Abstract
Background
Cancers with aberrant expression of Serine/threonine kinase 33 (STK33) has been reported to be particularly aggressive. However, its expression, clinical significance and biological functions in gastric cancer remain largely unknown. In the present study, we determined the expression and function of STK33 in gastric cancer and delineated the clinical significance of Krüppel-like factor 4 (KLF4) /STK33 signaling pathway.
Methods
STK33 expression and its association with multiple clinicopathologic characteristics were analyzed immunohistochemically in human gastric cancer specimens. STK33 knockdown and overexpression were used to dissect the underlying mechanism of its functions in gastric cancer cells. Regulation and underlying mechanisms of STK33 expression by KLF4 in gastric cancer cells were studied using cell and molecular biological methods.
Results
Drastically higher expression of STK33 was observed in gastric cancer and gastric intraepithelial neoplasia tissues compared to adjacent normal gastric tissues. Increased STK33 expression correlated directly with tumor size, lymph node and distant metastasis; and patients with low STK33 expression gastric cancer were predicted to have a favorable prognosis. Enforced expression of STK33 promoted gastric cancer cell proliferation, migration, and invasion in vitro and in vivo, whereas reduced STK33 did the opposite. Moreover, STK33 promoted epithelial–mesenchymal transition (EMT) in vitro. Mechanistically, KLF4 transcriptionally inhibited STK33 expression in gastric cancer cells. KLF4-mediated inhibition of gastric cancer cell invasion was reversed by upregulation of STK33 expression.
Conclusions
STK33 has pro-tumor function and is a critical downstream mediator of KLF4 in gastric cancer. STK33 may serve as a potential prognostic marker and therapeutic target for gastric cancer.
Keywords: STK33, KLF4, invasion, metastasis, gastric cancer
INTRODUCTION
Gastric cancer is a significant global health problem, being the fourth most common cancer and the second leading cause of cancer death worldwide (1,2). Despite important advances in the understanding of its epidemiology, pathology, and molecular mechanisms, gastric cancer still exhibits a poor 5 year overall survival rate of <25%, largely due to late detection and limited effectiveness of current treatment options (3–6). Although various genetic and molecular alterations have been found to be associated with the malignant transformation of gastric cancer, they may represent only the pathogenesis of this disease, and have not been identified as a specific sequence of changes leading to gastric carcinoma (7–9). Therefore, the role and detailed mechanisms of genetic and epigenetic changes in gastric cancer development and progression remain to be investigated.
Serine/threonine kinase 33 (STK33) is a serine/threonine kinase that belongs to the calcium/calmodulin-dependent family of kinases, discovered by sequencing the human chromosome 11 region 11p15 and mouse chromosome 7 (10,11). Prior studies have analyzed STK33 mRNA and protein expression in a variety of normal human adult and embryonic tissues and revealed that STK33 is mainly expressed in testis, fetal lung and heart, and is weakly expressed in most other organs including stomach (12). Recently, several studies found that STK33 is highly expressed in many solid tumors, including hepatocellular carcinoma, large cell lung cancer and hypopharyngeal squamous cell carcinoma (13–16). These studies suggest that STK33 have critical roles in promoting tumor growth and metastasis. Also, STK33 has both kinase-dependent and -independent regulatory effects in tumor development and progression (11,14,17). However, so far no study has evaluated the expression and biological function of STK33 in gastric cancer.
In the present study, we found that STK33 was highly expressed in gastric cancer and was positively correlated with tumor metastasis and worse prognosis. Enforced expression of STK33 induced more aggressive phenotype of gastric cancer cells. Furthermore, a cause-effect relationship was discovered that KLF4 suppressed STK33 expression, and STK33-meditated EMT and metastasis of gastric cancer cells.
MATERIALS AND METHODS
Patients and Tissue Microarray
With the approval from the Ethics Committee at Changhai Hospital, tissue microarrays (TMAs) containing gastric cancer, gastric intraepithelial neoplasia and normal gastric specimens were obtained from patients who underwent endoscopic or surgical resection after histopathologic diagnosis of gastric cancer at the institution. Twelve different tissue microarray blocks containing a total of 670 cores (25 normal mucosa cores, 20 gastric intraepithelial neoplasia cores and 625 gastric cancer cores) were finally constructed. All of the tissue specimens for this study were obtained with patient informed consent wherever necessary. To analyze the co-expression of STK33 and KLF4, TMAs with consecutive tissues containing 45 paired primary gastric cancer and matched normal adjacent gastric specimens were subsequently used (SuperChip Shanghai, China). The characteristics of the two cohorts of TMAs are listed in Supplementary Table S1 and S2. All analyses for human specimens were also approved by the institutional review board of The University of Texas MD Anderson Cancer Center and conducted in accordance with the Declaration of Helsinki.
Bioinformatic Analysis
Gene expression data and corresponding clinical data of 300 gastric cancer patients were downloaded from the publicly available Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) databases with accession numbers GSE 66229. To gain further insight into the biologic pathways involved in gastric cancer pathogenesis through STK33 pathway, a GSEA was performed by using TCGA database. The gene sets showing FDR of 0.25, a well-established cutoff for the identification of biologically relevant gene, were considered enriched between classes under comparison.
Immunohistochemistry (IHC)
Immunohistochemical analyses of gastric specimens were conducted with anti-STK33 (Cat. #ab206296, Abcam) and anti-KLF4 (Cat. #sc-166238, Santa Cruz) antibodies as described previously (18). Expression of STK33 and KLF4 were estimated based on the percentage and intensity of the stained tumor cells. The staining percentage and staining intensity were graded and calculated for the final staining score by 2 investigators under an Olympus CX31 microscope (Olympus, Center Valley, PA) as described previously (18).
Immunofluorescence (IF)
To evaluate the protein expression and cellular localization, gastric cancer cells were seeded onto glass slides, washed with phosphate-buffered saline, fixed with 4% paraformaldehyde for 15 minutes, permeabilized with 0.5% Triton X-100 in phosphate-buffered saline for 15 minutes at room temperature, and blocked for 1 hour with 5% bovine serum albumin in phosphate-buffered saline. The cells were stained with an anti-E-Cadherin (1:200 dilution, Cat. #20874-I-AP, Proteintech), anti-vimentin (1:100 dilute, Cat. #sc-6260, Santa Cruz), anti-Flag (1:200 dilute, Cat. #F1804, Sigma), anti-KLF4 (1:50, Santa Cruz) and anti-STK33 (1:200, Abcam) antibodies overnight at 4°C. Cells were mounted with 4’,6-diamidino-2-phenylindole (DAPI) Fluoromount-G medium and a DAPI nuclear stain (Southern Biotech). Images were pseudo-colored using an Olympus microscope.
RNA Extraction and Quantitative Real-time PCR
Total RNA was extracted from the gastric cancer cells using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. cDNA was synthesized by random primers and PrimeScript RT reagent Kit (TaKaRa, Shiga, Japan). Quantitative PCR was performed using SYBR-green mastermix (TaKaRa, Shiga, Japan) and run in a LightCycler 480II instrument (Roche, Mannheim, Germany). PCR primers used are indicated in Supplementary Table S3.
Protein Extraction and Western Blot
Standard Western blotting was carried out using whole-cell protein lysates; primary antibodies against STK33 (Cat. #ab206296, Abcam), KLF4 (Cat. #sc-166238, Santa Cruz), Flag (Sigma), Vimentin (Santa Cruz), E-cadherin (Proteintech), N-cadherin (Cat. #66219-I-LG, Proteintech), Snail (Cat. #ab53519, Abcam), Slug (Cat. #ab27568, Abcam), ZEB1 (Cat. # 3396,Cell Signaling Technology), ZEB2 (Cat. #271984, Santa Cruz), Twist (Cat. #ab50581, Abcam); and a secondary antibody (anti-rabbit IgG or anti-mouse IgG; Westang Biotechnology). Equal protein sample loading was monitored using an anti-GAPDH or anti-β-actin antibody.
Cell Culture and Transient Transfection
Non-malignant gastric cell GES-1, 293T and gastric cancer cell lines BCG-823, MKN-28, MKN-45, MGC-803, and SGC-7901 cells were purchased from the Committee of Type Culture Collection of Chinese Academy of Sciences or were purchased from the American Type Culture Collection (Manassas, VA). All of the cell lines were obtained in 2014 and authenticated in August 2016 using short tandem repeat analysis, routinely tested for mycoplasma contamination by using Hoechst staining and PCR, and the cell lines they provided were passaged in our laboratory for fewer than 6 months after reception. The plasmids pcDNA3.0-STK33 (pSTK33) and pcDNA3.0-Flag-KLF4 (pFlag-KLF4) were purchased from Addgene (Cambridge, MA, USA). Small interfering RNAs (siRNAs) for STK33 (siSTK33) and KLF4 (siKLF4) were synthesized by GenePharma as described in our previous studies (19). A negative control siRNA (Invitrogen) and control pcDNA3.0 vector were used. Transient transfection was performed using Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, USA) with plasmid or siRNAs for 48 hours before functional assays were performed.
Tumor-cell Migration/Invasion Assay
Both cell scratch-wound (horizontal migration) and modified Boyden chamber (vertical invasion) assays were performed to determine the migratory ability and invasiveness of gastric cancer cells with altered STK33 expression as described previously (31). For the cell scratch wound assays, the cells were incubated for 12–48 hours after the wound was generated with 10-μL tip of a pipette. The cells in the wounded monolayer were photographed at different time points, and cell migration was assessed by measuring gap sizes in multiple fields. For the Boyden chamber assay, 24-well tissue culture plates with 12 cell culture inserts (Millipore) were used. Each insert contained an 8-μm-pore-size polycarbonate membrane with a pre-coated thin layer of a basement membrane matrix (ECMatrix for the invasion assay) or without a coated matrix (for the migration assay). Ten percent fetal bovine serum-containing medium was placed in the lower chambers to act as a chemoattractant. Cells (5×105) in a 300-μL volume of serum-free medium were placed in the upper chambers and incubated at 37°C for 48 hours. Cells on the lower surface of the polycarbonate membrane, which had invaded the ECMatrix and migrated through the membrane, were stained, counted, and photographed under a microscope.
Cell Proliferation Assay
Cells were seeded at 2×103 cells/well in 96-well plates and cultured in 100 μl of culture medium. After 24 h, 10 μl Cell Counting Kit-8 (CCK-8, Dojindo, Japan) was added to each well and cells were then incubated at 37°C for 2 h. The absorbance was then read at 450 nm using a microplate reader according to the manufacturer’s instructions.
Animals
Male and female pathogen-free athymic nude mice were purchased from the National Cancer Institute. The animals were maintained in facilities approved by AAALAC International in accordance with the current regulations and standards of the U.S. Department of Agriculture and Department of Health and Human Services. All animal studies have been conducted in accordance with the Institutional Animal Care and Use Committee (IACUC) of Second Military Medical University and The University of Texas MD Anderson Cancer Center.
Tumor Growth and Metastasis
Gastric cancer cells (1×106) in 0.1 mL of Hank’s balanced salt solution were injected subcutaneously on the left thigh of nude mice. The resulting tumors were measured every week. Tumor-bearing mice were killed by cervical dislocation when they became moribund or on day 35 after injection, and their tumors were removed and weighed. Tumor volume was calculated as follows: V (volume) = (length × width2)/2.
To quantitatively measure lung metastasis, 2×106 tumor cells suspended in 0.2 ml serum-free DMEM were injected into the lateral tail vein of nude mice. After 6 or 10 weeks, all of mice were killed. Their lung tissues were dissected and fixed with 4% phosphate-buffered neutral formalin for at least 72 h. Lung tissues were analyzed by hematoxylin and eosin staining. To measure liver metastasis, 1×106 tumor cells were injected intravenously into another group of mice via the ileocolic vein. The mice were killed on day 28 and their liver surface metastases were counted.
Construction of STK33 Promoter Reporter Plasmids
A 1900-base pair (bp) DNA fragment containing the STK33 5’ sequence from −1904 to −4 relative to the transcription initiation site was subcloned into a pGL3-basic vector (Promega). The resulting full-length reporter plasmid, which contained multiple KLF4 binding sites (KBSs), was designated pGL3–1769 (the promoter reporter plasmid was named by the start site of the KBS). Deletion mutation reporters for this plasmid (pGL3–1009, pGL3–322, pGL3–283, and pGL3–67) were then generated. All of these constructs were verified by sequencing the inserts and flanking regions of the plasmids.
Promoter Reporter and Dual Luciferase Assay
Gastric cancer cells were transfected with STK33 promoter reporters, siRNAs, or specific gene expression plasmids. The STK33 promoter activity in these cells was normalized via cotransfection of a β-actin/Renilla luciferase reporter containing a full-length Renilla luciferase gene. The resulting luciferase activity in the cells was quantified using a dual luciferase assay system (Promega) 24 hours after transfection.
Chromatin Immunoprecipitation Assay
Gastric cancer cells (2×106) were prepared for a chromatin immunoprecipitation (ChIP) assay using a ChIP assay kit (Millipore) according to the manufacturer’s protocol. The resulting precipitated DNA specimens were analyzed using PCR to amplify fractions of the STK33 promoter. The PCR products were resolved electrophoretically on a 2% agarose gel and visualized using ethidium bromide staining.
Statistical Analysis
Associations between STK33 expression and clinicopathological variables were analyzed by nonparametric analysis, using the Mann-Whitney U test for dichotomization variables and the Kruskal-Wallis test for the others. The median overall survival (OS) and its 95% confidence intervals (95% CIs) were estimated by Kaplan-Meier method. Difference in survivals was analyzed using the log-rank test. All in vitro experiments were performed in triplicate and at least three times. The correlation of STK33 and KLF4 expression was examined by Spearman correlation test. Data were presented either as means ± standard deviation (SD) from one representative independent experiment of three with similar results or means ± standard error of the mean (SEM) from three independent experiments. In all of the tests, P values less than 0.05 were considered statistically significant. Statistical analyses were conducted using GraphPad Prism software (Intuitive Software for Science, San Diego, CA) or SPSS 17.0 statistical software (SPSS Inc, Chicago, Illinois, USA).
RESULTS
Elevated expression of STK33 in primary gastric cancer directly correlated with metastasis and poor survival
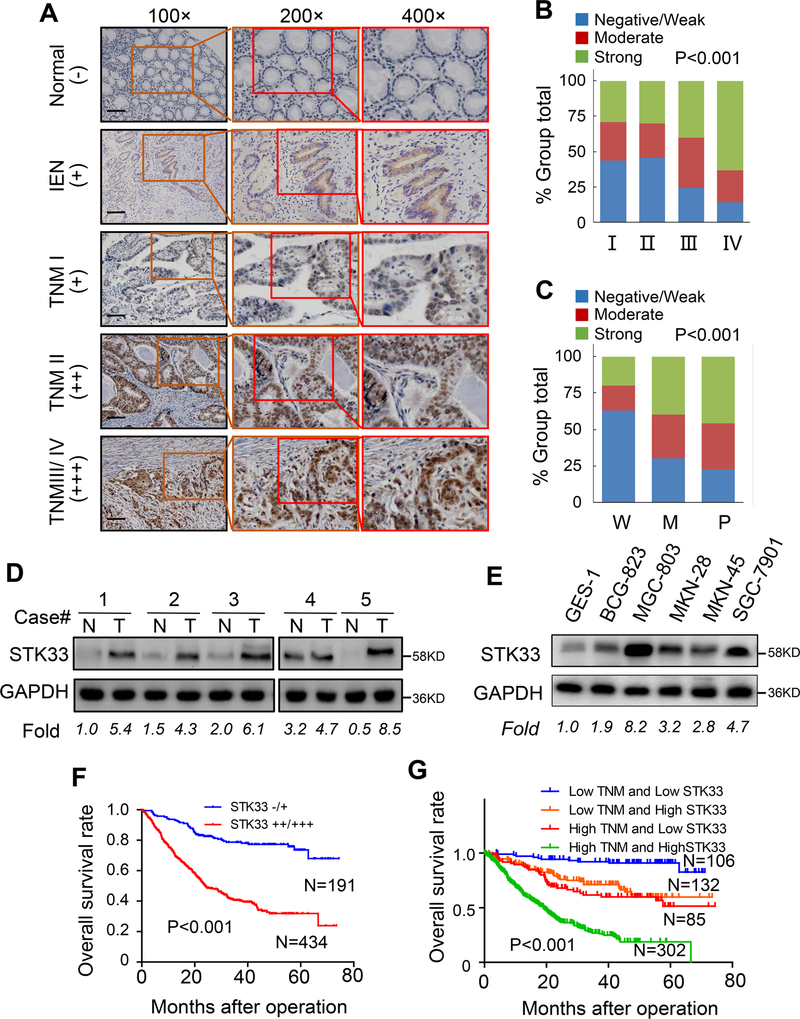
We first examined the STK33 expression profile in a TMA consisting of normal gastric, intraepithelial neoplasia, and gastric cancer tissues. As shown in Figure 1A and Table S4, STK33 expression gradually increased from normal gastric tissue through to gastric intraepithelial neoplasia and to gastric cancer, suggesting that STK33 accumulation expression is an early event in the multistep progression of gastric carcinogenesis. Next, we analyzed the relationship between clinicopathologic parameters and STK33 expression in gastric cancer specimens. Higher expression of STK33 was significantly associated with bigger tumor size (P=0.005), more lymph node (P<0.001) and distant site metastasis (P=0.033), higher TNM stage (P<0.001) (Figure 1A and 1B), and worse differentiation (P<0.001) (Figure 1C and Supplementary Figure S1). No significant association was observed between STK33 expression and patient sex, age, tumor location, Lauren classification, serum CEA level, or serum CA19–9 level (Table S5). Western blot analysis demonstrated that STK33 expression in gastric cancer tissue specimens and cell lines were higher than that in normal gastric tissues and cell lines (Figure1D and 1E). Furthermore, we observed that patients with high STK33 expression had shorter OS than did those with low STK33 (P < 0.0001) (Figure 1F) and STK33 predicted poor prognosis independent of TNM stage (Figure 1G). To further explore the role of STK33 in predicting gastric cancer prognosis, we performed univariate and multivariate analyses of clinical follow-up data. Our data showed STK33 expression was positively correlated with OS duration (Table S6 and S7). To validate our findings, we further analyzed STK33 mRNA expression and corresponding clinical data from the publicly available GEO database (GSE66229). Evaluation of STK33 in 300 patients with gastric cancer revealed that high STK33 mRNA expression was associated with decreased OS and DFS and advanced TNM stage (Supplementary Figure S2A–S2C). We then tested whether the prognostic value of STK33 mRNA was independent of TNM stage. Results showed STK33 remained to be significantly associated with OS and DFS in gastric cancer patients when adjusted by TNM stage (Supplementary Figure S2D and S2E). Taken together, these data indicated that STK33 plays a critical role in gastric cancer development and progression and might be a potential prognostic biomarker for this disease.
Figure 1. Close association with upregulated expression of STK33 and advanced tumor stage.
A, representative images of STK33 in TMAs of normal gastric tissue, gastric intraepithelial neoplasia (IEN) and gastric cancer with different tumor stages. Scale bars: 50 μm. B, association of elevated expression of STK33 with tumor stage in gastric cancer. C, association of elevated expression of STK33 with tumor differentiation in gastric cancer. D, Western blot analysis of STK33 expression in 5 paired human gastric cancer and adjacent normal gastric tissue specimens. STK33 protein expression were normalized according to GAPDH. E, Western blot showing the expression of STK33 protein in non-malignant and malignant gastric cell lines. F, association of STK33 expression with OS rate in patients with gastric cancer. The patients were stratified into two groups according to STK33 immunohistochemical staining score. Patients with low STK33 expression (−/+) had much longer OS durations than did patients with high STK33 expression (++/+++). G, STK33 predicted poorer OS in gastric cancer patients adjusted by TNM stages. *P<0.05.
STK33 promoted migration and invasion of gastric cancer cells in vitro and metastasis in animal models
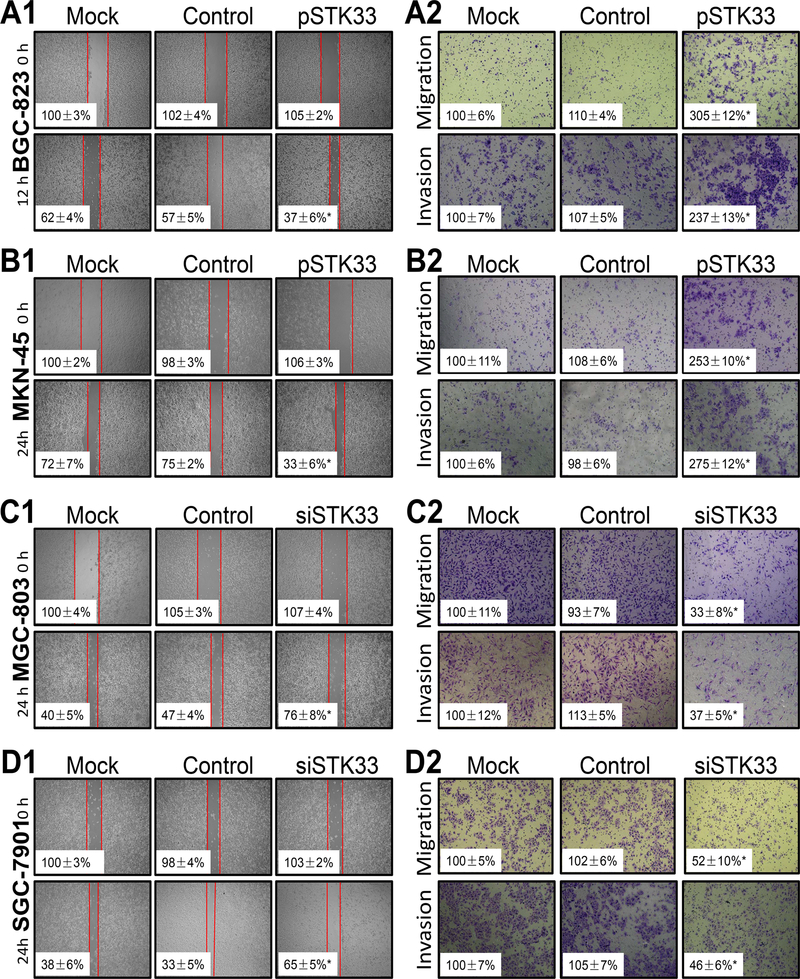
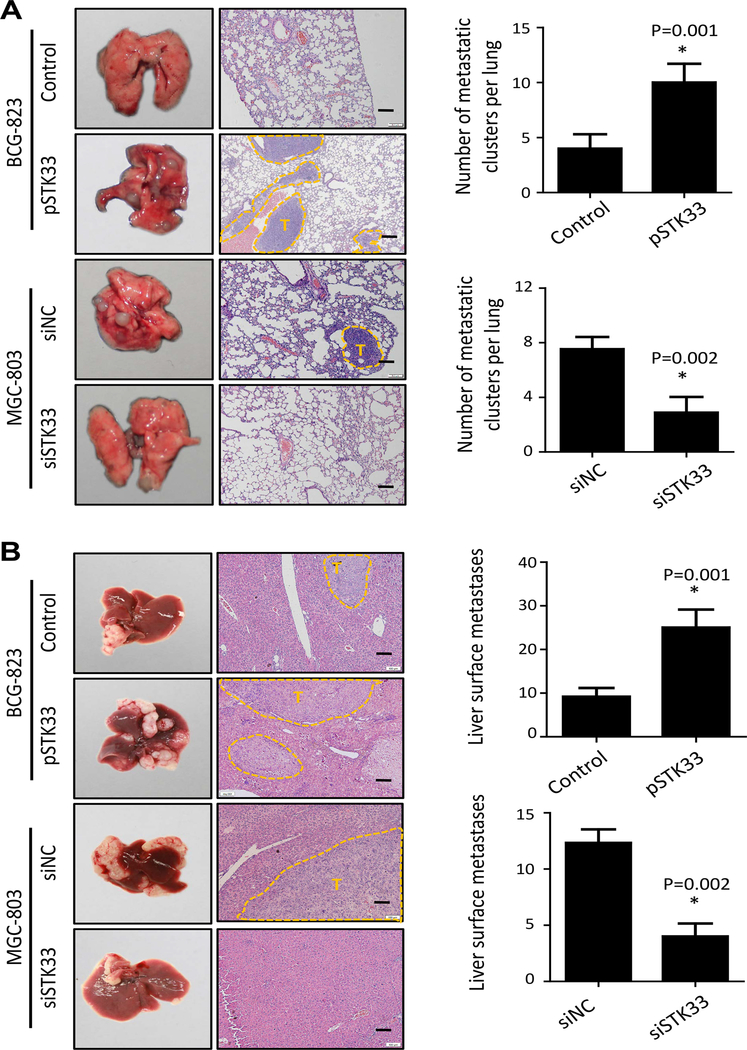
By IHC and IF analyses, we observed that STK33 was located in the cytoplasm and/or nucleus of gastric cancer cells (Supplementary Figure S3A and S3B). To explore the biologic pathways involved in gastric cancer pathogenesis stratified by the levels of STK33 expression, we performed GSEA analysis using TCGA dataset. Enrichment plots of GSEA showed that the gene signatures of tumor metastasis were more correlated with patients with higher STK33 expression than patients with lower STK33 expression (Supplementary Figure S4A). To validate the GSEA analysis on STK33, we genetically increased the ectopic expression of STK33 in two gastric cancer cell lines (BCG-823 and MKN-45) with relatively lower endogenous levels of STK33 expression. Wound-healing assay showed that migratory ability of BCG-823 and MKN-45 cell were significantly enhanced by STK33 (Figure 2A1 and 2B1). Transwell assays demonstrated that STK33 overexpression promoted the migration and invasion in both gastric cancer cells more so than that of control cells (Figure 2A2 and 2B2). Two gastric cancer cell lines with relatively higher endogenous STK33 expression (MGC-803 and SGC-7901) were then transfected with siRNAs-targeting STK33. Wound-healing assay suggested that STK33 depletion in MGC-803 and SGC-7901 cells reduced cell migratory ability (Figure 2C1 and 2D1). In Transwell assays, STK33 silencing inhibited the motility and invasiveness of both gastric cancer cells (Figure 2C2 and 2D2). Further, we confirmed the in vitro results in metastatic tumor models of gastric cancer, finding that enforced expression of STK33 markedly increased lung and hepatic metastasis of BCG-823 cells, whereas knockdown of expression of STK33 abrogated lung and hepatic metastasis of MGC-803 cells (Figure 3A and 3B). Transfection efficacy of STK33 cDNA in BGC-823 and MKN-45 cells and siRNA in MGC-803 and SGC-7901 cells were shown in Figure 4B. Consistent with the diagnostic and prognostic value of STK33 in gastric cancer patients, these data indicated that STK33 plays critical roles in gastric cancer cell metastasis.
Figure 2. Influence of STK33 expression on gastric cancer cell migration and invasion in vitro.
BCG-823 (A1 and A2), MKN-45 (B1 and B2), MGC-803 (C1 and C2), and SGC-7901 (D1 and D2) cells were transfected with pSTK33, siSTK33, control vectors, or siRNAs (mock) for 48 hours. For a cell scratch-wound assay, cells in each group were placed in six-well plates, wounded via scratching, and maintained at 37°C for at least 12 hours. Cell cultures were photographed, and cell migration was assessed by measuring the cell-free areas in multiple fields (the insert numbers indicate the percentage mean gap areas ± standard deviation in triplicate). The migration and invasion of gastric cancer cells were determined as described in Materials and Methods. The data represent the means ± standard error of the mean in triplicate from one representative experiment of three with similar results. *P < 0.05 in comparisons of pSTK33-treated, siSTK33-treated, mock, and control groups (Student t-test).
Figure 3. STK33-promoted gastric cancer cell metastasis in vivo.
Lung and liver metastasis mouse models were used to analyze the effect of STK33 on tumor metastasis in vivo. A, representative images of gross lung metastases and hematoxylin- and eosin-stained sections of lung of the mice from the indicated groups and their numbers of lung surface metastatic foci are shown. The number of lung metastatic foci revealed by hematoxylin and eosin staining is calculated microscopically, represented as the mean ± SEM. B, representative images of gross liver surface metastases and hematoxylin- and eosin-stained sections of liver from indicated groups (tumor areas are outlined with dashed lines; T, tumor). Scale bars: 100 μm. *P<0.05.
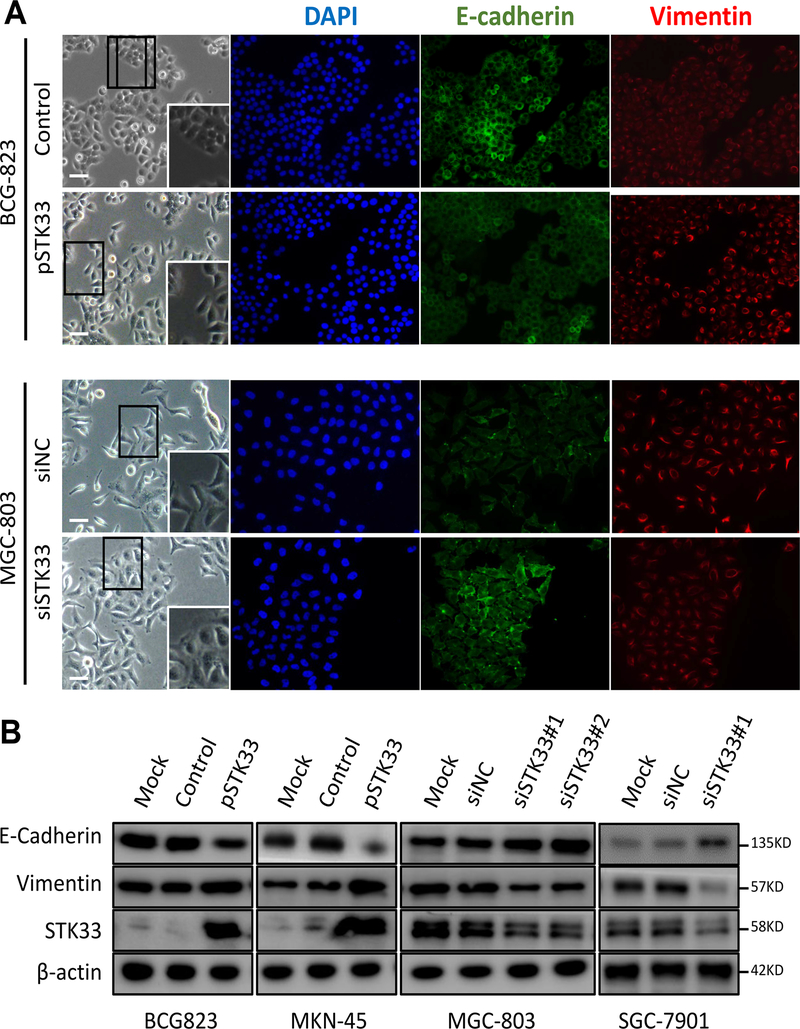
Figure 4. STK33-promoted EMT in gastric cancer cells.
A, left: shown are phase contrast microphotographs of BCG-823 cells infected with either empty vector or STK33-overexpressing vector and MGC-803 transfected with either non-targeting siRNA (siNC) or siSTK33. Higher magnification of the highlighted region is shown in the inset. Right: expression of E-cadherin and vimentin detected by immunofluorescence. Nuclear counterstaining with diamidino-2-phenylindole (DAPI) is shown separately. Scale bars: 50 μm. B, expression of STK33, E-cadherin and Vimentin detected by western blotting, in total lysates from BCG-823 and MGC-803 cells infected with the indicated constructs. β-actin was used as a loading control.
STK33 promoted epithelial-mesenchymal transition (EMT) in gastric cancer cells
Acquisition of EMT phenotype is a critical process for epithelial cells to gain ability to migration and invasion. We tested if STK33 is involved in EMT of gastric cancer cells. We observed that BCG-823 cells grew as compact colonies and, upon STK33 overexpression, acquired a spindle-like shape, showed increased scattering, downregulated E-cadherin and upregulated Vimentin protein expression, all features suggesting an EMT. Consistently, STK33-silenced MGC-803 displaying looser growth pattern-resulted in the formation of compact colonies, reduced scattering, upregulated E-cadherin and downregulated Vimentin, supporting a mesenchymal-epithelial transition (MET) (Figure 4A). E-cadherin, Vimentin and other EMT related markers in STK33-dysregulated gastric cancer cell lines were determined by Western blot and/or real-time PCR (Figure 4B and Supplementary Figure S4B and S4C). These results showed that STK33 might promote migration and invasion of gastric cancer cells through EMT.
STK33 promoted proliferation in vitro and tumor growth in vivo of gastric cancer cells
To determine the effect of STK33 expression on gastric cancer cell proliferation, BCG-823 and MKN-45 cells were transfected with pSTK33, whereas MGC-803 and SGC-7901 cells were transfected with siSTK33. Cell-counting kit-8 assays indicated that exogenous expression of STK33 resulted in a significant increase in the proliferation index of BCG-823 and MKN-45 cells, whereas knockdown of STK33 in MGC-803 and SGC-7901 cells did the opposite (Supplementary Figure S5A). In animal models, transfection with pSTK33 or siSTK33 consistently promoted (Supplementary Figure 5B1–5D1) or suppressed (Supplementary Figure S5B2–S5D2) the growth of tumor cells in the subcutis. Both in vitro and in vivo experimental evidence validated the oncogenic role of STK33 in gastric cancer.
Lost or reduced expression of KLF4 caused STK33 overexpression in gastric cancer cells
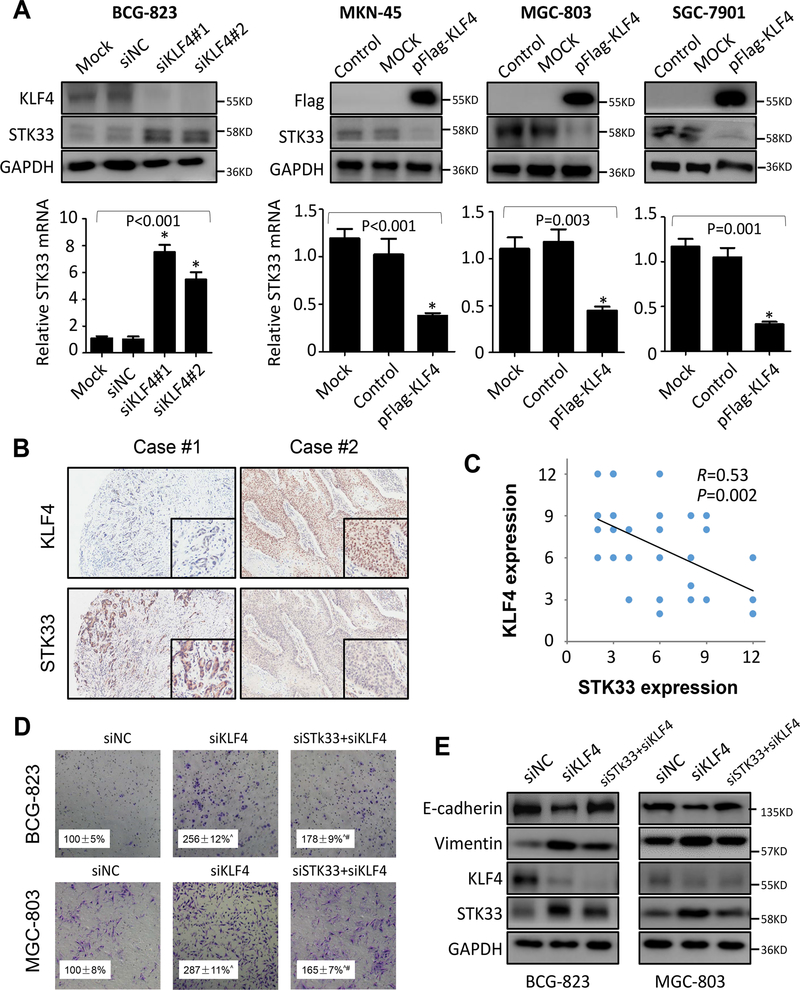
Previously, our group have demonstrated that KLF4, also known as gut-enriched KLF, serves as an important transcription factor involved in the multistep of gastrointestinal tumor development and progression, including EMT (20,21). To understand the molecular mechanisms underlying STK33-induced signaling leading to enhanced tumor cell aggressiveness, we examined the effect of altered KLF4 expression in gastric cancer cells by Western blot and immunofluorescence. As shown in Figure 5A and Supplementary Figure S6A, overexpression of KLF4 in BCG-823, MKN-45 and SGC-7901 cells markedly downregulated the expression of both STK33 mRNA and protein, whereas knockdown of expression of KLF4 upregulated them in MGC-803 cells. Subsequently, we performed correlation analysis of protein expression between KLF4 and STK33 using TMAs with consecutive tissue sections. Our results showed that the gastric cancer specimens in the TMAs, which gained high expression of KLF4, had low expression of STK33 (Figure 5B). Importantly, statistical analysis revealed that the expression of STK33 was negatively correlated with that of KLF4 in the gastric cancer specimens (R = 0.53; P = 0.002) (Figure 5C).
Figure 5. Negative regulation of STK33 by KLF4 in gastric cancer cells.
BCG-823 cells were transfected with siKLF4#1 and siKLF4#2 or control siRNA for 48 hours, and MKN45, MGC-803 and SGC-7901 cells were transfected with pFlag-KLF4 or control vector for 48 hours. A, total RNA and protein lysates were harvested, and the expression of STK33 and KLF4 in the lysates was determined using real-time PCR and Western blot. B, immunohistochemical stains for KLF4 and STK33 from TMAs with consecutive gastric cancer tissue sections. Representative images of gastric cancer sections with STK33 and KLF4 staining are shown (100× magnification in the main images, 200× magnification in the inserts). C, assessment of the negative correlation between KLF4 and STK33 expression in gastric cancer specimens (N = 45) using Pearson correlation coefficient analysis. Some of the dots on the graph represent more than one specimen. D, Boyden chamber analysis of effect of alerted expression of STK33 on KLF4-mediated invasion in gastric cancer cells. The experiments were performed independently three times. Ŝtatistically significant when compared with siNC group (P < 0.05); #statistically significant when compared with siKLF4 group (P < 0.05). E, transfection efficacy, and EMT markers were examined by Western blot.
To understand the role of KLF4 in STK33–mediated aggressive phenotype, we inhibited KLF4 expression in BCG-823 and MGC-803 cells with or without knockdown of STK33. As shown in Figure 5D and Supplementary Figure S6B, downregulation of KLF4 expression enhanced the invasion and migration of gastric cancer cells. However, knockdown of STK33 partially inhibited the promoting effect of KLF4 knockdown on gastric cancer cell invasion and migration, suggesting that STK33 was involved in KLF4-mediated cell invasion and migration. KLF4 was reported to be critical for progression and maintenance of phenotypic and cellular changes associated with EMT. Therefore, we evaluated the possibility of STK33 contributing to EMT by KLF4. Western blot analysis showed that KLF4 regulated expression of STK33, E-cadherin and Vimentin in a dose-dependent manner (Supplementary Figure S7A). Further analysis revealed that the inhibition of STK33 partially rescued the dysregulated-E-cadherin and -Vimentin induced by dysregulated KLF4 expression in both protein and mRNA level (Figure 5E and Supplementary Figure S7B). Moreover, the morphological changes of gastric cancer cells induced by siKLF4 was partially reverted by siSTK33 (Supplementary Figure S7C). Collectively, these data strongly demonstrated that STK33 expression and STK33-mediated EMT was negatively regulated by KLF4.
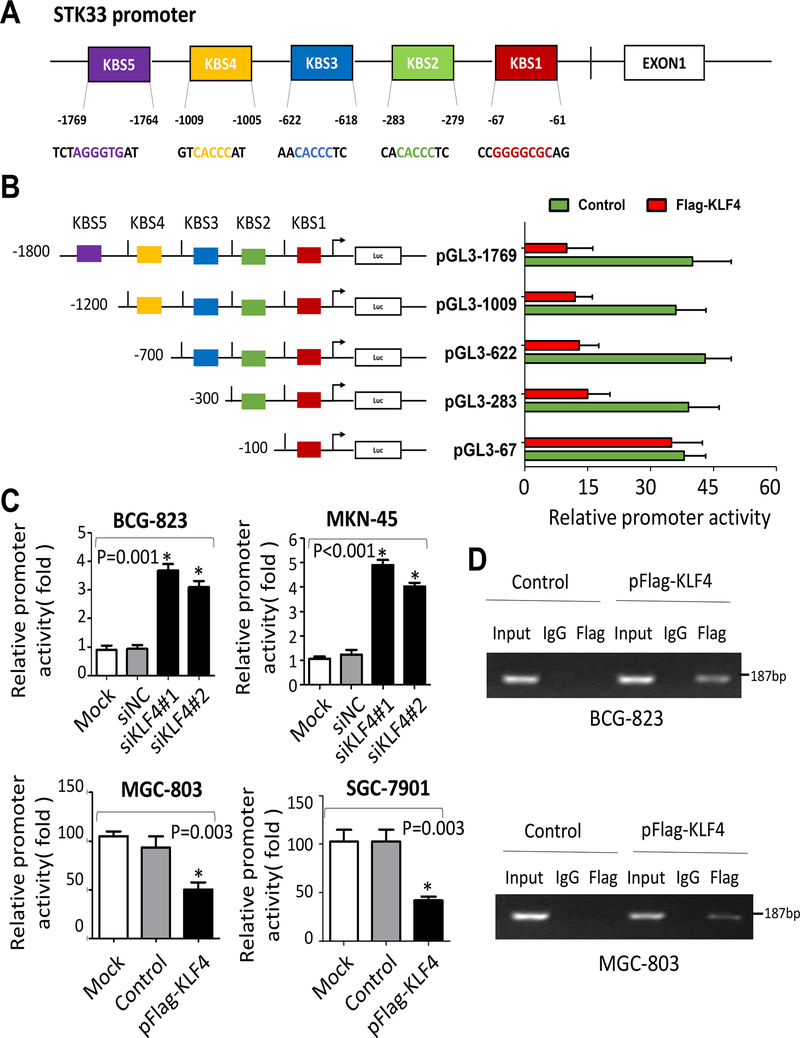
KLF4 transcriptionally inhibited STK33 expression in gastric cancer cells
To further understand the molecular mechanism of STK33 expression regulated by KLF4, we analyzed the STK33 promoter sequence for the canonical KLF4-bingding sites (KBSs) 5’-CACCC-3’ and 5’-(G/A)(G/A)GG(C/T)G(C/T)-3’, which were described previously (18). Sequence analysis of the STK33 promoter uncovered five putative KBSs located at −1769 (KBS5), −1009 (KBS4), −622(KBS3), −283 (KBS2) and −67 (KBS1) bp relative to the transcriptional start site of STK33 (Figure 6A). We then generated the full-length STK33 promoter pSTK33–1769 and deletion mutants of it. To determine whether KLF4 regulates STK33 expression at the transcriptional level, we co-transfected the deletion mutant reporters with or without KLF4 expression vectors into 293T cells. Luciferase reporter assay results demonstrated that deletion of the region from −402 to −211 bp, covering the KBS2, markedly increased the promoter activity of STK33 inhibited by KLF4 (Figure 6B). To further determine whether KLF4 regulates STK33 promoter transcriptional activity in gastric cancer cells, we co-transfected a full length reporter with KLF4 expression vectors or siKLF4 into gastric cancer cells. As shown in Figure 6C, increased KLF4 expression in BCG-823 and MKN-45 cells attenuated STK33 promoter activity, whereas knockdown of KLF4 expression in MGC-803 and SGC-7901 cells showed activating effect. Finally, to determine whether KLF4 directly interacts with the STK33 promoter, we performed a ChIP assay using specific PCR primers. Our results confirmed that KLF4 directly bound to the KBS2 site (Figure 6D) rather than other KBSs in the STK33 promoter in gastric cancer cells. To further confirm the negative regulatory effect of KLF4 on the expression of STK33, we downregulated KLF4 in pancreatic cancer cells (L3.6pl). In accordance with our previous findings, STK33 was transcriptionally inhibited by KLF4 and partially reverted KLF4-mediated EMT in pancreatic cancer cells (Supplementary Figure S8A–S8D). These data strongly suggested that KLF4 bound directly to the STK33 promoter and transcriptionally inhibited STK33 expression in gastric cancer cells.
Figure 6. Direct binding of KLF4 to the STK33 promoter.
A, five KBSs located at different sites in the STK33 promoter sequence. B, STK33 promoter reporters (pGL3–1769, −1009, −622, −283 and −67) were transfected into 239T cells in triplicate with KLF4 expression or control vectors for 24 hours. The STK33 promoter activity was examined by dual luciferase assay. The promoter activities of the treated groups relative to those of the control groups are shown. C, BCG-823 and MKN-45 cells were co-transfected with pGL3–1769, siKLF4 (#1 or #2), or a nontargeting siRNA. MGC-803 and SGC-7901 cells were co-transfected with pGL3–1769, a Flag-tagged KLF4, or control vector. The promoter activities were determined by dual luciferase assay. The experiments were performed independently three times. D, results of ChIP assay conducted using chromatins isolated from BCG-823 and MGC-803 cells. The immunoprecipitated DNA was analyzed by PCR followed by agarose gel electrophoresis. Genomic DNA input was 1%. The experiments were performed three times independently. *P < 0.05.
STK33 kinase inhibitor BRD-8899 did not influence STK33-induced cell proliferation, migration and invasion
BRD-8899 has been reported to be one of the most potent and selective kinase inhibitor for STK33 (22). To further explore the role of STK33 in gastric cancer cells, we used BRD-8899 to analysis its kinase activity. We found that BRD-8899 failed to inhibit the proliferation, migration and invasion of gastric cancer cells (Supplementary Figure S9A and S9B). Therefore, the kinase activation effect of STK33 does not appear to be necessary in gastric cancer cell proliferation, migration and invasion.
DISCUSSION
In the present study, we determined the expression of STK33 in gastric cancer and the role of KLF4/STK33 signaling in gastric tumorigenesis. We found STK33 expression was progressively upregulated from normal gastric tissues through to precancerous tissues to gastric cancer tissues. High levels of STK33 expression were directly correlated with increased lymph node and distant site metastasis, advanced TNM stage, poor differentiation and reduced OS duration. Additionally, STK33 played critical roles in regulating EMT and tumor metastasis, a novel mechanism in gastric cancer metastasis. Moreover, lost expression of KLF4, resulted in upregulated expression and enhanced function of STK33 in gastric cancer cells. Mechanistically, KLF4 bound directly to the STK33 promoter and transcriptionally inhibited the expression of STK33 in gastric cancer cells. Taken together, these data demonstrated that STK33 serves as an important regulator of progression and metastasis of gastric cancer, which is negatively regulated by KLF4.
Elevated expression of STK33 has been demonstrated in a variety of solid tumors, including hepatocellular carcinoma, and large cell lung cancer and hypopharyngeal squamous cell carcinoma (14–16). Recent evidence from our group identified a significantly elevated STK33 expression in pancreatic cancer tissues relative to adjacent normal pancreatic tissues specimens (23). Our current study for the first time established the frequent overexpression of STK33 in majority of gastric cancer, which was significantly correlated with advanced clinical stage and poorer survival, suggesting that upregulated STK33 in gastric cancer might facilitate the metastatic phenotype. These findings were further validated by analysis of another independent cohort from GEO database containing 300 gastric cancer patients (GSE66229). Importantly, we found STK33 expression in gastric intraepithelial neoplasia was higher than in normal gastric tissues but much lower than in gastric cancer, indicating a role of STK33 in the early stage of gastric carcinogenesis. Furthermore, univariate and multivariate Cox regression analysis demonstrated that STK33 was an independent prognostic factor for poorer outcome in gastric cancer patients, suggesting that STK33 expression could be used as a potential predictor as it is intimately involved in gastric cancer progression. These findings underscored a potentially important role of STK33 in the development and progression of gastric cancer.
Enforced expression of STK33 enhances the aggressiveness of many types of cancer (13,14,16). Overexpression of STK33 in pancreatic cancer could substantially promote cell proliferation, migration and invasiveness (23). STK33 is found to be capable of suppressing p53 gene, whose accumulation in the nucleus is associated with an increasing susceptibility to chemotherapy and radiotherapy (16). Therefore, overexpression of STK33 may define a subgroup of cancer cells that are less sensitive to antitumor treatment. To clarify the biological function of STK33 in regulating gastric cancer cell motility and invasiveness, we conducted a series of in vitro and in vivo assays and discovered that STK33 enhanced proliferation, migration and invasion of gastric cancer cells and tumor growth and metastasis in an orthotopic mouse model of gastric cancer.
Metastasis represented a multi-step cell-biological process including a key event, EMT. EMT plays a pivotal role in tumor invasion and metastasis during tumor progression by suppressing expression of epithelial markers and inducing expression of mesenchymal markers. In this study, we uncovered that an EMT mechanism was involved in the STK33-mediated aggressive phenotype. STK33 showed an anti-epithelial and pro-EMT function in gastric cancer through activating Vimentin, a mesenchymal marker, and the concomitant repression of E-cadherin, an important epithelial gene. This is the first study to identify STK33 as an EMT regulator in gastrointestinal tumors. However, the underlying mechanism how STK33 modulates the EMT-related genes is currently unknown and warrants further investigation.
Molecular basis of STK33 overexpression in cancer cells remains largely unclear. STK33 protein can be stabilized by HSP90 chaperone complex (24). We found that reduced expression of Transcription factor KLF4 caused STK33 overexpression in gastric cancer. Our previous studies found that KLF4 was downregulated in gastric cancer and regulated the expression of a vast number of oncogenes that are involved in many cellular functions, ranging from differentiation to proliferation and metastasis in gastrointestinal cancers (18,25–29). Loss of KLF4 expression contributes to human gastric cancer development and progression by regulating Sp1 (21). Also, we reported that KLF4-knockout in gastric progenitor cells resulted in the formation and progression of tumors in the antrum (20). Evidently, a number of regulators critical in the initiation and execution of EMT are transcriptionally modulated by KLF4 (30–32). Our promoter sequence analysis identified multiple KLF4 binding sites on STK33 promoter, suggesting that KLF4 involve in the regulation of SKT33 expression and STK33-mediated EMT in gastric cancer. We found four lines of evidence supporting a critical role of KLF4 as a transcriptional suppressor of STK33. First, the expression of STK33 was negatively correlated with that of KLF4 in gastric cancer specimens. Second, overexpression of KLF4 downregulated the expression of STK33 at both mRNA and protein level, whereas reduced KLF4 did the opposite. Third, knockdown of STK33 partially abrogated the enhanced gastric cancer cell invasion and reversed the morphologic changes of EMT induced by downregulation of KLF4. Fourth, KLF4 bound directly to the promoter region of STK33 and transcriptionally suppressed its promoter activity. Therefore, our findings provided clinical and mechanistic evidence supporting the existence of a novel KLF4/STK33 signaling pathway and its critical contribution to invasion and metastasis of gastric cancer cells.
There are limited literatures discussed about the role of KLF4 in regulating EMT of gastric cancer, but other discoveries about KLF4 regulating EMT in non-gastric cancer may bring inspiration to our understanding of KLF4. In a mouse model of breast cancer, KLF4 inhibited EMT through reducing the expression of Snail, a key mediator of EMT and metastasis (30). The expression of E-cadherin, an epithelial marker, is controlled by a balance between ZEB2 and KLF4 in cancer cell lines. KLF4 binds to the E-cadherin promoter in a region overlapping with a known ZEB2 binding site, but these two transcription factors exhibited opposite effects on the activity of E-cadherin promoter (32,33). A recent publication has shown that KLF4 was a transcriptional regulator of genes critical for EMT, and in addition, they revealed a series of key genes as direct transcriptional target of KLF4, including E-cadherin, N-cadherin, vimentin, β-catenin, VEGF-A, endothelin-1 and Jnk1 (34). Consistent with those previous studies, our study mechanistically showed that KLF4 was a regulator of E-cadherin and Vimentin, both critical for EMT. Arguably, these regulations could be partially mediated by altered expression of STK33. Therefore, KLF4 may impact EMT directly through the regulation of epithelial and mesenchymal genes and/or indirectly through the regulation anti-epithelial and pro-mesenchymal factors (e.g., STK33).
Finally, KLF4 has long been established as a key factor in carcinogenesis, while different studies show that KLF4 play different roles in different cancer types or different stages of cancer development and progression (25). Targeted activation of KLF4 could suppress the growth of late-stage cancer cells, but may predispose epithelial cells to the risk of transformation into precancerous lesions. In contrast to KLF4, STK33 is generally upregulated in various cancer types, including those precancerous lesions. Targeted inhibition of STK33 as a downstream mediator of KLF4 may not have that complication.
In summary, our clinical and experimental evidence strongly support that STK33 is oncogenic in gastric cancer. STK33 promoted gastric cancer cell EMT and metastasis. KLF4 inhibited STK33 expression by directly binding to the KBS of its promoter and attenuated its oncogenic function. Therefore, STK33 plays a critical role in promoting gastric cancer progression and metastasis, and the aberrant KLF4/STK33 signaling pathway might be a promising molecular target for designing new strategy to control metastatic gastric cancer.
Supplementary Material
Translational Relevance.
Gastric cancer has a high incidence of recurrence and metastasis. The mechanisms underlying these aggressive behaviors are poorly understood. In this study, the expression and role of Serine/threonine kinase 33 (STK33) in gastric cancer development and progression was determined. We found that STK33 was frequently overexpressed in gastric intraepithelial neoplasia and cancer, which was significantly associated with advanced tumor stage and metastasis. Moreover, STK33 was an independent prognostic predictor; gastric cancer patients with high STK33 expression had poorer survival than those with low STK33 expression. Mechanistically, STK33 promoted gastric cancer metastasis by inducing EMT and reduced expression of Krüppel-like factor 4 (KLF4) caused STK33 overexpression. Our findings suggested that STK33 serve as a new prognostic biomarker for gastric cancer patients and targeted inhibition of STK33 expression reverse EMT, reduce incidence of distant metastasis and improve prognosis of patients with gastric cancer. Therefore, our study may have a significant effect on clinical management of gastric cancer.
Acknowledgments
Financial Support: This work was supported by grants R01CA172233, R01CA195651, and R01CA198090 from the National Cancer Institute, National Institutes of Health (to K. Xie) and grant NSFC no. 81772640 (to X. Kong) from the National Natural Science Foundation of China.
Footnotes
Disclosure of Potential Conflicts of Interest: The authors declare no conflict of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: a cancer journal for clinicians 2016;66(1):7–30 doi 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet 2016;388(10060):2654–64 doi 10.1016/s0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 3.Tan P, Yeoh KG. Genetics and Molecular Pathogenesis of Gastric Adenocarcinoma. Gastroenterology 2015;149(5):1153–62 e3 doi 10.1053/j.gastro.2015.05.059. [DOI] [PubMed] [Google Scholar]
- 4.Badgwell B, Blum M, Estrella J, Ajani J. Personalised therapy for localised gastric and gastro-oesophageal adenocarcinoma. The Lancet Oncology 2016;17(12):1628–9 doi 10.1016/S1470-2045(16)30521-6. [DOI] [PubMed] [Google Scholar]
- 5.Shan LH, Sun WG, Han W, Qi L, Yang C, Chai CC, et al. Roles of fibroblasts from the interface zone in invasion, migration, proliferation and apoptosis of gastric adenocarcinoma. Journal of clinical pathology 2012;65(10):888–95 doi 10.1136/jclinpath-2012-200909. [DOI] [PubMed] [Google Scholar]
- 6.Eren OO, Sonmez OU, Oyan B. Is It Time for Maintenance Chemotherapy for Advanced Gastric Adenocarcinoma? J Clin Oncol 2016;34(17):2067 doi 10.1200/JCO.2015.65.8088. [DOI] [PubMed] [Google Scholar]
- 7.Werner M, Becker KF, Keller G, Hofler H. Gastric adenocarcinoma: pathomorphology and molecular pathology. Journal of cancer research and clinical oncology 2001;127(4):207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng L, Wang L, Ajani J, Xie K. Molecular basis of gastric cancer development and progression. Gastric cancer : official journal of the International Gastric Cancer Association and the Japanese Gastric Cancer Association 2004;7(2):61–77 doi 10.1007/s10120-004-0277-4. [DOI] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513(7517):202–9 doi 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mujica AO, Hankeln T, Schmidt ER. A novel serine/threonine kinase gene, STK33, on human chromosome 11p15.3. Gene 2001;280(1–2):175–81. [DOI] [PubMed] [Google Scholar]
- 11.Brauksiepe B, Mujica AO, Herrmann H, Schmidt ER. The Serine/threonine kinase Stk33 exhibits autophosphorylation and phosphorylates the intermediate filament protein Vimentin. BMC biochemistry 2008;9:25 doi 10.1186/1471-2091-9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mujica AO, Brauksiepe B, Saaler-Reinhardt S, Reuss S, Schmidt ER. Differential expression pattern of the novel serine/threonine kinase, STK33, in mice and men. Febs J 2005;272(19):4884–98 doi 10.1111/j.1742-4658.2005.04900.x. [DOI] [PubMed] [Google Scholar]
- 13.Scholl C, Frohling S, Dunn IF, Schinzel AC, Barbie DA, Kim SY, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell 2009;137(5):821–34 doi 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 14.Yang T, Song B, Zhang J, Yang GS, Zhang H, Yu WF, et al. STK33 promotes hepatocellular carcinoma through binding to c-Myc. Gut 2014. doi 10.1136/gutjnl-2014-307545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang L, Chen C, Zhang G, Ju Y, Zhang J, Wang H, et al. STK33 overexpression in hypopharyngeal squamous cell carcinoma: possible role in tumorigenesis. BMC cancer 2015;15(1):13 doi 10.1186/s12885-015-1009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang P, Cheng H, Wu J, Yan A, Zhang L. STK33 plays an important positive role in the development of human large cell lung cancers with variable metastatic potential. Acta biochimica et biophysica Sinica 2015. doi 10.1093/abbs/gmu136. [DOI] [PubMed] [Google Scholar]
- 17.Sun EL, Liu CX, Ma ZX, Mou XY, Mu XA, Ni YH, et al. Knockdown of human serine/threonine kinase 33 suppresses human small cell lung carcinoma by blocking RPS6/BAD signaling transduction. Neoplasma 2017. doi 10.4149/neo_2017_608. [DOI] [PubMed] [Google Scholar]
- 18.Guo K, Cui J, Quan M, Xie D, Jia Z, Wei D, et al. The Novel KLF4/MSI2 Signaling Pathway Regulates Growth and Metastasis of Pancreatic Cancer. Clin Cancer Res 2017;23(3):687–96 doi 10.1158/1078-0432.CCR-16-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi M, Cui J, Du J, Wei D, Jia Z, Zhang J, et al. A novel KLF4/LDHA signaling pathway regulates aerobic glycolysis in and progression of pancreatic cancer. Clin Cancer Res 2014;20(16):4370–80 doi 10.1158/1078-0432.CCR-14-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Q, Jia Z, Wang L, Kong X, Li Q, Guo K, et al. Disruption of Klf4 in villin-positive gastric progenitor cells promotes formation and progression of tumors of the antrum in mice. Gastroenterology 2012;142(3):531–42 doi 10.1053/j.gastro.2011.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanai M, Wei D, Li Q, Jia Z, Ajani J, Le X, et al. Loss of Kruppel-like factor 4 expression contributes to Sp1 overexpression and human gastric cancer development and progression. Clin Cancer Res 2006;12(21):6395–402 doi 10.1158/1078-0432.CCR-06-1034. [DOI] [PubMed] [Google Scholar]
- 22.Luo T, Masson K, Jaffe JD, Silkworth W, Ross NT, Scherer CA, et al. STK33 kinase inhibitor BRD-8899 has no effect on KRAS-dependent cancer cell viability. Proc Natl Acad Sci U S A 2012;109(8):2860–5 doi 10.1073/pnas.1120589109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong F, Kong X, Du Y, Chen Y, Deng X, Zhu J, et al. STK33 promotes growth and progression of pancreatic cancer as a critical downstream mediator of HIF-1alpha. Cancer Res 2017. doi 10.1158/0008-5472.can-17-0067. [DOI] [PubMed] [Google Scholar]
- 24.Azoitei N, Hoffmann CM, Ellegast JM, Ball CR, Obermayer K, Gossele U, et al. Targeting of KRAS mutant tumors by HSP90 inhibitors involves degradation of STK33. The Journal of experimental medicine 2012;209(4):697–711 doi 10.1084/jem.20111910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei D, Wang L, Yan Y, Jia Z, Gagea M, Li Z, et al. KLF4 Is Essential for Induction of Cellular Identity Change and Acinar-to-Ductal Reprogramming during Early Pancreatic Carcinogenesis. Cancer cell 2016;29(3):324–38 doi 10.1016/j.ccell.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei D, Gong W, Kanai M, Schlunk C, Wang L, Yao JC, et al. Drastic down-regulation of Kruppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res 2005;65(7):2746–54 doi 10.1158/0008-5472.CAN-04-3619. [DOI] [PubMed] [Google Scholar]
- 27.Wei D, Kanai M, Jia Z, Le X, Xie K. Kruppel-like factor 4 induces p27Kip1 expression in and suppresses the growth and metastasis of human pancreatic cancer cells. Cancer Res 2008;68(12):4631–9 doi 10.1158/0008-5472.CAN-07-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan Y, Li Z, Kong X, Jia Z, Zuo X, Gagea M, et al. KLF4-Mediated Suppression of CD44 Signaling Negatively Impacts Pancreatic Cancer Stemness and Metastasis. Cancer Res 2016;76(8):2419–31 doi 10.1158/0008-5472.CAN-15-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun H, Tang H, Xie D, Jia Z, Ma Z, Wei D, et al. Kruppel-like Factor 4 Blocks Hepatocellular Carcinoma Dedifferentiation and Progression through Activation of Hepatocyte Nuclear Factor-6. Clin Cancer Res 2016;22(2):502–12 doi 10.1158/1078-0432.CCR-15-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yori JL, Seachrist DD, Johnson E, Lozada KL, Abdul-Karim FW, Chodosh LA, et al. Kruppel-like factor 4 inhibits tumorigenic progression and metastasis in a mouse model of breast cancer. Neoplasia (New York, NY) 2011;13(7):601–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin ZS, Chu HC, Yen YC, Lewis BC, Chen YW. Kruppel-like factor 4, a tumor suppressor in hepatocellular carcinoma cells reverts epithelial mesenchymal transition by suppressing slug expression. Plos One 2012;7(8):e43593 doi 10.1371/journal.pone.0043593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koopmansch B, Berx G, Foidart JM, Gilles C, Winkler R. Interplay between KLF4 and ZEB2/SIP1 in the regulation of E-cadherin expression. Biochem Biophys Res Commun 2013;431(4):652–7 doi 10.1016/j.bbrc.2013.01.070. [DOI] [PubMed] [Google Scholar]
- 33.Li R, Liang J, Ni S, Zhou T, Qing X, Li H, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 2010;7(1):51–63 doi 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 34.Tiwari N, Meyer-Schaller N, Arnold P, Antoniadis H, Pachkov M, van Nimwegen E, et al. Klf4 is a transcriptional regulator of genes critical for EMT, including Jnk1 (Mapk8). Plos One 2013;8(2):e57329 doi 10.1371/journal.pone.0057329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.