Abstract
Background
Colorectal cancer is a successful model of genetic biomarker development in oncology. Currently, several predictive or prognostic genetic alterations have been identified and are used in clinical practice. The RAS gene family, which includes KRAS and NRAS act as predictors for anti-epithelial growth factor receptor treatment (anti-EGFR), and it has been suggested that NRAS mutations also play a role in prognosis: patients harboring NRAS alterations have a significantly shorter survival compared to those with wild type tumours. BRAF V600E mutations are rare and occur mostly in tumors located in the ascending colon in elderly female patients. BRAF is instrumental in establishing prognosis: survival is shorter by 10–16 months in BRAF-mutant patients, and BRAF may be a negative prognostic factor for patients who undergo hepatic or pulmonary metastasectomy. Moreover, this mutation is used as a negative predictive factor for anti-EGFR therapies. Two new biomarkers have recently been added to the metastatic colorectal cancer panel: HER2 and microsatellite instability. While HER2 is still being investigated in different prospective studies in order to validate its prognostic role, microsatellite instability already guides clinical decisions in substituted with advanced colorectal cancer.
Conclusions
There are current evidences that support using above mentioned genetic biomarkers to better identify the right medicine that is supposed to be used in the right patient. This approach contributes to a more individualized patient-oriented treatment in daily clinical practice.
Key words: metastatic colorectal cancer, KRAS, NRAS, BRAF, HER2, microsatellite instability
Introduction
Colorectal cancer (CRC) is the third most common cancer in men and the second most common in women.1 It is also one of the leading causes of death worldwide, accounting for 10% of all cancer deaths. Although screening, addressability and increased awareness have augmented the number of cases in the non-metastatic setting, approximately one in four individuals with CRC will be diagnosed in stage IV.1, 2 As such, a significant part of cancer research has focused on identifying novel therapies and therapeutic advances in the field of metastatic colorectal cancer (mCRC) over the last 20 years have significantly extended overall survival (OS) from 10 months to more than 20 months.3 A large part of this improvement is due to the approval of new molecular therapies (such as Bevacizumab, Cetuximab, Panitumumab, Aflibercept and Regorafenib) that are given in combination with different classical or modern cytotoxic agents (including Oxaliplatin, Irinotecan Capecitabine or Trifluridine/Tipiracil).4 Another cornerstone of improved patient management consists of identifying markers and tumor molecular anomalies that predict treatment response and can discriminate between different types of prognosis in such patients. Additionally, because this improvement in survival has also been associated with substantial health care financial burden, appropriate selection of patients for specific treatments is of utmost importance. Currently, there are several biomarkers that help clinicians in making the optimal treatment decision: KRAS, NRAS, BRAF mutations, human epidermal growth factor receptor 2 (HER2) amplification and microsatellite instability (MSI) or mismatch repair (MMR), they all play a significant role in the process, facilitating selection of the right treatment for the right patient. The aim of this review is to provide clinicians with an update on the particular features of these biomarkers. The correlations between demographic, clinical, pathological and molecular characteristics of KRAS, NRAS, BRAF, HER2 and MSI and patient outcome will be presented, together with the role of these assessments in determining mCRC prognosis and treatment personalization.
This paper shows the impact of such biomarkers analyzing the results of clinical trials and their outcomes from the perspective of routine clinical practice.
Methods
A search algorithm (Figure 1) based on a combination of the terms ‘‘metastatic colorectal cancer’’ AND “therapy” OR “treatment” AND “RAS” OR “KRAS” OR “NRAS” AND “BRAF” AND “HER2” AND “MSI” OR “MMR” was used for the search in PubMed and EMBASE, data was gathered from the beginning of the database PubMed and the search was updated until 30th of April 2019. All the studies which analyzed the biomarkers KRAS, NRAS, BRAF, HER2 and MSI, were considered eligible for inclusion in this review. Eligibility criteria for study selection included: 1) clinical studies/trials; and 2) reviews; 3) meta-analysis. The exclusion criteria were: a) articles not within the field of interest of this review: not discussing about clinical, pathological and molecular correlations, predictive factors, prognostic factors of the studied biomarkers; b) editorials, letters to the editor, commentaries, conference proceedings; c) case reports or small case series; d) articles not in English; e) studies not in humans.
Figure 1.
Flow chart of the search for the eligible studies.
Eligible studies reported on patients with metastatic colorectal cancer and included details on KRAS, NRAS, BRAF, HER2 and MSI biomarker status of the tumour, oncological outcomes and type of therapy implemented.
KRAS: an indispensable biomarker for anti-epidermal growth factor receptor treatment
RAS is a family of proteins expressed in all cell organs belonging to a class of protein called GTPase, and its role is to transmit signals within cells. These signals finally stimulate cell proliferation. RAS regulated signal pathways control processes like cell proliferation, cell differentiation, cell adhesion, apoptosis and cell migration. When they are mutated, the cell will have an increased potential of invasion and metastasis. The main members of the RAS family are KRAS and NRAS mutations.5, 6, 7 These are point mutations in which a single nucleotide base is changed, inserted or deleted from a DNA sequence. Moreover, these are frequently somatic mutations (acquired during lifetime).8 In the metastatic setting, KRAS mutations occur in approximately 40% of the cases, especially in exon 2, codons 12 (70–80%) and 13 (15–20%). Different KRAS mutations are mainly located in exon 3, codons 59–61, and in exon 4, codons 117 and 146. In exon 2, mutations are common in codons 12: G12D, G12V and G12C; in exon 3, the affected codons are Q61H and Q61R, while in exon 4 the codons involved are A146T and A146V.9
The geographic distribution of RAS mutations is uneven. The distribution of KRAS mutation among clinical trials was 44.7% in Western European countries, 35.8% in Eastern European countries, while 19.5% of the patients were from the Middle East. Also, in one Middle Eastern study by Zekri et al., the percentage of mutations ranged from 13% to 45% depending on country.10, 11, 12, 13, 14, 15 Most of the data suggest that geographical distribution is not a significant factor in how such mutations are positioned in the codons. As for racial distribution, certain studies indicated that African-American patients have RAS tumours (68%) more often than Caucasians. It is unclear if such geographic and racial variations are due to genetic background or environmental and lifestyle differences.16
Concerning gender, data has been inconclusive. In a meta-analysis by Kafatos et al, RAS mutations were distributed almost equally among men and women: 43.8% in men vs 43.3% in women, P = 0.006.17 In another study by Kwak et al., the rate of mutations was significantly higher in women than in men (46% vs 34.4%, P = 0.03). Codon 12 mutations were more prevalent in women than in men (73.4% vs 66.2%). In the same study, the occurrence of KRAS mutation subtypes did not appear to depend on gender (P = 0.592) and no significant differences were noticed regarding the codon 12 and codon 13 mutations in women versus in men (P = 0.166, P = 0.122).18
The full analysis of the RAS gene exons 2, 3 and 4 revealed that these mutations correlate with certain typical clinical, pathological and molecular features, depending on their exact location on exons and codons. For instance, the mutations of exon 2 and codon 12 are associated with the well/ moderately differentiated adenocarcinoma subtype and the mucinous subtype.19
In terms of where tumours with KRAS mutations are located, findings are inconsistent: some studies have shown that the KRAS mutation does not correlate with the location of the tumour, while others have found that tumours with KRAS mutations occur more frequently in the caecum area. These studies are limited by the lack of clinical and histological data, as well as by their retrospective observational methodology.18
Epidermal growth factor receptor (EGFR) is considered a very important component of initiation and progression in colorectal cancer. It is a membrane-bound receptor tyrosine kinase and became a key target for monoclonal antibodies which bind on the extracellular domain of the receptor. The KRAS status is critical for the medical oncologists because it guides the treatment. KRAS mutations have been considered a predictive feature for resistance to EGFR biological therapy, as confirmed by the results of the CRYSTAL and OPUS trials.20, 21
These indicated that adding Cetuximab to standard chemotherapy protocols such as FOLFOX (regimen of chemotherapy consisting of the next citotoxic agents: Oxaliplatin, 5-Fluorouracil and Folinic acid) and FOLFIRI (regimen of chemotherapy consisting of the next citotoxic agents: Irinotecan, 5-Fluorouracil and Folinic acid) has no benefit in patients with KRAS mutation. However, in patients with RAS wild-type (WT), the addition of Cetuximab resulted in improved prognosis.22, 23 The same conclusions were shown as well in clinical trials with high enrolment number like COIN, NORDIC or PRIME.24, 25, 26 Some trials investigated potential correlations between KRAS mutation and Bevacizumab efficacy, but with negative results.27 Another predictive element in relation to KRAS is the location of the tumour: left-side CRC tumours are more responsive to anti-EGFR treatment. This may be due to the more frequent activation of EGFR signalling in left-sided tumours compared to those on the right side.28 In contrast with anti-EGFR treatment, the effectiveness of Bevacizumab is not dependent on tumour location.29 Recent data indicates that different KRAS mutations exert different biological effects and their impact on EGFR resistance is not consistent throughout the mutational spectrum. Thus, not all tumours with KRAS mutation are resistant to EGFR-inhibiting treatment. This phenomenon is probably due to the heterogeneity of tumours. It should also be noted that patients with tumours on the right colon exhibit more frequent BRAF mutations, which result in relative resistance to anti-EGFR treatment.28, 29, 30
Available data is inconclusive regarding the prognostic role of KRAS. One study showed that patients with KRAS mutation who presented with stage IV CRC, synonymous with metastatic disease, had a higher mortality rate (34% versus 18.5%) and reduced OS (23.5 months versus 14 months) compared to patients without this mutation. Also, the presence of the KRAS mutation was found to be an independent risk factor for reduced survival.8 The mutation status of KRAS has not been associated significantly with OS in the case of patients receiving best supportive care (BSC) in a randomized phase III trial comparing Cetuximab with BSC.33 One explanation for such inconsistent results is that different mutations in different codons may lead to variable tumour activity of the KRAS protein.34
NRAS: the little brother of KRAS
Patients with NRAS mutation form a distinct mCRC subgroup from a clinical and molecular standpoint. 3–5% of CRC show a mutation in exons 2, 3 and 4 of the NRAS gene, but the impact of these genetic changes is less studied compared to KRAS and BRAF mutations due to the low number of patients.34
One study by Schiripa et al. found the NRAS mutation in 6% of the 785 included patients. Available literature indicates that the clinical and pathological features of patients with KRAS and NRAS mutation are similar. In another study, tumours with NRAS mutation were identified on the proximal colon in 33% of the cases, on the distal colon in 36% and at the rectal level in 31% of the cases. KRAS and NRAS mutant tumours exhibit similar metastasis patterns, namely their dissemination is frequently hepatic, pulmonary and peritoneal. Nevertheless, there are individual differences: low incidence of mucinous histology in patients with NRAS mutation (4% versus 26%, P = 0.012) and less frequent lung metastasis (30% versus 35%).34
Patients with NRAS mutation respond poorly to anti-EGFR treatment. De Roock et al. evaluated the role of the NRAS mutation in a cohort of patients treated with Cetuximab and chemotherapy. Only one of the 13 patients responded to treatment. A phase II trial that included patients with NRAS mutation found them non-responsive to anti-EGFR treatment. Similar results were obtained in a phase III trial testing Panitumumab against BSC: none of the patients with NRAS mutation responded to the treatment.35 Similar results were found in another study: only one of the 37 patients responded with an objective response rate of 2.7%. Given the ineffectiveness of anti-EGFR treatment in patients with NRAS mutation, the European Drug Agency does not recommend the use of anti-EGFR drugs in such patients.34
In Schiripa’s study, patients with NRAS mutation had a lower OS rate compared to WT patients – 25.6 months vs 42.7 months. There were no differences in the survival of patients with NRAS and those with KRAS mutations. OS depended on the position of the mutation on the exons. In mCRC patients with NRAS mutation, it was significantly shorter in exon 3 compared to mCRC RAS WT patients (HR 2.85; 95% CI 1.87–4.36, P < 0.01) and to patients with NRAS mutation in exon 2 (HR 2.0; 95% CI 1.04–4.0, P = 0.039).34, 35, 36
BRAF: rare, but important
BRAF plays a role in MAP-kinase (MAPK) pathway activation and contributes to cellular growth, proliferation and differentiation, as well as to other key cellular processes such as migration, apoptosis and cellular survival. Approximately 90% of BRAF mutations occur at the level of T1799 transversion in exon 15, which leads to the substitution of valine for glutamic acid (V600E).37 This substitution regulates phosphorylation, increasing BRAF activity by approximately ten times compared to WT.38
CRC patients with BRAF mutation are a small and unique group that make up 8%–12% of all the patients suffering from CRC.39, 40, 41, 42, 43, 44 Concerning the epidemiology of the BRAF mutation, several studies have revealed similar rates of occurrence across the world, with only minor variations between regions.45, 46, 47, 48 The BRAF mutation has been reported in multiple studies in association with various clinical and pathological parameters in mCRC patients. It is more common in women older than 70, and for tumours located in the right colon.49 BRAF mutations are less frequent if the left colon is affected (4%) and in rectal cancers (2%).44 In terms of aggressiveness, approximately 60% of the BRAF mutant tumours are poorly differentiated, and only 36% of them are well or moderately differentiated. Histologically, the mucinous subtype is more frequently associated with BRAF mutant cancers (22–67%).46, 47, 48, 49 Unlike most colorectal cancers, tumours with BRAF mutations metastasize more frequently in the peritoneum and less commonly in the lungs and liver.50
The relationship between BRAF mutations and certain molecular tumour characteristics has been investigated. The BRAF and KRAS mutations are mutually exclusive. The BRAF mutation coexists with the PIK3CA mutation in 13% of the patients and with the PTEN mutation in 22% of the patients.51, 52, 53
BRAF V600 mutations are significantly more common in patients with MSI high (38.9%) than in those with MSI-low (9.3%; OR = 8.18; 95% CI = 5.08–13.17).51 Patients with sporadic CRC and MSI-high carry the BRAF mutation in 91% of the cases.54, 55 Therefore, BRAF mutation testing is also useful in identifying patients with Lynch syndrome. Patients with MSI-high and the absence of BRAF mutation should undergo genetic polymerase chain reaction testing to confirm Lynch syndrome.56, 57
BRAF mutations are considered a biomarker for negative prognosis in mCRC. Several trials such as COIN, PRIME, CRYSTAL and OPUS, as well as a meta-analysis of 21 trials with patients in the metastatic stage, have found reduced survival and shorter progression-free survival (PFS) in mCRC patients with BRAF mutation. Thus, regardless of the approach to treatment, median survival is generally reported as 10–16 months shorter in CRC patients with BRAF mutation than in those without it.53, 58 In recent years, the role of the BRAF mutation in patients undergoing metastasectomy has been discussed. In a retrospective study of 309 patients whose secondary hepatic lesions were surgically removed, recurrence-free survival was 5.7 months for those with BRAF mutation compared to 11 and 14.4 months for RAS mutant and RAS WT without BRAF mutation, respectively (P = 0.003).59 Renaud et al. evaluated the pulmonary metastasectomies retrospectively in 180 patients with BRAF mutation, KRAS mutant and WT CRC. Patients with CRC and BRAF mutation had a lower survival rate following surgery compared to those with KRAS mutant tumours or WT (15, 55 and 98 months, respectively, P < 0.0001).60
Several post-hoc analyses of phase III randomized trials have assessed the predictive role of the BRAF V600E mutation concerning the effectiveness of anti-EGFR therapies. The results of these retrospective studies did not reach statistical significance and were insufficient for a definitive conclusion about the potential use of BRAF V600E as a biomarker for determining primary resistance to anti-EGFR agents in CRC.61, 62, 63, 64, 65 Therefore, the predictive role of the BRAF mutation for anti-EGFR agents after two meta-analyses is still unclear.66, 67 Pietrantonio et al concluded that the BRAF mutations could be a negative predictive factor for anti-EGFR agents, thus supporting the meta-analysis conducted by Yuan et al.68 However, another meta-analysis by Rowland et al. of 7 randomized controlled trials looking at OS and PFS concluded that the evidence is insufficient in order to justify the exclusion of anti-EGFR agents in the case of patients with BRAF mutation.53, 67, 68
It has been suggested that BRAF-mutant patients might benefit more from an intensive chemotherapy regimen, such as the FOLFOXIRI (regimen of chemotherapy consisting of the next citotoxic agents: Oxaliplatin, Irinotecan, 5-Fluorouracil and Folinic acid) and Bevacizumab protocol. Loupakis et al. obtained encouraging results in their study of 15 patients with BRAF mutation in a validation cohort with a median OS of 24.1 months and median PFS of 11.8 months.69 Although the results were confirmed in the phase 3 trial TRIBE, this strategy was not embraced by all oncologists due to the high toxicity profile in which FOLFOXIRI-Bevacizumab was compared with FOLFIRI-Bevacizumab as the first line of treatment in mCRC. Recently, the FDA has approved the combined use of Encorafenib (a small molecule which blocks BRAF by acting as a competitive RAF kinase inhibitor), Binimetinib (an inhibitor of the mitogen-activating kinase) and Cetuximab for the treatment of mCRC patients with BRAF V600E mutation who underwent one or two lines of chemotherapy for their metastatic disease based on the results of the BEACON trial, which found a 62% OS rate 1 year after the analysis. The median PFS for the patients treated with this triple combination was of 8 months (95% CI = 5.6–9.3) regardless of whether or not they had previously benefitted from one or two lines of treatment. The overall response rate was 48%, and in the case of patients who had previously undergone a single line of treatment it was 62%.70
Non-V600 BRAF mutations are a special and infrequent category (they occur in 2% of mCRC patients). Certain differences between patients with V600 and those with non-V600 BRAF mutations were noticed: the latter were found in younger patients (58 versus 68), mostly male (65% versus 46%), with well-differentiated tumours located less often in the right colon and which more frequently suffer concurrent RAS mutations. Survival was also much longer in this category of patients compared to mCRC patients with V600E BRAF mutation or RAS WT (60.7 months versus 11.4 and 43 months, respectively, P < 0.001). Non-V600 BRAF mutations define a distinct molecular subtype of mCRC with excellent prognosis.53
HER2: the stranger on the shore
HER2 oncogene is a member of the tyrosine kinase family similar to EGFR, HER-1, HER-3 and HER-4. HER-2 is located on chromosome 17 and codes a transmembrane protein of 185 kD which is activated through ligand binding. HER-2 activation initiates the signal pathways, including MAPK and PI3K/AKT, which are essential for cellular proliferation and differentiation. Meanwhile, the family of receptors is located on the normal cells; multiple studies have shown that they are overexpressed in multiple malignant tumors, including colorectal cancer.71 HER2 amplification is a relevant genetic alteration in mCRC. This fact was documented in the HERACLES and MyPathway clinical trials. This biomarker can be screened at diagnosis and has a prevalence of approximately 5% in patients with KRAS WT mCRC.72 Seo et al. investigated different correlations between different clinicopathological variables and the HER2 status. Overexpression of HER2 was not associated with gender and micro-satellite status but was correlated with an aggressive tumoural behaviour which includes profound invasion, lymphatic metastases, distant metastases, perineural invasion and distal colon location with the highest incidence in the rectum.73, 74, 75
HER2 can be a predictive factor for anti-EGFR therapies. Two retrospective clinical series supports the idea that HER2 signalling activation could determine cetuximab resistance.76, 77, 78, 79 Raghav et al. analysed the impact of HER2 amplification and the efficacy of monoclonal antibodies in RAS and BRAF WT mCRC in a cohort of 99 patients, which included 99 patients. 37 of 99 patients had HER2 amplification identified with next-generation sequencing. Median PFS with anti-EGFR treatment was significantly shorter for the patients with HER2 amplification compared to those without amplification (2.9 months versus 8.1 months, P < 0.0001).72 Yonesaka et al., evaluated the clinical impact of de novo HER2 amplification in 233 patients treated with cetuximab.78 Median PFS and OS were reduced in patients with amplified HER versus unamplified HER2, 3 months versus 5 months and OS was 10.2 months versus 30.5 months. (P < 0.0013).80 In the HERACLES-A clinical trial, which included only patients with HER2 positive mCRC patients who received previously Panitumumab and Cetuximab had resistance to Trastuzumab. However, these data are retrospective, and they must be carefully taken into consideration because they have to be validated in prospective clinical trials. HERACLES-A clinical trial opened new therapeutic perspectives in mCRC with the use of HER2 dual blockade with Lapatinib and Trastuzumab in patients with KRAS WT in exon 2 (codons 12 and
13) pretreated with four lines or more of chemotherapy and with resistance to Cetuximab and Panitumumab. The objective response rate was 35%, clinical benefit was 70%, median PFS was 5,5 months, and the safety profile was agreeable.72 The early trials proposed a negative prognostic impact of HER2 overexpression, but more recent trials did not confirm this fact.80, 81 Li’s meta-analysis indicated that HER2 overexpression probably has a minor impact on OS in patients with mCRC. The prognostic role of HER2 in mCRC remains uncertain due to few clinical trials which analyzed this problem.82
Microsatellite instability: the new player which brings hope
Microsatellites are repetitive sequences of coding, and non-coding DNA.83 MSI is the result of the inability of MMR gene to repeat the DNA errors appeared during replication. Insertions and deletions represent somatic mutations in these repetitive sequences of DNA, and they determine genomic instability. MMR genes inactivation is the result of MLH1 promoter hypermethylation or the germinal mutations of MLH1, MSH2, MSH6 and PMS2.84 Germinal anomalies of MSI represent the molecular basis of Lynch syndrome.85 MSI is detected in approximately 15% of patients with mCRC; only 3% of cases are associated with Lynch syndrome and the other 12% are caused by sporadic hyper-methylation of the MLH1 gene. The MSI prevalence is similar across different populations: 12% of Afro-Americans are positive, 12% of Hispanics are positive, and 12% of Caucasians are positive. CRC tumours are more frequently positive for MSI in stage II and III than in stage IV. Sporadic MSI positive tumours are located proximal, appear more frequent in older women, are poorly differentiated, have mucinous histology and have pronounced lymphocytic infiltration.86
Mismatch deficiency causes multiple somatic mutations that can produce multiple immunogenic neoantigens and antigens, which will increase the response to checkpoint inhibitors. Le et al. in a phase II study have shown that patients with deficient mismatch CRC were treated with anti-PD-1 pembrolizumab and they had an objective response rate of 62% compared with MSI-L tumours. Nivolumab monotherapy and the nivolumab ipilimumab combination in patients with pretreated MSI-H mCRC was studied. After a median follow-up of 21 months, patients treated with nivolumab had a response rate of 34%. The responses were durable; 64% of patients had at least a response for a year. After one year, 44% of patients did not progress, and 73% were alive. After a median follow-up of 13.4 months, the patients who received the combination of nivolumab and ipilimumab had a response rate of 55%, 71% did not have progression, and 85% were alive. The discovery of MSI in CRC widened the molecular landscape and created the premises for a new type of systemic treatment: immunotherapy. MSI-H became a predictive biomarker for immunotherapy in stage IV currently CRC.87
Figure 2.
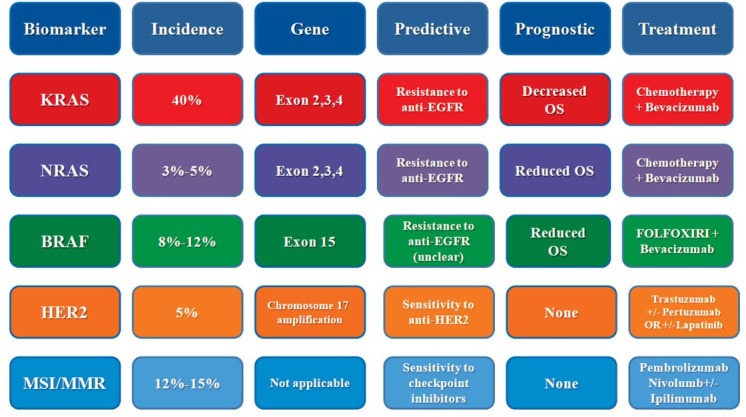
Summary features of biomarkers in stage IV colorectal cancer
EGFR = epithelial growth factor receptor; FOLFOXIRI = regimen of chemotherapy consisting of 5-fluorouracil, folinic acid, oxaliplatin and irinotecan; MMR = mismatch repair; MSI = microsatellite instability; OS = overall survival; WT = wild type
MSI represents a favourable prognostic factor in stage II and III of disease, but this is not applicable for metastatic disease. In a study of 2439 patients with mCRC, no significant survival differences were observed between patients with MSI-H and MSI-L tumours (3.82 versus 2.95 years; P = 0,76).88, 89
Conclusions
Biomarker testing in mCRC patients has become a routine in clinical practice. While the predictive role of KRAS and NRAS is well-known and widely used in treatment selection, other features of these biomarkers are only now being investigated in prospective studies. NRAS-mutated tumors are associated with reduced OS and resistance to anti-EGFR treatment. Similarly, the presence of a BRAF mutation in mCRC seems to also predict resistance to anti-EGFR treatment and identifies a very poor-prognosis subgroup of patients. More recently, HER2 overexpression has been linked to sensitivity to anti-Her2 treatment in mCRC patients and MSI/MMR status has been shown to predict tumor response to checkpoint inhibitors. All these information help identify personalized treatments for cancer patients, thus increasing overall survival and significantly decreasing drug-related toxicity.
Disclosure
No potential conflicts of interest were disclosed.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics. CA Cancer J Clin. 2002;2005:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Moghimi-Dehkordi B, Safaee A. An overview of colorectal cancer survival rates and prognosis in Asia. World J Gastrointest Oncol. 2012;4:71–5. doi: 10.4251/wjgo.v4.i4.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolpin BM, Mayer RJ. Systemic treatment of colorectal cancer. Gastroenterology. 2008;134:1296–310. doi: 10.1053/j.gastro.2008.02.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly H, Goldberg RM. Systemic therapy for metastatic colorectal cancer: current options, current evidence. J Clin Oncol. 2005;23:4553–60. doi: 10.1200/JCO.2005.17.749. [DOI] [PubMed] [Google Scholar]
- 5.Bos JL, Fearon ER, Hamilton SR, Verlaan-de Vries M, van Boom JH, van der Eb AJ. Prevalence of RAS mutations in human colorectal cancers. Nature. 1987;327:293–7. doi: 10.1038/327293a0. et al. [DOI] [PubMed] [Google Scholar]
- 6.Finkelstein SD, Sayegh R, Christensen S, Swalsky PA. Genotypic classification of colorectal adenocarcinoma. Cancer. 1993;71:3827–38. doi: 10.1002/1097-0142(19930615)71:12<3827::aid-cncr2820711207>3.0.co;2-n. 10.1002/1097-0142(19930615)71:12<3827::aid-cncr2820711207>3.0.co;2-n [DOI] [PubMed] [Google Scholar]
- 7.Boughdady IS, Kinsella AR, Haboubi NY, Schofield PF. K-ras gene mutations in adenomas and carcinomas of the colon. Surg Oncol. 1992;1:275–82. doi: 10.1016/0960-7404(92)90087-2. [DOI] [PubMed] [Google Scholar]
- 8.Heinemann V, Stintzing S, Kirchner T, Boeck S, Jung A. Clinical relevance of EGFR- and KRAS-status in colorectal cancer patients treated with monoclonal antibodies directed against the EGFR. Cancer Treat Rev. 2009;35:262–71. doi: 10.1016/j.ctrv.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Gong J, Cho M, Sy M, Salgia R, Fakih M. Molecular profiling of metastatic colorectal tumors using next-generation sequencing: a single-institution experience. Oncotarget. 2017;8:42198–213. doi: 10.18632/oncotarget.15030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ibrahim T, Saer-Ghorra C, Trak-Smayra V, Nadiri S, Yazbeck C, Baz M. Molecular characteristics of colorectal cancer in a Middle Eastern population in a single institution. Ann Saudi Med. 2018;38:251–9. doi: 10.5144/0256-4947.2018.251. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abubaker J, Bavi P, Al-Haqawi W, Sultana M, Al-Harbi S, Al-Sanea N. Prognostic significance of alterations in KRAS isoforms KRAS-4A/4B and KRAS mutations in colorectal carcinoma. J Pathol. 2009;219:435–45. doi: 10.1002/path.2625. et al. [DOI] [PubMed] [Google Scholar]
- 12.Zahrani A, Kandil M, Badar T, Abdelsalam M, Al-Faiar A, Ismail A. Clinico-pathological study of K-ras mutations in colorectal tumors in Saudi Arabia. Tumori. 2014;100:75–9. doi: 10.1700/1430.15819. [DOI] [PubMed] [Google Scholar]
- 13.Murtaza B, Bibi A, Rashid M, Khan Y, Chaudri M, Shakoori A. Spectrum of KRAS mutations in Pakistani colorectal cancer patients. Braz J Med Biol Res. 2014;47:35–41. doi: 10.1590/1414-431X20133046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segal G, Liebermann N, Klang S, Siegelmann-Daniel N, Beit-Or A, Klien B. Identification of KRAS mutations in colorectal cancer patients in Israel. Harefuah. 2011;150:447–50. et al. [PubMed] [Google Scholar]
- 15.Marchoudi N, Joutei HAH, Jouali F, Fekkak J, Rhaissi H. Distribution of KRAS and BRAF mutations in Moroccan patients with advanced colorectal cancer. Pathol Biol. 2013;61:273–6. doi: 10.1016/j.patbio.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Chaiyapan W, Duangpakdee P, Boonpipattanapong T, Kanngern S, Sangkhatthat S. Somatic mutations of K-RAS and BRAF in Thai colorectal cancer and their prognostic value. Asian Pac J Cancer Prev. 2013;14:329–32. doi: 10.7314/apjcp.2013.14.1.329. [DOI] [PubMed] [Google Scholar]
- 17.Kafatos G, Niepel D, Lowe K, Jenkins-Anderson S, Westhead H, Garawin T. RAS mutation prevalence among patients with metastatic colorectal cancer : a meta-analysis of real-world data. Biomark Med. 2017;11:751–60. doi: 10.2217/bmm-2016-0358. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwak MS, Cha JM, Cho YH, Kim SH, Yoon JY, Jeon JW. Clinical predictors for KRAS codon 13 mutations in patients with colorectal cancer. J Clin Gastroenterol. 2018;52:431–6. doi: 10.1097/MCG.0000000000000809. et al. [DOI] [PubMed] [Google Scholar]
- 19.Dobre M, Dinu DE, Panaitescu E, Birla RE, Iosif CI, Boeriu M. KRAS gene mutations – prognostic factor in colorectal cancer? Rom J Morphol Embryol. 2015;56(2 Suppl):671–8. et al. PMID: 26429158. [PubMed] [Google Scholar]
- 20.Van Cutsem, Köhne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–17. doi: 10.1056/NEJMoa0805019. et al. [DOI] [PubMed] [Google Scholar]
- 21.Bokemeyer C, Bondarenko I, Hartmann JT, de Braud F, Schuch G, Zubel A. Efficacy according to biomarker status of cetuximab plus FOLFOX-4 as first-line treatment for metastatic colorectal cancer: the OPUS study. Ann Oncol. 2011;22:1535–46. doi: 10.1093/annonc/mdq632. et al. [DOI] [PubMed] [Google Scholar]
- 22.Pentheroudakis G, Kotoula V, De Roock W, Kouvatseas G, Papakostas P, Makatsoris T. Biomarkers of benefit from cetuximab-based therapy in metastatic colorectal cancer: interaction of EGFR ligand expression with RAS/ RAF, PIK3CA genotypes. BMC Cancer. 2013;13:49. doi: 10.1186/1471-2407-13-49. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao C, Yang ZY, Hu XF, Chen Q, Tang JL. PIK3CA exon 20 mutations as a potential biomarker for resistance to anti- EGFR monoclonal antibodies in KRAS wild-type metastatic colorectal cancer: a systematic review and meta-analysis. Ann Oncol. 2012;23:1518–25. doi: 10.1093/annonc/mdr464. [DOI] [PubMed] [Google Scholar]
- 24.Brulé SY, Jonker DJ, Karapetis CS, O’Callaghan CJ, Moore MJ, Wong R. Location of colon cancer (right-sided versus left-sided) as a prognostic factor and a predictor of benefit from cetuximab in NCIC CO.17. Eur J Cancer. 2015;51:1405–14. doi: 10.1016/j.ejca.2015.03.015. et al. [DOI] [PubMed] [Google Scholar]
- 25.Petrelli F, Tomasello G, Borgonov K, Ghidini M, Turati L, Dallera P. Prognostic survival associated with left-sided vs right-sided colon cancer A systematic review and meta-analysis. JAMA Oncol. 2016;3:211–19. doi: 10.1001/jamaoncol.2016.4227. et al. [DOI] [PubMed] [Google Scholar]
- 26.Tejpar S, Celik I, Schlichting M, Sartorius U, Bokemeyer C, Van Cutsem E. Association of KRAS G13D tumor mutations with outcome in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. J Clin Oncol. 2012;30:3570–7. doi: 10.1200/JCO.2012.42.2592. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Ye Y, Sun H, Shi G. Association between KRAS codon 13 mutations and clinical response to anti-EGFR treatment in patients with metastatic colorectal cancer: results from a meta-analysis. Cancer Chemother Pharmacol. 2013;71:265–72. doi: 10.1007/s00280-012-2005-9. [DOI] [PubMed] [Google Scholar]
- 28.Peeters M, Douillard JY, Van Cutsem E, Siena S, Zhang K, Williams R. Mutant KRAS codon 12 and 13 alleles in patients with metastatic colorectal cancer: assessment as prognostic and predictive biomarkers of response to panitumumab. J Clin Oncol. 2013;31:759–65. doi: 10.1200/JCO.2012.45.1492. et al. [DOI] [PubMed] [Google Scholar]
- 29.Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten Ras mutations in patients with colorectal cancer: the multicenter ‘‘RASCAL” study. J Natl Cancer Inst. 1998;90:675–84. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 30.Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study onthe PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28:466–74. doi: 10.1200/JCO.2009.23.3452. et al. [DOI] [PubMed] [Google Scholar]
- 31.Ogino S, Meyerhardt JA, Irahara N, Niedzwiecki D, Hollis D, Saltz LB. KRAS mutation in stage III colon cancer and clinical outcome following intergroup trial CALGB 89803. Clin Cancer Res. 2009;15:7322–9. doi: 10.1158/1078-0432.CCR-09-1570. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–65. doi: 10.1056/NEJMoa0804385. et al. [DOI] [PubMed] [Google Scholar]
- 33.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–62. doi: 10.1016/S1470-2045(10)70130-3. et al. [DOI] [PubMed] [Google Scholar]
- 34.Schiripa M, Cremolini C, Loupakis F, Morvillo M, Bergamo F, Zoratto F. Role of NRAS mutations as prognostic and predictive markers in metastatic colorectal cancer. Int J Cancer. 2015;136:83–90. doi: 10.1002/ijc.28955. et al. [DOI] [PubMed] [Google Scholar]
- 35.Au HJ, Karapetis CS, O’Callaghan CJ, Tu D, Moore MJ, Zalcberg JR. Health-related quality of life in patients with advanced colorectal cancer treated with cetuximab: Overall and KRAS-specific results of the NCIC CTG and AGITG CO.17 Trial. J Clin Oncol. 2009;27:1822–8. doi: 10.1200/JCO.2008.19.6048. et al. [DOI] [PubMed] [Google Scholar]
- 36.Matallanas D, Birtwistle M, Romano D, Zebisch A, Rauch J, von Kriegsheim A. Raf family kinases: old dogs have learned new tricks. Genes Cancer. 2011;2:232–60. doi: 10.1177/1947601911407323. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM. Mechanism of activation of the RAF- ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–67. doi: 10.1016/s0092-8674(04)00215-6. et al. [DOI] [PubMed] [Google Scholar]
- 38.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. et al. [DOI] [PubMed] [Google Scholar]
- 39.Maughan TS, Adams RA, Smith CG, Meade AM, Seymour MT, Wilson RH. Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet. 2011;377:2103–14. doi: 10.1016/S0140-6736(11)60613-2. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Souglakos J, Philips J, Wang R, Marwah S, Silver M, Tzardi M. Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. Br J Cancer. 2009;101:465–72. doi: 10.1038/sj.bjc.6605164. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from theMRC FOCUS trial. J Clin Oncol. 2009;27:5931–7. doi: 10.1200/JCO.2009.22.4295. et al. [DOI] [PubMed] [Google Scholar]
- 42.Tran B, Kopetz S, Tie J, Gibbs P, Jiang ZQ, Lieu CH. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer. 2011;117:4623–32. doi: 10.1002/cncr.26086. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yokota T, Ura T, Shibata N, Takahari D, Shitara K, Nomura M. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br J Cancer. 2011;104:856–62. doi: 10.1038/bjc.2011.19. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tie J, Gibbs P, Lipton L, Christie M, Jorissen RN, Burgess AW. Optimizing targeted therapeutic develop- ment: analysis of a colorectal cancer patient population with the BRAF(V600E) mutation. Int J Cancer. 2011;128:207584. doi: 10.1002/ijc.25555. et al. [DOI] [PubMed] [Google Scholar]
- 45.Li HT, Lu YY, An YX, Wang X, Zhao QC. KRAS, BRAF and PIK3CA mutations in human colorectal cancer: relationship with metastatic colorectal cancer. Oncol Rep. 2011;25:1691–97. doi: 10.3892/or.2011.1217. [DOI] [PubMed] [Google Scholar]
- 46.Hsieh LL, Er TK, Chen CC, Hsieh JS, Chang JG, Liu TC. Characteristics and prevalence of KRAS, BRAF, and PIK3CA mutations in colorectal cancer by high-resolution melting analysis in Taiwanese population. Clin Chim Acta. 2012;413:1605–11. doi: 10.1016/j.cca.2012.04.029. [DOI] [PubMed] [Google Scholar]
- 47.Shen Y, Wang J, Han X, Yang H, Wang S, Lin D. Effectors of epidermal growth factor receptor pathway: the genetic profiling of KRAS, BRAF, PIK3CA, NRAS mutations in colorectal cancer characteristics and personalized medicine. PLoS One. 2013;8:e81628. doi: 10.1371/journal.pone.0081628. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lo L, Price T, Young J, Townsend A. BRAF mutation in colorectal cancer. https://www.intechopen.com/books/colorectal-cancer-from-pathogenesis-to-treatment/braf-mutation-in-colorectal-cancer [cited 10 Feb 2019]. Available at. [DOI]
- 49.Kambara T, Simms LA, Whitehall VL, Spring KJ, Wynter CV, Walsh MD. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137–44. doi: 10.1136/gut.2003.037671. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tran B, Kopetz S, Tie J, Gibbs P, Jiang ZQ, Lieu CH. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer. 2011;117:4623–32. doi: 10.1002/cncr.26086. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen D, Huang JF, Liu K, Zhang LQ, Yang Z, Chuai ZR. BRAF V600E mutation and its association with clinicopathological features of colorectal cancer: a systematic review and meta-analysis. PLoS One. 2014;9:e90607. doi: 10.1371/journal.pone.0090607. et al. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogino S, Nosho K, Kirkner GJ, Shima K, Irahara N, Kure S. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol. 2009;27:1477–84. doi: 10.1200/JCO.2008.18.654. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zarkavelis G, Boussiosa S, Papadakia A, Katsanos KH, Christodoulou DK, Pentheroudakis G. Current and future biomarkers in colorectal cancer. Ann Gastroenterol. 2017;30:613–21. doi: 10.20524/aog.2017.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Imai K, Yamamoto H. Carcinogenesis and microsatellite instability: the interrelation- ship between genetics and epigenetics. Carcinogenesis. 2008;29:673–80. doi: 10.1093/carcin/bgm228. [DOI] [PubMed] [Google Scholar]
- 55.Jensen LH, Lindebjerg J, Byriel L, Kolvraa S, Crüger DG. Strategy in clinical practice for classification of unselected colorectal tumours based on mismatch repair deficiency. Colorectal Dis. 2008;10:490–7. doi: 10.1111/j.1463-1318.2007.01378.x. [DOI] [PubMed] [Google Scholar]
- 56.Rahner N, Friedrichs N, Steinke V, Aretz S, Friedl W, Buettner R. Coexisting somatic promoter hypermethylation and pathogenic MLH1 germline mutation in Lynch syndrome. J Pathol. 2008;214:10–16. doi: 10.1002/path.2263. et al. [DOI] [PubMed] [Google Scholar]
- 57.Domingo E, Laiho P, Ollikainen M, Pinto L, Wang A, French J. BRAF screening as a low-cost effective strategy for simplifying HNPCC genetic testing. J Med Genet. 2004;41:664–8. doi: 10.1136/jmg.2004.020651. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cremolini C, Loupakis F, Antoniotti C, Lupi C, Sensi E, Lonardi S. FOLFOXIRI plus bevacizumab versus FOLFIRI plus bevacizumab as first-line treatment of patients with metastatic colorectal cancer: updated overall survival and molecular subgroup analyses of the open-label, phase 3 TRIBE study. Lancet Oncol. 2015;16:1306–15. doi: 10.1016/S1470-2045(15)00122-9. et al. [DOI] [PubMed] [Google Scholar]
- 59.Sanz- Garcia E, Argiles G, Elez E, Tabernero J. BRAF mutant colorectal cancer: prognosis, treatment, and new perspectives. Ann Oncol. 2017;28:2648–57. doi: 10.1093/annonc/mdx401. [DOI] [PubMed] [Google Scholar]
- 60.Renaud S, Romain B, Falcoz PE, Olland A, Santelmo N, Brigand C. KRAS and BRAF mutations are prognostic biomarkers in patients undergoing lung metastasectomy of colorectal cancer. Br J Cancer. 2015;112:720–8. doi: 10.1038/bjc.2014.499. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bokemeyer C, Van Cutsem E, Rougier P, Ciardiello F, Heeger S. Addition of cetuximab to chemotherapy as first-line treatment for KRAS wild-type metastatic colorectal cancer: pooled analysis of the CRYSTAL and OPUS randomised clinical trials. Eur J Cancer. 2012;48:1466–75. doi: 10.1016/j.ejca.2012.02.057. Schlichting, et al. [DOI] [PubMed] [Google Scholar]
- 62.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–34. doi: 10.1056/NEJMoa1305275. et al. [DOI] [PubMed] [Google Scholar]
- 63.Seymour MT, Brown SR, Middleton G, Maughan T, Richman S, Gwyther S. Panitumumab and irinotecan versus irinotecan alone for patients with KRAS wild-type, fluorouracil-resistant advanced colorectal cancer (PICCOLO): a prospectively stratified randomised trial. Lancet Oncol. 2013;14:749–59. doi: 10.1016/S1470-2045(13)70163-3. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karapetis CS, Jonker D, Daneshmand M, Hanson JE, O’Callaghan CJ, Marginean C. PIK3CA, BRAF, and PTEN status and benefit from cetuximab in the treatment of advanced colorectal cancer—results from NCIC CTG/AGITG CO.17. Clin Cancer Res. 2014;20:744–53. doi: 10.1158/1078-0432.CCR-13-0606. et al. [DOI] [PubMed] [Google Scholar]
- 65.Peeters M, Price TJ, Cervantes A, Sobrero AF, Ducreux M, Hotko Y. Final results from a randomized phase 3 study of FOLFIRI {+/-} panitumumab for second-line treatment of metastatic colorectal cancer. Ann Oncol. 2014;25:107–16. doi: 10.1093/annonc/mdt523. et al. [DOI] [PubMed] [Google Scholar]
- 66.Pietrantonio F, Petrelli F, Coinu A, Di Bartolomeo M, Borgonovo K, Maggi C. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: a meta-analysis. Eur J Cancer. 2015;51:587–94. doi: 10.1016/j.ejca.2015.01.054. et al. [DOI] [PubMed] [Google Scholar]
- 67.Rowland A, Dias MM, Wiese MD, Kichenadasse G, McKinnon RA, Karapetis CS. Meta-analysis of BRAF mutation as a predictive biomarker of benefit from anti-EGFR monoclonal antibody therapy for RAS wild-type metastatic colorectal cancer. Br J Cancer. 2015;112:1888–94. doi: 10.1038/bjc.2015.173. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yuan ZX, Wang XY, Qin QY, Chen DF, Zhong QH, Wang L. The prognostic role of BRAF mutation in metastatic colorectal cancer receiving anti-EGFR monoclonal antibodies: a meta-analysis. PLoS One. 2013;8:e65995. doi: 10.1371/journal.pone. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Loupakis F, Cremolini C, Salvatore L, Masi G, Sensi E, Schirripa M. FOLFOXIRI plus bevacizumab as first-line treatment in BRAF mutant metastatic colorectal cancer. Eur J Cancer. 2014;50:57–63. doi: 10.1016/j.ejca. et al. [DOI] [PubMed] [Google Scholar]
- 70.Van Cutsem E, Huijberts S, Grothey A, Yaeger R, Cuyle PJ, Elez E. BEACON CRC study safety lead-in (SLI) in patients with BRAF V600E metastatic colorectal cancer: Binimetinib, Encorafenib and Cetuximab Triplet Therapy for patients with BRAF V600E-mutant metastatic colorectal cancer: safety lead-in results from the phase III BEACON colorectal cancer study. J Clin Oncol. 2019;37:1460–9. doi: 10.1200/JCO.18.02459. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schuell B, Gruenberger T, Scheithauer W, Zielinski Ch, Wrba F. HER 2/neu protein expression in colorectal cancer. BMC Cancer. 2006;6:123. doi: 10.1186/1471-2407-6-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Siena S, Sartore-Bianchi A, Marsoni S, Hurwitz HI, McCall SJ, Penault-Llorca F. Targeting the human epidermal growth factor receptor 2 ( HER2) oncogene in colorectal cancer. Ann Oncol. 2018;29:1108–19. doi: 10.1093/annonc/mdy100. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seo AN, Kwak Y, Kim DW, Kang SB, Choe G, Kim WH. Her2 status in colorectal cancer: its clinical significance and the relationship between HER2 gene amplification and expression. PLoS One. 2014;9:e98528. doi: 10.1371/journal.pone. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ingold Heppner B, Behrens HM, Balschun K, Haag J, Krüger S, Becker T. HER2/neu testing in primary colorectal carcinoma. Br J Cancer. 2014;111:1977–84. doi: 10.1038/bjc.2014.483. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Missiaglia E, Jacobs B, D’Ario G, Di Narzo AF, Soneson C, Budinska E. Distal and proximal colon cancers differ in terms of molecular, pathological, and clinical features. Ann Oncol. 2014;25:1995–2001. doi: 10.1093/annonc/mdu275. et al. [DOI] [PubMed] [Google Scholar]
- 76.Nam SK, Yun S, Koh J, Kwak Y, Seo AN, Park KU. BRAF, PIK3CA, and HER2 oncogenic alterations according to KRAS mutation status in advanced colorectal cancers with distant metastasis. PLoS One. 2016;11:e0151865. doi: 10.1371/journal.pone. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bertotti A, Migliardi G, Galimi F, Sassi F, Torti D, Isella C. A molecularly annotated platform of patient-derived xenografts (“Xenopatient”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discovery. 2011;1:508–23. doi: 10.1158/2159-8290. et al. [DOI] [PubMed] [Google Scholar]
- 78.Yonesaka K, Zejnullahu K, Okamoto I, Satoh T, Cappuzzo F, Souglakos J. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3002442. et al. 99ra86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martin V, Landi L, Molinari F, Fountzilas G, Geva R, Riva A. HER2 gene copy number status may influence clinical efficacy to anti-EGFR monoclonal antibodies in metastatic colorectal cancer patients. Br J Cancer. 2013;108:668–75. doi: 10.1038/bjc.2013. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Osako T, Miyahara M, Uchino S, Inomata M, Kitano S, Kobayashi M. Immunohistochemical study of c-erbB-2 protein in colorectal cancer and the correlation with patient survival. Oncology. 1998;55:548–55. doi: 10.1159/000011911. [DOI] [PubMed] [Google Scholar]
- 81.Kapitanovic S, Radosevic S, Kapitanovic M, Andelinovic S, Ferencic Z, Tavassoli M. The expression of p185(HER-2/neu) correlates with the stage of disease and survival in colorectal cancer. Gastroenterology. 1997;112:1103–13. doi: 10.1016/S0016-5085(97)70120-3. et al. [DOI] [PubMed] [Google Scholar]
- 82.Li C, Liu DR, Ye LY, Huang LN, Jaiswai S, Li XW. HER-2 overexpression and survival in colorectal cancer: a meta-analysis. J Zhejiang Univ Sci B. 2014;15:582–9. doi: 10.1631/jzus.B1300258. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang L. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part II. The utility of microsatellite instability testing. J Mol Diagn. 2008;10:301–7. doi: 10.2353/jmoldx.2008.080062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ward R, Meagher A, Tomlinson I, O’Connor T, Norrie M, Wu R. Microsatellite instability and the clinicopathological features of sporadic colorectal cancer. Gut. 2001;48:821–9. doi: 10.1136/gut.48.6.821. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tiwari AK, Roy HK, Lynch HT. Lynch syndrome in the 21st century: clinical perspectives. QJM. 2016;109:151–8. doi: 10.1093/qjmed/hcv137. [DOI] [PubMed] [Google Scholar]
- 86.Ashktorab H, Ahuja S, Kannan L, Lior X, Ellis NA, Xicola RM. A meta-analysis of MSI frequency and race in colorectal cancer. Oncotarget. 2016;7:34546–57. doi: 10.18632/oncotarget.8945. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol. 2018;36:773–9. doi: 10.1200/JCO.2017.76.9901. et al. [DOI] [PubMed] [Google Scholar]
- 88.Fujiyoshi K, Yamamoto G, Takenoya T, Takahashi A, Arai Y, Yamada M. Metastatic pattern of stage IV colorectal cancer with high-frequency micro-satellite instability as a prognostic factor. Anticancer Res. 2017;37:239–47. doi: 10.21873/anticanres.11313. et al. [DOI] [PubMed] [Google Scholar]
- 89.Kawakami H, Zaanan A, Sinicrope FA. MSI testing and its role in management of colorectal cancer. Curr Treat Options Oncol. 2015;16:30. doi: 10.1007/s11864-015-0348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]


