Abstract
Background
To perform a radiomics analysis in local recurrence (LR) surveillance of limb soft tissue sarcoma (STS)
Patients and methods
This is a sub-study of a prospective multicenter study with Institutional Review Board approval supported by ESSR (European Society of Musculoskeletal Radiology). radiomics analysis was done on fast spin echo axial T1w, T2w fat saturated and post-contrast T1w (T1wGd) 1.5T MRI images of consecutively recruited patients between March 2016 and September 2018.
Results
N = 11 adult patients (6 men and 5 women; mean age 57.8 ± 17.8) underwent MRI to exclude STS LR: a total of 33 follow-up events were evaluated. A total of 198 data-sets per patients of both pathological and normal tissue were analyzed. Four radiomics features were significantly correlated to tumor size (p < 0.02) and four radiomics features were correlated with grading (p < 0.05). ROC analysis showed an AUC between 0.71 (95%CI: 0.55–0.87) for T1w and 0.96 (95%CI: 0.87–1.00) for post-contrast T1w.
Conclusions
radiomics features allow to differentiate normal tissue from pathological tissue in MRI surveillance of local recurrence of STS. radiomics in STS evaluation is useful not only for detection purposes but also for lesion characterization.
Key words: sarcoma, recurrence, magnetic resonance imaging, ROC curve
Introduction
radiomics is an advanced quantitative image features analysis defined as the conversion of clinical images to higher dimensional data and the subsequent mining of these data for improved decision support in research and clinical practice.1 The majority of clinically available medical images can potentially be evaluated with radiomics analysis. In this perspective, images of computed tomography, Magnetic Resonance Imaging (MRI), ultrasound, mammography or digital breast tomosynthesis and Fluorodesoxyglucose Positron-emission tomography-computed tomography (FDG PET/CT) include more data than what is visible on human eyes.1, 2, 3 Indeed, mathematical algorithms of radiomics examine hundreds of quantitative images considering medical images as data sources. The extracted imaging data could be the product of the mechanisms occurring at a genetic and molecular level linked to the genotypic and phenotypic characteristics of the tissue.4, 5, 6, 7 Among its applications, radiomics has been evaluated to differentiate normal and pathological tissue.1, 2, 3, 5 Hence, we thought about the potential use of radiomics in local recurrence surveillance of soft tissue sarcoma. The incidence of local recurrence (LR) of STS is about 6,5%–25% and is associated with poor outcome for patients.7 According to the American College of Radiology (ACR) Appropriateness Criteria guidelines, MRI is the most appropriate imaging test for LR surveillance of malignant or aggressive musculoskeletal soft-tissue tumors.8 However, data informing the appropriate use of MRI in the surveillance setting are conflicting.5,8,9 Indeed, MRI can differentiate local recurrence from post-surgical seroma, hematoma, inflammation and scarring, but some post-operative changes in the surgical bed can be similar to those of recurrence with conventional T1-weighted, T2- weighted, and post-contrast sequences posing diagnostic dilemmas especially when sarcoma recurrence has low signal intensity on fluid-sensitive images.1,8, 9, 10, 11 We hypothesized that radiomics analysis of MRI of patients undergoing follow-up for STS allows to differentiate normal tissue from pathological tissue of LR. Therefore, the aim of our study was to perform a radiomics analysis of MRI of patients undergoing local surveillance for Soft-tissue sarcoma recurrence.
Patients and methods
This is an exploratory study of an ongoing Italian prospective (blind) multicenter study with institutional review board approval (blind). Written informed consent was obtained from participants. This study is endorsed by ESSR (European Society of Musculoskeletal Radiology). Prospective recruitment of patients, as per protocol, includes MRI and US with commercially available equipment. MRI parameters of sequences included in the radiomics analysis are reported in Table 1.
Table 1.
MRI Parameters
| Manufacturer | Siemens Healthcare, Erlangen, Germany |
|---|---|
| T1-weighted MR imaging | |
| Repetition time / echo time (TR/TE) | 500/8 |
| Acquisition voxel size (mm3) | 0.6x0.7x3.0 |
| T2-weighted MR imaging* | |
| Repetition time / echo time (TR/TE) | 6200/110 |
| Acquisition voxel size (mm3) | 0.6x0.7x3.0 |
| T1-weighted MR imaging* with Gadolinium | |
| Repetition time / echo time (TR/TE) | 5/3 |
| Acquisition voxel size (mm3) | 0.6x0.7x3.0 |
* T2-weighted MR imaging and T1- weighted MR imaging with Gadolinium are acquired with fat-saturation
Patients
All consecutive MRI Images of follow-up events acquired between March 2016 to September 2018 were included. A follow-up event was considered a complete MRI assessment to exclude STT LR. Inclusion criteria were: patients included were 18 years and older operated on for localized soft tissue sarcomas of the limb. Exclusion criteria were patients unable to understand or execute written informed consent, unable or unwilling to agree to follow-up during observation period and patients with metastatic disease.
Experimental design
This study focused on finding if some radiomics features could discriminate on MRI patients who had confirmed significant disease at histology (LR) from those who did not. To ensure unbiased assessment, all MRI annotations were performed blinded to the biopsy findings. Only cases with matched locations (surgery, biopsy, reports, and radiologist’s delineations) were considered for inclusion in this study.
Two data-sets were created: pathological and control.
Each data set includes MRI images each with distinctive Regions of interest (ROIs). ROIs were positioned by two researchers (blind and blind; R1 and R2 respectively) expert in quantitative image analysis (8 and 4 years of experience) blindly one from each other. R1 and R2 ROIs data were used separately for intra- and inter-observer agreement estimation while the mean value was used for other estimations. Discrepancies higher that 15% between R1 ROI and R2 ROI were handled with arbitration.
Regions of interest including all the visible tumor or suspicious area represented the pathological data-set. Regions of interest in the same slices of tissue with no imaging evidence of recurrence according radiological assessment represented the control data-set. ROIs of control-data set were positioned not before 20 mm to the tumor to exclude inclusion of tumoral tissue. We did not include cancer cases with recurrences detected only with US. Image analysis was done per-lesion and not per-patient (Figure 1).
Figure 1.
Example of workflow.
Radiomics analysis
Radiomics analysis was performed on all MRI images included in pathological and control data-set within manually selected ROIs (Figure 1) as previously done for other 3D radiological techniques.2 From each image, we extracted 104 image features using an open-source software platform for medical image informatics, image processing, and three-dimensional visualization (3D Slicer 4.7; www.slicer.org) built over two decades through support from the National Institutes of Health and a worldwide developer community. 3D-Slicer can be employed for quantitative image feature extraction and image data mining research in large patient cohorts.6
Preprocessing
All patients were examined by using a 1.5-Tesla equipment (Magnetom Avanto, Siemens Heathcare) with a standard protocol. For each patient, T1-weighted (T1w), Gd-enhanced T1-weighted (T1WGd) with 0.1-mmol/kg doses of gadoteric acid and T2-weighted with fat saturation (T2w fs) volume images were resampled to an uniform pixel spacing of 0.5 x 0.5 x 3mm. Then they were cropped to the lesion region of interest as delineated by R1 and R2. Inhomogeneity correction was applied to T1w and T2w fat saturated images to account for the presence of bias field artifacts. Next, T1w and T2w fs images were corrected for inherent acquisition-to acquisition signal intensity variations (non standardness) using scale-based standardization. This procedure was applied to mitigate the inherent drift phenomenon that accompanies MRI intensities as previously done in literature.12
Statistical analysis
A) Comparison of radiomics features of normal and pathological tissue on MRI was done with non-parametric tests (Mann-Whitney U test for unpaired data with 1000 bootstraps samples to compare patients and controls) considering a p value of 0.05 as statistically significant. B) Accuracy was measured using receiver operating characteristic (ROC) analyses to estimate the area under the curve (AUC) and by estimating thresholds for sensitivity and specificity for the radiomics features that significantly differed between patients and controls, considering the mean value, to avoid over-fitting. Ninety-five percent confidence intervals (95%CIs) were calculated. Using statistical software, p values below 0.05 were considered statistically significant. C) Correlation analysis and univariate linear regression were performed to determine the association between the radiomics features and the corresponding clinical and prognostic parameters. Bonferroni correction was used to adjust for multiple comparisons. D) Mean time for single patient radiomics analysis was also calculated. Reading time was estimated with a commercially available stopwatch including the time to download images, perform image adjustment and analysis and finally data collection in the database. Statistical tests were done using statistical software (STATA MP, StataCorp, 4905 Lakeway Dr, College Station, TX, USA and MedCalc). E) Intra-observer agreement was estimated: for research purposes Cronbach’s alpha was considered acceptable if between 0.7 and 0.8.13
Results
Intra-observer agreement resulted to be 0.62 (95% CI: 0.52–0.67) for single measurements and 0.75 (95% CI: 0.69–0.80) for the average measure and was deemed acceptable for the purpose of the study. There were no discrepancies higher that 15% between R1 ROI and R2 ROI requesting arbitration.
N = 11 adult patients (6 men and 5 women) with suspicious STS LR were included for a total of 33 follow-up events on MRI. A total of 198 data-sets per patients of both pathological and normal tissue were analyzed (99 pathological and 99 control data-sets). Characteristics of the 19 pathological findings in 33 follow-up events are reported in Table 2. N = 3 patients had multiple lesions.
Table 2.
Distribution of the extremity soft tissue sarcoma patients’ clinical characteristics in 19 pathological findings of 11 patients in 33 follow-up events. N = 3 patients had multiple lesions
| Clinical Characteristic | |
|---|---|
| Age (years) | 57.8 ± 17.8 |
| Tumor size (mm) | 26,2 ± 16.9 |
| Grade (%) | |
| G1 | 4 (21) |
| G2 | 6 (37) |
| G3 | 8 (42) |
| Unassigned | 1 (5) |
| Depth (%) | |
| Superficial | 6 (32) |
| Deep | 13 (68) |
| Location (%) | |
| Upper extremity | 5 (26) |
| Lower extremity | 14 (74) |
| Histology (%) | |
| Pleomorphic liposarcoma | 6 (33) |
| Myxofibrosarcoma | 5 (27) |
| Myxoid liposarcoma | 2 (10) |
| Leiomyosarcoma | 2 (10) |
| Nerve sheath tumors | 2 (10) |
| Synovial sarcoma | 2 (10) |
After feature number reduction to avoid overfitting, Mann-Whitney U test identified n = 7, n = 13 and n = 12 features able to differentiate pathological tissue from normal tissue on T1w MRI images, T2w fat saturated MRI images and T1wGd respectively (p < 0.001). Table 3 shows feature domain according to different MRI sequences.
Table 3.
Feature domain according to different MRI sequences
| Feature | Description | Significance | T1-weighted MR imaging | T2-weighted MR imaging* | T1-weighted MR imaging with Gadolinium |
|---|---|---|---|---|---|
| Shape domain | descriptors of the three-dimensional size and shape of the ROI. | These features are independent from distribution the gray in level the ROI intensity and are therefore only calculated on the non-derived image and mask | 1 | 1 | 2 |
| First order | Mean, standard deviation, median, and range; first-order differentials computed using Sobel operators | Localize hypo- and hyperintense regions; gradients detect edges and quantify region boundaries | 1 | 1 | 1 |
| Gray level co-occurrence matrix (GLCM) | Localization of regions with significant intensity changes; gradients detect edges and quantify region boundaries | Localizes regions based on underlying heterogeneity of voxel intensities | 3 | 7 | 6 |
| Gray level run lenght matrix (glrlm) | quantifies gray level runs, which are defined as the length in number of pixels, of consecutive pixels that have the same gray level value. | In a gray level run length matrix the element describes the number of runs with gray level and length occur in the image (ROI) along angle | 2 | 3 | 6 |
| Gray level size zone matrix (glszm) domain | It is an advanced statistical matrix used for texture characterization. It estimates bivariate conditional probability density function of the image distribution values | represent the count of how many times a given size of given grey level occur | 0 | 0 | 0 |
Some radiomics features were significantly correlated to tumor size: 4 features (Compactness, MajorAxis, Flatness, Mean) with r = 0.75 with p < 0.02, on T1w MRI images, 2 features (MajorAxis, RootMeanSquared) on T2w fat saturated MRI images with r = 0.65 with p < 0.01, and 2 features (MajorAxis, Maximum) on T1wGd with r = 0.65 with p < 0.01. Four radiomics features (Sum entropy, difference entropy, energy, lmc2) were correlated with grading (4 features with r = 0.74 and p < 0.05) on T1w MRI images and none on T2w fat saturated MRI images and T1wGd respectively.
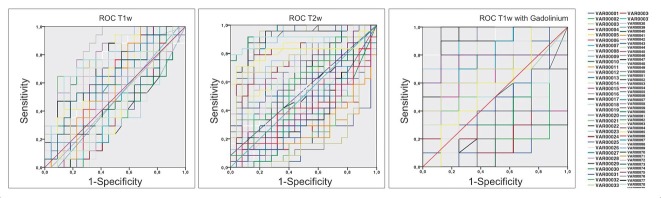
ROC analysis performed on T1w images, T2w fat saturated images and T1wGd showed an AUC between 0.71 (95%CI: 0.55–0.87) and 0.96 (95%CI: 0.87–1.00). Detailed results are reported in Table 4 and Figure 2. T2w fat saturated with fat-saturation and T1w post-Gadolinium showed a better performance than T1w images (p < 0.05).
Table 4.
ROC results according to different MRI sequences of the selected features.* T2-weighted MR imaging and T1-weighted MR imaging with Gadolinium are acquired with fat-saturation. Areas under the curve for differentiation of normal and pathological tissue (LR) had p < 0.05
| Sequence | Minimum AUC | 95%CI | Maximum AUC | 95%CI |
|---|---|---|---|---|
| T1-weighted MR imaging | 0.71 | 0.54–0.88 | 0.84 | 0.77–0.96 |
| T2-weighted MR imaging* | 0.81 | 0.67–0.95 | 0.91 | 0.83–1.00 |
| Twith 1-weighted Gadolinium MR imaging* | 0.87 | 0.69–1.00 | 0.96 | 0.87–1.00 |
Figure 2.
Examples of AUCs with a reduced number of features on T1w, T2w fat saturated with fat-saturation, and T1w post-Gadolinium showing a better performance for T2w fat saturated with fat-saturation and T1w post-Gadolinium (p < 0.05). Features from 1 to 26 belong to the shape domain; features (VAR00..) from 27 to 45 belong to the first order domain; features from 46 to 72 belong to the glcm (gray-level co-occurrence matrix) domain; features from 73 to 88 belong to gray level Run Lenght Matrix (glrlm) domain; features from 88 to 104 belong to the gray level size zone matrix (glszm) domain.
Mean time to perform radiomics analysis was at least 5 h per patient including the creation of a data set of 19 patients with suspicious cancer visible at MRI, therefore the total time to perform analysis, excluding the time to recruit and select patients was at least 100 h.
Discussion
In this study we performed a radiomics analysis on MRI images of patients suspected of having STS LR belonging to an ongoing prospective trial. The aim was to investigate whether radiomics features derived from standard clinical MRI sequences could be used to differentiate normal tissue from cancerous tissue of LR. In clinical practice, differentiating normal from pathological tissue on MRI in patients suspected of having LR is relevant especially when LR are not nodules, but plaque-like “tails” of tumor on MRI, especially for both undifferentiated pleomorphic sarcoma and myxofibrosarcoma.5,11 Despite the relatively low number of patients included in this study, we had a high percentage of pleomorphic sarcoma/liposarcoma and/or myxo-fibrosarcoma accounting for 11/19 of the lesions evaluated. The number of lesions evaluated in this study is similar to the number of pleomorphic sarcoma/liposarcoma and/or myxofibrosarcoma evaluted in the study by Corino et al.14 In the study by Corino et al.14, a radiomics approach with first order features performed on 13 pleomorphic sarcoma/liposarcoma and/or myxofibrosarcoma resulted in good accuracy and AUC. In the present study, the number of pleomorphic sarcoma/liposarcoma and/ or myxofibrosarcomas evaluated could be considered relatively high considering that patients underwent MRI evaluation for LR and not of a primary tumor. MRI could be challenging in these patients and the use of radiomics could enhance the role of MRI in STS local surveillance. Chou et al. 15 reported MRI sensitivity for LR detection ranging from 69%without contrast medium to 90% with contrast-enhanced sequences. AUCs of this study, especially for T1w post-gadolinium and T2w fat saturated are better that 90%. As reported by Fayad L. et al.16, postoperative inflammation and fibrosis in the surgical bed may share many of the same characteristics as tumor on conventional MR images. Indeed they can occasionally appear mass-like. Only in the absence of abnormal T2-weighted signal intensity the presence of recurrent tumor could be excluded. Very rarely, a sarcoma recurrence may be of low signal intensity on T2-weighted images and a T1-weighted study shows architectural distortions due to the tumoral presence. We believe that in these difficult radiological cases, radiomics may help even expert Radiologists in the detection of LR. A preliminary report showed that using a radiomics approach to characterize musculoskeletal tumors, machine learning performs even better than expert radiologists17 (Chhabra A, personal communication). Indeed, the main result of this study is that few radiomics features can differentiate LR from normal tissue on T1w MRI images, T2w fat saturated MRI images and T1w postgadolinium sequences. These features belonged to different classes. The majority of them belonged to the gray level co-occurrence matrix (GLCM) domain possibly reflecting underlying heterogeneity of voxel intensities. Although it is known that in soft-tissue tumors fluid-sensitive images should have a more favorable contrast between tumor and surrounding because skeletal muscle is of intermediate signal intensity, radiomics features were able to differentiate LR from normal tissues even on T1w sequences. This data reflects the great value of this application for STT survelliance. In addition, a very good AUC can be obtained with only few features for every MRI sequence. We acknowledge that, according to standard clinical practice, T2w fat saturated and post-contrast T1w sequences had the best AUCs. After feature reduction to avoid decrease the performance18 we found that a set of few radiomics features had an AUC generally better than data reported for conventional MRI. Indeed, radiomics based AUC for T1-weighted MR imaging with Gadolinium reached a best result of 0.96. These data support the design of future studies using of radiomics for STS surveillance. In addition, in spite of the small number of patients, some radiomics features correlated with tumor size and grading. These data support the hypothesis that few radiomics features could reflect tumoral biology and aggressiveness.14 We acknowledge that mean time to perform radiomics analysis was long and intra-observer agreement resulted acceptable for the purpose of the study. In addition, a different selection of the ROI could lead to a different measurement of the features even with good intra- and inter-observer agreement. Further improvements are needed to accelerate the workflow in clinical practice and to keep ROIs as stable as possible although we did not have any cases with discrepancies higher that 15%.
This study has several limitations. First, radiomics features were not extracted on ADC maps, that have been demonstrated to assess tumor cellularity even when different scanners are used.14 However, this is not a multicentric study and the MRI image sample was included from only one center. Second, the study population was relatively small. However, considering the incidence of LR in patients who underwent surgery for STT (reported rates of LR range from 6.5% to approximately 25%)5, the number of patients and images evaluated seem to be sufficient for the purpose of the study. The presence of a relatively high number of pleomorphic sarcoma and myxofibrosarcoma increases the clinical significance of the study because they are tumors difficult to be evaluated on MRI. Selection biases are excluded due to consecutive patient enrollment in the present study. Moreover, three MRI sequences per lesion were analyzed increasing data robustness. Finally, we also acknowledge that future software developments will reduce the necessity of freehand ROIs positioning.19 We also believe that further research will be critical to fully unveil the potential or radiomics in STT evaluation, indeed, it has been studied that radiomics features extracted from MR images are independently associated with survival when accounting for age and tumor grade and helps in differential diagnosis.20,21
In conclusion, radiomics features allow to detect LR on MRI images in STS local surveillance. radiomics in STS evaluation is useful not only for detection purposes but also for lesion biological characterization.
Disclosure
No potential conflicts of interest were disclosed.
References
- 1.Gillies RJ, Kinahan PE, Hricak H. radiomics: images are more than pictures, They are data. Radiology. 2016;278:563–77. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tagliafico AS, Valdora F, Mariscotti G, Durando M, Nori J, La Forgia D. An exploratory radiomics analysis on digital breast tomosynthesis in women with mammographically negative dense breasts. Breast. 2018;40:92–6. doi: 10.1016/j.breast.2018.04.016. et al. [DOI] [PubMed] [Google Scholar]
- 3.Valdora F, Houssami N, Rossi F, Calabrese M, Tagliafico AS. Rapid review: radiomics and breast cancer. Breast Cancer Res Treatm. 2018;169:217–29. doi: 10.1007/s10549-018-4675-4. [DOI] [PubMed] [Google Scholar]
- 4.Limkin EJ, Sun R, Dercle L, Zacharaki EI, Robert C, Reuzé S. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol. 2017;28:1191–206. doi: 10.1093/annonc/mdx034. et al. [DOI] [PubMed] [Google Scholar]
- 5.Ezuddin NS, Pretell-Mazzini J, Yechieli RL, Kerr DA, Wilky BA, Subhawong TK. Local recurrence of soft-tissue sarcoma: issues in imaging surveillance strategy. Skeletal Radiol. 2018;47:1595–606. doi: 10.1007/s00256-018-2965-x. [DOI] [PubMed] [Google Scholar]
- 6.Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging. 2012;30:1323–41. doi: 10.1016/j.mri.2012.05.001. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abatzoglou S, Turcotte RE, Adoubali A, Isler MH, Roberge D. Local recurrence after initial multidisciplinary management of soft tissue sarcoma: is there a way out? Clin Orthop Relat Res. 2010;468:3012–8. doi: 10.1007/s11999-010-1481-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheney MD, Giraud C, Goldberg SI, Rosenthal DI, Hornicek FJ, Choy E. MRI surveillance following treatment of extremity soft tissue sarcoma. J Surg Oncol. 2014;109:593–6. doi: 10.1002/jso.23541. et al. [DOI] [PubMed] [Google Scholar]
- 9.Noebauer-Huhmann IM, Weber M-A, Lalam RK, Trattnig S, Bohndorf K, Vanhoenacker F. Soft tissue tumors in adults: ESSR-approved guidelines for diagnostic imaging. Semin Musculoskelet Radiol. 2015;19:475–82. doi: 10.1055/s-0035-1569251. et al. [DOI] [PubMed] [Google Scholar]
- 10.Roberts CC, Kransdorf MJ, Beaman FD, Adler RS, Amini B, Appel M. ACR Appropriateness criteria follow-up of malignant or aggressive musculoskeletal tumors. J Am Coll Radiol. 2016;13:389–400. doi: 10.1016/j.jacr.2015.12.019. et al. [DOI] [PubMed] [Google Scholar]
- 11.Del Grande F, Subhawong T, Weber K, Aro M, Mugera C, Fayad LM. Detection of soft-tissue sarcoma recurrence: added value of functional MR imaging techniques at 3.0 T. Radiology. 2014;271:499–511. doi: 10.1148/radiol.13130844. [DOI] [PubMed] [Google Scholar]
- 12.Algohary A, Viswanath S, Shiradkar R, Ghose S, Pahwa S, Moses D. radiomic features on MRI enable risk categorization of prostate cancer patients on active surveillance: Preliminary findings. J Magn Reson Imaging JMRI. 2018;48:818–28. doi: 10.1002/jmri.25983. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bland JM, Altman DG. Cronbach’s alpha. BMJ. 1997;314:572. doi: 10.1136/bmj.314.7080.572. http://www.ncbi.nlm.nih.gov/pubmed/9055718 Available at. Accessed October 22, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corino VDA, Montin E, Messina A, Casali PG, Gronchi A, Marchianò A. radiomic analysis of soft tissues sarcomas can distinguish intermediate from high-grade lesions. J Magn Reson Imaging. 2018;47:829–40. doi: 10.1002/jmri.25791. et al. [DOI] [PubMed] [Google Scholar]
- 15.Chou S-HS, Hippe DS, Lee AY, Scherer K, Porrino JA, Davidson DJ. Gadolinium contrast enhancement improves confidence in diagnosing recurrent soft tissue sarcoma by MRI. Acad Radiol. 2017;24:615–22. doi: 10.1016/j.acra.2016.12.010. et al. [DOI] [PubMed] [Google Scholar]
- 16.Fayad LM, Jacobs MA, Wang X, Carrino JA, Bluemke DA. Musculoskeletal tumors: How to use anatomic, functional, and metabolic MR techniques. Radiology. 2012;265:340–56. doi: 10.1148/radiol.12111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Annual Scientific Meeting Abstracts of the International Skeletal Society (ISS) Berlin, Germany. Skeletal Radiol. 2018;2018;47:1315–25. doi: 10.1007/s00256-018-2994-5. [DOI] [PubMed] [Google Scholar]
- 18.Damper RI, MacDonald SL. Statistical clustering procedures applied to low-cost speech recognition. J Biomed Eng. 1984;6:265–71. doi: 10.1016/0141-5425(84)90072-4. http://www.ncbi.nlm.nih.gov/pubmed/6239064 Available at. Accessed October 22, 2018. [DOI] [PubMed] [Google Scholar]
- 19.Fanizzi A, Losurdo L, Basile TMA, Bellotti R, Bottigli U, Delogu P. Fully automated support system for diagnosis of breast cancer in contrast-enhanced spectral mammography images. J Clin Med. 2019;8:E891. doi: 10.3390/jcm8060891. et al. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie H, Hu J, Zhang X, Ma S, Ma S, Liu Y, Wang X. Preliminary utilization of radiomics in differentiating uterine sarcoma from atypical leiomyoma: Comparison on diagnostic efficacy of MRI features and radiomic features. Eur J Radiol. 2019;115:39–45. doi: 10.1016/j.ejrad.2019.04.004. et al. [DOI] [PubMed] [Google Scholar]
- 21.Spraker MB, Wootton LS, Hippe DS, Ball KC, Peeken JC, Macomber MW. MRI radiomic features are independently associated with overall survival in soft tissue sarcoma. 23; Adv Radiat Oncol. 2019;4:413–21. doi: 10.1016/j.adro.2019.02.003. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]