Significance
The extent to which variants with genome regulatory and evolutionary roles affect mammalian phenotypes is unclear. We systemically analyzed large datasets covering genomics, transcriptomics, epigenomics, metabolomics, and 34 phenotypes in over 44,000 cattle. This allowed us to provide a framework to rank over 17.7 million sequence variants based on their contribution to gene regulation, evolution, and variation in 34 complex traits. Validated in independent datasets with over 7,500 cattle, our sequence-variant ranking showed consistent performances in genomic prediction of phenotypes. Our study provides methods and an analytical framework to quantify the functional importance of sequence variants. By providing public data of biological priors on genomic markers, our work can make the global selection of animals efficient and accurate.
Keywords: gene regulation, evolution, quantitative traits, animal breeding, cattle
Abstract
Many genome variants shaping mammalian phenotype are hypothesized to regulate gene transcription and/or to be under selection. However, most of the evidence to support this hypothesis comes from human studies. Systematic evidence for regulatory and evolutionary signals contributing to complex traits in a different mammalian model is needed. Sequence variants associated with gene expression (expression quantitative trait loci [eQTLs]) and concentration of metabolites (metabolic quantitative trait loci [mQTLs]) and under histone-modification marks in several tissues were discovered from multiomics data of over 400 cattle. Variants under selection and evolutionary constraint were identified using genome databases of multiple species. These analyses defined 30 sets of variants, and for each set, we estimated the genetic variance the set explained across 34 complex traits in 11,923 bulls and 32,347 cows with 17,669,372 imputed variants. The per-variant trait heritability of these sets across traits was highly consistent (r > 0.94) between bulls and cows. Based on the per-variant heritability, conserved sites across 100 vertebrate species and mQTLs ranked the highest, followed by eQTLs, young variants, those under histone-modification marks, and selection signatures. From these results, we defined a Functional-And-Evolutionary Trait Heritability (FAETH) score indicating the functionality and predicted heritability of each variant. In additional 7,551 cattle, the high FAETH-ranking variants had significantly increased genetic variances and genomic prediction accuracies in 3 production traits compared to the low FAETH-ranking variants. The FAETH framework combines the information of gene regulation, evolution, and trait heritability to rank variants, and the publicly available FAETH data provide a set of biological priors for cattle genomic selection worldwide.
Understanding how mutations lead to phenotypic variation is a fundamental goal of genomics. With a few exceptions, complex traits with significance in evolution, medicine, and agriculture are determined by many mutations and environmental effects. Genome-wide association studies (GWASs) have been successful in finding associations between single-nucleotide polymorphisms (SNPs) and complex traits (1). Usually, there are many variants, each of small effect, which contribute to trait variation. Consequently, very large sample size is needed to find significant associations that explain most of the observed genetic variation. In humans, the sample size has reached over 1 million (2).
To test the generality of the findings in humans, it is desirable to have another species with very large sample size, and cattle is a possible example. There are over 1.46 billion cattle worldwide (3), and millions are being genotyped or sequenced as well as phenotyped (4, 5). Cattle have been domesticated from 2 subspecies of the humpless taurine (Bos taurus) and humped zebu (Bos indicus), which diverged ∼0.5 million years ago from extinct wild aurochs (Bos primigenius) (6). The increasing amount of genomic data and an outbred genome make cattle the only comparable GWAS model to humans. In addition, cattle have a very different demographic history than humans. While humans went through an evolutionary bottleneck about 10,000 to 20,000 y ago and then expanded to a population of billions, cattle have declined in effective population size due to domestication and breed formation, leading to a different pattern of linkage disequilibrium (LD) to humans. Insights into the genome–phenome relationships from cattle provide a valuable addition to the knowledge for other mammals. The knowledge of cattle genomics is also of direct practical value as rearing cattle is a major agricultural industry worldwide.
Despite the huge sample sizes used in human GWASs, identification of the causal variants for a complex trait is still difficult. This is due to the small effect size of most causal variants and the LD between variants. Consequently, there are usually many variants in high LD, any one of which could be the cause of the variation in phenotype. Prioritization of these variants can be aided by functional information on genomic sites. For instance, mutations that change an amino acid are more likely to affect phenotype than synonymous mutations.
Many mutations affecting complex traits regulate gene transcription-related activities. This has been demonstrated in many studies of human genomics, including but not limited to the analysis of intermediate trait quantitative trait loci (QTLs), such as metabolic QTLs (mQTLs) (7) and expression QTLs (eQTLs) (8) and analysis of regulatory elements, such as promoters (9) and enhancers (10), which can be identified with chromatin immunoprecipitation sequencing (ChIP-seq). In animals, the Functional Annotation of Animal Genomes (FAANG) project has started (11), and animal functional data have been accumulating (12–14). However, it is unclear which types of functional information improve the identification of causal mutations.
Mutations affecting complex traits may be subject to natural or artificial selection, which leaves a “signature” in the genome (15, 16). Given the unique evolutionary path of cattle, which has been significantly shaped by human domestication (17), it is attractive to test whether variants showing signatures of selection contribute to variation in complex traits. Mutations within genomic sites that are conserved across species may also affect complex traits. A previous study in humans showed that among a number of functional annotations, conserved sites across 29 mammals had the strongest enrichment of heritability in 17 complex traits (18).
We aim to determine which of several possible indicators of function are most useful for predicting sequence variants that are most likely to affect 34 traits in B. taurus dairy cattle. The indicators considered fall into 3 groups: 1) functional annotations of the bovine genome based, for instance, on ChIP-seq experiments; 2) evolutionary data, such as a site being under selection; and 3) GWAS data from traits that are relatively close to the primary action of the mutation, such as gene expression. Using these indicators of function, we define 30 sets of variants and estimate the variance explained by each set across 34 traits in 44,270 cattle. We then combine the estimates of heritability per variant across traits and across functional and evolutionary categories to define a Functional-And-Evolutionary Trait Heritability (FAETH) score that ranks variants on variance explained in complex traits. We validate the FAETH score in an independent dataset of 7,551 Danish cattle. The FAETH score of over 17 million variants with detailed user instructions is publicly available at https://doi.org/10.26188/5c5617c01383b (19). A tutorial demonstrating the calculation of the FAETH score along with demo data and R scripts can be found at https://ruidongxiang.com/2019/07/19/calculation-of-faeth-score-2/.
Results
Analysis Overview.
Our approach was to estimate the trait variance explained by a set of variants defined by some external data, such as the mapping of the gene expression QTLs (geQTLs), RNA splicing QTLs (sQTLs), or genome annotation, for 34 traits measured in dairy cattle. Sequence variants available to this study included over 17 million SNPs and indels. Any large set of variants can explain almost all of the genetic variance due to the LD between surrounding and causal variants. Therefore, we fitted each externally defined set of variants in a model together with a standard set of 630,000 SNPs from the bovine high-density (HD) SNP array. We combined the results from all 34 traits and all sets of variants to derive a score for each variant based on its expected contribution to the genetic variance in these 34 traits and tested the validity of this score in an independent cattle dataset.
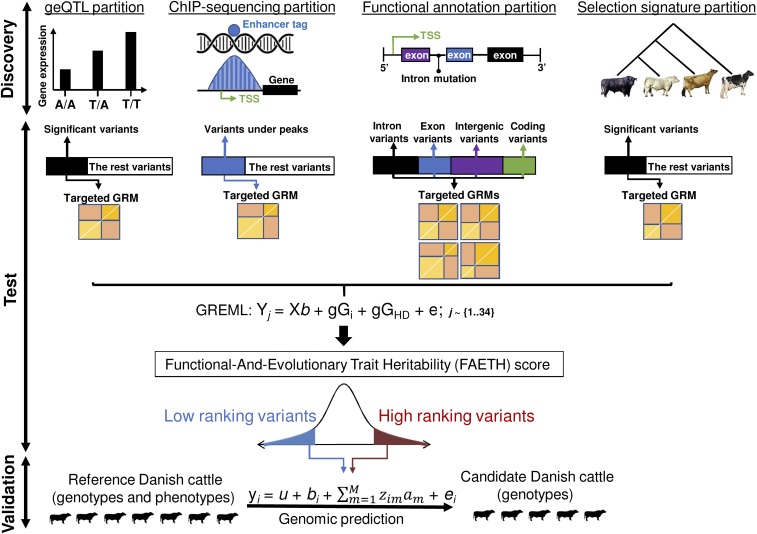
Our analysis had 4 major steps (Fig. 1).
-
1)
The 17 million sequence variants (1000 Bull Genomes Run6) (20) were classified according to external information from the discovery analysis of the function and evolution of each genomic site. The basis for this classification was either publicly available data or our own data as described in Materials and Methods. The genome was partitioned 15 different ways as listed in Table 1. For example, the category of geQTL partitioned the genome variants into a set of targeted variants with geQTL P value < 0.0001 and a set of remaining variants (i.e., the “rest” of the variants). Another partition, e.g., variant annotation, based on a publicly available annotation of the bovine genome, divided variants into several nonoverlapping sets, such as “intergenic,” “intron,” and “splice sites.”
-
2)
For each set of variants in each partition of the genome, separate genomic relationship matrices (GRMs) were calculated among the 11,923 bulls or 32,347 cows. Where a partition included only 2 sets (e.g., geQTL and the rest), a GRM was calculated only for the targeted set (e.g., geQTL).
-
3)
For each of the 34 traits, the variance explained by random effects described by each GRM was estimated using restricted maximum likelihood (this analysis is referred to as a genomic REML or GREML). Each GREML analysis fitted a random effect described by the targeted GRM and a random effect described by the GRM calculated from the HD SNP chip (630,002 SNPs). Each GREML analysis estimated the proportion of genetic variance, , explained by the targeted GRM in each of the 34 decorrelated traits (Cholesky orthogonalization) (ref. 21 and Materials and Methods) in each sex. The explained by each targeted set of variants was divided by the number of variants in the set to calculate the per variant, i.e., per-variant , and this was averaged for each variant across the 34 decorrelated traits.
-
4)
The FAETH score of all variants was calculated by averaging the per-variant across traits and informative partitions (13 out of 15). Two partitions determined as not informative were not included in the FAETH score computation. Variance explained and the accuracy of genomic predictions (using an independent dataset of 7,551 Danish cattle with 3 milk production traits) was compared between variants of high and low FAETH score.
Fig. 1.
Overview of the analysis. The discovery analysis involved the selection of variants from functional and evolutionary datasets; this figure shows examples of some of the datasets used. In the test analysis, each of the variant sets was used to make GRMs. Then, each one was analyzed in the GREML (gGi), together with the high-density SNP chip GRM (gGHD) for each of the 34 traits (Yj, ). Once the heritability, , of each gGi was calculated, it was averaged across traits and adjusted for the number of variants used to build the gGi to calculate the per-variant . The FAETH scoring of each variant was derived based on their memberships to differentially partitioned sets and the per-variant . In the validation analysis, variants with high and low FAETH ranking were tested in a Danish cattle dataset for GREML and genomic prediction of 3 production traits. The Australian test dataset contained 9,739 bulls and 22,899 cows of Holstein breed, 2,059 bulls and 6,174 cows of Jersey, 2,850 cows of mixed breeds, and 125 bulls and 424 cows of Australian Red. The Danish reference set contained 4,911 Holstein, 957 Jersey, and 745 Danish Red bulls, and the Danish validation population contained 500 Holstein, 517 Jersey, and 192 Danish Red bulls.
Table 1.
Variant sets selected from functional and evolutionary partitions
| Partitions | Targeted variant sets (no. of variants) | Animal no. |
| Gene expression QTLs | geQTLs with metaanalysis P < 1e−4 from blood and milk cells, liver, and muscle (110,200) | 209 |
| Exon expression QTLs | eeQTLs with metaanalysis P < 1e−4 from blood and milk cells, liver, and muscle (945,832) | 209 |
| Splicing QTLs | sQTLs with metaanalysis P < 1e−4 from blood and milk cells, liver, and muscle (1,112,324) | 209 |
| Allele specific expression QTLs | aseQTLs with metaanalysis P < 1e−4 from blood and milk cells (1,100,446) | 112 |
| Polar lipid metabolite QTLs | mQTLs with metaanalysis P < 1e−4 from 19 types of milk metabolites (5,365) | 338 |
| ChIP-seq peaks | Under H3K4Me3 and H3K27Ac peaks from liver, muscle, and mammary gland (1,166,795) | 15 |
| Variant annotation | Annotated as UTR (42,350), intergenic (11,869,145), gene end (1,007,214), intron (4,629,025), splice.sites (11,080), coding.related (105,969), and noncoding.related (4,589) | na |
| Predicted CTCF sites | Variants tagged by mapped CTCF-binding motifs from humans, mice, dogs, and macaques as published in ref. 32 (252,234) | na |
| HPRS | Genome sites within the top 1% gkm SVM score from the HPRS as published in ref. 31 (169,773) | na |
| Conserved 100 species | Bovine genome sites lifted over from human sites with PhastCon score (34) > 0.9 calculated using genomes of 100 vertebrate species (378,301) | na |
| Selection signature | GWAS P < 1e−4 between 7 beef and 8 dairy breeds, 1000 Bull Genome (6,218) | 1,370 |
| Young variants | Ranked within the bottom 1% of the proportion of positive correlations (PPRR) with rare variants, 1000 Bull Genome (893,986) | 2,330 |
| LD score quartiles | First quartile (4,417,033/4,416,205), second quartile (4,418,731/4,419,930), third quartile (4,415,633/4,415,481), and fourth quartile (4,417,975/4,417,756) | 44,270 |
| Variant density quartiles | First quartile (4,429,833), second quartile (4,414,996), third quartile (4,427,220), and fourth quartile (4,397,323) | |
| MAF quartiles | First quartile (4,414,292/4,417,036), second quartile (4,421,093/4,417,428), third quartile (4,416,834/4,418,157), and fourth quartile (4,417,153/4,418,157) |
For the 3 categories of quartiles, the numbers of variants on the left and right side of the slash were for the bulls and cows, respectively. LD score indicates the sum of linkage disequilibrium correlation between a variant and all variants in the surrounding 50-kb region, GCTA-LDS (38). The details of the variant annotations can be found in SI Appendix, Table S1. The animal numbers are the sample size in each discovery analysis. Fourth quartile scores > third quartile > second quartile > first quartile. na, not applicable.
Characteristics of Variant Sets with Regulatory and Evolutionary Significance.
Based on the 15 partitions of the genome in Table 1, we defined 30 sets of variants. The details of the discovery analysis defining these sets can be found in Materials and Methods. Briefly, regulatory variant sets including geQTLs, sQTLs, and allele-specific expression QTLs (aseQTLs) were discovered from multiple tissues, including white blood and milk cells, liver, and muscle. The milk cells were dominated by immune cells. However, they also contained mammary epithelial cells and had high transcriptomic similarity to the mammary gland tissue (13, 22). The polar lipid metabolites mQTLs were discovered using a multitrait metaanalysis (23) of 19 metabolite profiles, such as phosphatidylcholine, phosphatidylethanolamine, and phosphatidylserine (24), from bovine milk fat. The ChIP-seq data used in our analysis contained previously published H3K27Ac and H3K4me3 marks in liver and muscle tissues (25, 26) and newly generated H3K4Me3 marks from the mammary gland.
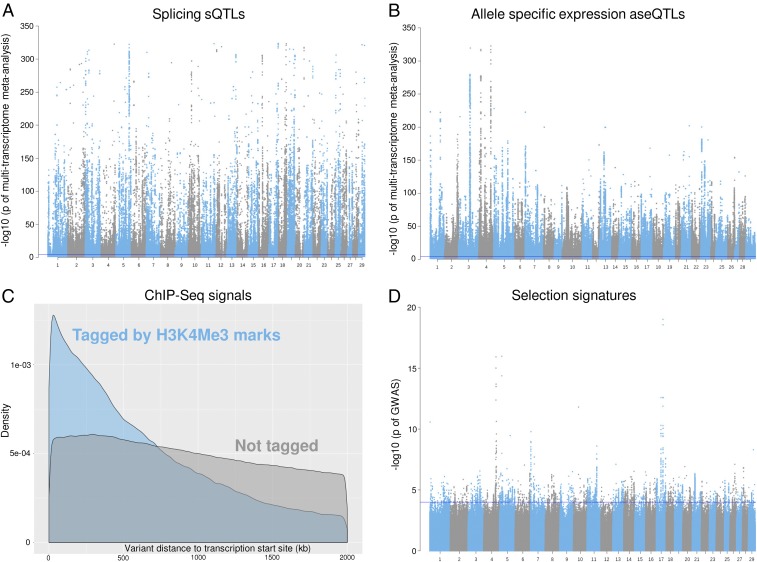
Fig. 2 illustrates some of the properties of these variant sets. Many sQTLs with strong effects on the intron excision ratio (27) were discovered in a metaanalysis of sQTLs mapped in white blood and milk cells, liver, and muscle (13) (Fig. 2A). Many significant aseQTLs were discovered using a gene-wise metaanalysis of the effects of the driver variant (dVariant) on the transcript variant (tVariant) at exonic heterozygous sites (28) from white blood and milk cells (Fig. 2B). Fig. 2C shows that variants tagged by the marks of H3K4Me3, a marker for promoters, were closer to the transcription start site than other variants.
Fig. 2.
Examples of regulatory and evolutionary signals from the discovery analysis. (A) A Manhattan plot of the metaanalysis of sQTLs from white blood and milk cells and liver and muscle tissues. (B) A Manhattan plot of the metaanalysis of aseQTLs in the white blood cells. (C) A distribution density plot of variants tagged by H3K4Me3 ChIP-seq mark from mammary gland within 2 Mb of gene transcription start site. (D) Artificial selection signatures between 8 dairy and 7 beef cattle breeds with the linear mixed-model approach using the 1000 Bull Genome database. The blue line indicates −log10(P value) = 4.
The variant annotation partition had 7 merged sets (Table 1 and SI Appendix, Table S1) based on the Variant Effect Prediction of Ensembl (29) and NGS-SNP (30). Additional information of variant function annotation was obtained from the Human Projection of Regulatory Regions (HPRS) as published in ref. 31 and predicted CCCTC-binding factor (CTCF) sites as published in ref. 32.
The evolutionary variant sets were discovered from across- and within-species genome analyses. Variants within cross-species conserved sites were lifted over from human genome sites (hg38), those with the PhastCon score >0.9 calculated using genome sequences of 100 vertebrate species. The LiftOver (https://genome.ucsc.edu/cgi-bin/hgLiftOver) rate from human conserved sites to bovine was 92.3%, which was higher than the LiftOver rate using the human sites with the PhastCon score >0.9 across 29 mammalian species (33, 34). Detailed results of the analysis of conserved sites can be found in SI Appendix, Note S1.
The within-species evolutionary analysis used the whole-genome sequence variants from Run 6 of the 1000 Bull Genomes project (35). Those variants with higher frequency in dairy than in beef breeds (“selection signature”; Table 1, Fig. 2D, and SI Appendix, Fig. S1) were detected from a GWAS where the breed type was modeled as a binary phenotype in the linear mixed model (36) of 15 beef and dairy breeds.
With the 1000 Bull Genomes data, we used a statistic to identify variants possibly subject to recent artificial and/or natural selection, PPRR (the proportion of positive correlations [r] with rare variants). SI Appendix, Fig. S2A illustrates a coalescence where a mutation has been positively selected, i.e., is relatively young and has increased in frequency rapidly. In this coalescence, the selected mutation was seldom on the same branch as rare mutations, and so the LD r between the selected mutation and rare alleles was typically negative. This was similar to the logic employed by ref. 37. In this partition of the genome, the 1% of variants with the lowest PPRR, after correcting for the variants’ own allele frequency (SI Appendix, Fig. S2 and Materials and Methods), were defined as young variants.
The quartile categories partitioned the genome variants into 4 sets of variants of similar size based on either their LD score (sum of LD r2 between a variant and all of the variants in the surrounding 50-kb region, GCTA-LDS) (38) or the number of variants within a 50-kb window (variant density) or their minor allele frequency (MAF) (38) (Table 1). Note that the fourth quartile had the highest value, and the first quartile had the lowest value for LD score, MAF, and SNP density.
The Proportion of the Genetic Variance for 34 Traits Explained by Each Set of Variants.
In the test datasets of 11,923 bulls and 32,347 cows, common variants (MAF ≥ 0.001) of the sets described above were used to make GRMs (36). Each of these GRMs was then fitted together with the high-density variant chip GRM (variant number = 632,002) in the GREML analysis to estimate the proportion of additive genetic variance explained by each functional and evolutionary set of variants, , in each of the 34 decorrelated traits, separately in bulls and cows (Table 2). Overall, the ranking of the averaged across 34 traits, , was highly consistent between bulls and cows (r = 0.94). All of the estimates, except that of the intergenic variants, were higher for bull traits than cow traits, consistent with the higher heritability of phenotypic records in bulls than in cows (39) because bull phenotypes are actually the average of many daughter phenotypes of the bull. When the HD variants were fitted alone, they explained on average 17.8% (±2.7%) of the variance in bulls and 4.7% (±1.4%) in cows (SI Appendix, Table S2). The estimates of mQTLs and the conserved sites across 100 species (termed as “conserved 100 species” in Table 2 and the following text) were much larger than their genome fractions in both sexes (Table 2). For other variant sets, the estimates generally increased with the number of variants in the set. For example, eQTLs, including exon expression QTLs (eeQTLs), sQTLs, and aseQTLs, which included around 5% of the total variants, explained 11 to ∼15% of trait variance in bulls and 2.5 to ∼4% of trait variance in cows. The young variants inferred by the statistic PPRR, which accounted for 0.54% of the total number of variants, explained 0.78% of the trait variance in bulls and 0.12% of the trait variance in cows.
Table 2.
The relative proportion of selected variant in sets compared to the total number of variants analyzed (genome fraction) and their averaged heritability in bulls and cows, across 34 traits
| Category | Genome fraction, % | in bulls, % | in cows, % |
| eeQTLs | 4.77 | 14.52 (2.2) | 3.96 (1.2) |
| sQTLs | 5.57 | 15.08 (2.5) | 3.88 (1.2) |
| aseQTLs | 5.21 | 11.0 (2.0) | 2.47 (0.7) |
| mQTLs | 0.03 | 0.71 (0.2) | 0.12 (0.04) |
| geQTLs | 0.53 | 1.54 (0.4) | 0.19 (0.06) |
| ChIP-seq | 6.60 | 4.21 (0.8) | 0.90 (0.3) |
| Noncoding.related | 0.03 | 0.06 (0.02) | 0.013 (0.004) |
| Splice.sites | 0.06 | 0.08 (0.02) | 0.02 (0.005) |
| UTR | 0.24 | 0.18 (0.03) | 0.03 (0.01) |
| Coding.related | 0.60 | 0.26 (0.06) | 0.04 (0.012) |
| Geneend | 5.70 | 3.76 (0.8) | 0.80 (0.2) |
| Intron | 26.2 | 5.56 (0.7) | 1.53 (0.3) |
| Intergenic | 67.2 | 10.3 (1.3) | 17.3 (2.2) |
| Predicted CTCF sites | 1.43 | 0.36 (0.08) | 0.046 (0.02) |
| HPRS | 0.96 | 0.31 (0.08) | 0.045 (0.02) |
| Conserved 100 species | 2.1 | 41.4 (2.6) | 17.4 (2.3) |
| Selection signatures | 0.02 | 0.011 (0.004) | 0.002 (0.0008) |
| Young variants | 0.54 | 0.78 (0.2) | 0.12 (0.05) |
| LD score q1 | 25 | 4.57 (0.6) | 1.18 (0.3) |
| LD score q2 | 25 | 5.56 (0.7) | 1.45 (0.3) |
| LD score q3 | 25 | 6.38 (0.8) | 1.75 (0.4) |
| LD score q4 | 25 | 6.94 (0.9) | 2.01 (0.5) |
| Variant density q1 | 25 | 5.59 (0.7) | 1.49 (0.3) |
| Variant density q2 | 25 | 5.42 (0.7) | 1.45 (0.3) |
| Variant density q3 | 25 | 5.72 (0.7) | 1.55 (0.3) |
| Variant density q4 | 25 | 5.99 (0.7) | 1.65 (0.4) |
| MAF q1 | 25 | 1.36 (0.2) | 0.35 (0.08) |
| MAF q2 | 25 | 11.5 (1.3) | 3.51 (0.7) |
| MAF q3 | 25 | 29.2 (2.4) | 10.3 (1.8) |
| MAF q4 | 25 | 40.5 (2.8) | 15.6 (2.4) |
SEs are in parenthesis. q1 ∼ q4 were the genome partitions based on the first, second, third, and fourth quartiles of MAF, LD score, and the number of variants (variant density) per 50-kb windows. Fourth quartile > third quartile > second quartile > first quartile.
The increased greatly from MAF quartiles 1 to 4. However, the dramatically low estimates for the first MAF quartile may be associated with the reduced imputation accuracy for low MAF variants. By contrast, increased only slightly with LD score and even less with variant density.
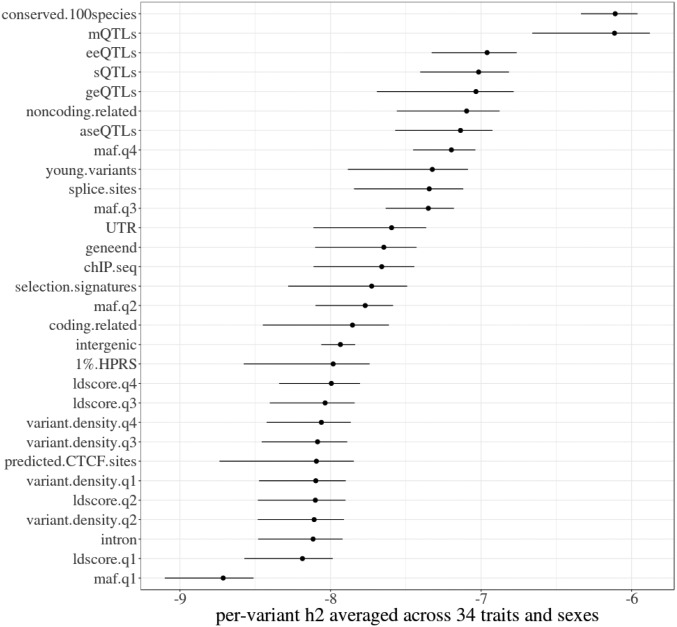
Estimates of were divided by the number of variants in the set to calculate the per-variant allowing comparison of the genetic importance of variant sets made with a varied number of variants. Since the per-variant was estimated independently in bulls and cows and yet showed high consistency between sexes (SI Appendix, Fig. S3), the average per-variant across sexes was used to rank each variant set (Fig. 3). Conserved 100 species and mQTLs made the top of the rankings (Fig. 3), due to their highly concentrated (41.4% in bulls and 17.4% in cows for conserved 100 species, and 0.71% in bulls and 0.12% in cows for mQTLs; Table 2) in a relatively small genome fraction (2.2% and 0.03%, respectively; Table 2). These 2 top sets were followed by several expression QTL sets, including eeQTLs, sQTLs, geQTLs, and aseQTLs (Fig. 3). Similar rankings were achieved by the “non.coding related” set (0.03% of genome variants) that included variants annotated as “non_coding_transcript_exon_variant” and “mature_miRNA_variant” (SI Appendix, Table S1), the “splice.site” set (0.06% of genome variants, including all of the variants annotated as associated with splicing functions), and the set of young variants (0.54% of genome variants). The “UTR” set, which included variants annotated as within 3′ and 5′ untranslated regions of genes, and the “geneend” set, which included variants annotated as downstream and upstream of genes, both had modest rankings along with the ChIP-seq and selection signatures sets. The “coding.related” set, dominated by variants annotated as synonymous and missense (SI Appendix, Table S1), ranked higher than the top 1% HPRS, intergenic variants, and predicted CTCF sites. Intron and the first quartile MAF set had the lowest per variant h2.
Fig. 3.
The proportion of genetic variances explained by sets of variants selected from functional and evolutionary categories. The ranking of variant sets based on the log10 scale of per-variant , averaged across bulls (left error bar) and cows (right error bar).
The impact of MAF on the ranking of variant sets was examined by calculating, for each set, the per-variant expected from the number of variants in a set belonging to each MAF quartile. This MAF expected per-variant was then subtracted from the observed per-variant to calculate the MAF adjusted per-variant (SI Appendix, Note S2). Excluding the sets based on MAF quartiles, the ranking of the unadjusted per-variant was well correlated (r = 0.9) with their ranking on the MAF adjusted per-variant . These results suggested an overall small impact of MAF on the variant set ranking of per-variant .
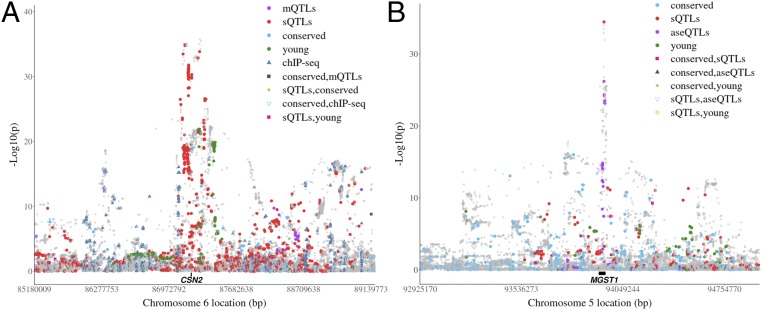
Variants from sets highly ranked for per-variant were highlighted in important QTL regions with the multitrait GWAS results (Fig. 4). In the expanded region of beta-casein (CSN2), a major but complex QTL for milk protein due to the existence of multiple QTL with strong LD, different high-ranking variant sets tended to tag variants with strong effects from multiple locations (Fig. 4A). Many variants with the strongest effects and close to CSN2 were tagged by sQTLs. Several clusters of variants from up- and downstream of CSN2 with slightly weaker effects were tagged by sets of ChIP-seq marks, young variant, and mQTLs. Conversely, for the expanded region of microsomal GST 1 (MGST1), a major QTL for milk fat, variants from high-ranking sets were more enriched in 2 major locations (Fig. 4B). The top variant within the MGST1 gene was again a sQTL, confirming previous results that regulatory variants are enriched in this region (13). Although not enriched in the MGST1 peak region, conserved sites tagged many variants that were not tagged by other top sets. The young variant sets appear to have tagged a different variant cluster around 0.7 Mb downstream from MGST1 (Fig. 4B).
Fig. 4.
Examples of top-ranked variant sets in important bovine trait QTL. (A) Manhattan plot of the metaanalysis of GWAS of 34 traits in the ±2 Mb region surrounding the beta casein (CSN2) gene, a major QTL for milk protein yield. (B) Manhattan plot of the metaanalysis of GWAS of 34 traits in the ±1 Mb region of the microsomal GST 1 (MGST1) gene, a major QTL for milk fat yield. The dots are colored based on their set memberships. The black bar between the gray dots and the X-axis indicates the gene locations.
The FAETH Score of Sequence Variants.
To quantify the relative importance of variants using a combination of functionality, evolutionary significance as well as their trait heritability, a framework was introduced to score variants based on their memberships to the sets of variants. Each time the genome variants were partitioned into nonoverlapping sets, each variant was a member of only one set and was assigned the per-variant of that variant. Therefore, all variants were assigned the same number (13 partitions) of per-variant , and the average of these 13 partitions was calculated for each variant and called the FAETH score. A criterion of per-variant > per-variant was also imposed to determine whether the variant set was informative. This criterion determined that 2 variant sets (HPRS and predicted CTCF sites) were not informative, and they were not included in the FAETH scoring (Materials and Methods). The FAETH score of 17,669,372 sequence variants for their genetic contribution to complex traits has been made publicly available at https://doi.org/10.26188/5c5617c01383b (19). A tutorial of the calculation of FAETH scores after was obtained can be found at https://ruidongxiang.com/2019/07/19/calculation-of-faeth-score-2/.
Variants with High FAETH Score Have Consistent Effects.
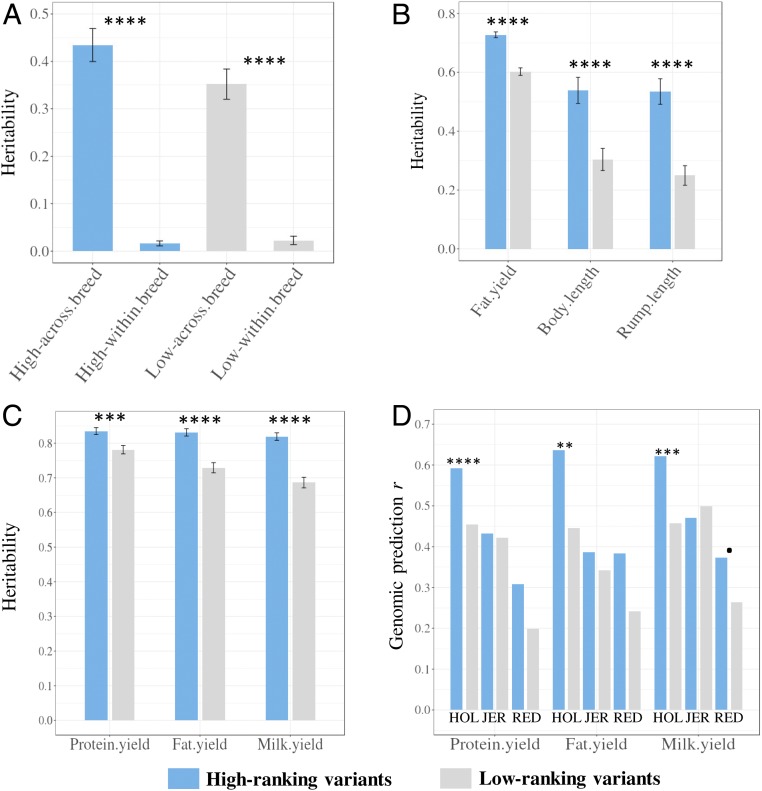
In the above analyses, the effect of a variant was estimated across all breeds. However, it is possible to fit a nested model in which both the main effect and an effect of the variant nested within a breed are included. If a variant is causal or in high LD with a causal variant, we might expect the effect to be similar in all breeds. Whereas if the variant is merely in LD with the causal variant, the effect might vary between breeds. Based on the FAETH score, the top 1/3 and bottom 1/3 ranked sequence variants in the Australian data were selected as “high” and “low” ranking variants, respectively. Fig. 5A shows the estimates of across-breed and within-breed variances for both high- and low-ranking variants. In both cases, the within-breed variance was small, but the high-ranking variants had a larger across-breed variance and a smaller within-breed variance than the low-ranking variants. This implied that the FAETH score identified variants with consistent phenotypic effects across breeds.
Fig. 5.
Further tests of the variant FAETH score. (A) The heritability of high and low FAETH ranking variants for the multibreed GRM and the within-breed GRM (2 GRMs fitted together) estimated across 34 traits in the Australian data. The error bars are the SE of heritability calculated across 34 traits. (B) The heritability of high and low FAETH ranking variants for 3 additional traits to the 34 traits in the Australian data used to calculate the FAETH score. (C) The multibreed heritability of high and low FAETH variants for 3 production traits in Danish data. The error bars are the SEs of the heritability of each GREML analysis. (D) Prediction accuracy of gBLUP of 3 production traits in Danish data using high and low FAETH variants (averaged between bulls and cows). The genomic predictors were trained in multiple breeds and predicted into single breeds (HOL, Holstein; JER, Jersey). P values of significant difference based on Z-score test: •P < 0.1; **P < 0.01; ***P < 0.001; ****P < 0.0001. Note that for the prediction accuracy r, the significance of difference was based on the sample sizes of the Danish candidate subset where there were 500 Holstein, 517 Jersey, and 192 Danish Red (Materials and Methods).
Additional data were obtained to test the FAETH score. Table 3 highlights the FAETH annotation of several causal or putative causal mutations where all of them were categorized as high FAETH ranking. Fig. 5B showed that the high-ranking variants had significantly (Z-score test: P < 0.0001) higher heritability estimates than the low-ranking ones for fat yield, body length, and rump length (original traits, not the Cholesky-transformed traits) that were not part of the Australian dairy 34 traits used to calculate the FAETH score. Also, as a proof of concept, high FAETH-ranking variants had significant enrichment (P = 4.5e−35), with pleiotropic SNPs significantly associated with 32 traits in beef cattle containing B. taurus and B. indicus subspecies (SI Appendix, Fig. S4). The enrichment of the low FAETH-ranking variants in these significant beef cattle pleiotropic SNPs was not different from random (SI Appendix, Fig. S4). These results supported the generality of the FAETH variant ranking in different traits, breeds, and subspecies.
Table 3.
FAETH annotation of previously identified causal or putative causal mutations for dairy cattle complex traits using the top variant sets
| Loci | Causal candidates | Annotation | Tagging variant sets | FAETH ranking |
| SLC37A1 | Chr1:144377960 (45) | Intron | aseQTL | High |
| DGAT1 | Chr14:1802266 (41) | Coding.related | mQTL, eeQTL, sQTL, aseQTL, ChIP-seq | High |
| FASN | Chr19:51386735 (71) | Intron | mQTL, eeQTL, sQTL, ChIP-seq | High |
| GHR | Chr20:31909478 (71) | Coding.related | Conserved 100 species | High |
“High” means that the variant was ranked within the top 1/3 of the FAETH score.
Validation of the FAETH Score in Danish Cattle.
An independent dataset of 7,551 Danish cattle of multiple breeds was used to test the FAETH score. The Australian high- and low-ranking variants were mapped in the Danish data. In the GREML analysis of Danish data, the high-ranking variants had significantly higher heritability than the low-ranking variants across three production traits (Z-score test: P < 0.001 for protein yield and P < 0.0001 for fat and milk yield) (Fig. 5C). The genomic best linear unbiased prediction (gBLUP) of Danish traits was also evaluated where the models were trained in the multiple-breed reference data to predict 3 production traits in each of 3 breeds (3 × 3 = 9 scenarios; Fig. 5D). Out of these 9 scenarios, high-ranking variants had higher accuracies than the low-ranking variants in 8 scenarios. Based on the sample sizes of the Danish candidate subset (500 Holstein, 517 Jersey, and 192 Danish Red), the significance levels of the increase in prediction accuracy for the high-ranking variants for these 8 scenarios are specified in Fig. 5D.
Discussion
GWASs have been very successful in finding variants associated with complex traits, but they have been less successful in identifying the causal variants because often there are a large group of variants, in high LD with each other (particularly in livestock) that are all associated with the trait. To distinguish among these variants, it would be useful to have information, external to the traits being analyzed, that points to variants that are likely to have an effect on phenotype. In this paper, we have evaluated 30 sources of external information based on genome annotation, evolutionary data, and intermediate traits such as gene expression and milk metabolites. Then, we assessed the variance that each set of variants explained when they were included in a statistical model that also included a constant set of 600,000 SNPs from the bovine HD SNP array. The purpose of this method is to find sets of variants that add to the variance explained by the HD SNPs, presumably because they are in higher LD with the causal variants than the HD SNPs are. Since the causal variants themselves are likely to be among the sequence variants analyzed, this method is a filter for classes of variants that are enriched for causal variants or variants in high LD with them. Although developed in cattle, the general framework of estimating FAETH score by combining the information of functionality, evolution, and complex trait heritability can be directly applied to other species. Additional tests of FAETH outside of the analyzed 34 traits and multiple beef cattle traits and the positive validation results in the Danish data support the across-breed, across-subspecies, and across-country usage of the FAETH score. Further, FAETH score not only contains a ranking of millions of variants that can be used as biological priors for genomic prediction (e.g., BayesRC) (40) but also includes the information of the variant membership to different functional and evolutionary categories. This additional information can be used by other researchers to annotate their variants of interests (e.g., Table 3).
Our results agreed with the report in humans (18) that the conserved sites had very strong enrichment of trait heritability. Interestingly, our analysis showed that genomic sites with conservation across a larger number of species appeared to have tagged variants with stronger enrichment of heritability, compared to the sites conserved across a smaller number of species (SI Appendix, Note S1). It may be worth studying the impact of the extent of the cross-species conservation on the amount of trait variation explained by the tagged variants in the future.
Our analysis also highlights the importance of intermediate trait QTL, including QTLs for metabolic traits and gene expression (mQTLs, geQTLs, eeQTLs, sQTLs, and aseQTLs). This is not a surprising result as the significant contribution of different intermediate trait QTLs to complex trait variations have been reported in humans (7, 27, 41–43) and cattle (13, 44–46). An advantage of these intermediate traits over conventional phenotypes is that individual QTL explain a larger proportion of the variance. For instance, cis eQTL tend to have a large effect on gene expression. This increases the signal-to-noise ratio and so increases power to distinguish causal variants from variants in partial LD with them. However, an intermediate QTL mapping study requires a large number of resources, especially when considering different metabolic profiles and tissues with large sample size. In the current analysis, we utilized several methods to combine results from individual studies of intermediate QTL mapping (21, 23, 28) (Materials and Methods and SI Appendix, Eqs. 1, 2, 3, and 5 and Note S3). This could reduce the noise from individual analyses, and this is likely to increase the chance of finding causal mutations.
To our knowledge, no study has systematically compared the genetic importance of mQTLs with eQTLs. The high ranking of mQTLs over eQTLs in our study might be related to the fact that the mQTLs were discovered from the milk fat, and the analyzed phenotype in the test data contained several milk-production traits. However, out of the 5,365 chosen mQTL variants, 961 variants were from the ±2 Mb region of DGAT1, while no mQTLs were from chromosome 5, which harbors MGST1 (SI Appendix, Table S3 and Fig. 4B), both of which are known major milk fat QTL. This suggests that many variants from the mQTL set not only influence milk fat production but may have other functions, including contributing to variation in the general process of fat synthesis, which is active in many mammalian tissues. Several large-scale human studies have highlighted the importance of mQTLs in various complex traits (7, 47).
Consistent with previous studies in cattle and humans (13, 27, 43), splicing sQTLs and the related eeQTLs ranked slightly higher than other eQTLs (Fig. 3). Cattle aseQTLs and geQTLs were found to have a similar magnitude of enrichment with trait QTL (28) and this is consistent with the current observation.
We proposed a method to identify variants that are young but at a moderate frequency and found this set was enriched for effects on quantitative traits (Figs. 3 and 4). However, Kemper et al. (48) showed that variants identified by selection signatures using traditional methods, such as fixation index (49) and integrated haplotype score (50) had little contribution to complex traits in cattle. In the current study, the selection signatures between beef and dairy cattle (“selection signature” set as shown in Table 1) explained some genetic variation in complex traits, although its contribution is relatively small (Table 2 and Fig. 3). It is possible that the inclusion of many nonproduction traits in the current study increased the chance of finding the trait-related sequence variants that are under artificial selection. Also, the use of sequence variants in the current study may have increased power compared to the study conducted by Kemper et al. (48), which used HD chip variants.
The set of variants with low PPRR (“young variants”) had a higher ranking of genetic importance to the complex traits than the other artificial selection signatures (Fig. 3). The identification of relatively young variants is based on the theory that very recent selection will increase the frequency of the favored alleles (37). Thus, the young variant set could contain variants that were either under artificial selection and/or recently appeared, and this may be the reason that it explained more trait variation than the artificial selection signatures. As shown in Fig. 4, many young variants can be found in major production trait QTL.
Genome-regulatory elements such as enhancers and promoters are important regulators of gene expression, and they can be identified by ChIP-seq assays. In humans, ChIP-seq–tagged binding QTLs (bQTLs) showed significant enrichments in complex and disease traits (51). We did not have enough individuals with ChIP-seq data to identify bQTLs. However, with only a limited amount of ChIP-seq data, variants tagged by H3K4me3 ChIP-seq showed a closer distance to the transcription start sites (Fig. 2C), and H3K4me3 and H3K27ac together tagged variants that had some contribution to complex trait variation (Fig. 3). Also, the FAETH ranking of the ChIP-seq–tagged variant set was similar to the ranking of variant annotation sets of gene end (variants within regions up- and downstream of genes) and UTR (variants within 3′ and 5′ UTR). It is logical that variants with the potential to affect promoters and/or enhancers are annotated as close to genes or located in gene-regulatory regions.
The variant annotation sets of noncoding-related and splice sites ranked relatively high for their contribution to trait variation (Fig. 3). Previously, variants annotated as splice sites had a high ranking of genetic importance to cattle complex traits (52). The majority of the variants from the noncoding-related set are “non_coding_transcript_exon_variant” (SI Appendix, Table S1), which is “a sequence variant that changes noncoding exon sequence in a noncoding transcript” according to VEP (29). This group of variants can be associated with long noncoding RNAs, and they are found to contribute to complex traits in humans (53) and cattle (54). Variants annotated as coding-related, of which the majority of variants are missense and synonymous (SI Appendix, Table S1), had a relatively low ranking of genetic importance to complex traits (Fig. 3). It seems a surprising result, but Koufariotis et al. (52) also reported similar observations in cattle. Perhaps coding variants that influence phenotype are subject to purifying selection and hence have low heterozygosity and hence low contribution to variance.
The contribution of variants with different LD properties to complex traits is an ongoing debate in humans (55–57). In our analysis of cattle, a domesticated species with strong LD between variants, variant LD differences had negligible influence on complex traits (Table 2). Also, variants within regions that have more variants (variant density) did not explain more trait variation. Common variants, as expected (58), had a substantial contribution to complex traits (Table 2 and Fig. 3).
Based on the variant membership to differentially partitioned genome sets and the value of the per-variant , the FAETH score of sequence variants combined the information of evolutionary and functional significance and heritability estimates across multiple complex traits for each variant. This analytical framework provides a simple but effective and comprehensive ranking for each variant that entered the analysis. Additional information on functional and/or evolutionary datasets can be easily integrated and linked to the variant contributions for multiple complex traits. A single score for each variant also makes the potential use of FAETH score easy and straightforward. For example, variants can be categorized as high and low FAETH ranking to create biological priors to inform Bayesian modeling for genomic selection (40). Additionally, different genome partitions of the variant sets in the FAETH data can be used to annotate interesting variants such as finding conserved sites that are also eQTLs. For example, we used FAETH data to annotate some causal or potential causal mutations for dairy cattle complex traits (Table 3). These results could improve our understanding of the biology behind the variant contribution to complex traits.
The FAETH score was further tested using Australian data. By building the within-breed GRM and comparing it with the multibreed GRM in the Australian data (Fig. 5A) using a method proposed by Khansefid et al. (59), our analysis implied that the variants with the high FAETH ranking contained variants with consistent effects across different breeds. Although estimated using 34 traits, our results show that FAETH ranking of variants can distinguish informative and uninformative variants beyond these 34 traits (Fig. 5B). Also, FAETH ranking of variants showed signs of being able to identify informative genetic markers for multiple traits in beef cattle including B. indicus subspecies (SI Appendix, Fig. S4). All of these results support the general use of FAETH variant scoring across different traits and breeds.
The FAETH score based on GREML using multiple Australian breeds was first tested with GREML using multiple Danish breeds (Fig. 5C). In this test, variants with high FAETH ranking explained significantly more genetic variance in protein, fat, and milk yield than the low-ranking variants. When the genomic predictors were trained in multiple Danish breeds and used to predict into single breeds (Fig. 5D), significant increases in prediction accuracies for the high-FAETH variants were mostly seen in the Holstein breed, and the increases for the Jersey breed were not significant. Several reasons contributed to this, including the most noticeable fact that the Holstein breed, which is genetically distant from the Jersey (60, 61), dominated both Danish (Holstein: Jersey = 5:1) and Australian (Holstein: Jersey = 4:1) populations. The relatively small sample size of the Danish validation population (519 for Jersey and 192 for Danish Red) reduced the power of Z-score test of significance of difference between correlations (i.e., prediction accuracies) of high- and low-FAETH variants. Also, since the Jersey breed has the smallest effective population size (62), it is expected that the advantage of a dense set of selected sequence variants is lowest (or absent) in that breed (63). Future tests in larger populations with increased breed diversities will provide better evaluation of the performance of the FAETH-ranked variants in multibreed analyses. Increasing the breed diversity, sample size, and tissues types in the functional genomic data may also improve the genomic prediction performances of FAETH ranking in specific cattle breeds. Nevertheless, the test results of the FAETH score in additional dairy traits and in beef cattle GWASs support that the FAETH ranking can prioritize informative variants in different populations.
In humans, Finucane et al. (18) combined many sources of data to calculate a prior probability that a variant affects a phenotype. Our approach is different from theirs in some respects. They used GWAS summary data and stratified LD score regression, whereas we used raw data and GREML. They fitted all sources of information simultaneously, whereas we fitted one variant set at a time in competition with the HD variants. We were unable to fit all sources at once with GREML for computational reasons but also because the extensive LD in cattle makes it harder than in humans to separate the effects of multiple variant sets. On the other hand, GREML is more powerful than LD score regression (64).
Our study demonstrates that the increasing amount of genomic and phenotypic data makes the cattle model a robust and critical resource for testing genetic hypotheses for large mammals. A recent large-scale study for cattle stature also supports the general utility of the cattle model in GWASs (5). In the current study, we highlight the contribution of the variants associated with intermediate QTLs and noncoding RNAs to complex traits, and this is consistent with many observations in human studies (8, 9, 27). However, we also provide contrasting evidence to results from humans. We found LD property of variants (e.g., variants from genomic regions with high LD) had negligible influences on trait heritability, contrasting with the recent evidence for the strong influence of LD property on human complex traits (55). In addition, variants under artificial selection had limited contributions to bovine complex traits, while in humans (where artificial selection is absent), natural selection clearly operates on complex traits (65). While the reasons for these contrasting results are yet to be studied, our findings from cattle add valuable insights into the ongoing discussions of the genetics of complex traits.
Our study has limitations. While some discovery analyses of the intermediate QTLs used relatively large sample size, the number of tissues and/or types of “omics” data included for discovering expression QTLs and mQTLs is yet to be increased. Also, in the discovery analysis, the selection criteria for informative variants to be included for building GRMs were relatively simple. In the test analysis, the heritability estimation for different GRMs used the GREML approach, which has been under some debate because of its potential bias (56, 66). Analysis of functional categories by the genomic feature models with BLUP has been previously tested (67), although this method can be computationally intensive. We aimed to treat each discovery dataset as equal as possible, and all GRMs were analyzed in the test dataset in the same systematic way. The positive results from the validation analysis suggest that informative variants have been well captured in the discovery and test analyses. The current version of FAETH score is based on included functional and evolutionary datasets. The FAETH score will be updated as more functional and evolutionary datasets become available.
Conclusions
We provide an extensive evaluation of the contribution of sequence variants with functional and evolutionary significance to multiple bovine complex traits. While developed using genomic and phenotypic data in the cattle model, the analytical approaches for the functional and evolutionary datasets and the FAETH framework of variant ranking can be applied equally well in other species. With their utility demonstrated, the publicly available FAETH score will provide functional and evolutionary annotation for sequence variants and effective and simple-to-implement biological priors for advanced genome-wide mapping and prediction.
Materials and Methods
Discovery Analysis.
Discovery data availability is detailed in SI Appendix, Table S4. A total of 360 cows from a 3-y experiment at the Ellinbank research facility of Agriculture Victoria in Victoria, Australia, were used to generate RNA-seq and milk fat metabolite datasets. Animal use was approved by Agriculture Victoria Animal Ethics Committee Application 2013-23.
The geQTLs, eeQTLs, and sQTLs in white blood and milk cells in a total of 131 Holstein and Jersey cows previously published (13) were used. The geQTLs, eeQTLs, and sQTLs in liver and semitendinosus muscle samples from Angus steers were also used (13). The aseQTLs were discovered using RNA-seq data from white blood and milk cells in a total of 112 Holstein cows (5). The metaanalysis of these 4 types of eQTLs, including SI Appendix, Eqs. 1–3 (published in refs. 13 and 68), are detailed in SI Appendix, Note S3.
The discovery of polar lipid metabolite mQTLs in bovine milk fat was based on the mass spectrometry-quantified concentration of 19 polar lipids from 338 Holstein cows. The lipid extraction description and the multitrait metaanalysis of single-trait GWASs including SI Appendix, Eqs. 4 and 5 (23) can be found in SI Appendix, Note S3.
ChIP-seq marks indicative of enhancers and promoters were discovered from a combination of experimental and published datasets. ChIP-seq peak data of trimethylation at lysine 4 of histone 3 (H3K4me3) from 9 bovine muscle samples (26) and H3K4me3 and acetylation at lysine 27 of histone 3 (H3K27ac) from 4 bovine liver samples (25) were downloaded. The generation of mammary H3K4me3 ChIP-seq peaks from 2 lactating Holstein cows (collected with the approval of Agriculture Victoria Animal Ethics Committee Application 2014-23) is detailed in SI Appendix, Note S3.
The discovery of variant sets with evolutionary significance was based on the whole-genome sequences of Run 6 of the 1000 Bull Genomes project (35). The analysis used a subset of 1,370 cattle of 15 dairy and beef breeds with a linear mixed-model method (SI Appendix, Eq. 6 and Note S3).
To fully utilize the 1000 Bull Genomes data, the metric PPRR (MAF <0.01), was developed to infer the variant age. PPRR was then calculated as (Eq. 7), where was the PPRR; was the count (N) of all of the positive correlations (r) between the genotypes of common variants and the genotypes of rare variants in a given window with a size of k (k = 50 kb for this study for computational efficiency). was the count of all correlations regardless of the sign. The calculation of can be easily and effectively performed using plink1.9 (www.cog-genomics.org/plink/1.9/). The rationale of PPRR computation is detailed in SI Appendix, Note S3.
Conserved genome sites in cattle were based on the lifted over (https://genome.ucsc.edu/cgi-bin/hgLiftOver) human sites with PhastCon score (34) > 0.9 computed across 100 vertebrate species. The analysis is detailed in SI Appendix, Note S1.
The variant annotation category was based on Ensembl variant Effect Predictor (29) and NGS-variant (30). Several variant annotations were merged from the original annotations to achieve reasonable sizes for GREML (SI Appendix, Table S1). The gkm SVM score of predicted regulatory potential for bovine genome sites was obtained from the HPRS (31). Variants in our study that overlapped with HPRS and within the top 1% of the SVM score (169,773 variants) were selected. The predicted CTCF sites were obtained from Wang et al. (32) and variants that overlapped these predicted bovine CTCF sites from ref. 32 were selected (252,234 variants).
Variant sets based on their distribution of LD score, density, and MAF were created using the GCTA-LDS method (38) based on imputed genome sequences of the test dataset of 11,923 bulls and 32,347 cows (detailed below). Over 17.6 million genome variants were partitioned into 4 quartiles of LD score per region (region size = 50 kb), the number of variants per window (window size = 50 kb), and MAF sets of variants that were used to make GRMs. The quartile partitioning of sequence variants followed the default setting of the GCTA-LDS. As a byproduct of GCTA LD score calculation, the number of variants per 50 kb window was computed, and the quartiles of the value of variant number per region for each variant was used to generate the variant density sets.
Test Analysis.
The test analysis with Australian data, including model SI Appendix, Eqs. 8 and 9, are detailed in SI Appendix, Note S3. Briefly, a total of 11,923 bulls (data provided by DataGene, http://www.datagene.com.au/ and CRV, https://www.crv4all-international.com/) and 32,347 cows (only provided by DataGene) from Holstein (9,739 ♂/22,899 ♀), Jersey (2,059 ♂/6,174 ♀), mixed breed (0 ♂/2,850 ♀) and Red dairy breeds (125 ♂/424 ♀) with 34 phenotypic traits (deviations for cows and daughter trait deviations for bulls [20]) were used (SI Appendix, Table S2). The trait decorrelation followed the procedure of Cholesky factorization (21). A total of 17,669,372 imputed sequence variants with Minimac3 imputation accuracy (69) R2 > 0.4 were used as genotype data. The construction of GRM used GCTA (36) and the heritability analysis with 2-GRM REML used MTG2 (70). An online tutorial for calculating FAETH score after the heritability estimation is available at https://ruidongxiang.com/2019/07/19/calculation-of-faeth-score-2/.
Validation Analysis.
The validation used variants within the top 1/3 (high) and bottom 1/3 (low) ranking from the Australian analysis to make GRMs in a total of 7,551 Danish bulls of Holstein (5,411), Jersey (1,203), and Danish Red (937), with a total of 8,949,635 imputed sequence variants in common between the Danish and Australian datasets, with a MAF ≥ 0.002 and imputation accuracy measured by the info score provide by IMPUTE2 ≥ 0.9 in the Danish data (62). Deregressed proofs (DRPs) were available for all animals in the Danish dataset for milk, fat, and protein yield. The Danish dataset was divided into a reference and validation set, where the reference set included 4,911 Holstein, 957 Jersey, and 745 Danish Red bulls, and the candidate set included 500 Holstein, 517 Jersey, and 192 Danish Red bulls. Over 1.25 million high-ranking variants and over 1.25 million low-ranking variants were used to make the high- and low-ranking GRMs. For the individuals in the reference set, each trait of protein, milk, and fat yield was analyzed with the GREML model (Eq. 10) using GCTA (36), where yDan was the vector of DRP of analyzed Danish individuals; β was the vector of fixed effects (breeds); Χ was a design matrix relating phenotypes to their fixed effects; u was the vector of animal effects where , was the genomic relationship matrix between Danish individuals, was the incidence matrix, and e was the vector of residual. This allowed the estimate of of high- and low-ranking variants in the Danish data.
To test the variant ranking, genomic prediction with gBLUP was performed by dividing the Danish individuals into reference and validation datasets. The –blup-variant option in GCTA (36) was used to obtain variant effects from the GREML analyses, which were used to predict genomic estimated breeding value (GEBV) in the validation population. Prediction accuracies were computed for each of the breeds in the validation population, as the correlation between GEBV and DRP. More tests of the FAETH score using additional Australian dairy and beef cattle data are detailed in SI Appendix, Note S3.
Supplementary Material
Acknowledgments
Australian Research Council’s Discovery Projects (DP160101056) supported R.X. and M.E.G. Dairy Futures Cooperative Research Centre supported the generation of the Holstein and Jersey transcriptome data. DairyBio (a joint venture project between Agriculture Victoria and Dairy Australia) funded the generation of the mammary ChIP-seq data. I.v.d.B. was supported by the Center for Genomic Selection in Animals and Plants, funded by Innovation Fund Denmark Grant 0603-00519B. No funding bodies participated in the design of the study; nor the collection, analysis, or interpretation of data; nor the writing of the manuscript. We thank DataGene and CRV for access to data used in this study and Gert Nieuwhof, Kon Konstantinov, Timothy P. Hancock (Datagene) and Chris Schrooten (CRV) for preparation and provision of data. We thank partners from the 1000 Bull Genomes project for the data access. We thank Dr. Majid Khansefid for the discussion of aseQTL analysis.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The Functional-And-Evolutionary Trait Heritability (FAETH) score with its user guide are publicly available at https://doi.org/10.26188/5c5617c01383b.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904159116/-/DCSupplemental.
References
- 1.Visscher P. M., et al. , 10 years of GWAS discovery: Biology, function, and translation. Am. J. Hum. Genet. 101, 5–22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nielsen J. B., et al. , Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat. Genet. 50, 1234–1239 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Food and Agriculture Organisation of the United Nations, FAOSTAT http://www.fao.org/faostat/en/#search/Cattle. Accessed 31 August 2018. [Google Scholar]
- 4.Taylor J. F., Taylor K. H., Decker J. E., Holsteins are the genomic selection poster cows. Proc. Natl. Acad. Sci. U.S.A. 113, 7690–7692 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouwman A. C., et al. , Meta-analysis of genome-wide association studies for cattle stature identifies common genes that regulate body size in mammals. Nat. Genet. 50, 362–367 (2018). [DOI] [PubMed] [Google Scholar]
- 6.MacHugh D. E., Shriver M. D., Loftus R. T., Cunningham P., Bradley D. G., Microsatellite DNA variation and the evolution, domestication and phylogeography of taurine and zebu cattle (Bos taurus and Bos indicus). Genetics 146, 1071–1086 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yousri N. A., et al. , Whole-exome sequencing identifies common and rare variant metabolic QTLs in a Middle Eastern population. Nat. Commun. 9, 333 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Battle A., Brown C. D., Engelhardt B. E., Montgomery S. B.; GTEx Consortium; Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration &Visualization—EBI; Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz; Lead analysts; Laboratory, Data Analysis &Coordinating Center (LDACC); NIH program management; Biospecimen collection; Pathology; eQTL manuscript working group , Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).29022597 [Google Scholar]
- 9.Lizio M., et al. ; FANTOM consortium , Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 16, 22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersson R., et al. , An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersson L., et al. ; FAANG Consortium , Coordinated international action to accelerate genome-to-phenome with FAANG, the Functional Annotation of Animal Genomes project. Genome Biol. 16, 57 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark E. L., et al. , A high resolution atlas of gene expression in the domestic sheep (Ovis aries). PLoS Genet. 13, e1006997 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiang R., et al. , Genome variants associated with RNA splicing variations in bovine are extensively shared between tissues. BMC Genomics 19, 521 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giuffra E., Tuggle C. K.; FAANG Consortium , Functional annotation of animal genomes (FAANG): Current achievements and roadmap. Annu. Rev. Anim. Biosci. 7, 65–88 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Zeng J., et al. , Signatures of negative selection in the genetic architecture of human complex traits. Nat. Genet. 50, 746–753 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Yang J., et al. , Genetic signatures of high-altitude adaptation in Tibetans. Proc. Natl. Acad. Sci. U.S.A. 114, 4189–4194 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu L., et al. , Genomic signatures reveal new evidences for selection of important traits in domestic cattle. Mol. Biol. Evol. 32, 711–725 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finucane H. K., et al. ; ReproGen Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium; RACI Consortium , Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiang R., et al. , The functional and evolutionary trait heritability (FAETH) score of over 17 million cattle sequence variants. University of Melbourne. 10.26188/5c5617c01383b. Deposited 28 August 2019. [DOI]
- 20.Hayes B. J., Daetwyler H. D., 1000 Bull genomes project to map simple and complex genetic traits in cattle: Applications and outcomes. Annu. Rev. Anim. Biosci. 7, 89–102 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Xiang R., MacLeod I. M., Bolormaa S., Goddard M. E., Genome-wide comparative analyses of correlated and uncorrelated phenotypes identify major pleiotropic variants in dairy cattle. Sci. Rep. 7, 9248 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cánovas A., et al. , Comparison of five different RNA sources to examine the lactating bovine mammary gland transcriptome using RNA-Sequencing. Sci. Rep. 4, 5297 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolormaa S., et al. , A multi-trait, meta-analysis for detecting pleiotropic polymorphisms for stature, fatness and reproduction in beef cattle. PLoS Genet. 10, e1004198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z., Moate P., Cocks B., Rochfort S., Comprehensive polar lipid identification and quantification in milk by liquid chromatography-mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 978–979, 95–102 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Villar D., et al. , Enhancer evolution across 20 mammalian species. Cell 160, 554–566 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao C., et al. , Genome-wide H3K4me3 analysis in Angus cattle with divergent tenderness. PLoS One 10, e0115358 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y. I., et al. , Annotation-free quantification of RNA splicing using LeafCutter. Nat. Genet. 50, 151–158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khansefid M., et al. , Comparing allele specific expression and local expression quantitative trait loci and the influence of gene expression on complex trait variation in cattle. BMC Genomics 19, 793 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLaren W., et al. , The Ensembl variant effect predictor. Genome Biol. 17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grant J. R., Arantes A. S., Liao X., Stothard P., In-depth annotation of SNPs arising from resequencing projects using NGS-SNP. Bioinformatics 27, 2300–2301 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen Q. H., et al. , Mammalian genomic regulatory regions predicted by utilizing human genomics, transcriptomics, and epigenetics data. Gigascience 7, 1–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang M., et al. , Putative bovine topological association domains and CTCF binding motifs can reduce the search space for causative regulatory variants of complex traits. BMC Genomics 19, 395 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollard K. S., Hubisz M. J., Rosenbloom K. R., Siepel A., Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20, 110–121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siepel A., et al. , Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daetwyler H. D., et al. , Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat. Genet. 46, 858–865 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Yang J., Lee S. H., Goddard M. E., Visscher P. M., GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Field Y., et al. , Detection of human adaptation during the past 2000 years. Science 354, 760–764 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J., et al. ; LifeLines Cohort Study , Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat. Genet. 47, 1114–1120 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kemper K. E., et al. , Improved precision of QTL mapping using a nonlinear Bayesian method in a multi-breed population leads to greater accuracy of across-breed genomic predictions. Genet. Sel. Evol. 47, 29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacLeod I. M., et al. , Exploiting biological priors and sequence variants enhances QTL discovery and genomic prediction of complex traits. BMC Genomics 17, 144 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ongen H., et al. ; GTEx Consortium , Estimating the causal tissues for complex traits and diseases. Nat. Genet. 49, 1676–1683 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Consortium G.; GTEx Consortium , Human genomics. The genotype-tissue expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348, 648–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhernakova D. V., et al. , Identification of context-dependent expression quantitative trait loci in whole blood. Nat. Genet. 49, 139–145 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Kemper K. E., et al. , Leveraging genetically simple traits to identify small-effect variants for complex phenotypes. BMC Genomics 17, 858 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanchez M.-P., et al. , Within-breed and multi-breed GWAS on imputed whole-genome sequence variants reveal candidate mutations affecting milk protein composition in dairy cattle. Genet. Sel. Evol. 49, 68 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Littlejohn M. D., et al. , Sequence-based association analysis reveals an MGST1 eQTL with pleiotropic effects on bovine milk composition. Sci. Rep. 6, 25376 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin S.-Y., et al. ; Multiple Tissue Human Expression Resource (MuTHER) Consortium , An atlas of genetic influences on human blood metabolites. Nat. Genet. 46, 543–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kemper K. E., Saxton S. J., Bolormaa S., Hayes B. J., Goddard M. E., Selection for complex traits leaves little or no classic signatures of selection. BMC Genomics 15, 246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Depaulis F., Veuille M., Neutrality tests based on the distribution of haplotypes under an infinite-site model. Mol. Biol. Evol. 15, 1788–1790 (1998). [DOI] [PubMed] [Google Scholar]
- 50.Voight B. F., Kudaravalli S., Wen X., Pritchard J.K., A map of recent positive selection in the human genome. PLoS Biol 4(3):e72 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tehranchi A. K., et al. , Pooled ChIP-seq links variation in transcription factor binding to complex disease risk. Cell 165, 730–741 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koufariotis L. T., Chen Y.-P. P., Stothard P., Hayes B. J., Variance explained by whole genome sequence variants in coding and regulatory genome annotations for six dairy traits. BMC Genomics 19, 237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan J. Y., et al. , Cis-acting complex-trait-associated lincRNA expression correlates with modulation of chromosomal architecture. Cell Rep. 18, 2280–2288 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Cai W., et al. , Genome wide identification of novel long non-coding RNAs and their potential associations with milk proteins in Chinese Holstein cows. Front. Genet. 9, 281 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Speed D., Cai N., Johnson M. R., Nejentsev S., Balding D. J.; UCLEB Consortium , Reevaluation of SNP heritability in complex human traits. Nat. Genet. 49, 986–992 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang J., Zeng J., Goddard M. E., Wray N. R., Visscher P. M., Concepts, estimation and interpretation of SNP-based heritability. Nat. Genet. 49, 1304–1310 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Evans L. M., et al. ; Haplotype Reference Consortium , Comparison of methods that use whole genome data to estimate the heritability and genetic architecture of complex traits. Nat. Genet. 50, 737–745 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang J., et al. , Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–569 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khansefid M., et al. , Estimation of genomic breeding values for residual feed intake in a multibreed cattle population. J. Anim. Sci. 92, 3270–3283 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Lund M. S., Su G., Janss L., Guldbrandtsen B., Brøndum R. F., Genomic evaluation of cattle in a multi-breed context. Livest. Sci. 166, 101–110 (2014). [Google Scholar]
- 61.van den Berg I., Boichard D., Lund M. S., Sequence variants selected from a multi-breed GWAS can improve the reliability of genomic predictions in dairy cattle. Genet. Sel. Evol. 48, 83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gibbs R. A., et al. ; Bovine HapMap Consortium , Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science 324, 528–532 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.MacLeod I. M., Hayes B. J., Goddard M. E., The effects of demography and long-term selection on the accuracy of genomic prediction with sequence data. Genetics 198, 1671–1684 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ni G., Moser G., Wray N. R., Lee S. H.; Schizophrenia Working Group of the Psychiatric Genomics Consortium , Estimation of genetic correlation via linkage disequilibrium score regression and genomic restricted maximum likelihood. Am. J. Hum. Genet. 102, 1185–1194 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo J., et al. , Global genetic differentiation of complex traits shaped by natural selection in humans. Nat. Commun. 9, 1865 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krishna Kumar S., Feldman M. W., Rehkopf D. H., Tuljapurkar S., Limitations of GCTA as a solution to the missing heritability problem. Proc. Natl. Acad. Sci. U.S.A. 113, E61–E70 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fang L., et al. , Use of biological priors enhances understanding of genetic architecture and genomic prediction of complex traits within and between dairy cattle breeds. BMC Genomics 18, 604 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khansefid M., et al. , Gene expression analysis of blood, liver, and muscle in cattle divergently selected for high and low residual feed intake. J. Anim. Sci. 95, 4764–4775 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Das S., et al. , Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee S. H., van der Werf J. H., MTG2: An efficient algorithm for multivariate linear mixed model analysis based on genomic information. Bioinformatics 32, 1420–1422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pausch H., et al. , Meta-analysis of sequence-based association studies across three cattle breeds reveals 25 QTL for fat and protein percentages in milk at nucleotide resolution. BMC Genomics 18, 853 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.