Significance
Aminoacyl-transfer RNA (tRNA) synthetases (aaRSs) are ubiquitously expressed enzymes catalyzing the charging of tRNAs with their cognate amino acids to provide key building blocks for protein synthesis. Currently, aaRSs represent the largest family of proteins implicated in the etiology of neurodegenerative Charcot–Marie–Tooth (CMT) disease. Using an assay that enables detection of aminoacylation within the human cell environment, this work investigates charged tRNA in CMT disease patients. The results suggest histidyl-tRNA synthetase (HisRS)–linked CMT disease is not caused by a loss-of-function mechanism. Further biochemical and biophysical analyses of several CMT disease-linked mutant HisRS proteins go on to show that disease severity is correlated with the degree of mutation-induced structural opening, thus consistent with a conformation-driven gain-of-function mechanism.
Keywords: histidyl-tRNA synthetase, Charcot–Marie–Tooth disease, structural rearrangement, gain-of-function mechanism
Abstract
Aminoacyl-transfer RNA (tRNA) synthetases (aaRSs) are the largest protein family causatively linked to neurodegenerative Charcot–Marie–Tooth (CMT) disease. Dominant mutations cause the disease, and studies of CMT disease-causing mutant glycyl-tRNA synthetase (GlyRS) and tyrosyl-tRNA synthetase (TyrRS) showed their mutations create neomorphic structures consistent with a gain-of-function mechanism. In contrast, based on a haploid yeast model, loss of aminoacylation function was reported for CMT disease mutants in histidyl-tRNA synthetase (HisRS). However, neither that nor prior work of any CMT disease-causing aaRS investigated the aminoacylation status of tRNAs in the cellular milieu of actual patients. Using an assay that interrogated aminoacylation levels in patient cells, we investigated a HisRS-linked CMT disease family with the most severe disease phenotype. Strikingly, no difference in charged tRNA levels between normal and diseased family members was found. In confirmation, recombinant versions of 4 other HisRS CMT disease-causing mutants showed no correlation between activity loss in vitro and severity of phenotype in vivo. Indeed, a mutation having the most detrimental impact on activity was associated with a mild disease phenotype. In further work, using 3 independent biophysical analyses, structural opening (relaxation) of mutant HisRSs at the dimer interface best correlated with disease severity. In fact, the HisRS mutation in the severely afflicted patient family caused the largest degree of structural relaxation. These data suggest that HisRS-linked CMT disease arises from open conformation-induced mechanisms distinct from loss of aminoacylation.
Charcot–Marie–Tooth (CMT) disease is the most common inherited disorder of the peripheral nervous system with a prevalence of 1 in 2,500 individuals. The disease is characterized by progressive muscle weakness, atrophy, and sensory loss in the distal extremities (1) and is associated with broad genetic heterogeneity. So far, mutations in more than 90 genes are causatively linked to the disease (2, 3). However, the mechanism(s) involved in the disease for most genes are still poorly understood. The largest gene family implicated in CMT disease encodes aminoacyl-transfer RNA (tRNA) synthetase (aaRS), with 6 members (i.e., glycyl-, tyrosyl-, tryptophanyl-, alanyl-, histidyl-, methionyl-tRNA synthetase) identified to date (4–9), highlighting the importance of understanding the etiology of aaRS-linked CMT diseases (10).
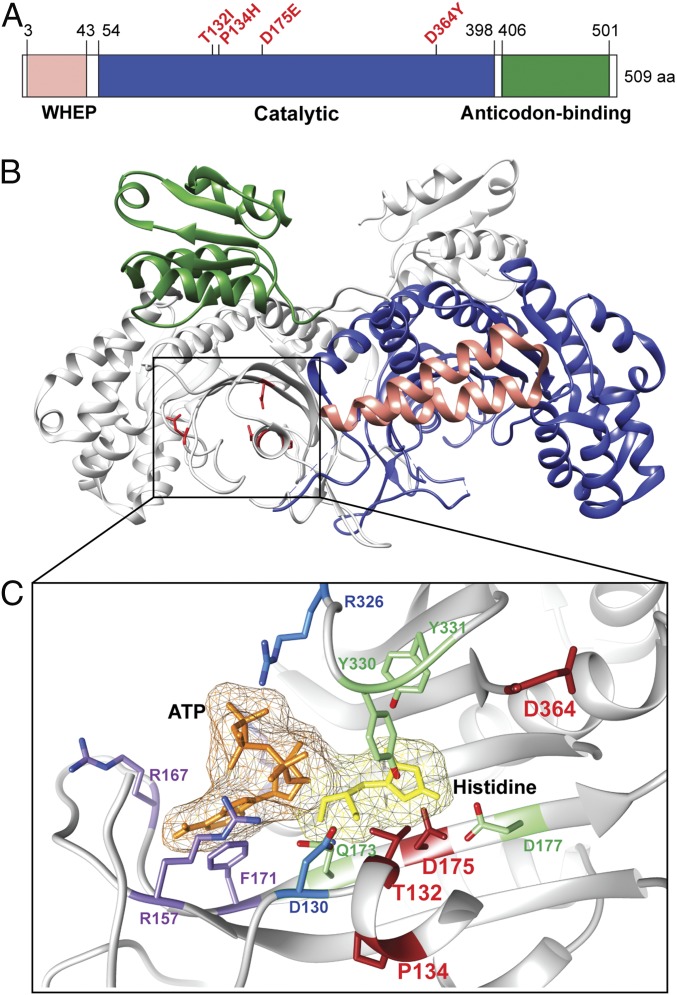
The aaRSs drive cellular messenger RNA translation by catalyzing the attachment of amino acids to their cognate tRNAs for use in ribosomal protein synthesis (11). The aminoacylation reaction is carried out in 2 steps. In the first step, aaRSs bind to amino acid and adenosine 5′-triphosphate (ATP) to catalyze the formation of an enzyme-bound aminoacyl-adenylate. The reaction liberates pyrophosphate (PPi). In the second step, the activated amino acid subsequently reacts with tRNA to yield aminoacyl-tRNA. The histidyl-tRNA synthetase (HisRS or HARS1) is one of the 6 aaRSs associated with CMT disease, designated as subtype CMT2W with predominant axonal phenotypes and autosomal dominant inheritance (7, 12). The subtype is also known as AD-CMTax-HARS based on a new CMT disease classification system (13). The human HisRS protein is constituted of 3 distinct domains: the N-terminal WHEP domain, catalytic domain, and C-terminal anticodon-binding domain (Fig. 1A). Interestingly, CMT disease-associated HisRS mutations are located in the catalytic domain surrounding the active site (Fig. 1).
Fig. 1.
Structure of HisRS and location of CMT2W-causing mutations. (A) Domain structure of human HisRS labeled with the 4 CMT2W-causing mutations studied in this paper. aa, amino acids. (B) Crystal structure of dimeric human HisRS (PDB ID code 6O76), which is obtained by reprocessing the deposited data of PDB ID code 4X5O to reveal the WHEP domain for one of the 2 subunits of the dimeric HisRS. (C) Close-up view of the histidine- and ATP-binding sites in human HisRS. Histidine and ATP were modeled into the structure based on the human HisRS: histidine cocrystal (PDB ID code 4PHC) and E. coli HisRS/ATP cocrystal (PDB ID code 1KMN), respectively. The residues involved in histidine and ATP binding, but not both, are labeled in green and purple, respectively, whereas the residues involved in the binding of both ATP and histidine are labeled in blue.
Four CMT2W families were initially identified, with each linked to a distinct mutation (i.e., T132I, P134H, D175E, D364Y) through strong genetic evidence (7). Relatively speaking, patients in the family that carry a P134H mutation exhibit the most severe and consistent phenotypes, with onset in childhood or (late) adolescence. Patients carrying a D364Y mutation show relatively severe disease (although the mutation has incomplete penetrance). Patients with a T132I or D175E mutation are either mildly affected or have variable disease presentations. The disease severity was assessed by a combination of different clinical factors, including the average age at disease onset of the patients carrying the mutation and the severity of the symptoms. To understand the impact of these mutations on tRNA aminoacylation activity within the components of a cell, functional activity was evaluated using a genetic complementation assay in haploid yeast (7). Because 3 of the 4 mutations (T132I, P134H, and D364Y) caused a complete loss of function in this yeast model, whereas D175E HisRS had partial activity, the results could not correlate disease severity with activity (7).
In 2 other aaRS-linked CMT disease subtypes—CMT2D and DI-CMTC (linked to glycyl-tRNA and tyrosyl-tRNA synthetases [GlyRS and TyrRS], respectively), a toxic gain-of-function mechanism became evident (14–22). At the molecular structure level, this gain of function is supported by mutation-induced conformational opening to expose new surfaces to mediate aberrant interactions with other proteins (23, 24). Moreover, the yeast complementation assay was carried out using haploid cells and is not a straightforward mimic of the situation in diploid human cells. Considering the dominant nature of the CMT disease-linked aaRS mutations, and the existence of a wild-type (WT) allele of the tRNA synthetase in patients, activities need examination at the patient level to better understand the impact of a dominant mutation on tRNA aminoacylation.
Here, we obtained samples from the most severely affected P134H patient family and subjected the samples to several analyses, including a Northern blot protocol that interrogates the endogenous tRNA aminoacylation status. No difference in the aminoacylation level of tRNAHis and other tRNAs was observed between P134H patients and their unaffected family members. Further analysis of aminoacylation using purified recombinant HisRS and the 4 CMT disease-causing mutants confirmed the lack of correlation between enzymatic activity and disease severity. In contrast, 3 independent biophysical analyses on HisRS proteins, including small-angle X-ray scattering (SAXS), hydrogen/deuterium exchange (HDX), and switchSENSE hydrodynamic sizing, consistently revealed that the largest conformational change and structural opening were induced by P134H HisRS when compared with the other mutant proteins, and strongly correlated the opening up of the structure with CMT disease severity.
Results and Discussion
CMT Disease Patients Carrying the P134H Mutation in HisRS Do Not Have a Defect in tRNA Aminoacylation.
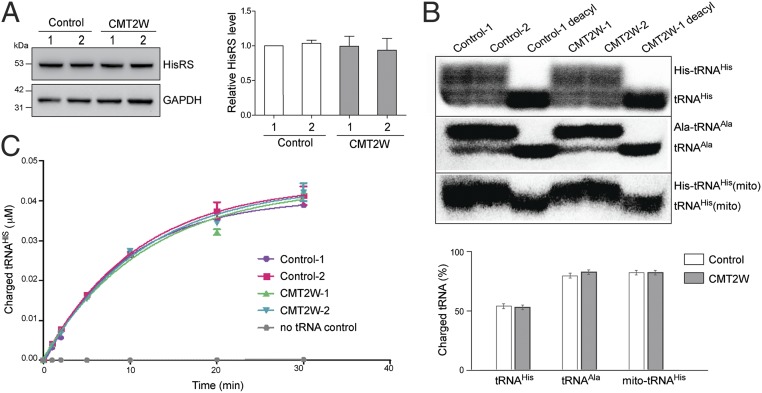
In the yeast complementation assay, HisRSP134H was shown to be completely unable to support cell growth (7), suggesting the mutant protein lost the enzymatic activity for charging tRNAHis with histidine. To understand whether the WT HARS allele in P134H patients can provide sufficient activity to compensate for the loss-of-function mutant allele, we analyzed HisRS activity in the CMT2W patient family. Immortalized lymphocytes from 2 P134H patients and 2 unaffected family members not bearing the mutation were first compared for the level of HisRS protein. No difference was identified (Fig. 2A), thus indicating the mutation does not affect HisRS expression and stability. To evaluate HisRS activity, we performed Northern blot analysis to examine the aminoacylation status in situ of endogenous tRNAs in these samples. Total RNAs were extracted and analyzed on gels under acidic conditions to preserve the charged tRNAs from deacylation. The charged fraction of the cognate cytosolic tRNAHis was around 50% in these cells, and these levels were the same among patients and their unaffected family members (Fig. 2B). For controls, we examined a noncognate cytoplasmic tRNA (tRNAAla) and the mitochondrial tRNAHis, and again found no difference in their charging status between patient and control samples (Fig. 2B). Next, to confirm the lack of activity loss in patients, we carried out an in vitro aminoacylation assay with cell lysates and compared their capacities for aminoacylating tRNAHis. We found no decrease in aminoacylation activity with the 2 CMT disease patient samples versus the control samples (Fig. 2C). Apparently, the presence of the HisRSP134H mutant with HisRSWT does not create a (dominant) negative effect to compromise the cellular charging capacity. Thus, we concluded that the levels of charged tRNAHis are unperturbed in the cells of the most severely afflicted patients.
Fig. 2.
CMT2W patients carrying the P134H mutation do not have a defect in tRNA aminoacylation. (A, Left) Western blot analysis of 4 PBMC-derived lymphoblast samples from 1 family: 2 isolated from CMT2W patients (CMT2W 1 and 2) and 2 from healthy members (control 1 and 2). (A, Right) Normalized relative levels of HisRS protein among all family members. (B, Upper) Northern blot analysis of total RNA extracted from PBMC-derived lymphoblast samples probed against nuclear-encoded tRNAHis, nuclear-encoded tRNAAla, and mitochondrial (mito)-encoded tRNAHis. Total RNA was loaded onto a denaturing sequencing gel to separate charged from uncharged tRNAs. deacyl, deacylated samples included as controls. (B, Lower) Charged fraction of investigated tRNAs was quantified as a ratio of charged-to-total tRNA. Quantification data are presented as mean ± SD (n = 2 biological replicates per group). (C) In vitro tRNA aminoacylation assay with lysates of PBMCs to detect HisRS activity. Quantification data are presented as mean ± SD (n = 3 technical replicates).
No Correlation between Enzymatic Activity of CMT2W Mutations and Disease Severity.
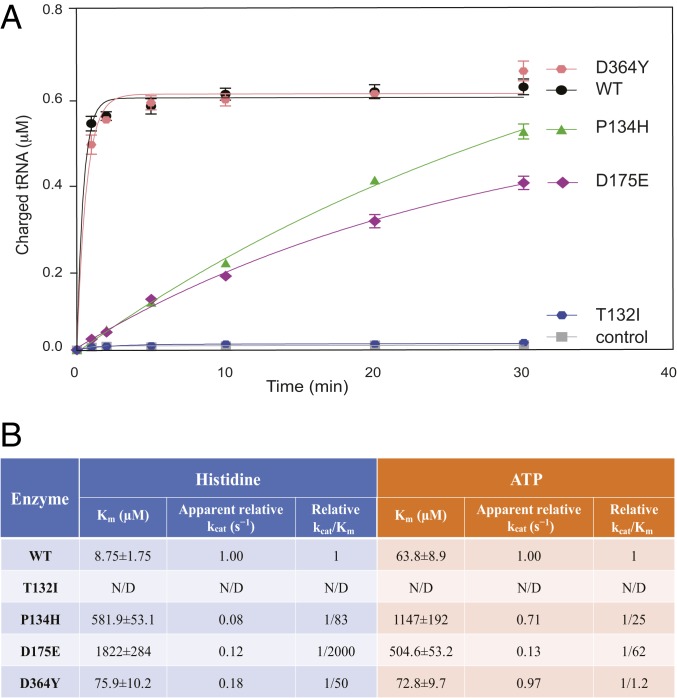
The apparent lack of an aminoacylation defect in CMT2W patients with the most severe forms of the disease does not support a loss-of-function mechanism (7). This prompted us to compare the aminoacylation activity of HisRSP134H with other CMT2W-linked mutants associated with milder phenotypes, using the in vitro aminoacylation assay. The recombinant human HisRS WT (HisRSWT) protein and the 4 CMT2W mutants reported in the original study (7) were overexpressed and purified to allow accurate analysis (SI Appendix, Fig. S1A). (The human HisRS is dimeric, and all CMT2W mutations do not appear to affect dimer formation, as shown in SI Appendix, Fig. S1B.) Interestingly, in contrast to complete loss-of-function phenotypes in yeast (7), HisRSP134H had substantial activity, with an initial aminoacylation rate at ∼40% of the WT protein, whereas HisRSD364Y had full aminoacylation activity. Consistent with the yeast assay, HisRST132I had a complete loss of function and HisRSD175E had partial activity slightly below that of HisRSP134H (Fig. 3A). Given that the T132I mutation caused the most severe loss of aminoacylation function but induced a relatively mild disease phenotype, a lack of correlation between enzymatic activity and disease severity is indicated.
Fig. 3.
Enzymatic analyses of HisRS and its CMT2W mutants. (A) In vitro tRNA aminoacylation assay with purified HisRS proteins. A reaction with WT HisRS, but no tRNA, was included as a control. (B) Steady-state kinetics of WT and mutant HisRS proteins for histidine and ATP determined by ATP/PPi exchange analysis. Data are presented as mean ± SD (n = 3 replicates per group). kcat, catalytic rate constant; N/D, not determined.
A recent study reported additional mutations linked to CMT2W (i.e., V155G, Y330C, S356N) and examined their impact on enzyme kinetics (12). All 3 mutants reportedly have a defect in binding to substrates, especially ATP and histidine (12). To investigate whether this defect pertains to the 4 earlier mutants described above, we carried out a kinetic analysis using an ATP/PPi exchange assay that monitors the first step of the 2-step aminoacylation reaction (25). In this assay, like in the overall aminoacylation assay, HisRST132I has a complete loss of function, and thus does not allow for kinetic analysis. Other mutants (HisRSP134H, HisRSD175E, and HisRSD364Y), to different extents, have an increased Michaelis constant (Km) for histidine and ATP (Fig. 3B), indicating some deficiency in substrate binding, consistent with the findings on the other 3 CMT2W mutants (12). However, because the first step of the tRNA aminoacylation reaction is not rate-limiting, a mild deficiency in histidine and ATP binding may not affect the efficiency of the overall aminoacylation reaction. Similar to the tRNA aminoacylation assay (Fig. 3A), HisRSD364Y showed the least amount of activity defect in the ATP/PPi exchange assay (Fig. 3B). The same order of enzymatic deficiency is obtained from tRNA aminoacylation and ATP/PPi exchange assays (T132I > D175E > P134H > D364Y), which has no correlation to the disease severity associated with each mutation.
Crystal Structure of HisRSD175E Shows No Conformational Change at the Active Site.
The increased Km for histidine and ATP for the mutant proteins indicate potentially altered active site structures. We set out to determine the crystal structures of the CMT2W mutants. [The crystal structure of human HisRSWT has been previously solved (26, 27).] Crystals were obtained for 3 mutants (HisRST132I, HisRSD175E, and HisRSD364Y). However, only HisRSD175E crystals were diffracted to high resolution (3.1 Å). We solved the crystal structure of this mutant using molecular replacement with the HisRSWT structure (Protein Data Bank [PDB] ID code 4X5O) as a template. Interestingly, the crystal structure of HisRSD175E is similar to the HisRSWT structure with a root-mean-square deviation for 376 Cα atoms of 0.97 Å (SI Appendix, Fig. S2). The crystallization conditions used for this mutant were almost identical to the conditions used for the HisRSWT (26), and the crystals have the same space group (P41212). We did not observe any significant differences around the ATP- or histidine-binding sites, as we would expect from the kinetic studies (Fig. 3B). We attributed the lack of conformational change to the constraints imposed by the crystal lattice, as we observed for other CMT disease-linked tRNA synthetase mutants (23, 24), and moved on to analyzing the structural characteristics of those proteins in solution.
SAXS Shows an Extended Conformation for CMT2W Mutants.
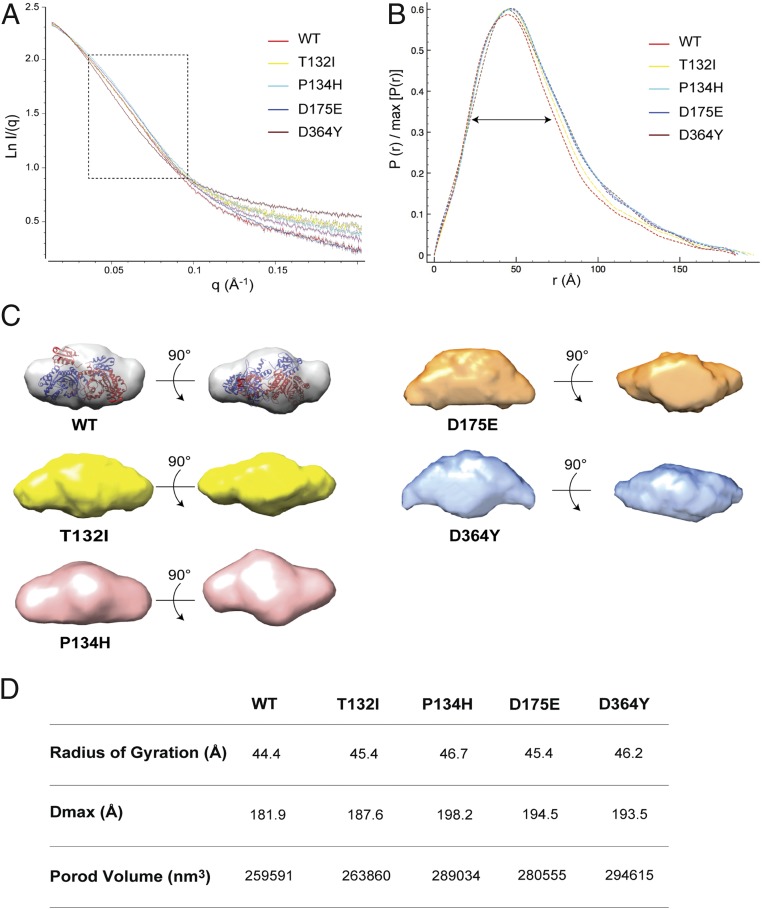
SAXS experiments were conducted for the 4 CMT2W mutants and HisRSWT. For each protein, the shapes of the scattering curves are independent of the protein concentration in the range of 25 to 215 μM (SI Appendix, Fig. S3A) and the corresponding Guinier plots show parallel and linear fits (SI Appendix, Fig. S3B), indicating the absence of significant aggregation during the measurements. The molecular masses extrapolated from the scattering curves indicate a dimeric organization of all proteins, consistent with estimates from gel filtration chromatography (SI Appendix, Fig. S1B).
A close look at the scattering curves (Fig. 4A) within the medium-resolution region (q values from 0.035 to 0.1 Å−1) shows a clear separation between the different spectra, indicating conformational differences between the proteins. The pairwise distance distribution functions [P(r)s] of the 4 CMT2W mutants display a shift toward higher r values (paired set of distances between all of the electrons within the protein structure), indicating more relaxed structures for the mutants (Fig. 4B). Consistently, ab initio shape reconstruction analyses show a more elongated shape for all of the mutants compared with HisRSWT (Fig. 4C). To obtain a quantitative comparison of the proteins, we derived the radius of gyration, maximum dimension (Dmax), and Porod volume from the scattering data. Interestingly, all of the parameters are larger for the mutants than for HisRSWT. Among the mutants, HisRSP134H exhibits the largest, whereas HisRST132I exhibits the smallest, conformational change, based on radius of gyration and Dmax parameters (Fig. 4D).
Fig. 4.
SAXS analysis of WT HisRS and its CMT disease mutants. (A) Experimental solution scattering data for the HisRS proteins. The dotted square shows the medium-resolution region of the scattering spectra. (B) P(r) of the SAXS data determined during data reduction with GNOM. (C) Two different views of the ab initio envelopes generated with DAMMIF for the HisRS proteins. The structure of the dimeric HisRS was manually docked into the envelope of HisRSWT. (D) Summary table of main SAXS parameters for the proteins tested.
The switchSENSE Technology Confirms the Increased Size for CMT2W Mutants.
In order to confirm the observations that all 4 CMT disease mutations induce structural relaxation, we used the switchSENSE technology (28) to assess the hydrodynamic radius of those proteins. Small double strands of DNA are tethered to the gold surface of the analytical chip on one side and allow the binding to proteins of interest on the other end (SI Appendix, Fig. S4A). The motion of the DNA levers is then triggered by alternating the voltage across the electrode surface and is recorded in real time through the fluorescence probe attached to the DNA. Upon conjugation of the HisRS proteins, the hydrodynamic friction of the DNA double strand is affected, as well as the movement of the DNA. The movements were recorded (SI Appendix, Fig. S4B) and used to estimate the hydrodynamic radius of the proteins (SI Appendix, Fig. S4C). We found all mutants, to different extents, have a larger hydrodynamic radius than the HisRSWT protein. The relative radius increase for the mutants ranges between 4% and 20%. HisRST132I is the smallest of the mutants recorded, with only a 4% increase observed in comparison to HisRSWT. The recorded hydrodynamic radii for HisRSP134H, HisRSD175E, and HisRSD364Y are similar, with 18%, 20%, and 16% increases, respectively. The results are consistent with those from SAXS in suggesting structural relaxation for all mutants and in characterizing HisRST132I with less pronounced structural opening than the other CMT2 mutants.
HDX Analyses Suggest Disease-Correlated Conformational Change.
In order to pinpoint the site of the conformational opening, we recorded HDX for HisRSWT and the 4 CMT2W mutants. HDX is a powerful analytical technique for studying protein dynamics in solution and revealing conformational changes when mutants are compared with the WT protein. After each exchange period in a D2O buffer, the protein is rapidly proteolyzed into fragments and analyzed using high-resolution proteomics methods to gain insights on the solvent accessibility to different regions of the protein.
For all of the proteins, HDX/mass spectrometry (MS) analysis gave excellent peptide coverage (>90%) (SI Appendix, Fig. S5A). Compared with HisRSWT, HisRSP134H and HisRSD364Y show an overall increase (6% and 3%, respectively, after 10,000 s of exchange), whereas HisRST132I and HisRSD175E show an overall decrease (4% and 5%, respectively, after 10,000 s of exchange) in deuteration level (SI Appendix, Fig. S5B). Interestingly, T132 and D175 residues are located near the histidine-binding pocket, and both T132I and D175E mutations substitute a smaller side chain with a larger one, which would stabilize the pocket in the absence of the amino acid (Fig. 1C). The stabilization effect would reduce the dynamics of the structure and explain the overall decrease in deuterium uptake for these 2 mutants. Combining the SAXS and hydrodynamic sizing data (Fig. 4D and SI Appendix, Fig. S4C) with the HDX analysis, we conclude that the structures of HisRST132I and HisRSD175E are enlarged and extended but, at the same time, become less flexible than that of HisRSWT.
In contrast, P134 and D364 are located on the periphery and with the side chains pointed away from the active site (Fig. 1C). The distal location of the P134H and D364Y mutations (relative to the T132I and D175E mutations) to the active site also explains their lesser impact on the enzymatic activity of HisRS (Fig. 3). The overall increase in deuterium uptake induced by P134H and D364Y mutations and their SAXS and hydrodynamic sizing data (Fig. 4D and SI Appendix, Fig. S4C) indicate an extended and relaxed structure for HisRSP134H and HisRSD364Y. Clinically, the P134H and D364Y CMT2W patients have more severe disease than the T132I and D175E patients (7). Therefore, based on these 3 biophysical analyses (SAXS, switchSENSE, and HDX), we conclude that all 4 CMT2W mutants have a more extended structure than the WT HisRS and that the extent of the size increase and the flexibility of the structure positively correlates with the disease severity.
Modeling the Conformational Change Induced by CMT2W Mutations in HisRS.
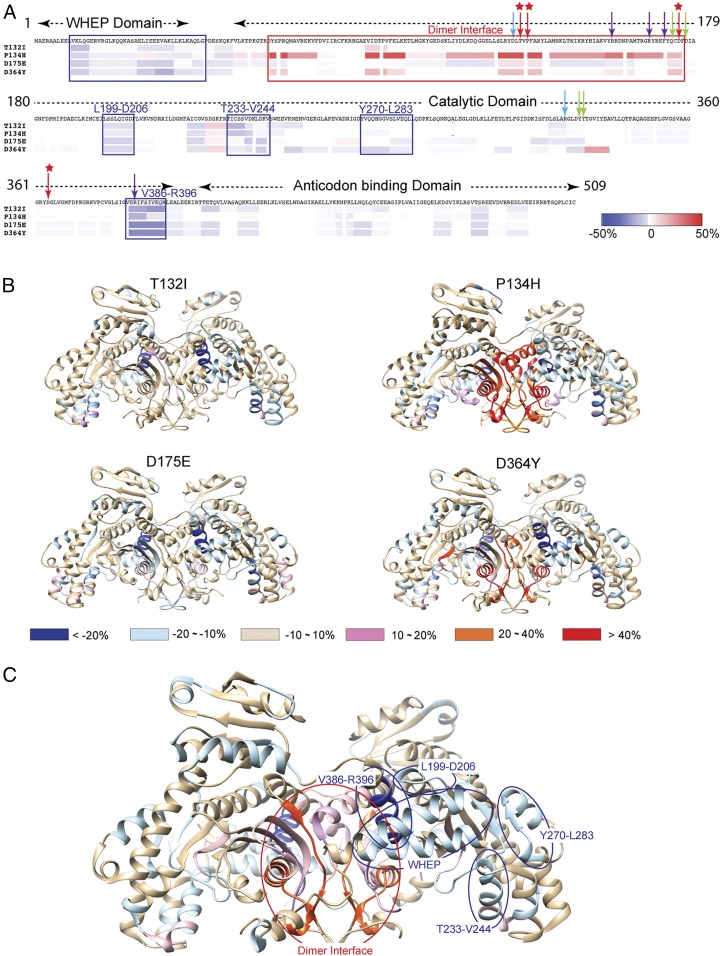
By aligning the differential deuterium uptake data of the 4 mutants together (Fig. 5A), we made 2 observations. First, albeit differences in overall change, the N-terminal region of the catalytic domain (D64 to F176), which constitutes the dimer interface (Fig. 5), shows a general increase in HDX for CMT2W mutants compared with HisRSWT. This observation indicates that the mutation-induced structural extension occurs at the dimer interface. Importantly, this increase in deuterium incorporation is unlikely due to mutation-induced dimer dissociation, as we found that the mutant proteins form stable dimers even at a concentration as low as 200 nM, substantially lower than the concentration we used for HDX experiments (6 μM).
Fig. 5.
HDX analysis. (A) Differential deuterium incorporation induced by each of the 4 CMT2W mutations after 10,000 s of HDX mapped onto the HisRS sequence. Blue rectangles indicate consensus regions that show a decrease in deuterium incorporation compared with the WT, while the red rectangle indicates a region with increased deuterium incorporation. The arrows indicate the CMT2W-causing mutations (red) and the residues involved in histidine binding (green), ATP binding (purple), or both (blue). (B) Mapping differential deuterium incorporation induced by each CMT2W mutation onto the HisRS dimer crystal structure. (C) Average levels of deuterium incorporation change induced by the 4 CMT2W mutations mapped onto the dimeric HisRS crystal structure.
Second, all 4 mutants share a decrease in deuterium uptake for the WHEP domain and for 4 regions in the C-terminal part of the catalytic domain: L199-D206, T233-V244, Y270-L283, and V386-R396 (Fig. 5A). Interestingly, the WHEP domain and these regions are contiguous in structure (Fig. 5C), suggesting that the conformational change induced by a CMT2W mutation strengthens the interactions between the WHEP domain and this part of the catalytic domain. Moreover, the areas associated with decreased deuterium uptake are also adjacent to the dimerization interface (Fig. 5C). This suggests that the enhanced interaction between the WHEP domain and the C-terminal part of the catalytic domain may help to open up the dimerization interface, and do so more effectively in HisRSP134H and HisRSD364Y than in HisRSD175E and HisRST132I (Fig. 5B).
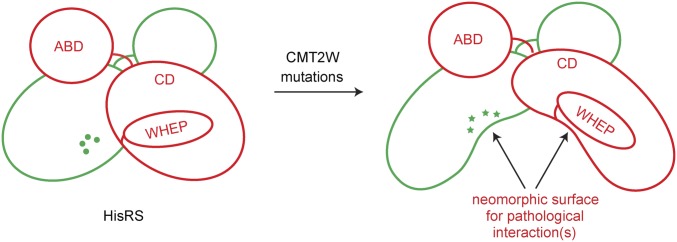
Based on these considerations, we created a cartoon to illustrate the conformational change induced by CMT2W mutants (Fig. 6). The conformational change opens up the dimer interface area of HisRS and is likely to expose neomorphic surfaces that may engender aberrant interactions with CMT2W mutants but not with WT HisRS. This could form the structural basis for potential toxic gain-of-function effects of CMT2W mutants. Such toxic gain-of-function effects, due to aberrant interactions with other partners, have been demonstrated for CMT disease-linked GlyRS and TyrRS mutants (18, 21, 24).
Fig. 6.
Schematic illustration of the conformational change induced by CMT2W mutations on HisRS. The conformational change involves the opening and relaxation at the dimer interface without disrupting dimer formation. The relaxation at the dimer interface is accompanied by decreased flexibilities at the WHEP domain and part of the catalytic domain (CD), possibly through strengthened interactions between them. The conformational opening creates neomorphic surfaces that may engender aberrant interactions with other cellular molecules, leading to a diseased state. ABD, anticodon-binding domain.
It is interesting to note that many CMT disease-associated aaRSs contain a WHEP domain. These include GlyRS, HisRS, TrpRS, and MetRS. In fact, all single-WHEP domain-containing aaRSs are implicated in CMT disease, suggesting the relevance of this appended domain for CMT disease. Indeed, Cader and coworkers (17) demonstrated that the toxicity of GlyRSP234KY in the fly model is WHEP domain-dependent. The WHEP domain in both HisRS and GlyRS is associated with a high level of deuterium update during HDX analysis, indicating a dynamic and flexible conformation. In case of HisRS, as discussed above, the WHEP domain seems to play a role in stabilizing the opening conformation of HisRS CMT disease mutants. Such a role was not observed in GlyRS, where no obvious change in deuterium update occurred in the WHEP domain of the CMT disease mutants (23), suggesting that facilitating conformational opening may not be the key role of the WHEP domain for its CMT disease association.
By measuring the activities of individual mutant HisRSs we were able to rule out a correlation between activity and disease severity and, instead, go on to show that a correlation can be found with the degree of conformational change associated with each mutation. Those neomorphic surfaces exposed by the conformational change may mediate aberrant interactions and toxic gain-of-function effects, leading to a diseased state (Fig. 6). In conclusion, this study has provided an examination of tRNA aminoacylation in CMT disease patient cells. Both the lack of aminoacylation defect in CMT disease patients and the lack of correlation between the level of aminoacylation defect caused by different CMT2W-causing mutations and disease severity suggest strongly that CMT2W is not caused by a loss-of-function mechanism or a dominant negative effect of the mutations affecting tRNA aminoacylation in patients. Moreover, this study successfully links CMT2W phenotypes with physical alternations of the HisRS protein, opening doors for future molecular mechanistic studies.
Materials and Methods
Expression and Purification of HisRS and the CMT2W Mutants.
The Escherichia coli strain Rosetta [DE3] pLysS (Novagen) was used for expression of HisRS and all 4 recombinant CMT2W mutant proteins. Cultures were grown overnight to saturation in Luria-Bertani (LB) medium containing 100 μg/mL ampicillin and 34 μg/mL chloramphenicol. An aliquot of the overnight culture was diluted 1/25 in Super Broth (SB) medium and grown at 37 °C. At an optical density of 600 nm of 0.7, isopropyl β-d-thiogalactopyranoside was added to a final concentration of 0.2 mM and the cells were grown at 37 °C for 3 h. The induced cells were harvested, washed, and collected by centrifugation (5,000 × g, 10 min). The resulting bacterial pellets were frozen at −20 °C (24).
The pellet was resuspended in 5 volumes (vol/wt) of buffer A (50 mM Tris⋅HCl [hydrogen ion concentration (pH) 8], 300 mM NaCl, 20 mM imidazole, 1 mM phenyl-methyl-sulphonyl-fluoride supplemented with 0.1 mg/mL lysozyme, 10 μg/mL DNase I [Zymo Research], 20 mM MgSO4, protease inhibitor mixture [Sigma] [50 μL/g of cells]). After a 20-min incubation with gentle agitation, the cells were disrupted by sonication (using a 750-W sonicator and 5 cycles of 30 s each at 60% power output). The lysate was clarified by centrifugation at 35,000 × g for 30 min. Starting from a 1-L culture, the clarified supernatant was injected onto a 5-mL HisTrap FF column (GE Healthcare), previously equilibrated in buffer A supplemented with 1 M NaCl. Elution was carried out using a gradient of imidazole in buffer A supplemented with 1 M NaCl. The fractions containing the protein were combined and loaded onto a Superdex 200 HR 16/60 column (GE Healthcare) and eluted in 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes), 150 mM NaCl, and 1 mM Tris(2-carboxyethyl)phosphine (pH 7.5). The protein was concentrated using Centricon Plus-20 (molecular cutoff of 10,000 Da; Millipore) and then stored at −20 °C (24).
All purification steps were carried out at 4 °C. The apparent molecular mass of proteins eluted from the gel filtration column was deduced from a calibration carried out with low-molecular-weight and high-molecular-weight calibration kits (Amersham Pharmacia Biotech). Protein concentrations were calculated using the theoretical absorption coefficient ε (milligrams per milliliter per centimeter) at 280 nm as obtained using the program ProtParam at the EXPASY server (https://www.expasy.org/tools).
In Vitro Aminoacylation Assay.
As described previously (25), the aminoacylation assays were performed at room temperature with 50 mM Hepes (pH 7.5), 20 mM KCl, 5 mM MgCl2, 4 mM ATP, 2 mM dithiothreitol (DTT), 4 µg/mL pyrophosphatase, 20 μM cold l-histidine, and 1.34 μM [3H]-histidine (1 mCi/mL) as the assay solution. Total yeast tRNA (1 μg/μL) was mixed with the assay solution, and the reaction was initiated by adding 200 nM HisRS protein or 1 μg/μL cell lysate into the mixture. The cell lysate was obtained by resuspension of the centrifuged cells in ice-cold lysis buffer supplemented with proteases inhibitors and followed by a 30-min incubation. Then, the lysate was clarified after a 10-min centrifugation at 12,000 rpm, and the enzyme concentration was measured by bicinchoninic acid (BCA). At varying time intervals, 5-μL aliquots were applied to a MultiScreen 96-well filter plate (0.45-μm pore size hydrophobic, low-protein-binding membrane; Millipore), which is prewetted with quench solution containing 0.5 mg/mL DNA and 100 mM ethylenediaminetetraacetic acid (EDTA) in 300 mM NaOAc (pH 3.0). After all time points were collected, 100 μL of 20% trichloroacetic acid (TCA) was added to precipitate the nucleic acids. The plate was then washed 4 times with 200 μL of 5% TCA containing 100 mM cold histidine, followed once with 95% ethanol. On drying after completion of the washing steps, 70 μL of 100 mM NaOH was added to elute the tRNAs, which was then centrifuged into a 96-well flexible PET microplate (PerkinElmer) with 150 μL of Supermix scintillation mixture (PerkinElmer). After mixing, the radioactivity in each well of the plate is counted in a 1450 LSC & Luminescence Counter (PerkinElmer).
Patient Lymphocyte Immortalization, Cell Culture, and Growth.
For each patient, 20 mL of peripheral blood was collected for generating immortalized lymphocytes. Within 48 h of the blood draw, peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation using Histopaque-1077 (Sigma–Aldrich), followed by removing red blood cells using RBC Lysis Buffer (Thermo Fisher Scientific), and cultured in RPMI-1640 media supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Immortalization was accomplished by adding Epstein–Barr virus and cyclosporin (1:1,000) to the resuspended PBMCs, followed by incubation at 37 °C for 1 mo. Immortalized lymphocytes were maintained in RPMI-1640 media with 10% FBS and 1% penicillin/streptomycin. When collecting the lymphocytes, the cells were separated from the media by centrifugation at 500 × g for 5 min.
The Ethical Review Board of the University of Antwerp approved the study, and all patients or their legal representatives signed an informed consent form.
Northern Blot Analysis.
Oligonucleotide probes for Northern blots were 5′ 32P-labeled with T4 polynucleotide kinase (PNK) and gel-purified. The probe sequences are as follows: 5′TGCCGTGACTCGGATTCGAACCGAGGTTGCTGCGGCCACAACGCAGAGTACTAACCACTATACGATCACGGC for nuclear-encoded tRNAHis,
5′TGGAGAATGYGGGCGTCGATCCCRCTACCTCTYGCATGCTAAGCRAGCGCTCTACCRCCTGAGCTAATTCCCC (R = A/G; Y = C/T) for nuclear-encoded tRNAAla (IGC), and 5′GGTAAATAAGGGGTCGTAAGCCTCTGTTGTCAGATTCACAATCTGATGTTTTGGTTAAACTATATTTAC for mitochondrial-encoded tRNAHis.
Total RNA (2.5 μg) in 10 mM NaOAc/HOAc (pH 4.8) was mixed with an equal volume of 8 M urea, 0.1 M NaOAc/HOAc (pH 4.8), 0.05% Bromophenol blue, and 0.05% Xylene cyanol. Total RNA (2.5 μg) was deacylated in Tris⋅HCl (pH 9.0) at 37 °C for 45 min and included as a control. Samples were run on an 8% denaturing sequencing gel (8 M urea, 0.1 M NaOAc/HOAc [pH 5.0]) at 500 V for 24 h at 4 °C. RNA was transferred and fixed to a Hybond-XL membrane (GE Healthcare) using a gel dryer at 80 °C for 2 h. The membrane was prehybridized twice at room temperature for 30 min in 20 mM sodium phosphate (pH 7.0), 300 mM NaCl, and 1% sodium dodecyl sulfate (SDS). Hybridization of radiolabeled oligo (7 pmol) was performed in 20 mM sodium phosphate (pH 7.0), 300 mM NaCl, and 1% SDS at 60 °C for 16 h. Membranes were washed 3 times in 20 mM sodium phosphate (pH 7.0), 300 mM NaCl, 0.1% SDS, and 2 mM EDTA at 60 °C for 20 min. The dried membranes were exposed to imaging plates and quantified using a phosphoimager.
ATP/PPi Exchange Assays.
As described previously (25), the ATP/PPi exchange assays were performed at room temperature with 50 mM Hepes (pH 7.5), 20 mM KCl, 10 mM MgCl2, 2 mM DTT, and 4 μg/mL pyrophosphatase as the assay solution. To determine the kinetic parameters of histidine, 0 to 2 mM histidine, 2 mM ATP, 1 mM NaPPi, and 0.001 μCi/μL [32P] NaPPi were mixed with the assay solution, and the reaction was initiated by adding 200 nM HisRS protein into the mixture. To determine the kinetic parameters of ATP, 1 mM histidine, 0 to 1 mM ATP, 0 to 1 mM NaPPi, and the adjusted amounts of [32P] NaPPi were mixed with the assay solution, and the reaction was initiated by adding 200 nM HisRS protein into the mixture. At varying time intervals, 15-μL aliquots were applied to a MultiScreen 96-well filter plate (0.45-μm pore size hydrophobic, low-protein-binding membrane; Millipore), which is prewetted with quench solution containing 1 M HCl, 200 mM sodium PPi, and 4% charcoal. The plate was washed 4 times with 200 μL of wash solution (1 M HCl, 200 mM sodium PPi) and then dried for ∼30 min under a heat lamp. The wells of the plate were punched into small scintillation vials with 3 mL of scintillation fluid and allowed to sit for at least 30 min. The radioactivity in each vial was counted in the scintillation counter. The amount of charcoal-bound radioactivity was converted into reaction velocity by using the specific activity of the 32PPi. A nonlinear regression fit of the plot of velocity versus substrate concentration provided the maximum velocity (Vmax) and Km values.
Crystal Structure Analysis.
Crystallization experiments were performed immediately after protein purification. Screening experiments were carried out using the sitting-drop vapor diffusion method at 20 °C in 96-well Medical Research Council (MRC) crystallization plates using a nanodrop-dispensing robot (TTP Labtech’s mosquito Crystal) and various commercial crystallization kits (Molecular Dimensions Limited, Emerald BioStructures, and GenaBiosciences). Reservoir solutions were 30 μL in volume, and crystallization drops were composed of 100, 200, or 300 nL of protein solution at 10 mg/mL and 100 nL of reservoir solution. Crystallization plates were sealed with transparent film after setup of the drops and transferred to a storage cabinet at 20 °C (24).
The HisRSD175E crystals grew typically within 72 h to a size of 0.2 × 0.1 × 0.1 mm3 by mixing 300 nL of protein at 15 mg/mL with 100 nL of the same condition used for the HisRSWT crystals (26). Single crystals were harvested from the drop and flash-frozen in liquid nitrogen at 100 K with 2-methyl-2,4-pentanediol as a cryoprotectant. Diffraction datasets were collected at the Stanford Synchrotron Radiation Lightsource (SSRL).
SAXS and Ab Initio Shape Reconstruction.
Data reduction was performed using the established procedure available at beamline BL4-2, and buffer background runs were subtracted from sample runs. The forward scattering intensity was calibrated using bovine serum albumin (BSA) as a reference. All SAXS measurements were carried out at the SSRL on beamline BL4-2 at a working energy of 12.5 KeV corresponding to λ = 0.992 Å. The sample-to-detector distance of the X-rays was 2.847 m, leading to scattering vectors q ranging from 0.028 to 4.525 nm−1. The scattering vector is defined as q = 4π/λ sinθ, where 2θ is the scattering angle. The exposure time was optimized to reduce radiation damage. Data reduction was performed using the established procedure available at the beamline, and buffer background runs were subtracted from sample runs. The radius of gyration (Rg) and forward intensity at zero angle [I(0)] were determined with the program PRIMUS (29) according to the Guinier approximation at low values, in a Q⋅Rg range up to 1.3: Ln[I(Q)] = Ln[I0] − ().
The forward scattering intensity was calibrated using BSA as a reference. The Rg and P(r) were calculated with the program GNOM. The Dmax value was adjusted such that the Rg value obtained from GNOM agreed with that obtained from the Guinier analysis.
SAXS data were collected at 10 °C using 30 μL of 5 different protein concentrations ranging from 25 to 215 μM in 10 mM Hepes (pH 7.5) buffer with 5 mM EDTA, and loaded in a fully automated sample charger. Ten exposures, each of 10 s, were made for each protein concentration and were combined to give the average scattering curve for each measurement (24). Any data points affected by aggregation, possibly induced by radiation damage, were excluded. The profiles obtained at 5 different protein concentrations had the same shape and were flat at low q values, indicating the absence of significant aggregation. Then, we used the higher concentration to have the maximum information at high resolution.
Three-dimensional bead models were built by fitting the scattering data with the program DAMMIF (30). Thirty independent models were generated with DAMMIF assuming P2 symmetry. The models resulting from independent runs were superimposed using the DAMAVER suite (31). This yielded an initial alignment of structures based on their axes of inertia, followed by minimization of the normalized spatial discrepancy, which is 0 for identical objects and larger than 1 for systematically different objects. The aligned structures were then averaged, giving an effective occupancy to each voxel in the model, and filtered at half-maximal occupancy to produce models of the appropriate volume that were used for all subsequent analyses.
HDX Analysis.
Before carrying out HDX experiments, the optimal proteolysis conditions that produced the best sequence coverage map of HisRSWT were established as described (24, 32). Briefly, 3 μL of HisRSWT protein (1 mg/mL) that quenched with 18 μL of 0.08 M guanidine hydrochloride (GuHCl) and digested on an immobilized pepsin column for 40 s generated the most unique peptides (n = 302), which covered 100% of the WT protein sequence. To initiated H/D exchange reactions, 3 μL of protein stock solution (WT or mutants) was mixed with 9 μL of D2O buffer (8.3 mM Tris, 50 mM NaCl in D2O [deuterium ion concentration (pD) 7.6]) and incubated for 10, 100, 1,000, 10,000, and 100,000 s at 0 °C. At the designated times, the HDX reaction was quenched by adding 18 μL of ice-cold quench buffer (0.08 M GuHCl, 0.8% formic acid, 16.6% glycerol) and samples were immediately frozen at −80 °C. In addition, nondeuterated and fully deuterated samples were prepared as previously described (33). The 30-μL quenched samples were thawed at 4 °C and immediately passed over the immobilized pepsin column (1 × 20 mm, 40 mg/mL porcine pepsin; Sigma). The digested peptides were collected on a trap column (0.2 × 2 mm, Magic C18AQ; Michrom) for desalting and separated with a Magic C18 column (0.2 × 50 mm, 3 μm, 200 Å; Michrom) by a 30-min linear acetonitrile gradient of 6.4 to 38.4%. The accurate peptide mass measurement was performed with an OrbiTrap Elite mass spectrometer (Thermo Fisher Scientific), which was set up for optimal HDX performance with a minimum back-exchange rate (34, 35). Data acquisition was completed in either data-dependent tandem MS or MS1 profile mode, and peptide identification was done using Proteom Discoverer software (Thermo Fisher Scientific). Deuterium incorporation levels of each peptide at each time point were determined by HDXaminer (Sierra Analytics, Inc.) with back-exchange correction use above control samples (36).
switchSENSE Analysis.
Kinetic measurements were performed using a DRX series instrument (Dynamic Biosensors) with a MPC-48-2-Y1 chip (Dynamic Biosensors). All HisRS proteins were dialyzed against the T40 buffer (10 mM Tris⋅HCl [pH 7.4], 40 mM NaCl, 0.05% Tween 20), which is optimized for switchSENSE measurements. For the determination of hydrodynamic radius in solution, a reporter fluorophore has been attached to the DNA’s distal end, which is read out by an epifluorescence setup. Sizing experiments were performed in T40 buffer with an association time of 20 min at a flow rate of 50 μL⋅min−1 and a HisRS concentration of 150 nM to saturate with proteins the RNA molecules attached to the biochip. We verified that no interaction was observed with scrambled equivalent RNAs (×1 to ×4) at this protein concentration. For sizing, the time-resolved fluorescence intensity signal corresponding to the upward motion of the DNA nanolevers was evaluated as described by Langer et al. (28) using switchANALYSIS software (Dynamic Biosensors GmbH). The size analysis allowed estimation of an apparent size value for all of the proteins tested.
Supplementary Material
Acknowledgments
We thank all the patients and family members who participated in this study for their contribution. We also thank the staff at beamline 12-2 of the SSRL for assistance with X-ray diffraction data collection and the staff at beamline 4-2 for assistance with SAXS data collection. This work was supported by US NIH Grant R01 GM088278 (to X.-L.Y.) and a fellowship from National Foundation for Cancer Research. J.B. is supported by a senior clinical researcher mandate of the Research Fund-Flanders under Grant Agreement 1805016N.
Footnotes
Conflict of interest statement: T.W. is employed by the company Dynamic Biosensors GmbH.
Data deposition: The crystal structure coordinates of human wild-type (WT) and D175E histidyl-transfer RNA synthetase (HisRS) have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank, www.pdb.org (PDB ID codes 6O76 and 5W6M, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908288116/-/DCSupplemental.
References
- 1.Skre H., Genetic and clinical aspects of Charcot-Marie-Tooth’s disease. Clin. Genet. 6, 98–118 (1974). [DOI] [PubMed] [Google Scholar]
- 2.Rossor A. M., Tomaselli P. J., Reilly M. M., Recent advances in the genetic neuropathies. Curr. Opin. Neurol. 29, 537–548 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gutmann L., Shy M., Update on Charcot-Marie-Tooth disease. Curr. Opin. Neurol. 28, 462–467 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Antonellis A., et al. , Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am. J. Hum. Genet. 72, 1293–1299 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jordanova A., et al. , Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat. Genet. 38, 197–202 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Latour P., et al. , A major determinant for binding and aminoacylation of tRNA(Ala) in cytoplasmic Alanyl-tRNA synthetase is mutated in dominant axonal Charcot-Marie-Tooth disease. Am. J. Hum. Genet. 86, 77–82 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Safka Brozkova D., et al. , Loss of function mutations in HARS cause a spectrum of inherited peripheral neuropathies. Brain 138, 2161–2172 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai P. C., et al. , A recurrent WARS mutation is a novel cause of autosomal dominant distal hereditary motor neuropathy. Brain 140, 1252–1266 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez M., et al. ; Inherited Neuropathy Consortium , Exome sequencing identifies a significant variant in methionyl-tRNA synthetase (MARS) in a family with late-onset CMT2. J. Neurol. Neurosurg. Psychiatry 84, 1247–1249 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei N., Zhang Q., Yang X. L., Neurodegenerative Charcot-Marie-Tooth disease as a case study to decipher novel functions of aminoacyl-tRNA synthetases. J. Biol. Chem. 294, 5321–5339 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibba M., Soll D., Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69, 617–650 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Abbott J. A., et al. , Substrate interaction defects in histidyl-tRNA synthetase linked to dominant axonal peripheral neuropathy. Hum. Mutat. 39, 415–432 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathis S., et al. , Charcot-Marie-Tooth diseases: An update and some new proposals for the classification. J. Med. Genet. 52, 681–690 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Storkebaum E., et al. , Dominant mutations in the tyrosyl-tRNA synthetase gene recapitulate in Drosophila features of human Charcot-Marie-Tooth neuropathy. Proc. Natl. Acad. Sci. U.S.A. 106, 11782–11787 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Motley W. W., et al. , Charcot-Marie-Tooth-linked mutant GARS is toxic to peripheral neurons independent of wild-type GARS levels. PLoS Genet. 7, e1002399 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niehues S., et al. , Impaired protein translation in Drosophila models for Charcot-Marie-Tooth neuropathy caused by mutant tRNA synthetases. Nat. Commun. 6, 7520 (2015). Correction in: Nat. Commun. 7, 10497 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grice S. J., et al. , Dominant, toxic gain-of-function mutations in gars lead to non-cell autonomous neuropathology. Hum. Mol. Genet. 24, 4397–4406 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He W., et al. , CMT2D neuropathy is linked to the neomorphic binding activity of glycyl-tRNA synthetase. Nature 526, 710–714 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grice S. J., Sleigh J. N., Zameel Cader M., Plexin-semaphorin signaling modifies neuromuscular defects in a Drosophila model of peripheral neuropathy. Front. Mol. Neurosci. 11, 55 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sleigh J. N., et al. , Trk receptor signaling and sensory neuron fate are perturbed in human neuropathy caused by Gars mutations. Proc. Natl. Acad. Sci. U.S.A. 114, E3324–E3333 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mo Z., et al. , Aberrant GlyRS-HDAC6 interaction linked to axonal transport deficits in Charcot-Marie-Tooth neuropathy. Nat. Commun. 9, 1007 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benoy V., et al. , HDAC6 is a therapeutic target in mutant GARS-induced Charcot-Marie-Tooth disease. Brain 141, 673–687 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He W., et al. , Dispersed disease-causing neomorphic mutations on a single protein promote the same localized conformational opening. Proc. Natl. Acad. Sci. U.S.A. 108, 12307–12312 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blocquel D., et al. , Alternative stable conformation capable of protein misinteraction links tRNA synthetase to peripheral neuropathy. Nucleic Acids Res. 45, 8091–8104 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beebe K., Waas W., Druzina Z., Guo M., Schimmel P., A universal plate format for increased throughput of assays that monitor multiple aminoacyl transfer RNA synthetase activities. Anal. Biochem. 368, 111–121 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Z., et al. , Internally deleted human tRNA synthetase suggests evolutionary pressure for repurposing. Structure 20, 1470–1477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koh C. Y., Wetzel A. B., de van der Schueren W. J., Hol W. G., Comparison of histidine recognition in human and trypanosomatid histidyl-tRNA synthetases. Biochimie 106, 111–120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langer A., et al. , Protein analysis by time-resolved measurements with an electro-switchable DNA chip. Nat. Commun. 4, 2099 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konarev P. V., et al. , PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 (2003). [Google Scholar]
- 30.Franke D., Svergun D. I., DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Volkov V. V., Svergun D. I., Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li S., et al. , Mechanism of intracellular cAMP sensor Epac2 activation: cAMP-induced conformational changes identified by amide hydrogen/deuterium exchange mass spectrometry (DXMS). J. Biol. Chem. 286, 17889–17897 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mendillo M. L., et al. , A conserved MutS homolog connector domain interface interacts with MutL homologs. Proc. Natl. Acad. Sci. U.S.A. 106, 22223–22228 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walters B. T., Ricciuti A., Mayne L., Englander S. W., Minimizing back exchange in the hydrogen exchange-mass spectrometry experiment. J. Am. Soc. Mass Spectrom. 23, 2132–2139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marsh J. J., et al. , Structural insights into fibrinogen dynamics using amide hydrogen/deuterium exchange mass spectrometry. Biochemistry 52, 5491–5502 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z., Smith D. L., Determination of amide hydrogen exchange by mass spectrometry: A new tool for protein structure elucidation. Protein Sci. 2, 522–531 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.