Significance
Programmed ribosomal frameshifting (PRF) is important to many viruses, but how structures in mRNA stimulate PRF remains uncertain. By studying a frameshift signal stimulating very high PRF in West Nile virus (WNV), we identify features that differ from other structures stimulating more modest PRF levels. Using single-molecule force spectroscopy to mimic the effects of tension applied to the RNA by the ribosome during translation, we find that the WNV frameshift signal exhibits exceptionally heterogeneous conformational dynamics, forming several different pseudoknots and multiple combinations of hairpins, all connected on 2 mutually exclusive pathways. This work supports a role for conformational heterogeneity in PRF, suggesting that dynamic fluctuations rather than static properties play a key role in stimulating PRF efficiently.
Keywords: programmed ribosomal frameshifting, RNA folding, pseudoknots, force spectroscopy, West Nile virus
Abstract
Specific structures in mRNA can stimulate programmed ribosomal frameshifting (PRF). PRF efficiency can vary enormously between different stimulatory structures, but the features that lead to efficient PRF stimulation remain uncertain. To address this question, we studied the structural dynamics of the frameshift signal from West Nile virus (WNV), which stimulates −1 PRF at very high levels and has been proposed to form several different structures, including mutually incompatible pseudoknots and a double hairpin. Using optical tweezers to apply tension to single mRNA molecules, mimicking the tension applied by the ribosome during PRF, we found that the WNV frameshift signal formed an unusually large number of different metastable structures, including all of those previously proposed. From force-extension curve measurements, we mapped 2 mutually exclusive pathways for the folding, each encompassing multiple intermediates. We identified the intermediates in each pathway from length changes and the effects of antisense oligomers blocking formation of specific contacts. Intriguingly, the number of transitions between the different conformers of the WNV frameshift signal was maximal in the range of forces applied by the ribosome during −1 PRF. Furthermore, the occupancy of the pseudoknotted conformations was far too low for static pseudoknots to account for the high levels of −1 PRF. These results support the hypothesis that conformational heterogeneity plays a key role in frameshifting and suggest that transitions between different conformers under tension are linked to efficient PRF stimulation.
Programmed ribosomal frameshifting (PRF) involves a recoding of the translation of mRNA by ribosomes owing to a shift in the reading frame, resulting in the production of an alternate polypeptide chain (1–4). PRF is triggered by a stimulatory structure in the mRNA, most often a pseudoknot formed when nucleotides within a hairpin loop base pair with complementary ones outside of the loop (5), in conjunction with a slippery sequence located 6 to 8 nt upstream, where the shift in reading frame takes place (2, 3, 6). PRF is particularly notable in viruses (7), many of which use a programmed shift into the −1 frame (−1 PRF) to produce 2 different polypeptides in a defined ratio. The expression ratio of the frameshifted proteins can affect essential aspects of viral function, such as genome replication and packaging or viral invasiveness (8–13), making it an attractive target for drugs aiming to attenuate viruses by altering PRF efficiency levels (14, 15).
The level of −1 PRF stimulated by different structures can vary enormously, from a few percent up to ∼70% to 80% (16–21). However, the features that contribute to efficient stimulation remain under debate (2), in part because the mechanisms driving frameshifting remain incompletely understood. Given that tension in the mRNA applied by the ribosome during translocation (22, 23) is a key feature of several proposed frameshifting mechanisms (2, 24, 25), it has been suggested that PRF efficiency is linked to the mechanical properties of the stimulatory structure (17, 24, 26). This hypothesis has motivated studies of stimulatory structures under mechanical tension using single-molecule force spectroscopy (SMFS), a powerful tool for studying conformational dynamics whereby force is applied to the ends of a molecule by a probe, such as optical tweezers, and the molecular extension is monitored as the structure changes in response to the tension (27). Such measurements applying force across the stimulatory structure can be used to mimic the tension applied by the translocating ribosome (22, 23) and characterize tension-induced dynamics in the stimulatory structure.
SMFS studies have uncovered interesting trends in the mechanical properties of frameshift stimulatory structures. Work examining pseudoknots containing mutations abolishing key tertiary contacts has suggested a correlation between PRF efficiency and the unfolding force for stimulatory pseudoknots (28, 29). However, a survey of numerous different pseudoknots showed that the mechanical resistance was uncorrelated to −1 PRF efficiency; instead, −1 PRF efficiency was found to correlate with conformational plasticity or heterogeneity (16). This correlation was supported by subsequent work extending SMFS studies of −1 PRF to other pseudoknots (30), different types of stimulatory structures such as hairpins (31), and the effects of antiframeshifting ligands (32). Evidence supporting the importance of conformational plasticity has also been found from single-molecule fluorescence experiments of ribosomes translocating through pseudoknots (33) and ensemble structural studies of stimulatory structures using such methods as selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) (21, 34). However, structures stimulating extremely high levels of frameshifting, which should pose the most stringent test of any hypotheses relating stimulatory structure properties and −1 PRF efficiency, have not yet been studied to probe their mechanical properties or dynamics.
Here we report SMFS measurements of the frameshift signal from West Nile virus (WNV), a mosquito-borne flavivirus causing neurologic disease that uses −1 PRF to control the relative expression levels of structural and nonstructural proteins encoded within a single open reading frame (9). Whereas most viral frameshift signals stimulate −1 PRF with an efficiency in the range of 5% to 30% (16, 17), the WNV frameshift signal has been reported to do so at up to 70% efficiency (21), among the highest levels measured to date. The WNV frameshift signal is also notable because the nature of the stimulatory structure is unclear. Initial studies of a 75-nt segment of the WNV mRNA suggested that, based on bioinformatic predictions, the stimulatory structure is a 61-nt pseudoknot (35). However, subsequent work extending the RNA by ∼50 nt and applying SHAPE analysis suggested that the frameshift signal forms competing tandem stem loop and 109-nt pseudoknot structures (21), with the tandem stem loops sharing stem 1 in common with the 61-nt pseudoknot proposed previously but the 109-nt pseudoknot being mutually exclusive with the other structures.
Using optical tweezers to measure the unfolding and refolding of individual mRNA molecules held under tension, we aimed not only to test the link between −1 PRF stimulation efficiency and conformational heterogeneity in the case of an extremely efficient frameshift signal, but also to confirm which, if any, of the structures proposed for the WNV frameshift signal were formed. We found that the conformational dynamics of the WNV frameshift signal were much more complex than those of structures stimulating −1 PRF less efficiently, confirming the hypothesis. Indeed, the WNV frameshift signal formed all the structures proposed previously, as well as several others: analysis of the unfolding and refolding trajectories revealed 2 parallel pathways, each containing multiple intermediates. Suggestively, the number of transitions between these states was highest in the range of forces expected to be applied by the ribosome during −1 PRF, supporting the notion that dynamic conformational changes under tension rather than static properties play a key role in stimulating −1 PRF efficiently.
Results
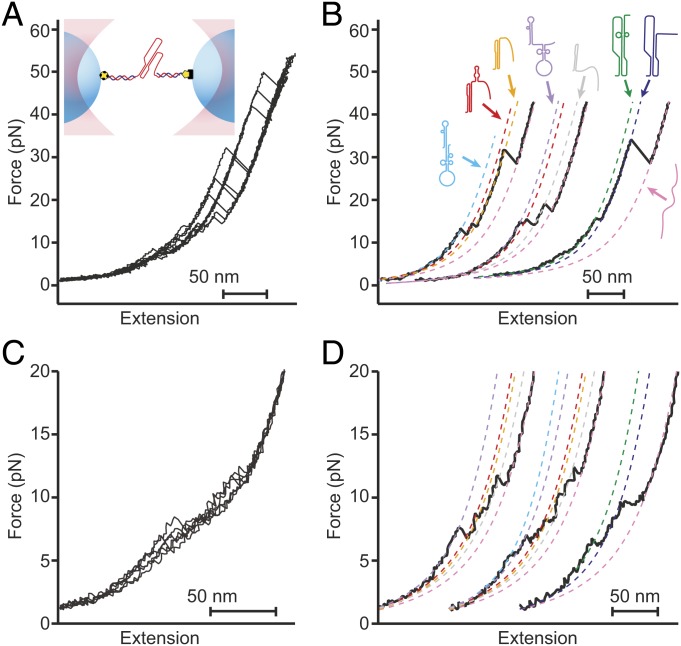
We studied the 111 nucleotides downstream of the WNV slippery sequence and linker (SI Appendix, Table S1). We first used a dual-luciferase assay of frameshifting to confirm that this frameshift signal stimulated very high −1 PRF levels, finding a stimulation efficiency of 78 ± 8% (all errors represent standard error of the mean, SEM). For SMFS measurements, we next transcribed a RNA construct containing these 111 nucleotides flanked on each side by kilobase-long handle sequences, annealing the transcripts to single-stranded DNA complementary to the handle regions and attached to beads held in optical traps (Fig. 1 A, Inset) (16). The tethered RNA was held near zero force for 3 s to allow time for folding, after which the traps were separated at constant speed to ramp up the force before bringing them together to ramp the force back down. These force ramps mimicked the situation during −1 PRF, where the ribosome makes repeated attempts to resolve the stimulatory structure before finally translocating through it (33, 36), causing rapid, nonequilibrium fluctuations in tension in the mRNA.
Fig. 1.
Force spectroscopy measurements of the WNV frameshift signal. (A) Representative unfolding FECs from the same molecule showing multiple “rips” where the extension increases when different parts of the structure unfold as the force is ramped up. (Inset) The frameshift signal is held under tension via duplex handles connected to beads held in optical traps. (B) Fitting unfolding FECs (black) to WLC models containing different amounts of unfolded RNA (dashed lines) reveals multiple different conformations of the frameshift signal. Cartoons show proposed structures for the different states based on observed length changes and unfolding forces. (C) Representative refolding FECs showing heterogeneous behavior as in unfolding curves. (D) WLC fits (dashed lines) to refolding curves (black) reveal the same set of states (color-coded as in B) as seen in unfolding curves.
Changes in structure were monitored by plotting the force as a function of the molecular extension, generating force-extension curves (FECs). FECs characteristically displayed regions in which the force rose nonlinearly with extension, reflecting the stretching of the handles during parts of the unfolding trajectory where the structure remained constant, separated by “rips” in the curve where the extension increased abruptly and the force dropped, indicating the unfolding of some part of the RNA (Fig. 1A). Notably, repeated unfolding of the same molecule revealed different patterns of rips of different length in the FECs (Fig. 1 A and B, black), indicating the presence of a heterogeneous mixture of conformational states. Similarly heterogeneous behavior was also seen in refolding curves (Fig. 1 C and D, black).
We characterized the structural transitions occurring in these FECs by fitting the curves to worm-like chain (WLC) polymer-elasticity models (37) before and after each rip (Fig. 1 B and D, dashed lines). Using one WLC for the duplex handles in series with a second WLC for the variable amount of unfolded RNA present in each conformation, we determined the contour length of unfolded RNA, LcU, before and after each transition. We identified at least 6 conformational states with different LcU values (Table 1), indicating a minimum of 6 different structures in the frameshift signal. Some of these states (Fig. 1B, orange and dark blue) unfolded with the broad distribution of generally high forces characteristic of tertiary structures such as pseudoknots (16, 38), whereas the others unfolded with a narrower distribution of forces in the range of 10 to 20 pN, more characteristic of simple duplexes (39, 40), suggesting the presence of 2 distinct kinds of pseudoknots as well as various helix and/or hairpin structures. The same set of LcU values from the WLC fits were seen for refolding as for unfolding (SI Appendix, Table S2), indicating that the same set of intermediate structures was sampled.
Table 1.
Unfolding lengths and forces of observed and predicted structures
| Observed LcU, nm | Proposed state | Expected LcU, nm | Observed FU, pN |
| 0 | DHP | 0 | 10.8 ± 0.3 |
| 9 ± 1 | DHP− | 10.0 | 12.0 ± 0.3 |
| PKD | 9.8 | 11.4 ± 0.2 | |
| 12.1 ± 0.8 | PKD− | 12.4 | 21 ± 1 |
| 20.4 ± 0.6 | PKA+HP | 20.5 | 13.0 ± 0.4 |
| 31 ± 1 | PKA | 31.5 | 24 ± 1 |
| 40 ± 1 | S1A | 40.9 | 16.7 ± 0.2 |
| 60.7 ± 0.4 | U | 61.3 |
Errors represent SEM.
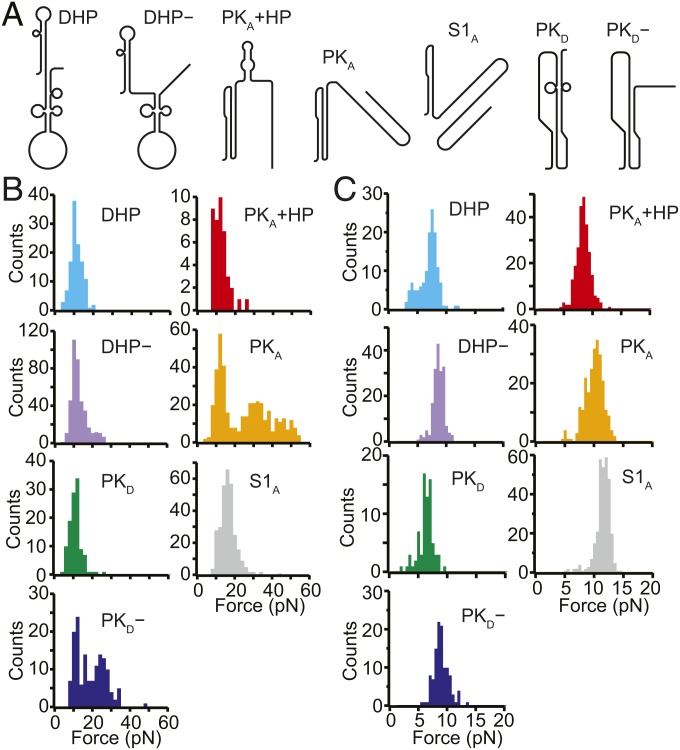
To identify the structures corresponding to each state in the FECs, we matched the observed LcU values to the expectations for the pseudoknot structures proposed in the literature as well as to the stem-loop structures predicted by mfold (41). Seven structures were found to match the observed LcU values within experimental error (Fig. 2A and SI Appendix, Fig. S1): the pseudoknot proposed by Firth and Atkins (35), denoted by PKA, which unfolded over a broad range of forces (Fig. 2B, orange); a pseudoknot similar to that proposed by Moomau et al. (21), denoted by PKD, which unfolded partially at low force (Fig. 2B, green); a partially folded version of PKD, denoted by PKD−, which unfolded over a broad range of forces (Fig. 2B, blue); fully and partially folded versions of the double-hairpin also proposed by Moomau et al. (21), denoted by DHP and DHP−, unfolding over a narrow range of low forces (Fig. 2B, cyan and purple, respectively); a combination of PKA and a short hairpin, denoted by PKA+HP, unfolding partially over a narrow range of low forces (Fig. 2B, red); and an extended version of stem 1 from PKA, denoted by S1A, similar to one of the hairpins in DHP, which unfolded over a narrow range of low forces (Fig. 2B, gray). The correspondence of each structure to the observed lengths and unfolding forces is shown in Table 1.
Fig. 2.
Different structures formed by the WNV frameshift signal in FECs. (A) Secondary structure models for the different conformations matching the states observed in the FECs (see also SI Appendix, Fig. S1). (B) Unfolding force distributions for the different states. The unfolding forces for DHP, DHP−, S1A, PKD, and PKA+HP were in the range of ∼5 to 20 pN, characteristic of secondary structures. (For PKD, and PKA+HP, a duplex/hairpin unfolded rather than the more stable pseudoknot.) Most unfolding forces for PKA and PKD− were in the range of 20 to 60 pN, characteristic of fully formed pseudoknots, although some transitions occurred at lower force, suggesting incomplete tertiary contact formation. (C) Refolding force distributions for each state. Refolding forces were only slightly smaller than unfolding forces for DHP, DHP−, S1A, PKD, and PKA+HP but were considerably smaller for PKA and PKD−.
These unfolding forces matched the expectation that tertiary structure unfolding generally involves broader force distributions and higher forces than secondary structure unfolding (16, 39). The sole exception was PKD, whose unusually low unfolding force reflected the low-stability contacts at the 3′ end of stem 2, as revealed in SHAPE data (21). The forces observed in refolding were generally lower than the unfolding forces for the corresponding structures (Fig. 2C), with small differences for states where only secondary structure was broken/formed during unfolding/refolding (DHP, DHP−, S1A, PKD, PKA+HP) but larger differences in the cases of PKD− and PKA, which involved breaking/forming tertiary structures. Thus, transitions involving secondary structures alone were not far from equilibrium, leading to the observation of occasional quasi-equilibrium reversible hopping between states (SI Appendix, Fig. S2), whereas those involving tertiary interactions were generally farther from equilibrium, as expected based on previous work (38, 42–44).
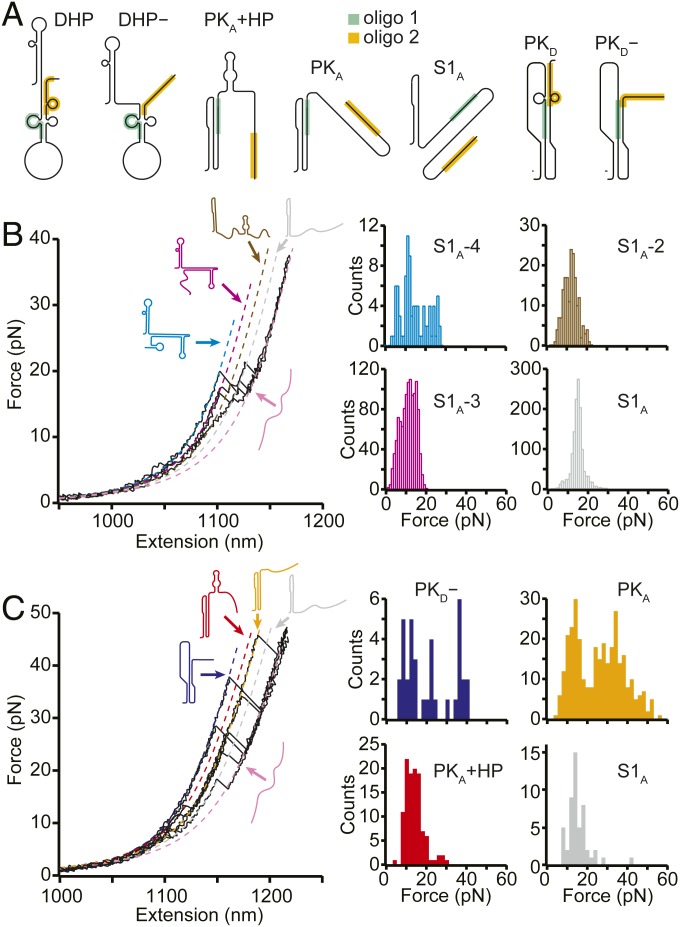
To confirm the structural assignments for these states, we remeasured the FECs in the presence of antisense oligonucleotides (SI Appendix, Table S1) binding to specific regions of the sequence at which they block RNA base pair formation (Fig. 3A, shaded regions). Considering first the oligo that blocked formation of stem 2 in both PKA and PKD (Fig. 3A and SI Appendix, Fig. S1, green), denoted by oligo 1, we found that unfolding events had forces in the range of 10 to 20 pN (Fig. 3B). This result indicates that only secondary structures were present, confirming that oligo 1 blocked pseudoknot formation. Indeed, not only were PKA, PKD, and PKD− prevented from forming, but, based on the LcU values from WLC fits (Fig. 3A, dashed lines) to the different states observed in these FECs (SI Appendix, Table S3), so were all the other states in Fig. 2A with the exception of S1A. In their stead, various combinations of S1A with nonnative helices that are energetically disfavored in the absence of antisense oligomer binding (SI Appendix, Fig. S3) were observed, matching predictions from structure-prediction tools (41, 45, 46).
Fig. 3.
FECs with antisense oligos. (A) Structural models showing where antisense oligos 1 (green) and 2 (orange) bind on each state. (B) With oligo 1 present, FECs showed only the low unfolding forces characteristic of secondary structures, indicating that the pseudoknots were prevented from forming, as expected from the structural models in A. Several unfolding transitions not observed without the oligo were seen, matching predictions for structures formed under the constraints imposed by the antisense oligo. (C) With oligo 2 present, high-force unfolding events corresponding to PKA and PKD− were observed, but transitions corresponding to DHP and DHP− were not, consistent with the structural models in A.
A second antisense oligonucleotide (oligo 2) was used to block base-pairing at the 3′ end of the frameshift signal (Fig. 3A and SI Appendix, Fig. S1, orange), disrupting DHP, DHP−, and PKD while permitting the formation of PKA, PKA+HP, PKD−, and S1A. High-force unfolding events were indeed seen with oligo 2 present, corresponding to unfolding of PKA and PKD− (Fig. 3B, orange and dark blue, respectively), as were lower-force events corresponding mainly to unfolding of PKA+HP and S1A (Fig. 3B, red and gray, respectively). The contour lengths for unfolding these states matched the values observed without oligos and those expected from the structural models of each state (SI Appendix, Table S3) in all cases but one: PKD−, which was slightly shorter (∼1.5 nm) than without oligo 2. This difference is attributed to the fact that oligo 2 can invade stem 2 of PKD− by 2 nt, shortening the expected length change on unfolding by 1.3 nm. States with the LcU and unfolding force values expected for DHP, DHP−, and PKD were not observed in the presence of oligo 2, as expected; PKD− was present, but its occupancy was reduced by roughly 2-fold.
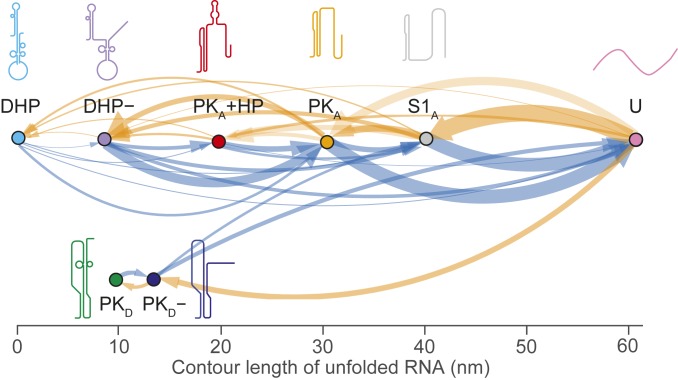
Finally, we examined the sequence in which the states formed, cataloging all pairwise transitions between states during unfolding and refolding curves to reveal all possible pathways through them (47). The resulting transition maps for unfolding (Fig. 4, blue) and refolding (Fig. 4, orange) show 2 distinct pathways for both unfolding and refolding the WNV frameshift signal. Most unfolding and refolding FECs (80 ± 2% of each) involved transitions along a pathway involving S1A, PKA, and DHP (Fig. 4, Top), whereas a minority (20 ± 2%) were on a second pathway involving PKD and PKD− (Fig. 4, Bottom). The observed transitions on the 2 pathways were consistent with the properties of the structures identified for each state: on the most common pathway, DHP, DHP−, PKA, and S1A all share a similar stem structure (S1A) and thus can be interconverted readily, whereas the pseudoknots on the minority pathway have a stem structure inconsistent with DHP or PKA and thus cannot interconvert with the latter structures. Note that even though PKD and DHP− had similar unfolding forces and contour lengths, PKD unfolded only via PKD−, which could be readily identified by its distinctive LcU value and unfolding force (Table 1).
Fig. 4.
Transition map of unfolding and refolding pathways. All pairwise transitions between states observed in unfolding (blue) and refolding (orange) FECs are shown, with the thickness of each arrow proportional to the probability of observing the associated transition. The same 2 distinct pathways are seen for both unfolding and refolding.
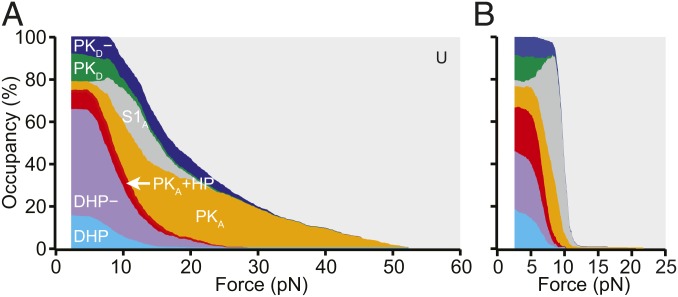
To complement the transition maps, we also analyzed how the occupancy of each state changed as a function of force during unfolding (Fig. 5A) and refolding (Fig. 5B). All the different structures that could be formed by the WNV frameshift signal were occupied at low force, with the exception of S1A: roughly 16% of unfolding curves started in DHP, 50% started in DHP−, 9% started in PKA+HP, 4% started in PKA, 13% started in PKD, and 8% started in PKD−. The occupancies were similar when examining only the first pulls for each molecule (SI Appendix, Table S4), where the RNA had folded before it was tethered to the beads, suggesting that the distribution of states was not significantly affected by waiting only 3 s between successive pulls, and, moreover, that refolding under tension when tethered to beads was not significantly different from folding in the absence of force before tethering. However, the occupancies of PKA and PKA+HP were considerably higher at the end of refolding FECs than at the start of unfolding FECs, and the occupancy of DHP− was correspondingly lower (SI Appendix, Table S4), indicating that a significant reconfiguration of PKA into the double-hairpin structures occurred between pulls. The low occupancy of DHP compared with DHP− could reflect the presence of transitions into DHP occurring at forces sufficiently low so as to preclude their ready identification; attempts to detect such transitions using an analysis sensitive to low-force transitions (48) were unsuccessful.
Fig. 5.
Force-dependent conformational heterogeneity. The occupancy of the different conformations as a function of the force applied during unfolding (A) and refolding (B) highlights the extreme conformational heterogeneity of the WNV frameshift sequence. Differences between unfolding and refolding reflect the nonequilibrium nature of the transitions.
Discussion
Comparing the behavior of the WNV stimulatory structure with that of structures stimulating −1 PRF at more moderate levels, the most obvious difference is the extreme conformational plasticity of the WNV frameshift signal. Previous SMFS studies of stimulatory structures inducing −1 PRF with efficiencies in the range of 15% to 20%, like those from the severe acute respiratory syndrome coronavirus, sugar cane yellow leaf virus, and mouse mammary tumor virus (19, 20, 49), found that in each case, only 2 structures formed: the native pseudoknot and an alternate structure (16). In contrast, the WNV frameshift signal formed 3 different pseudoknot structures based on 2 mutually incompatible configurations, as well as several different stem-loop structures. Consequently, the folding of this RNA is remarkably complex, involving 2 largely disjunct pathways, each with multiple intermediate states. In fact, the SMFS measurements likely understate the full complexity of the pathways: H-type pseudoknots such as PKA, PKD, and PKD− are known to have at least 2 parallel pathways involving stem 1 or stem 2 as intermediates, with the flux between the pathways determined by the relative stability of the 2 stems (50). However, the unfolding force of pseudoknots is often sufficiently high so as to make hairpin intermediates very short-lived and thus not always observed, as here; only the stem 1 intermediate (S1A) of PKA was seen, and none of the hairpin intermediates for PKD or PKD− were observed.
The pathways defined in Fig. 4 have some interesting properties. From the transition maps, although the refolding pathways are completely disjunct, the unfolding pathways appear to be connected through S1A: PKD− never transitioned to PKA, but it did sometimes appear to change directly into S1A. In principle, such a change should not be seen, because S1A is incompatible with both stems in PKD−. However, in every case, these transitions occurred in the same force range at which S1A refolds (∼10 to 14 pN) (Fig. 2C, gray), so that S1A would be expected to form quite rapidly after unfolding of PKD−, resulting in an apparent transition between PKD− and S1A. A more intriguing observation is that in the pathway starting at DHP, PKA was able to form after the double-hairpin conformations unfolded, requiring the formation of new tertiary contacts during unfolding. This behavior is contrary to the standard hierarchy of events seen during unfolding, where typically tertiary structures unfold before secondary structures (38, 42–44), as seen in the pathway involving PKD. However, such a conversion of a double hairpin to a pseudoknot has been reported previously in SMFS measurements of the rpsO operator from Escherichia coli (51); here it is enabled by the similarity between the refolding force for PKA (Fig. 2C, orange) and the unfolding forces for DHP and DHP− (Fig. 2B, cyan and purple). The existence of direct transitions from the double hairpin to PKA suggests that unfolding of the 3′ hairpin in DHP− is likely coordinated cooperatively with stem 2 formation in PKA.
Turning to the various states observed by SMFS, the length changes and sequential transitions identified from the FECs help refine and expand on the results from previous studies of the WNV frameshift signal. The apparently incompatible structures proposed on the basis of bioinformatic predictions (35) and SHAPE analysis (21) were in fact all present, with their incompatibility resolved by their existence on separate, mutually exclusive pathways. The structure of the pseudoknot proposed by Moomau et al. (21) was refined based on the observed length changes, revealing 2 variants: one with a long stem 2 and the other with a truncated stem 2 (Fig. 2A and SI Appendix, Fig. S1). An additional variant of the double hairpin proposed by Moomau et al. (21) (DHP) was also observed, with one stem, DHP−, partially unfolded. Moreover, the unfolding force distributions for PKA and PKD− (Fig. 2B, orange and blue) suggested the presence of 2 populations: one without properly formed tertiary contacts that unfolded in the ∼10 to 15 pN range characteristic of secondary structures and the other with a broader unfolding force distribution in the 20 to 50 pN range characteristic of pseudoknots with a fully formed tertiary structure. Importantly, all these structures were sufficiently stable (or metastable) so as to persist during the 3-s waiting period between successive FECs, such that significant fractions of the unfolding FECs started out in them.
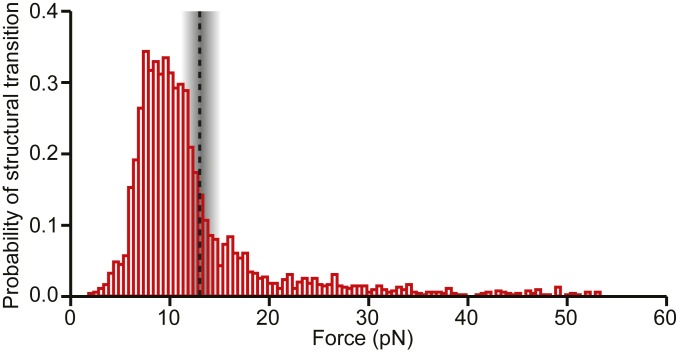
The occupancies of the diverse conformational states formed by the WNV frameshift signal were not static, however: a considerable number of transitions between structures were observed under tension, including both quasi-equilibrium fluctuations between conformations and nonequilibrium switches from one structure to another. Looking at the probability of undergoing some form of structural transition in the FECs as a function of force (Fig. 6), we found that it was highly peaked in the range of 7 to 13 pN. Intriguingly and suggestively, this maximum is in the range of forces just below the force at which ribosomes have been observed to stall during translation, 13 ± 2 pN (52). Given that the ribosome ramps the force on the stimulatory structure up and down in multiple attempts to unfold it during −1 PRF (33, 36), at loading rates estimated to be even higher than those used in the SMFS measurements (SI Appendix), the stimulatory structure would be expected to pass through the force range most likely to induce conformational transitions multiple times, analogous to the conditions present in the SMFS measurements.
Fig. 6.
Force-dependent probability of structural transitions. The probability of the WNV stimulatory structure changing conformation under tension during unfolding and refolding pulls peaks at ∼7 to 13 pN, just below the maximum force applied by the ribosome during translocation before it stalls (dashed line).
The results presented here suggest that it is primarily the dynamic properties of stimulatory structures, rather than their static properties, that are important for inducing −1 PRF. Pseudoknots are considered the canonical stimulatory structure and are often assumed to be the active element inducing −1 PRF (6, 21, 53, 54). Such an assumption seems unlikely to apply here, however: the prevalence of the pseudoknot structures PKA, PKD, and PKD−, which combined were present only 34 ± 2% of the time at zero force, is insufficient to account for the −1 PRF efficiency of ∼80% observed for this frameshift signal, even if each of the pseudoknot structures stimulated −1 PRF with 100% efficiency. Thus, some other factor(s) presumably must be involved. Indeed, the diversity of structures formed by the WNV frameshift signal at zero force raises the question of whether it is meaningful to speak of a single “native” structure stimulating frameshifting. We note that these considerations reflect the value of studying a frameshift signal with very high efficiency: it helps clarify the factors that must be important for stimulating −1 PRF.
The fact that the WNV frameshift signal—one of the highest-efficiency −1 PRF stimulators studied to date—exhibits the most complex, heterogeneous dynamics yet observed in any frameshift signal, along with the observation that the probability of structural transitions is most pronounced in the force range that the RNA is expected to experience during −1 PRF, suggest that the key factor for stimulating −1 PRF at high levels is most likely the propensity of the frameshift signal to undergo conformational changes when under tension from ribosomal attempts to translocate through it. We speculate that mechanistically, such conformational changes (whether near-equilibrium fluctuations or out-of-equilibrium switches) could trigger frameshifting by communicating abrupt tension fluctuations to the tRNA-mRNA complex, possibly in concert with other regulatory mechanisms yet to be fully elucidated. Such a picture is consistent with previous work connecting −1 PRF to pseudoknot conformational flexibility or dynamics (16, 18, 21, 30) and the ability to inhibit late-stage translocation during ribosome-induced unfolding of the first few base pairs (33). Notably, this perspective embeds the dynamic mechanical unfolding of the frameshift signal at the crux of the mechanisms that control −1 PRF efficiency. Future work extending single-molecule and kinetic assays of frameshifting (36, 55–57) to measure simultaneously the dynamics of both the ribosomal complex and the frameshift stimulatory structure in the act of frameshifting may help clarify the role of mRNA conformation dynamics in −1 PRF through more direct observation.
Methods
SMFS Measurements and Analysis.
Samples were prepared, measured, and analyzed as reported previously (16) and described in detail in SI Appendix.
Dual-Luciferase Frameshift Assay.
RNA transcripts were made containing the Renilla luciferase gene in the 0 frame upstream of the firefly luciferase in the −1 frame and separated by the WNV slippery sequence, linker, and stimulatory structure. Frameshifting efficiency was then measured from the luciferase luminescence ratio as described previously (58) and compared with values for negative and positive controls. Details are provided in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by the Canadian Institutes of Health Research, the National Research Council Canada, and Alberta Innovates.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.V.R. is a guest editor invited by the Editorial Board.
See Commentary on page 19225.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905258116/-/DCSupplemental.
References
- 1.Ketteler R., On programmed ribosomal frameshifting: The alternative proteomes. Front. Genet. 3, 242 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brierley I., Gilbert R. J. C., Pennell S., “Pseudoknot-dependent programmed —1 ribosomal frameshifting: Structures, mechanisms and models” in Recoding: Expansion of Decoding Rules Enriches Gene Expression, Nucleic Acids and Molecular Biology, Atkins J. F., Gesteland R. F., Eds. (Springer, New York, 2010), pp. 149–174. [Google Scholar]
- 3.Dinman J. D., Mechanisms and implications of programmed translational frameshifting. Wiley Interdiscip. Rev. RNA 3, 661–673 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atkins J. F., Loughran G., Bhatt P. R., Firth A. E., Baranov P. V., Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 44, 7007–7078 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dam E., Pleij K., Draper D., Structural and functional aspects of RNA pseudoknots. Biochemistry 31, 11665–11676 (1992). [DOI] [PubMed] [Google Scholar]
- 6.Giedroc D. P., Cornish P. V., Frameshifting RNA pseudoknots: Structure and mechanism. Virus Res. 139, 193–208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plant E. P., “Ribosomal frameshift signals in viral genomes” in Viral Genomes - Molecular Structure, Diversity, Gene Expression Mechanisms and Host-Virus Interactions, Garcia M. L., Romanowski V., Eds. (Intech Open, 2012), pp. 93–122. [Google Scholar]
- 8.Kendra J. A., et al. , Ablation of programmed −1 ribosomal frameshifting in Venezuelan equine encephalitis virus results in attenuated neuropathogenicity. J. Virol. 91, e01766-e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melian E. B., et al. , Programmed ribosomal frameshift alters expression of West Nile virus genes and facilitates virus replication in birds and mosquitoes. PLoS Pathog. 10, e1004447 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melian E. B., et al. , NS1′ of flaviviruses in the Japanese encephalitis virus serogroup is a product of ribosomal frameshifting and plays a role in viral neuroinvasiveness. J. Virol. 84, 1641–1647 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brierley I., Dos Ramos F. J., Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Res. 119, 29–42 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dulude D., Berchiche Y. A., Gendron K., Brakier-Gingras L., Heveker N., Decreasing the frameshift efficiency translates into an equivalent reduction of the replication of the human immunodeficiency virus type 1. Virology 345, 127–136 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Shehu-Xhilaga M., Crowe S. M., Mak J., Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J. Virol. 75, 1834–1841 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belew A. T., Dinman J. D., Cell cycle control (and more) by programmed -1 ribosomal frameshifting: Implications for disease and therapeutics. Cell Cycle 14, 172–178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gareiss P. C., Miller B. L., Ribosomal frameshifting: An emerging drug target for HIV. Curr. Opin. Investig. Drugs 10, 121–128 (2009). [PubMed] [Google Scholar]
- 16.Ritchie D. B., Foster D. A., Woodside M. T., Programmed −1 frameshifting efficiency correlates with RNA pseudoknot conformational plasticity, not resistance to mechanical unfolding. Proc. Natl. Acad. Sci. U.S.A. 109, 16167–16172 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao S., Chen S.-J., Predicting ribosomal frameshifting efficiency. Phys. Biol. 5, 016002 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y., et al. , Comparative studies of frameshifting and nonframeshifting RNA pseudoknots: A mutational and NMR investigation of pseudoknots derived from the bacteriophage T2 gene 32 mRNA and the retroviral gag-pro frameshift site. RNA 8, 981–996 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornish P. V., Hennig M., Giedroc D. P., A loop 2 cytidine-stem 1 minor groove interaction as a positive determinant for pseudoknot-stimulated −1 ribosomal frameshifting. Proc. Natl. Acad. Sci. U.S.A. 102, 12694–12699 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plant E. P., et al. , A three-stemmed mRNA pseudoknot in the SARS coronavirus frameshift signal. PLoS Biol. 3, e172 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moomau C., Musalgaonkar S., Khan Y. A., Jones J. E., Dinman J. D., Structural and functional characterization of programmed ribosomal frameshift signals in West Nile virus strains reveals high structural plasticity among cis-acting RNA elements. J. Biol. Chem. 291, 15788–15795 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wen J.-D., et al. , Following translation by single ribosomes one codon at a time. Nature 452, 598–603 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qu X., et al. , The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature 475, 118–121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen T. M., Reihani S. N. S., Oddershede L. B., Sørensen M. A., Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting. Proc. Natl. Acad. Sci. U.S.A. 104, 5830–5835 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Namy O., Moran S. J., Stuart D. I., Gilbert R. J. C., Brierley I., A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature 441, 244–247 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plant E. P., et al. , The 9-A solution: How mRNA pseudoknots promote efficient programmed −1 ribosomal frameshifting. RNA 9, 168–174 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ritchie D. B., Woodside M. T., Probing the structural dynamics of proteins and nucleic acids with optical tweezers. Curr. Opin. Struct. Biol. 34, 43–51 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen G., Chang K.-Y., Chou M.-Y., Bustamante C., Tinoco I. Jr, Triplex structures in an RNA pseudoknot enhance mechanical stability and increase efficiency of −1 ribosomal frameshifting. Proc. Natl. Acad. Sci. U.S.A. 106, 12706–12711 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong Z., et al. , Mechanical unfolding kinetics of the SRV-1 gag-pro mRNA pseudoknot: Possible implications for −1 ribosomal frameshifting stimulation. Sci. Rep. 6, 39549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Messieres M., et al. , Single-molecule measurements of the CCR5 mRNA unfolding pathways. Biophys. J. 106, 244–252 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ritchie D. B., et al. , Conformational dynamics of the frameshift stimulatory structure in HIV-1. RNA 23, 1376–1384 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie D. B., Soong J., Sikkema W. K. A., Woodside M. T., Anti-frameshifting ligand reduces the conformational plasticity of the SARS virus pseudoknot. J. Am. Chem. Soc. 136, 2196–2199 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Wu B., et al. , Translocation kinetics and structural dynamics of ribosomes are modulated by the conformational plasticity of downstream pseudoknots. Nucleic Acids Res. 46, 9736–9748 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuhlmann M. M., Chattopadhyay M., Stupina V. A., Gao F., Simon A. E., An RNA element that facilitates programmed ribosomal readthrough in Turnip crinkle virus adopts multiple conformations. J. Virol. 90, 8575–8591 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Firth A. E., Atkins J. F., A conserved predicted pseudoknot in the NS2A-encoding sequence of West Nile and Japanese encephalitis flaviviruses suggests NS1′ may derive from ribosomal frameshifting. Virol. J. 6, 14 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan S., Wen J. D., Bustamante C., Tinoco I. Jr, Ribosome excursions during mRNA translocation mediate broad branching of frameshift pathways. Cell 160, 870–881 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang M. D., Yin H., Landick R., Gelles J., Block S. M., Stretching DNA with optical tweezers. Biophys. J. 72, 1335–1346 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen G., Wen J.-D., Tinoco I. Jr, Single-molecule mechanical unfolding and folding of a pseudoknot in human telomerase RNA. RNA 13, 2175–2188 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liphardt J., Onoa B., Smith S. B., Tinoco I. Jr, Bustamante C., Reversible unfolding of single RNA molecules by mechanical force. Science 292, 733–737 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Woodside M. T., et al. , Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc. Natl. Acad. Sci. U.S.A. 103, 6190–6195 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuker M., Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Onoa B., et al. , Identifying kinetic barriers to mechanical unfolding of the T. thermophila ribozyme. Science 299, 1892–1895 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neupane K., Yu H., Foster D. A. N., Wang F., Woodside M. T., Single-molecule force spectroscopy of the add adenine riboswitch relates folding to regulatory mechanism. Nucleic Acids Res. 39, 7677–7687 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greenleaf W. J., Frieda K. L., Foster D. A. N., Woodside M. T., Block S. M., Direct observation of hierarchical folding in single riboswitch aptamers. Science 319, 630–633 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bellaousov S., Mathews D. H., ProbKnot: Fast prediction of RNA secondary structure including pseudoknots. RNA 16, 1870–1880 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kato Y., Sato K., Asai K., Akutsu T., Rtips: Fast and accurate tools for RNA 2D structure prediction using integer programming. Nucleic Acids Res. 40, W29–W34 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sen Mojumdar S., et al. , Partially native intermediates mediate misfolding of SOD1 in single-molecule folding trajectories. Nat. Commun. 8, 1881 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Solanki A., Neupane K., Woodside M. T., Single-molecule force spectroscopy of rapidly fluctuating, marginally stable structures in the intrinsically disordered protein α-synuclein. Phys. Rev. Lett. 112, 158103 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Chamorro M., Parkin N., Varmus H. E., An RNA pseudoknot and an optimal heptameric shift site are required for highly efficient ribosomal frameshifting on a retroviral messenger RNA. Proc. Natl. Acad. Sci. U.S.A. 89, 713–717 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roca J., et al. , Monovalent ions modulate the flux through multiple folding pathways of an RNA pseudoknot. Proc. Natl. Acad. Sci. U.S.A. 115, E7313–E7322 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu Y.-J., Wu C.-H., Yeh A. Y.-C., Wen J.-D., Folding a stable RNA pseudoknot through rearrangement of two hairpin structures. Nucleic Acids Res. 42, 4505–4515 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaiser C. M., Tinoco I. Jr, Probing the mechanisms of translation with force. Chem. Rev. 114, 3266–3280 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang X., Cheng Q., Du Z., A genome-wide analysis of RNA pseudoknots that stimulate efficient −1 ribosomal frameshifting or readthrough in animal viruses. BioMed Res. Int. 2013, 984028 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Houck-Loomis B., et al. , An equilibrium-dependent retroviral mRNA switch regulates translational recoding. Nature 480, 561–564 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caliskan N., Katunin V. I., Belardinelli R., Peske F., Rodnina M. V., Programmed −1 frameshifting by kinetic partitioning during impeded translocation. Cell 157, 1619–1631 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen C., et al. , Dynamics of translation by single ribosomes through mRNA secondary structures. Nat. Struct. Mol. Biol. 20, 582–588 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen J., et al. , Dynamic pathways of −1 translational frameshifting. Nature 512, 328–332 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grentzmann G., Ingram J. A., Kelly P. J., Gesteland R. F., Atkins J. F., A dual-luciferase reporter system for studying recoding signals. RNA 4, 479–486 (1998). [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.