Significance
The viruses from the Nucleo-Cytoplasmic Large DNA Virus (NCLDV) assemblage regularly draw the attention of the scientific community for their surprising features, from the gigantism of some viruses’ particles to their genome content. They shook the very definition of viruses and shed new light on the debate over their nature and putative role in the evolution of the cellular domains. Their origin(s) and evolution remain to be elucidated, however. Our phylogenetic analyses reveal the evolutionary relationships between the NCLDV families and their origin from a common ancestor, from which they diversified before the origin of modern eukaryotes. Our results also point to their likely role in the emergence of multiple DNA-dependent RNA polymerases in proto-eukaryotes.
Keywords: evolution, giant viruses, NCLDV, RNA polymerase, proto-eukaryotes
Abstract
Giant and large eukaryotic double-stranded DNA viruses from the Nucleo-Cytoplasmic Large DNA Virus (NCLDV) assemblage represent a remarkably diverse and potentially ancient component of the eukaryotic virome. However, their origin(s), evolution, and potential roles in the emergence of modern eukaryotes remain subjects of intense debate. Here we present robust phylogenetic trees of NCLDVs, based on the 8 most conserved proteins responsible for virion morphogenesis and informational processes. Our results uncover the evolutionary relationships between different NCLDV families and support the existence of 2 superclades of NCLDVs, each encompassing several families. We present evidence strongly suggesting that the NCLDV core genes, which are involved in both informational processes and virion formation, were acquired vertically from a common ancestor. Among them, the largest subunits of the DNA-dependent RNA polymerase were transferred between 2 clades of NCLDVs and proto-eukaryotes, giving rise to 2 of the 3 eukaryotic DNA-dependent RNA polymerases. Our results strongly suggest that these transfers and the diversification of NCLDVs predated the emergence of modern eukaryotes, emphasizing the major role of viruses in the evolution of cellular domains.
The discovery of giant viruses in the early 21st century has revived the debate on the nature of viruses and their role in evolution (1–12). The 1-µm-long particles of pithoviruses (13) can be seen under a light microscope, and the 2.5 Mb-long genomes of pandoraviruses, larger than those of many cellular organisms, encode for more than 2,000 proteins, mostly ORFans (14). However, these unexpected features notwithstanding, giant viruses are a bona fide part of the virosphere, relying on the infected cells for the production of energy and protein synthesis. Phylogenetic and comparative genomics analyses have shown that giant viruses together with smaller eukaryotic dsDNA viruses form a supergroup, dubbed the Nucleo-Cytoplasmic Large DNA Viruses (NCLDV) (15, 16). This assemblage encompasses families of large and giant viruses, including Poxviridae, Iridoviridae, Ascoviridae, Asfarviridae, Marseilleviridae, Mimiviridae, and Phycodnaviridae, as well as several lineages of as-yet unclassified viruses, such as pithoviruses, pandoraviruses, molliviruses, and faustoviruses (17). Altogether, the NCLDVs are associated with diverse eukaryotic phyla, from phagotrophic protists to insects and mammals, and some cause devastating diseases, such as smallpox (Poxviridae) or African swine fever (Asfarviridae), and play important ecological roles, such as termination of algal blooms (Phycodnaviridae) (18).
The origin and evolution of the NCLDVs remain subjects of controversy. It is still unclear if these viruses form a monophyletic group, if proteins conserved in most NCLDVs had a congruent evolutionary history, or if some of them were acquired several times independently from their hosts. Most phylogenetic analyses performed up to now have been based on individual proteins or various subsets of conserved proteins (19, 20). These analyses usually recovered the monophyly of various NCLDV families but often offered contradicting results, and the relationships between the families remained debated. For instance, it has been proposed that the giant pandoraviruses are related to members of the Phycodnaviridae family (21), but this grouping was not recovered in a recent phylogeny based on their DNA polymerases (22). According to some studies, the different families of the NCLDVs emerged during the diversification of modern eukaryotes (23), whereas other studies suggest that NCLDVs form a monophyletic group branching between Archaea and Eukarya (19). Some authors have even suggested that several families of giant viruses could have originated independently from extinct cellular lineages, possibly even before the last universal common ancestor of Archaea, Bacteria, and Eukarya (10, 24).
To study the relationships between NCLDVs and the 3 cellular domains, it is first necessary to have a robust phylogeny of NCLDVs themselves. Constant improvements of the phylogenetic tools and substantial expansion of the collection of large and giant viruses prompted us to perform an updated and in-depth phylogenetic analysis of the NCLDVs. We mined available genomes for homologous genes, built clusters of orthologous genes, and performed extensive phylogenetic analyses on the 8 most conserved ones, separately and in concatenations. In addition, we investigated the relationships between NCLDVs and eukaryotes through the phylogeny of the DNA-dependent RNA polymerases (RNAPs). Unlike in previous analyses, we included in our study the 3 eukaryotic RNAPs (RNAP-I, -II, and -III) and concatenated the 2 largest subunits. The robust phylogenies that we obtained show that core genes involved in virion morphogenesis, genome transcription, and replication have coevolved in the entire NCLDV lineage. Furthermore, our results reveal the existence of 2 superclades of NCLDVs that diverged after the separation of the archaeal and eukaryotic lineages but before the emergence of the last eukaryotic common ancestor (LECA). Surprisingly, our data suggest that eukaryotic RNAP-III is the actual cellular ortholog of the archaeal and bacterial RNAP, while eukaryotic RNAP-II and possibly RNAP-I were transferred between 2 viral families and proto-eukaryotes. Overall, our results reveal that the diversification of NCLDVs predates the emergence of LECA, and that the ancestors of contemporary NCLDVs have played important roles in the emergence and diversification of modern eukaryotes.
Results
Identification of the Core Genes.
Many new NCLDV genomes have been published following the latest comprehensive comparative genomics analyses (20, 25), substantially increasing their known diversity and enriching families that were previously poorly represented. As a result, the list of the most conserved genes among the NCLDVs could have changed drastically since the last estimation, prompting us to reanalyze this list. To identify NCLDV orthologs, we designed a pipeline based on the best bidirectional BLAST hit combined with manual curation to remain as exhaustive as possible while avoiding the inclusion of paralogs (Methods). The sets of conserved proteins classified according to their conservation among NCLDVs are summarized at https://zenodo.org/record/3368642 (26).
Our results show that only 3 proteins are strictly conserved among the 73 selected NCLDV genomes (SI Appendix, Table S1): family B DNA polymerase (DNApol B), the D5-like primase-helicase (primase hereinafter), and homologs of the Poxvirus Late Transcription Factor VLTF3 (VLTF3-like). Acknowledging various reasons that may preclude detection of homologous genes (e.g., high divergence or genuine loss in a taxon), we decided to lower our conservation threshold to include genes found in at least 95% of the genomes. This alteration increased our set of core genes by 3: the transcription elongation factor II-S (TFIIS), the genome packaging ATPase (pATPase), and the major capsid protein (MCP). Notably, no homolog of the MCP has been found in pandoraviruses (14), whereas pATPases are apparently lacking in Pithovirus (13), Cedratvirus (27), and Orpheovirus (28). Conservation of the NCLDV genes is discussed further in SI Appendix.
To this set of 6 proteins we added the 2 largest RNAP subunits (RNAP-a and -b) despite their notable absence in members of all genera of the Phycodnaviridae family except for the Coccolithovirus genus. Indeed, these 2 proteins are otherwise highly conserved among the NCLDVs (present in 92% of the genomes) and are the largest universal markers (found in all members of the 3 cellular domains), which makes them perfectly suited for reconstructing the evolutionary relationships between NCLDVs and cellular organisms. The subsequent analyses, from alignments to phylogenetic reconstruction, were performed on this set of 8 markers.
Phylogenies of NCLDVs.
Using a maximum-likelihood (ML) framework, we obtained the monophyly of known NCLDV families in most of the 8 single-protein phylogenetic trees, except for the Phycodnaviridae. However, these trees lacked resolution, particularly for the 2 shortest markers (TFIIS and VLTF3-like), and we noticed several incongruences in relationships between families. [Trees are listed in Additional data at https://zenodo.org/record/3368642 (26); further details are provided in SI Appendix.] The Poxviridae and Aureococcus anophagefferens virus consistently formed long branches and displayed the most unstable positions; thus, we removed these taxa from our subsequent analyses to avoid potential artifacts, such as the long-branch attraction artifact. Remarkably, phylogenetic analyses of the resultant datasets resulted in much more congruent single-protein trees with higher supports for most nodes (SI Appendix, Fig. S1). This congruence suggested that the 8 core proteins selected for this study share the same phylogenetic signal. Thus, we decided to build NCLDV phylogenetic trees based on these 8 core genes using 2 independent approaches: concatenation and subtree prune-and-regraft (SPR) supertree reconstruction (Methods).
As a first step in the concatenation approach, we performed comparative phylogenetic analyses of differential concatenations using a homemade pipeline to check the congruence between the markers (Methods and SI Appendix, Table S2). This test did not reveal any major incongruences and strongly supported the absence of conflicting signals that would have prevented concatenating them. We then performed Bayesian inferences with the CAT-GTR model (Methods) on the 8-core concatenation. After reaching a good convergence (maxdiff <0.1), we obtained a phylogenetic tree with all nodes but 2 minor ones with maximal support (posterior probability 1). Strikingly, we obtained the same phylogenetic tree topology using the SPR supertree reconstruction, which is independent of any concatenation (SI Appendix, Fig. S2). The congruent topology obtained using different approaches very strongly suggests that it represents the vertical evolutionary history of the NCLDV core genes. This notably implies that NCLDV informational proteins have coevolved with proteins involved in virion formation (SI Appendix, Figs. S3 and S4; details in SI Appendix).
The Bayesian tree and the SPR supertree confidently position a number of recently identified viruses. The family Mimiviridae includes the unclassified Klosneuvirus, Indivirus, Catovirus, Hokovirus (29), and Tupanvirus (30) and is associated with unclassified viruses with smaller genomes often referred to as the “extended Mimiviridae” (20) or, more recently, the “Mesomimivirinae” (31). We refer to this grouping as the putative order “Megavirales” (justification in SI Appendix). Pithovirus sibericum, Cedratvirus A11, and Orpheovirus IHUMI-LCC2 represent a new distinct family that we herein refer to as a Pitho-like clade, whose exact position remains to be investigated considering their still-limited representation. Faustovirus (32, 33), Pacmanvirus (34), and Kaumoebavirus (35) form a well-supported clade with the African swine fever virus (ASFV-1) of the Asfarviridae, as previously suggested (36). The family Phycodnaviridae encompasses pandoraviruses and Mollivirus sibericum. As often observed in published NCLDV phylogenies (25), Ascoviridae were nested within the Iridoviridae.
To tentatively root the NCLDV phylogeny, we performed a ML tree of the MCP-pATPase concatenation, using the polintoviruses, which could be the closest outgroup to the NCLDVs (37, 38), as an outgroup. Notably, the MCP-pATPase tree rooted using polintovirus sequences (SI Appendix, Fig. S5) was almost identical to that obtained with the NCLDVs alone (SI Appendix, Fig. S4), and the number of positions was not dramatically reduced (601 with polintoviruses, 625 without). With this root, NCLDVs are split into 2 superclades corresponding to a bipartition already observed in all single-gene trees and in the Bayesian tree (Fig. 1) and the SPR supertree. The first superclade includes the Marseilleviridae with the Ascoviridae, the Pitho-like virus clade, and the Iridoviridae (hereinafter referred to as the MAPI superclade), whereas the second includes the Phycodnaviridae with the Asfarviridae and the Megavirales (hereinafter referred to as the PAM superclade).
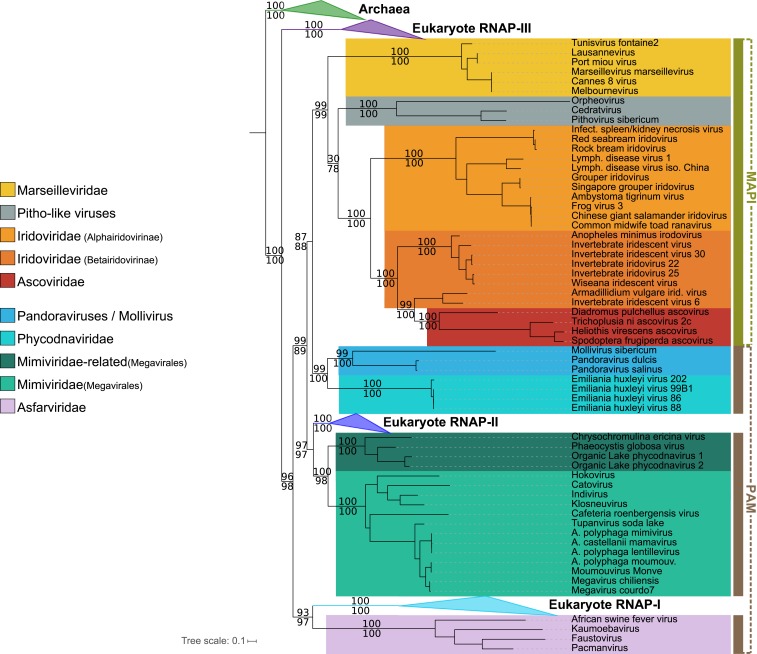
Fig. 1.
Phylogenetic tree of the NCLDVs. Bayesian inference (CAT-GTR model) of the concatenated 8 core proteins from the NCLDVs after removal of Poxviridae and A. anophagefferens. Genome sizes (in bp) are represented next to each virus name. The scale bar indicates the average number of substitutions per site. The values at branches represent Bayesian posterior probabilities. Nodes without maximum support are indicated in red. The tree has been rooted between the MAPI and the PAM superclades [unrooted tree in Additional data at https://zenodo.org/record/3368642 (26)].
Relationships between NCLDVs and the 3 Cellular Domains.
The RNA and DNA polymerases of NCLDV have homologs in the 3 domains of life (Archaea, Bacteria, and Eukarya), making it a priori possible to investigate their evolutionary relationships with cellular organisms. However, the family B DNA polymerase, often used to tentatively affiliate new NCLDV genomes to known taxa (39), cannot be used for this task, since they are absent from most Bacteria and their phylogenetic analyses produce very complex scenarios (40). In contrast, the phylogeny of the 2 largest RNAP subunits, which are also the largest universal markers, allows positioning the ancestors of the 3 cellular domains (41).
Most phylogenetic analyses of RNAPs performed until now included only the eukaryotic RNA polymerase II (RNAP-II), which is the most widely studied and usually considered the most similar to the archaeal RNAPs (42). Here we decided to include all 3 eukaryotic RNAPs (RNAP-I, RNAP-II, and RNAP-III); we used a normalized RNAP nomenclature (SI Appendix). Importantly, these 3 multisubunit RNAPs are present in all eukaryotes, indicating that all were already present in the LECA. Therefore, their inclusion in our dataset should produce 3 universal eukaryotic phylogenies and thus 3 positions for LECA in the viral/cellular RNAP tree.
We previously obtained a robust phylogenetic RNAP tree with a concatenation of the 2 largest RNAP subunits (in ML and Bayesian frameworks) using a balanced dataset (i.e., the same number of species for each domain) devoid of known fast-evolving species to prevent long-branch attraction artifacts and using RNAP-II as the eukaryotic representative (41, 43). Here we added the eukaryotic RNAP-I and RNAP-III to this dataset. Importantly, the 3 eukaryotic RNAPs displayed globally congruent phylogenies, corroborating their presence in the LECA. As in our previous study, Archaea and Eukarya form 2 monophyletic sister groups in our new phylogeny of concatenated RNAP subunits when the tree is rooted with Bacteria as the outgroup (SI Appendix, Fig. S6).
We included the sequences of NCLDVs into this new dataset (except for Poxviridae and A. anophagefferens) to investigate the timeline of NCLDV diversification in the context of cellular evolution. The ML phylogenetic analysis of concatenated RNAP subunits yielded the 3-domain topology (SI Appendix, Fig. S7) in which NCLDVs branch after the divergence of the archaeal and eukaryotic lineages. We then removed Bacteria to increase the phylogenetic resolution and used the Archaeal branch as the outgroup (single-protein trees in SI Appendix, Fig. S8; concatenation in SI Appendix, Fig. S9). The trees were highly similar, and the supports for several nodes indeed became stronger. Considering the systematic monophyly of each cellular clade (the Archaea and the 3 eukaryotic RNAP homologs), we decided to use an independent constraint for each of them during the alignment process (Methods) to improve the resolution by limiting misalignments. The resulting concatenation of the 2 subunits switched from 1,683 positions to 1,595, and the highly supported reconstructed tree obtained in the ML framework (LG+C60 model; Fig. 2) was strictly identical to the tree without any constraint.
Fig. 2.
ML phylogenetic tree of the concatenated 2 largest RNAP subunits from Archaea, Eukaryotes, and NCLDVs, with Archaea serving as the outgroup. The scale bar indicates the average number of substitutions per site. Values atop and below the branches represent support calculated by the SH-like aLRT (1,000 replicates) and UFBoot (1,000 replicates), respectively.
Surprisingly, RNAP-III, rather than RNAP-II, appears to be the closest eukaryotic RNAP to the archaeal outgroup with strong supports, suggesting that it could be the actual ortholog of the archaeal enzyme. The most significant feature of this tree is that the LECA, despite being a single time point in the history of Eukaryotes, is represented 3 times. Notably, 2 of these positions are nested within the diversity of NCLDVs, indicating that NCLDVs predated the emergence of the LECA. As a consequence, NCLDVs form 3 monophyletic subgroups well separated from the 3 eukaryotic RNAPs. To validate this result, we performed an approximately unbiased (AU) tree topology test, in which the likelihood of the unconstrained tree was compared with those of 2 alternative topologies obtained by constraining either the monophyly of NCLDVs or the monophyly of cellular organisms (Methods and SI Appendix). The AU test rejected the 2 alternative trees with P values <1e-3. Remarkably, the relative positions of the NCLDV families and superclades in the concatenated RNAP tree are completely congruent with the NCLDV topology in the Bayesian and SPR Supertree trees previously obtained with the 8 core proteins (Fig. 1). This was not due to the fact that the RNAPs are the longest markers in these analyses, since we obtained highly similar trees with and without the RNAPs in comparative phylogenetics tests (SI Appendix, Table S2 and Fig. S10). This is remarkable, since the RNAP proteins represent nearly one-half of the total positions (47%) in the global concatenation. The congruence of the NCLDV topology, notably between the RNAP phylogenetic trees before (SI Appendix, Fig. S11) and after the addition of cellular taxa (Fig. 2), is schematically represented in SI Appendix, Fig. S12.
Three clades of the NCLDVs are distinguishable in the viral/cellular RNAP tree and correspond to the monophyletic MAPI superclade next to the Phycodnaviridae, the Megavirales, and the Asfarviridae (Fig. 2). The PAM superclade is indeed not monophyletic in this tree, with the Phycodnaviridae ambiguously branching as a sister group to the MAPI superclade (a position to consider with caution; SI Appendix) and, even more notably, the eukaryotic RNAP-I and -II branching within this clade (discussion in SI Appendix). The eukaryotic RNAP-II is a sister group to the Megavirales, whereas the eukaryotic RNAP-I is a sister group to the Asfarviridae. To assess the robustness of these groupings, we reconstructed a consensus bootstrap tree of the concatenated RNAP subunits. In parallel, we also performed a phylogenetic analysis based on reconstructed ancestral sequences to replace the 3 eukaryotic RNAP clades (Methods). Both methods support the relationships between the Megavirales and the eukaryotic RNAP-II, as well as between the Asfarviridae and the eukaryotic RNAP-I (SI Appendix, Fig. S13). However, the single-subunit trees (SI Appendix, Fig. S8) suggest a more complex scenario for the Asfarviridae and the eukaryotic RNAP-I, whose position differs in the 2 trees: the Asfarviridae are a sister group to the RNAP-I in the individual a subunit tree, as in the tree based on concatenated RNAP subunits (Fig. 2), whereas they branch within the Megavirales in the b subunit tree, with the RNAP-I and -III being sister clades. This suggests that the 2 RNAP subunits of the Asfarviridae were involved in 2 separate transfer events with proto-eukaryotes, which could explain their long branch in the RNAP trees.
The branching of NCLDVs as a sister group to the eukaryotic RNAP-III indicates that they have probably obtained their RNAP from proto-eukaryotes after their divergence from the archaeal lineage. The unexpected evolutionary relationships between RNAP-I and -II and NCLDVs suggest that these 2 eukaryotic RNAPs were either recruited from NCLDVs or transferred from proto-eukaryotes to the ancestors of the Asfarviridae family and Megavirales order. Transfers from cells to viruses seem unlikely in this case, because replacements of the 2 largest core genes in 2 major NCLDV families by their cellular counterparts would have most certainly resulted in substantial alterations in the NCLDV topologies obtained during the comparative phylogenetics tests, which is not the case (SI Appendix, Table S2 and Fig. S12). In particular, replacements of the ancestral NCLDV RNAP in 2 viral families by transfers from the eukaryotic RNAP-I and -II likely would have led to specific topological features not observed in the phylogenetic trees (SI Appendix and SI Appendix, Fig. S14).
These data strongly suggest that the transfers of the RNAP-encoding genes were directed from viruses to cells after the diversification of these RNAPs within the NCLDVs. However, one should remain cautious regarding the direction of transfer between the eukaryotic RNAP-I and the Asfarviridae, owing to the length of the corresponding branches and the fact that the 2 subunits apparently had different origins. Moreover, the number of representatives in the Asfarviridae clade is still limited, and future analyses with more genomes could change our conclusions. Nevertheless, the virus-to-cell transfer scenario of the eukaryotic RNAP-II is not impacted by these limitations.
Based on this observation and our previous results on the MAPI and PAM superclades (see SI Appendix for discussions about the root), we postulate a possible, hypothetical scenario depicted in Fig. 3 for the evolution of NCLDVs and their RNAPs that parsimoniously explain the distribution of our data. According to this hypothesis, the ancestral eukaryotic RNAP (at least the 2 largest subunits), more similar to RNAP-III, was first transferred to the ancestor of NCLDVs. After the divergence between the MAPI and the PAM superclades, this viral RNAP diverged in the common ancestor of the Megavirales and Asfarviridae following the emergence of the Phycodnaviridae and was subsequently transferred to proto-eukaryotes to give rise to the RNAP-II. Separately, a duplication of the ancestral RNAP-III in proto-eukaryotes occurred, and the a subunit of this newly formed RNAP was exchanged between proto-eukaryotes and the Asfarviridae. Alternatively, only the b subunit of the RNAP-III could be duplicated and directly coupled with a partnering a subunit from the Asfarviridae. Either way, this new complex, partly viral and partly cellular from duplication, resulted in the RNAP-I. Although this scenario remains hypothetical and to be further tested, every alternative scenario would still imply that NCLDVs diversified before the emergence of modern eukaryotes.
Fig. 3.
Schematic representation of the hypothetical scenario for the transfers of RNAP between cells and NCLDVs. An ancestral RNAP that later gave rise to the eukaryotic RNAP-III, an actual ortholog of the archaeal RNAP, was transferred from proto-eukaryotes to the ancestor of modern NCLDVs. Following the emergence of the Phycodnaviridae (with an RNAP whose origin requires further investigation), a significantly divergent RNAP was transferred from the common ancestor of the Asfarviridae and Megavirales to proto-eukaryotes. A new eukaryotic RNAP also emerged from a duplication event from RNAP-III, possibly followed by replacement of the largest subunit with that of Asfarviridae; further investigations are needed to confirm the direction of this transfer. These events occurred before the LECA that marked the emergence of modern eukaryotes.
Discussion
From our investigation of the NCLDV genomes, including those of most recently identified giant and large dsDNA viruses, we can reconstruct a robust phylogenetic tree that likely represents their vertical evolutionary history. Our results provide a solid framework for proposed and sometimes debated positions of different NCLDV families. Notably, Pithovirus and related viruses form a separate, yet to be named family most closely related to the Marseilleviridae. Pandoraviruses and the Mollivirus branch within the Phycodnaviridae as a sister group to Coccolithovirus, confirming the results of Yutin and Koonin (21). Our results reveal 2 robust clusters: the MAPI, comprising the Marseilleviridae, the Ascoviridae, the Pitho-like clade, and the Iridoviridae, and the PAM, which includes the Phycodnaviridae, the Asfarviridae, and the Megavirales. The monophyly of the 2 superclades is supported with an external outgroup in the MCP-pATPase concatenated tree, and the monophyly of the MAPI cluster is further supported in the RNAP trees.
These results call for reassessment of the taxonomy of large and giant dsDNA viruses included in the NCLDV assemblage, as recently suggested by Koonin and Yutin (44). In particular, the expansion of the Mimiviridae family and the discovery of associated but more distantly related viruses suggest that a family-level taxon might not be adequate to encompass this diversity. A new order, the Megavirales, might be more appropriate. Furthermore, the Asfarviridae clade includes Faustovirus (32, 33), Kaumoebavirus (35), and Pacmanvirus (34), which have been suggested to represent separate families (35), and thus an order-level taxon would be needed for their classification. Similarly, the placement of the pandoraviruses and Mollivirus within the Phycodnaviridae indicates that this family might not be monophyletic and should be revised. Ascoviridae regularly branch within Iridoviridae, advocating for a reconsideration of these 2 families. The elusive position of the Poxviridae, which were removed from most of our analyses, and their actual association with NCLDVs remain to be investigated.
The monophyly of NCLDVs is not recovered in the NCLDV/cellular RNAP tree. NCLDVs do not form a fourth domain of life, as has been proposed by some authors (19), nor do they nest among eukaryotes (23). While some genes in the NCLDV genomes might have been recruited from different sources, including their modern hosts and bacteria, we have shown that a congruent vertical evolutionary history of the NCLDV core genes is traceable and sound. The selected core genes indeed shared a similar global vertical evolution and were inherited from a common ancestor, which was likely smaller, as hypothesized previously (45), and possibly related to polintoviruses (11). Notably, these core genes are involved in both genome replication and virion formation, key features of viruses, supporting their evolution from a viral ancestor that emerged either shortly after the divergence between Archaea and proto-eukaryotes or before this divergence. In this latter case, viruses related to the ancestors of NCLDVs might have been lost in Archaea or still have infected archaeal lineages, such as the Asgard archaea, which have not yet been explored for their viruses. Notably, although our RNAP phylogeny supports the 3-domain topology for the tree of life (Archaea and Eukaryotes being sister clades), our scenario for NCLDV evolution is also compatible with the 2-domain hypothesis for eukaryogenesis (i.e., Eukaryotes emerging from within Archaea).
Interestingly, giant viruses do not cluster together in the NCLDV trees. Most of them are present in the PAM superclade (the Mimiviridae in the “Megavirales” and pandoraviruses/Mollivirus in the Phycodnaviridae), whereas Orpheovirus is present in the Pitho-like clade within the MAPI superclade (Fig. 1). The scattered distribution of giant viruses within the diversity of NCLDVs strongly opposes a giant—viral or cellular—ancestor scenario as proposed previously (10, 24). Indeed, this would suggest a parallel genome reduction in many families and subsets of families, which is not parsimonious, especially when considering a common ancestry of NCLDV with smaller viruses infecting bacteria and archaea (17). In contrast, the occurrence of several independent and massive increases in the genome size in different virus groups along the evolution of NCLDVs, potentially through successive steps of reduction and expansion of their genomes (46, 47), seems more likely.
Our analyses of the 2 largest subunits of the RNAP, including the 3 eukaryotic polymerases, revealed that the genuine ortholog of the archaeal and bacterial RNAP might actually be the eukaryotic RNAP-III. In agreement with this unexpected result, homologs of the eukaryotic RNAP-III–specific subunit RPC34 are present in most archaeal lineages (48, 49). Importantly, the inclusion in our analyses of the 3 eukaryotic RNA polymerases, which emerged before the emergence of modern eukaryotes, provided a relative time frame for NCLDV evolution. Our RNAP trees indeed strongly imply that the diversification of NCLDVs occurred after the divergence between Archaea and proto-eukaryotes but predated the evolutionary bottleneck that marked the emergence of modern eukaryotes. Several authors have suggested that NCLDVs have played a central role in the origin of eukaryotes (6, 8). Our results indeed suggest that modern eukaryotes obtained their RNAP-II from NCLDVs, and possibly their RNAP-I as well, during the proto-eukaryotic stage. Our results indicate that further investigation into the diversity and molecular biology of NCLDVs will probably have a major impact on our understanding of the origin and early evolution of eukaryotes.
Methods
Datasets.
We initially collected a total of 96 NCLDV genomes from public databases (SI Appendix, Table S1) that we used to build the core genome. This dataset comprises 17 Mimiviridae, 6 Marseilleviridae, 30 Iridoviridae, 4 Ascoviridae, 14 Poxviridae, 4 Asfarviridae, 15 Phycodnaviridae, 3 unclassified viruses (referred to as Pitho-like viruses), 2 pandoraviruses, and 1 mollivirus.
Preliminary phylogenetic analyses showed high redundancy within some groups already comprising many members compared to others. We thus decided to remove some genomes to obtain a more balanced sampling (SI Appendix, Table S1): 14 Iridoviridae, 2 Phycodnaviridae, and 4 Mimiviridae. These analyses also revealed that the Poxviridae on the one hand, and a single virus (A. anophagefferens) on the other hand always produce long branches and tend to change position in the tree depending on the considered proteins or concatenation of proteins. Thus, we decided to remove these viruses (14 Poxviridae and A. anophagefferens) from subsequent analyses, leading to the dataset of 61 genomes used in the phylogenetic analyses.
Ten polintovirus sequences were collected from the Repbase collection (50) (https://www.girinst.org/repbase/update/index.html): Polinton-1_HM, Polinton-3_TC, Polinton-5_NV, Polinton-2_NV, Polinton-1_DY, Polinton-1_TC, Polinton-1_SP, Polinton-2_SP, Polinton-2_DR, and Polinton-1_DR.
The cellular taxa included in some analyses were selected based on previous work performed by some of our group (41). The list of selected taxa is presented in SI Appendix, Table S3.
Core Genome Building.
Because of the high divergence level of NCLDV genomes, we were not able to directly identify genes shared among all of them. This is why we first started from 2 subsets of NCLDVs, both of which were sufficiently coherent and comprising enough members. Those 2 subsets were the viruses annotated as Mimiviridae on the one hand and as Marseilleviridae on the other hand.
For each subset of genomes, we proceeded as follows. We defined groups of orthologous genes by blasting 1 proteome against all of the others. We only considered hits that had an E-value <1 e−10. We then identified pairwise reciprocal best hits with at least 20% similarity and at least 40% alignment coverage. We finally identified the union of all the sets of orthologs and retained those present in more than one-half of the members of the subset.
The result was 2 sets of orthologs, 1 set for each subset of NCLDV genomes. We compared these 2 sets by identifying the matching proteins using BLAST and HMM profiles and obtained orthologs found in both Mimiviridae and Marseilleviridae. Using the aforementioned BLAST criteria, we checked for the presence of these orthologs in other NCLDV proteomes. When a protein was missing, we checked the presence of a corresponding gene using TBLASTN to account for incomplete annotations of the genomes, and also used HMM profiles to account for high sequence divergence. This process resulted in a set of putative orthologous proteins found in all NCLDV families.
To detect errors, typically different proteins assigned to the same group, we used HMMer (51) to find a matching HMM profile in the PFAM database (http://pfam.xfam.org/) for each group and discarded those significantly matching more than 1 PFAM profile (after checking that these profiles were not from the same protein family). Finally, we aligned the remaining orthologs and visually inspected the alignments as a last control.
We obtained a list of orthologs that we ordered according to their presence in NCLDV genomes to define different categories of core proteins.
Phylogenetic Analyses.
Alignments.
All alignments were performed using MAFFT v7.397 and the E-INS-i algorithm (52), which is designed to align sequences that are susceptible to containing large insertions. For 1 RNA polymerase analysis (see Relationships between NCLDVs and the 3 Cellular Domains), constraints in the alignments were used with the seed option; independent alignments of each cellular clade (Archaea and the 3 eukaryotic RNA polymerases) performed separately served as constraints for the global alignment. For the viral phylogenies, we trimmed each alignment of the positions containing more than 20% of gaps. For the RNA polymerase phylogenies with cellular sequences, the alignments were trimmed with BMGE, with the -m BLOSUM30 and -b 1 options (53).
ML phylogenies.
Single-protein and concatenated protein phylogenies were conducted within the ML framework using IQ-TREE v1.6.3 (54). We first performed a model test with the Bayesian information criterion by including protein mixture models (55). For mixture model analyses, we used the PMSF models (56). The support values were computed either from 100 bootstrap replicates in the case of the nonparametric bootstrap or from 1,000 replicates for the Shimodaira–Hasegawa (SH)-like approximation likelihood ratio test (aLRT) (57) and ultrafast bootstrap approximation (UFBoot) (58).
Comparative phylogenetic analyses.
To detect potential incongruences within the signal carried by core proteins (after removal of Poxviridae and A. anophagefferens) that could prevent their global concatenation, we performed comparative phylogenetic analyses of every possible combination of 6 out of 8 core proteins through the ML framework. The 36 ML trees generated were carefully analyzed for reference features estimated from the Bayesian phylogenetic tree (Fig. 1), as well as from most phylogenetic trees obtained throughout this study. The presence or absence of these features were counted, and accordingly each feature was scored for its observed frequency among the trees, and each tree was scored according to the number of observed reference features (SI Appendix, Table S2).
Supermatrix analysis.
We obtained a supermatrix by concatenating the 8 amino acid alignments of the core genes. For supermatrices containing more characters, we computed ML trees using the aforementioned method and performed Bayesian analyses using phyloBayes MPI v1.5a (59) and the CAT-GTR model (60). The other parameters were set on default. Four independent chains were run until at least 2 reached convergence with a maximum difference value <0.1. The tree presented in Fig. 1 was obtained from the convergence (maxdiff value 0.097) of 2 chains of 3,426 and 3,276 generations. The first 25% of trees were removed as burn-in. The consensus tree was obtained by selecting 1 out of every 2 trees. To account for composition bias, we also applied 2 different character recodings, using 4 bins according to 2 different binnings: the adaptation of the 6 Dayhoff groups (61) to 4 bins proposed by Lartillot in phyloBayes manual and the one proposed by Susko and Rogers (62). For these analyses, a GTR+Γ4+I model was used.
Supertree analysis.
Horizontal gene transfers can deeply impact tree reconstruction when using alignment-based methods. Supertree methods aim to reconciliate sets of phylogenetic trees, typically gene/protein trees, into an organismal tree even when such evolutionary phenomena occur. Among the different proposed criteria for supertree methods, the SPR distance has proven to lead to more accurate tree reconstructions (63). We used SPR Supertree v1.2.1 (63) from the 8 single protein phylogenies that we previously inferred, after collapsing the clades for which the support was <95%.
Ancestral sequence reconstruction.
In an attempt to reduce the risk of long branch attraction, we replaced the eukaryotic clades in the RNAP tree by their ancestral sequences. These sequences were inferred using IQ-TREE. We selected sites with a posterior probability >0.7 and replaced the other sites by gaps.
Topology test.
IQ-TREE v1.6.3 was used to perform AU tree topology tests (64) for comparing the tree obtained with the concatenated RNAP genes (Fig. 2) with 2 other trees that we built using the same methodology but constraining the monophyly of the NCLDVs and the monophyly of the cellular organisms. The AU tests rejected these 2 new trees with P values <1 e-3.
Visualization.
The phylogenetic trees were visualized with FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/) and iTOL (65).
Data Availability.
All the trees presented in this study are included as Newick files within Additional data, together with the alignments in FASTA format (https://zenodo.org/record/3368642; see ref. 26). This folder also contains a table listing the proteins conserved among the NCLDV families. In addition, an online platform has been developed to grant readers easy access to the multiple sequence alignments and position-specific scoring matrix (PSSM) files (http://giphy.pasteur.fr/PhyloM/NCLDV/), with usage information.
Supplementary Material
Acknowledgments
We thank Alexis Criscuolo for his expert support in computational analyses and the development of the online platform, and Violette Da Cunha for assistance. This work was supported by a European Research Council grant from the European Union’s Seventh Framework Program (FP/2007-2013)/Project EVOMOBIL-ERC Grant Agreement 340440.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: FASTA and tree files are accessible at https://zenodo.org/record/3368642.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912006116/-/DCSupplemental.
References
- 1.La Scola B., et al. , A giant virus in amoebae. Science 299, 2033 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Raoult D., Forterre P., Redefining viruses: Lessons from Mimivirus. Nat. Rev. Microbiol. 6, 315–319 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Moreira D., Brochier-Armanet C., Giant viruses, giant chimeras: The multiple evolutionary histories of Mimivirus genes. BMC Evol. Biol. 8, 12 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filée J., Chandler M., Gene exchange and the origin of giant viruses. Intervirology 53, 354–361 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Forterre P., Giant viruses: Conflicts in revisiting the virus concept. Intervirology 53, 362–378 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Nasir A., Forterre P., Kim K. M., Caetano-Anollés G., The distribution and impact of viral lineages in domains of life. Front. Microbiol. 5, 194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takemura M., Yokobori S., Ogata H., Evolution of eukaryotic DNA polymerases via interaction between cells and large DNA viruses. J. Mol. Evol. 81, 24–33 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Forterre P., Gaïa M., Giant viruses and the origin of modern eukaryotes. Curr. Opin. Microbiol. 31, 44–49 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Forterre P., To be or not to be alive: How recent discoveries challenge the traditional definitions of viruses and life. Stud. Hist. Philos. Biol. Biomed. Sci. 59, 100–108 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Claverie J.-M., Abergel C., Giant viruses: The difficult breaking of multiple epistemological barriers. Stud. Hist. Philos. Biol. Biomed. Sci. 59, 89–99 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Koonin E. V., Krupovic M., Polintons, virophages and transpovirons: A tangled web linking viruses, transposons and immunity. Curr. Opin. Virol. 25, 7–15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mihara T., et al. , Taxon richness of “Megaviridae” exceeds those of bacteria and archaea in the ocean. Microbes Environ. 33, 162–171 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Legendre M., et al. , Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc. Natl. Acad. Sci. U.S.A. 111, 4274–4279 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Philippe N., et al. , Pandoraviruses: Amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341, 281–286 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Iyer L. M., Aravind L., Koonin E. V., Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 75, 11720–11734 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koonin E. V., Yutin N., “Nucleo-cytoplasmic large DNA viruses (NCLDV) of eukaryotes” in eLS (Wiley, Chichester, UK, 2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koonin E. V., Krupovic M., Yutin N., Evolution of double-stranded DNA viruses of eukaryotes: From bacteriophages to transposons to giant viruses. Ann. N. Y. Acad. Sci. 1341, 10–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brussaard C., Kempers R., Kop A., Riegman R., Heldal M., Virus-like particles in a summer bloom of Emiliania huxleyi in the North Sea. Aquat. Microb. Ecol. 10, 105–113 (1996). [Google Scholar]
- 19.Boyer M., Madoui M.-A., Gimenez G., La Scola B., Raoult D., Phylogenetic and phyletic studies of informational genes in genomes highlight existence of a 4 domain of life including giant viruses. PLoS One 5, e15530 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yutin N., Colson P., Raoult D., Koonin E. V., Mimiviridae: Clusters of orthologous genes, reconstruction of gene repertoire evolution and proposed expansion of the giant virus family. Virol. J. 10, 106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yutin N., Koonin E. V., Pandoraviruses are highly derived phycodnaviruses. Biol. Direct 8, 25 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Legendre M., et al. , Diversity and evolution of the emerging Pandoraviridae family. Nat. Commun. 9, 2285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreira D., López-García P., Evolution of viruses and cells: Do we need a fourth domain of life to explain the origin of eukaryotes? Philos. Trans. R. Soc. Lond. B. Biol. Sci. 370, 20140327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Claverie J.-M., Abergel C., Open questions about giant viruses. Adv. Virus Res. 85, 25–56 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Yutin N., Koonin E. V., Hidden evolutionary complexity of nucleo-cytoplasmic large DNA viruses of eukaryotes. Virol. J. 9, 161 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guglielmini J., Woo A. C., Krupovic M., Forterre P., Gaia M., Additional data. Zenodo. https://zenodo.org/record/3368642. Deposited 14 August 2019.
- 27.Andreani J., et al. , Cedratvirus, a double-cork structured giant virus, is a distant relative of pithoviruses. Viruses 8, E300 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andreani J., et al. , Orpheovirus IHUMI-LCC2: A new virus among the giant viruses. Front. Microbiol. 8, 2643 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulz F., et al. , Giant viruses with an expanded complement of translation system components. Science 356, 82–85 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Abrahão J., et al. , Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat. Commun. 9, 749 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallot-Lavallée L., Blanc G., Claverie J.-M., Comparative genomics of Chrysochromulina ericina virus and other microalga-infecting large DNA viruses highlights their intricate evolutionary relationship with the established mimiviridae family. J. Virol. 91, e00230-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reteno D. G., et al. , Faustovirus, an asfarvirus-related new lineage of giant viruses infecting amoebae. J. Virol. 89, 6585–6594 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klose T., et al. , Structure of faustovirus, a large dsDNA virus. Proc. Natl. Acad. Sci. U.S.A. 113, 6206–6211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andreani J., et al. , Pacmanvirus, a new giant icosahedral virus at the crossroads between Asfarviridae and Faustoviruses. J. Virol. 91, e00212-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bajrai L. H., et al. , Kaumoebavirus, a new virus that clusters with Faustoviruses and Asfarviridae. Viruses 8, E278 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oliveira G. P., de Aquino I. L. M., Luiz A. P. M. F., Abrahão J. S., Putative promoter motif analyses reinforce the evolutionary relationships among Faustoviruses, Kaumoebavirus, and Asfarvirus. Front. Microbiol. 9, 1041 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krupovic M., Bamford D. H., Koonin E. V., Conservation of major and minor jelly-roll capsid proteins in Polinton (Maverick) transposons suggests that they are bona fide viruses. Biol. Direct 9, 6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krupovic M., Koonin E. V., Polintons: A hotbed of eukaryotic virus, transposon and plasmid evolution. Nat. Rev. Microbiol. 13, 105–115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fischer M. G., Giant viruses come of age. Curr. Opin. Microbiol. 31, 50–57 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Filée J., Forterre P., Sen-Lin T., Laurent J., Evolution of DNA polymerase families: Evidences for multiple gene exchange between cellular and viral proteins. J. Mol. Evol. 54, 763–773 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Da Cunha V., Gaia M., Gadelle D., Nasir A., Forterre P., Lokiarchaea are close relatives of Euryarchaeota, not bridging the gap between prokaryotes and eukaryotes. PLoS Genet. 13, e1006810 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Werner F., Grohmann D., Evolution of multisubunit RNA polymerases in the three domains of life. Nat. Rev. Microbiol. 9, 85–98 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Da Cunha V., Gaia M., Nasir A., Forterre P., Asgard archaea do not close the debate about the universal tree of life topology. PLoS Genet. 14, e1007215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koonin E. V., Yutin N., “Evolution of the large nucleocytoplasmic DNA viruses of eukaryotes and convergent origins of viral gigantism” in Advances in Virus Research, Kielian M., Mettenleiter T. C., Roossinck M. J., Eds. (Academic Press, New York, 2019), pp. 167–202. [DOI] [PubMed] [Google Scholar]
- 45.Yutin N., Wolf Y. I., Koonin E. V., Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life. Virology 466-467, 38–52 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filée J., Route of NCLDV evolution: The genomic accordion. Curr. Opin. Virol. 3, 595–599 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Filée J., Giant viruses and their mobile genetic elements: The molecular symbiosis hypothesis. Curr. Opin. Virol. 33, 81–88 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Eme L., Spang A., Lombard J., Stairs C. W., Ettema T. J. G., Archaea and the origin of eukaryotes. Nat. Rev. Microbiol. 15, 711–723 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Blombach F., et al. , Identification of an ortholog of the eukaryotic RNA polymerase III subunit RPC34 in Crenarchaeota and Thaumarchaeota suggests specialization of RNA polymerases for coding and non-coding RNAs in Archaea. Biol. Direct 4, 39 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jurka J., et al. , Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110, 462–467 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Eddy S. R., Accelerated profile HMM searches. PLOS Comput. Biol. 7, e1002195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Criscuolo A., Gribaldo S., BMGE (Block Mapping and Gathering with Entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 10, 210 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen L.-T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., Jermiin L. S., ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang H.-C., Minh B. Q., Susko E., Roger A. J., Modeling site heterogeneity with posterior mean site frequency profiles accelerates accurate phylogenomic estimation. Syst. Biol. 67, 216–235 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Guindon S., et al. , New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Minh B. Q., Nguyen M. A. T., von Haeseler A., Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lartillot N., Lepage T., Blanquart S., PhyloBayes 3: A Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25, 2286–2288 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Lartillot N., Philippe H., A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 21, 1095–1109 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Embley T. M., van der Giezen M., Horner D. S., Dyal P. L., Foster P., Mitochondria and hydrogenosomes are two forms of the same fundamental organelle. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 191–201; discussion 201–202 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Susko E., Roger A. J., On reduced amino acid alphabets for phylogenetic inference. Mol. Biol. Evol. 24, 2139–2150 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Whidden C., Zeh N., Beiko R. G., Supertrees based on the subtree prune-and-regraft distance. Syst. Biol. 63, 566–581 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shimodaira H., An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51, 492–508 (2002). [DOI] [PubMed] [Google Scholar]
- 65.Letunic I., Bork P., Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the trees presented in this study are included as Newick files within Additional data, together with the alignments in FASTA format (https://zenodo.org/record/3368642; see ref. 26). This folder also contains a table listing the proteins conserved among the NCLDV families. In addition, an online platform has been developed to grant readers easy access to the multiple sequence alignments and position-specific scoring matrix (PSSM) files (http://giphy.pasteur.fr/PhyloM/NCLDV/), with usage information.