Abstract
Although antisense oligonucleotides (ASOs) are well tolerated preclinically and in the clinic, some sequences of ASOs can trigger an inflammatory response leading to B cell and macrophage activation in rodents. This prompted our investigation into the contribution of genetic architecture to the ASO-mediated inflammatory response. Genome-wide association (GWA) and transcriptomic analysis in a hybrid mouse diversity panel (HMDP) were used to identify and validate novel genes involved in the acute and delayed inflammatory response to a single 75 mg/kg dose of an inflammatory 2′-O-methoxyethyl (2′MOE) modified ASO. The acute response was measured 6 h after ASO administration, via evaluation for increased plasma production of interleukin 6 (IL6), IL10, monocyte chemoattractant protein 1 (MCP-1) and macrophage inflammatory protein-1β (MIP-1β). Delayed inflammation was evaluated by spleen weight increases after 96 h. We identified single nucleotide polymorphisms (SNPs) on chromosomes 16 and 17 associated with plasma MIP-1β, IL6, and MCP-1 levels, and one on chromosome 8 associated with increases in spleen weight. Systems genetic analysis utilizing transcriptomic data from HMDP strain macrophages determined that the acute inflammatory SNPs were expression quantitative trait locis (eQTLs) for CCAAT/enhancer-binding protein beta (Cebpb) and salt inducible kinase 1 (Sik1). The delayed inflammatory SNP was an eQTL for Rho guanine nucleotide exchange factor 10 (Arhgef10). In vitro assays in mouse primary cells and human cell lines have confirmed the HMDP finding that lower Sik1 expression increases the acute inflammatory response. Our results demonstrate the utility of using mouse GWA study (GWAS) and the HMDP for detecting genes modulating the inflammatory response to pro-inflammatory ASOs in a pharmacological setting.
Keywords: inflammation, GWAS, oligonucleotide, eQTL
Introduction
Antisense oligonucleotides (ASOs) are a well-recognized, potent therapeutic platform used in the clinic to treat a variety of human conditions, including cardiometabolic, infectious, inflammatory, and neurological disease [1–7]. By utilizing short synthetic (12–24mer) chemically modified DNA-like oligonucleotides to bind complimentary mRNA sequences, ASOs are able to modulate the degradation, splicing, and regulation of mRNA in a highly selective, sequence-specific manner [8]. In the most commonly applied mechanism, an ASO binds complementary mRNA and induces RNAseH1-mediated degradation, preventing translation and thus reducing the overall quantity of the target protein [9]. Iterative platform modifications throughout the past 30 years have increased stability, potency, specificity, and reduced toxicity. In particular, the modification of the phosphodiester backbone to 2′-O-methoxyethyl (2′MOE) has not only significantly increased stability and potency but also reduced ASO-mediated inflammation [10–15].
The past several decades have seen rapid expansion in our understanding of the cell surface and endosome-associated toll-like receptors (TLRs), of which TLR 3, 7, 8, and 9 recognize nucleic acids [16–19]. Early structure–activity relationship studies determined that unmethylated cytosine-guanine (CpG) motifs, common in bacteria and DNA viruses, have reduced frequency and increased methylation in vertebrates, thus acting as a ligand for TLR9 [20,21]. Activation of TLR9 in the endosome signals via myeloid differentiation of primary response genes 88 (Myd88), triggering the activation of mitogen-activated protein kinase (MAPK) family members and nuclear factor kappa-light-chain-enhances of activated B cells (NF-κB) signaling pathways [19,21,22]. In addition, receptor for advance glycation end product (RAGE) has been shown to promote inflammatory responses on interaction with CpG-containing ligands at the cell surface, provoking endosomal uptake and further contact with TLR9, leading to enhanced NF-κB signaling [23].
Previous work has demonstrated that non-CpG containing 2′MOE ASOs are able to activate splenic B cells and macrophage populations, and this response is dependent on MyD88 and TLR9 function [24]. Further, the RAGE receptor is involved in mediating the inflammatory response to systemic administration of non-CpG containing sequences [23]. Aside from TLR9-mediated stimulation, ASOs are able to initiate TLR9 and Myd88 independent proinflammatory responses as well [15,25].
Therefore, although ASOs are widely accepted, effective therapeutic agents and are generally well tolerated in the clinic, a small number of sequences are capable of triggering an inflammatory response, resulting in B cell and macrophage activation in rodents in vivo [15,24]. These sequences are not utilized as clinical candidates. However, an understanding of the mechanism of the proinflammatory effects of these sequences is critical to the development of future clinical candidates. To date, there has been no in vivo systematic interrogation of the role of genetic contribution in the ASO-mediated inflammatory response.
The Hybrid Mouse Diversity Panel (HMDP) is a powerful tool for carrying out the investigation of complex traits in vivo. It consists of 30 classical inbred mouse strains, in addition to more than 70 recombinant inbred strains, resulting in a panel of more than 100 genetically unique inbred mouse strains. These recombinant inbred strains were derived from intercrosses of eight “founder strains,” allowing important insights into genetically derived phenotypic differences among mice. All of the mice in the HMDP are genotyped at 140,000 high-quality single nucleotide polymorphisms (SNPs), with sufficient power to detect traits contributing to 10% of overall phenotypic variance [26–28]. Using the sequenced strains of the HMDP, we can carry out a mouse genome-wide association study (GWAS) to detect SNPs associated with causal variants, contributing to a complex phenotype in a hypothesis-free manner. GWAS generally has been used to identify hundreds of genes contributing to human pathophysiology, including cancer, cardiovascular, metabolic, inflammatory, and neurological diseases [29–32]. Using mouse models in genetic association studies, though not exactly replicating human genetics, provides multiple advantages over human populations, including cost effectiveness, reproducibility of results, and reduced impact of environmental factors.
To better understand the genomic variants that are associated with ASO-mediated inflammation, we employed GWAS and transcriptomic analysis of the HMDP by using an inflammatory ASO (ION 421856) that has previously been shown to induce acute and delayed inflammation in vivo [24]. The acute inflammatory response was measured in plasma collected 6 h after a single dose of 75 mg/kg inflammatory or control ASO. Plasma was evaluated for interleukin 6 (IL6), IL10, MCP-1, and MIP-1β production in comparison to mice dosed with a control, noninflammatory ASO in 100 HMDP strains. Delayed response was measured after 96 h by euthanizing the mice and observing increases in spleen weight. We observed significant variability between the HMDP strains for changes in spleen weight and all four markers of acute inflammation, and all markers positively correlated with each other (Supplementary Fig. S1). These results are summarized in Table 1 and Supplementary Table S1. Using Factored Spectrally Transformed Linear Mixed Model (FaST-LMM), we identified a region on chromosome 8 associated with spleen weight increase. Systems genetic analysis suggested this locus contributed to the expression of Rho guanine nucleotide exchange factor 10 (Arhgef10). In addition, we identified a locus on chromosome 16 associated with increased MIP-1β plasma levels, and 2 loci on chromosome 17 associated with IL6 and MCP-1 plasma levels. CCAAT/enhancer-binding protein beta (Cebpb) was identified as a gene likely to be regulated in trans by the region on chromosome 16, whereas both chromosome 17 loci likely regulated salt inducible kinase 1 (Sik1). Additional in vitro studies confirmed that Sik1 contributes to the ASO-mediated acute inflammatory response. Our results demonstrated that genetic variation impacts the in vivo inflammatory response to ASOs and validates our use of the HMDP for toxicological evaluation of antisense drugs.
Table 1.
Peak Single Nucleotide Polymorphisms Identified by Genome-Wide Association Study for the Indicated Trait in the Hybrid Mouse Diversity Panel Strains After Inflammatory Antisense Oligonucleotide Administration
| Trait | Chromosome | Peak SNP | Position | P | LD block | Genes within LD block |
|---|---|---|---|---|---|---|
| Spleen weight | 8 | rs13479604 | 9757872 | 7.64E-07 | 9,530,463–11,385,428 | 7 |
| MIP-1β | 16 | rs48685261 | 73451453 | 2.06E-06 | 73,258,917–74,084,707 | 1 |
| IL6 | 17 | rs33060525 | 35595846 | 7.80E-07 | 35,561,965–35,615,192 | 1 |
| MCP-1 | 17 | rs33441142 | 34175992 | 9.69E-07 | 34,187,385–34,179,669 | 0 |
IL, interleukin; LD, linkage disequilibrium; MCP-1, monocyte chemoattractant protein 1; MIP-1β, macrophage inflammatory protein-1β; SNP, single nucleotide polymorphism.
Materials and Methods
Antisense oligonucleotides
2′MOE ASOs were synthesized at Ionis Pharmaceuticals (Carlsbad, CA) as previously described [33]. The inflammatory ASO (ION 421856) and control (ION 141923) ASOs were formulated in saline and were delivered via subcutaneous injection into the mice.
Mice
An institutional animal care and use committee approved all procedures and protocols for the mouse pharmacology studies. Mice were obtained from The Jackson Laboratory and housed at Ionis Pharmaceuticals, maintained on a chow diet, and entered into studies before they exceeded 7 weeks of age. At 6 h postdose, mice were anesthetized with a controlled flow of isoflurane/oxygen mixture and whole blood (∼100 μL) was collected via orbital bleed with heparinized catheters and EDTA tubes. At 96 h postdose, mice were humanely anesthetized with isoflurane and euthanized by cervical dislocation. Whole blood was immediately collected via cardiac puncture with a 1 cc syringe with a 23G needle. Whole blood was centrifuged in EDTA tubes at 5,000 RPM for 10 min, and plasma was transferred to a 96-well plate and stored −20°C. At necropsy, only spleens and livers were harvested and collected. Spleen and liver samples were separated into several Eppendorf tubes, immediately snap frozen in liquid nitrogen, and stored at −80°C.
Cell lines and reagents
Mouse splenocytes were harvested from naive 7-week-old male C57BL/6 mice, which were anesthetized and euthanized via cervical dislocation. Spleens were pushed through a 0.4 μM cell strainer in 5 mL cold PBS, then slowly pipetted onto 5 mL Lympholyte-M Cell Separation Media (Cedarlane Labs). This mixture was spun at 1,500g for 20 min, and the splenocyte layer was harvested and centrifuged at 800g for 10 min. The splenocytes were resuspended in RPMI1 supplemented with 10% FBS and 1% penstrep. Live cells were counted by using a TC20 Automated Cell Counter (Biorad) after mixing with Trypsin Blue (Fisher Scientific). Cells were plated at 50,000 cells/well in 96-well round-bottom plates. Inflammatory and control ASOs, lipopolysaccharide (LPS), and HG-9-91-01 were added to the media at the indicated concentrations.
Cytokine/chemokine analysis
Plasma was collected at 6 and 96 h postdose, stored in 96-well plates, and placed at −20°C until use. The main analytes of interest were IL6, IL10, MCP-1, and MIP-1β. MesoScale Discovery developed a custom 4-plex cytokine and chemokine mouse plate. Assay plates were prepared as per the manufacturer's instructions. Twenty-five microliter of plasma per sample was added to 25 μL Meso Scale Discovery (MSD) buffer, and the plates were placed on a shaker plate for 2 h. Each plate was washed three times between each step. Antibody conjugations were prepared per instructions, and the plate was developed and read on a MSD QuickPlex SQ 120. Conjugate levels were normalized to a scale solution on each plate.
RNA analysis
Total mRNA was isolated by using a QIAGEN RNAeasy kit (QIAGEN, Valencia, CA). Target mRNA expression was determined by quantitative polymerase chain reaction (qPCR) using StepOne RT-PCR machines (Applied Biosystems), as previously described. Relative levels of Sik1 were normalized to cyclophilin.
Association analysis
We identified genetic associations by using the FaST-LMM, which is a reformulated mixed-model analysis that performs linearly in run time and memory footprint for GWAS in very large datasets. We retrieved the genotypes from the University of California, Los Angeles (UCLA) systems genetics database along with chromosomal locations of linkage disequilibrium (LD) blocks. The global gene expression microarrays that were generated from microarrays of chow-fed male mice from 95 HMDP strains were obtained from the UCLA systems genetics database. These data were used to perform the cis and trans-analysis, as previously described. FaST-LMM on ASO accumulation and activity properties was performed with a genome-wide significance threshold of 4.1 × 10−6 as described.
Statistics
Data are reported as means ± S.E.M. The statistical tests and significance are described in the figure legends.
Results
Systems genetics analysis of delayed inflammation-associated SNPs
To identify regions associated with delayed ASO-mediated inflammation, 6-week-old male mice from the HMDP strains were administered one 75 mg/kg dose of either the 2′MOE inflammatory ASO (ION 421856) or the noninflammatory control (ION 141923) ASO (Supplementary Fig. S2). Ninety-six hours after dosing, mice were euthanized and the spleens were removed and weighed (Fig. 1A and Supplementary Table S1).
FIG. 1.
Systems genetic analysis identifies Arhgef10 as a cis-eQTL for increased spleen weight after inflammatory ASO administration. (A) Six-week-old male mice from 100 HMDP strains (n = 4/strain/condition) were dosed with a single 7.5 mg/kg dose of either control ASO (ION 421923) or inflammatory ASO (ION 421856). Spleens were harvested after 96 h, weighed, and recorded as mean percent increase. Results are presented as mean ± S.E.M. (Supplementary Table S1) (B) Manhattan plot showing the −log10 of the association P values [−log(P)] for increased spleen weight in the HMDP strains. Each chromosome is plotted on the x-axis in alternating light and dark colors. Genome-wide significance threshold line is shown in gray (−logP ≤ 5.39). Arrow indicates the location of the significant peak. (C) Distribution of percent spleen weight increase based on the genotype of the associated SNP on chromosome 8. Mean ± S.E.M. Two tailed t-test, P ≤ 0.0096 (D) Distribution of Arhgef10 expression by microarray, relative to C57Bl6/J expression, based on the genotype of the associated SNP on chromosome 8. Mean ± S.E.M. Two-tailed t-test, P ≤ 0.0024. (E) Percent increase in the spleen weight after inflammatory ASO administration was correlated with Arhgef10 expression, with the increase significantly lower in strains with high expression by microarray. Mean ± S.E.M. Two-tailed t-test, P ≤ 0.0008. Arhgef10, Rho guanine nucleotide exchange factor 10; ASO, antisense oligonucleotide; eQTL, expression quantitative trait loci; HMDP, hybrid mouse diversity panel; MCP-1, monocyte chemoattractant protein-1; MIP-1β, macrophage inflammatory protein-1β; SNP, single nucleotide polymorphism.
The association analysis was accomplished via FaST-LMM, where adjusted association P values were calculated for 108,064 SNPs with a minor allele frequency of >5% (P < 0.05, the genome-wide equivalent for GWA using FaST-LMM in the HMDP is P ≤ 4.1 × 10−6, −log10P ≤ 5.39). At this threshold, a genome-wide significant locus associated with variation in increased spleen weight was identified on chromosome 8 (rs13479604, P ≤ 7.63 × 10−7) (Fig. 1B, C). This locus contained seven genes within the LD block, four of which are expressed in the spleen (Table 2).
Table 2.
Genes Identified as Located Within the Linkage Disequilibrium Block for Peak Single Nucleotide Polymorphism rs13479604, Associated with Increases in Spleen Weight
| Gene symbol | Gene name | Localization |
|---|---|---|
| Lig4 | Ligase IV, DNA, ATP-dependent | Splenic |
| Irs2 | Insulin receptor substrate 2 | Splenic |
| Col4a1 | Collagen, type IV, alpha 1 | Splenic |
| Col4a2 | Collagen, type IV, alpha 2 | Splenic |
| Fam155a | Family with sequence similarity 155, member A | Nonsplenic |
| Myo16 | Myosin XVI | Nonsplenic |
| B930025P03Rik | RIKEN cDNA B930025P03 gene | Nonsplenic |
Most SNPs sequenced in the HMDP occur in noncoding regions, making identification of a causal coding mutation unlikely. Therefore, it is important to identify genes within the LD block that also demonstrate expression regulated by the identified SNPs. Isolated macrophages from chow-fed male mice were subjected to global gene expression microarray analysis [34]. These data were used in combination with the SNP data to generate a list of expression quantitative trait loci (eQTL), leading to the identification of cis- (within 1 Mb of the peak SNP) and trans- (farther than 1 Mb from the peak SNP) regulated genes corresponding to the identified SNP at chromosome 8. Arhgef10 was found to be regulated in cis by this SNP (Supplementary Table S2). This gene has been reported to play a role in inflammation, oxidative stress, thrombus formation, and fibrosis [35]. Further analysis demonstrated that Arhgef10 expression varied significantly depending on the genotype at SNP rs13479604 (Fig. 1D). Importantly, the basal level of macrophage Arhgef10 expression across the HMDP was strongly correlated with the increase in spleen weight seen after ASO administration, as mice with higher levels of basal macrophage Arhgef10 expression demonstrated significantly lower increases in spleen weight (P ≤ 0.0008) (Fig. 1E and Supplementary Fig. S3).
Systems genetics analysis of acute inflammation-associated SNPs
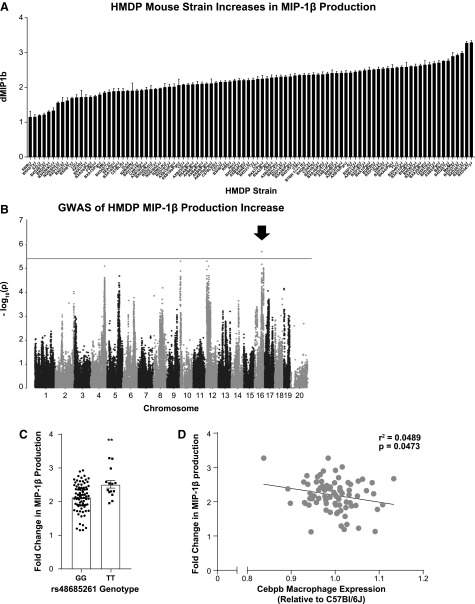
To identify regions associated with acute ASO-mediated inflammation, 6-week-old male mice from the HMDP strains were administered one 75 mg/kg dose of either the 2′MOE inflammatory ASO (ION 421856) or the noninflammatory control (ION 141923) ASO. As previous work has identified peak cytokine production 6 h postdose [24], plasma was isolated and evaluated for IL6, IL10, MCP-1, and MIP-1β concentrations at this time-point (Supplementary Table S1). A wide spectrum in plasma levels for these cytokines was observed. FaST-LMM analysis identified one peak SNP associated with changes in MIP-1β plasma levels in response to ASO administration on chromosome 16 (rs4868521, P ≤ 2.055 × 10−6) (Fig. 2B, C). One gene, Robo2, was located in the LD block for this locus (Table 1).
FIG. 2.
Systems genetic analysis identifies Cebpb as a trans-eQTL for MIP-1β production after inflammatory ASO administration. (A) Plasma MIP-1β after control and inflammatory ASO exposure are presented as mean fold increase+S.E.M. for all strains tested (Supplementary Table S1). (B) Manhattan plot showing the −log(10) of the association P values [−log(P)] for fold increase in MIP-1β expression in the HMDP strains. Each chromosome is plotted on the x-axis in alternating light and dark colors. Genome-wide significance threshold is shown in gray [−log(P) = 5.39]. Arrow indicates the location of the significant peak. (C) Distribution of the fold change in MIP-1β production based on the genotype of the associated SNP on chromosome 16. Mean ± S.E.M. Two tailed t-test, P ≤ 0.0021. (D) Fold change in MIP-1β production after inflammatory ASO administration was correlated with Cebpb expression measured by microarray relative to C57Bl6J. Linear regression, P ≤ 0.0473. Cebpb, CCAAT/enhancer-binding protein beta.
As described for the delayed inflammatory phenotype, we performed transcriptomic (eQTL) analysis by using macrophage microarray expression data, accessible on NCBI GEO GSE97207. This effort produced a list of genes that were regulated in cis and trans by the peak region on chromosome 16. Systems genetics analysis utilizing the macrophage microarray expression data identified 3 genes potentially regulated in cis by this peak, and 13 regulated in trans (Supplementary Tables S2 and S3). Unusually none of the genes identified in cis was found to correlate with the MIP-1β response phenotype. However, macrophage expression Cebpb, regulated in trans, was found to correlate with the increase in MIP-1β production after administration of the inflammatory ASO (Fig. 2D). Its inclusion in this screen is not surprising as Cebpb is a transcription factor expressed in macrophages and has long been known to be responsible for the expression of pro-inflammatory response genes [36].
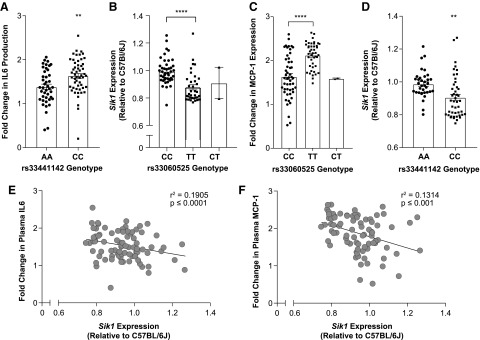
The IL6 and MCP-1 plasma responses to the inflammatory ASO most closely overlapped out of all the measured endpoints in this study (Supplementary Fig. S1). FaST-LMM analysis of both IL6 and MCP-1 responses identified overlapping peaks on chromosome 17 (Fig. 3), although each had a separate peak SNP (rs33060525, P ≤ 7.8 × 10−7 and rs33441142, P ≤ 9.69 × 10−7) (Figs. 3D, E, and 4A, B). The rs33060525 locus contained one gene in the LD block, 2300002M23Rik, which has a putative function in extracellular matrix. The rs33441142 loci contained no coding regions within the LD block (Table 1). eQTL analysis performed on these peak SNPs utilizing the macrophage expression microarray data determined that Sik1 was strongly regulated in cis by both SNPs identified in chromosome 17 (Fig. 4C, D and Supplementary Tables S4, S5, S6, S7, S8, S9). Importantly, the peak SNP genotypes strongly correlate with the level of basal macrophage Sik1 expression. Further, basal Sik1 expression is significantly negatively correlated with the level of increase seen in plasma IL6 and MCP-1 (Fig. 4E, F).
FIG. 3.
IL6 and MCP-1 production after inflammatory ASO administration are closely correlated. (A). Plasma IL6 after control and inflammatory ASO exposure are presented as mean fold increase+S.E.M. for all strains tested (Supplementary Table S1). (B) As with IL6, plasma was analyzed by MSD plate ELISA for MCP-1 production and recorded as fold increase (Supplementary Table S1). (C) IL6 and MCP-1-fold increase were the most closely correlated endpoints out of all inflammatory endpoints in this study. Linear regression, P < 0.0001. (D) Manhattan plot showing the −log(10) of the association P values [−log(P)] for fold increase in IL6 expression in the HMDP strains. Each chromosome is plotted on the x-axis in alternating light and dark colors. Genome-wide significance threshold is shown in gray [−log(P) = 5.39]. Arrow indicates the location of the significant peak. (E) As with IL6, Manhattan plot for MCP-1 production following the same parameters. IL, interleukin; MSD, Meso Scale Discovery.
FIG. 4.
Systems genetic analysis identifies Sik1 as a cis-eQTL for IL6 and MCP-1 production after inflammatory ASO administration. (A) Distribution of the fold change in IL6 production based on the genotype of the associated SNP on chromosome 17. Mean ± S.E.M. Two-tailed t-test, P ≤ 0.0016. (B) Distribution of Sik1 expression by microarray relative to C57Bl/6J expression based on the genotype of the associated SNP on chromosome 17. Mean ± S.E.M. Two-tailed t-test, P ≤ 0.0032. (C) Distribution of the fold change in MCP-1 production based on the genotype of the associated SNP on chromosome 17. Mean ± S.E.M. Two-tailed t-test, P < 0.0001. (D) Distribution of Sik1 expression by microarray relative to C57Bl/6J based on the genotype of the associated SNP on chromosome 17. Mean ± S.E.M. Two-tailed t-test, P < 0.0001. (E) Fold change in IL6 production after inflammatory ASO administration was correlated with Sik1 expression measured by microarray relative to C57Bl/6J. Linear regression, P < 0.0001. (F) Fold change in MCP-1 production after inflammatory ASO administration was correlated with Sik1 expression measured by microarray relative to C57Bl/6J. Linear regression, P ≤ 0.0009. Sik1, salt inducible kinase 1.
Validation of Sik1's role in ASO-associated acute inflammation
Since Sik1 plays a key role in the acute inflammatory response, we investigated its putative role in ASO-associated cytokine and chemokine release. Systems analysis of the peak SNPs rs33060525 and rs33441142 demonstrated a significant difference in plasma MCP-1 and MIP-1β levels based on the genotype distribution at those SNPs. Further, our analysis determined that HMDP strains with lower Sik1 expression had significantly higher IL6 and MCP-1 plasma levels after ASO administration.
To experimentally validate the role of Sik1, we utilized the pan-SIK inhibitor HG-9-91-01 in mouse primary splenocytes isolated from healthy C57Bl6/J mice [37]. These splenocytes were pretreated with HG-9-91-01 for 3 h and subsequently exposed to either a control ASO, an inflammatory ASO, or LPS for 24 h. MSD analysis of the cell culture media demonstrated that splenocytes treated with HG-9-91-01 displayed a significantly increased release of MCP-1 and IL6, but not IL10 or MIP-1β (Fig. 5). Further, this correlation between Sik1 inhibition and increased IL6 and MCP-1 release was seen across multiple strains of mice, validating Sik1 as an important mediator of ASO-associated acute inflammation (Supplementary Fig. S4).
FIG. 5.
Validation of the role of Sik1 in acute inflammation in response to inflammatory ASO administration. Splenocytes were harvested from 7-week-old male mice as described. They were pretreated with either HG-9-91-01 or DMSO for 3 h; then, they were treated with either control ASO (ION 421923), an inflammatory ASO (ION 421856). or 4 μg LPS. Significant increases in production of IL6, MCP-1, and MIP-1β were observed in response to the inflammatory ASO when pretreated with the SIK inhibitor. LPS, lipopolysaccharide.
Discussion
Innovations in medicinal chemistry have drastically improved ASO activity and target tissue specificity, allowing increasingly lower doses of drug to be administered in the clinic. Although antisense therapeutics are well tolerated in the clinic, ongoing research efforts seek to identify factors that allow some sequences to induce a proinflammatory response in the murine models. Although these problematic sequences are quickly identified and removed well before reaching the clinic, more efforts are needed to better understand the mechanism of this response.
Here, we investigate the role of genetics in modulating the response to an inflammatory ASO sequence in vivo. Our data show that, after a single dose of a proinflammatory 2′MOE ASO, the plasma levels of IL6, IL10, MCP-1, and MIP-1β vary significantly among the 100 genetically unique HMDP strains. In a delayed response, spleen weights also vary significantly between the mice. The variability in these parameters allowed us to perform GWAS and led to the identification of inflammatory ASO-associated eQTLs Arhgef10, associated with increased spleen weight, Cebpb, associated with increased MIP-1β, and Sik1, associated with production of IL6 and MCP-1. No significant peaks were seen in the GWAS for IL10 production (Supplementary Fig. S5). These results are particularly noteworthy as Cebpb and Sik1 are known to be involved with TLR signaling, which has been previously shown to be crucial for the acute ASO-mediated inflammatory process.
Systems genetics analysis identified Cepbp as a trans-eQTL, concordant with the peak SNP identified in GWAS analysis of MIP-1β increase. Further analysis showed that its expression correlated with MIP-1β plasma levels after inflammatory ASO administration. Cebpb, a member of the CCAAT/enhancer-binding protein family, is a transcription factor strongly regulated by the TLRs and NF-κB pathway and is known to mediate the expression of a variety of inflammatory factors, including MIP-1β [36,38]. The gene encoding MIP-1β protein contains a Cebpb regulatory motif in the promoter [39]. The identification of Cebpb is not surprising in a study examining factors involved in inflammation and its inclusion serves as an important control validating the use of the HMDP.
Systems genetics analysis identified Sik1 as a gene with expression concordant with the identified SNPs rs33060525 and rs33441142. Further analysis showed that increasing levels of Sik1 expression among the HMDP mice led to decreased production of IL6 and MCP-1 in plasma after inflammatory ASO administration. The genetic data were further confirmed in vivo by the use of the SIK inhibitor HG-9-91-01 in mouse primary splenocytes, which showed increased IL6 and MCP-1 production when treated with an inflammatory ASO in conjunction with the SIK inhibitor.
Sik1 is a member of a family of related serine-threonine kinases that have been implicated in controlling liver glucose homeostasis, hepatic lipogenesis, steroidogenesis, adipogenesis, and TLR-mediated inflammation [40–43]. Canonically, the SIKs control the phosphorylation and subcellular localization of class IIa histone deacetylases (HDACs) and cAMP-regulated transcriptional coactivators (CRTCs) [44,45]. Notably, Sik1 expression has been found to inhibit NF-κB activation in response to TLR signaling, resulting in decreased expression and production of proinflammatory cytokines both in vitro and in vivo [43,46], and Sik3 has been shown to negatively regulate the production of IL6, nitric oxide, and IL12p40 in vivo [47]. Our data are in line with these published reports, confirming the role of Sik1 as a mediator of ASO-induced NF-κB activation.
Arhgef10 was identified as a cis-eQTL regulated by a peak SNP strongly associated with increases in spleen weight. Systems genetics analysis of this gene showed that its expression was further correlated with the delayed inflammatory phenotype. SNPs in Arhgef10 have been linked with the incidence of thrombotic stroke in humans; however, the function of this gene is not well understood [35]. It is a member of the family of guanidine nucleotide exchange factors, which catalyze the exchange of bound GDP by GTP, thereby regulating the activity of small Rho-GTPases [48]. The best-known function of Arhgef10 is to regulate the activity of RhoA kinase (ROCK); Arhgef10 upregulation has been found to increase the activity of ROCK in vivo [35]. Importantly, Rock2 has been found by our group to be involved in the activity of ASOs in vivo, suggesting that Arhgef10 is a possible link between ASO activity and inflammation [49]. Further, the RhoA-Rho kinase pathway has been shown to regulate a wide variety of functions, including endothelial dysfunction, inflammation, and apoptosis, and can be induced by LPS exposure [50–52]. Specifically, it has been demonstrated that ROCK1 negatively regulates splenic erythropoiesis via regulation of P53, important in the development of the delayed inflammatory phenotype in response to ASO [53].
Surprisingly, although TLR9 is a complex pathway, other intermediaries of its downstream activation were not identified. Closer analysis of HMDP macrophage expression found that TLR9, similar to many, though expressed, did not sufficiently vary in its expression across the HMDP strains (Supplementary Fig. S6), making identification using the HMDP unlikely.
In summary, genetic factors have been shown for the first time to be capable of altering the ASO-mediated inflammatory response in vivo. Here, we establish the utility of the HMDP as a method for detecting genes modulating the inflammatory response to ASOs in an in vivo setting. Future work will focus on translating these findings to human systems and applying this understanding to more efficiently screen ASOs preclinically. Further studies evaluating these factors will provide us with a better understanding of ASO response, potential effects across patient populations and help design drugs with improved therapeutic benefits.
Supplementary Material
Acknowledgments
Ionis Pharmaceuticals, Inc. approved and sponsored the study design, data collection, analysis, and preparation of this article. Supported authors E.P., P.C., W.F., S.R., J.H., S.A.B., P.N., R.M.C., and R.G.L. The National Institute of Health sponsored data collection. HL28481, HL30568, HL114437, and HL123295 supported authors C.P. and A.J.L. The authors would like to thank the vivarium staff, preclinical toxicity group (Todd Machemer), bioinformatics group, the members of the cardiovascular research group (Amy Wei, Steve Yeh, PhD, Adam Mullick, PhD, Stan Riney), and Tracey Riegle who have provided crucial help with animal housing, experimental techniques, and discussions during preparation of this article.
Declarations
Ethics Statement
Ionis is AAALAC accredited and follows the 8th Ed. Of the Guide for the Care and Use of Laboratory Animals and the 2013 AVMA guidelines for the euthanasia of animals. All animals in this study were anesthetized with Isoflurane and euthanized via cervical dislocation. The Ionis IACUC-approved protocol is No. P-0225. This protocol was approved on May 28, 2014.
Availability of data and material
All macrophage microarray expression data are available at NCBI GEO GSE97207 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97207).
Author Disclosure Statement
E.P., P.C., W.F., S.R., J.H., S.A.B., P.N., R.M.C., and R.G.L. were employees at Ionis Pharmaceuticals when these studies were completed.
Supplementary Material
References
- 1. Raal FJ, Santos RD, Blom DJ, Marais AD, Charng MJ, Cromwell WC, Lachmann RH, Gaudet D, Tan JL, et al. (2010). Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: a randomised, double-blind, placebo-controlled trial. Lancet 375:998–1006 [DOI] [PubMed] [Google Scholar]
- 2. Jaschinski F, Rothhammer T, Jachimczak P, Seitz C, Schneider A. and Schlingensiepen KH. (2011). The antisense oligonucleotide trabedersen (AP 12009) for the targeted inhibition of TGF-beta2. Curr Pharm Biotechnol 12:2203–2213 [DOI] [PubMed] [Google Scholar]
- 3. Al-Asaaed S. and Winquist E. (2013). Custirsen (OGX-011): clusterin inhibitor in metastatic prostate cancer. Curr Oncol Rep 15:113–118 [DOI] [PubMed] [Google Scholar]
- 4. Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, et al. (2013). Treatment of HCV infection by targeting microRNA. N Engl J Med 368:1685–1694 [DOI] [PubMed] [Google Scholar]
- 5. Jones NR, Pegues MA, McCrory MA, Singleton W, Bethune C, Baker BF, Norris DA, Crooke RM, Graham MJ. and Szalai. AJ. (2012). A selective inhibitor of human C-reactive protein translation is efficacious in vitro and in C-reactive protein transgenic mice and humans. Mol Ther Nucleic Acids 1:e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hua Y, Sahashi K, Rigo F, Hung G, Horev G, Bennett CF. and Krainer AR. (2011). Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature 478:123–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kordasiewicz HB, Stanek LM, Wancewicz EV, Mazur C, McAlonis MM, Pytel KA, Artates JW, Weiss A, Cheng SH, et al. (2012). Sustained therapeutic reversal of huntington's disease by transient repression of huntingtin synthesis. Neuron 74:1031–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crooke ST. (2004). Antisense strategies. Curr Mol Med 4:465–487 [DOI] [PubMed] [Google Scholar]
- 9. Lee RG, Crosby J, Baker BF, Graham MJ. and Crooke RM. (2013). Antisense technology: an emerging platform for cardiovascular disease therapeutics. J Cardiovasc Transl Res 6:969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krieg AM. (1996). An innate immune defense mechanism based on the recognition of CpG motifs in microbial DNA. J Lab Clin Med 128:128–133 [DOI] [PubMed] [Google Scholar]
- 11. Monteith DK, Henry SP, Howard RB, Flournoy S, Levin AA, Bennett CF. and Crooke ST. (1997). Immune stimulation—a class effect of phosphorothioate oligodeoxynucleotides in rodents. Anticancer Drug Des 12:421–432 [PubMed] [Google Scholar]
- 12. Roberts TL, Sweet MJ, Hume DA. and Stacey KJ. (2005). Cutting edge: species-specific TLR9-mediated recognition of CpG and non-CpG phosphorothioate-modified oligonucleotides. J Immunol 174:605–608 [DOI] [PubMed] [Google Scholar]
- 13. Burel SA, Machemer T, Ragone FL, Kato H, Cauntay P, Greenlee S, Salim A, Gaarde WA, Hung G, et al. (2012). Unique O-methoxyethyl ribose-DNA chimeric oligonucleotide induces an atypical melanoma differentiation-associated gene 5-dependent induction of type I interferon response. J Pharmacol Exp Ther 342:150–162 [DOI] [PubMed] [Google Scholar]
- 14. Henry S, Stecker K, Brooks D, Monteith D, Conklin B. and Bennett CF. (2000). Chemically modified oligonucleotides exhibit decreased immune stimulation in mice. J Pharmacol Exp Ther 292:468–479 [PubMed] [Google Scholar]
- 15. Younis HS, Vickers T, Levin AA. and Henry SP. (2006). CpG and non-CpG oligodeoxynucleotides induce differential proinflammatory gene expression profiles in liver and peripheral blood leukocytes in mice. J Immunotoxicol 3:57–68 [DOI] [PubMed] [Google Scholar]
- 16. Akira S. and Hemmi H. (2003). Recognition of pathogen-associated molecular patterns by TLR family. Immunol Lett 85:85–95 [DOI] [PubMed] [Google Scholar]
- 17. Akira S. and Takeda K. (2004). Toll-like receptor signalling. Nat Rev Immunol 4:499–511 [DOI] [PubMed] [Google Scholar]
- 18. Kawai T. and Akira S. (2010). The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat Immunol 11:373–384 [DOI] [PubMed] [Google Scholar]
- 19. Kawai T. and Akira S. (2011). Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34:637–650 [DOI] [PubMed] [Google Scholar]
- 20. Krieg AM, Yi AK, Matson S, Waldschmidt TJ, Bishop GA, Teasdale R, Koretzky GA. and Klinman DM. (1995). CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 374:546–549 [DOI] [PubMed] [Google Scholar]
- 21. Bauer S, Kirschning CJ, Hacker H, Redecke V, Hausmann S, Akira S, Wagner H. and Lipford GB. (2001). Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc Natl Acad Sci U S A 98:9237–9242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Blasius AL. and Beutler B. (2010). Intracellular toll-like receptors. Immunity 32:305–315 [DOI] [PubMed] [Google Scholar]
- 23. Sirois CM, Jin T, Miller AL, Bertheloot D, Nakamura H, Horvath GL, Mian A, Jiang J, Schrum J, et al. (2013). Rage is a nucleic acid receptor that promotes inflammatory responses to DNA. J Exp Med 210:2447–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paz S, Hsiao J, Cauntay P, Soriano A, Bai L, Machemer T, Xiao X, Guo S, Hung G, et al. (2017). The distinct and cooperative roles of toll-like receptor 9 and receptor for advanced glycation end products in modulating in vivo inflammatory responses to select CpG and non-CpG oligonucleotides. Nucleic Acid Ther 27:272–284 [DOI] [PubMed] [Google Scholar]
- 25. Senn JJ, Burel S. and Henry SP. (2005). Non-CpG-containing antisense 2′-methoxyethyl oligonucleotides activate a proinflammatory response independent of toll-like receptor 9 or myeloid differentiation factor 88. J Pharmacol Exp Ther 314:972–979 [DOI] [PubMed] [Google Scholar]
- 26. Ghazalpour A, Rau CD, Farber CR, Bennett BJ, Orozco LD, van Nas A, Pan C, Allayee H, Beaven SW, et al. (2012). Hybrid mouse diversity panel: a panel of inbred mouse strains suitable for analysis of complex genetic traits. Mamm Genome 23:680–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bennett BJ, Farber CR, Orozco L, Kang HM, Ghazalpour A, Siemers N, Neubauer M, Neuhaus I, Yordanova R, et al. (2010). A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res 20:281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rau CD, Parks B, Wang Y, Eskin E, Simecek P, Churchill GA. and Lusis AJ. (2015). High-density genotypes of inbred mouse strains: improved power and precision of association mapping. G3 (Bethesda) 5:2021–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hui ST, Parks BW, Org E, Norheim F, Che N, Pan C, Castellani LW, Charugundla S, Dirks DL, et al. (2015). The genetic architecture of NAFLD among inbred strains of mice. Elife 4:e05607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Farber CR, Bennett BJ, Orozco L, Zou W, Lira A, Kostem E, Kang HM, Furlotte N, Berberyan A, et al. (2011). Mouse genome-wide association and systems genetics identify Asxl2 as a regulator of bone mineral density and osteoclastogenesis. PLoS Genet 7:e1002038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parks BW, Sallam T, Mehrabian M, Psychogios N, Hui ST, Norheim F, Castellani LW, Rau CD, Pan C, et al. (2015). Genetic architecture of insulin resistance in the mouse. Cell Metab 21:334–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rau CD, Wang J, Avetisyan R, Romay MC, Martin L, Ren S, Wang Y, and Lusis (2015). Mapping genetic contributions to cardiac pathology induced by beta-adrenergic stimulation in mice. Circ Cardiovasc Genet 8:40–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Watts LM, Manchem VP, Leedom TA, Rivard AL, McKay RA, Bao D, Neroladakis T, Monia BP, Bodenmiller DM, et al. (2005). Reduction of hepatic and adipose tissue glucocorticoid receptor expression with antisense oligonucleotides improves hyperglycemia and hyperlipidemia in diabetic rodents without causing systemic glucocorticoid antagonism. Diabetes 54:1846–1853 [DOI] [PubMed] [Google Scholar]
- 34. Sallam T, Jones M, Thomas BJ, Wu X, Gilliland T, Qian K, Eskin A, Casero D, Zhang Z, et al. (2018). Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat Med 24:304–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Matsushita T, Ashikawa K, Yonemoto K, Hirakawa Y, Hata J, Amitani H, Doi Y, Ninomiya T, Kitazono T, et al. (2010). Functional SNP of ARHGEF10 confers risk of atherothrombotic stroke. Hum Mol Genet 19:1137–1146 [DOI] [PubMed] [Google Scholar]
- 36. Tsukada J, Yoshida Y, Kominato Y. and Auron PE. (2011). The CCAAT/enhancer (C/EBP) family of basic-leucine zipper (bZIP) transcription factors is a multifaceted highly-regulated system for gene regulation. Cytokine 54:6–19 [DOI] [PubMed] [Google Scholar]
- 37. Sundberg TB, Choi HG, Song JH, Russell CN, Hussain MM, Graham DB, Khor B, Gagnon J, O'Connell DJ, et al. (2014). Small-molecule screening identifies inhibition of salt-inducible kinases as a therapeutic strategy to enhance immunoregulatory functions of dendritic cells. Proc Natl Acad Sci U S A 111:12468–12473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hirata M, Kugimiya F, Fukai A, Ohba S, Kawamura N, Ogasawara T, Kawasaki Y, Saito T, Yano F, et al. (2009). C/EBPbeta promotes transition from proliferation to hypertrophic differentiation of chondrocytes through transactivation of p57. PLoS One 4:e4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang Z, Bryan JL, DeLassus E, Chang LW, Liao W. and Sandell LJ. (2010). CCAAT/enhancer-binding protein β and NF-κB mediate high level expression of chemokine genes CCL3 and CCL4 by human chondrocytes in response to IL-1β. J Biol Chem 285:33092–33103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Qu C. and Qu Y. (2017). Down-regulation of salt-inducible kinase 1 (SIK1) is mediated by RNF2 in hepatocarcinogenesis. Oncotarget 8:3144–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yoon YS, Seo WY, Lee MW, Kim ST. and Koo SH. (2009). Salt-inducible kinase regulates hepatic lipogenesis by controlling SREBP-1c phosphorylation. J Biol Chem 284:10446–10452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nixon M, Stewart-Fitzgibbon R, Fu J, Akhmedov D, Rajendran K, Mendoza-Rodriguez MG, Rivera-Molina YA, Gibson M, Berglund ED, Justice NJ. and Berdeaux R. (2016). Skeletal muscle salt inducible kinase 1 promotes insulin resistance in obesity. Mol Metab 5:34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yong Kim S, Jeong S, Chah KH, Jung E, Baek KH, Kim ST, Shim JH, Chun E. and Lee KY. (2013). Salt-inducible kinases 1 and 3 negatively regulate toll-like receptor 4-mediated signal. Mol Endocrinol 27:1958–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wein MN, Foretz M, Fisher DE, Xavier RJ. and Kronenberg HM. (2018). Salt-inducible kinases: physiology, regulation by camp, and therapeutic potential. Trends Endocrinol Metab 29:723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Clark K, MacKenzie KF, Petkevicius K, Kristariyanto Y, Zhang J, Choi HG, Peggie M, Plater L, Pedrioli PG, et al. (2012). Phosphorylation of CRTC3 by the salt-inducible kinases controls the interconversion of classically activated and regulatory macrophages. Proc Natl Acad Sci U S A 109:16986–16991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Y, Gao W, Yang K, Tao H. and Yang H. (2018). Salt-inducible kinase 1 (SIK1) is induced by alcohol and suppresses microglia inflammation via NF-κB signaling. Cell Physiol Biochem 47:1411–1421 [DOI] [PubMed] [Google Scholar]
- 47. Sanosaka M, Fujimoto M, Ohkawara T, Nagatake T, Itoh Y, Kagawa M, Kumagai A, Fuchino H, Kunisawa J, Naka T. and Takemori H. (2015). Salt-inducible kinase 3 deficiency exacerbates lipopolysaccharide-induced endotoxin shock accompanied by increased levels of pro-inflammatory molecules in mice. Immunology 145:268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rossman KL, Der CJ. and Sondek J. (2005). GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol 6:167–180 [DOI] [PubMed] [Google Scholar]
- 49. Pirie E, Ray S, Pan C, Fu W, Powers AF, Polikoff D, Miller CM, Kudrna KM, Harris EN, et al. (2018). Mouse genome-wide association studies and systems genetics uncover the genetic architecture associated with hepatic pharmacokinetic and pharmacodynamic properties of a constrained ethyl antisense oligonucleotide targeting malat1. PLoS Genet 14:e1007732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Loirand G, Scalbert E, Bril A. and Pacaud P. (2008). Rho exchange factors in the cardiovascular system. Curr Opin Pharmacol 8:174–180 [DOI] [PubMed] [Google Scholar]
- 51. Mong PY. and Wang Q. (2009). Activation of Rho kinase isoforms in lung endothelial cells during inflammation. J Immunol 182:2385–2394 [DOI] [PubMed] [Google Scholar]
- 52. Wang C, Song S, Zhang Y, Ge Y, Fang X, Huang T, Du J. and Gao. J. (2015). Inhibition of the Rho/Rho kinase pathway prevents lipopolysaccharide-induced hyperalgesia and the release of TNF-alpha and IL-1beta in the mouse spinal cord. Sci Rep 5:14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vemula S, Shi J, Mali RS, Ma P, Liu Y, Hanneman P, Koehler KR, Hashino E, Wei L. and Kapur R. (2012). Rock1 functions as a critical regulator of stress erythropoiesis and survival by regulating p53. Blood 120:2868–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All macrophage microarray expression data are available at NCBI GEO GSE97207 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97207).