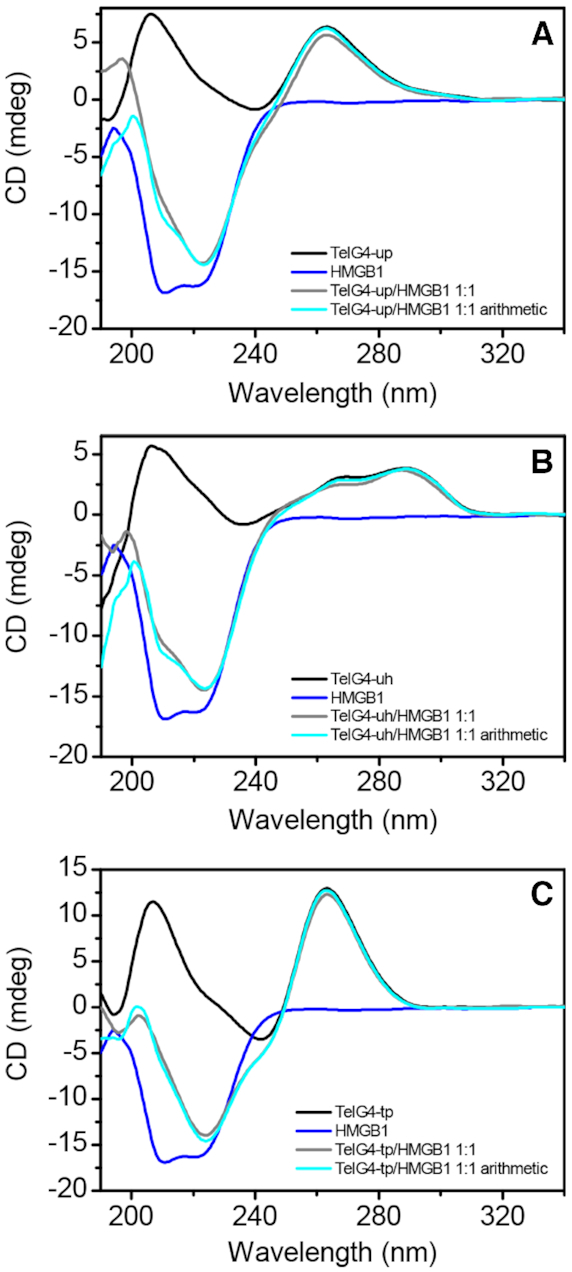
CD spectra of (A) TelG4-up, (B) TelG4-uh and (C) TelG4-tp G4s (1 μM) (black lines), HMGB11–166 (1 μM) (blue lines), and 1:1 G4/HMGB11–166 mixtures (gray lines). The spectra derived from the arithmetical sum of the individual spectra of G4s and HMGB11-166 are reported as well (cyan lines).
HMGB11–166 and box-A do not induce the unfolding of telomeric G4s
The CD spectra of the proteins (HMGB11–166 or box-A) alone, as well as those of G4/protein (1:1 molar equiv.) mixtures, were also recorded and are shown in Figure 2 and Supplementary Figure S7A–C, along with the spectra resulting from the arithmetic sum of the spectra of the single components. In all cases, no significant changes were observed in the spectral region where only the G4 chromophores absorb (λ > 245 nm), clearly indicating that no unfolding or structural alterations of DNA secondary structures occur in the presence of the HMGB11-166 protein or the box-A domain. Within the 190–245 nm wavelength range, despite no considerable variations between the experimental and the corresponding calculated DNA/protein spectra, such spectra differ to a certain extent from each other, thus suggesting the DNA–protein interaction.
Furthermore, to evaluate any possible stabilizing/destabilizing effect of HMGB11–166 and box-A on the investigated G4s, CD melting experiments were carried out (Supplementary Figure S7D–F, Supplementary Material). Upon HMGB11-166 addition to the G4 solutions, CD melting curves showed a slight increase in the thermal stability of the DNA structures (ΔTm = 2.9 (±0.7)°C and 2.3 (±0.7)°C for TelG4-uh and TelG4-tp, respectively). It is to be noted that the melting profile of TelG4-up in the presence of HMGB11–166 exhibits two inflection points (probably because of the formation of some aggregates besides the DNA/protein complex), a first one at around 53°C, and a second one at around 72°C (with a ΔTm of ∼6.5°C compared to the G4 alone). Quite different effects on the thermal stability of G4s were observed in the presence of box-A. When it was added to TelG4-uh and TelG4-tp solutions, no changes in the thermal stability of these DNA structures were observed. On the other hand, a significant increase of the stability of TelG4-up was observed in the presence of box-A (ΔTm = 4.7 (±1.0)°C), thus suggesting a probable pivotal role of box-A in the interaction with this G4.
NMR investigation of HMGB11-166 binding to TelG4-up, TelG4-uh and TelG4-tp
The region of the HMGB11–166 protein surface involved in the interaction with G4 molecules has been investigated by monitoring the changes in cross-peak intensity ratio (I/I0), as well as the chemical shift variations (Δδ) occurring in the 2D 1H–15N HSQC spectrum of the uniformly 15N-labeled protein upon the addition of increasing amounts (to reach concentrations of 0.25, 0.5, 1, 2, 25 μM) of each G4-forming DNA (I), versus the free protein in solution (I0). The concentration of the protein used for the NMR titrations was relatively low (25 μM) because, at higher protein concentrations, protein-DNA aggregation caused extensive precipitation upon addition of DNA. To be sure that our results were not affected by non-specific aggregation phenomena, we analyzed in detail the effects measured at low concentrations of the DNA molecules, i.e. upon the addition of 1 μM TelG4-up (Figure 3A and Supplementary Figure S8, Supplementary Material). The affected residues (Table 1) were distributed on both domains.
Figure 3.
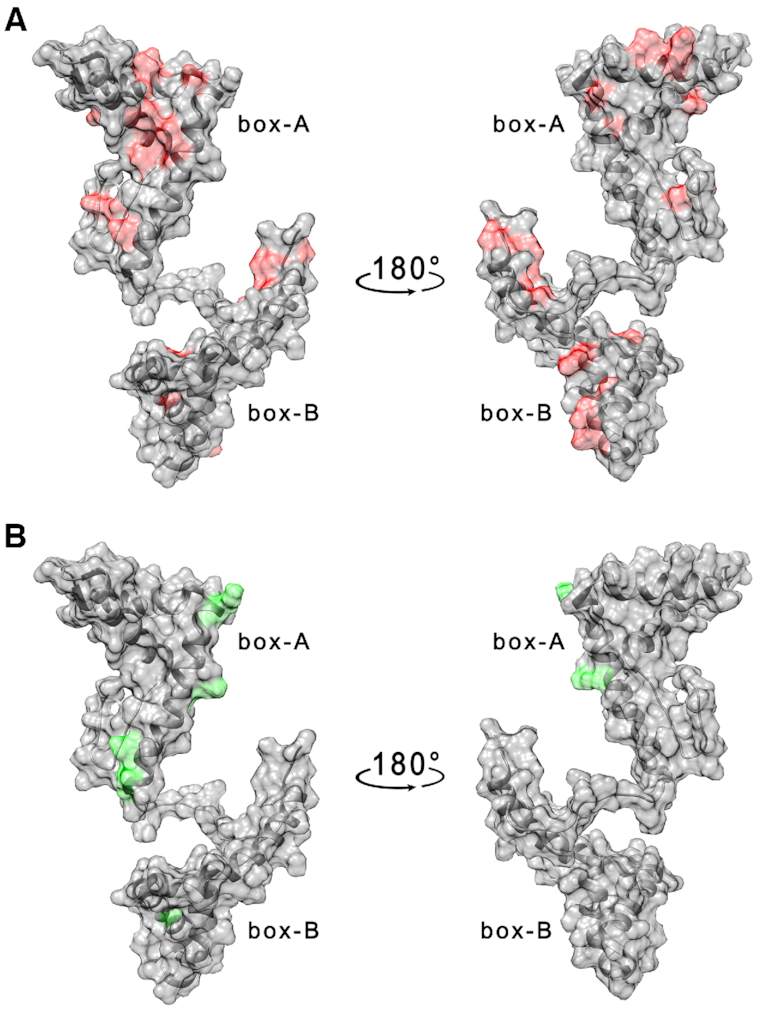
(A) Per residues intensity changes and (B) chemical shift perturbation of HMGB11-166 protein (25 μM) in the presence of 1 μM TelG4-up DNA. The residues exhibiting the largest effects have been colored in red (A) and green (B) on the structure of free HMGB1 (pdb: 2YRQ).
Table 1.
Residues showing a decrease in signal intensity or a chemical shift perturbation upon addition of 1 μM G4 DNA (see table header) to 25 μM HMGB11–166
| TelG4-up | TelG4-uh | TelG4-tp | ||||
|---|---|---|---|---|---|---|
| Residue* | Intensity decrease | Chemical shift perturbation | Intensity decrease | Chemical shift perturbation | Intensity decrease | Chemical shift perturbation |
| G4 | X | |||||
| K8 | X | |||||
| R10 | X | |||||
| M13 | X | |||||
| S14 | X | X | ||||
| S15 | X | X | ||||
| Y16 | X | X | ||||
| F18 | X | X | ||||
| V20 | X | X | ||||
| T22 | X | X | ||||
| C23 | X | X | X | X | ||
| R24 | X | X | ||||
| E25 | X | |||||
| V36 | X | |||||
| E40 | X | X | ||||
| F41 | X | X | X | |||
| S42 | X | |||||
| K43 | X | X | X | X | ||
| K44 | X | |||||
| S46 | X | X | ||||
| E47 | X | X | ||||
| R48 | X | X | ||||
| K50 | X | |||||
| A54 | X | |||||
| K55 | X | |||||
| E56 | X | |||||
| K57 | X | X | ||||
| F60 | X | |||||
| M63 | X | X | X | |||
| K65 | X | |||||
| D67 | X | X | ||||
| E72 | X | X | X | |||
| E74 | X | X | ||||
| T77 | X | X | X | |||
| Y78 | X | X | X | |||
| I79 | X | |||||
| G83 | X | |||||
| F89 | X | |||||
| K90 | X | |||||
| D91 | X | |||||
| A94 | X | |||||
| R97 | X | X | ||||
| S100 | X | |||||
| F102 | X | |||||
| F103 | X | X | ||||
| F105 | X | X | X | |||
| K114 | X | |||||
| I122 | X | |||||
| V125 | X | |||||
| A126 | X | |||||
| K127 | X | |||||
| G130 | X | X | ||||
| W133 | X | |||||
| K141 | X | |||||
| K154 | X | |||||
| I159 | X | X | X | |||
| A160 | X | |||||
| R163 | X | |||||
| A164 | X | X | X | |||
| K165 | X | X | ||||
*Residues of box-A and box-B domains are in bold. Significantly decreased residues displayed a change in intensity deviating more than 1 σ from the average variation observed over the entire sequence.
Analogous titrations were performed for TelG4-uh and TelG4-tp, whose effects on the HMGB11–166 spectrum are shown in Supplementary Figures S9 and S10 (Supplementary Material), respectively. Also for these systems, we measured the effect of DNA on the protein NMR spectrum at 1 μM G4 concentration. The observed changes in signal intensity and/or chemical shift variations, indicating an interaction in the intermediate regime on the NMR timescale, are summarized in Table 1 and Supplementary Figures S9 and S10 in the Supplementary Material.
Overall, these results clearly demonstrated that HMGB11-166 interacts with the three subtypes of telomeric G4s. In particular, it seems that box-A involvement in the interaction is more extensive than that of box-B (Table 1). While a single interaction region could not be identified, the sequence stretches from residue 20 to 25, and from residue 35 to 45 were the major perturbed sites.
Braco-19 affects but does not prevent the binding of HMGB11-166 to TelG4-up, TelG4-uh and TelG4-tp
To shed light on the structural determinants of G4 recognition by HMGB1, competitive NMR binding studies in the presence of the well-known G4-binding ligand Braco-19 were carried out. The trisubstituted acridine Braco-19 is known to bind to the external G-tetrad planes of G4s through stacking interactions with a Kd of about 3.2 × 10−8 M (31,48), so it was expected to affect or even prevent the binding of HMGB1 if occurring at the G-tetrads. Preliminarily, Braco-19 was added to the protein to evaluate any possible interaction with HMGB1. In this respect, only minor effects were observed even at a protein/ligand ratio of 1:100 (Supplementary Figure S11, Supplementary Material).
HMGB11-166 (25 μM) was titrated with increasing amounts of TelG4-up, TelG4-uh and TelG4-tp (from 0.25 up to 25 μM) in the presence of Braco-19 (2.5 mM). For all systems, the presence of Braco-19 considerably diminished the widespread effect of G4s on the cross-peaks of HMGB1 (I/I0 values were on average closer to 1), thus indicating a significant reduction of the protein-DNA interaction, likely preventing concatenate protein/DNA aggregation. This was particularly evident for TelG4-up where the residues mainly affected by DNA interaction were all in the box-A domain (F19, E25, K28, K30, H31, E40, E47, M52, A54, K55, E56, K57, G58, A64, E74, M75) (Figure 4A and Supplementary S12A, Supplementary Material), with some closely located residues exhibiting chemical shift perturbations (V20, S35, K59, Figure 4B and Supplementary Figure S12B, Supplementary Material). However, an interaction in the intermediate regime on the NMR timescale is substantially maintained.
Figure 4.
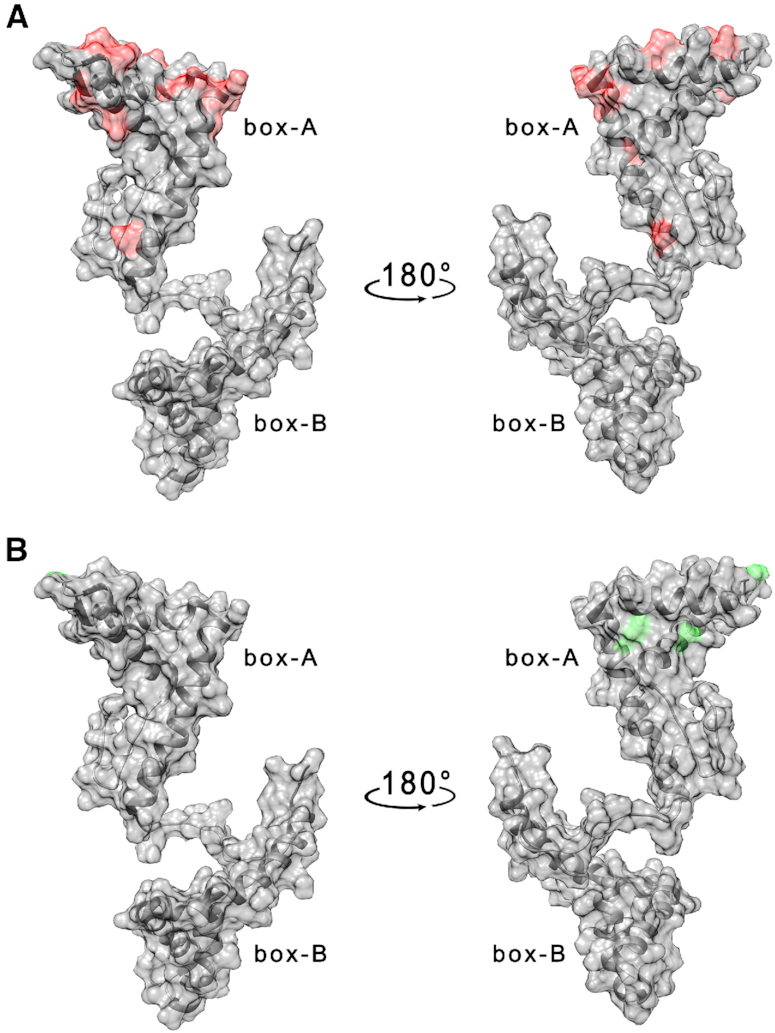
(A) Per residues intensity changes and (B) chemical shift perturbation of HMGB11-166 protein (25 μM) in the presence of 1 μM TelG4-up DNA and Braco-19 (2.5 mM). The residues exhibiting the largest effects have been colored in red (A) and green (B) on the structure of free HMGB1 (pdb: 2YRQ).
As far as TelG4-uh and TelG4-tp are concerned, even though the general decrease in the intensity of HMGB1 cross-peaks was reduced in the presence of Braco-19, the perturbed few residues were scattered along the whole protein sequence, suggesting a non-specific interaction.
Overall, these results show that Braco-19 affects but does not prevent the binding of HMGB1 to telomeric G4s in vitro, and suggest a major role for box-A, at least in the interaction with TelG4-up.
To further investigate the role of the G-tetrad planes of G4s in the binding to HMGB1, the effects of a second G4-binding ligand, RHPS4, on HMGB11-166/G4s interaction were evaluated performing new competitive NMR binding studies. HMGB11-166 (25 μM) was thus titrated with increasing amounts of TelG4-up, TelG4-uh and TelG4-tp (from 0.25 up to 25 μM) in the presence of the same high excess of ligand (2.5 mM). In these conditions, the effects of telomeric G4s on the protein NMR signals were weaker than those observed when Braco-19 was present in solution and became significant only at higher DNA concentrations. The perturbed residues, also for TelG4-up, were scattered along the whole protein sequence, even if with larger engagement of box-A (Supplementary Figure S13, Supplementary Material).
NMR investigation of the interaction of isolated box-A and box-B domains with TelG4-up in the absence and presence of Braco-19
To better define the regions of HMGB1 involved in the interaction and shed light on the mechanism of G4 recognition, we performed NMR titrations of the two isolated box domains (A and B) with TelG4-up.
As in the case of the HMGB11–166 protein, the effects of the DNA G4 on box-A were already visible after the addition of very low amounts of nucleic acid molecule (1 μM). The residues with the highest decrease in signal intensity are placed in the short arm of the L-shaped structure (Table 2, Figure 5A and Supplementary Figure S14A, Supplementary Material) with few neighboring residues exhibiting a small chemical shift perturbation (Figure 5B and Supplementary Figure S14B, Supplementary Material). At higher concentrations of TelG4-up (2 < [C] < 25 μM) the protein signals completely disappeared because of extensive line broadening, possibly due to aggregation processes occurring also between the isolated box-A and the DNA.
Table 2.
Residues showing a decrease in signal intensity or a chemical shift perturbation upon addition of 1 μM TelG4-up DNA to 25 μM of protein (see table header)
| Box-A | Box-B | ||||
|---|---|---|---|---|---|
| Residue* | Intensity decrease | Chemical shift perturbation | Residue* | Intensity decrease | Chemical shift perturbation |
| M13 | X | F103 | X | ||
| S14 | X | F105 | X | ||
| Y16 | X | S107 | X | ||
| F18 | X | E108 | X | ||
| F19 | X | Y109 | X | ||
| V20 | X | R110 | X | ||
| Q21 | X | K112 | X | ||
| T22 | X | K114 | X | ||
| E26 | X | E116 | X | ||
| H27 | X | G119 | X | ||
| K30 | X | K128 | X | ||
| H31 | X | Y144 | X | ||
| D33 | X | E145 | X | ||
| A34 | X | L151 | X | ||
| F41 | X | E153 | X | ||
| S42 | X | K154 | X | ||
| K43 | X | E156 | X | ||
| C45 | X | X | D158 | X | |
| S46 | X | A160 | X | ||
| W49 | X | Y162 | X | ||
| M52 | X | ||||
| A54 | X | ||||
| K55 | X | ||||
| G58 | X | ||||
| K59 | X | ||||
| F60 | X | ||||
| M63 | X | X | |||
| D67 | X | ||||
*Significantly decreased residues displayed a change in intensity deviating more than 1 σ from the average variation observed over the entire sequence.
Figure 5.
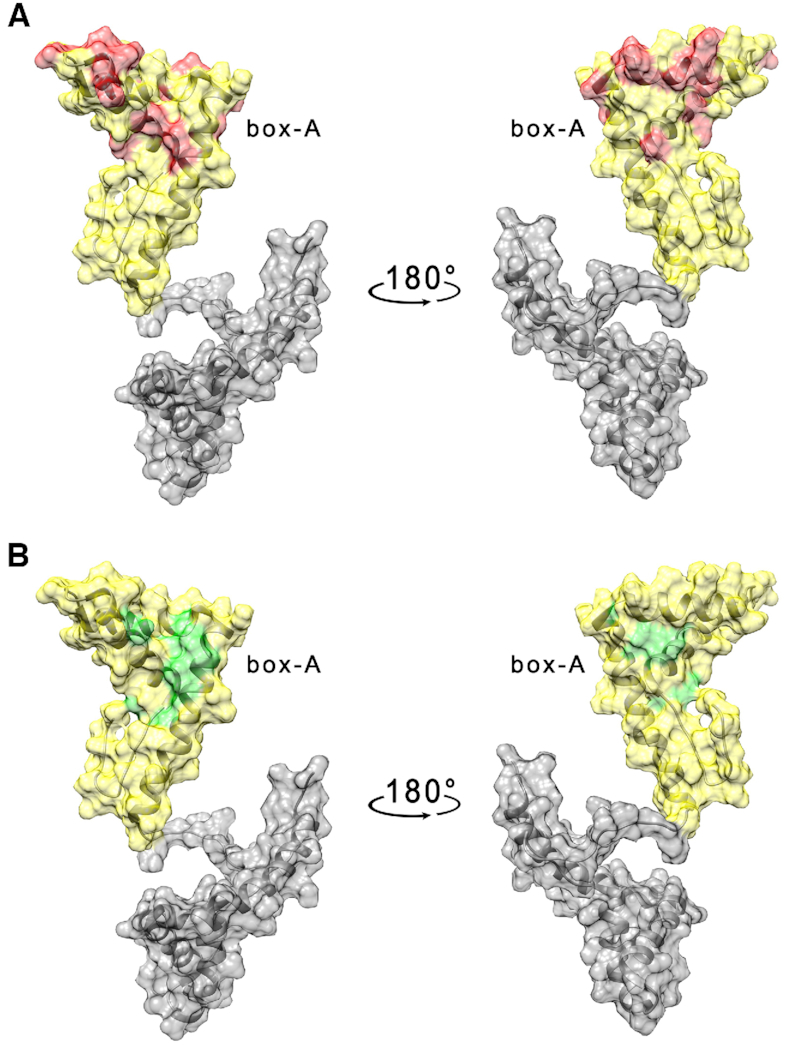
(A) Per residues intensity changes and (B) chemical shift perturbation of box-A domain of HMGB1 (25 μM) in the presence of 1 μM TelG4-up DNA. The residues exhibiting the largest effects have been colored in red (A) and green (B) on the structure of free HMGB1 (pdb: 2YRQ).
On the other hand, the effect of TelG4-up on the isolated box-B domain was found to be less significant than those observed for box-A and HMGB11-166, and higher concentration of G4 were allowed in solution without having the complete disappearance of the signals from the 2D 1H–15N HSQC spectrum. The residues with the highest decrease in signal intensity at equimolar concentration (25 μM) of TelG4-up and box-B (Table 2) are spread on a wide protein surface, preventing the identification of a specific binding region (Supplementary Figure S15, Supplementary Material). Moreover, no significant chemical shift perturbations were observed. Intriguingly, after adding the isolated box-A domain to the solution containing the TelG4-up/box-B mixture, an extensive further decrease of box-B signal intensities was observed (Figure 6 and Supplementary Figure S16, Supplementary Material), thus indicating that the interaction of box-B with the G4 was greatly favored by the presence of box-A, possibly because of a cooperative binding of the two domains and/or by aggregation induced phenomena.
Figure 6.
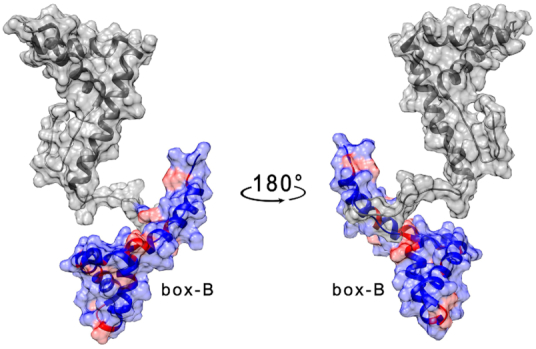
Per residues intensity changes of box-B after the addition of box-A (25 μM) to the complex box-B/TelG4-up (25 μM). The residues exhibiting the largest effects have been colored in red on the structure of free HMGB1 (pdb: 2YRQ).
In addition, we also investigated the interaction of the box-A domain with TelG4-up in the presence of Braco-19 (2.5 mM). As observed for the HMGB11-166 protein (Figure 4A and Supplementary Figure S12A, Supplementary Material), the overall effects are weakened in the presence of the G4 ligand and the residues with the highest decrease in signal intensity (T22, H27, K28, K30, H31, E40, K43, K50) were located on a smaller region of the protein surface (Figure 7A and Supplementary Figure S17A, Supplementary Material), with some residues exhibiting a chemical shift perturbation (E26, K55, F60, T77, Figures 7B and S17B, Supplementary Material). The interaction of box-A with TelG4-up was investigated also in the presence of RHPS4 (2.5 mM). As previously observed with HMGB11-166, the effects of TelG4-up on the box-A NMR signals were weaker than those observed when Braco-19 was present in solution. (Supplementary Figure S18, Supplementary Material).
Figure 7.
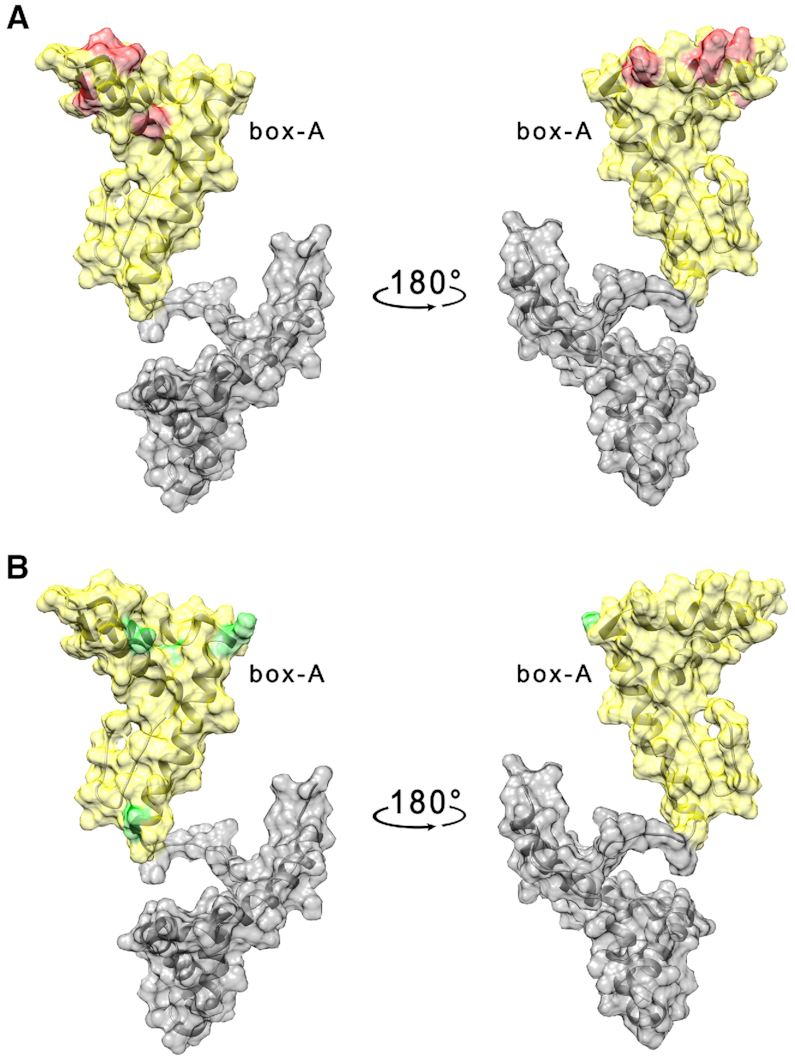
(A) Per residues intensity changes and (B) chemical shift perturbation of box-A domain protein (25 μM) in the presence of 1 μM TelG4-up DNA and Braco-19 (2.5 mM). The residues exhibiting the largest effects have been colored in red (A) and green (B) on the structure of free HMGB1 (pdb: 2YRQ).
Evaluation of binding affinity by fluorescence titration and SPR experiments
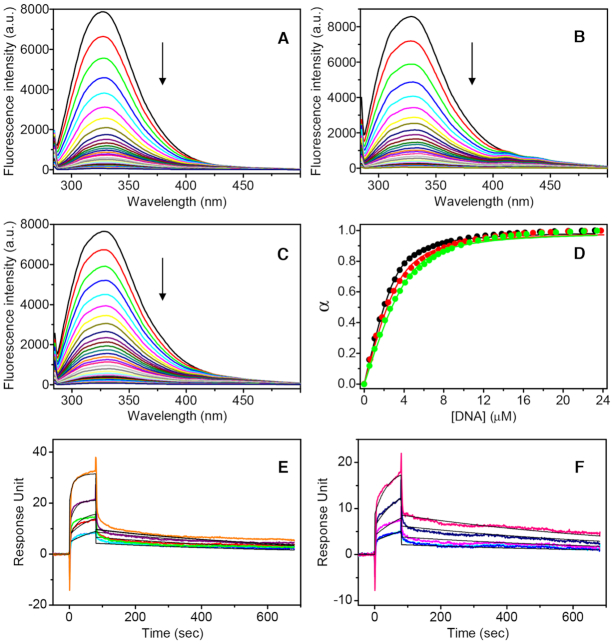
The binding of HMGB11-166 to the telomeric G4s was examined by fluorescence titration and SPR experiments. Fluorescence experiments were carried out by monitoring the changes in the intrinsic fluorescence of the protein upon binding to DNA (16). Fluorescence emission spectra of HMGB11-166 were recorded upon addition of increasing amounts of each G4. Titrations were performed in the presence of Braco-19 (1:1 ligand/DNA), after excluding protein/drug interactions, to avoid protein-DNA aggregation phenomena in solution that would be detrimental to the experiments. As clearly shown in Figure 8A–C, a significant decrease in fluorescence intensity on increasing the concentration of G4s was observed. The titration curves (Figure 8D) were fitted by means of nonlinear regression algorithm using an independent and equivalent binding sites model (35), to get the equilibrium dissociation constants (Kd). From these fittings, Kd values of 4.2 (±0.3) × 10−7, 6.0 (±0.7) × 10−7 and 7.0 (±0.9) × 10−7 M were determined for TelG4-up, TelG4-uh and TelG4-tp, respectively. These results indicate that HMGB11-166 interacts with all three G4s, showing a preference for the unimolecular parallel-stranded TelG4-up. For comparison, fluorescence titrations were also carried out in the presence of the G4 binder RHPS4 (after excluding protein/drug interactions). As in the first set of experiments, increasing the concentration of G4s resulted in a gradual decrease in the fluorescence intensity of HMGB11-166 until saturation was reached (Supplementary Figure S19, Supplementary Material), thus indicating the formation of protein/DNA complexes also in this case. The Kd values obtained from fitting (5.7 (±0.5) × 10−7, 6.8 (±0.7) × 10−7 and 7.2 (±0.8) × 10−7 M for TelG4-up, TelG4-uh and TelG4-tp, respectively) suggest that RHPS4 slightly decreases the affinity of HMGB11–166 for the G4s.
Figure 8.
(A–C) Fluorescence emission spectra of HMGB11–166 (3 μM) in the absence and presence of stepwise addition (5 μl) of (A) TelG4-up, (B) TelG4-uh, and (C) TelG4-tp G4s at 25°C. (D) Titration curves obtained by plotting the fraction of bound protein (α) versus the DNA concentration for TelG4-up (black), TelG4-uh (red), and TelG4-tp (green).The circles represent the experimental data, the lines are the best fit obtained with the theoretical model. (E, F) Time evolution SPR sensorgrams obtained at 25°C by injections of (E) TelG4-up and (F) TelG4-uh at various concentrations on the chip-immobilized HMGB11-166 with a contact time of 80 s, a dissociation time of 600 s, and a flow rate of 30 μl/min. The sensorgrams are shown as colored lines and their respective fits, based on the 1:1 kinetic interaction model, as black lines.
To get further insights into the kinetics and affinity of these interactions, SPR experiments were performed. Representative SPR sensorgrams obtained for the interaction of immobilized HMGB11-166 with TelG4-up and TelG4-uh are shown in Figure 8E and F, whereas no reliable SPR data were obtained for TelG4-tp, probably due to the occurrence of non-specific interactions. On the other hand, SPR experiments on TelG4-up and TelG4-uh showed a response proportional to DNA concentration, indicative of a specific interaction with the protein. The association (kon) and dissociation (koff) rate constants were derived from the fitting, and the binding dissociation constants (Kd) were kinetically determined. Kd values of 4.3 (±0.2) × 10−7 and 8.2 (±0.3) × 10−7 M were obtained for TelG4-up and TelG4-uh, respectively, in close agreement with fluorescence data. The comparison of kinetic constants obtained for TelG4-up and TelG4-uh reveals that koff values are similar for the two systems (1.62 (±0.03) × 10−3 and 1.26 (±0.02) × 10−3 s−1, respectively), while kon value was greater for TelG4-up (3.81 (±0.08) × 103 M−1 s−1) than for TelG4-uh (1.54 (±0.03) × 103 M−1 s−1), thus suggesting that the difference in affinity arises from the association rates.
Calculation of the structural models of HMGB1/TelG4-up complex
Docking calculations were performed using the software HADDOCK to obtain 3D structural models describing the interaction mechanism of HMGB1 and TelG4-up. The two isolated domains were first separately docked in distinct calculations on the DNA G4 structure. Since the interaction of box-A is not inhibited in the presence of the G4 binder Braco-19, two possible binding regions on TelG4-up were hypothesized for this domain: (i) with the G-tetrad plane and (ii) with the loops. Conversely, since the interaction of box-B is completely hindered by the presence of Braco-19, the G-tetrad plane was considered as the only binding region for this protein domain.
Two different sets of docking calculations were performed for the isolated box-A: (i) on the loops of the G4 structure (residues T6, T7, A8, T12, T13, A14), considering as interacting residues the amino acids experiencing a decrease in signal intensity during the titration performed in the presence of Braco-19; (ii) on the G-tetrad plane (residues G5, G11, G17, G23), considering as interacting residues the amino acids experiencing a decrease in signal intensity during the titration performed in the absence of Braco-19, but those interacting in the presence of Braco-19. In the first set (i), three different calculations (one for each loop of the TelG4-up interacting with box-A) were performed (Supplementary Figure S20A, panels I-III, and Supplementary Table S1, Supplementary Material). Immediately after, the case of box-A interacting at the same time with two loops of TelG4-up was analyzed by HADDOCK. The analysis of docking calculations where box-A interacts with loops I–II and II–III, respectively is summarized in Supplementary Figure S20A, panels IV–V, and Supplementary Table S1.
Conversely, a single docking calculation was performed to investigate the interaction of box-A with the G-tetrad plane of TelG4-up (set ii). The calculation provided multiple clusters with equivalent HADDOCK-statistics (see Supplementary Figure S20B and Supplementary Table S2, Supplementary Material). A deeper inspection of the models shows that each cluster (Supplementary Figure S20B-II-III-IV), except one (Supplementary Figure S20B-I), differs from the other only for a symmetric rotation of box-A around the DNA structure, while maintaining the protein interface region.
Similarly, the docking of box-B on the G-tetrad plane (residues G5, G11, G17, G23) provided many different clusters which, however, can be grouped in two distinct binding modes (Supplementary Figure S20C and Supplementary Table S3). The first (panels I and II, Table S3, I–II) exhibits a lower HADDOCK-score. The second binding mode shows a higher HADDOCK-score but with a lower violation of the experimental restraints (panels III and IV, Table S3, III–IV). In all the models the hydrophobic residues of the short arm of the box-B form stacking interactions with the G-tetrad plane. At the same time, the C-terminal residues of the protein interact with the DNA phosphate groups. It is interesting to point out that the hydrophobic residues F103 and I122, belonging to box-B and crucial for the interaction with duplex DNA (24), here interact with the G4-forming DNA.
Firstly, to investigate the binding mode of the HMGB11-166 protein to TelG4-up, the simultaneous binding of the two boxes to the G4 was analyzed. This is in agreement with our experimental data where the interaction of box-B with TelG4-up occurs only in the presence of box-A. Therefore, multibody docking calculations were performed considering: i) the interaction of the isolated box-A and box-B with two different G-tetrad planes of the same DNA molecule (model A), ii) the interaction of the isolated box-A with the loop region of TelG4-up and of isolated box-B with one G-tetrad plane (model B). Each of the two distinct docking calculations provided many different clusters, which were selected taking into account the experimental NMR data and the length of the linker connecting box-A and box-B in the HMGB1 protein, which was generated with Modeller program (39) after each docking calculation. The models of the HMGB1/TelG4-up complexes obtained by Modeller, starting from the HADDOCK clusters, were further refined with HADDOCK. The models with the best HADDOCK scores are shown in Supplementary Figures S21–S22 (Supplementary Material) and the docking statistics reported in Table S4. The docking complexes with the lowest HADDOCK-score (for model A and model B, respectively) are shown in Figure 9. The best calculated complex for model A, where HMGB1 sandwiches the DNA construct, has a lower HADDOCK-score than the best complex calculated for model B.
Figure 9.
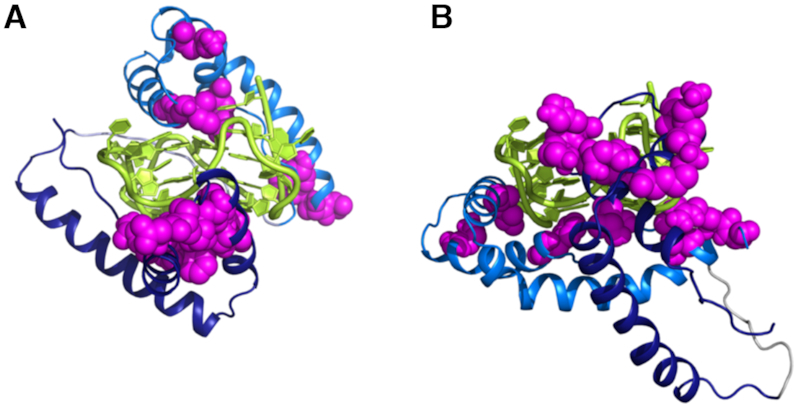
Best first models of HMGB11-166 and TelG4-up obtained performing multi-body docking calculations, with the software HADDOCK (see main text), and considering in one case the interaction of box-A with the G-tetrad plane (model A) and in the other case the interaction with the loop (model B). The interaction of box-B is with the G-tetrad plane in both cases. Box-A is in blue, box-B in marine, TelG4-up in yellow, and the ‘active’ residues in the two domains as magenta spheres.
Biological analyses
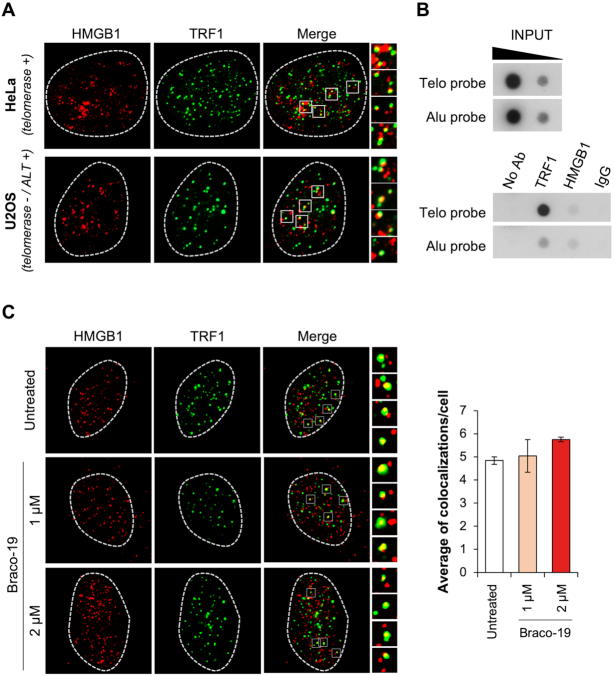
Finally, to define the biological relevance of the obtained results, we extended our study to tumor cell lines. Based on the data described above, we first evaluated whether HMGB1 localizes at telomeres in both telomerase positive cells and in cells lacking telomerase activity (telomerase negative cells), in which the telomere length is maintained through homologous recombination dependent mechanisms (ALT cells). To this aim, HeLa cells—a telomerase positive cervical carcinoma cell line with short telomeres—and U2OS—an ALT osteosarcoma cell line with longer telomers – underwent to immunofluorescence (IF) microscopy. Notably, we found that, in both the cell lines analyzed, HMGB1 colocalize with TRF1—an effective marker for interphase telomeres—indicating that, independently from telomere length and from the mechanism of telomere maintenance, HMGB1 is largely associated with telomeres (Figure 10A). In parallel with the IF experiments, the telomeric localization of HMGB1 was further confirmed in HeLa cells by chromatin immunoprecipitation (ChIP) assay by using a telomeric probe (Figure 10B). The enrichment of TRF1 protein at telomeres was used as positive control of Telo-ChIP. Moreover, additional IF experiments performed in U2OS evidenced that the treatment of the cells with Braco-19, at doses able to induce DNA damage (Supplementary Figure S23, Supplementary Material), does not affect significantly the amount of HMGB1 localized at telomeres (Figure 10C), consistent with the results of the biophysical studies.
Figure 10.
HMGB1 localizes at telomeres. (A) To establish whether HMGB1 colocalizes with telomeres, telomerase-positive HeLa cells and telomerase-negative/ALT-positive U2OS cells were processed for immunofluorescence using antibodies against the telomeric protein TRF1 (green) and HMGB1 (red). Nuclei are indicated by dotted lines. The images were acquired with a Leica Deconvolution microscope (magnification 63×). Enlarged views of co-localization spots are reported. (B) Formaldehyde-cross-linked chromatin fragments, obtained from HeLa cells, were immunoprecipitated with antibodies against HMGB1 and TRF1, as positive control for telomeric sequences. Chromatin immunoprecipitation with Rabbit immunoglobulins (IgG) and without antibody were used as negative controls. To verify that an equivalent amount of chromatin was used in the immunoprecipitates, 0.5 and 0.05 μg of the total chromatin (Input) were included in the blot. Specific (Telo) and nonspecific (Alu) probes were used. (C) U2OS cells were treated with Braco-19 at the indicated doses for 24 h and processed for IF analysis as in (A). Left panel: Representative immunofluorescence images. Right panel: Quantitative analysis of HMGB1/TRF1 colocalization, expressed as the number of HMGB1/TRF1 colocalizations for cell. Histograms show the mean values ± S.D. of three independent experiments.
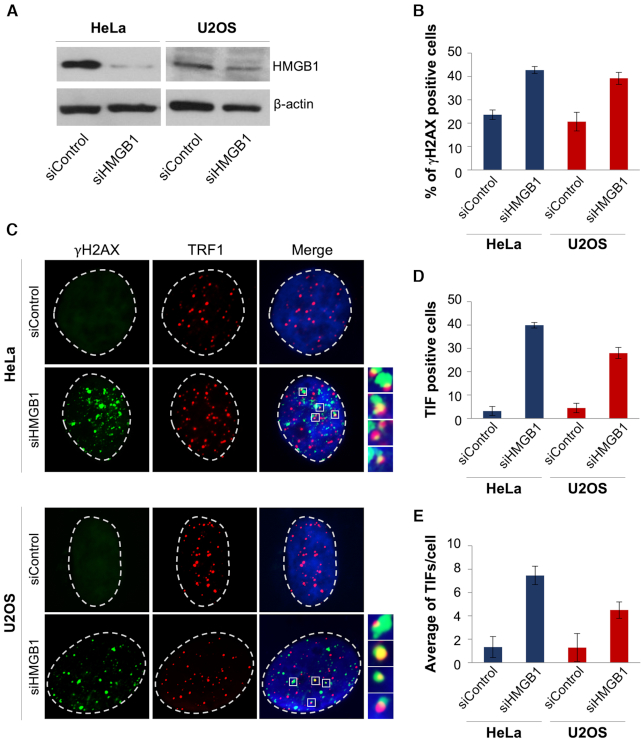
Next, moved by the observation of the telomere localization of HMGB1, we questioned whether this protein could play a role in the maintenance of telomere integrity. To address this point, HeLa and U2OS cells were transfected with a siRNA against HMGB1 (siHMGB1), or a non-targeting control (siControl), and the effect of gene silencing on DNA damage response (DDR) was evaluated by IF analyses (Figure 11). Interestingly, the down-regulation of HMGB1, verified by western blotting (Figure 11A), determined a substantial increase of the phosphorylated form of H2AX (γH2AX), a hallmark of DNA double-strand breaks (Figure 11B). Moreover, several of the damage spots induced by the silencing of HMGB1 were found to co-localize with TRF1, indicating that HMGB1 plays a critical role in telomere maintenance (Figure 11C). In particular, the quantitative analysis performed by measuring the number of telomere-induced foci (TIFs) deriving from the colocalization between γH2AX and TRF1, revealed that silencing of HMGB1 determines a significant increase in both the percentage of TIF positive cells (cells with at least four co-localization spots) and the average number of TIFs per cell (Figure 11D and E). Notably, a number of non-telomeric γH2AX foci were also observed in HMGB1 silenced cells, suggesting that this protein, probably interacting with other G4 structures present in the genome (49), may play a more general role in the maintenance of DNA integrity.
Figure 11.
Knocking-down of HMGB1 induces telomere instability. HeLa and U2OS cells were transfected with a siRNA against HMGB1 (siHMGB1) or a non-targeting siRNA (siControl) and assayed. (A) Representative western blot showing the expression levels of HMGB1 evaluated in HeLa and U2OS cells, silenced or not (siControl) for HMGB1. Analysis of β-actin was reported as loading control. (B–E) The indicated cell lines were processed for immunofluorescence (IF) using antibodies against γH2AX and TRF1 to visualize the DNA damage and telomeres, respectively. (B) Histogram showing the percentage of γH2AX-positive cells. (C) Representative images of IF microscopy experiments. γH2AX spots are visualized in green, TRF1 in red and nuclei are stained in blue. Enlarged views of Telomere Induced Foci (TIFs) are reported. The images were acquired with a Leica Deconvolution microscope (magnification 63x). (D, E) Quantitative analysis of TIFs. The graphs represent the percentages of (D) TIF-positive cells and (E) the mean number of TIFs per cell in the indicated samples. Cells with at least four γH2AX/TRF1 foci were scored as TIF positive. Histograms show the mean values ± S.D.
DISCUSSION
The G4s represent one of the most significant nucleic acid secondary structures involved in the regulation of various cellular processes. For appropriate biological function, the formation, stabilization, and resolution of G4s need to be regulated in a tight spatiotemporal manner (50). In this frame, G4 recognition and processing by nucleic acid directed proteins represent a key event to activate or deactivate physiological or pathological pathways (51,52). In the telomeric region, where G4s have been shown to have key regulatory roles in telomere extension and maintenance (53), an ensemble of interacting proteins is involved in the modulation of the telomere end-conformation and, thus, in the modification of its homeostasis (50,52).
In the present study, to get insights into the telomeric G4 DNA recognition by HMGB1 protein, we performed detailed biophysical studies complemented with biological analyses. Since there is controversy as to the exact conformation of human telomeric G4 under physiological conditions, and it has become increasingly apparent that the human telomeric sequence may be structurally heterogeneous, we investigated both the parallel-stranded and the hybrid [3+1] G4 structures formed by the d[(TTAGGG)4TT] truncation of human telomeric DNA. In addition, to evaluate a possible role of loops in the G4/protein recognition, we analysed the tetramolecular G4 formed by the short d(TTAGGGT) sequence that has no loops. CD experiments, performed to further investigate the protein/DNA interaction and monitor the unfolding or any possible change in the G4 structure upon binding, showed that no unfolding or structural alterations of DNA secondary structures occur upon binding to HMGB1 or box-A. 3D models of HMGB1/TelG4-up complex obtained by docking calculations driven by the experimental NMR data, shed light on the possible interaction mechanism and highlighted the role of the G4 shape in protein–DNA recognition.
All the biophysical methodologies employed here (CD, fluorescence, and NMR spectroscopy as well as SPR) provided evidence that HMGB1 interacts with the three subtypes of telomeric G4s, but with some differences. The NMR data provided evidence of a better interaction with the unimolecular parallel-stranded TelG4-up and suggested a major role of the box-A domain. Interestingly, the G4-binding region on box-A seems to be located mainly on the concave surface of the short arm of the L-shaped structure of the protein, on the same side of that involved in the interaction with four-way junction DNA (54), but on the opposite side to that of DNA duplex binding (23). The binding studies in the presence of the G4 binders Braco-19 and RHPS4 indicated that the ligands affect but do not prevent the G4/protein interaction in vitro. This is in agreement with the results of immunofluorescence analysis showing that telomeric localization of HMGB1 is not dramatically affected by the G4-binding compounds. Altogether, our data suggest that the protein interacts with G4 DNA also (but not only) in a region not involved in the binding with Braco-19 and RHPS4. Considering that such ligands bind to this DNA structure by stacking on the external G-tetrad(s), it is plausible that HMGB1 also interacts with the loops of G4. In summary, the experimental and computational studies reported here show that (i) the G4 architecture is important for the HMGB1 recognition and (ii) the structural determinants for a better interaction may also be represented by the presence and specific conformation of loops.
To gain insights into the energetic aspects of binding, the interaction of HMGB1 with the three G4s was examined by fluorescence titration and SPR experiments. Fluorescence experiments—performed by examining the protein/DNA interaction in the presence of Braco-19 or RHPS4—confirmed the results obtained by NMR: (i) the G4 ligands do not prevent the interaction and (ii) HMGB1 has a preference for the unimolecular parallel TelG4-up. SPR results indicated a specific protein interaction for TelG4-up and TelG4-uh, with a lower Kd for the former, in agreement with fluorescence data. Overall, the biophysical data showed that the G4 structural variability may have significant implications in protein recognition. Since the investigated G4 structures mainly differ in presence and conformation of loops, these results suggest an important role for the double-chain-reversal loops of unimolecular parallel G4 conformation in the interaction with HMGB1. Interestingly, the Kd observed for HMGB1/TelG4-up interaction is comparable to that found for the binding of this protein to another parallel-stranded G4, namely the KRAS promoter G4 (Kd of about 5 × 10−7 M) (16), thus suggesting that this protein might have a more global effect as G4-interactor, being not selective for telomeric G4s. In addition, the affinity for TelG4-up was also found to be similar to that previously observed for the binding of HMGB1 to cisplatin-modified DNA (Kd of 3.7 × 10−7 M) (55), and two orders of magnitude higher than that reported for the nonspecific binding to B-form DNA (Kd of about 5 × 10−5 M) (56). The quantitative estimate of on/off rates of binding revealed that the difference in affinity arises from the association rates. Note that kon generally determines the binding affinity to a higher extent than koff for proteins that bind with large conformational changes, whereas, for rigid complexes, koff is the major determinant for binding affinity (57).
Regarding the biological relevance of such interactions, a couple of studies have focused on the roles of HMGB1 in telomere biology. Polanska et al. showed that knockout of the HMGB1 gene in mouse embryonic fibroblasts (MEFs) resulted in a decline in telomerase activity and telomere dysfunction, while overexpression of HMGB1 enhanced telomerase activity (18). Ke et al. demonstrated that the decreasing HMGB1 levels promote telomere dysfunction and confer radiosensitivity in human breast cancer cells (17). Our biological data identified HMGB1 as a telomere-associated protein in both telomerase-positive (HeLa) and telomerase-negative (U2OS) tumor cell lines and showed that the silencing of HMGB1 encoding gene in such cells induces telomere DNA damage foci. Even if we cannot exclude a broad role of HMGB1 in maintenance of DNA integrity through interaction with G4 structures interspersed in the genome (49,58), our findings evidence that HMGB1 is indispensable for telomere homeostasis and suggest that this protein could actually represent a new target for cancer therapy. Overall, our data indicate that the relationship between HMGB1 and telomere biology, remained unclear until now, may de facto be the G4 DNA structures.
Supplementary Material
ACKNOWLEDGEMENTS
S.I. and P.Z. were recipient of a fellowship from the Umberto Veronesi Foundation (FUV).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Italian Association for Cancer Research [16730 to B.P., 18695 to A.R., 21579 to A.B.]; Regione Campania-POR Campania FESR 2014/2020 [B61G18000470007]. Funding for open access charge: Department of Pharmacy, University of Naples Federico II.
Conflict of interest statement. None declared.
REFERENCES
- 1. Blackburn E.H., Epel E.S., Lin J.. Human telomere biology: a contributory and interactive factor in aging, disease risks, and protection. Science. 2015; 350:1193–1198. [DOI] [PubMed] [Google Scholar]
- 2. Blasco M.A. Telomere length, stem cells and aging. Nat. Chem. Biol. 2007; 3:640–649. [DOI] [PubMed] [Google Scholar]
- 3. Hänsel-Hertsch R., Di Antonio M., Balasubramanian S.. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017; 18:279–284. [DOI] [PubMed] [Google Scholar]
- 4. Neidle S. Quadruplex nucleic acids as targets for anticancer therapeutics. Nat. Rev. Chem. 2017; 1:0041. [Google Scholar]
- 5. Moye A.L., Porter K.C., Cohen S.B., Phan T., Zyner K.G., Sasaki N., Lovrecz G.O., Beck J.L., Bryan T.M.. Telomeric G-quadruplexes are a substrate and site of localization for human telomerase. Nat. Commun. 2015; 6:7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim N., Piatyszek M., Prowse K., Harley C., West M., Ho P., Coviello G., Wright W., Weinrich S., Shay J.. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994; 266:2011–2015. [DOI] [PubMed] [Google Scholar]
- 7. de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005; 19:2100–2110. [DOI] [PubMed] [Google Scholar]
- 8. McEachern M.J., Krauskopf A., Blackburn E.H.. Telomeres and their control. Annu. Rev. Genet. 2000; 34:331–358. [DOI] [PubMed] [Google Scholar]
- 9. Zaug A.J., Podell E.R., Cech T.R.. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:10864–10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang F., Tang M.-l., Zeng Z.-x., Wu R.-y., Xue Y., Hao Y.-h., Pang D.-W., Zhao Y., Tan Z.. Telomere- and telomerase-interacting protein that unfolds telomere G-quadruplex and promotes telomere extension in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:20413–20418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pagano B., Margarucci L., Zizza P., Amato J., Iaccarino N., Cassiano C., Salvati E., Novellino E., Biroccio A., Casapullo A. et al.. Identification of novel interactors of human telomeric G-quadruplex DNA. Chem. Commun. 2015; 51:2964–2967. [DOI] [PubMed] [Google Scholar]
- 12. Kang R., Chen R., Zhang Q., Hou W., Wu S., Cao L., Huang J., Yu Y., Fan X., Yan Z. et al.. HMGB1 in health and disease. Mol. Aspects Med. 2014; 40:1–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Štros M. HMGB proteins: Interactions with DNA and chromatin. Biochim. Biophys. Acta - Gene Regul. Mech. 2010; 1799:101–113. [DOI] [PubMed] [Google Scholar]
- 14. Kang R., Zhang Q., Zeh H.J., Lotze M.T., Tang D.. HMGB1 in Cancer: Good, Bad, or Both. Clin. Cancer Res. 2013; 19:4046–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kang R., Xie Y., Zhang Q., Hou W., Jiang Q., Zhu S., Liu J., Zeng D., Wang H., Bartlett D.L. et al.. Intracellular HMGB1 as a novel tumor suppressor of pancreatic cancer. Cell Res. 2017; 27:916–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Amato J., Madanayake T.W., Iaccarino N., Novellino E., Randazzo A., Hurley L.H., Pagano B.. HMGB1 binds to the KRAS promoter G-quadruplex: a new player in oncogene transcriptional regulation. Chem. Commun. 2018; 54:9442–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ke S., Zhou F., Yang H., Wei Y., Gong J., Mei Z., Wu L., Yu H., Zhou Y.. Downregulation of high mobility group box 1 modulates telomere homeostasis and increases the radiosensitivity of human breast cancer cells. Int. J. Oncol. 2015; 46:1051–1058. [DOI] [PubMed] [Google Scholar]
- 18. Polanská E., Dobšáková Z., Dvořáčková M., Fajkus J., Štros M.. HMGB1 gene knockout in mouse embryonic fibroblasts results in reduced telomerase activity and telomere dysfunction. Chromosoma. 2012; 121:419–431. [DOI] [PubMed] [Google Scholar]
- 19. Wang J., Tochio N., Takeuchi A., Uewaki J., Kobayashi N., Tate S.. Redox-sensitive structural change in the A-domain of HMGB1 and its implication for the binding to cisplatin modified DNA. Biochem. Biophys. Res. Commun. 2013; 441:701–706. [PubMed] [Google Scholar]
- 20. Rowell J.P., Simpson K.L., Stott K., Watson M., Thomas J.O.. HMGB1-Facilitated p53 DNA binding occurs via HMG-Box/p53 transactivation domain interaction, regulated by the acidic tail. Structure. 2012; 20:2014–2024. [DOI] [PubMed] [Google Scholar]
- 21. Weir H.M., Kraulis P.J., Hill C.S., Raine A.R., Laue E.D., Thomas J.O.. Structure of the HMG box motif in the B-domain of HMG1. EMBO J. 1993; 12:1311–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grasser K.D., Teo S.-H., Lee K.-B., Broadhurst R.W., Rees C., Hardman C.H., Thomas J.O.. DNA-binding properties of the tandem HMG boxes of high-mobility-group protein 1 (HMG1). Eur. J. Biochem. 1998; 253:787–795. [DOI] [PubMed] [Google Scholar]
- 23. Sánchez-Giraldo R., Acosta-Reyes F.J., Malarkey C.S., Saperas N., Churchill M.E.A., Campos J.L.. Two high-mobility group box domains act together to underwind and kink DNA. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015; 71:1423–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stott K., Tang G.S.F., Lee K.-B., Thomas J.O.. Structure of a complex of tandem HMG boxes and DNA. J. Mol. Biol. 2006; 360:90–104. [DOI] [PubMed] [Google Scholar]
- 25. Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S.. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006; 34:5402–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xue Y., Kan Z., Wang Q., Yao Y., Liu J., Hao Y., Tan Z.. Human telomeric DNA forms Parallel-Stranded intramolecular G-Quadruplex in K+ solution under molecular crowding condition. J. Am. Chem. Soc. 2007; 129:11185–11191. [DOI] [PubMed] [Google Scholar]
- 27. Amato J., Morigi R., Pagano B., Pagano A., Ohnmacht S., De Magis A., Tiang Y.P., Capranico G., Locatelli A., Graziadio A. et al.. Toward the development of specific G-Quadruplex binders: synthesis, biophysical, and biological studies of new hydrazone derivatives. J. Med. Chem. 2016; 59:5706–5720. [DOI] [PubMed] [Google Scholar]
- 28. Cantor C.R., Warshaw M.M., Shapiro H.. Oligonucleotide interactions. III. Circular dichroism studies of the conformation of deoxyoligonucleolides. Biopolymers. 1970; 9:1059–1077. [DOI] [PubMed] [Google Scholar]
- 29. Amato J., Iaccarino N., Pagano B., Morigi R., Locatelli A., Leoni A., Rambaldi M., Zizza P., Biroccio A., Novellino E. et al.. Bis-indole derivatives with antitumor activity turn out to be specific ligands of human telomeric G-quadruplex. Front. Chem. 2014; 2:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pagano B., Amato J., Iaccarino N., Cingolani C., Zizza P., Biroccio A., Novellino E., Randazzo A.. Looking for efficient G-quadruplex ligands: Evidence for selective stabilizing properties and telomere damage by drug-like molecules. ChemMedChem. 2015; 10:640–649. [DOI] [PubMed] [Google Scholar]
- 31. Campbell N.H., Parkinson G.N., Reszka A.P., Neidle S.. Structural Basis of DNA Quadruplex Recognition by an Acridine Drug. J. Am. Chem. Soc. 2008; 130:6722–6724. [DOI] [PubMed] [Google Scholar]
- 32. Gavathiotis E., Heald R.A., Stevens M.F.G., Searle M.S.. Drug recognition and stabilisation of the parallel-stranded DNA quadruplex d(TTAGGGT)4 containing the human telomeric repeat. J. Mol. Biol. 2003; 334:25–36. [DOI] [PubMed] [Google Scholar]
- 33. Keller R. The Computer Aided Resonance Assignment Tutorial. 2004; Goldau: CANTINA Verlag. [Google Scholar]
- 34. Ulrich E.L., Akutsu H., Doreleijers J.F., Harano Y., Ioannidis Y.E., Lin J., Livny M., Mading S., Maziuk D., Miller Z. et al.. BioMagResBank. Nucleic Acids Res. 2008; 36:D402–D408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Giancola C., Pagano B.. Energetics of ligand binding to G-quadruplexes. Top. Curr. Chem. 2013; 330:211–242. [DOI] [PubMed] [Google Scholar]
- 36. van Zundert G.C.P., Rodrigues J.P.G.L.M., Trellet M., Schmitz C., Kastritis P.L., Karaca E., Melquiond A.S.J., van Dijk M., de Vries S.J., Bonvin A.M.J.J.. The HADDOCK2.2 Web Server: user-Friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016; 428:720–725. [DOI] [PubMed] [Google Scholar]
- 37. Wassenaar T.A., van Dijk M., Loureiro-Ferreira N., van der Schot G., de Vries S.J., Schmitz C., van der Zwan J., Boelens R., Giachetti A., Ferella L. et al.. WeNMR: structural biology on the grid. J. Grid Comput. 2012; 10:743–767. [Google Scholar]
- 38. Heddi B., Phan A.T.. Structure of human telomeric DNA in crowded solution. J. Am. Chem. Soc. 2011; 133:9824–9833. [DOI] [PubMed] [Google Scholar]
- 39. Fiser A., Šali A.. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 2003; 374:461–491. [DOI] [PubMed] [Google Scholar]
- 40. Biroccio A., Amodei S., Antonelli A., Benassi B., Zupi G.. Inhibition of c-Myc oncoprotein limits the growth of human melanoma cells by inducing cellular crisis. J. Biol. Chem. 2003; 278:35693–35701. [DOI] [PubMed] [Google Scholar]
- 41. Salvati E., Leonetti C., Rizzo A., Scarsella M., Mottolese M., Galati R., Sperduti I., Stevens M.F.G., D’Incalci M., Blasco M. et al.. Telomere damage induced by the G-quadruplex ligand RHPS4 has an antitumor effect. J. Clin. Invest. 2007; 117:3236–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dai J., Carver M., Yang D.. Polymorphism of human telomeric quadruplex structures. Biochimie. 2008; 90:1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Renčiuk D., Kejnovská I., Školáková P., Bednářová K., Motlová J., Vorlíčková M.. Arrangements of human telomere DNA quadruplex in physiologically relevant K+ solutions. Nucleic Acids Res. 2009; 37:6625–6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dai J., Carver M., Punchihewa C., Jones R.A., Yang D.. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007; 35:4927–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gavathiotis E., Searle M.S.. Structure of the parallel-stranded DNA quadruplex d(TTAGGGT)4 containing the human telomeric repeat: evidence for A-tetrad formation from NMR and molecular dynamics simulations. Org. Biomol. Chem. 2003; 1:1650–1656. [DOI] [PubMed] [Google Scholar]
- 46. Randazzo A., Spada G.P., Webba Da Silva M. Circular dichroism of quadruplex structures. Top. Curr. Chem. 2013; 330:67–86. [DOI] [PubMed] [Google Scholar]
- 47. Salvati E., Botta L., Amato J., Di Leva F.S., Zizza P., Gioiello A., Pagano B., Graziani G., Tarsounas M., Randazzo A. et al.. Lead discovery of Dual G-Quadruplex stabilizers and Poly(ADP-ribose) polymerases (PARPs) Inhibitors: A new avenue in anticancer treatment. J. Med. Chem. 2017; 60:3626–3635. [DOI] [PubMed] [Google Scholar]
- 48. Moore M.J.B., Schultes C.M., Cuesta J., Cuenca F., Gunaratnam M., Tanious F.A., Wilson W.D., Neidle S.. Trisubstituted acridines as G-quadruplex telomere targeting agents. Effects of extensions of the 3,6- and 9-Side chains on quadruplex binding, telomerase activity, and cell proliferation. J. Med. Chem. 2006; 49:582–599. [DOI] [PubMed] [Google Scholar]
- 49. Hänsel-Hertsch R., Beraldi D., Lensing S. V., Marsico G., Zyner K., Parry A., Di Antonio M., Pike J., Kimura H., Narita M. et al.. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016; 48:1267–1272. [DOI] [PubMed] [Google Scholar]
- 50. Rhodes D., Lipps H.J.. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015; 43:8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zyner K.G., Mulhearn D.S., Adhikari S., Martínez Cuesta S., Di Antonio M., Erard N., Hannon G.J., Tannahill D., Balasubramanian S.. Genetic interactions of G-quadruplexes in humans. Elife. 2019; 8:e46793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sissi C., Gatto B., Palumbo M.. The evolving world of protein-G-quadruplex recognition: a medicinal chemist's perspective. Biochimie. 2011; 93:1219–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bochman M.L., Paeschke K., Zakian V.A.. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012; 13:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Webb M., Thomas J.O.. Structure-specific binding of the two tandem HMG boxes of HMG1 to four-way junction DNA is mediated by the A domain. J. Mol. Biol. 1999; 294:373–387. [DOI] [PubMed] [Google Scholar]
- 55. Pil P., Lippard S.. Specific binding of chromosomal protein HMG1 to DNA damaged by the anticancer drug cisplatin. Science. 1992; 256:234–237. [DOI] [PubMed] [Google Scholar]
- 56. Lange S.S., Vasquez K.M.. HMGB1: the jack-of-all-trades protein is a master DNA repair mechanic. Mol. Carcinog. 2009; 48:571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kastritis P.L., Bonvin A.M.J.J.. On the binding affinity of macromolecular interactions: daring to ask why proteins interact. J. R. Soc. Interface. 2012; 10:20120835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S.. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015; 33:877–881. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.