Significance
In humans, essential iron-sulfur (Fe-S) cluster cofactors are synthesized using an assembly complex that depends on the protein frataxin (FXN). The physiological function of FXN, which is linked to the fatal neurodegenerative disease Friedreich’s ataxia (FRDA), is still under debate. Here, we show that FXN accelerates Fe-S cluster formation by positioning a mobile loop cysteine of the assembly complex to interact with the substrate and function as an acid, nucleophile, and sulfur carrier during the reaction. Our architectural switch model suggests that FXN controls Fe-S cluster biosynthesis by inducing an unusual rearrangement of protein subunits in the assembly complex. These results provide mechanistic insights into this critical biological process and establish a foundation for the design of new FRDA treatments.
Keywords: Friedreich’s ataxia, allostery, enzyme mechanism, bioinorganic, iron-sulfur
Abstract
The function of frataxin (FXN) has garnered great scientific interest since its depletion was linked to the incurable neurodegenerative disease Friedreich’s ataxia (FRDA). FXN has been shown to be necessary for iron-sulfur (Fe-S) cluster biosynthesis and proper mitochondrial function. The structural and functional core of the Fe-S cluster assembly complex is a low-activity pyridoxal 5′-phosphate (PLP)–dependent cysteine desulfurase enzyme that consists of catalytic (NFS1), LYRM protein (ISD11), and acyl carrier protein (ACP) subunits. Although previous studies show that FXN stimulates the activity of this assembly complex, the mechanism of FXN activation is poorly understood. Here, we develop a radiolabeling assay and use stopped-flow kinetics to establish that FXN is functionally linked to the mobile S-transfer loop cysteine of NFS1. Our results support key roles for this essential cysteine residue in substrate binding, as a general acid to advance the Cys-quinonoid PLP intermediate, as a nucleophile to form an NFS1 persulfide, and as a sulfur delivery agent to generate a persulfide species on the Fe-S scaffold protein ISCU2. FXN specifically accelerates each of these individual steps in the mechanism. Our resulting architectural switch model explains why the human Fe-S assembly system has low inherent activity and requires activation, the connection between the functional mobile S-transfer loop cysteine and FXN binding, and why the prokaryotic system does not require a similar FXN-based activation. Together, these results provide mechanistic insights into the allosteric-activator role of FXN and suggest new strategies to replace FXN function in the treatment of FRDA.
Friedreich’s ataxia (FRDA) is an autosomal recessive neurodegenerative disease that has no cure. It is primarily caused by the down-regulation of the protein frataxin (FXN) that results from a GAA triplet repeat expansion in the first intron of the FXN gene (1). In the search for the function of FXN, early studies led to many different proposals that include the control of reactive oxygen stress, in iron storage, as an iron chaperone, and as an iron sensor (2). FXN was later demonstrated to bind to the human iron-sulfur (Fe-S) assembly complex and promote Fe-S cluster biosynthesis in eukaryotes (3–8). However, mechanistic details and the precise role of FXN in Fe-S cluster assembly remain elusive, thereby limiting FXN replacement strategies for the treatment of FRDA.
Fe-S clusters are protein cofactors that are necessary for many critical biochemical processes (9, 10). Fe-S clusters have a variety of enzymatic roles, including electron transfer, substrate binding and activation, and the initiation of radical chemistry (11, 12). Different Fe-S cluster stoichiometries, such as the [2Fe-2S] and [4Fe-4S] forms, are synthesized and distributed to apo target proteins by conserved biosynthetic pathways. A key enzyme in these pathways is the cysteine desulfurase, which uses a pyridoxal 5′-phosphate (PLP) cofactor to convert l-cysteine to l-alanine and provide the sulfur for cluster synthesis. In contrast to the prokaryotic cysteine desulfurase IscS, the human Fe-S assembly subcomplex requires interactions with both a LYRM superfamily protein (ISD11) and an acyl carrier protein (ACP) for the stability and function of the NFS1 catalytic subunit (13–18). Both NFS1 and IscS generate a persulfide intermediate on a conserved cysteine residue of a mobile S-transfer loop; the sulfur is then transferred to a cysteine residue on the Fe-S scaffold protein (19). In humans, the scaffold protein ISCU2 combines the sulfane sulfur with Fe2+ and electrons, possibly from a ferredoxin (20, 21), to generate [2Fe-2S]2+ cluster intermediates (5, 22, 23). These clusters are then transferred intact to a carrier protein to complete the catalytic cycle for the assembly complex.
Identifying the role of FXN in Fe-S cluster biosynthesis has been complicated by the existence of FXN forms with different lengths in vivo. The longer form, FXNΔ1–55, can undergo an iron-dependent oligomerization to produce a species reminiscent of ferritin that may function in iron storage (24–28). The truncated form, FXNΔ1–80, has a modest binding affinity for iron (3 to 55 μM), which led to the proposal that FXN accelerates Fe-S cluster synthesis by functioning as a chaperone that donates iron (29–38). More recently, multiple studies provide evidence that FXN enhances both the cysteine desulfurase activity (increasing the catalytic constant [kcat] by ∼12-fold) and the subsequent Fe-S cluster assembly rate on the scaffold protein (5, 22, 39–42). Remarkably, both FXN and its prokaryotic homolog, CyaY, stimulate the in vitro Fe-S assembly reactions by the human Fe-S biosynthetic complex but inhibit the equivalent reaction by the prokaryotic system (43, 44). FXN has been proposed to affect the activity of the cysteine desulfurase enzyme by enhancing substrate binding to the PLP cofactor (39) and to increase the rate of the sulfur transfer reaction to the scaffold protein (40, 41) and small-molecule thiols (5, 41). However, the underlying biochemical logic for why FXN is required to synthesize Fe-S clusters in eukaryotes, but not prokaryotes, and whether FXN affects specific chemical steps in the PLP-based mechanism of the cysteine desulfurase are currently unknown.
In this study, a newly developed acid-quench radiolabeling assay and stopped-flow kinetics were used to probe how FXN affects the rates of individual steps in the cysteine desulfurase reaction and subsequent intermolecular sulfur transfer to ISCU2. Our results are consistent with the essential cysteine of the mobile S-transfer loop functioning in substrate binding, as a general acid, as a nucleophile, and as a sulfur carrier during different steps of Fe-S cluster biosynthesis. Intriguingly, we show that FXN accelerates each of these steps linked to the mobile S-transfer loop. The mechanistic details and resulting model described in this paper provide insights into the function of FXN and have potential implications for the treatment of FRDA.
Results
FXN Accelerates Persulfide Formation on NFS1 and ISCU2.
Our first objective was to test whether FXN modulates the rates of persulfide formation on NFS1 and the interprotein sulfur transfer to ISCU2. The human NFS1–ISD11 complex was recombinantly purified with Escherichia coli ACP (previously named the SDAec complex) (18) and mixed with ISCU2 (referred to as the SDAecU complex), ISCU2 and FXN (referred to as the SDAecUF complex), or ISCU2 and the E. coli FXN homolog CyaY (referred to as the SDAecUCec complex). A rapid acid-quench assay was developed that couples high-performance liquid chromatography (HPLC) with radiolabeled sulfur detection. In this assay, Fe-S assembly complexes were reacted with l-[35S]cysteine substrate, quenched with acid after different reaction times, and applied to a reversed-phase HPLC column for an analysis of protein content (absorbance) and persulfide label (radioactivity). The abilities to rapidly protonate thiolate nucleophiles, which inhibits both enzymatic and nonenzymatic sulfur transfer reactions for persulfide species (45), and analyze proteins for 35S incorporation under quench conditions are critical aspects of this assay. Control experiments established conditions to separate proteins in the SDAecUF complex that contain cysteine residues (NFS1 and ISCU2; SI Appendix, Figs. S1 and S2). The effectiveness of acid in quenching sulfur transfer from NFS1 to ISCU2 was established by reactions under normal and quench conditions (SI Appendix, Fig. S3). Initial studies with excess l-[35S]cysteine substrate resulted in labeling of multiple cysteine residues on NFS1 and ISCU2 (SI Appendix, Fig. S4). Some of these labeled cysteine residues, especially on NFS1 (C158, C163, C353, and C426), are likely due to side reactions of the SDAecU complex that occur with excess cysteine substrate, long incubation times, and no additional reductant or iron; these conditions likely permit sulfur transfer reactions from persulfide species to excess cysteine in solution and result in the subsequent scrambling of the radiolabel on the protein. The ability of cysteine desulfurases to readily generate free l-cysteine persulfide has been previously reported (46). To minimize nonspecific labeling, stoichiometric amounts of the substrate were used to determine the rates of persulfide formation on NFS1 and ISCU2.
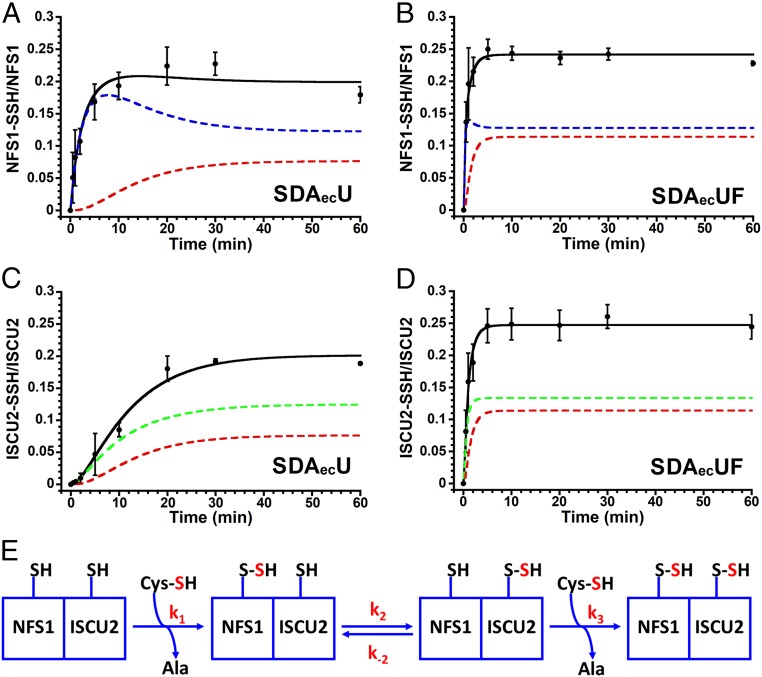
The SDAecU and SDAecUF complexes were combined with l-[35S]cysteine and quenched after different reaction times, and the proteins were separated under quench conditions. Plotting the amount of radiolabel as a function of time revealed saturation between 0.20 and 0.25 labels per NFS1 (Fig. 1 A and B) and ISCU2 (Fig. 1 C and D) for the SDAecU and SDAecUF complexes. Interestingly, the amount of label on NFS1 did not decrease at longer reaction times as one might expect for sequential irreversible reactions using limited substrate. Persulfide formation on NFS1 and ISCU2, which were fit simultaneously, matched well with an equilibrium sulfur transfer model with a second cysteine turnover (Fig. 1E and SI Appendix, Fig. S5). Models that lack the equilibrium step (k-2) or second cysteine turnover (k3) did not adequately fit the data. The rates of persulfide formation on NFS1 were constrained to be the same for the SDAecU (or SDAecUF) complex with or without a persulfide-bound ISCU2 (Fig. 1E; k1 = k3). Under these limiting substrate conditions, the slow step for both the SDAecU and SDAecUF complexes was the formation of the persulfide species on NFS1 (Fig. 1 and Table 1). The kinetic model that best fit the data suggests that FXN accelerates both the rate of persulfide formation on NFS1 (6.5-fold greater) and sulfur transfer from NFS1 to ISCU2 (30-fold greater), and that the persulfide transfer reaction from NFS1 to ISCU2 is reversible with an equilibrium constant near 1 for both the SDAecU and SDAecUF complexes (Table 1).
Fig. 1.
FXN accelerates the persulfide formation kinetics on both NFS1 and ISCU2. The SDAecU and SDAecUF complexes were reacted with a stoichiometric amount of l-[35S]cysteine and quenched with acid, and the proteins were separated using a reversed-phase column. Formation of [35S]-persulfide was quantitated and plotted for NFS1 (SDAecU) (A), NFS1 (SDAecUF) (B), ISCU2 (SDAecU) (C), and ISCU2 (SDAecUF) (D). (E) Best-fit kinetic model (k1 = k3) for persulfide accumulation on cysteine residues of NFS1 and ISCU2. The data for the SDAecU or SDAecUF complex were fit simultaneously with KinTek, and the fits were validated using FitSpace (SI Appendix, Fig. S5). Blue, green, and red dashed lines represent the simulated amounts of persulfide on NFS1, ISCU2, or both NFS1 and ISCU2, respectively. Black lines indicate the total amount of simulated persulfide on NFS1 or ISCU2. Replicate errors (n = 3) are indicated by error bars.
Table 1.
Persulfide formation kinetics for Fe-S assembly complexes
| Experiments | Parameters | SDAecU | SDAecUF | FXN rate enhancement |
| [35S]-Cys sulfur transfer reactions (min−1 × 10−2) | Persulfide formation on NFS1 (k1 and k3) | 0.8 (0.75 to 0.81)* | 5.2 (5.1 to 5.2)* | 6.5 |
| Persulfide transfer: NFS1 to ISCU2 (k2) | 8.6 (8.5 to 9.6)* | 266 (252 to 282)* | 30.9 | |
| Persulfide back transfer: ISCU2 to NFS1 (k-2) | 8.4 (8.2 to 10.0)* | 254 (241 to 70)* | 30.2 | |
| ISCU2/NFS1 equilibrium constant (k2/k-2) | 1.0 | 1.1 | ||
| Pulse–chase experiments (min−1 × 10−2) | Persulfide decay for NFS1 | 8 ± 1 | 42 ± 7 | 5.3 |
| Persulfide decay for ISCU2 | 6 ± 2 | 0.048 ± 0.0015† | 0.008 |
The values in parentheses denotes the lower and upper limits of the best fit value of the rate constant. These values were calculated at 99% and 99.7% confidence contours for the SDAecU and SDAecUF complexes, respectively.
Determined with a linear fit.
Evidence for a Reversible Sulfur Transfer Reaction Between NFS1 and ISCU2.
Next, we extended this radiolabeling assay to (1) test whether the sulfur transfer reaction between NFS1 and ISCU2 is indeed reversible and (2) determine whether FXN affects the susceptibility of the persulfide species to thiol-based cleavage using a pulse–chase experiment. The SDAecU and SDAecUF complexes were first reacted with l-[35S]cysteine for 40 min to allow persulfide labeling of NFS1 and ISCU2. The quantitation of the radiolabel indicated that both the SDAecU and SDAecUF complexes had similar amounts of persulfide associated with NFS1 (47%) and ISCU2 (53%) (SI Appendix, Fig. S6 A and D). Spiking a portion of the labeled SDAecU sample with unlabeled ISCU2 or SDAec shifted the [35S]-persulfide population toward ISCU2 (35% NFS1, 65% ISCU2; SI Appendix, Fig. S6B) or NFS1 (62% NFS1, 38% ISCU2; SI Appendix, Fig. S6C), respectively. Similarly, adding unlabeled ISCU2 or SDAec to the labeled SDAecUF sample shifted the population of radiolabel toward ISCU2 (33% NFS1, 67% ISCU2; SI Appendix, Fig. S6E) or NFS1 (55% NFS1, 45% ISCU2; SI Appendix, Fig. S6F), respectively. The observation that the total radioactivity remained the same (SI Appendix, Fig. S6) but the label distribution shifted upon the addition of unlabeled proteins is consistent with a reversible interprotein sulfur transfer reaction between NFS1 and ISCU2 in the absence of iron, as suggested by our kinetic model (Fig. 1E).
Next, we determined the susceptibility of the persulfide species to reductive cleavage using a pulse–chase experiment. A stoichiometric amount of l-[35S]cysteine was first incubated with the individual SDAecU and SDAecUF complexes. The labeled complexes were then combined with 1 mM nonradioactive l-cysteine for various times, quenched with acid, and analyzed as described above. For the SDAecU complex, spiking the sample with nonradioactive cysteine resulted in the loss of most of the labeled sulfur on both NFS1 and ISCU2, which occurred at similar rates (SI Appendix, Fig. S7 A and B and Table 1). Intriguingly, the addition of FXN resulted in an ∼5-fold increased rate of sulfur loss from NFS1, but almost no loss of label from ISCU2. These data, coupled to the evidence for a reversible sulfur transfer reaction (SI Appendix, Fig. S6), are consistent with a model in which the persulfide loss occurs primarily from NFS1, which is then regenerated by either enzymatic turnover using a nonlabeled cysteine or back-transfer of the sulfur from ISCU2 (SI Appendix, Fig. S7C). For the SDAecU complex, we hypothesize that after 35S cleavage, the persulfide on NFS1 is replaced through interprotein sulfur transfer from ISCU2, explaining the similar [35S]-persulfide depletion rates for NFS1 and ISCU2 (Table 1). In contrast, we suggest that the SDAecUF complex regenerates the persulfide species through rapid enzymatic turnover at the high cysteine concentration of the chase experiment, thereby inhibiting the back-transfer of label from ISCU2 to NFS1 and reconciling the apparent protection of the label on ISCU2. Notably, the kcat for the cysteine desulfurase reaction is ∼12-fold greater for the SDAecU complex in the presence of FXN (18). Together, these radiolabeling experiments strongly support the proposed role of FXN as an allosteric activator that accelerates sulfur chemistry (5, 40, 41), more specifically the rate of both persulfide formation on NFS1 and interprotein sulfur transfer to ISCU2.
FXN Accelerates the Formation and Decay of PLP Intermediates for the Human Cysteine Desulfurase Complex.
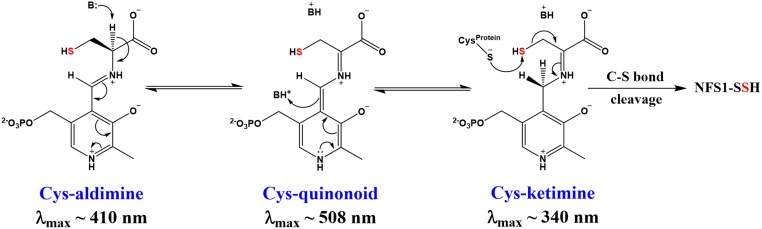
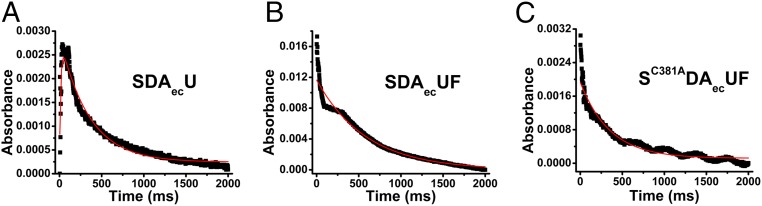
To determine how FXN accelerates persulfide formation on NFS1, the PLP chemistry associated with the cysteine desulfurase reaction was directly probed using stopped-flow kinetics. Previously, Bollinger and coworkers (47, 48) used stopped-flow kinetic and isotope effect experiments to dissect the mechanism of formation of the persulfide species, identify 7 accumulating intermediates, and map the intermediates to the mechanism of cysteine desulfurases from Synechocystis sp. PCC 6803. According to the currently accepted cysteine desulfurase mechanism (47, 49, 50) (SI Appendix, Fig. S8), the addition of cysteine leads to the sequential formation of Cys-aldimine, Cys-quinonoid, and Cys-ketimine intermediates prior to the C-S bond cleavage step and formation of the persulfide species (Fig. 2). We monitored the Cys-aldimine at 410 nm, the Cys-quinonoid at 508 nm, and the Cys-ketimine at 340 nm (Materials and Methods). The SDAecU and SDAecUF complexes were combined with l-cysteine, and the changes in absorbance were monitored over the first 2,000 ms by stopped-flow kinetics. For the SDAecU complex, we observed both the development and decay of the Cys-aldimine intermediate (Fig. 3A). In contrast, the build-up of the Cys-aldimine species was not observed for the SDAecUF complex, possibly due to its development occurring during the dead time of the instrument, and the intermediate accumulated at an ∼8-fold higher level than for the equivalent experiment with the SDAecU complex (Fig. 3 A and B). Despite the different formation kinetics, the Cys-aldimine intermediates for the 2 complexes decay at similar rates (∼7% difference; Table 2). This result suggests that FXN promotes l-cysteine interactions with the PLP cofactor to generate the aldimine intermediate and initiate catalytic turnover for the human Fe-S assembly complex.
Fig. 2.
Scheme showing key intermediates in the cysteine desulfurase reaction that were monitored by stopped-flow kinetics. In the currently accepted mechanism, the enzyme goes through sequential Cys-aldimine, Cys-quinonoid, and Cys-ketimine intermediates prior to C-S bond cleavage and generation of the NFS1 persulfide species (NFS1-SSH).
Fig. 3.
Formation and decay of the Cys-aldimine intermediate monitored by stopped-flow kinetics. The SDAecU, SDAecUF, and SC381ADAecUF complexes (100 μM) were individually mixed with 10 mM l-cysteine, and the Cys-aldimine intermediate was tracked by changes in absorbance at 410 nm as a function of time. The traces are an average of 3 independent experiments. The rates for the development and decay of the Cys-aldimine intermediate were obtained by nonlinear regression analysis (solid red lines).
Table 2.
Rates for individual steps in the cysteine desulfurase reaction for Fe-S assembly complexes
| Enzyme or complex | Rate of Cys-aldimine formation (s−1) | Rate of Cys-aldimine decay (s−1) | Rate of Cys-quinonoid formation (s−1) | Rate of Cys-quinonoid decay (s−1) | Relative rate of Cys-quinonoid decay |
| SDAecU | 96 ± 15 | 1.82 ± 0.05 | 19.5 ± 0.5 | 1.19 ± 0.03 | 1 |
| SDAecUF | ND | 1.70 ± 0.01 | 17.5 ± 0.4 | 4.80 ± 0.07 | 4.0 |
| SDAecUCec | ND | ND | 5.6 ± 0.2 | 2.9 ± 0.1 | 2.44 |
| SC381ADAecUF | ND | 2.84 ± 0.005 | 59 ± 2 | 0.099 ± 0.004 | 0.08 |
| IscS | ND | ND | 88 ± 1 | 9.77 ± 0.06 | 8.21 |
| IscSC328A | ND | ND | 96 ± 20 | 0.08 ± 0.01 | 0.067 |
ND, not determined.
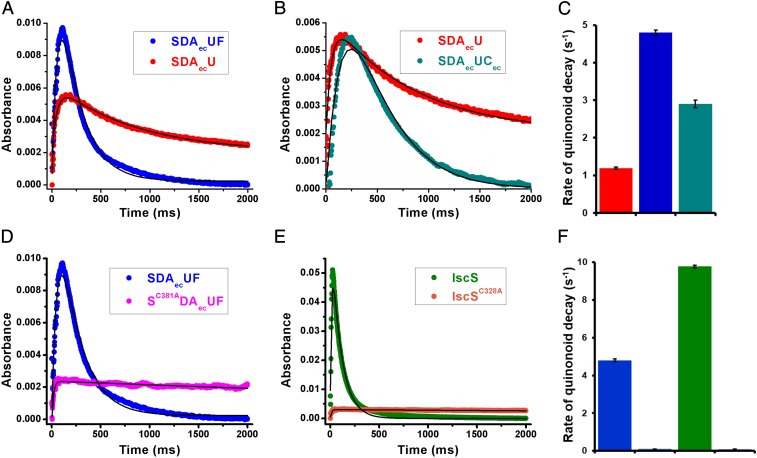
We next examined the effect of FXN on the formation and decay of the Cys-quinonoid intermediate. The SDAecU and SDAecUF complexes exhibited similar (∼11% difference) kinetics for the formation of the Cys-quinonoid species (Fig. 4A and Table 2), consistent with the comparable rates of decay for the Cys-aldimine intermediate. Notably, the addition of FXN to the SDAecU complex dramatically increased the decay kinetics (4-fold faster) for the Cys-quinonoid intermediate (Fig. 4 A and C and Table 2). Next, we tested if the FXN homolog CyaY, which is known to also activate the cysteine desulfurase activity of the SDAecU complex (44), also has the ability to accelerate this step of the reaction. Similar to the effect with FXN, the addition of CyaY to the SDAecU complex increased the rate of Cys-quinonoid decay (2.4-fold faster) (Fig. 4 B and C and Table 2). The conversion of the Cys-quinonoid to the Cys-ketimine species requires protonation at the C4′ position (Fig. 2). Thus, these results support a role for FXN in accelerating the protonation of the Cys-quinonoid intermediate, which prompts its conversion to the Cys-ketimine species, during the catalytic cycle of the human cysteine desulfurase complex (Fig. 2 and SI Appendix, Fig. S8).
Fig. 4.
Decay of the Cys-quinonoid intermediate is accelerated by the presence of FXN and the mobile S-transfer loop cysteine. The different complexes (100 μM) were combined with 10 mM l-cysteine, and the absorbance at 508 nm was followed as a function of time. Stopped-flow kinetic traces are displayed for the Cys-quinonoid intermediate of the SDAecU (red) and SDAecUF (blue) complexes (A), SDAecU (red) and SDAecUCec (cyan) complexes (B), SDAecUF (blue) and SC381ADAecUF (pink) complexes (D), and IscS (green) and IscSC328A (orange) variants (E). A comparison of the Cys-quinonoid decay rates for complexes (using the same color scheme) for A and B (C) and D and E (F) is shown. Note that the SC381ADAecUF and IscSC328A variants have extremely slow decay kinetics. All kinetic traces are an average of 3 independent experiments, and the rates for the development and decay of the Cys-quinonoid intermediate were obtained by nonlinear regression analysis (solid lines).
We then examined the kinetics for the development of the Cys-ketimine intermediate. An increase in absorbance at 340 nm was observed but could not be adequately fit to obtain rate constants (SI Appendix, Fig. S9). This may be due to contributions from additional intermediates after the carbon-sulfur (C-S) bond cleavage event (SI Appendix, Fig. S8) that have similar absorbance properties to the Cys-ketimine species. However, we do observe that there is a larger amount of absorbance at 340 nm for SDAecUF than for SDAecU (SI Appendix, Fig. S9 A and B), which corresponds to the faster kinetics for the Cys-quinonoid decay for the SDAecUF complex. Overall, the observation of the decay of a 410-nm species, the formation and decay of a 508-nm species, and the formation of a 340-nm species is compatible with the current mechanism and the assignment of sequential Cys-aldimine, Cys-quinonoid, and Cys-ketimine intermediates. Together, these stopped-flow experiments are consistent with the radiolabeling studies and indicate that FXN binding accelerates PLP-based chemistry that occurs prior to the C-S bond cleavage and persulfide formation reaction on the mobile S-transfer loop.
The Mobile S-Transfer Loop Cysteine Is the Proton Donor for Quinonoid Decay.
The mobile S-transfer loop cysteine has a critical role in C-S bond cleavage of the Cys-ketimine intermediate to generate the NFS1 persulfide species (Fig. 2). Here, we tested whether the mobile S-transfer loop cysteine also provides the proton necessary to advance the Cys-quinonoid intermediate. The NFS1 S-transfer loop cysteine was converted to alanine, and the resulting SC381ADAec complex was purified and combined with ISCU2 and FXN. Stopped-flow kinetics for the SC381ADAecUF complex revealed a decrease in the accumulation, but similar decay kinetics, for the Cys-aldimine intermediate compared with the SDAecUF complex (Fig. 3C). Interestingly, the Cys-quinonoid intermediate for the SC381ADAecUF complex does not appear to decay (Fig. 4 D and F and Table 2), consistent with a proton donation role for C381. We then tested whether this general acid role of the mobile S-transfer loop cysteine is a shared feature among the type I cysteine desulfurases (51). Native IscS and the IscSC328A variant were purified and analyzed using stopped-flow kinetics. Native IscS showed faster Cys-quinonoid formation (5-fold) and decay (2-fold) kinetics than the SDAecUF complex (Fig. 4 E and F and Table 2). The IscSC328A variant exhibited extremely slow decay of the Cys-quinonoid intermediate (Fig. 4 E and F and Table 2), analogous to the result from the SC381ADAecUF complex. These results are consistent with the mobile S-transfer loop cysteine providing the proton to advance the Cys-quinonoid intermediate. Interestingly, both the presence of FXN and the mobile S-transfer loop cysteine have a role in advancing the Cys-quinonoid intermediate, suggesting that these processes are functionally linked.
Discussion
A direct connection between the FXN gene and the neurodegenerative disease FRDA was established in a seminal 1996 study (1). Typical FRDA patients have lower than wild-type FXN expression levels (4 to 36% of controls) (52–56), suggesting that a threshold level of FXN is required for normal function. FXN depletion results in the loss of activity for Fe-S cluster enzymes, increased iron accumulation in mitochondria, and enhanced sensitivity to oxidative stress (57). Strategies to treat FRDA have focused on iron chelators, antioxidants, and mechanisms to increase FXN levels (58, 59). Unfortunately, FRDA remains incurable. An alternate approach to generating an FRDA treatment is to first determine the biological role of FXN and then replace that function.
Early studies focused on a possible role of FXN in iron metabolism. A function in iron storage was proposed based on the ability of the larger form of FXN (FXNΔ1–55) to undergo an iron-dependent oligomerization to produce a species reminiscent of ferritin (24–28, 60). However, this iron-induced oligomerization appears to be dispensable in vivo (61). The truncated form of FXN (FXNΔ1–80), which is widely considered to be the functional form in vivo, is monomeric even in the presence of iron (62), interacts with the NFS1–ISD11–ISCU2 complex (5, 7, 22, 31, 63), and rescues cells challenged with FXN depletion (64). The truncated form of FXN was proposed to function as a chaperone that delivers iron for Fe-S cluster assembly. This proposal was based on the observations that (1) FXN depletion disrupts iron homeostasis and promotes iron accumulation in the mitochondria, (2) FXN contains an iron-binding carboxylate patch, (3) FXN has the ability to reactivate the [4Fe-4S]–containing aconitase, and (4) the Fe-S assembly pathway lacks a designated iron chaperone (29, 33). However, truncated FXN has a weak binding affinity toward ferrous iron (55 μM), which is the substrate for Fe-S cluster biosynthesis, and the iron-binding carboxylate patch residues are only semiconserved (33, 34, 36, 37). Moreover, a similar iron overload phenotype is generated upon depletion of other members of the mitochondrial Fe-S assembly system (65), indicating that this phenotype is connected to the loss of Fe-S clusters and not necessarily to the ability to donate iron for this pathway. Furthermore, FRDA clinical variants for FXN have been identified in a region distinct from the carboxylate patch. These FRDA variants are not iron-binding residues and exhibit diminished ability to bind and activate the Fe-S assembly complex (66–68). Finally, the presence of a ∼150 μM labile iron pool in mitochondria could function as a feedstock for iron-containing proteins and raises the question as to whether a designated iron chaperone is even required for the Fe-S cluster biosynthetic pathway (69). Despite these concerns, many investigators still champion iron storage or donor roles for FXN.
More recently, evidence supporting a role for FXN in promoting sulfur-based chemistry associated with eukaryotic Fe-S cluster biosynthesis has emerged (5, 22, 39–41, 70). In this study, we have further interrogated and tested this role of FXN using 2 types of experiments. First, we developed a radiolabeling assay that couples an acid-quench step with HPLC-based separation and analysis of the labeled proteins. Reactions were performed with stoichiometric amounts of cysteine in the absence of iron to minimize loss of the protein-bound label. Using this assay, we found that FXN accelerates the rate of persulfide formation on NFS1 as well as the interprotein sulfur transfer reaction from NFS1 to ISCU2 (Fig. 1 and Table 1).
Second, we applied a stopped-flow kinetic approach to examine which chemical step or steps FXN accelerates in the formation of the persulfide species on NFS1. The addition of either FXN or its prokaryotic homolog CyaY to the SDAecU complex dramatically increased the decay kinetics for the Cys-quinonoid intermediate, which correlates well with the rate enhancement of the eukaryotic cysteine desulfurase complex upon the addition of either FXN or CyaY (44). Advancement of the quinonoid to ketimine intermediate requires protonation at the C4′ position. Substitution of the mobile S-transfer loop cysteine with an alanine for either the human SDAecUF complex or E. coli IscS enzyme inhibited the decay of the Cys-quinonoid intermediate (Fig. 4), consistent with previous mechanistic proposals (47, 50) and implying that this is a common feature of type I cysteine desulfurases. These data strongly support a role for the mobile S-transfer loop cysteine as a general acid that protonates the Cys-quinonoid intermediate.
Interestingly, we found that type I cysteine desulfurases with substitutions for the mobile S-transfer loop cysteine, which therefore lack the proposed proton donor for formation of the Cys-ketimine species, did not accumulate higher levels of earlier catalytic intermediates, as one might predict. Moreover, there appears to be a positive correlation between how much of the Cys-quinonoid accumulates for different complexes and the rate at which the intermediate decays (SI Appendix, Fig. S10), which is counterintuitive. Such an effect can be readily explained if the mobile S-transfer loop cysteine also has a role in a step prior to the formation of the Cys-ketimine intermediate. The SC381ADAecUF complex that lacks the mobile S-transfer loop cysteine shows markedly diminished levels of Cys-aldimine, suggesting the S-transfer loop cysteine is also involved in the formation of the Cys-aldimine intermediate. The mobile S-transfer loop cysteine has been previously proposed to participate in substrate binding (46); possible roles include stabilizing the substrate through noncovalent interactions or functioning as a general base that deprotonates the amine of the substrate to promote Cys-aldimine formation (Fig. 5). Moreover, Cys-aldimine accumulation for SDAecU was very similar to that of SC381ADAecUF, but significantly lower than that of SDAecUF, suggesting the loss of the mobile S-transfer loop cysteine or the loss of FXN has an equivalent detrimental effect on substrate binding and Cys-aldimine formation.
Fig. 5.
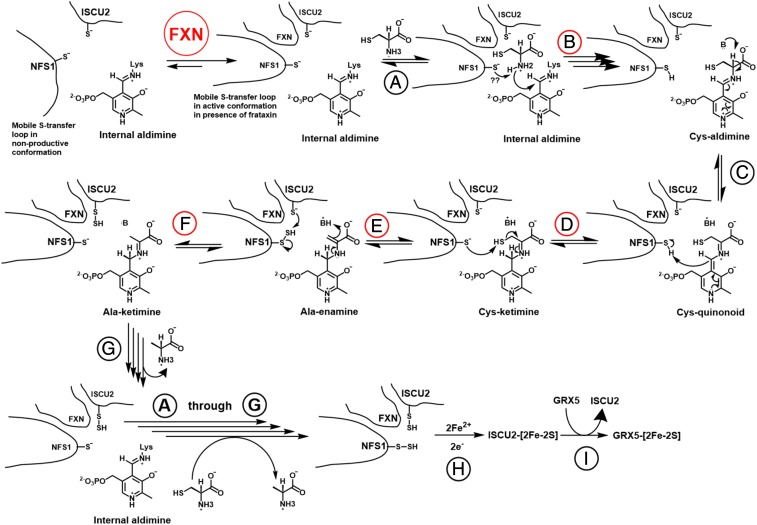
Model for FXN activation of eukaryotic Fe-S cluster biosynthesis. In the absence of FXN, the NFS1 mobile S-transfer loop exists primarily in a nonproductive open conformation. FXN binding favors the active closed conformation. In step A, cysteine substrate enters the active site channel and forms a noncovalent Michaelis complex. In step B, catalytic cysteine of the mobile S-transfer loop either provides noncovalent interactions or acts as a base and deprotonates the amine (shown in the figure) of the cysteine substrate to facilitate the formation of Cys-aldimine. In step C, the Cα proton of the substrate is abstracted to generate the Cys-quinonoid intermediate. In step D, S-transfer loop cysteine acts as an acid and provides the proton for Cys-ketimine formation. In step E, S-transfer loop cysteine now acts as a nucleophile and cleaves the C-S bond to generate a persulfide intermediate. In step F, S-transfer loop cysteine then functions as a sulfur carrier and delivers the sulfane sulfur to ISCU2. Our results show that this step is reversible in the absence of iron. In step G, the product alanine leaves, regenerating the internal aldimine (resting state). Steps A to G take place again with another turnover of cysteine to alanine and generation of persulfide on NFS1. In step H, the complex with 2 persulfides then reacts with ferrous iron and electrons to synthesize a [2Fe-2S] cluster on ISCU2. In step I, the [2Fe-2S] cluster is transferred to apo-GRX5. Steps accelerated by FXN are labeled in red. Note that the steps accelerated by FXN involved the S-transfer loop cysteine.
Overall, the mobile S-transfer loop cysteine is responsible for the critical steps in the cysteine desulfurase mechanism (Fig. 5): facilitating substrate binding (step B), functioning as a general acid to accelerate the decay of the Cys-quinonoid intermediate (step D), acting as a nucleophile to generate the persulfide species on NFS1 (step E), and delivering the sulfane sulfur to the acceptor protein ISCU2 (step F). Our results show that each of these roles assigned to the mobile S-transfer loop cysteine is enhanced or accelerated by FXN. Notably, the conversion of the Cys-aldimine to the Cys-quinonoid species (Fig. 5, step C), which is thought to be independent of the mobile S-transfer loop cysteine and involve the PLP-binding lysine functioning as a base, did not show slower kinetics when either the mobile S-transfer loop cysteine or FXN was absent. Together, these data support the conclusion that the mobile S-transfer loop and FXN are functionally linked for the eukaryotic Fe-S assembly system.
Model for the Role of FXN in Fe-S Cluster Biosynthesis.
The eukaryotic SDAecU complex has unusually low activity for a cysteine desulfurase enzyme (kcat < 10% of prokaryotic IscS) and requires FXN binding to stimulate activity and approach the catalytic turnover rate of IscS (5, 18). In 2017, the first crystal structure of a mitochondrial cysteine desulfurase (SDAec) complex surprisingly revealed an incomplete substrate-binding site, a solvent-exposed PLP cofactor, a disordered mobile S-transfer loop, and an overall “open” α2β2γ2 architecture in which ISD11 molecules mediate interactions between NFS1 subunits (18). Quaternary interactions between NFS1 subunits are therefore quite different from those in its prokaryotic homolog IscS, which have contributions from each subunit of the homodimer to the active site channel of the other subunit (71, 72). A subsequent structure revealed the SDAec complex can also adopt a “closed” α2β2γ2 architecture exhibiting an NFS1–NFS1 interface with similar subunit interactions as IscS (73). Overlaying the 2 SDAec structures reveals that the NFS1-ISD11-ACP units superimpose but use distinct protein–protein interfaces to form the open and closed quaternary structures. Electron microscopy studies revealed that the predominant form of the SDAec complex in solution is the open form (18). This open architecture provides a rationale for the essential nature of ISD11 in eukaryotes, the low basal levels of cysteine desulfurase activity for the SDAec complex, and the requirement of an allosteric effector to enhance the activity of the enzyme.
We propose that the SDAec complex exists as an equilibrium mixture of open and closed forms, with the open architecture being the dominant species in solution (18). In the open architecture, the incomplete active site structure and solvent-exposed PLP likely hinder substrate binding. Moreover, the mobile S-transfer loop is not constrained by the other NFS1 subunit in the open form and probably adopts largely nonproductive conformations (Fig. 5), which results in a low probability of the catalytically essential cysteine residue sampling the active site and thereby limits cysteine desulfurase activity. In our model, FXN addition induces an open-to-closed architectural rearrangement for the complex. This structural switch repositions the second NFS1 subunit to complete the channel to the active site and excludes nonproductive conformations of the mobile S-transfer loop. As a consequence, specific chemical steps involving the mobile S-transfer loop cysteine that include substrate binding, advancement of the Cys-quinonoid to the Cys-ketimine intermediate, NFS1 persulfide formation, and interprotein sulfur transfer to ISCU2 are accelerated. It is also possible, but less likely, that FXN binds to the open form of the complex, mimicking the second NFS1 subunit in the closed form, and directly excludes nonproductive conformations to activate the cysteine desulfurase complex. We suggest that CyaY, despite a mere 20% sequence identity to FXN, activates the human Fe-S assembly complex by inducing a similar open-to-closed structural transition. In contrast, prokaryotic IscS already exists in a high activity closed form that cannot be further activated by the binding of an allosteric effector that induces an open-to-closed architectural switch, explaining the dissimilar effects upon FXN/CyaY binding to the different cysteine desulfurases (44).
This stimulation of the sulfur transfer chemistry explains the previously reported FXN-based acceleration of [2Fe-2S] cluster formation on ISCU2 (23). Notably, the role of FXN in directly assembling the [2Fe-2S] cluster intermediate on ISCU2 (Fig. 5, step H) has not been evaluated. Once the cluster is synthesized, the final step of [2Fe-2S] cluster biosynthesis is the transfer of the intact Fe-S cluster to a carrier protein such as GRX5 (Fig. 5, step I). This step was recently shown to be unaffected by FXN but accelerated for variants of ISCU2 (42). Interestingly, the FXN-dependent stimulation of [2Fe-2S] cluster synthesis and this FXN-independent acceleration of cluster transfer were shown to be additive, and both increase the overall Fe-S cluster flux through the pathway (42). Finally, we did not evaluate if Fe2+ affects the kinetics/equilibrium of any of the steps in this study. Considering Fe2+ has been shown to further accelerate the cysteine desulfurase activity in the presence of FXN (5), a future study evaluating the effect of Fe2+ on these reactions might provide additional mechanistic insight.
Overall, our results strongly support a role for FXN as an allosteric modulator that stimulates PLP-based chemistry and sulfur delivery to ISCU2 for [2Fe-2S] cluster synthesis. We provide an architectural switch model that explains how FXN binding completes the active site for NFS1 and controls the S-transfer loop trajectory. The incorporation of FXN, along with ISD11/ACP, into the eukaryotic cysteine desulfurase is likely a mechanism to either control the sulfur flux through different biosynthetic pathways or tune the level of Fe-S cluster assembly. Consistent with this idea, a recent study discovered that FXN was dispensable in mammalian systems under anoxic conditions (74), which would have a lower demand for Fe-S clusters. Now that the FXN activation mechanism is better understood, new approaches such as the development of molecules that exclude nonproductive mobile S-transfer loop conformations or stabilize the closed form of the cysteine desulfurase architecture might have potential as FXN replacement strategies and FRDA therapeutics.
Materials and Methods
Protein Preparations.
Plasmids containing human NFS1 (Δ1–55) and ISD11 (pZM4) were generously provided by S. Leimkühler, University of Potsdam, Potsdam, Germany (75). The NFS1 and ISD11 plasmids were transformed into E. coli strain BL21(DE3) cells and the proteins were copurified with the bacterial ACP (ACPec) as the SDAec complex (18). Human ISCU2 (Δ1–35) and FXN (Δ1–55) were separately expressed without their mitochondrial targeting sequence and purified as previously described (5). The spontaneous conversion of Δ1–55 FXN to the truncated form was confirmed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (5). E. coli IscS was purified as previously described (43). The QuikChange protocol (Agilent) was used to introduce the C381A point mutation into the NFS1-pET15b plasmid and C328A point mutation into a plasmid containing native IscS (43, 75). To remove any residual persulfide, the SDAec, IscS, and ISCU2 samples were treated with 3 mM dithiothreitol (DTT) prior to a final gel filtration purification step that was performed in an anaerobic glove box. Protein concentrations were estimated using the following extinction coefficients: SDAec and IscS using 6.6 mM−1⋅cm−1 at 388 nm (in 0.1 M NaOH) (76), ISCU2 using 8,490 M−1⋅cm−1 at 280 nm, FXN using 26,030 M−1⋅cm−1 at 280 nm, and CyaY using 28,990 M−1⋅cm−1 at 280 nm (5, 77). Protein variants were purified using the same protocol used for native proteins and assumed to have the same extinction coefficient as the native proteins. Cysteine desulfurase activities (kcat) for the IscS, SDAecU, and SDAecUF complexes were 8.07 ± 0.13 min−1, 0.82 ± 0.03 min−1, and 10.1 ± 0.2 min−1, respectively. Unless otherwise stated, all protein manipulations and reactions were carried out in an anaerobic glove box (MBRAUN; maintained at ∼14 °C with O2 < 1 ppm).
Protein Separation and 35S-Labeling Reactions.
Proteins and protein complexes were injected (150-μL volume; SI Appendix, Fig. S1) into a reversed-phase C4 column (WAT011807; Waters) attached to an HPLC system (1260 Infinity; Agilent Technologies) and separated using a gradient from 30 to 70% of buffer B (60% CH3CN, 40% isopropanol, 0.1% trifluoroacetic acid [TFA]), with the remainder of the composition made up of buffer A (0.1% TFA in water [pH 2.0]). Both the absorbance (diode array detector) and, after mixing with scintillation mixture (BioCount 111182), the [35S] signal (model 5C β-RAM radio-HPLC detector; LabLogic) were recorded for the eluted proteins.
An l-[35S]cysteine stock solution was generated by diluting 20 µL of l-[35S]cysteine (10.2 μM, 1.00796 mCi; PerkinElmer) 50 times with nonradioactive l-cysteine to generate 1 mL of a 1-mM stock solution. The final concentration of l-[35S]cysteine in the stock solution was 204 nM. The retention time of NFS1 was determined by reacting a 25-μL aliquot of the SDAec complex (37.5 μM) with 25 μL of l-[35S]cysteine (final concentration of 500 μM) for 30 min at 37 °C (SI Appendix, Fig. S2). The reaction was stopped by addition of 50 μL of a quenching solution, and the proteins were separated under low pH (quench) conditions. The quenching solution was made by mixing 1 volume of concentrated HCl with 4 volumes of 6 M guanidine hydrochloride (pH < 1). Note that ISD11, ACPec, and FXN lack cysteine residues needed to form a persulfide species and are not labeled by l-[35S]cysteine.
To demonstrate that the quench solution inhibits sulfur transfer, labeled NFS1 was first generated by reacting the SDAec complex (30 μM) with l-[35S]cysteine (400 μM final concentration, 45-μL total reaction volume) for 30 min at 37 °C. The labeled NFS1 was either (1) incubated with ISCU2 (80 μM, 5 μL) for 30 min, followed by the addition of 50 μL of the quenching solution, or (2) first combined with 50 μL of the quenching solution and then incubated with ISCU2 (80 μM, 5 μL) for 30 min. Samples were analyzed by reversed-phase chromatography under quench conditions (SI Appendix, Fig. S3). Data collection and analysis were performed with Laura software (LabLogic). To quantitate the amount of persulfide associated with the peak, the area under the scintillation curve (in counts per minute) was converted into a [35S] concentration using a standard curve generated from known amounts of l-[35S]cysteine and then multiplied by the dilution factor (unlabeled/labeled l-cysteine = 4,901.96).
Assays for Monitoring Persulfide Formation and Decay.
For the persulfide formation and decay assays, 30 μM SDAec and 30 μM ISCU2 were combined to generate the SDAecU complex, which was converted to the SDAecUF complex by the addition of 75 μM FXN. The SDAecU and SDAecUF complexes were reacted with either 30, 200, or 600 μM cysteine (0.0204% [35S]cysteine), incubated at 37 °C for different lengths of time (30 s to 1 h), and diluted 4-fold into the quenching solution (Fig. 1 and SI Appendix, Fig. S4). The amount of persulfide associated with NFS1 and ISCU2 was calculated as described above, plotted against time, and fit to a reaction scheme (discussed below) using KinTek software (KinTek Corporation). To determine which cysteine residues were labeled with persulfide species, the SDAecU complex was combined with 300 µM l-cysteine, incubated at 37 °C for 30 min, and quenched with concentrated HCl. The sample was digested with pepsin (100:1 sample/pepsin based on concentration in milligrams per milliliter) at 22 °C for 30 min and subjected to liquid chromatography/tandem mass spectrometry analysis (Center for Mass Spectrometry at Texas A&M University).
Validation of Data Fitting for Persulfide Formation Kinetics.
The data were fitted to a simulated mechanism according to the reaction scheme (Fig. 1E) using KinTek software. The estimation of error was determined by FitSpace confidence contour analysis. FitSpace determines the goodness of the fit by measuring the dependence of the sum of square errors on each pair of parameters, while all other parameters were varied (SI Appendix, Fig. S5). The confidence interval of all of the fitted parameters was constrained by χ2 threshold values of 0.99 and 0.997 for the SDAecU and SDAecUF data, respectively. These thresholds indicate that varying the parameter within the confidence interval would not increase the χ2 value more than 1% (SDAecU) and 0.3% (SDAecUF) from the minimum χ2 value, which represents the best fit. Confidence contour plots are provided that indicate well-defined regions determined by χ2 threshold values (red-colored patches in SI Appendix, Fig. S5).
Reversibility of Sulfur Transfer Reaction between NFS1 and ISCU2.
The SDAecU (30 μM SDAec and 30 μM ISCU2) and SDAecUF (SDAecU plus 75 μM FXN) complexes were reacted with 30 μM l-[35S]cysteine for 40 min at 37 °C. The amount of label associated with NFS1 and ISCU2 (SI Appendix, Fig. S6) was determined as described above. Portions of the SDAecU and SDAecUF samples were then spiked with either unlabeled ISCU2 (60 μM) or SDAec (60 μM) and incubated for an additional 30 min, and the reaction was then diluted 4-fold into the quenching solution at various times. NFS1 and ISCU2 were separated by HPLC as described above, and the radioactivity was measured using a scintillation counter for the samples before and after spiking with unlabeled ISCU2 or SDAec.
Pulse–Chase Experiments.
Pulse–chase experiments were conducted by first reacting the SDAecU and SDAecUF complexes (30 μM SDAec and 30 μM ISCU2, with or without 75 μM FXN) with 30 μM [35S]cysteine for 40 min (the amount of label maximized after 10 to 20 min) to generate radiolabeled NFS1 and ISCU2. Next, a final concentration of 1 mM nonradioactive l-cysteine was added to the samples and then diluted 4-fold into the quenching solution at various times. The extent of residual persulfide labeling on NFS1 and ISCU2 was determined and fit to a linear equation (persulfide decay from ISCU2 for the SDAecUF sample) or an exponential decay equation [y = y0 + A0*exp (−k*x)] using Kaleidagraph, where the k value gives the apparent rate of persulfide loss (Table 1 and SI Appendix, Fig. S7).
Stopped-Flow Kinetics for the Cysteine Desulfurase Reactions.
Individual 100 μM samples of the different enzyme or enzyme complexes, SDAecU (100 μM SDAec + 100 μM ISCU2), SDAecUF (100 μM SDAec + 100 μM ISCU2 + 100 μM FXN), SDAecUCec (100 μM SDAec + 100 μM ISCU2 + 100 μM CyaY), SC381ADAecUF (100 μM SC381ADAec + 100 μM ISCU2 + 100 μM FXN), IscS, or IscSC328A, in assay buffer [50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 250 mM NaCl (pH 7.5)] were placed in one of the syringes of the stopped-flow apparatus (KinTek Corporation). The other syringe contained 10 mM l-cysteine. The samples were mixed by simultaneously pressing both syringes. Formation of the aldimine, quinonoid, and ketimine intermediates was followed by monitoring changes in absorbance at 410 nm, 508 nm, and 340 nm (discussed below), respectively. For the aldimine and quinonoid kinetics, traces monitored at 410 nm and 508 nm with time were fitted with Origin software (OriginLab) to a consecutive B equation [y = y0 + (k1*[A]0/(k2 − k1))*(exp(−k1*t) − exp(−k2*t))], where k1 and k2 are rate constants of the formation and decay of intermediates respectively. The ketimine kinetics could not be adequately fit using a consecutive B equation.
Determination of Wavelength of Maximum Absorbance for PLP-Associated Intermediates in the Cysteine Desulfurase Reactions.
The addition of 5 mM l-cysteine to IscS (50 μM) in a stopped-flow experiment resulted in an absorbance spectrum with a wavelength of maximum absorbance (λmax) values of 410 nm and 508 nm after 20 ms (SI Appendix, Fig. S11) that were assigned to the aldimine and quinonoid intermediates, respectively. Changes in the PLP absorbance spectra were also monitored as a function of cysteine concentration for the IscSC328A and SC381ADAecUF variants, which lack the mobile S-transfer loop cysteine and are incapable of cleaving the C-S bond to generate protein-bound persulfide species. At higher cysteine concentrations, the IscSC328A variant developed peaks at ∼350 nm and ∼420 nm (SI Appendix, Fig. S12) similar to those previously reported for the C326A variant of a cyanobacterial cysteine desulfurase (CD0387), which were assigned to the Cys-ketimine and Cys-aldimine species, respectively (47). Wavelength scans for stopped-flow experiments with the native SDAecU and SDAecUF complexes did not result in a change in the spectrum with sufficient signal to noise to allow the assignment of λmax for particular intermediates. The addition of different cysteine concentrations to the SC381ADAecUF variant resulted in the development of a stable peak at ∼340 nm at the highest cysteine concentration (SI Appendix, Fig. S12), which was assigned as the Cys-ketimine species. Overall, we monitored the kinetics of formation and decay of the aldimine, quinonoid, and ketimine intermediates for the human and E. coli cysteine desulfurases at a single wavelength by their changes in absorbance (discussed above) at 410 nm, 508 nm, and 340 nm, respectively, which are similar to wavelengths assigned in previous cysteine desulfurase mechanistic studies (46–50).
Supplementary Material
Acknowledgments
We thank Professor Tadhg Begley for the generous use of his radio-HPLC and stopped-flow apparatus. We also thank Dr. Jamison P. Huddleston and Dr. Rung-yi Lai for their assistance with the KinTek software and radio-HPLC, respectively. We thank Dr. Deepika Das, Seth Cory, and Dr. Chris Putnam for helpful discussions. This work was supported, in part, by NIH Grant R01GM096100, National Science Foundation Grant CHE 1508269, and Welch Foundation Grant A-1647.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.R.D. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909535116/-/DCSupplemental.
References
- 1.Campuzano V., et al. , Friedreich’s ataxia: Autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271, 1423–1427 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Lane D. J., Richardson D. R., Frataxin, a molecule of mystery: Trading stability for function in its iron-binding site. Biochem. J. 426, e1–e3 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Rouault T. A., Mammalian iron-sulphur proteins: Novel insights into biogenesis and function. Nat. Rev. Mol. Cell Biol. 16, 45–55 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Stehling O., Lill R., The role of mitochondria in cellular iron-sulfur protein biogenesis: Mechanisms, connected processes, and diseases. Cold Spring Harb. Perspect. Biol. 5, a011312 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai C. L., Barondeau D. P., Human frataxin is an allosteric switch that activates the Fe-S cluster biosynthetic complex. Biochemistry 49, 9132–9139 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Stehling O., Elsässer H. P., Brückel B., Mühlenhoff U., Lill R., Iron-sulfur protein maturation in human cells: Evidence for a function of frataxin. Hum. Mol. Genet. 13, 3007–3015 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Gerber J., Mühlenhoff U., Lill R., An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep. 4, 906–911 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mühlenhoff U., Richhardt N., Ristow M., Kispal G., Lill R., The yeast frataxin homolog Yfh1p plays a specific role in the maturation of cellular Fe/S proteins. Hum. Mol. Genet. 11, 2025–2036 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Lill R., Function and biogenesis of iron-sulphur proteins. Nature 460, 831–838 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Johnson D. C., Dean D. R., Smith A. D., Johnson M. K., Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74, 247–281 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Py B., Barras F., Building Fe-S proteins: Bacterial strategies. Nat. Rev. Microbiol. 8, 436–446 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Holliday G. L., et al. , Atlas of the radical SAM superfamily: Divergent evolution of function using a “plug and play” domain. Methods Enzymol. 606, 1–71 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiedemann N., et al. , Essential role of Isd11 in mitochondrial iron-sulfur cluster synthesis on Isu scaffold proteins. EMBO J. 25, 184–195 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Vranken J. G., et al. , The mitochondrial acyl carrier protein (ACP) coordinates mitochondrial fatty acid synthesis with iron sulfur cluster biogenesis. eLife 5, e17828 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adam A. C., Bornhövd C., Prokisch H., Neupert W., Hell K., The Nfs1 interacting protein Isd11 has an essential role in Fe/S cluster biogenesis in mitochondria. EMBO J. 25, 174–183 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi Y., Ghosh M. C., Tong W. H., Rouault T. A., Human ISD11 is essential for both iron-sulfur cluster assembly and maintenance of normal cellular iron homeostasis. Hum. Mol. Genet. 18, 3014–3025 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terali K., Beavil R. L., Pickersgill R. W., van der Giezen M., The effect of the adaptor protein Isd11 on the quaternary structure of the eukaryotic cysteine desulphurase Nfs1. Biochem. Biophys. Res. Commun. 440, 235–240 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Cory S. A., et al. , Structure of human Fe-S assembly subcomplex reveals unexpected cysteine desulfurase architecture and acyl-ACP-ISD11 interactions. Proc. Natl. Acad. Sci. U.S.A. 114, E5325–E5334 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith A. D., et al. , Sulfur transfer from IscS to IscU: The first step in iron-sulfur cluster biosynthesis. J. Am. Chem. Soc. 123, 11103–11104 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Webert H., et al. , Functional reconstitution of mitochondrial Fe/S cluster synthesis on Isu1 reveals the involvement of ferredoxin. Nat. Commun. 5, 5013 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Cai K., Tonelli M., Frederick R. O., Markley J. L., Human mitochondrial ferredoxin 1 (FDX1) and ferredoxin 2 (FDX2) both bind cysteine desulfurase and donate electrons for iron-sulfur cluster biosynthesis. Biochemistry 56, 487–499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colin F., et al. , Mammalian frataxin controls sulfur production and iron entry during de novo Fe4S4 cluster assembly. J. Am. Chem. Soc. 135, 733–740 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Fox N. G., Das D., Chakrabarti M., Lindahl P. A., Barondeau D. P., Frataxin accelerates [2Fe-2S] cluster formation on the human Fe-S assembly complex. Biochemistry 54, 3880–3889 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adamec J., et al. , Iron-dependent self-assembly of recombinant yeast frataxin: Implications for Friedreich ataxia. Am. J. Hum. Genet. 67, 549–562 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schagerlöf U., et al. , Structural basis of the iron storage function of frataxin from single-particle reconstruction of the iron-loaded oligomer. Biochemistry 47, 4948–4954 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gakh O., et al. , Physical evidence that yeast frataxin is an iron storage protein. Biochemistry 41, 6798–6804 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Park S., Gakh O., Mooney S. M., Isaya G., The ferroxidase activity of yeast frataxin. J. Biol. Chem. 277, 38589–38595 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nichol H., et al. , Structure of frataxin iron cores: An X-ray absorption spectroscopic study. Biochemistry 42, 5971–5976 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Bulteau A. L., et al. , Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science 305, 242–245 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y., et al. , Mrs3p, Mrs4p, and frataxin provide iron for Fe-S cluster synthesis in mitochondria. J. Biol. Chem. 281, 22493–22502 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Zaidi A., et al. , Interaction of frataxin, an iron binding protein, with IscU of Fe-S clusters biogenesis pathway and its upregulation in AmpB resistant Leishmania donovani. Biochimie 115, 120–135 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Li H., Gakh O., Smith D. Y. 4th, Isaya G., Oligomeric yeast frataxin drives assembly of core machinery for mitochondrial iron-sulfur cluster synthesis. J. Biol. Chem. 284, 21971–21980 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cook J. D., et al. , Monomeric yeast frataxin is an iron-binding protein. Biochemistry 45, 7767–7777 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoon T., Cowan J. A., Iron-sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. J. Am. Chem. Soc. 125, 6078–6084 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Yoon T., Dizin E., Cowan J. A., N-terminal iron-mediated self-cleavage of human frataxin: Regulation of iron binding and complex formation with target proteins. J. Biol. Inorg. Chem. 12, 535–542 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Huang J., Dizin E., Cowan J. A., Mapping iron binding sites on human frataxin: Implications for cluster assembly on the ISU Fe-S cluster scaffold protein. J. Biol. Inorg. Chem. 13, 825–836 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Kondapalli K. C., Kok N. M., Dancis A., Stemmler T. L., Drosophila frataxin: An iron chaperone during cellular Fe-S cluster bioassembly. Biochemistry 47, 6917–6927 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rötig A., et al. , Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat. Genet. 17, 215–217 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Pandey A., et al. , Frataxin directly stimulates mitochondrial cysteine desulfurase by exposing substrate-binding sites, and a mutant Fe-S cluster scaffold protein with frataxin-bypassing ability acts similarly. J. Biol. Chem. 288, 36773–36786 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bridwell-Rabb J., Fox N. G., Tsai C. L., Winn A. M., Barondeau D. P., Human frataxin activates Fe-S cluster biosynthesis by facilitating sulfur transfer chemistry. Biochemistry 53, 4904–4913 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parent A., et al. , Mammalian frataxin directly enhances sulfur transfer of NFS1 persulfide to both ISCU and free thiols. Nat. Commun. 6, 5686 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Das D., Patra S., Bridwell-Rabb J., Barondeau D. P., Mechanism of frataxin “bypass” in human iron-sulfur cluster biosynthesis with implications for Friedreich’s ataxia. J. Biol. Chem. 294, 9276–9284 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adinolfi S., et al. , Bacterial frataxin CyaY is the gatekeeper of iron-sulfur cluster formation catalyzed by IscS. Nat. Struct. Mol. Biol. 16, 390–396 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Bridwell-Rabb J., Iannuzzi C., Pastore A., Barondeau D. P., Effector role reversal during evolution: The case of frataxin in Fe-S cluster biosynthesis. Biochemistry 51, 2506–2514 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bailey T. S., Zakharov L. N., Pluth M. D., Understanding hydrogen sulfide storage: Probing conditions for sulfide release from hydrodisulfides. J. Am. Chem. Soc. 136, 10573–10576 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tirupati B., Vey J. L., Drennan C. L., Bollinger J. M. Jr, Kinetic and structural characterization of Slr0077/SufS, the essential cysteine desulfurase from Synechocystis sp. PCC 6803. Biochemistry 43, 12210–12219 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Behshad E., Bollinger J. M. Jr, Kinetic analysis of cysteine desulfurase CD0387 from Synechocystis sp. PCC 6803: Formation of the persulfide intermediate. Biochemistry 48, 12014–12023 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Behshad E., Parkin S. E., Bollinger J. M. Jr, Mechanism of cysteine desulfurase Slr0387 from Synechocystis sp. PCC 6803: Kinetic analysis of cleavage of the persulfide intermediate by chemical reductants. Biochemistry 43, 12220–12226 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Zheng L., White R. H., Cash V. L., Jack R. F., Dean D. R., Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 90, 2754–2758 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng L., White R. H., Cash V. L., Dean D. R., Mechanism for the desulfurization of L-cysteine catalyzed by the nifS gene product. Biochemistry 33, 4714–4720 (1994). [DOI] [PubMed] [Google Scholar]
- 51.Mihara H., Kurihara T., Yoshimura T., Soda K., Esaki N., Cysteine sulfinate desulfinase, a NIFS-like protein of Escherichia coli with selenocysteine lyase and cysteine desulfurase activities. Gene cloning, purification, and characterization of a novel pyridoxal enzyme. J. Biol. Chem. 272, 22417–22424 (1997). [DOI] [PubMed] [Google Scholar]
- 52.Campuzano V., et al. , Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum. Mol. Genet. 6, 1771–1780 (1997). [DOI] [PubMed] [Google Scholar]
- 53.Willis J. H., Isaya G., Gakh O., Capaldi R. A., Marusich M. F., Lateral-flow immunoassay for the frataxin protein in Friedreich’s ataxia patients and carriers. Mol. Genet. Metab. 94, 491–497 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deutsch E. C., et al. , A rapid, noninvasive immunoassay for frataxin: Utility in assessment of Friedreich ataxia. Mol. Genet. Metab. 101, 238–245 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steinkellner H., Scheiber-Mojdehkar B., Goldenberg H., Sturm B., A high throughput electrochemiluminescence assay for the quantification of frataxin protein levels. Anal. Chim. Acta 659, 129–132 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Saccà F., et al. , A combined nucleic acid and protein analysis in Friedreich ataxia: Implications for diagnosis, pathogenesis and clinical trial design. PLoS One 6, e17627 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmucker S., Puccio H., Understanding the molecular mechanisms of Friedreich’s ataxia to develop therapeutic approaches. Hum. Mol. Genet. 19, R103–R110 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Erwin G. S., et al. , Synthetic transcription elongation factors license transcription across repressive chromatin. Science 358, 1617–1622 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marmolino D., Friedreich’s ataxia: Past, present and future. Brain Res. Brain Res. Rev. 67, 311–330 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Park S., et al. , Yeast frataxin sequentially chaperones and stores iron by coupling protein assembly with iron oxidation. J. Biol. Chem. 278, 31340–31351 (2003). [DOI] [PubMed] [Google Scholar]
- 61.Aloria K., Schilke B., Andrew A., Craig E. A., Iron-induced oligomerization of yeast frataxin homologue Yfh1 is dispensable in vivo. EMBO Rep. 5, 1096–1101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gakh O., et al. , Normal and Friedreich ataxia cells express different isoforms of frataxin with complementary roles in iron-sulfur cluster assembly. J. Biol. Chem. 285, 38486–38501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmucker S., et al. , Mammalian frataxin: An essential function for cellular viability through an interaction with a preformed ISCU/NFS1/ISD11 iron-sulfur assembly complex. PLoS One 6, e16199 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmucker S., Argentini M., Carelle-Calmels N., Martelli A., Puccio H., The in vivo mitochondrial two-step maturation of human frataxin. Hum. Mol. Genet. 17, 3521–3531 (2008). [DOI] [PubMed] [Google Scholar]
- 65.Lill R., et al. , The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim. Biophys. Acta 1823, 1491–1508 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Tsai C. L., Bridwell-Rabb J., Barondeau D. P., Friedreich’s ataxia variants I154F and W155R diminish frataxin-based activation of the iron-sulfur cluster assembly complex. Biochemistry 50, 6478–6487 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bridwell-Rabb J., Winn A. M., Barondeau D. P., Structure-function analysis of Friedreich’s ataxia mutants reveals determinants of frataxin binding and activation of the Fe-S assembly complex. Biochemistry 50, 7265–7274 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H., Gakh O., Smith D. Y. 4th, Ranatunga W. K., Isaya G., Missense mutations linked to friedreich ataxia have different but synergistic effects on mitochondrial frataxin isoforms. J. Biol. Chem. 288, 4116–4127 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lindahl P. A., Moore M. J., Labile low-molecular-mass metal complexes in mitochondria: Trials and tribulations of a burgeoning field. Biochemistry 55, 4140–4153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turowski V. R., Busi M. V., Gomez-Casati D. F., Structural and functional studies of the mitochondrial cysteine desulfurase from Arabidopsis thaliana. Mol. Plant 5, 1001–1010 (2012). [DOI] [PubMed] [Google Scholar]
- 71.Kaiser J. T., et al. , Crystal structure of a NifS-like protein from Thermotoga maritima: Implications for iron sulphur cluster assembly. J. Mol. Biol. 297, 451–464 (2000). [DOI] [PubMed] [Google Scholar]
- 72.Cupp-Vickery J. R., Urbina H., Vickery L. E., Crystal structure of IscS, a cysteine desulfurase from Escherichia coli. J. Mol. Biol. 330, 1049–1059 (2003). [DOI] [PubMed] [Google Scholar]
- 73.Boniecki M. T., Freibert S. A., Mühlenhoff U., Lill R., Cygler M., Structure and functional dynamics of the mitochondrial Fe/S cluster synthesis complex. Nat. Commun. 8, 1287 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ast T., et al. , Hypoxia rescues frataxin loss by restoring iron sulfur cluster biogenesis. Cell 177, 1507–1521.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marelja Z., Stöcklein W., Nimtz M., Leimkühler S., A novel role for human Nfs1 in the cytoplasm: Nfs1 acts as a sulfur donor for MOCS3, a protein involved in molybdenum cofactor biosynthesis. J. Biol. Chem. 283, 25178–25185 (2008). [DOI] [PubMed] [Google Scholar]
- 76.Peterson E. A., Sober H. A., Preparation of crystalline phosphorylated derivatives of vitamin B6. J. Am. Chem. Soc. 76, 169–175 (1954). [Google Scholar]
- 77.Gill S. C., von Hippel P. H., Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182, 319–326 (1989). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.