Abstract
Small endonucleolytic ribozymes promote the self-cleavage of their own phosphodiester backbone at a specific linkage. The structures of and the reactions catalysed by members of individual families have been studied in great detail in the past decades. In recent years, bioinformatics studies have uncovered a considerable number of new examples of known catalytic RNA motifs. Importantly, entirely novel ribozyme classes were also discovered, for most of which both structural and biochemical information became rapidly available. However, for the majority of the new ribozymes, which are found in the genomes of a variety of species, a biological function remains elusive. Here, we concentrate on the different approaches to find catalytic RNA motifs in sequence databases. We summarize the emerging principles of RNA catalysis as observed for small endonucleolytic ribozymes. Finally, we address the biological functions of those ribozymes, where relevant information is available and common themes on their cellular activities are emerging. We conclude by speculating on the possibility that the identification and characterization of proteins that we hypothesize to be endogenously associated with catalytic RNA might help in answering the ever-present question of the biological function of the growing number of genomically encoded, small endonucleolytic ribozymes.
INTRODUCTION
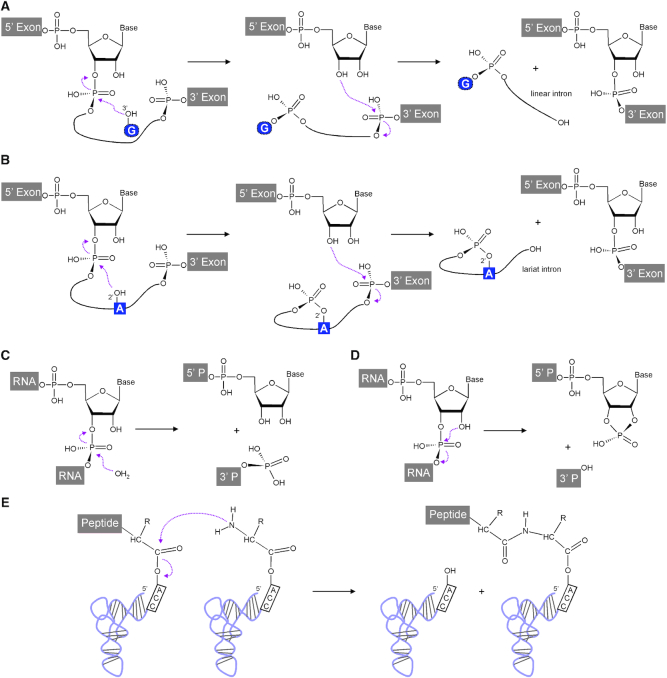
In the current year, we look back to 30 years of continued discoveries on catalytic RNA, after the Nobel Prize in Chemistry was awarded to Tom Cech and Sid Altman in 1989 ‘for their discovery of catalytic properties of RNA' (http://www.nobelprize.org/prizes/chemistry/1989/summary/). The distinguished discoveries were made on self-splicing Group I introns (1) and on the RNA subunit of bacterial RNase P (2). The reactions of both these catalytic RNAs target RNA 3′,5′-phosphodiester bonds, either in a series of two autocatalytic transesterification reactions in cis, or by means of a hydrolysis reaction carried out by the catalytic RNA component M1 of bacterial RNase P (Figure 1A, C). The majority of currently known natural ribozymes, short for ribonucleic acid enzymes, act on RNA 3′,5′-phosphodiester bonds, with further examples being group II introns (3,4) and the small nucleolytic ribozymes (5), whose reactions are shown in Figure 1B, D. In contrast, the peptidyl transferase centre of the ribosome consists of an RNA that catalyses the formation of peptide bonds (6; Figure 1E).
Figure 1.
Reactions naturally catalysed by RNA. Two sequential transesterification reactions catalysed by group I. (A) and group II (B) introns in cis. These result in joined exons and linear and lariat introns, respectively. RNA hydrolysis catalysed in trans by the M1 RNA subunit of bacterial RNase P. (C) results in a phosphate containing 5′ end of the mature tRNA as the 3′ cleavage product (3′ P) and a 3′ hydroxyl group at the 5′ cleavage product (5′ P). Small nucleolytic ribozymes undergo transesterification reactions in cis (D), in which a specific 2′-hydroxyl attacks the neighbouring 3′,5′-phosphodiester bond. This results in a 2′,3′-cyclic phosphate and a 5′ hydroxyl at the 5′ and 3′ cleavage products, respectively. (E) Peptide bond formation catalysed in the ribosomal peptidyl transferase centre. This figure was adapted from (163).
Early observations made by Harry Noller et al. already pointed strongly towards the possibility that proteins are made in an RNA-catalysed fashion, as translation proved to be resistant to mild proteolytic treatment (7). Final proof for ribosomal RNAs being the catalyst in protein formation, rather than ribosomal proteins, came from high-resolution crystal structures that revealed that the peptidyl transferase centre is made up exclusively of RNA constituents (6,8). In all likelihood, RNA-catalysed reactions had a broader range of substrate classes during evolution; however, due to the greater chemical diversity of amino acid side chains, proteinaceous enzymes would take over in the transition from the RNA World (9) to the current protein-dominated life. Such a shift in mode of operation is not easily conceivable for the peptidyl transferase centre in the ribosome and indeed this most central reaction in the generation of proteins remained RNA-catalysed. To make up for the limited chemical variability of the RNA nucleobases, in comparison to proteins, the efficiency of the essential, non-replaceable peptidyl transferase reaction was improved by other means. Examples of these improvements include not only the obvious ribosomal and auxiliary proteins, but also the more than one hundred chemical modifications that are known to occur in tRNA molecules (10). These modifications allow for an optimized translation as a consequence of altered binding energies between codon and anticodon, as highlighted recently in a seminal contribution (11). While most tRNA modifications are not absolutely essential, the absence of individual chemical modifications on tRNAs can have dramatic effects on protein translation (12), which results in protein aggregation (13), further highlighting their importance in and necessity for the optimization of the peptidyl transferase reaction.
By contrast, the four building blocks of RNA appear well suited to catalyse reactions at 3′,5′-phosphodiester bonds, often with the help of divalent metal ions and, occasionally, a small-molecule catalytic co-factor in the glmS ribozyme (14, and see below). Beyond the self-splicing introns and the M1 RNA mentioned above, an increasing number of ribozyme classes catalysing cleavage reactions of phosphodiester bonds was discovered in the past 30 years. Together, these RNA motifs form the family of the self-cleaving ribozymes, as they naturally cleave distinct positions in their own phosphodiester backbone (Figure 2). As the reaction is reversible, the ribozymes can in principle also accelerate the reverse, ligation reaction (Figure 2). This is, however, observed only in some cases due to instability of the products of the cleavage reaction. A significant body of data has accumulated on the structures of these catalytic RNA motifs and the varying catalytic strategies that they apply in their reactions. In this review, we summarize different approaches that were used to discover entirely new self-cleaving ribozyme motifs, or to identify novel examples of known motifs in various genetic contexts. Compared to their structures and mechanisms, the biological functions—particularly of the newly discovered, genomically encoded ribozyme motifs—have attracted less attention. We summarize the current understanding for those ribozymes where functions are known or emerging.
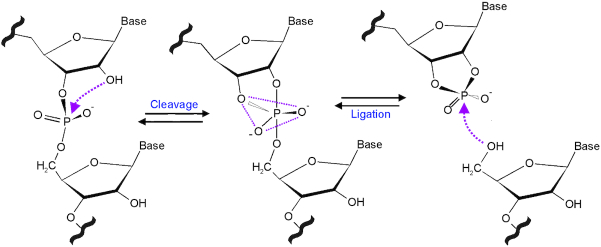
Figure 2.
The cleavage and ligation mechanism of small nucleolytic ribozymes. The 2′ OH attacks nucleophilically the neighbouring 3′,5′-phosphodiester bond. Upon passing through a trigonal bipyramidal transition state, the cleavage reaction yields a 5′ product with a 2′,3′-cyclic phosphate and a 3′ product with a 5′ OH. In the reverse ligation reaction, this 5′ hydroxyl group attacks the 2′,3′-cyclic phosphate, and, passing through the same transition state, the two RNAs are joined by a conventional 3′, 5′ phosphodiester.
DISCOVERY OF SELF-CLEAVING RIBOZYMES
The first structural class of self-cleaving ribozyme to be discovered was the hammerhead ribozyme (Figure 3A). It was found in the minus polarity strand of satellite RNAs of the tobacco ringspot virus (15) after a significant body of work investigating the replication mechanisms of satellite RNAs. Additional self-cleaving ribozyme classes were also discovered as a result of studying biological systems in which they participate. The plus polarity strand of the tobacco ringspot virus satellite RNA revealed the hairpin ribozyme (16; Figure 3B), and only two other examples of hairpin ribozymes are known to date (17). This research and observations that hepatitis δ virus (HDV) has similar properties to those of plant satellite RNAs spurred the identification of the HDV ribozyme (Figure 3C; 18). Finally, the Varkud satellite (VS) ribozyme (Figure 3D) was found in transcripts of DNA satellites associated with mitochondria in certain Neurospora isolates (19).
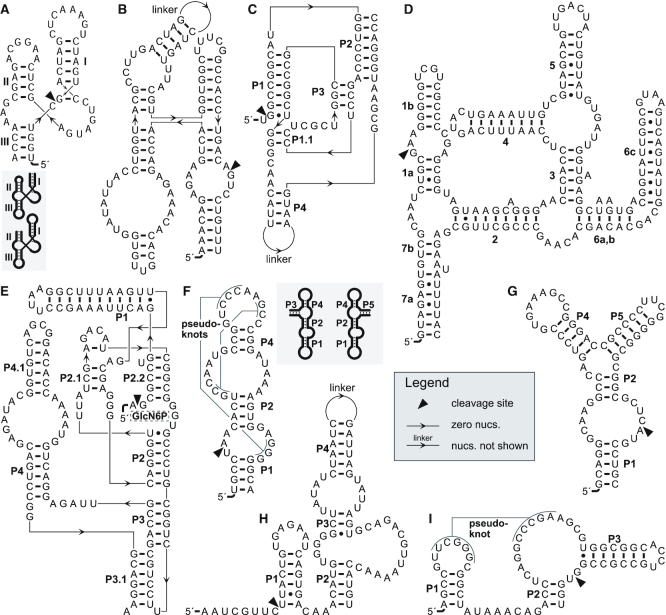
Figure 3.
Example secondary structures of the small nucleolytic ribozyme motifs. (A) The hammerhead ribozyme Ara2 (37), a type-III motif, featuring an open helix III. The inset shows the shapes of circularly permuted type-I (left) and type-II (right) hammerhead ribozymes with open helices I and II, respectively. (B) The hairpin ribozyme of tobacco ringspot virus satellite RNA (164). (C) The genomic hepatitis δ virus ribozyme (165). (D) The Varkud satellite ribozyme (166). (E) The glmS ribozyme (167) with the binding position of its cofactor glucosamine 6-phosphate (GlcN6P). (F) A type-P1 twister ribozyme from rice. The inset shows type-P3 (left) and type-P5 (right) permuted forms (23). Examples of a twister sister (G), hatchet (H) and a pistol (I) ribozymes (27). For each motif the helices are named or numbered in black according to the most-established nomenclature for that ribozyme class. This figure was drawn with R2R (168) and Adobe Illustrator.
DISCOVERY BY BIOINFORMATICS
The remaining self-cleaving ribozyme classes, among those currently known, were found bioinformatically. The glmS ribozyme (Figure 3E) was uncovered in a search for novel classes of riboswitches (20). Riboswitches are RNAs that regulate genes based on their ability to detect a specific small molecule metabolite or ion ligand (21). Since riboswitches usually occur in the 5′ UTRs of bacterial genes, the search compared InterGenic Regions (IGRs) in the bacterium Bacillus subtilis against those of other Firmicute species. Conserved IGRs upstream of metabolic genes were investigated further. The glmS ribozyme is found in the 5′ UTRs of glmS mRNAs, whose product synthesizes glucosamine 6-phosphate (GlcN6P). The ribozyme cleaves itself only in the presence of high concentrations of GlcN6P (14), and, in a negative feedback loop, this cleavage dramatically reduces gene expression of glmS (22).
Roughly 10 years later, a general search for all types of conserved RNA structures in bacteria revealed the twister ribozyme (Figure 3F; (23). This search analysed multiple lineages of bacteria and archaea (24). In brief, for each lineage, it grouped together similar IGRs based on BLAST (25) comparisons. For each group of similar IGRs, the process determined a possible secondary structure using the CMfinder software (26) and automatically provided a score to indicate the likelihood that each IGR group exhibits an evolutionarily conserved secondary structure. This search uncovered many RNAs of various types (24), including a conserved RNA motif now known as the twister ribozyme (Figure 3F).
The initial clue that this RNA might function as a ribozyme was that the genes frequently found near to bacterial twister ribozymes are also commonly found nearby to bacterial hammerhead ribozymes (23). This similarity in associated genes suggested a similar function, a hypothesis that was confirmed experimentally (23). The reason for the gene associations that twister and hammerhead ribozymes share has not yet been determined. Regardless of the biological role of the ribozyme in these genomic contexts, the fact that two structurally unrelated self-cleaving ribozyme classes are frequently found in these locations suggests both that there is some biological need for highly efficient self-cleavage activity, and that the exact RNA motif is unimportant. These facts, in turn, suggest that there might be additional self-cleaving ribozyme classes in these locations that serve the same, as-yet unknown purpose (27).
This hypothesis was explored (27) by first enumerating genetic elements commonly associated with self-cleaving ribozymes in bacteria, and then extracting IGRs near to these genetic elements. These IGRs were expected to be enriched for self-cleaving ribozymes, and were therefore analysed with the above mentioned method (24) to find conserved RNA structures. RNA structures in these IGRs were considered to be candidate self-cleaving ribozymes.
Three novel classes of self-cleaving ribozymes were found in this manner (27): twister sister (27), hatchet (28) and the pistol (29) ribozymes (Figure 3G–I). Additional RNA structures were discovered that did not cleave in experiments (27,30), and the functions of these RNAs are thus far unknown. It is possible that some of them function as ribozymes, but were not active in the experimental conditions tested.
DISCOVERY BY HIGH-THROUGHPUT EXPERIMENTAL METHODS
Non-natural self-cleaving ribozyme classes have been discovered by an in vitro selection strategy, e.g. (31,32). In such a strategy, randomly synthesized RNA molecules are subjected to a selection procedure in a laboratory such that self-cleaving RNAs can survive and be replicated, while other RNAs generally fail to be propagated. After multiple rounds of repeated selection, the pool of RNAs becomes heavily enriched for self-cleaving ribozymes.
Such a strategy is also capable, with adjustments, of finding natural self-cleaving ribozymes (33). By using RNA transcribed from a library derived from the human genome as starting material (instead of random RNA molecules), the selection would amplify any self-cleaving ribozymes present in humans. Although this work did not result in the discovery of a novel structural class, it uncovered previously unknown variants of HDV ribozymes in humans and other mammals (33). Thus, experimental methods are also capable of discovering self-cleaving ribozymes.
SIMILARITY SEARCH
Computational methods to find sequences that are similar to known examples can find new examples of self-cleaving ribozymes, although these approaches are by definition not capable of finding new structural classes. (We avoid the term ‘homology search’, because there is good reason to believe that some structurally similar ribozymes might result from convergent evolution (34). Thus, we say that ribozymes belong to the same structural class, although homology is less certain.) Most notably, similarity searches were fundamental to the discovery of the widespread nature of HDV (35) and hammerhead (36–40) ribozymes, which earlier were thought to be in a much less diverse group of organisms. (One work, however, found widespread hammerhead ribozymes using the previously discussed method to discover conserved RNA structures de novo, where one such RNA structure was found to correspond to bacterial hammerhead ribozymes (41)).
Similarity search methods can be divided into two main categories: structural profiling and pattern matching. Structural-profiling methods exploit statistical models of an RNA’s sequence and structural conservation. The most popular statistical model for structural profiling is the covariance model (42,43). Covariance models use the frequencies of nucleotides and base pairs in the columns of an RNA’s multiple-sequence alignment in order to find statistically similar sequences. A fast and highly accurate implementation that provides statistical significance of predicted homologs (in the form of E-values) is found in the Infernal software (44). The only input needed by a covariance model is a multiple-sequence alignment annotated with a conserved secondary structure, and the sequences to be searched. Covariance models allow for highly accurate searches of ribozymes and other RNAs.
The other similarity search technique popular with self-cleaving ribozymes is based on a user-defined search pattern that specifies the conserved sequence and structural features of a particular self-cleaving ribozyme class. For example, many studies (35–37,40) have used RNABOB (45; http://eddylab.org/software.html), and several similar computer programs have also been successfully applied. Defining a good search pattern can be much more difficult than working with covariance models (46), and covariance models automatically exploit even slight biases in nucleotide frequencies to detect similar sequences. However, most known self-cleaving ribozymes have certain positions that exhibit extremely high conservation, and, in practice, pattern-based searches have been successful for these RNAs, e.g. (37).
PRINCIPLES OF RIBOZYME CATALYSIS
The thorough investigation of the small nucleolytic ribozymes in vitro has addressed important aspects of the chemical mechanisms by which the transesterification reactions (Figure 2) are accelerated. Together with extensive structural studies, these investigations have resulted for most ribozymes in a rather complete picture of their physicochemical behaviour, as detailed in several excellent articles, e.g. (47–51). The most recent comprehensive review contains a detailed discussion of biochemical and other aspects of all self-cleaving ribozymes discovered up to and including the twister ribozyme (51). However, several new ribozyme motifs were subsequently uncovered (27–29) and their analysis allows us to deduce commonalities of catalysis for all self-cleaving ribozyme classes, as summarized below.
All natural small nucleolytic ribozymes feature helical segments that are connected by formally unpaired, often highly conserved nucleotides (Figure 3). The latter are usually involved in the formation of tertiary interactions with other parts of the ribozyme, leading to a compaction of the RNA structure and notably the formation of the catalytic centre that is required for the rate enhancement of the cleavage reaction. For this, four classical principles (α-δ) have been coined (Figure 4): in-line arrangement for the nucleophilic attack by the 2′ OH and departure of the leaving group (α), neutralization of the negative charge at the two non-bridging oxygens (β), deprotonation of the attacking 2′ OH (γ) and neutralization of the negative charge at the 5′ oxygen atom (δ; 52,53). A recent study compared more than 80 high-resolution structures of four small endonucleolytic ribozymes, to determine the extent by which the four principles contribute to catalysis in individual catalytic RNA motifs (54). This study further uncovered two additional principles, termed γ' and γ'', which also contribute to the activation of the 2′ OH nucleophile by a guanine. In brief, both lead to an acidification of the 2′ OH: in the γ' principle by hydrogen bond donation, and in the γ'' principle by the release of the 2′ OH from inhibitory interactions through competitive hydrogen bonding (54).
Figure 4.
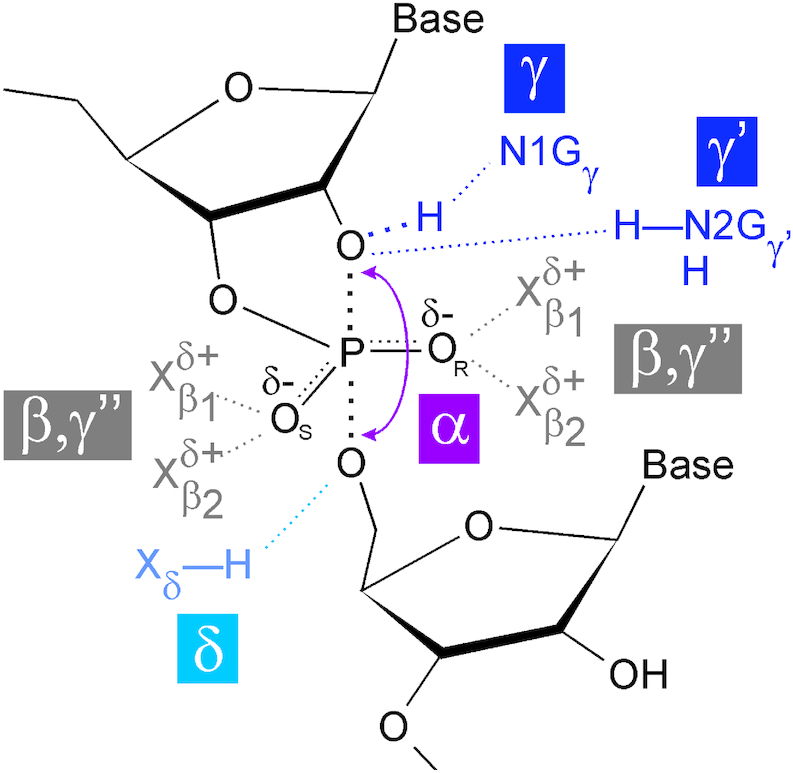
Catalytic strategies of the small nucleolytic ribozymes. To accelerate self-cleavage, the ribozyme motifs employ, to varying degrees, in-line arrangement (α), neutralization of the non-bridging pro-RP (OR) and pro-SP (OS) oxygen atoms (β), deprotonation of the attacking 2′ OH (γ) and neutralization of the negative charge at the 5′ oxygen atom (δ) (52,53). Additionally, the 2′ OH can be acidified by hydrogen bond donation to it (γ') or by preventing inhibitory interactions (γ''). Frequently, the N1 atom of a guanine contributes to the γ strategy, and the exocyclic amine of (sometimes the same) guanine to the γ' principle, while the functional groups involved in the realization of the β, γ'' and δ principles are more variable. For details see (54).
For an in-line attack, the ideal 180° arrangement of the 2′ oxygen, the central phosphate atom and the departing 5′ oxygen (Figure 4) is expected to be realized only in the short-lived transition state of the reaction, if at all. Consequently, a ground state crystal structure could deviate from this arrangement. The closer the arrangement is to 180° in the ground state, the higher the contribution of the α strategy is presumed to be, for a given ribozyme. This is exemplified by the hammerhead, glmS and pistol ribozymes, which deviate by less than 30° from the ideal arrangement (54).
For the neutralization of negative charges at the non-bridging oxygens (β strategy, Figure 4), divalent metal ions or hydrogen atoms bound to either oxygen or nitrogen atoms are in place in the different ribozyme motifs, and often, the β principle employs intricate hydrogen bonding networks (54). The hammerhead ribozyme, for example, employs hydrogens from either a water molecule or a 2′-hydroxyl, and a divalent metal ion (54). These interactions in the hammerhead ribozyme appear fewer in number compared to other ribozymes, as the glmS, twister, pistol and hairpin ribozymes employ complex hydrogen bond networks contributing to the β strategy. In these ribozymes, exo- and endocyclic amines, ordered water molecules or exocyclic hydroxyl groups serve as hydrogen bond donors. Notably, for the glmS ribozyme, two such interactions are contributed by the GlcN6P cofactor (54).
Arguably most important for the overall catalytic rate enhancement of the nucleolytic ribozymes is the use of general acid-base catalysis (55), frequently performed by dedicated nucleobases. For example, in the hairpin, twister and VS ribozymes, elegant pH-dependent kinetic analyses of the cleavage reactions have identified specific guanosines as base and adenines as acid in the realization of the γ and δ principles of catalysis (55–61). As a variation to this “G/A" mechanism, the glmS, the hammerhead and the pistol ribozymes also employ the N1 atom of guanines as the general base, however, they all differ in the nature of the general acid, a role that is fulfilled by the GlcN6P cofactor, the 2′ hydroxyl and a divalent metal ion, respectively (55,62). The general acid-base mechanism of these ribozymes thus can be described as “G/X", or more generally as “N/X", with N for nucleotide and X for a variable chemical entity. As a variation of an “N/X" general acid-base mechanism, the HDV and the twister sister ribozymes use a nucleotide for general acid-base catalysis: in the HDV ribozyme, a cytosine acts as the general acid and a metal ion bound water molecule as the general base, and the same chemical entities are also thought to act in the mechanism of the twister sister ribozyme (63–66).
In summary, the principles of RNA catalysis, as described above, are rather well understood, thanks to a substantial body of kinetic and structural studies (summarized in 51,54). By contrast, comparatively little is known about the biological functions of ribozymes. However, some principles are emerging, and these will be summarized next.
BIOLOGICAL ROLES OF SELF-CLEAVING RIBOZYMES
Although much remains to be learnt about the biological significance of self-cleaving ribozymes, their abundance and wide distribution suggest that they fulfil important functions in nature. Some of these functions have been deciphered, but for many more ribozyme examples, they remain to be elucidated. To date, self-cleaving RNAs are known to play crucial roles in rolling circle replication of some circular RNA entities, such as viroids or plant virus satellite RNAs, have been found as parts of numerous retrotransposons, and in isolated cases have been shown to play roles in mRNA biogenesis and gene regulation. In the following subsections we will briefly summarize these ribozyme functions.
SELF-CLEAVING RIBOZYMES IN REPLICATION OF CIRCULAR MOLECULES
For viroids, plant virus satellite RNAs, HDV RNAs and in transcripts of the mitochondrial Varkud plasmid in Neurospora crassa, self-cleaving ribozymes play an integral role in replication.
Viroids are circular RNAs that do not code for a protein (67,68). They use their hosts’ transcription and processing machinery in concert with their own RNA elements to propagate (69). Viroids can be divided into two distinct families: the Avsunviroidae, which are named after the AvocadoSunblotchViroid (ASBVd), and the Pospiviroidae named after the PotatoSpindleTuberViroid (PSTVd). Members of both families undergo rolling circle replication. For Pospiviroidae, this is performed in the nucleus and in an asymmetric fashion that relies, however, fully on the plant host's enzymes without involvement of ribozymes (reviewed in 70).
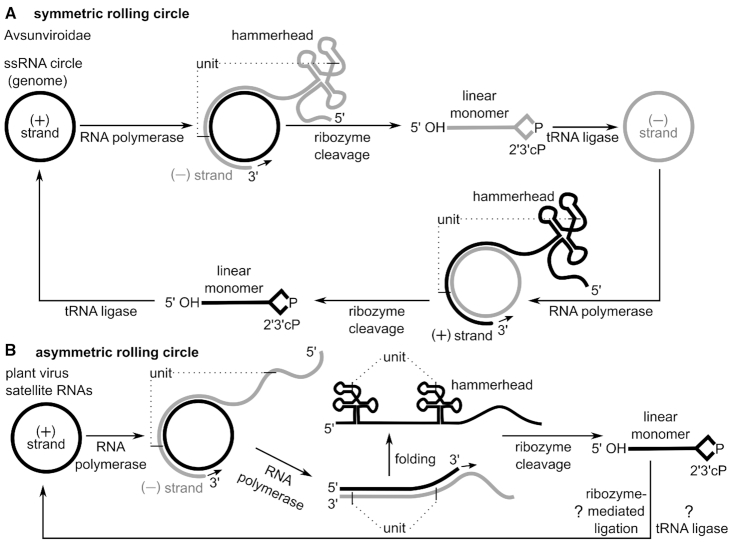
Avsunviroidae replicate their single-stranded, circular RNA genomes in chloroplasts through the symmetric rolling circle mechanism. Integral steps of this mechanism are carried out by hammerhead ribozymes, which have been found in the strands of both polarities (71,72). In this process, the single-stranded, circular RNA referred to as (+) strand is transcribed by the hosts’ DNA-dependent RNA polymerase (Pol) into an RNA, referred to as the (−) strand (Figure 5A). Although not typical, some DNA-dependent RNA polymerases, including RNA Pol II can use certain RNA molecules as templates (73–75). The previously mentioned (−) strand RNA contains multiple reverse-complement copies of the (+) strand that are concatenated together, making it ‘oligomeric’ or ‘multimeric’. In the multimeric (−) strand, hammerhead ribozymes can fold and become active. Their cleavage generates monomeric units, which host enzymes circularize. For example, plant tRNA ligases could catalyse the connection of RNAs with a 2′,3′-cyclic phosphate and 5′-hydroxyl group generated by ribozyme cleavage (76,77); for Avsunviroidae, the chloroplastic isoform of tRNA ligase indeed was shown to be involved in their circularization (78). The resulting circular (−) strand RNA serves as a template for the synthesis of a new (+) strand via analogous (symmetrical) transcription, cleavage and ligation steps (reviewed in 68,70).
Figure 5.
Self-cleaving ribozymes support rolling circle replication mechanisms. (A) Symmetric rolling circle mechanism in viroids of the Avsunviroidae family and plant virus satellite RNAs. Circular (+) strand RNA is transcribed by DNA-dependent RNA polymerase into oligomeric (−) strand RNAs. Dotted lines indicate cleavage sites that define a single unit within the oligomeric RNA. Units are separated by hammerhead ribozyme cleavage and circularized by host enzymes, such as tRNA ligase. The circular (−) strand RNA is used for a second round of amplification yielding the (+) strand genome. In some plant virus satellite RNAs a hairpin instead of a hammerhead ribozyme could catalyse the cleavage of (+) strand oligomeric transcript, as well as potentially the ligation of the (+) strand linear monomer into a circular RNA. (B) Asymmetric rolling circle mechanism in plant virus satellite RNAs. First, the (+) strand RNA is transcribed by the host RNA polymerase into a long oligomeric transcript, the (−) strand RNA. Then the (−) strand RNA serves as template for a second transcription resulting in an oligomeric (+) strand. Hammerhead ribozymes, which are encoded in the (+) strand, cleave the oligomeric transcript into linear monomers. These unit-length transcripts are circularized either by ribozyme-mediated or enzymatic ligation.
Other types of circular RNAs also use ribozymes as part of their rolling circle replication scheme. Plant virus satellite RNAs (Figure 5B), for example, can replicate only in the presence of a helper RNA virus, whose proteins also perform necessary tasks for the satellite RNA (68,79). As with viroids, ribozymes contribute to replication by cleaving the multimeric linear transcripts into monomers. These monomers are ligated such that they form circles, a reaction often carried out by cytoplasmic tRNA ligases (76). However, unlike the ribozymes in viroids, some ribozymes in satellite RNAs can, at least in vitro, catalyse self-ligation, in addition to self-cleavage. All known satellite RNAs that use a catalytic RNA during rolling circle replication use hammerhead ribozymes, and three of these satellite RNAs additionally employ hairpin ribozymes encoded in their minus polarity strand (15,80,81). These three satellite RNAs are the only known natural occurrences of the hairpin ribozyme (17). Indeed the hairpin ribozyme is an efficient RNA self-ligase (Figure 2), which suggests that it —and not a proteinaceous enzyme— might also perform the circularization step in vivo (80,82).
Satellite RNAs can use the symmetric or asymmetric rolling circle pathway. Satellites that use asymmetric rolling circle replication, where no circular RNA intermediate is formed (Figure 5B), employ hammerhead ribozymes only in the (+) strand. However, satellites using the symmetric rolling circle mechanism use ribozymes on both strands (+ and −, Figure 5A). In known examples, these ribozymes either both belong to the hammerhead ribozyme class or there is a hammerhead on one strand and a hairpin ribozyme on the other (80,83,84).
A subviral entity closely related to viroids is the HDV RNA. Similarly to plant virus satellite RNAs, HDV RNA depends on a helper virus (the Hepatitis B Virus) for its transmission (68). The HDV RNA has been found in higher eukaryotes, including humans, and has a genome size of about 1,700 nucleotides (85). The HDV RNA is divided into two distinct domains, one of which contains the coding sequence for a protein, the delta-antigen (δAg; 86–88). The other genomic region corresponds to a ∼350 nt viroid-like sequence that folds into a rod-like secondary structure for protection against cellular RNases and contains self-cleaving ribozymes necessary for replication in its terminal part (88–90). The HDV RNA replicates via a symmetric rolling circle mechanism catalysed by host enzymes such as a DNA-dependent RNA polymerase and progresses through subsequent co-transcriptional self-cleavage of HDV ribozymes (91–93). The resulting RNA fragments can be ligated by other host-specific enzymes, which are presumed to resemble the tRNA ligases used for this purpose in plants (94).
In the fungus Neurospora crassa, self-cleaving ribozymes are part of a rolling circle mechanism that includes a DNA stage. In the Varkud isolate of Neurospora, the mitochondria contain an 881-nt DNA plasmid (Varkud plasmid) whose transcription product is an oligomeric RNA containing self-cleaving ribozymes (19,95). These VS ribozymes perform cleavage and ligation reactions in vitro, which are both necessary for the replication of the satellite RNA (96–99). The oligomeric transcript is cleaved into monomers by the VS ribozyme, which, in turn, ligate these monomeric RNAs to generate circular RNA intermediates. These circular RNAs are thought to serve as templates for reverse transcription by an RT encoded on the Varkud plasmid. After displacement or degradation of the RNA template from the (−) strand cDNA, synthesis of the (+) strand DNA and ligation to generate a closed circular DNA presumably occur (97).
SELF-CLEAVING RIBOZYMES AS PART OF RETROTRANSPOSONS
There are several self-cleaving ribozyme classes that are found as parts of retrotransposons. A retrotransposon is a type of mobile genetic element that can move copies of itself to different locations within the genome. In this process, the retrotransposon is first copied from the genomic locus by transcription into RNA. Then this RNA intermediate is reverse transcribed into cDNA and inserted back into the genome at a new location. To facilitate this process, retrotransposons usually encode proteins such as reverse transcriptases (RTs) or endonucleases (ENs; 100). Here, we will briefly discuss the self-cleaving ribozyme-containing R2 elements, the L1Tc retrotransposon found in Trypanosoma cruzi, short interspersed nuclear elements (SINEs) in Schistosomes, Penelope-like elements and retrozymes.
A very well-studied retrotransposon subclass, which contains HDV-like ribozymes, are so-called R2 elements. These non-long terminal repeat (non-LTR) retrotransposons insert site-specifically into ribosomal DNA (rDNA). First discovered in Drosophila melanogaster (101,102), examples of these elements are now known in other insect species such as the silkmoth Bombyx mori (103,104), in arthropods, nematodes, birds and tunicates (105). R2 elements can only integrate at one position within 28S rRNA genes, defined by sequence features at this location (Figure 6A). This restriction in genomic location of R2 elements made them relatively easy to study (105).
Figure 6.
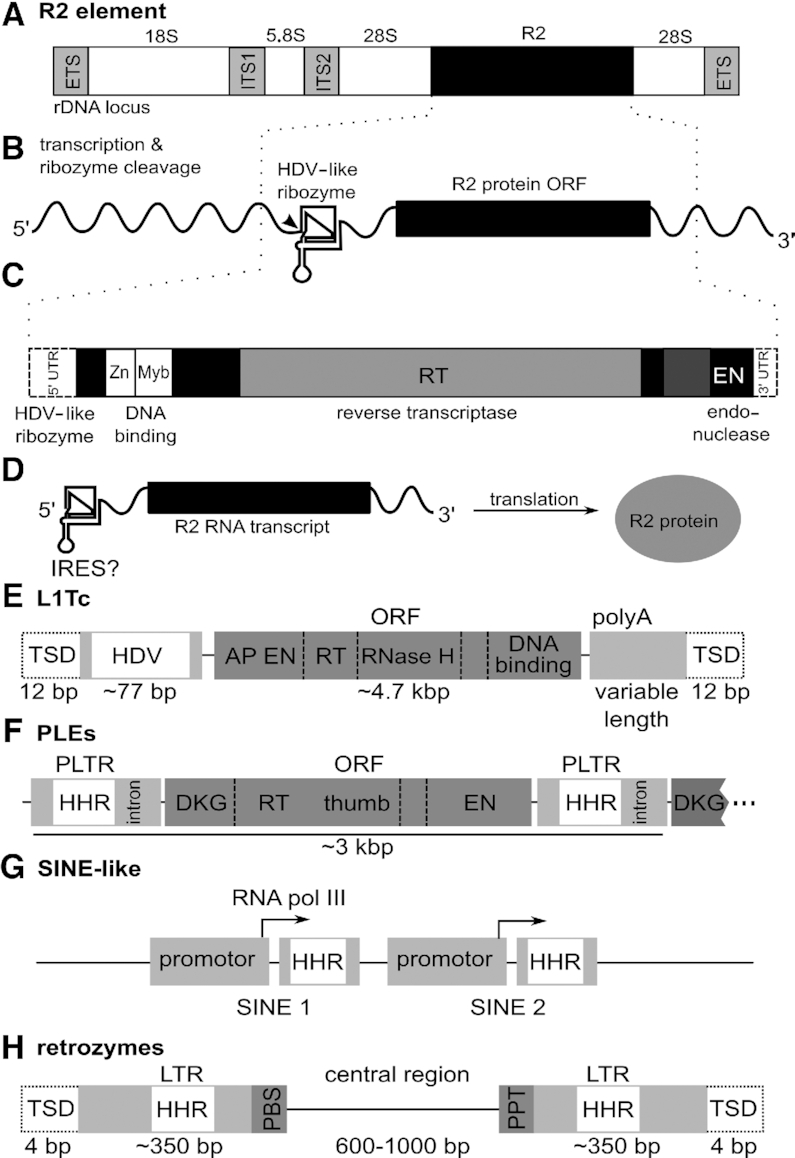
Several retrotransposons contain self-cleaving ribozymes. (A–D) The R2 element in Bombyx mori. (A) Organization of an rDNA unit with an R2 element inserted into a specific site within the 28S rDNA is shown. The external transcribed spacers (ETS) and internal transcribed spacers (ITS1 and ITS2) found in the precursor rRNA are depicted as light grey boxes. (B) Transcription of the R2 element in B. mori yields an RNA consisting of the HDV-like ribozyme in its 5′ UTR, the open reading frame (ORF) for the R2 protein and a 3′ UTR. (C) An expanded view of the R2 ORF illustrates a (simplified) composition of protein domains, which include zinc-finger and Myb-like nucleic acid binding motifs, the reverse transcriptase (RT) domain and the endonuclease domain (EN). Dotted lines designate untranslated regions. (D) Translation of the open reading frame generates R2 protein. Translation initiation likely occurs through IRES-like structure of the 5′ UTR. R2 proteins can bind the 5′ and 3′ end of the R2 element RNA. (E) Schematic representation of the simplified composition of L1Tc retrotransposons from Trypanosoma cruzi. The element is flanked by target site duplications (TSD) of usually 12 bp and it encodes a protein with an apurinic/apyrimidinic endonuclease (AP EN) domain, a reverse transcriptase (RT) and RNase H domain and a DNA binding domain. The first 77 nt of L1Tc harbor an HDV ribozyme (HDV). (F) Schematic representation of the composition of Penelope-like elements (PLEs). PLEs occur as tandem or multi-copy repeats in which an open reading frame (ORF) is flanked by Penelope long terminal repeats (PLTRs). The ORF encodes a protein with an RT and EN domain. The PLTRs contain a hammerhead ribozyme (HHR) and have been shown to also contain an intron in some PLEs. (G) Schematic representation of small interspersed nuclear element-like retrotransposons in Schistosoma often found in repetitive sequences. They consist of a promoter followed by a hammerhead ribozyme (HHR). All promoters could initiate transcription. (H) Schematic representation of the composition of retrozymes. Retrozymes are flanked by target site duplications (TSDs) and LTRs, which contain hammerhead ribozymes (HHR). The central region does not contain an open reading frame and is flanked by primer binding site (PBS) and poly-purine tract (PPT) elements needed for priming of DNA synthesis from the RNA element.
R2 elements are composed of an open reading frame (ORF), which encodes the R2 protein and is flanked by 5′ and 3′ untranslated regions (UTRs, Figure 6B). The R2 protein is a multi-domain protein that harbours DNA-binding domains, a reverse transcriptase domain and an endonuclease domain (Figure 6C; (106–109). The R2 protein and its UTRs are able to achieve retrotransposition, although additional host factors are likely also involved. As part of the R2 element, the HDV-like ribozyme appears to be important for several aspects of retrotransposition. First, as the full-length R2 element is co-transcribed with the 28S rRNA by RNA polymerase I (110), the ribozyme, which is present in the first ∼184 nt of R2 (111,112), cleaves this transcript and thus separates it from the 28S rRNA. Second, to allow translation of the uncapped R2 protein transcript, the HDV-like ribozyme is thought to function as an internal ribosomal entry site (IRES, Figure 6D; 113–116). Third, the R2 protein can bind both of its transcript's UTRs, which facilitates the integration process (117,118). In the 5′ UTR, the R2 protein binds to a pseudoknot that is part of an extended sequence within the conserved HDV ribozyme (111,119). Thus, R2 protein binding supports genome integration of the retrotransposon by a mechanism independent from self-cleavage. Lastly, a by-product of ribozyme-mediated processing of the 28S co-transcript is the propagation of several non-autonomous parasites of R2, so-called SIDEs (short internally deleted elements). These truncated R2 elements have lost their ORF, but are able to replicate non-autonomously using R2 proteins provided in trans by complete R2 elements (120).
Apart from retrotransposons that insert site-specifically into the host genome, there are other self-cleaving ribozymes found in association with retroelements that insert with little specificity, and can be found essentially anywhere in the genome. In Trypanosoma cruzi the retrotransposon L1Tc (Figure 6E) occurs in disparate genomic locations often nearby to repetitive DNA, and forms tandem repeats (121). L1Tc is a type of long interspersed nuclear element (LINE). Like all LINEs, L1Tc carries all genetic elements required for its transposition, and as such is termed ‘autonomous’. In its 5′ UTR, L1Tc contains an HDV-like ribozyme, which is active in co-transcriptional cleavage assays (122). Thus, in this example as well, the ribozyme can trigger the release of the transposable element, in this case from long polycistronic transcripts, which are common in Trypanosomes (122). In eukaryotes, a 5′ cap and 3′ poly(A) tail are normally required for an efficient start of translation. In certain cases where these essential features are absent, HDV-like ribozymes likely support translation initiation instead. As in the case of R2 retrotransposons, it has been proposed that HDV-like ribozymes can function as IRESes (122). In vitro investigations and in vivo translation assays revealed translation efficiencies similar to or higher than that of the Hepatitis C Virus IRES positive control (114). Therefore, it seems likely that the HDV-like ribozyme in L1Tc and R2 elements acts similarly to an IRES, presumably by binding the translation machinery and enabling translation initiation.
In the aforementioned secondary structure-based computational searches, HDV-like ribozymes were found to be widespread in nature (35). Among the uncovered examples, there were several HDV-like ribozymes found in close proximity to genes coding for RT-like proteins, and these proteins are typical of retrotransposons. Also, many genomic regions were found in which hundreds of intergenic HDV-like ribozyme copies were each located between conserved downstream sequences and different upstream sequences (35). In these cases, the downstream regions could reflect conserved retrotransposon elements, while the differing upstream regions are consistent with varying integration sites. Thus, these ribozyme examples could be parts of retrotransposons.
In the same computational search, additional examples were identified, which also contain HDV-like ribozymes and belong to so-called baggins retrotransposons and retrotransposon-like elements (RTEs). These elements do not carry their own promoters, but are transcribed because they occur in introns or immediately downstream of LTR retrotransposons (114,123). Some LINEs and non-autonomous SINEs also make use of hammerhead or HDV-like ribozymes, and these are known in a diverse variety of metazoans (114,124). In all these cases, it is likely that the self-cleaving ribozymes function to liberate the mobile genetic element from its co-transcript, as in the examples described earlier.
Retrotransposons can be grouped into different subclasses that are distinguished by the presence of LTRs that usually serve as transcriptional promoters (125), and/or their capacity to code for reverse transcriptases. One family of retrotransposons that does not exactly fit these distinctions, and therefore forms its own subclass, is that of Penelope-like elements (PLEs; 126,127). Penelope-like elements have been identified in different phyla of eukaryotes. They are massively widespread in metazoans, can be found in vertebrates, but not in mammalian genomes and their distribution includes genomes of fungi, plants and protists (126,128,129).
PLEs code for reverse transcriptase and endonuclease enzymes and carry Penelope-like LTRs (PLTRs) at their ends (Figure 6F; 130). PLEs are about 3 kb on average and the PLTRs always contain hammerhead ribozymes, as shown recently (131). PLEs are often found adjacent to one-another in a ‘tandem’ arrangement, as they tend to insert into or adjacent to pre-existing PLE copies. This tandem arrangement is likely needed for the element to be expressed (132). PLEs typically contain transcriptional promoters in their PLTRs. From this promoter, downstream PLEs within a tandem arrangement (Figure 6F) can be transcribed, as confirmed by studies of tandem PLE elements demonstrating that the downstream PLE’s transcript start site lies in the PLTR of its upstream partner (129).
Additionally, the tandem arrangement of PLEs also appears to be necessary for efficient ribozyme cleavage. The hammerhead ribozymes in PLEs have a palindromic, extremely short loop in stem III and are mostly of the type-I secondary structure, where the 5′ and 3′ end of the ribozyme come together in stem I (Figure 3A). Hammerhead ribozymes with short stem III structures have been previously hypothesized to be thermodynamically unstable (133) and subsequent analysis showed that it is likely that these RNAs do not cleave as monomers, but form dimers (131,133,134). Thus, the PLE’s frequent tandem layout might help adjacent hammerhead ribozyme pairs to form dimers so that they can self-cleave.
As in vitro self-cleavage rates for PLE hammerhead ribozymes are very low, even as dimers, it is plausible that these RNAs rely in addition on other factors, such as RNA chaperones, to increase cleavage speed in vivo (131). Cleavage of the PLE transcript by the ribozyme would generate RNA ends that could be ligated, either by the ribozyme's intrinsic ligation function (Figure 2) or by proteinaceous ligases present in the cells (as mentioned above for the replication of circular RNAs). The resulting circular RNAs could serve as template for a rolling circle-like amplification step, similar to those shown in Figure 5. Oligomeric transcripts, that have not been cleaved by the ribozyme to form monomers, can then be used by the PLE RT and EN enzymes to support genomic insertion of the retrotransposon (135). This would automatically lead to a sequence of several adjacent PLEs, which is indeed how they are often observed in the genome. Such a repeat architecture would allow efficient retrotransposition, because a downstream PLE can be transcribed by the promoter in the upstream PLE (126).
Other elements that use hammerhead ribozymes are the subtype of SINE-like retrotransposons occurring in Schistosoma mansoni and related organisms (124,136). These retrotransposons are often found as part of repetitive sequences and appear to be transcribed by RNA polymerase III. In vitro cleavage assays have shown that the hammerhead ribozyme is capable of liberating SINE copies from multimeric transcripts by cleavage in cis or trans (124). Once liberated, the RNA is copied into a DNA by reverse transcriptase, and this DNA integrates elsewhere in the host genome. However, as the RNA polymerase III promoter is located only at the 3′ end of cleaved multimeric transcripts, single copies of the SINEs lose their ability to propagate (Figure 6G; 124). Thus, SINEs often occur in tandem repeats of at least two elements, where the 5′-most element transcribes the following SINEs (124). As a variation of the theme, transcripts of satellite DNA in different newt species (137,138) self-cleave by their embedded hammerhead ribozyme, but these transcripts appear to be generated by RNA polymerase II (137).
In plants, the recently identified transposable elements termed retrozymes contain hammerhead ribozymes. Retrozymes are mostly found in eudicots, with some examples in ferns, monocots and algae. These roughly 1- to 1.5-kb-long elements are delimited by 4-bp target-site duplications (TSDs) and LTRs of about 350 bp that each harbour a hammerhead ribozyme (Figure 6H; 139). The central region of the element has a variable length of about 600–1000 bp and does not seem to encode a protein. While retrozymes are thought to replicate through rolling circle amplification (Figure 5), they require proteins expressed from autonomous elements to insert new copies into the genome. Their central region is flanked by two conserved domains, the primer binding site (PBS) and a poly-purine tract (PPT), which are also typical for Ty3-gypsy long terminal repeat retrotransposons (139). These regions are required for the mobilization of LTR-retrotransposons, as they serve to prime DNA synthesis from the linear RNA transcript (125,140,141).
In summary, self-cleaving ribozymes in retrotransposons enable propagation through self-cleavage and self-ligation. However, in studying self-cleaving ribozyme functions we should consider additional possible activities that these catalytic RNA can achieve, e.g. functioning as an IRES or interaction partner for proteins.
SELF-CLEAVING RIBOZYMES IN EUKARYOTIC GENE REGULATION AND mRNA BIOGENESIS
Several self-cleaving ribozymes have been implicated to play roles in gene regulation and mRNA biogenesis. HDV-like ribozymes in CPEB3 genes in mammals (33) and hammerhead ribozymes in amniotes (39,142) are each found in introns. This genomic location suggests a possible ribozyme function on pre-mRNA processing and/or alternative splicing, even though definitive experimental proof has yet to be brought forward. Another possibility for ribozyme involvement in mRNA biogenesis includes an unusual hammerhead ribozyme that was described in some mammalian C-type lectin (Clec2) and Clec-like genes (143). The hammerhead structure in this RNA is separated by an insertion of several hundred nucleotides. The ribozyme is only active when these flanking regions come together, allowing them to fold into the ribozyme structure. In vitro, cleavage for such a bipartite hammerhead ribozyme was observed. Interestingly, the ribozyme cleavage site lies exactly between the translation termination and polyadenylation signal within the 3′ UTR. This means that upon cleavage the polyadenylation signal is removed from the 3′ end of the mRNA, which leads to a reduction in protein expression in vivo (143). The discovery and characterization of this unusual ribozyme example has fuelled the search for trans-cleaving ribozymes (144).
SELF-CLEAVING RIBOZYME BIOLOGY IN BACTERIA
The recent discovery of new self-cleaving ribozyme classes by comparative sequence analysis is based on the observation that, in bacteria, self-cleaving ribozymes are often found in close proximity to each other or to certain types of genes (see ribozyme discovery section). This observation, however, not only provides the means to discover more self-cleaving ribozymes, but also opens up the possibility to decipher their biological roles in bacteria, which so far are largely elusive.
To date the only well-understood example of bacterial self-cleaving ribozyme function is the afore-mentioned glmS ribozyme class (14). Here, the fact that this RNA is only found in the 5′ UTR of the glmS mRNA immediately suggested a role in gene regulation similar to that of riboswitches. Distinct to the majority of those motifs (145), however, the specific metabolite GlcN6P does not induce a conformational change, but is directly involved in the self-cleavage of the mRNA, as detailed above. Hence, the GlcN6P-induced cleavage of the glmS mRNA followed by the degradation by cellular RNases (22) provides the means for negative feedback regulation in many Gram-positive and some Gram-negative bacteria (146).
An HDV-like ribozyme discovered in Faecalibacterium prausnitzii is located upstream of the glmM gene coding for phosphoglucosamine mutase. This location suggests a possible involvement in metabolite-dependent gene regulation (147) as has been proven in the case of glmS self-cleaving ribozymes (14).
In addition to the glmS ribozyme, a biological role for some bacterial hammerhead (41) and twister ribozyme examples (148) has been suggested in the processing of mRNAs. These ribozymes have the potential to influence gene expression in the 3′ UTR as illustrated by reporter constructs which were engineered to contain twister ribozymes in their 3′ UTR (148). It has also been suggested that self-cleaving ribozymes can process polycistronic transcripts and that these processing events could lead to transcripts with different stabilities or translation efficiencies (41,148), but additional work is needed to concretely define the specific purpose of these ribozymes. Due to their genomic location near phage genes, it is also likely that these ribozyme functions could be important in bacteriophages as well as in bacteria or their interactions (23,27).
OUTLOOK
While the principles of ribozyme catalysis are comparably well understood, the biological function of most genomically encoded catalytic RNAs remains to be deciphered. The use of bioinformatics has in recent years significantly increased the number of both ribozyme families and examples of old and new members thereof. Most of these, however, are functionally orphans, and only hypothetical functions of such motifs have been discussed in the context of the genomically encoded hammerhead ribozymes (149). One aspect that has not been in the limelight of ribozyme research is the ubiquitous formation of RNA-protein complexes (RNPs) in cellular systems, and in particular for both group II introns and RNase P, the functional importance of the respective interacting proteins on RNA catalysis is well documented (e.g. (150,151). Although RNPs have been reported for some nucleolytic ribozymes, or a positive contribution of proteins to their catalysis has been reported, e.g. (152–158) or hypothesized (27), there is little known about the nature of the endogenously interacting protein(s), nor their general influence on ribozyme function in vivo. In recent years, several unbiased experimental approaches have been established that allow for the identification of RNPs from cellular systems (159–162). Given that virtually all RNA molecules in cellular systems interact with proteins, we propose that the application of such methods to ribozymes might lead to the identification of their proteinaceous interaction partners. Such endogenous proteins interacting with ribozymes (EPIRs) might bind their catalytic RNA specifically or non-specifically. In either case, their nature and characterization in vivo or in vitro could shed light on the long-standing question (137) of the biological function of genomically encoded ribozymes.
ACKNOWLEDGEMENTS
We thank Dr Benedikt Beckmann for helpful discussions and Drs Timothy J. Wilson, Monica Hagedorn and Claudia Gerlich for their helpful comments on the manuscript.
FUNDING
German Research Foundation (DFG) [LU1889/4-1 to C.E.W., CH3459/10-1 to C.H. and WE6322/1-1 to Z.W.]; Tönjes-Vagt-Stiftung [TVSXXXIV to C.H.]. Funding for open access charge: DFG and Leipzig University within the program of Open Access Publishing.
Conflict of interest statement. None declared.
REFERENCES
- 1. Cech T.R., Zaug A.J., Grabowski P.J.. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell. 1981; 27:487–496. [DOI] [PubMed] [Google Scholar]
- 2. Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S.. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983; 35:849–857. [DOI] [PubMed] [Google Scholar]
- 3. Peebles C.L., Perlman P.S., Mecklenburg K.L., Petrillo M.L., Tabor J.H., Jarrell K.A., Cheng H.L.. A self-splicing RNA excises an intron lariat. Cell. 1986; 44:213–223. [DOI] [PubMed] [Google Scholar]
- 4. van der Veen R., Arnberg A.C., van der Horst G., Bonen L., Tabak H.F., Grivell L.A.. Excised group II introns in yeast mitochondria are lariats and can be formed by self-splicing in vitro. Cell. 1986; 44:225–234. [DOI] [PubMed] [Google Scholar]
- 5. Lilley D.M. Structure, folding and mechanisms of ribozymes. Curr. Opin. Struct. Biol. 2005; 15:313–323. [DOI] [PubMed] [Google Scholar]
- 6. Nissen P., Hansen J., Ban N., Moore P.B., Steitz T.A.. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000; 289:920–930. [DOI] [PubMed] [Google Scholar]
- 7. Noller H.F., Hoffarth V., Zimniak L.. Unusual resistance of peptidyl transferase to protein extraction procedures. Science. 1992; 256:1416–1419. [DOI] [PubMed] [Google Scholar]
- 8. Korostelev A., Trakhanov S., Laurberg M., Noller H.F.. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006; 126:1065–1077. [DOI] [PubMed] [Google Scholar]
- 9. Cech TR, Atkins JF, Gesteland RF. The RNA World. 2005; 3rd ednCold Spring Harbor: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 10. Helm M., Alfonzo J.D.. Posttranscriptional RNA Modifications: playing metabolic games in a cell's chemical Legoland. Chem. Biol. 2014; 21:174–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grosjean H., Westhof E.. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 2016; 44:8020–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ranjan N., Rodnina M.V.. tRNA wobble modifications and protein homeostasis. Translation. 2016; 4:e1143076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nedialkova D.D., Leidel S.A.. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell. 2015; 161:1606–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Winkler W.C., Nahvi A., Roth A., Collins J.A., Breaker R.R.. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004; 428:281–286. [DOI] [PubMed] [Google Scholar]
- 15. Prody G.A., Bakos J.T., Buzayan J.M., Schneider I.R., Bruening G.. Autolytic processing of dimeric plant virus satellite RNA. Science. 1986; 231:1577–1580. [DOI] [PubMed] [Google Scholar]
- 16. Buzayan J.M., Gerlach W.L., Bruening G.. Satellite tobacco ringspot virus RNA: A subset of the RNA sequence is sufficient for autolytic processing. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:8859–8862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bajaj P., Steger G., Hammann C.. Sequence elements outside the catalytic core of natural hairpin ribozymes modulate the reactions differentially. Biol. Chem. 2011; 392:593–600. [DOI] [PubMed] [Google Scholar]
- 18. Sharmeen L., Kuo M.Y., Dinter-Gottlieb G., Taylor J.. Antigenomic RNA of human hepatitis delta virus can undergo self-cleavage. J. Virol. 1988; 62:2674–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saville B.J., Collins R.A.. A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell. 1990; 61:685–696. [DOI] [PubMed] [Google Scholar]
- 20. Barrick J.E., Corbino K.A., Winkler W.C., Nahvi A., Mandal M., Collins J., Lee M., Roth A., Sudarsan N., Jona I. et al.. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:6421–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Breaker R.R. Prospects for riboswitch discovery and analysis. Mol. Cell. 2011; 43:867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Collins J.A., Irnov I., Baker S., Winkler W.C.. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev. 2007; 21:3356–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roth A., Weinberg Z., Chen A.G., Kim P.B., Ames T.D., Breaker R.R.. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat. Chem. Biol. 2014; 10:56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weinberg Z., Wang J.X., Bogue J., Yang J., Corbino K., Moy R.H., Breaker R.R.. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 2010; 11:R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J.. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yao Z., Weinberg Z., Ruzzo W.L.. CMfinder–a covariance model based RNA motif finding algorithm. Bioinformatics. 2006; 22:445–452. [DOI] [PubMed] [Google Scholar]
- 27. Weinberg Z., Kim P.B., Chen T.H., Li S., Harris K.A., Lünse C.E., Breaker R.R.. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat. Chem. Biol. 2015; 11:606–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li S., Lünse C.E., Harris K.A., Breaker R.R.. Biochemical analysis of hatchet self-cleaving ribozymes. RNA. 2015; 21:1845–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Harris K.A., Lünse C.E., Li S., Brewer K.I., Breaker R.R.. Biochemical analysis of pistol self-cleaving ribozymes. RNA. 2015; 21:1852–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weinberg Z., Lünse C.E., Corbino K.A., Ames T.D., Nelson J.W., Roth A., Perkins K.R., Sherlock M.E., Breaker R.R.. Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions. Nucleic Acids Res. 2017; 45:10811–10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Williams K.P., Ciafré S., Tocchini-Valentini G.P.. Selection of novel Mg(2+)-dependent self-cleaving ribozymes. EMBO J. 1995; 14:4551–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tang J., Breaker R.R.. Structural diversity of self-cleaving ribozymes. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:5784–5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Salehi-Ashtiani K., Lupták A., Litovchick A., Szostak J.W.. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006; 313:1788–1792. [DOI] [PubMed] [Google Scholar]
- 34. Salehi-Ashtiani K., Szostak J.W.. In vitro evolution suggests multiple origins for the hammerhead ribozyme. Nature. 2001; 414:82–84. [DOI] [PubMed] [Google Scholar]
- 35. Webb C.H., Riccitelli N.J., Ruminski D.J., Luptak A.. Widespread occurrence of self-cleaving ribozymes. Science. 2009; 326:953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seehafer C., Kalweit A., Steger G., Gräf S., Hammann C.. From alpaca to zebrafish: hammerhead ribozymes wherever you look. RNA. 2011; 17:21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Przybilski R., Graf S., Lescoute A., Nellen W., Westhof E., Steger G., Hammann C.. Functional hammerhead ribozymes naturally encoded in the genome of Arabidopsis thaliana. Plant Cell. 2005; 17:1877–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. de la Peña M., García-Robles I.. Ubiquitous presence of the hammerhead ribozyme motif along the tree of life. RNA. 2010; 16:1943–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. de la Peña M., García-Robles I.. Intronic hammerhead ribozymes are ultraconserved in the human genome. EMBO Rep. 2010; 11:711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jimenez R.M., Delwart E., Lupták A.. Structure-based search reveals hammerhead ribozymes in the human microbiome. J. Biol. Chem. 2011; 286:7737–7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Perreault J., Weinberg Z., Roth A., Popescu O., Chartrand P., Ferbeyre G., Breaker R.R.. Identification of hammerhead ribozymes in all domains of life reveals novel structural variations. PLoS Comput. Biol. 2011; 7:e1002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sakakibara Y., Brown M., Mian I.S., Underwood R., Haussler D.. Proceedings of the Twenty-seventh IEEE Hawaii International Conference of System Sciences (HICSS 94). 1994; 284–294. [Google Scholar]
- 43. Eddy S.R., Durbin R.. RNA sequence analysis using covariance models. Nucleic Acids Res. 1994; 22:2079–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nawrocki E.P., Eddy S.R.. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013; 29:2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Eddy S. RNABOB 2.1. 2005; http://eddylab.org/software.html. [Google Scholar]
- 46. Menzel P., Gorodkin J., Stadler P.F.. The tedious task of finding homologous noncoding RNA genes. RNA. 2009; 15:2075–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wilson T.J., Lilley D.M.. Biochemistry. The evolution of ribozyme chemistry. Science. 2009; 323:1436–1438. [DOI] [PubMed] [Google Scholar]
- 48. Cochrane J.C., Strobel S.A.. Catalytic strategies of self-cleaving ribozymes. Acc. Chem. Res. 2008; 41:1027–1035. [DOI] [PubMed] [Google Scholar]
- 49. Lau M.W., Ferre-D’Amare A.R.. Many activities, one structure: functional plasticity of ribozyme folds. Molecules. 2016; 21:E1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lilley D.M.J. How RNA acts as a nuclease: some mechanistic comparisons in the nucleolytic ribozymes. Biochem. Soc. Trans. 2017; 45:683–691. [DOI] [PubMed] [Google Scholar]
- 51. Jimenez R.M., Polanco J.A., Lupták A.. Chemistry and biology of self-cleaving ribozymes. Trends Biochem. Sci. 2015; 40:648–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Breaker R.R., Emilsson G.M., Lazarev D., Nakamura S., Puskarz I.J., Roth A., Sudarsan N.. A common speed limit for RNA-cleaving ribozymes and deoxyribozymes. RNA. 2003; 9:949–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Emilsson G.M., Nakamura S., Roth A., Breaker R.R.. Ribozyme speed limits. RNA. 2003; 9:907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Seith D.D., Bingaman J.L., Veenis A.J., Button A.C., Bevilacqua P.C.. Elucidation of catalytic strategies of small nucleolytic ribozymes from comparative analysis of active sites. ACS Catal. 2018; 8:314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wilson T.J., Liu Y., Li N.S., Dai Q., Piccirilli J.A., Lilley D.M.J.. Comparison of the structures and mechanisms of the pistol and hammerhead ribozymes. J. Am. Chem. Soc. 2019; 141:7865–7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wilson T.J., McLeod A.C., Lilley D.M.. A guanine nucleobase important for catalysis by the VS ribozyme. EMBO J. 2007; 26:2489–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wilson T.J., Li N.S., Lu J., Frederiksen J.K., Piccirilli J.A., Lilley D.M.. Nucleobase-mediated general acid-base catalysis in the Varkud satellite ribozyme. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:11751–11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pinard R., Hampel K.J., Heckman J.E., Lambert D., Chan P.A., Major F., Burke J.M.. Functional involvement of G8 in the hairpin ribozyme cleavage mechanism. EMBO J. 2001; 20:6434–6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lafontaine D.A., Wilson T.J., Norman D.G., Lilley D.M.. The A730 loop is an important component of the active site of the VS ribozyme. J. Mol. Biol. 2001; 312:663–674. [DOI] [PubMed] [Google Scholar]
- 60. Kath-Schorr S., Wilson T.J., Li N.S., Lu J., Piccirilli J.A., Lilley D.M.. General acid-base catalysis mediated by nucleobases in the hairpin ribozyme. J. Am. Chem. Soc. 2012; 134:16717–16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wilson T.J., Liu Y., Domnick C., Kath-Schorr S., Lilley D.M.. The novel chemical mechanism of the twister ribozyme. J. Am. Chem. Soc. 2016; 138:6151–6162. [DOI] [PubMed] [Google Scholar]
- 62. Han J., Burke J.M.. Model for general acid-base catalysis by the hammerhead ribozyme: pH-activity relationships of G8 and G12 variants at the putative active site. Biochemistry. 2005; 44:7864–7870. [DOI] [PubMed] [Google Scholar]
- 63. Nakano S., Chadalavada D.M., Bevilacqua P.C.. General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science. 2000; 287:1493–1497. [DOI] [PubMed] [Google Scholar]
- 64. Liu Y., Wilson T.J., Lilley D.M.J.. The structure of a nucleolytic ribozyme that employs a catalytic metal ion. Nat. Chem. Biol. 2017; 13:508–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Das S.R., Piccirilli J.A.. General acid catalysis by the hepatitis delta virus ribozyme. Nat. Chem. Biol. 2005; 1:45–52. [DOI] [PubMed] [Google Scholar]
- 66. Gaines C.S., York D.M.. Model for the functional active state of the TS ribozyme from molecular simulation. Angew. Chem. Int. Ed. Engl. 2017; 56:13392–13395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Flores R., Gas M.E., Molina-Serrano D., Nohales M.A., Carbonell A., Gago S., de la Peña M., Daròs J.A.. Viroid Replication: Rolling-Circles, enzymes and ribozymes. Viruses-Basel. 2009; 1:317–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Flores R., Grubb D., Elleuch A., Nohales M.-Á., Delgado S., Gago S.. Rolling-circle replication of viroids, viroid-like satellite RNAs and hepatitis delta virus: Variations on a theme. RNA Biol. 2014; 8:200–206. [DOI] [PubMed] [Google Scholar]
- 69. Flores R., Minoia S., Carbonell A., Gisel A., Delgado S., López-Carrasco A., Navarro B., Di Serio F.. Viroids, the simplest RNA replicons: How they manipulate their hosts for being propagated and how their hosts react for containing the infection. Virus Res. 2015; 209:136–145. [DOI] [PubMed] [Google Scholar]
- 70. Hammann C., Steger G.. Viroid-specific small RNA in plant disease. RNA Biol. 2012; 9:809–819. [DOI] [PubMed] [Google Scholar]
- 71. Daròs J.A., Marcos J.F., Hernandez C., Flores R.. Replication of avocado sunblotch viroid - evidence for a symmetrical pathway with 2 rolling circles and hammerhead ribozyme processing. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:12813–12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hutchins C.J., Rathjen P.D., Forster A.C., Symons R.H.. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986; 14:3627–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mühlbach H.P., Sänger H.L.. Viroid replication is inhibited by alpha-amanitin. Nature. 1979; 278:185–188. [DOI] [PubMed] [Google Scholar]
- 74. Navarro J.A., Vera A., Flores R.. A chloroplastic RNA polymerase resistant to tagetitoxin is involved in replication of avocado sunblotch viroid. Virology. 2000; 268:218–225. [DOI] [PubMed] [Google Scholar]
- 75. Schindler I., Mühlbach H.P.. Involvement of nuclear DNA-dependent RNA-polymerases in potato spindle tuber viroid replication - a reevaluation. Plant Sci. 1992; 84:221–229. [Google Scholar]
- 76. Englert M., Beier H.. Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res. 2005; 33:388–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Englert M., Latz A., Becker D., Gimple O., Beier H., Akama K.. Plant pre-tRNA splicing enzymes are targeted to multiple cellular compartments. Biochimie. 2007; 89:1351–1365. [DOI] [PubMed] [Google Scholar]
- 78. Nohales M.-Á., Molina-Serrano D., Flores R., Daròs J.A.. Involvement of the chloroplastic isoform of tRNA ligase in the replication of viroids belonging to the family Avsunviroidae. J. Virol. 2012; 86:8269–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Flores R., Gago-Zachert S., Serra P., Sanjuán R., Elena S.F.. Viroids: Survivors from the RNA world. Annu. Rev. Microbiol. 2014; 68:395–414. [DOI] [PubMed] [Google Scholar]
- 80. Buzayan J.M., Gerlach W.L., Bruening G.. Non-enzymatic cleavage and ligation of RNAs complementary to a plant-virus satellite RNA. Nature. 1986; 323:349–353. [Google Scholar]
- 81. Forster A.C., Symons R.H.. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987; 49:211–220. [DOI] [PubMed] [Google Scholar]
- 82. Fedor M.J. Structure and function of the hairpin ribozyme. J. Mol. Biol. 2000; 297:269–291. [DOI] [PubMed] [Google Scholar]
- 83. Flores R., Hernandez C., de la Peña M., Vera A., Daròs J.A.. Hammerhead ribozyme structure and function in plant RNA replication. Ribonucleases. 2001; 341:540–552. [DOI] [PubMed] [Google Scholar]
- 84. Song S.I., Silver S.L., Aulik M.A., Rasochova L., Mohan B.R., Miller W.A.. Satellite cereal yellow dwarf virus-RPV (satRPV) RNA requires a double hammerhead for self-cleavage and an alternative structure for replication. J. Mol. Biol. 1999; 293:781–793. [DOI] [PubMed] [Google Scholar]
- 85. Makino S., Chang M.F., Shieh C.K., Kamahora T., Vannier D.M., Govindarajan S., Lai M.M.. Molecular cloning and sequencing of a human hepatitis delta (delta) virus RNA. Nature. 1987; 329:343–346. [DOI] [PubMed] [Google Scholar]
- 86. Taylor J., Pelchat M.. Origin of hepatitis delta virus. Future Microbiol. 2010; 5:393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Taylor J.M. Structure and Replication of Hepatitis Delta Virus RNA. Curr. Top Microbiol. Immunol. 2006; 307:1–23. [DOI] [PubMed] [Google Scholar]
- 88. Wang K.S., Choo Q.L., Weiner A.J., Ou J.H., Najarian R.C., Thayer R.M., Mullenbach G.T., Denniston K.J., Gerin J.L., Houghton M.. Structure, sequence and expression of the hepatitis delta (delta) viral genome. Nature. 1986; 323:508–514. [DOI] [PubMed] [Google Scholar]
- 89. Chen P.J., Kalpana G., Goldberg J., Mason W., Werner B., Gerin J., Taylor J.. Structure and replication of the genome of the hepatitis delta virus. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:8774–8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Rizzetto M., Hoyer B., Canese M.G., Shih J.W., Purcell R.H., Gerin J.L.. delta Agent: Association of delta antigen with hepatitis B surface antigen and RNA in serum of delta-infected chimpanzees. Proc. Natl. Acad. Sci. U.S.A. 1980; 77:6124–6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ferre-D’Amare A.R., Zhou K.H., Doudna J.A.. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998; 395:567–574. [DOI] [PubMed] [Google Scholar]
- 92. Kuo M.Y., Sharmeen L., Dinter-Gottlieb G., Taylor J.. Characterization of self-cleaving RNA sequences on the genome and antigenome of human hepatitis delta virus. J. Virol. 1988; 62:4439–4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chadalavada D.M., Cerrone-Szakal A.L., Bevilacqua P.C.. Wild-type is the optimal sequence of the HDV ribozyme under cotranscriptional conditions. RNA. 2007; 13:2189–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Reid C.E., Lazinski D.W.. A host-specific function is required for ligation of a wide variety of ribozyme-processed RNAs. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Saville B.J., Collins R.A.. RNA-mediated ligation of self-cleavage products of a Neurospora mitochondrial plasmid transcript. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:8826–8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Collins R.A. The Neurospora Varkud satellite ribozyme. Biochem. Soc. Trans. 2002; 30:1122–1126. [DOI] [PubMed] [Google Scholar]
- 97. Kennell J.C., Saville B.J., Mohr S., Kuiper M.T., Sabourin J.R., Collins R.A., Lambowitz A.M.. The VS catalytic RNA replicates by reverse transcription as a satellite of a retroplasmid. Genes Dev. 1995; 9:294–303. [DOI] [PubMed] [Google Scholar]
- 98. Lilley D.M. The Varkud satellite ribozyme. RNA. 2004; 10:151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ouellet J., Byrne M., Lilley D.M.J.. Formation of an active site in trans by interaction of two complete Varkud Satellite ribozymes. RNA. 2009; 15:1822–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Craig NL, Craigie R, Gellert M, Lambowitz AM. Mobile DNA II. 2002; American Society of Microbiology. [Google Scholar]
- 101. Dawid I.B., Rebbert M.L.. Nucleotide-sequences at the boundaries between gene and insertion regions in the rDNA of Drosophila melanogaster. Nucleic Acids Res. 1981; 9:5011–5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Roiha H., Miller J.R., Woods L.C., Glover D.M.. Arrangements and rearrangements of sequences flanking the 2 types of rDNA insertion in Drosophila melanogaster. Nature. 1981; 290:749–753. [DOI] [PubMed] [Google Scholar]
- 103. Eickbush T.H., Robins B.. Bombyxmori 28S ribosomal genes contain insertion elements similar to the Type I and II elements of Drosophila melanogaster. EMBO J. 1985; 2281–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Fujiwara H., Ogura T., Takada N., Miyajima N., Ishikawa H., Meakawa H.. Introns and their flanking sequences in Bombyxmori rDNA. Nucleic Acids Res. 1984; 12:6861–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Eickbush T.H., Eickbush D.G.. Integration, regulation, and long-term stability of R2 retrotransposons. Microbiol. Spectrum. 2015; 3:MDNA3-0011-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Bibillo A., Eickbush T.H.. The reverse transcriptase of the R2 non-LTR retrotransposon: Continuous synthesis of cDNA on non-continuous RNA templates. J. Mol. Biol. 2002; 316:459–473. [DOI] [PubMed] [Google Scholar]
- 107. Burke W.D., Calalang C.C., Eickbush T.H.. The site-specific ribosomal insertion element type-II of Bombyxmori (R2Bm) contains the coding sequence for a reverse transcriptase-like enzyme. Mol. Cell Biol. 1987; 7:2221–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kurzynska-Kokorniak A., Jamburuthugoda V.K., Bibillo A., Eickbush T.H.. DNA-directed DNA polymerase and strand displacement activity of the reverse transcriptase encoded by the R2 retrotransposon. J. Mol. Biol. 2007; 374:322–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Yang J., Malik H.S., Eickbush T.H.. Identification of the endonuclease domain encoded by R2 and other site-specific, non-long terminal repeat retrotransposable elements. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:7847–7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Eickbush D.G., Ye J., Zhang X., Burke W.D., Eickbush T.H.. Epigenetic regulation of retrotransposons within the nucleolus of Drosophila. Mol. Cell Biol. 2008; 28:6452–6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Eickbush D.G., Eickbush T.H.. R2 retrotransposons encode a self-cleaving ribozyme for processing from an rRNA cotranscript. Mol. Cell Biol. 2010; 30:3142–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Eickbush D.G., Burke W.D., Eickbush T.H.. Evolution of the R2 retrotransposon ribozyme and its self-cleavage site. PLoS One. 2013; 8:e66441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. George J.A., Eickbush T.H.. Conserved features at the 5 ' end of Drosophila R2 retrotransposable elements: Implications for transcription and translation. Insect Mol. Biol. 1999; 8:3–10. [DOI] [PubMed] [Google Scholar]
- 114. Ruminski D.J., Webb C.H., Riccitelli N.J., Luptak A.. Processing and translation initiation of non-long terminal repeat retrotransposons by hepatitis delta virus (HDV)-like self-cleaving ribozymes. J. Biol. Chem. 2011; 286:41286–41295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kieft J.S. Viral IRES RNA structures and ribosome interactions. Trends Biochem. Sci. 2008; 33:274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Berry K.E., Waghray S., Doudna J.A.. The HCV IRES pseudoknot positions the initiation codon on the 40S ribosomal subunit. RNA. 2010; 16:1559–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Christensen S.M., Ye J., Eickbush T.H.. RNA from the 5 ' end of the R2 retrotransposon controls R2 protein binding to and cleavage of its DNA target site. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:17602–17607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Christensen S.M., Eickbush T.H.. R2 target-primed reverse transcription: Ordered cleavage and polymerization steps by protein subunits asymmetrically bound to the target DNA. Mol. Cell Biol. 2005; 25:6617–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Moss W.N., Eickbush D.G., Lopez M.J., Eickbush T.H., Turner D.H.. The R2 retrotransposon RNA families. RNA Biol. 2011; 8:714–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Eickbush D.G., Eickbush T.H.. R2 and R2/R1 hybrid non-autonomous retrotransposons derived by internal deletions of full-length elements. Mob DNA. 2012; 3:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Bringaud F., Bartholomeu D.C., Blandin G., Delcher A., Baltz T., El-Sayed N.M.A., Ghedin E.. The Trypanosomacruzi L1Tc and NARTc non-LTR retrotransposons show relative site specificity for insertion. Mol. Biol. Evol. 2006; 23:411–420. [DOI] [PubMed] [Google Scholar]
- 122. Sanchez-Luque F.J., Lopez M.C., Macias F., Alonso C., Thomas M.C.. Identification of an hepatitis delta virus-like ribozyme at the mRNA 5′-end of the L1Tc retrotransposon from Trypanosomacruzi. Nucleic Acids Res. 2011; 39:8065–8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Tay W.T., Behere G.T., Batterham P., Heckel D.G.. Generation of microsatellite repeat families by RTE retrotransposons in lepidopteran genomes. BMC Evol. Biol. 2010; 10:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Ferbeyre G., Smith J.M., Cedergren R.. Schistosome satellite DNA encodes active hammerhead ribozymes. Mol. Cell Biol. 1998; 18:3880–3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Thompson P.J., Macfarlan T.S., Lorincz M.C.. Long terminal Repeats: From parasitic elements to building blocks of the transcriptional regulatory repertoire. Mol. Cell. 2016; 62:766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Evgen’ev M.B., Arkhipova I.R.. Penelope-like elements–a new class of retroelements: distribution, function and possible evolutionary significance. Cytogenet. Genome Res. 2005; 110:510–521. [DOI] [PubMed] [Google Scholar]
- 127. Evgen’ev M.B., Zelentsova H., Shostak N., Kozitsina M., Barskyi V., Lankenau D.H., Corces V.G.. Penelope, a new family of transposable elements and its possible role in hybrid dysgenesis in Drosophila virilis. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Arkhipova I.R. Distribution and phylogeny of Penelope-like elements in eukaryotes. Syst. Biol. 2006; 55:875–885. [DOI] [PubMed] [Google Scholar]
- 129. Arkhipova I.R., Pyatkov K.I., Meselson M., Evgen’ev M.B.. Retroelements containing introns in diverse invertebrate taxa. Nat. Genet. 2003; 33:123–124. [DOI] [PubMed] [Google Scholar]
- 130. Dalle Nogare D.E., Clark M.S., Elgar G., Frame I.G., Poulter R.T.. Xena, a full-length basal retroelement from tetraodontid fish. Mol. Biol. Evol. 2002; 19:247–255. [DOI] [PubMed] [Google Scholar]
- 131. Cervera A., de la Peña M.. Eukaryotic penelope-like retroelements encode hammerhead ribozyme motifs. Mol. Biol. Evol. 2014; 31:2941–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Schostak N., Pyatkov K., Zelentsova E., Arkhipova I., Shagin D., Shagina I., Mudrik E., Blintsov A., Clark I., Finnegan D.J. et al.. Molecular dissection of Penelope transposable element regulatory machinery. Nucleic Acids Res. 2008; 36:2522–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Forster A.C., Davies C., Sheldon C.C., Jeffries A.C., Symons R.H.. Self-cleaving viroid and newt RNAs may only be active as dimers. Nature. 1988; 334:265–267. [DOI] [PubMed] [Google Scholar]
- 134. Lünse C.E., Weinberg Z., Breaker R.R.. Numerous small hammerhead ribozyme variants associated with Penelope-like retrotransposons cleave RNA as dimers. RNA Biol. 2016; 14:1499–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Pyatkov K.I., Arkhipova I.R., Malkova N.V., Finnegan D.J., Evgen’ev M.B.. Reverse transcriptase and endonuclease activities encoded by Penelope-like retroelements. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:14719–14724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Laha T., McManus D.P., Loukas A., Brindley P.J.. Sj alpha elements, short interspersed element-like retroposons bearing a hammerhead ribozyme motif from the genome of the Oriental blood fluke Schistosoma japonicum. Biochim. Biophys. Acta-Gene Struct. Expr. 2000; 1492:477–482. [DOI] [PubMed] [Google Scholar]
- 137. Cremisi F., Scarabino D., Carluccio M.A., Salvadori P., Barsacchi G.. A newt ribozyme: a catalytic activity in search of a function. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:1651–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Epstein L.M., Gall J.G.. Self-cleaving transcripts of satellite DNA from the newt. Cell. 1987; 48:535–543. [DOI] [PubMed] [Google Scholar]
- 139. Cervera A., Urbina D., de la Peña M.. Retrozymes are a unique family of non-autonomous retrotransposons with hammerhead ribozymes that propagate in plants through circular RNAs. Genome Biol. 2016; 17:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Hu W.-S., Hughes S.H.. HIV-1 reverse transcription. Cold Spring Harbor Perspect. Med. 2012; 2:a006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Hughes S.H. Reverse transcription of retroviruses and LTR retrotransposons. Microbiol. Spectrum. 2015; 3:MDNA3-0027-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Garcia-Robles I., Sanchez-Navarro J., de la Peña M.. Intronic hammerhead ribozymes in mRNA biogenesis. Biol. Chem. 2012; 393:1317–1326. [DOI] [PubMed] [Google Scholar]
- 143. Martick M., Horan L.H., Noller H.F., Scott W.G.. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature. 2008; 454:899–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Webb C.-H.T., Lupták A.. Kinetic parameters of trans scission by extended HDV-like ribozymes and the prospect for the discovery of genomic trans-Cleaving RNAs. Biochemistry. 2018; 57:1440–1450. [DOI] [PubMed] [Google Scholar]
- 145. Barrick J.E., Breaker R.R.. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007; 8:R239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. McCown P.J., Roth A., Breaker R.R.. An expanded collection and refined consensus model of glmS ribozymes. RNA. 2011; 17:728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Passalacqua L.F.M., Jimenez R.M., Fong J.Y., Lupták A.. Allosteric modulation of the faecalibacterium prausnitzii hepatitis delta Virus-like ribozyme by glucosamine 6-Phosphate: The substrate of the adjacent gene product. Biochemistry. 2017; 56:6006–6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Felletti M., Bieber A., Hartig J.S.. The 3′-untranslated region of mRNAs as a site for ribozyme cleavage-dependent processing and control in bacteria. RNA Biol. 2017; 14:1522–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Hammann C., Luptak A., Perreault J., de la Peña M.. The ubiquitous hammerhead ribozyme. RNA. 2012; 18:871–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Marquez S.M., Chen J.L., Evans D., Pace N.R.. Structure and function of eukaryotic Ribonuclease P RNA. Mol. Cell. 2006; 24:445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Aizawa Y., Xiang Q., Lambowitz A.M., Pyle A.M.. The pathway for DNA recognition and RNA integration by a group II intron retrotransposon. Mol. Cell. 2003; 11:795–805. [DOI] [PubMed] [Google Scholar]
- 152. Bertrand E.L., Rossi J.J.. Facilitation of hammerhead ribozyme catalysis by the nucleocapsid protein of HIV-1 and the heterogeneous nuclear ribonucleoprotein A1. EMBO J. 1994; 13:2904–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Daròs J.A., Flores R.. A chloroplast protein binds a viroid RNA in vivo and facilitates its hammerhead-mediated self-cleavage. EMBO J. 2002; 21:749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Denti M.A., Martínez de Alba A.E., Sagesser R., Tsagris M., Tabler M.. A novel RNA-binding protein from Triturus carnifex identified by RNA-ligand screening with the newt hammerhead ribozyme. Nucleic Acids Res. 2000; 28:1045–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Luzi E., Eckstein F., Barsacchi G.. The newt ribozyme is part of a riboprotein complex. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:9711–9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Tsuchihashi Z., Khosla M., Herschlag D.. Protein enhancement of hammerhead ribozyme catalysis. Science. 1993; 262:99–102. [DOI] [PubMed] [Google Scholar]
- 157. Sioud M., Jespersen L.. Enhancement of hammerhead ribozyme catalysis by glyceraldehyde-3-phosphate dehydrogenase. J. Mol. Biol. 1996; 257:775–789. [DOI] [PubMed] [Google Scholar]
- 158. Herschlag D., Khosla M., Tsuchihashi Z., Karpel R.L.. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 1994; 13:2913–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Urdaneta E.C., Vieira-Vieira C.H., Hick T., Wessels H.H., Figini D., Moschall R., Medenbach J., Ohler U., Granneman S., Selbach M. et al.. Purification of cross-linked RNA-protein complexes by phenol-toluol extraction. Nat. Commun. 2019; 10:990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Trendel J., Schwarzl T., Horos R., Prakash A., Bateman A., Hentze M.W., Krijgsveld J.. The human RNA-Binding proteome and its dynamics during translational arrest. Cell. 2019; 176:391–403. [DOI] [PubMed] [Google Scholar]
- 161. Huang R., Han M., Meng L., Chen X.. Transcriptome-wide discovery of coding and noncoding RNA-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E3879–E3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Bao X., Guo X., Yin M., Tariq M., Lai Y., Kanwal S., Zhou J., Li N., Lv Y., Pulido-Quetglas C. et al.. Capturing the interactome of newly transcribed RNA. Nat. Methods. 2018; 15:213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Hammann C., Westhof E.. Searching genomes for ribozymes and riboswitches. Genome Biol. 2007; 8:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Butcher S.E., Burke J.M.. Structure-mapping of the hairpin ribozyme - Magnesium-dependent folding and evidence for tertiary interactions within the ribozyme-substrate complex. J. Mol. Biol. 1994; 244:52–63. [DOI] [PubMed] [Google Scholar]
- 165. Shih I.H., Been M.D.. Catalytic strategies of the hepatitis delta virus ribozymes. Annu. Rev. Biochem. 2002; 71:887–917. [DOI] [PubMed] [Google Scholar]
- 166. Suslov N.B., DasGupta S., Huang H., Fuller J.R., Lilley D.M.J., Rice P.A., Piccirilli J.A.. Crystal structure of the Varkud satellite ribozyme. Nat. Chem. Biol. 2015; 11:840–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Klein D.J., Ferre-D’Amare A.R.. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006; 313:1752–1756. [DOI] [PubMed] [Google Scholar]
- 168. Weinberg Z., Breaker R.R.. R2R–software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics. 2011; 12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]