Abstract
tRNA-derived RNA fragments (tRFs) have emerged as a new class of functional RNAs implicated in cancer, metabolic and neurological disorders, and viral infection. Yet our understanding of their biogenesis and functions remains limited. In the present study, through analysis of small RNA profile we have identified a distinct set of tRFs derived from pre-tRNA 3′ trailers in the hepatocellular carcinoma cell line Huh7. Among those tRFs, tRF_U3_1, which is a 19-nucleotide-long chr10.tRNA2-Ser(TGA)-derived trailer, was expressed most abundantly in both Huh7 and cancerous liver tissues, being present primarily in the cytoplasm. We show that genetic loss of tRF_U3_1 does not affect cell growth and it is not involved in Ago2-mediated gene silencing. Using La/SSB knockout Huh7 cell lines, we demonstrate that this nuclear-cytoplasmic shuttling protein directly binds to the 3′ U-tail of tRF_U3_1 and other abundantly expressed trailers and plays a critical role in their stable cytoplasmic accumulation. The pre-tRNA trailer-derived tRFs capable of sequestering the limiting amounts of La/SSB in the cytoplasm rendered cells resistant to various RNA viruses, which usurp La/SSB with RNA chaperone activity for their gene expression. Collectively, our results establish the trailer-derived tRF-La/SSB interface, regulating viral gene expression.
INTRODUCTION
Since the discovery of tRNA degradation products in sera and urine of cancer patients (1,2), numerous tRNA fragments (tRFs) were reported to be present in diverse organisms (3). Recent deep sequencing of small RNA libraries from various human cancer tissues and cell lines has identified diverse types of tRFs derived from precursor tRNAs (pre-tRNAs) and mature tRNAs (4–8). As evidenced by precise terminal sequences of some of those small RNAs (smRNAs), including tRFs [tRFs representing the 5′-end (tRF_5) or 3′-end (tRF_3) region of mature tRNA] and tRNA-derived stress-induced RNAs (also named tiRNAs or tRNA-halves generated by angiogenin-mediated cleavage at the tRNA anticodon region), and by the presence of distinct sets of tRFs in different cell types, these tRFs appear to be produced by controlled cellular processes (9).
Recently, mature tRNA-derived tRFs, which were previously regarded as merely degradation intermediates, have emerged as potential effectors implicated in cancer development, regulation of translation and target gene expression, metabolic and neurological disorders, epigenetic inheritance and ribosome biogenesis (9–17). For instance, some of tiRNAs and tRFs derived from 5′ end of tRNA were shown to possess translation suppressing activity (8,18).
Despite diverse unexpected functions of tRFs uncovered over the last 10 years, their roles in viral life cycle are largely unknown. tRFs derived from the 3′ end of certain mature tRNAs were proposed to act as a primer for reverse transcriptase of T-cell leukemia virus type 1 and to target the primer-binding site of HIV-1 to suppress viral replication (19,20). Wang et al. (21) demonstrated antiviral activity of angiogenin-cleaved tRFs accumulating in respiratory syncytial virus-infected cells. Yet, little is known about the potential roles of pre-tRNA trailers (also called tRF-1 or tRF_U3 family tRFs in this study), which are supposed to be degraded after being released during pre-tRNA processing (13), in viral life cycle.
In the present study, we analyzed the profiles of small RNAs in the human hepatocellular carcinoma (HCC) cell line Huh7 and human liver biopsies and identified various tRFs including a distinct set of pre-tRNA 3′ trailer-derived tRFs. Regulatory functions of the trailer-derived tRFs in viral and cellular gene expression were assessed by molecular biological approaches and further evaluated in virus-infected cells or using target tRNA and La/SSB knockout cell lines. We show that the trailer-derived tRFs negatively regulate La/SSB-dependent viral gene expression by sequestering the RNA chaperone La/SSB. Overall, our results establish the trailer-La/SSB interface, which regulates viral gene expression, and demonstrate a critical role of La/SSB in stabilization of the trailer released from pre-tRNA in the cytoplasm.
MATERIALS AND METHODS
Cell culture
The human hepatocellular carcinoma cell line Huh7, the human cervical carcinoma cell line HeLa, the murine macrophage cell line RAW264.7, human embryonic kidney 293 (HEK293) or 293T (HEK293T), and human colon fibroblast CCD-18Co cells were cultivated in DMEM supplemented with 10% FBS. Normal human primary hepatocytes (HH, #5200) were purchased from ScienCell Research Laboratories (Carlsbad, CA, USA) and cultivated according to manufacturer's recommendations. Human peripheral blood mononuclear cells (hPBMCs) were grown in DMEM supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin.
Small RNA deep sequencing and bioinformatic analysis
The small RNA high throughput sequencing data for human non-cancerous (n = 22) and cancerous (n = 14) human liver cancer tissues, and HCC lines (n = 5) were collected from in-house small RNA libraries and public databases. Small RNA sequences of 16−30 nt from small RNA libraries and human liver biopsies were retrieved after removal of Illumina adapters using the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). The reads (≥ 10 reads) were mapped to the human genome reference sequence (assembly hg19) in the University of California Santa Cruz (UCSC) Genome Browser (22) using the Bowtie (version 1.0.1) software. Short sequencing reads (≥16 nt) from the La PAR-CLIP (photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation) assay datasets (GSE95683) were aligned using the Bowtie software (23) to the tRF_U3 family tRFs with the allowance of two mismatches.
Establishment of tRF_U3_1 and La/SSB knockout cell lines
tRF_U3_1 and La/SSB knockout Huh7 cell lines were generated using the CRISPR-Cas9 genome editing system as described previously (24). Candidate colonies were screened by sequencing and target gene deletion was verified by northern blotting or immunoblotting.
Virus infection
Huh7 cells were infected with cell culture-derived hepatitis C virus (HCVcc) by adsorption for 12 h as previously described (25). Murine norovirus 1 (MNV-1) strain CW1 (kindly provided by Prof. Herbert Virgin IV, Washington University School of Medicine, St. Louis, MO, USA) was propagated in RAW264.7 cells (26). Plaque forming assay for MNV-1 was performed as described previously (27).
Northern blot analysis
Total RNA was subjected to electrophoresis on a 15% polyacrylamide–8 M urea gel and electrically transferred to positively charged nylon membranes (Roche). The membrane was UV-crosslinked and then probed with DNA oligonucleotide probes end-labeled with [γ-32P] ATP using T4 polynucleotide kinase. The sequences of probes used for northern blot analysis are shown in Supplementary Table S1.
Quantification of viral RNAs, mRNAs, and small RNAs
Total RNA was extracted with TRIzol (Invitrogen) and purified according to the manufacturer's procedures. Viral RNA titers and cellular mRNA levels were quantified by real-time RT-qPCR using TaqMan probes and SYBR green, respectively. MicroRNA (miRNA) expression was quantified by TaqMan probe-based real-time PCR assays (Applied Biosystems, Foster, CA, USA) as described previously (28). pre-tRNA trailers, which were recovered by gel elution following electrophoresis of total RNA on a denaturing polyacrylamide gel, were quantified by qPCR following cDNA synthesis using stem-loop primers specific to trailers.
Dual luciferase reporter assay
Bicistronic dual luciferase vector allowing cap-dependent translation of a Renilla luciferase (Rluc) reporter and HCV or poliovirus (PV) internal ribosome entry site (IRES)-mediated translation of a firefly luciferase (Fluc) reporter were transfected into cells. At 18−24 h post-transfection, luciferase activities in cell lysates were quantified using the Dual-Glo luciferase assay system (Promega, Madison, WI, USA).
Ethics statement
Liver biopsy specimens were obtained from the National Biobank of Korea (Pusan National University Hospital, Busan Korea, IRB Approval No. 2011-3) and from the Biobank, Severance Hospital, Yonsei University College of Medicine, Seoul, Korea (IRB Approval Nos. 7001988-201608-BR-239-02E and 7001988-201807-BR-420-01E), and written informed consent was obtained from all patients. The experiment with hPBMCs was approved by the Institutional Review Board of Yonsei University (Approval No. YUIRB-201207-967).
Additional methods
Additional detailed information and experimental procedures are described in online Supplementary Materials and Methods with associated references in the Supplementary Information.
RESULTS
Profile of tRFs derived from pre-tRNA 3′ trailers in the HCC cell line Huh7
By deep-sequencing analysis of smRNAs from the HCC cell line Huh7, we found that tRFs represent 62% of total reads (Figure 1A). Notably, ≥28-nt-long RNA species were identified mainly as tRNA-derived tRFs, whereas the majority of the 20–25-nt-long RNAs identified were miRNA species (Supplementary Figure S1A). Further analysis of the composition of tRFs derived from a selected set of isodecoder tRNA groups (tRNAs that have the same anticodon but variable degrees of sequence variation in other regions) revealed that each isodecoder tRNA group expresses a unique set of tRFs (Figure 1B, top). The majority of identified tRF_5, tRF_3, and i-tRF family tRFs, which are derived from mature tRNA (Supplementary Figure S1B–D), were mapped to multiple tRNA-coding loci (Figure 1B, bottom). In contrast, most tRF_U3 family tRFs, representing pre-tRNA 3′ trailers, were derived from a single genetic locus, thus allowing their unambiguous classification, specific detection by northern blotting or PCR-based analysis, and genetics-based functional studies.
Figure 1.
Profiles of pre-tRNA 3′ trailer-derived tRFs in the HCC cell line Huh7. (A) Pie chart representing the proportion of small RNAs in Huh7 cells (based on average RPM values from the two small RNA datasets GSM3666020 and GSM3666021). mt, mitochondria. (B) Proportion of tRFs (detected in the above-described datasets) derived from the selected isodecoder tRNA groups (top) and number of tRNA-coding genes for the indicated tRFs (bottom). tRFs in Huh7 cells were derived from a total of 54 isodecoder tRNA-coding genes (out of >270 isodecoder genes in human genome). After analysis of the proportion of tRFs for each isodecoder tRNA group, the ones showing > 0.1 in at least one proportion of each tRF family are presented. (C) The trailer-derived tRF_U3 family tRFs (> 100 average RPM values) sorted in descending order according to their read numbers. The enzymes involved in pre-tRNA processing are indicated. (D) Northern blot analysis for tRF_U3_1. Probes specific to the indicated trailers were used for analysis of total RNA (20 μg) from the indicated cell lines and non-cancerous human liver tissues along with synthetic tRF_U3_1 (0.1 ng). (E) Box-and-whisker plots showing tRF_U3_1 levels (read counts normalized using DESeq2) in human liver tissues [NC, normal (n = 22); C, cancer (n = 14)]. Middle bar, median; box, inter-quartile range (25–75%); bars extend to 1.5× the interquartile range. (F) Northern blot and immunoblot analyses of total RNA and proteins from the indicated subcellular fractions derived from an equal number (4.5 × 106) of Huh7 cells. Cellular fractionation was verified by immunoblotting (IB) for the indicated subcellular organelle markers. Nucl, nuclear fraction; Cyto, cytoplasmic fraction. (G) Major tRF_U3_1 isomers. (H) U-tail length variance in the trailer-derived tRFs in Huh7 cells.
Intriguingly, we found that the trailers were originated from a limited number of pre-tRNAs. The reads were derived from only 3.7% of total tRNA genes (625 tRNA-coding loci in the human genome). Among a total of 23 different tRFs with ≥ 10 reads (Supplementary Table S2), only 15 tRF_U3 family tRFs had >100 RPM counts with the tRF_U3_1 (1761 RPM) from chr10.tRNA2-Ser(TGA) being most abundantly detected trailer-derived tRF (Figure 1C). tRF_U3_1 copy number in a single Huh7 cell was 8311 ± 1116 (mean ± SEM), as determined by northern blot analysis (Supplementary Figure S2A). Its expression level was comparable to miR-122 copy number determined by RT-PCR (1.5 × 104 copies/cell) (28) or RNase protection assay (1.6 × 104 copies/cell) (29) in Huh7.
The tRF_U3_1 copy number was only 31% lower (albeit not statistically significant) than that of mature tRNA (10,914 ± 864). Notably, the average ratio of tRF to pre-tRNA was 5.9, suggesting stable maintenance of tRF_U3_1 processed out from its precursor (Supplementary Figure S2A–C). Since the pre-tRNA copy number (1,420 ± 188/cell) was 7.7-fold lower than that of mature tRNA, it was barely detected by the 3′-end region specific probe (probe #1, 18 nt). Nevertheless, by longer exposure of the blot hybridized with the probe #1 or improving detection sensitivity using longer probes (probe #2 and #3; 19 nt and 20 nt, respectively), we could detect pre-tRNA well separated from mature tRNA on a 10% denaturing polyacrylamide gel (Supplementary Figure S2D and E). The ratio of mature tRNA to pre-tRNA (6.9–9.9 with an average ratio of 8.7) was very close to the value estimated using their copy numbers determined with the blots hybridized with the 3′-end probe or the trailer probe (Supplementary Figure S2C). As evidenced by its slower migration than the in vitro transcript of pre-tRNA lacking the 5′-end leader, the detected endogenous pre-tRNA appeared to have both the leader and 3′-end trailer. Pre-tRNA processing intermediates bearing one of these flanking sequences were not detectable, suggesting rapid processing of pre-tRNA. Abundant expression of tRF_U3_1 was similarly observed in Huh7-derived cell lines as well as Huh7 (Supplementary Figure S3A). In the human primary hepatocytes (hPHs) used in this study, both tRF_U3_1 and its pre-tRNA levels were lower than in Huh7, whereas mature tRNA expression was rather increased (Supplementary Figure S3B), as assessed by northern blot analysis.
tRF_U3_1 as well as tRF_U3_2 (628 RPM) from chr10.tRNA6-Val(TAC) and tRF_U3_5 (235 RPM) from chr15.tRNA10-Ser(GCT) could be detected by northern blotting, thus verifying our deep-sequencing results, whereas the less abundantly expressed one tRF_U3_20 (39 RPM) from chr14.tRNA6-Pro(TGG) was below the limit of detection (Supplementary Figure S3C). These trailer-derived tRFs along with their precursors were not detectable in the CCD-18Co normal human colon fibroblasts, while the level of mature tRNA-Ser(TGA) was similar to that observed in Huh7 cells (Figure 1D, left). tRF_U3_1 was undetectable in non-cancerous human liver tissues (Figure 1D, right), as assessed by northern blot analysis.
Through analysis of small RNA datasets for human liver [non-cancerous (n = 22) and cancerous (n = 14) liver tissues] (Supplementary Table S3), we observed significantly increased tRF_U3_1 levels (5.32 log2-fold increase, Padj = 2.34E–04) in human liver cancer tissues (Figure 1E; see also Supplementary Table S4). We also observed multiple miRNAs (miR-221-3p, miR-200c-3p, miR-215-5p and miR-203a) upregulated >3 log2-fold in these cancer tissues (Supplementary Figure S3D). Of these miRNAs found to be upregulated in liver cancer tissues, miR-221-3p is widely accepted as a liver cancer miRNA marker (30). tRF_U3_1 was the only tRF_U3 family tRF differentially expressed with a baseMean value of >1000 (Supplementary Table S4), indicating that this trailer constitutes a unique small RNA marker abundantly present in liver cancer cells. Notably, tRF_U3_1 was found to be present in both the cytoplasm and the nucleus along with its precursor, albeit more abundantly in the former compartment (Figure 1F). Similarly, we also observed predominant cytoplasmic localization of the second most abundant trailer, tRF_U3_2, in Huh7 cells (Supplementary Figure S3E).
Sequence profile of pre-tRNA-Ser(TGA) 3′ trailer-derived tRFs
Although the mature sequences of tRNAs with a Ser(TGA) anticodon are highly conserved in humans and mice, their trailer sequences are variable in sequence and length (Supplementary Table S5). Even from a total of 32 tRNA-Ser species encoded by human chromosomes sequences (Supplementary Table S6), only 3 tRNA-Ser species from chr10.tRNA2-Ser(TGA), chr15.tRNA10-Ser(GCT), and chr17.tRNA7-Ser(GCT) loci yielded trailer-derived tRFs, tRF_U3_1, tRF_U3_5 and tRF_U3_16, respectively, which all have unique sequences.
The 3′-end of trailer sequences would have an oligo(U)-tail of variable length depending on RNA polymerase III termination efficiency at a short oligo(dT) tract on the non-template strand of the DNA. We found that in addition to tRF_U3_1, its two major isomers bearing UUUU-3′OH or UU-3′OH at their 3′-ends were also produced from the chr10.tRNA2-Ser(TGA) (Figure 1G), which has the T6 RNA polymerase III termination signal. Further isomer profile analysis for the 23 different tRF_U3 family tRFs (with ≥10 reads) revealed that the majority of abundant tRF_U3 family tRFs carries 3-4U 3′-end tails (Figure 1H and Supplementary Figure S3F).
La/SSB binds and stabilizes tRF_U3_1
Considering that tRF_U3_1, unlike other tRFs, is derived from a unique genetic locus and detected most abundantly, this trailer-derived tRF was chosen for further analysis. First, we performed affinity chromatography experiments using 5′-biotinylated tRF_U3_1 immobilized onto streptavidin beads to address whether tRF_U3_1 exerts its function by forming an active complex with cellular proteins. We detected a protein with an apparent molecular mass of ∼50-kDa specifically pulled-down by the tRF_U3_1 (Figure 2A). The bound protein was identified as the La/SSB (Sjögren syndrome antigen B, also known as LARP3 and La autoantigen) (31,32) by nano LC-MS/MS analysis (Supplementary Figure S4A).
Figure 2.
Identification of La/SSB as a tRF_U3_1-interacting protein. (A) Cellular proteins interacting with tRF_U3_1 were pulled down from Huh7 cell lysates using 5′-biotin-labeled tRF_U3_1 immobilized onto streptavidin beads, resolved by SDS-PAGE, and visualized by Coomassie blue staining. (B) A PAR-CLIP experiment was carried out to verify the specific interaction of tRF_U3_1 with La/SSB in the Huh7-derived cell line R-1, in which an HCV subgenomic replicon stably replicates. From the La/SSB crosslinked to RNA (marked with an arrowhead), RNA was recovered for TaqMan RT-qPCR quantification of the indicated small RNAs. ND, not detected. (C) Interaction between tRF_U3_1 and La/SSB assessed by a pull-down assay followed by immunoblotting analysis. Pull-down experiments were performed in the absence (−) or presence (+) of the indicated competitors (10-fold molar excess of 5′-biotin-labeled tRF_U3_1). (D) EMSA with radiolabeled tRF_U3_1 (0.1 nM) and La/SSB protein (100 nM) in the presence or absence of the indicated competitors (10 nM). C, complex; P, probe. (E) Detection of tRF_U3_1 in HEK293 cells by northern blotting. (F and G) Bioinformatic analysis of La/SSB-bound tRF_U3 family tRFs identified by PAR-CLIP experiments. Proportion of Flag-hLa-crosslinked tRF_U3 family tRFs in HEK293 cells was analyzed using the dataset from the small RNA cDNA library generated after digestion of non-crosslinked RNA with RNase A (GSM2521600). Shown in (F) are the tRF_U3 family tRFs, which occupy >2% in the proportion of the dataset, sorted in descending order according to their read numbers. Proportion of T-to-C conversion at each nucleotide of tRF_U3_1 reads is shown in (G).
We next confirmed this specific interaction between tRF_U3_1 and La/SSB in cells using a photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) assay (33) (Figure 2B). tRF_U3_1 was abundantly detected in the UV-crosslinked RNA-La/SSB complexes, whereas miR-122, a miRNA with highest abundance in human liver tissue (28) and three other miRNAs (miR-221, miR-23b and miR-26a) were not found in the complexes, indicating that tRF_U3_1 interacts with La/SSB with a high degree of specificity. We further confirmed the interaction between tRF_U3_1 and La/SSB by a pull-down experiment using a biotinylated tRF_U3_1 (Figure 2C). This interaction was inhibited by the addition of tRF_U3_1, whereas its variant tRF_U3_1_MT(3′-CC) carrying a cytidylate dinucleotide substitution for the 3′-terminal two uridylate residues failed to compete with tRF_U3_1 for interaction with La/SSB. Similar results were observed in a competitive electrophoretic mobility shift assay (EMSA). The tRF_U3_1-La/SSB interaction was not blocked by tRF_U3_1_MT(3′-CC) (Figure 2D). In contrast, tRF_U3_1 and the two other tRF_U3 family tRFs, tRF_U3_2 and tRF_U3_5, which have different sequences upstream of the 3′-U tail, effectively disrupted the interaction. These results together demonstrate the importance of 3′-end uridylate residues in the interaction between tRF_U3_1 and La/SSB.
Using a similar PAR-CLIP assay in HEK293 cells transiently expressing the Flag-tagged La/SSB, Gogakos et al. (34) have recently identified several La/SSB-associated RNAs. Following detection of tRF_U3_1 in this cell line (Figure 2E), we subsequently analyzed the deep-sequencing datasets generated by Gogakos et al. (34) and found that tRF_U3_1 comprises the most abundant tRF_U3 family tRFs among the reads in the dataset derived from the RNaseA-treated La/SSB-RNA crosslinked sample (Figure 2F). Moreover, in the reads matching to the tRF_U3_1, the T-to-C conversion, which is a hallmark of crosslinking between the thio-uridine-labeled RNA and the target protein, was found to occur in the U-rich 3′-end region of tRF_U3_1 (Figure 2G), providing additional evidence for the direct interaction of tRF_U3_1 with La/SSB.
Interestingly, some of these reads were even mapped to the region spanning from 65 to 82 nt, downstream of the RNase Z-mediated pre-tRNA processing site [83th nt of chr10.tRNA2-Ser(TGA)] (Supplementary Figure S4B). These reads, mainly starting from the 73rd or 75th nt, appeared to be generated by RNase A-digestion of non-protected RNA (Supplementary Figure 4C). All of these reads were in fact extended over the RNase Z-cleavage site, suggesting that La/SSB binding to this region might influence processing or its binding to this region might be transient. The high proportion of T-to-C conversion-bearing reads (at +75 and +79 nt) further supported the possibility of direct binding of La/SSB to this region (Supplementary Figure S4D). Notably, majority of the La/SSB-crosslinked RNA species were exactly 5′ co-terminal at nt 83 (G), indicating that these reads represent the free trailer that was either bound to La/SSB after processing or released from pre-tRNA as a La/SSB-bound form.
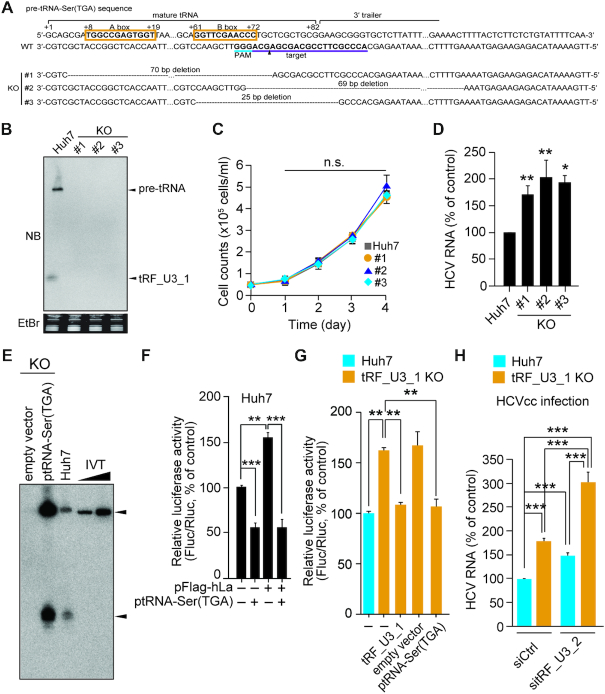
Figure 4.
Impact of tRF_U3_1 knockout on cell proliferation and HCV infection. (A) CRISPR-Cas9-mediated generation of tRF_U3_1 KO Huh7 cell lines. tRNA sequences were numbered according to the standardized numbering system (53). The sequences in bold type in the orange box are the A and B boxes that correspond to internal promoter regions of RNA polymerase III. The gRNA targeting sequence (purple) and protospacer adjacent motif (PAM, sky blue) are underlined. The putative cleavage site is indicated by an arrowhead. (B) Northern blot analysis for tRF_U3_1 in the indicated KO cell lines. (C) Growth curves of the tRF_U3_1 KO cell lines and its parental cell line Huh7. (D) RT-PCR quantification of HCV genome titers at two days post-infection of Huh7 and tRF_U3_1 KO cell lines with HCV. (E) Rescue of tRNA-Ser(TGA) expression was verified by northern blotting analysis of total RNA (20 μg) from the tRF_U3_1 KO cell line #1 ectopically expressing tRNA-Ser(TGA) precursor. IVT, in vitro transcript (100 and 500 pg) of tRNA-Ser(TGA) carrying its 3′ trailer sequence. (F and G) Effect of pre-tRNA-Ser(TGA) overexpression or synthetic tRF_U3_1 (100 nM) transfection on HCV IRES activity was assessed as described in Figure 3C at 24 h post-transfection of the dual-luciferase reporter plasmid into Huh7 or tRF_U3_1 KO cells. (H) Effect of tRF_U3_2 depletion by RNAi was assessed at two days post-infection of Huh7 or tRF_U3_1 KO cells with HCV. Where shown, error bars are standard deviations of at least three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; n.s., not significant; by unpaired two-tailed Student's t-test.
More direct experimental evidence of robust interaction between tRF_U3_1 and La/SSB was obtained by an RNA immunoprecipitation experiment wherein RNA bound to La/SSB was analyzed by northern blotting. While both the free trailer tRF_U3_1 and its precursor were coimmunoprecipitated with La protein, the copy number ratio of tRF_U3_1 to pre-tRNA in the immunoprecipitated La protein increased 2.4-fold compared to that in the input RNA in Huh7 cell lysates (9.9 in the La IP versus 4.1 in the input), suggesting La binding was significantly biased to the free trailer (Supplementary Figure S5). The mature tRNA lacking the trailer was not detected in the immunoprecipitated La/SSB, demonstrating specific binding of La protein to pre-tRNA and tRF.
Finally, as shown in Supplementary Figure S6A, tRF_U3_1 did not interact with Ago2. Analysis of smRNAs in Argonaute (Ago2) immunoprecipitates also revealed that tRF_U3_1 was not selectively bound by Ago2 (Supplementary Figure S6B). Furthermore, its miRNA-like function was not observed toward various mRNAs that carry a potential binding site for tRF_U3_1 (Supplementary Figure S6C–F).
tRF_U3_1 negatively regulates HCV IRES-mediated translation
The RNA chaperone La/SSB was previously demonstrated to bind to the stem-loop IV of HCV internal ribosome entry site (IRES) to promote IRES-mediated translation initiation (35). We tested whether tRF_U3_1 interferes with the La/SSB–HCV IRES interaction. The results of competitive EMSAs showed that tRF_U3_1, but not miR-122-5p, inhibits La/SSB binding to HCV IRES (Figure 3A; compare lanes 4 and 5 and lanes 6 and 7). We then tested its impact on HCV IRES-mediated translation using a dual-luciferase reporter assay system. All assays with tRF_U3_1 were performed at ≤ 100 nM, in which <10% reduction in cell viability was observed in HCV-replicating cells (R-1 cell line harboring an HCV subgenomic replicon in Huh7) and CCD-18Co cells (Figure 3B). As shown in Figure 3C, tRF_U3_1, but not miR-122-5p, significantly suppressed HCV IRES-mediated translation. Similar degrees of HCV translation inhibition were also obtained with two other 3′-U-tailed pre-tRNA trailer-derived tRFs (tRF_U3_2 and tRF_U3_5), which also interacted with La/SSB (Figure 2D). We also found that tRF_U3_1(3′-4U), an isomer of tRF_U3_1 bearing four uridylate residues at its 3′-end, exhibited inhibitory activity similar to that of tRF_U3_1, whereas tRF_U3_1_MT(3′-CC), which does not bind to La/SSB, and tRF-3002, which lacks the 3′ U-tail (Supplementary Table S7), had no effect on IRES-mediated translation (Figure 3D). These results together highlighted the important role of the 3′ U-tail of tRF_U3 family tRFs in regulating HCV IRES activity. Further, the importance of terminal two U residues was demonstrated by showing the inhibitory activity using tRF_U3_1(3′-UU), a tRF_U3_1 derivative with two U residues at its 3′-end (Supplementary Figure S7). In contrast, as_IRES (an antisense RNA oligonucleotide targeting the pseudoknot I region of HCV IRES), tRF-5001 (a tRF_5 family tRF), and tRF_U3_1(ΔU17–19) (a tRF_U3_1 derivative), all of which lack the U-tail at their 3′-ends, failed to suppress the IRES-mediated translation.
Figure 3.
Negative regulation of La/SSB-dependent HCV IRES-mediated translation by pre-tRNA 3′ trailer-derived tRFs. (A) EMSA was performed with radiolabeled HCV IRES RNA (nt 1-361 of HCV 5′-UTR) and La/SSB protein in the absence or presence of the indicated competitor RNAs. miR-122-5p, guide strand of miR-122 used as a single-strand RNA control. C, complex; P, probe. (B) Cell viability was measured by MTS assay at 2 days post-transfection of tRF_U3_1 into CCD-18Co or R-1 cells. (C) Effect of tRF_U3_1 and other trailer-derived tRFs or miR-122-5p (0.1 nM each) on HCV IRES-mediated translation in CCD-18Co cells transfected with a dual-luciferase reporter plasmid (top). Shown are the relative firefly luciferase (Fluc) and Renilla luciferase (Rluc) activities representing the IRES- and cap-dependent translation, respectively. (D) Effect of tRF_U3_1 derivatives on HCV IRES activity was assessed as in (C). (E and F) RT-PCR quantification of HCV RNA titers at two days post-infection of human primary hepatocytes (hPHs) or Huh7 cells with HCVcc (E). In (F), infected hPHs were transfected with the indicated tRFs. (G) IFN-β promoter activation activity of tRF_U3_1 was assessed in Huh7 cells transfected with an IFN-β promoter assay plasmid and the pRL-TK plasmid used for normalization of transfection efficiency. The transfected cells were treated with tRF_U3_1 for 24 h prior to determination of relative Fluc/Rluc activity. IRF3_5D, a plasmid expressing a constitutively active IRF3_5D used as a control. 5′ppp-tRF_U3_1, tRF_U3_1 carrying 5′-triphosphate. (H) IFN-β mRNA levels in HEK293T cells transfected with tRF_U3_1 or two other IFN-inducing RNA ligands (1 μg/ml). Shown are the GAPDH-normalized IFN-β mRNA levels determined at 24 h post-transfection, with PCR-amplified products visualized by EtBr staining. (I) IFN-α levels in hPBMCs transfected with trailer-derived tRFs (1 μM) or HCV 3′-UTR (5 μg/ml) were measured by ELISA at 16 h post-transfection. In (B–H), error bars are standard deviations from three independent experiments. Data in (I) are presented as mean ± SD, n = 3 technical replicates. Statistical significance of difference between groups was determined via unpaired two-tailed Student's t-test. *P < 0.05; **P < 0.01; ***P < 0.001; n.s., not significant; ND, not detected.
Huh7 cells were less competent in supporting HCV propagation than hPHs (Figure 3E), possibly due to the presence of numerous HCV IRES-inhibiting tRF_U3 family tRFs in Huh7. In fact, northern blot analysis showed that both tRF_U3_1 and its pre-tRNA levels in hPHs were substantially lower than those in Huh7, while mature tRNA level was increased (Supplementary Figure S3B). Accordingly, tRF_U3_1 transfection indeed resulted in a significant reduction in HCV RNA titers in HCVcc-infected hPHs (Figure 3F).
The inhibitory activity of tRF_U3_1 was further validated in the R-1 cells where HCV subgenomic replicon constitutively replicates. The results showed that tRF_U3_1, but not single-stranded miR-122-5p, substantially reduced HCV RNA abundance (53 ± 1% of untreated control) and consequently the abundance of HCV nonstructural proteins NS3 and NS5B (Supplementary Figure S8A). The duplex form of miR-122 increased subgenomic RNA levels, which was consistent with previous findings (36). The antiviral activity of tRF_U3_1 against HCV was attenuated specifically by a tRF_U3_1-targeting antisense peptide nucleic acid (PNA), whereas the PNA-SL3-17 targeting the HCV X-RNA at the 3′-end of the viral genome (37) decreased HCV subgenomic replicon RNA and NS5B protein levels (Supplementary Figure S8B).
To ensure that the inhibitory effect of tRF_U3_1 was not attributed to induction of type I interferon (IFN) production, we assessed whether it activates the IFN-β promoter using a reporter responding to IRF3, a transcription factor inducing IFN-β production. The results shown in Figure 3G indicate that the 5′-monophosphate form tRF_U3_1 has no intrinsic IFN-β-inducing nature. By contrast, its 5′-triphosphate form, which is expected to be recognized by RIG-I, a cytoplasmic pattern recognition receptor for 5′-triphosphate single strand RNA (38,39), and the constitutively active IRF3, IRF3_5D used as a positive control increased the reporter expression. Similarly, there was no upregulation of IFN-β mRNA by tRF_U3_1 in HEK293T cells (Figure 3H). Furthermore, we observed no substantial increases in IFN-α levels in hPBMCs in response to tRF_U3_1 or another tRF_U3 family tRF tRF_U3_16 (Figure 3I). These observations altogether indicated that the HCV inhibitory activity of the pre-tRNA trailer-derived tRFs was not attributed to IFN pathway activation.
tRF_U3_1 modulates La/SSB-dependent viral gene expression
To specifically determine to what extent HCV IRES-mediated translation is suppressed by the tRF_U3_1 alone, we generated tRF_U3_1 knockout (KO) Huh7 cell lines using the CRISPR-Cas9 system. In the three established KO cell lines with deletions in the tRNA-Ser(TGA) gene (Figure 4A), depletion of tRF_U3_1 and its precursor was verified by northern blotting using a probe complementary to tRF_U3_1 (Figure 4B). Further northern blotting analyses using probes targeting the 3′-end and 5′-end regions of mature tRNA-Ser(TGA) also showed no expression of mature form of tRNA-Ser(TGA) in the KO cell lines. Of note, the 5′-end targeting probe detected other tRNA-Ser(TGA) isodecoders due to their sequence similarity at the 5′-end region (Supplementary Figure S9A and B). These tRNA-Ser(TGA) (chr10.tRNA2) KO cell lines displayed a growth rate similar to that of the parental cell line Huh7 (Figure 4C), suggesting that accumulation or loss tRF_U3_1 alone has no effect on cell proliferation. In these tRF_U3_1 KO cell lines, HCV RNA titers increased significantly, confirming the HCV inhibitory activity of tRF_U3_1 (Figure 4D).
By using a vector expressing pre-tRNA-Ser(TGA), which was thus capable of rescuing tRF_U3_1 and its precursor biogenesis (Figure 4E), we further verified the HCV IRES regulatory activity of tRF_U3_1. As shown in Figure 4F, ectopic expression of pre-tRNA-Ser(TGA) under the control of its own RNA polymerase III promoter suppressed HCV IRES activity in both Huh7 and La/SSB-overexpressed Huh7 cells. As expected, tRF_U3_1 KO significantly increased HCV IRES activity, which could be reduced to the levels in Huh7 cells by synthetic tRF_U3_1 transfection or by ectopic expression of the tRNA-Ser(TGA) precursor (Figure 4G). Notably, depletion of tRF_U3_2 by RNAi in this KO cell line (Supplementary Figure S9C–E) further increased HCV RNA titers (Figure 4H).
We also found that tRF_U3_1 displayed a similar inhibitory effect on poliovirus (PV) IRES. La/SSB overexpression in HeLa cells, a susceptible host of PV, led to enhanced PV IRES activity, which was in turn significantly repressed upon ectopic expression of tRF_U3_1 using a tRNA-Ser(TGA) expression vector or by tRF_U3_1 transfection (Figure 5A). tRF_U3_1 expression in the tRF_U3_1 KO Huh7 cells also led to a significant reduction in the IRES-activity (Figure 5B). These results are in agreement with previous reports demonstrating the La/SSB-dependent PV IRES-mediated translation in rabbit reticulocyte lysates or La/SSB-depleted HeLa cells (40,41). Moreover, tRF_U3_1 exerted its antiviral activity against norovirus whose RNA genome interacts with La/SSB at its 5′ and 3′ ends (42). In the mouse macrophage cell line RAW264.7, which lacks this tRF (as the mouse genome does not have the corresponding tRNA-coding gene), both tRF_U3_1 and its isomer tRF_U3_1(3′-4U) significantly reduced intracellular viral RNA and the VP1 (major capsid protein) levels as well as infectious virus titers (Figure 5C and D). Collectively, our results highlight an unprecedented regulatory function of the trailer-derived tRFs in La/SSB-dependent viral gene expression.
Figure 5.
Inhibition of PV IRES-mediated translation and norovirus propagation by tRF_U3_1. (A and B) HeLa (A) or tRF_U3_1 KO Huh7 (B) cells were transfected with a dual luciferase reporter plasmid to monitor PV IRES-mediated translation at 6 h post-transfection of ptRNA-Ser(TGA) or tRF_U3_1 (100 nM), with or without pFlag-hLa. Reporter activity was determined as described in Figure 4F. (C and D) RAW264.7 cells were infected with mouse norovirus (MNV-1) at a multiplicity of infection of 0.005 and transfected with the indicated tRFs (100 nM). After 36 h, intracellular viral genome copy number (C) and infectious virus titer in culture media (D) were determined by RT-qPCR and plaque-forming assay, respectively. VP1, viral capsid protein. In all panels, error bars are standard deviations of three independent experiments. P values were calculated by unpaired two-tailed Student's t-test. **P < 0.01; ***P < 0.001; n.s., not significant.
Functional interplay between tRF_U3_1 and La/SSB
To further explore the functional implication of tRF_U3_1 binding to La/SSB, we used La/SSB KO Huh7 cell lines established using the CRISPR-Cas9 system (Supplementary Figure S10A). The two independently isolated La/SSB KO cell lines showed impaired growth kinetics (approximately 3-fold decreases in growth rate) (Supplementary Figure S10B), suggesting the critical albeit not indispensable role of La/SSB in cells. Surprisingly, northern blot analysis revealed that tRF_U3_1 accumulation was completely abolished in the absence of La/SSB (Figure 6A), which was accompanied with reduced levels of its precursor and mature tRNA, whereas miR-122 level remained unchanged. Similar results were also observed for two other trailer-derived tRFs, tRF_U3_2 and tRF_U3_5 (Supplementary Figure S11A and B). Transient depletion of La/SSB by RNAi also resulted in a substantial decrease of tRF_U3_1 level (Supplementary Figure S12A). Furthermore, an in vitro decay assay showed that tRF_U3_1 decayed faster in La/SSB-depleted Huh7 lysates (Supplementary Figure S12B). These results suggested that trailer-derived tRFs are stabilized by their interaction with La/SSB. This hypothesis was supported by our findings that tRF_U3_1, which was barely detectable even when its precursor was overexpressed in La/SSB KO cell lines, became accumulated to detectable levels upon rescue of La/SSB expression (Figure 6B). Similarly, ectopic expression of La/SSB in Huh7 cells induced a marked increase in tRF_U3_1 level. A similar stabilization effect was also observed upon La overexpression in the Huh7-derived tRF_U3_1 KO cell line transiently expressing pre-tRNA-Ser(TGA) (Supplementary Figure S13A–C). As in Huh7, both tRF_U3_1 and its precursor levels were increased by ectopic expression of the Flag-tagged human La protein. tRF_U3_1 bound more preferentially to the La protein than its precursor (Supplementary Figure S13D), an observation which was consistent with the results from Huh7 (Supplementary Figure S5),
Figure 6.
Effect of La/SSB knockout on cytoplasmic accumulation of the trailers and HCV and PV IRES-mediated translation. (A and B) Northern blot analysis for tRF_U3_1 and its precursor in La/SSB KO cell lines and their parental cell line Huh7 using the indicated probes. MiR-122 was used as a loading control (A). In (B), cells were transfected with plasmids to express hLa and/or pre-tRNA-Ser(TGA) prior to northern blot analysis of total RNA resolved by PAGE on a small (10 × 10-cm)- or medium (20 × 20-cm; bottom panels)-size denaturing acrylamide gel. (C) (Top) Schematic diagram showing the NLS and the three RNA-binding motifs of La/SSB, including the highly conserved La/SSB motif (LaM), the canonical RNA recognition motif (RRM1), and the atypical RRM2 motif. La/SSB KO cells were transfected with a plasmid expressing pre-tRNA-Ser(TGA) along with expression vectors for the indicated La proteins prior to northern blot and immunoblot analyses at 2 days post-transfection. (D) Confocal microscopy for the indicated hLa proteins ectopically expressed in La/SSB KO Huh7 cells. DAPI, nuclear staining. Scale bar, 20 μm. (E and F) Effect of ectopic expression of tRF_U3_1 on HCV IRES (E) or PV IRES (F)-mediated translation in La/SSB KO cell lines transiently expressing hLa. (G) Reduction of HCV RNA titers in La/SSB knockout Huh7 cell lines. HCV RNA titer was determined by RT-qPCR at two days post-infection. Where shown, error bars are standard deviations of at least three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; n.s., not significant; by unpaired two-tailed Student's t-test.
Ectopic expression of pre-tRNA-Ser(TGA) in these KO cell lines allowed us to detect both the precursor, which bears the trailer (tRF_U3_1) and possibly the 3-nt long 5′-end leader, and the mature tRNA lacking these parts (see the blots hybridized with the tRF_3 family-specific probe binding to the 3′-end region of mature tRNA). Strikingly, northern blot analysis revealed that pre-tRNA processing to the mature form could occur irrespective of the presence of La/SSB (compare lane 2 with lanes 6 and 10). Even in the absence of La/SSB, removal of the 5′-end leader and 3′-end trailer appeared not to be affected as evidenced by no detectable accumulation of pre-tRNA processing intermediates bearing one of these flanking sequences.
Next, we used a La/SSB derivative with a deletion of amino acids 375–408 including the nuclear localization signal (NLS) (Figure 6C, top) to ask whether this derivative, which localized only in the cytoplasm unlike the wild-type La/SSB predominantly residing in the nucleus (Figure 6D), can rescue tRF_U3_1 accumulation in La/SSB KO cells. The results showed that tRF_U3_1 became detectable following expression of the cytoplasm-residing La/SSB (Figure 6C, northern blot). Our results suggest that the fate and functions of the trailer-derived tRFs can be controlled by cytoplasmic abundance of La/SSB. In fact, we noticed that the inhibitory activity of tRF_U3_1 on HCV IRES disappeared in the absence of La/SSB (Figure 6E). Importantly, its inhibitory activity in the La/SSB KO cell lines was restored upon ectopic expression of La/SSB. Similar La/SSB-dependent regulatory function of tRF_U3_1 was observed with PV IRES as well (Figure 6F). In addition, as observed following ectopic expression of pre-tRNA-Ser(TGA) generating both tRF_U3_1 and its pre-tRNA, PV IRES-mediated translation enhanced by hLa expression was suppressed by synthetic tRF_U3_1 transfected into the La/SSB KO cells (Supplementary Figure S14). These results further support our conclusion that tRF_U3_1 inhibits HCV IRES-mediated translation by sequestering the limiting amounts of La/SSB available in the cytoplasm. Finally, taking advantage of these La/SSB KO cell lines, we were able to provide robust genetic evidence supporting the proviral activity of this IRES-trans-acting factor (Figure 6G).
DISCUSSION
Despite the accumulating evidence for the presence of diverse tRFs in cells, their regulatory roles in gene expression is just beginning to emerge. In particular, potential roles of pre-tRNA-derived tRFs during the course of viral infection are largely unknown. Unlike other tRFs representing various portions of mature tRNA, the trailer-derived tRFs, even the ones from isodecoder tRNAs, unambiguously map to specific genetic loci due to their sequence uniqueness. In addition, these trailer sequences, unlike tRFs derived from mature tRNAs, can be correctly annotated to corresponding pre-tRNA-coding genes because base modifications, which occur in the D-loop or TψC loop and thereby can introduce unwanted mutations during cDNA synthesis, are not present on this tail part of pre-tRNA (13). Based on these properties, we focused our investigation on the trailers highly abundant in liver cancer cells. Our studies show for the first time that a unique array of trailers are stabilized by La/SSB binding to their 3′ oligo(U) tract and negatively regulate La/SSB-dependent viral gene expression.
Our genetic and biochemical studies with tRF_U3_1 and La/SSB KO cell lines showed that the trailer-derived tRFs, which accumulate in the cytoplasm by yet an unknown mechanism, exert their antiviral activity by sequestering limiting amounts of cytoplasmic La/SSB from cis-acting RNA elements on non-coding regions of viral genomes, which are required for viral gene expression and/or replication. Our results recapitulated the positive regulatory role of La/SSB in HCV and PV IRES-mediated translation (35,43,44). We also demonstrated that tRF_U3_1 and its isomers have the potential to inhibit the propagation of norovirus. Thus, the inhibitory activity of trailer-derived tRFs is not likely limited to IRES-mediated viral gene expression. Rather, these tRFs might control various viruses that exploit La/SSB as a proviral factor. For example, La/SSB binding to a specific hepatitis B virus (HBV) cis-acting element was shown to enhance HBV RNA stability (45). For HIV, trans-activation response element (TAR) RNA-La/SSB complex formation was shown to relieve translational repression by the 5′ leader sequence of the TAR (46). Hence, pre-tRNA trailer-derived tRFs with La/SSB-sequestering activity may constitute an additional molecular network that regulates various steps in the life cycles of viruses.
Nowadays, it is widely accepted that La/SSB helps to ensure proper folding of pre-tRNA by binding to the 3′-end trailer and 5′-end region that facilitates tRNA processing in yeast (47,48). It may also act as a transcription termination factor for RNA polymerase III (49). However, crucial genetics studies on the roles of La/SSB in mammalian cells are still lacking. Although La/SSB mice were embryonic lethal (50), we could successfully establish the La/SSB KO Huh7 cell lines. Surprisingly, using these cell lines we found that pre-tRNA processing to mature tRNA can proceed independently of La/SSB. These unexpected data raise a possibility that other La/SSB-related proteins may also participate in pre-tRNA processing. The KO cell lines also enabled us to provide for the first time the genetic evidence for the proviral role of La/SSB in HCV IRES-mediated translation. Our results also show that La/SSB has an unappreciated role in stabilizing a set of trailers derived from the pre-tRNAs exported to the cytoplasm by an as yet unknown transport mechanism.
One such trailer tRF_U3_1, as well as its precursor, was more abundant in liver cancer cells as well as in Huh7 compared to normal liver tissues and hPHs. Steady state expression levels of these RNAs can be controlled by pre-tRNA transcription and processing capacity and their stabilization pathways. Mature tRNA levels in noncancerous liver tissues were slightly higher than in Huh7 cells whereas pre-tRNA, which was much less than mature tRNA and thus absent normally, was barely detectable in Huh7 cells. Consequently, the sum of pre-tRNA and mature tRNA amount appeared similar in Huh7 and non-cancerous human liver tissues or hPHs. Thus, it is unlikely that pre-tRNA-Ser(TGA) transcription upregulation is largely responsible for tRF_U3_1 and its precursor accumulation in liver cancer cells. It is plausible that elevation of pre-tRNA level in Huh7 might be instigated by slightly debilitated pre-tRNA processing and/or through its stabilization. When pre-tRNA processing becomes a bottle neck in tRNA maturation process possibly due to overall pre-tRNA overloading in rapidly growing cancer cells, La/SSB-binding to nascent pre-tRNA would prevent its degradation and might help its proper folding until the trailer separation occurs.
We found that the free trailer tRF_U3_1 was more preferentially associated with endogenous La/SSB than its precursor in Huh7 cells. In both Huh7 and pre-tRNA expressed tRF KO cell line, the tRF/pre-tRNA ratio increased (2.4-fold in Huh7 and 1.7-fold in the pre-tRNA overexpressed cells) in immunoprecipitated La protein compared to that in the inputs (Supplementary Figures S5 and S13). Such an increased ratio may reflect tRF_U3_1’s higher affinity to the La protein. Alternatively, this might be due to steric hindrance of La binding imposed by tRNA processing proteins such as RNase P and RNase Z (47,48). The molecular mechanism(s) for this free trailer-binding preference of La/SSB is unclear. Also, it remains an open question whether such a preference binding holds for other trailer tRFs. Given that La/SSB mainly resides in the nucleus, its use as a trailer tRF-stabilizing factor in the cytoplasm would be modulated by its abundance and subcellular localization in liver cancer cells. It is also plausible that besides La/SSB, an active stabilization pathway might be programmed in liver cancer cells for cytoplasmic accumulation of trailer tRF, which may require additional factors as yet undiscovered (Supplementary Figure S15).
Absence of mature tRNA in the La immunoprecipitates suggests that the T-to-C conversion sites in the 3′-end region of mature tRNA, which were detected in the La-crosslinked reads (Supplementary Figure S4C), might be the secondary contact sites of La where it transiently binds following recognition of the trailer. It appears that the 3′-end region alone is not sufficient for high affinity La binding. Because La binds more preferentially to tRF_U3_1 than to its precursor, there must be a mechanism for clearing La from the tRF separated from pre-tRNA and recycling into the pre-tRNA processing in the nucleus as well as in the cytoplasm.
Previously, tRF_U3_1 was also identified, albeit with different names, in various human transformed cell lines [tRF-1001 from prostate cancer cell lines (5), 3′-trailer of tRNA-Ser(TGA) from the human nasopharyngeal carcinoma 5–8F cell line (7), Cand45 from HEK293 cells (6), and Eym65 from the clonal lymphoma cell line BCP-1 (4)]. Aside from its impact on viral gene expression through a direct interaction with La/SSB, tRF_U3_1 abundantly expressed in various cell types may alter miRNA expression by binding and inhibiting La/SSB, because La/SSB is known to facilitate the biogenesis of miRNAs by binding to pre-miRNA stem-loop structures (32). Over the last 10 years, numerous studies have demonstrated regulatory roles of miRNAs in virus replication and pathogenesis (51). Thus, trailer-derived tRFs have the potential to control viral life cycle through regulation of miRNA expression. Further study will be required to characterize the indirect and/or direct interplays between trailer-derived tRFs and miRNAs. Finally, given that there would be several potential trailer-binding targets in host RNAs, these small RNAs may also be implicated in post-transcriptional regulation of cellular gene expression. Previously, some of tRF_3 or tRF_5 family tRFs were shown to be associated with various Ago proteins (17,52). tRF_U3_1, however, did not bind Ago2, and consequently did not display regulatory activity through the miRNA pathway (Supplementary Figure S6). Nevertheless, it remains to be explored whether other trailer-derived tRFs can participate in this regulatory pathway differently from tRF_U3_1.
As illustrated in Supplementary Figure S15, several unanswered questions remain. First, it is important to identify the mechanisms by which pre-tRNA is exported to the cytoplasm. While we showed that the two abundantly observed trailer tRFs, tRF_U3_1 and tRF_U3_2 are mainly present in the cytoplasm of Huh7, it is not clear how a selected set of pre-tRNAs are exported to the cytoplasm to dysregulate La/SSB per se and/or by the free trailers processed out from them. Second, it remains to be elucidated if there is an as yet undiscovered trailer stabilizing pathways besides La/SSB in liver cancer cells. Third, since our finding of the dispensable role of La/SSB in pre-tRNA processing does not reproduce the main function of this RNA chaperone, another future challenge is to track alternative pathways of tRNA biogenesis and their roles in various physiological and pathological conditions. Despite these unanswered questions, our results reveal an unprecedented viral gene expression regulation mechanism through a pre-tRNA trailer-derived tRF-La/SSB interface, which would play a role in determining host susceptibility to La/SSB-exploiting viruses and potentially in post-transcriptional cellular gene expression regulation as well. Our findings also indicate that the trailer-derived tRFs could represent broad-spectrum antiviral drugs targeting the La/SSB protein, which is exploited by various pathogenic RNA viruses.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Gue Su Chang (The Genome Institute, Washington University, School of Medicine, St. Louis, MO, USA) for helpful discussions. We also thank Sung Key Jang (POSTECH, Pohang, Korea), Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan), John Hiscott (McGill University, Montreal, Quebec, Canada), Christoph Seeger (Institute for Cancer Research, Fox Chase Cancer Center, Philadelphia, PA, USA), Herbert W. Virgin (Washington University School of Medicine, St. Louis, MO, USA), and Seung-Woo Cho (Yonsei University, Seoul, Korea) for reagents.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea funded by the Korean government (MSIP) [2014R1A2A2A01005522, 2016R1A5A1004694, 2018R1A2B6000985, 2019R1H1A2078176]; H.C. was partially supported by the Graduate School of Yonsei University Research Scholarship Grants in 2018; W.L. was the recipient of a postdoctoral fellowship [2017-12-0035] from Yonsei University. Funding for open access charge: Brain Korea 21 (BK21) PLUS program.
Conflict of interest statement. None declared.
REFERENCES
- 1. Borek E., Baliga B.S., Gehrke C.W., Kuo C.W., Belman S., Troll W., Waalkes T.P.. High turnover rate of transfer RNA in tumor tissue. Cancer Res. 1977; 37:3362–3366. [PubMed] [Google Scholar]
- 2. Speer J., Gehrke C.W., Kuo K.C., Waalkes T.P., Borek E.. tRNA breakdown products as markers for cancer. Cancer. 1979; 44:2120–2123. [DOI] [PubMed] [Google Scholar]
- 3. Kumar P., Mudunuri S.B., Anaya J., Dutta A.. tRFdb: a database for transfer RNA fragments. Nucleic Acids Res. 2015; 43:141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li Z., Kim S.W., Lin Y., Moore P.S., Chang Y., John B.. Characterization of viral and human RNAs smaller than canonical microRNAs. J. Virol. 2009; 83:12751–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee Y.S., Shibata Y., Malhotra A., Dutta A.. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009; 23:2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haussecker D., Huang Y., Lau A., Parameswaran P., Fire A.Z., Kay M.A.. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010; 16:673–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liao J.Y., Ma L.M., Guo Y.H., Zhang Y.C., Zhou H., Shao P., Chen Y.Q., Qu L.H.. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3 ' Trailers. PLoS One. 2010; 5:e10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ivanov P., Emara M.M., Villen J., Gygi S.P., Anderson P.. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 2011; 43:613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhu L., Liu X., Pu W., Peng Y.. tRNA-derived small non-coding RNAs in human disease. Cancer Lett. 2018; 419:1–7. [DOI] [PubMed] [Google Scholar]
- 10. Kuscu C., Dutta A.. A Pro-metastatic tRNA Pathway. Cell. 2016; 165:1314–1315. [DOI] [PubMed] [Google Scholar]
- 11. Venkatesh T., Suresh P.S., Tsutsumi R.. tRFs: miRNAs in disguise. Gene. 2016; 579:133–138. [DOI] [PubMed] [Google Scholar]
- 12. Kim H.K., Fuchs G., Wang S., Wei W., Zhang Y., Park H., Roy-Chaudhuri B., Li P., Xu J., Chu K. et al.. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. 2017; 552:57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol. 2018; 19:45–58. [DOI] [PubMed] [Google Scholar]
- 14. Kumar P., Kuscu C., Dutta A.. Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem. Sci. 2016; 41:679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sharma U., Conine C.C., Shea J.M., Boskovic A., Derr A.G., Bing X.Y., Belleannee C., Kucukural A., Serra R.W., Sun F. et al.. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016; 351:391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goodarzi H., Liu X., Nguyen H.C., Zhang S., Fish L., Tavazoie S.F.. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015; 161:790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maute R.L., Schneider C., Sumazin P., Holmes A., Califano A., Basso K., Dalla-Favera R.. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:1404–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sobala A., Hutvagner G.. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013; 10:553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ruggero K., Guffanti A., Corradin A., Sharma V.K., De Bellis G., Corti G., Grassi A., Zanovello P., Bronte V., Ciminale V. et al.. Small noncoding RNAs in cells transformed by human T-cell leukemia virus type 1: a role for a tRNA fragment as a primer for reverse transcriptase. J. Virol. 2014; 88:3612–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yeung M.L., Bennasser Y., Watashi K., Le S.Y., Houzet L., Jeang K.T.. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 2009; 37:6575–6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Q., Lee I., Ren J., Ajay S.S., Lee Y.S., Bao X.. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol. Ther. 2013; 21:368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meyer L.R., Zweig A.S., Hinrichs A.S., Karolchik D., Kuhn R.M., Wong M., Sloan C.A., Rosenbloom K.R., Roe G., Rhead B. et al.. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013; 41:64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee W., Lee S.H., Kim M., Moon J.S., Kim G.W., Jung H.G., Kim I.H., Oh J.E., Jung H.E., Lee H.K. et al.. Vibrio vulnificus quorum-sensing molecule cyclo(Phe-Pro) inhibits RIG-I-mediated antiviral innate immunity. Nat. Commun. 2018; 9:1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wobus C.E., Karst S.M., Thackray L.B., Chang K.O., Sosnovtsev S.V., Belliot G., Krug A., Mackenzie J.M., Green K.Y., Virgin H.W.. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2004; 2:e432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee W., Kim M., Lee S.H., Jung H.G., Oh J.W.. Prophylactic efficacy of orally administered Bacillus poly-gamma-glutamic acid, a non-LPS TLR4 ligand, against norovirus infection in mice. Sci. Rep. 2018; 8:8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim G.W., Lee S.H., Cho H., Kim M., Shin E.C., Oh J.W.. Hepatitis C virus core protein promotes miR-122 destabilization by inhibiting GLD-2. PLoS Pathog. 2016; 12:e1005714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chang J., Nicolas E., Marks D., Sander C., Lerro A., Buendia M.A., Xu C., Mason W.S., Moloshok T., Bort R. et al.. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004; 1:106–113. [DOI] [PubMed] [Google Scholar]
- 30. Rong M., Chen G., Dang Y.. Increased miR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer. 2013; 13:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bousquet-Antonelli C., Deragon J.M.. A comprehensive analysis of the La-motif protein superfamily. RNA. 2009; 15:750–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liang C., Xiong K., Szulwach K.E., Zhang Y., Wang Z., Peng J., Fu M., Jin P., Suzuki H.I., Liu Q.. Sjogren syndrome antigen B (SSB)/La promotes global microRNA expression by binding microRNA precursors through stem-loop recognition. J. Biol. Chem. 2013; 288:723–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M. Jr, Jungkamp A.C., Munschauer M. et al.. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010; 141:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gogakos T., Brown M., Garzia A., Meyer C., Hafner M., Tuschl T.. Characterizing expression and processing of precursor and mature human tRNAs by hydro-tRNAseq and PAR-CLIP. Cell Rep. 2017; 20:1463–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pudi R., Abhiman S., Srinivasan N., Das S.. Hepatitis C virus internal ribosome entry site-mediated translation is stimulated by specific interaction of independent regions of human La autoantigen. J. Biol. Chem. 2003; 278:12231–12240. [DOI] [PubMed] [Google Scholar]
- 36. Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P.. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005; 309:1577–1581. [DOI] [PubMed] [Google Scholar]
- 37. Ahn D.G., Shim S.B., Moon J.E., Kim J.H., Kim S.J., Oh J.W.. Interference of hepatitis C virus replication in cell culture by antisense peptide nucleic acids targeting the X-RNA. J. Viral Hepat. 2011; 18:e298–306. [DOI] [PubMed] [Google Scholar]
- 38. Hornung V., Ellegast J., Kim S., Brzozka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K.K., Schlee M. et al.. 5′-triphosphate RNA is the ligand for RIG-I. Science. 2006; 314:994–997. [DOI] [PubMed] [Google Scholar]
- 39. Takeuchi O., Akira S.. Pattern recognition receptors and inflammation. Cell. 2010; 140:805–820. [DOI] [PubMed] [Google Scholar]
- 40. Costa-Mattioli M., Svitkin Y., Sonenberg N.. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol. Cell Biol. 2004; 24:6861–6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Meerovitch K., Svitkin Y.V., Lee H.S., Lejbkowicz F., Kenan D.J., Chan E.K., Agol V.I., Keene J.D., Sonenberg N.. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 1993; 67:3798–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vashist S., Urena L., Chaudhry Y., Goodfellow I.. Identification of RNA-protein interaction networks involved in the norovirus life cycle. J. Virol. 2012; 86:11977–11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee K.M., Chen C.J., Shih S.R.. Regulation mechanisms of viral IRES-driven translation. Trends Microbiol. 2017; 25:546–561. [DOI] [PubMed] [Google Scholar]
- 44. King H.A., Cobbold L.C., Willis A.E.. The role of IRES trans-acting factors in regulating translation initiation. Biochem. Soc. Trans. 2010; 38:1581–1586. [DOI] [PubMed] [Google Scholar]
- 45. Ehlers I., Horke S., Reumann K., Rang A., Grosse F., Will H., Heise T.. Functional characterization of the interaction between human La and hepatitis B virus RNA. J. Biol. Chem. 2004; 279:43437–43447. [DOI] [PubMed] [Google Scholar]
- 46. Svitkin Y.V., Pause A., Sonenberg N.. La autoantigen alleviates translational repression by the 5′ leader sequence of the human immunodeficiency virus type 1 mRNA. J. Virol. 1994; 68:7001–7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yoo C.J., Wolin S.L.. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell. 1997; 89:393–402. [DOI] [PubMed] [Google Scholar]
- 48. Teplova M., Yuan Y.R., Phan A.T., Malinina L., Ilin S., Teplov A., Patel D.J.. Structural basis for recognition and sequestration of UUU(OH) 3′ temini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol. Cell. 2006; 21:75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wolin S.L., Cedervall T.. The La protein. Annu. Rev. Biochem. 2002; 71:375–403. [DOI] [PubMed] [Google Scholar]
- 50. Park J.M., Kohn M.J., Bruinsma M.W., Vech C., Intine R.V., Fuhrmann S., Grinberg A., Mukherjee I., Love P.E., Ko M.S. et al.. The multifunctional RNA-binding protein La is required for mouse development and for the establishment of embryonic stem cells. Mol. Cell Biol. 2006; 26:1445–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Trobaugh D.W., Klimstra W.B.. MicroRNA regulation of RNA virus replication and pathogenesis. Trends Mol. Med. 2017; 23:80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kumar P., Anaya J., Mudunuri S.B., Dutta A.. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014; 12:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Juhling F., Morl M., Hartmann R.K., Sprinzl M., Stadler P.F., Putz J.. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009; 37:159–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.