The variety of structures available to an individual RNA molecule through Watson/Crick and nonclassical interactions causes them to be conformationally dynamic; that is, any single RNA may exist in and transition among multiple configurations. Recently, single-molecule methodologies have made it possible to determine the number of RNA structures present in a sample and their relative distributions, folding pathways, and conformational dynamics. In PNAS, Halma et al. (1) used single-molecule force spectroscopy (SMFS), or “molecular tweezers,” to dissect the dynamic folding and unfolding pathways of a sequence in the West Nile virus (WNV) messenger RNA (mRNA) that directs ribosomes to shift reading frame with extremely high frequency. In addition to structures identified by prior studies (2, 3), they find metastable intermediates and variants that fold along 2 mutually exclusive pathways. The finding that transitions between different conformations are maximized in the range of forces applied by elongating ribosomes makes a significant contribution to our understanding of how RNA structural dynamics can influence biological function.
Structural elements in mRNAs present both problems and opportunities to ribosomes. They can act as “roadblocks,” causing elongating ribosomes to arrest. If the goal is to translate the entire mRNA, such barriers are not desirable, and ribosomes that are stopped for too long are rescued at the expense of the mRNA (4). However, if mRNA structural elements are resolvable, they can have regulatory functions by virtue of altering the kinetic parameters of translation elongation, for example, by adding time for recruitment of trans-acting factors or enabling noncanonical elongation events to proceed. One such event, called programmed −1 ribosomal frameshifting (−1 PRF), capitalizes on the ability of mRNA structures to pause ribosomes over a heptameric “slippery sequence,” whereupon a fraction of them can slip one nucleotide in the 5′ (or −1) direction (5). The elements that direct these slippage events range from simple stem loops to complex pseudoknots (stem loops in which the 3′ sequence can base pair with sequences in the loop sections). Many RNA viruses use −1 PRF as a genome condensation mechanism, enabling them to encode multiple proteins from a single mRNA, and −1 PRF rates typically range from ∼5 to ∼30%. Altering −1 PRF rates changes the stoichiometric ratios of the protein products, interfering with virus particle assembly and virus propagation, thus identifying −1 PRF as a potential target for antivirals (5). Although more-stable structures are assumed to promote greater rates of −1 PRF by providing stalled ribosomes with more time to slip, recent studies suggest that −1 PRF efficiency correlates better with conformational plasticity rather than mechanical resistance (e.g., see refs. 6 and 7).
The WNV element promotes ∼80% −1 PRF efficiency. A bioinformatics analysis first identified a 61-nucleotide (nt) pseudoknot in this sequence (3). A subsequent biophysical study expanded this to a 109-nt-long sequence capable of folding into mutually exclusive tandem stem-loop and mRNA pseudoknot structures (2). The current work (1) used a 111-nt WNV-derived RNA sequence that was flanked on each side by long “handle” regions. The handles were annealed to complementary single-stranded DNAs attached to beads suspended in optical traps. Force ramps were generated by separating the beads, enabling measurements of RNA unwinding (molecular extension) as a function of force to produce force-extension curves (FECs). Interpretation of the FECs is intuitive. Imagine stretching a spring: At first, very little force is required to lengthen it, but, as the process continues, increasingly more force is needed to continue stretching; that is, the resulting FEC follows an exponential-like function. Next, imagine that glue was used to connect 2 noncontiguous regions of the spring together. As the ends are pulled, at some point, enough force will be applied to break the adhesive. At that instant, the FEC shows a rapid drop in force accompanied by a sudden extension in length. In this study, this indicates that a double-stranded region of the RNA has been ripped apart. The reverse setup enabled measurements of RNA refolding. The same molecule can be repeatedly unfolded and refolded, revealing different FEC patterns indicative of multiple conformational states.
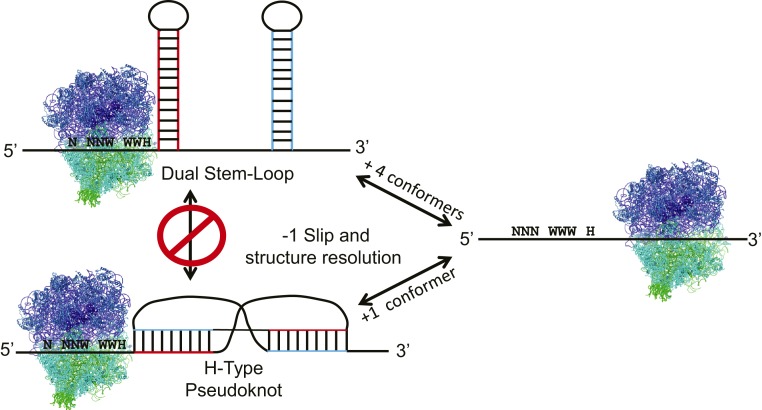
The contour length of each RNA molecule was determined before and after each transition by fitting the data into a worm-like chain polymer-elasticity model. Two distinct pseudoknot types were identified based on their broadly distributed and high-energy unfolding/refolding patterns, while the stem loops tended to unfold/refold in a lower energy range that was more tightly distributed. Matching the contour lengths with the previously proposed structures and computational predictions resulted in identification of 7 distinct metastable structures. These included 2 versions of the dual hairpin structure and another 2 variations of the extended pseudoknot first proposed in ref. 2, and 2 varieties of the shorter pseudoknot discussed in ref. 3. In a clever set of experiments, these structures were validated by measuring FECs in the presence of saturating concentrations of antisense oligonucleotides designed to inhibit the formation of structure-specific base pairs. Two distinct folding and unfolding pathways were then mapped by cataloguing all of the pairwise transitions. Approximately 80% of these folded along a pathway starting with the completely unfolded RNA to either the simple stem loop or to the smaller pseudoknot, and then either to the small pseudoknot with an additional hairpin or to the dual hairpin. In contrast, the remaining ∼20% folded directly from the linear RNA to the 2 versions of the larger pseudoknot (Fig. 1). None of the larger pseudoknots could be generated from the conformers generated from the majority pathway; that is, the 2 folding routes were independent of one another. The unfolding pathways were similarly independent, with the exception that a small fraction of the large pseudoknots could deform into the stem loop. Analyses of changes in occupancy state as a function of force during unfolding and refolding confirmed the 80:20 ratio, with the majority of the conformers occupying 1 of the 2 dual hairpins at steady state.
Fig. 1.
The WNV −1 PRF signal can assume 7 different conformations, 5 of which fold/unfold along the dual stem-loop pathway, while 2 follow the pseudoknot course. The structures along the 2 pathways are mutually incompatible, and the pathways only interconvert through the unfolded state. The tension required to induce ribosome stalling, which allows ribosomes to slide from the incoming reading frame (N NNW WWH) to the −1 frame (NNN WWW H), is in the same range as the force at which the RNA conformational changes occur. The high degree of conformational heterogeneity and complex dynamics are proposed to enable extremely high rates of −1 PRF.
These findings enhance our understanding of the biophysics of −1 PRF by correlating the extent of mRNA structural dynamism with −1 PRF efficiency. That this element promotes −1 PRF at an efficiency of ∼80%, coupled with the 80:20 occupancy status of the 2 mutually exclusive folding/unfolding pathways may or may not be coincidental. It is tempting to speculate that the structural elements that promote −1 PRF lie in the majority 80% occupancy pathway, despite the fact that this is dominated by stem-loop−based structures (<20% of the structures in this pathway are pseudoknots). This calls into question the assumption that mRNA pseudoknots are the canonical stimulators of −1 PRF. Indeed pseudoknots pose a topological problem. As described in the torsional restraint model (8), unwinding from the base of stem structure imparts rotational movement upon its more-distal region. A simple stem loop is not restrained, and the loop can rotate freely. In contrast, in a pseudoknot, the “loop” is part of the downstream stem (stem 2), and thus it is rotationally constrained. In essence, the proximal stem 1 cannot be fully unwound until stem 2 is first denatured. This poses a paradox because the intrinsic helicase activity of the ribosome cannot access stem 2 until stem 1 has been fully unwound. While trans-acting helicases may be available to solve this problem in live cells, in vitro −1 PRF assays using purified core components of the translational apparatus have been used for many years, demonstrating that the ribosome does not require help to elongate through a pseudoknot. The solution to this paradox may lie in the observation made by Halma et al. (1) that the energy required for transitions between FECs is in the same range as the force at which ribosomes stall (9). In vitro experiments have shown that EF-G (the bacterial homolog of eEF2) undergoes multiple rounds of guanosine triphosphate (GTP) hydrolysis as it attempts to translocate the ribosome through a −1 PRF signal (7, 10). This suggests that the hydrolysis of GTP by eEF2 initiates a power stroke which transfers energy into the mRNA structure (11). This may enable the −1 PRF-promoting mRNA structure to transition through multiple conformations as the ribosome is both slipping and attempting to resolve the structural roadblock. By this model, greater conformational plasticity may make it more likely that a topologically resolvable structure will be sampled. Additionally, the dynamic contributions of the entire translational apparatus in the −1 PRF process should considered. For example, the energy released by eEF2 can also be transduced up the peptidyl transfer RNA into the nascent peptide. We previously demonstrated that deletion of components of the ribosome-associated chaperone complex specifically inhibited −1 PRF, suggesting that cotranslational folding of nascent peptides may also influence −1 PRF efficiency (12).
One potential question is whether the use of molecular tweezers truly mimics an elongating ribosome. Pulling apart 2 beads to which the ends of the RNA are attached means that the unwinding force is vectored outward from both ends of the RNA molecule. In contrast, during translation, the force is internally vectored in the 5′ → 3′ direction by a ribosome trapped at the base of the structured element inside the mRNA. Thus, the critique here is that the SMFS approach is “frameshifting without the ribosome.” Despite this concern, this approach represents a significant contribution to the field. At best, the molecular genetics approaches that have been used for the past 30 y are indirect measures of the bulk fraction of ribosomes that are able to slip and then escape the highly structured stimulatory elements. At their worst, they leave a big black box around the events occurring during this process, and they can be influenced by unforeseen artifacts and unknown factors [e.g., the newly discovered Shiftless protein (13)]. We are beginning to see the work of a new generation of biophysicists who are building instruments capable of measuring the behavior of individual ribosomes at the near-quantum level. The coming decade promises to more fully illuminate the black box of −1 PRF.
Acknowledgments
My research is supported by the National Institute of General Medical Sciences of the National Institutes of Health (R01 GM117177).
Footnotes
The author declares no conflict of interest.
See companion article on page 19500.
References
- 1.Halma M. T. J., Ritchie D. B., Cappellano T. R., Neupane K., Woodside M. T., Complex dynamics under tension in a high-efficiency frameshift stimulatory structure. Proc. Natl. Acad. Sci. U.S.A. 116, 19500–19505 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moomau C., Musalgaonkar S., Khan Y. A., Jones J. E., Dinman J. D., Structural and functional characterization of programmed ribosomal frameshift signals in West Nile virus strains reveals high structural plasticity among cis-acting RNA elements. J. Biol. Chem. 291, 15788–15795 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Firth A. E., Atkins J. F., A conserved predicted pseudoknot in the NS2A-encoding sequence of West Nile and Japanese encephalitis flaviviruses suggests NS1′ may derive from ribosomal frameshifting. Virol. J. 6, 14 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuller A. P., Green R., Roadblocks and resolutions in eukaryotic translation. Nat. Rev. Mol. Cell Biol. 19, 526–541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dinman J. D., Mechanisms and implications of programmed translational frameshifting. Wiley Interdiscip. Rev. RNA 3, 661–673 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ritchie D. B., Foster D. A. N., Woodside M. T., Programmed −1 frameshifting efficiency correlates with RNA pseudoknot conformational plasticity, not resistance to mechanical unfolding. Proc. Natl. Acad. Sci. U.S.A. 109, 16167–16172 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu B., et al. , Translocation kinetics and structural dynamics of ribosomes are modulated by the conformational plasticity of downstream pseudoknots. Nucleic Acids Res. 46, 9736–9748 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plant E. P., Dinman J. D., Torsional restraint: A new twist on frameshifting pseudoknots. Nucleic Acids Res. 33, 1825–1833 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaiser C. M., Tinoco I. Jr, Probing the mechanisms of translation with force. Chem. Rev. 114, 3266–3280 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan S., Wen J.-D., Bustamante C., Tinoco I. Jr, Ribosome excursions during mRNA translocation mediate broad branching of frameshift pathways. Cell 160, 870–881 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C., et al. , Elongation factor G initiates translocation through a power stroke. Proc. Natl. Acad. Sci. U.S.A. 113, 7515–7520 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muldoon-Jacobs K. L., Dinman J. D., Specific effects of ribosome-tethered molecular chaperones on programmed −1 ribosomal frameshifting. Eukaryot. Cell 5, 762–770. (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X., et al. , Regulation of HIV-1 Gag-Pol expression by Shiftless, an inhibitor of programmed −1 ribosomal frameshifting. Cell 176, 625–635.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]