Significance
The ability of Mycobacterium tuberculosis to survive during prolonged treatment has been attributed to either transient stress responses or fixed heritable drug-resistance–producing mutations. We show that phase-variation in the M. tuberculosis glpK gene represents a third type of resistance mechanism. The ability of these glpK mutants to grow slowly and then rapidly revert suggests that these transiently heritable changes may also explain how a hidden population of drug-tolerant bacteria develops during tuberculosis treatment. As a genetically trackable cause of drug tolerance, M. tuberculosis glpK mutants provides a unique opportunity to study these phenomena at a cellular and mechanistic level. These mutants could also be used for developing drugs that target tolerant populations, leading to more rapid and effective tuberculosis treatments.
Keywords: glpK, Mycobacterium tuberculosis, reversible drug tolerance, phase variation, small colony variant
Abstract
The length and complexity of tuberculosis (TB) therapy, as well as the propensity of Mycobacterium tuberculosis to develop drug resistance, are major barriers to global TB control efforts. M. tuberculosis is known to have the ability to enter into a drug-tolerant state, which may explain many of these impediments to TB treatment. We have identified a mechanism of genetically encoded but rapidly reversible drug tolerance in M. tuberculosis caused by transient frameshift mutations in a homopolymeric tract (HT) of 7 cytosines (7C) in the glpK gene. Inactivating frameshift mutations associated with the 7C HT in glpK produce small colonies that exhibit heritable multidrug increases in minimal inhibitory concentrations and decreases in drug-dependent killing; however, reversion back to a fully drug-susceptible large-colony phenotype occurs rapidly through the introduction of additional insertions or deletions in the same glpK HT region. These reversible frameshift mutations in the 7C HT of M. tuberculosis glpK occur in clinical isolates, accumulate in M. tuberculosis-infected mice with further accumulation during drug treatment, and exhibit a reversible transcriptional profile including induction of dosR and sigH and repression of kstR regulons, similar to that observed in other in vitro models of M. tuberculosis tolerance. These results suggest that GlpK phase variation may contribute to drug tolerance, treatment failure, and relapse in human TB. Drugs effective against phase-variant M. tuberculosis may hasten TB treatment and improve cure rates.
Despite decades of control efforts, tuberculosis (TB) remains the leading cause of death from an infectious disease (1). The length and complexity of TB therapy is a major barrier to TB control. Drug-susceptible TB must be treated with multiple drugs, usually for 6 mo, and multidrug-resistant TB must be treated for at least 9 mo (2, 3). Relapses remain fairly common despite these regimens (4–7). Many of these clinical phenomena can likely be attributed to the ability of Mycobacterium tuberculosis to enter into a tolerant state when exposed to drugs, hypoxia, nutritional deprivation, or host defense mechanisms during human infections that renders them temporarily drug-resistant (8–10). This transient “phenotypic” drug resistance is not thought to be caused by genetic changes associated with new heritable drug resistance but has instead been attributed to reversible transcriptional and metabolic changes. A number of in vitro studies have confirmed that M. tuberculosis can become transiently drug-resistant when cultured under growth-limiting conditions, including drug treatment (11–13). Evidence for the development of reversible phenotypic drug resistance during human TB treatment includes the observation that most patients who relapse after treatment for drug-susceptible TB remain infected with the same fully drug-susceptible strain (14–16). M. tuberculosis that has been cultured from apparently latent, closed, and encapsulated lesions of patients undergoing TB treatment has also been found to be drug susceptible (17).
Phase variation is an adaptive mechanism that mediates reversible switching “on” and “off” of a gene by genotypic changes, such as DNA methylation, homologous recombination, DNA rearrangement, or insertions/deletions in short sequence repeats or homopolymeric tracts (HTs) located within the promoter region or the coding sequence of a gene (18). Reversible frameshift mutations in HTs are a result of slipped-strand mispairing errors during replication and are well documented in many bacterial species (19), with a high rate of HT variants observed in species with DNA mismatch repair (MMR) deficiency (20). M. tuberculosis also lacks a recognizable MMR system (21, 22). This suggests that the poly-G:C and poly-A:T tracts identified in the M. tuberculosis genome (21) may be susceptible to reversible insertion and deletion mutations during replication.
Here, we describe a mechanism of drug tolerance that is caused by genetically encoded but rapidly reversible mutations in the 7 cytosines (7C) HT of the glpK gene in M. tuberculosis. These mutations produce small colony and morphological variants that have reduced susceptibility to drugs, but unlike classic drug tolerance or persistence, are expressed as a transiently heritable trait. We propose that GlpK phase variability may account for much of the clinical and microbiological observations associated with persistence and relapse TB in humans.
Results
Small Colonies with Altered Morphology Are Detectable in Clinical M. tuberculosis Strains.
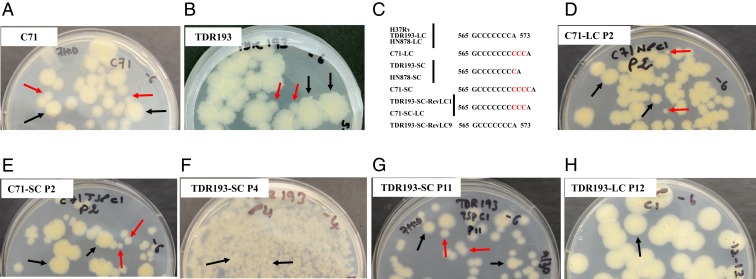
We examined cultures of 26 drug-susceptible (23) and 8 drug-resistant (1 isoniazid- [INH] and ethambutol- [EMB] resistant, 1 rifampicin- [RIF] and INH-resistant, and 6 RIF-INH-EMB–resistant) (24) clinical M. tuberculosis strains. Of these, we noted that 9 susceptible and 4 RIF-INH-EMB–resistant isolates contained a subpopulation of small colonies (SCs) with a smooth morphology mixed in with the large colonies (LCs) that predominated in the culture. We selected LCs and SCs of 1 pan-susceptible clinical isolate (C71) (Fig. 1A) and 1 multidrug-resistant clinical isolate (TDR193) (Fig. 1B). Whole-genome sequencing (WGS) of the LC (C71-LC) and SC (C71-SC) isolates showed that they differed only at the M. tuberculosis glpK gene (Rv3696c, a glycerol kinase) after exclusion of the highly repetitive PE/PPE/PE-PGRS genes from the analysis. Compared to the M. tuberculosis H37Rv reference sequence (GenBank ID AL123456.3) (21) (Fig. 1C and Table 1), C71-LC had a 3C insertion in a 7C HT (M. tuberculosis, nucleotide 566 to 572) of glpK. In contrast, the C71-SC sequence contained both 3C and 4C insertions in the same 7C HT, indicating a substrain mixture within the colony. The 3C insertion resulted in a GlpK192Gly insertion that preserved the glpK ORF; however, the 4C insertion increased the size of the 7C HT to 11C and produced a glpK frameshift. We noted similar mutational differences between the LCs and SCs of the multidrug-resistant strain TDR193. WGS of the LC (TDR193-LC) and the SC (TDR193-SC) sequences of strain TDR193 only identified mutations in glpK and rpoC (Table 1). TDR193-LC had WT glpK and a nonsynonymous mutation in rpoC. Mutations in rpoC have been previously described as compensatory for the rpoB mutations associated with RIF-resistance (25) and this mutation was not investigated further. TDR193-SC had a single C insertion in the glpK 7C HT causing a frameshift (Fig. 1C and Table 1). We then sequenced this glpK hotspot in 94 additional M. tuberculosis clinical isolates randomly selected from the TDR-TB strain bank to represent a broad range of susceptibility profiles as well as geographic diversity (24) (SI Appendix, Table S1). The 7C HT was WT in 68, had a 1C insertion in 10, and showed a mixture of WT and 1C insertions in 16.
Fig. 1.
Clinical M. tuberculosis isolates with mixtures of small smooth and large rough colonies. Initial cultures of (A) pan susceptible M. tuberculosis strain C71 and (B) multidrug-resistant M. tuberculosis strain TDR193 were plated on 7H10 agar containing glycerol. (C) The glpK slippage site sequences of LCs and SCs are shown. (D) C71-LC showed emergence of SCs at passage 2. (E) C71-SC showed emergence of LCs at passage 2. (F) Emergence at passage 4 and accumulation of LCs at (G) passage 11 of a TDR193-SC culture. (H) TDR193-LC phenotype was stable after 12 passages. Examples of SCs are indicated by red arrows and LCs by black arrows.
Table 1.
Genotypic and phenotypic characteristics of large and SCs of clinical M. tuberculosis strains
| Strain | embB | inhA | rpoB | rpoC | gyrA | gyrB | glpK | Colony size |
| TDR193-SC | Met306Val; Met423Thr | Ile194Thr | Ser531Leu | Ala542Ala | Glu21Gln; Ser95Thr; Gly668Asp | Val340Leu | Ins573C (7C → 8C) (frameshift) | Small |
| TDR193-LC | Met306Val; Met423Thr | Ile194Thr | Ser531Leu | Ala542Ala; Val775Ala | Glu21Gln; Ser95Thr; Gly668Asp | Val340Leu | WT (7C) | Large |
| TDR193-SC-RevLC1 | Met306Val; Met423Thr | Ile194Thr | Ser531Leu | Ala542Ala | Glu21Gln; Ser95Thr; Gly668Asp | Val340Leu | Ins573CCC (7C → 10C) (Gly192 insertion) | Large |
| TDR193-SC-RevLC9 | Met306Val; Met423Thr | Ile194Thr | Ser531Leu | Ala542Ala | Glu21Gln; Ser95Thr; Gly668Asp | Val340Leu | WT (7C) | Large |
| C71-SC | Ala630Val | WT | WT | WT | Glu21Gln | WT | Ins573CCCC (66%)/Ins573CCC (34%) (7C → 11C/10C) (Mix of frameshift and Gly192 insertion) | Small and Large |
| C71-LC | Ala630Val | WT | WT | WT | Glu21Gln | WT | Ins573CCC (7C → 10C) (Gly192 insertion) | Large |
| C71-SC- RevLC | Ala630Val | WT | WT | WT | Glu21Gln | WT | Ins573CCC (7C → 10C) (Gly192 insertion) | Large |
High-Frequency Reversions among Clinical glpK Mutants.
To determine whether clinical glpK SC frameshift mutants revert to normal colony size, we cultured TDR193-SC to stationary phase and plated the culture on glycerol-containing agar to identify LCs. TDR193-SC cultures reverted to LC morphology at high frequency both in the presence or absence of glycerol in liquid medium (2.9 × 10−2 ± 3.0 × 10−2 in 7H9 without glycerol vs. 1.7 × 10−2 ± 1.1 × 10−2 in 7H9 with glycerol). We also performed serial passage of C71-SC, C71-LC, TDR193-SC, and TDR193-LC single colonies in the absence of glycerol. Both C71-LC and C71-SC showed a mixture of SCs and LCs in just 2 passages (Fig. 1 D and E), perhaps because of the instability of the large 10C and 11C glpK HT regions in these strains. In contrast, for TDR193, the SC (TDR193-SC) reverted to LC in 4 passages (Fig. 1 F and G) but the LC (TDR193-LC) was stable for 12 passages (Fig. 1H). WGS of 1 C71-SC colony that reverted to a LC phenotype (C71-SC-RevLC) revealed a deletion of 1C in the glpK HT, reducing the 11C HT to 10C. WGS of 2 TDR193-SC colonies that reverted to an LC phenotype revealed either a deletion of 1C in the glpK HT (TDR193-SC-RevLC9), reducing the 8C HT back to 7C as exists in the H37Rv reference strain, or the insertion of an additional 2C in the glpK HT (TDR193-SC-RevLC1) increasing the HT to 10C (Fig. 1C and Table 1). Thus, each SC to LC mutant reestablished the correct glpK gene ORF. No other new mutations were detected in the genomes of the revertants.
Due to the high frequency of C71-SC and C71-LC reversions, compared to TDR193 variants, we did not perform any further experiments with C71 variants. Given the role of GlpK in glycerol metabolism, we cultured TDR193-SC, TDR193-LC, and TDR193-SC-RevLCs in media with or without glycerol supplementation. All strains had similar growth rates in liquid media (SI Appendix, Fig. S1A) and equal colony sizes on solid media (SI Appendix, Fig. S1B) in the absence of glycerol. However, in the presence of glycerol, strains with a functional glpK (TDR193-LC, TDR193-SC-RevLC1, and TDR193-SC-RevLC9) had improved growth in liquid media (SI Appendix, Fig. S1A) and produced larger colonies on solid media than TDR193-SC (SI Appendix, Fig. S1B). These results demonstrate that clinical M. tuberculosis strains frequently generate glpK 7C HT frameshift-variants that have a slower growth capacity under certain culture conditions and have the ability to rapidly revert to higher growth patterns by reestablishing a correct ORF.
M. tuberculosis Acquires Frameshift Mutations in the 7C HT of glpK during Murine Infections.
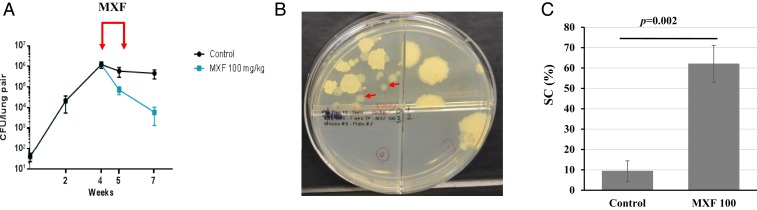
We studied glpK mutant generation in vivo by infecting C56BL/6 mice with a low dose of M. tuberculosis strain HN878 via aerosol. One group of mice were not treated with any antituberculars, and a second group received 100 mg/kg of Moxifloxacin (MXF) to simulate a human dose equivalent for 1 wk (Fig. 2A). SC cells appeared to spontaneously arise or to be selected for during infection and treatment (Fig. 2B). SCs were undetectable in preinfection cultures of HN878. However, by week 7 postinfection, SCs comprised 9% (±5%) of the colony forming units (CFU) recovered from the lungs of untreated control mice and 62% (±9%) of the CFU recovered from the lungs of MXF-treated mice (P = 0.002) (Fig. 2C). WGS of 1 SC (named HN878-SC) randomly picked from lung cultures of untreated mice and 3 LCs (HN878-XM4-LC1, HN878-XM4-LC2, and HN878-XM4-LC3) and 3 SCs (HN878-XM4-SC4, HN878-XM4-SC5, and HN878-XM4-SC6) randomly picked from lung cultures of MXF-treated mice was performed. Compared to the parental HN878 strain, no mutations were detected in all 3 LC genome sequences. In contrast, all 4 SCs had a 1C insertion in the 7C HT of glpK (Fig. 1C), except for 1 SC (HN878-XM4-SC4) that also had a ΔC44 frameshift mutation in Rv0452 and a synonymous Gly157Gly mutation in purM. To confirm the presence of glpK frameshift mutations in other SCs, we sequenced the glpK 7C HT in another 27 randomly picked SCs (12 from untreated and 15 from the MXF-treated group) and 20 LCs (11 from untreated and 9 from the MXF-treated group). All 20 LCs had a WT glpK sequence while all 27 SCs had the same 1C insertion in the glpK 7C HT, leading to a frameshift. These results strongly suggest that glpK frameshift mutants arise during infection and have an improved ability to persist in vivo, particularly during antibiotic treatment.
Fig. 2.
Frameshift glpK mutants develop in M. tuberculosis-infected mice and are less susceptible to antituberculosis treatment. (A) BALB/c mice were infected by aerosol with M. tuberculosis HN878 strain and treated with MXF (100 mg/kg) over 1 wk, as indicated by red arrows. Shown are the averages and SDs of CFU per lung of 4 to 5 mice killed at the indicated time points. (B) Small smooth colonies isolated from an infected mouse lung after 1 wk of MXF treatment. Examples of SCs are indicated by red arrows. (C) Frequency, in percentage (%), of SC in the control and MXF-treated groups of 4 mice each. Significant difference in frequency was determined by 2-tailed Student’s t test.
SC glpK Frameshift Mutants Are Less Susceptible to Subminimal Inhibitory Concentrations of Antituberculosis Drugs.
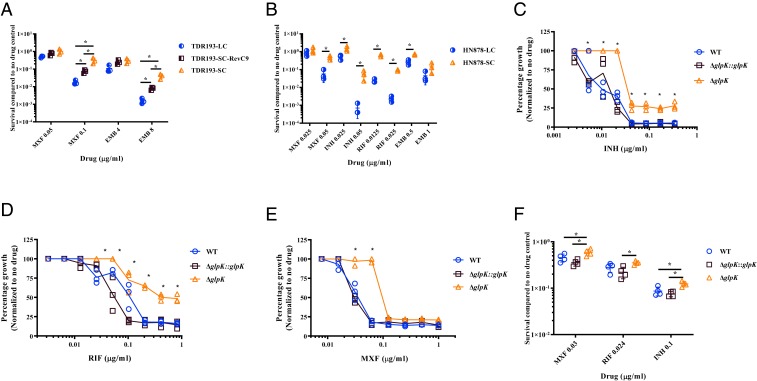
Noting the accumulation of SCs during mouse infection and treatment, we tested SCs and LCs from various M. tuberculosis genetic backgrounds: a clinical TDR193 isolate, an HN878 isolate identified after in vivo selection in mice, and SCs generated by performing clean glpK gene deletions in M. tuberculosis H37Rv, for alterations in drug susceptibility patterns. WGS analysis of the clinical multidrug-resistant TDR193-SC, TDR193-LC, and TDR193-SC-RevLC9 strains identified no other mutations except as previously noted (Table 1). Despite these similarities, the INH minimal inhibitory concentration (MIC) of TDR193-SC was 2-fold higher (5 µg/mL) than the MIC of TDR193-LC and TDR193-SC-RevLC9 (2.5 µg/mL) (Table 2). On the other hand, all variants had the same MICs for MXF, RIF, and EMB. However, the SC TDR193-SC had a 5- and 22-fold relative survival advantage measured by colony counts on sub-MIC concentrations of either MXF (0.1 µg/mL) or EMB (8 µg/mL), respectively, compared to LCs TDR193-SC-RevLC9 and TDR193-LC (Fig. 3A). The SC to LC revertant strain, TDR193-SC-RevLC9, had an intermediate survival advantage between the TDR193-LC and TDR193-SC. These differences at sub-MIC concentrations were only observed in the presence of glycerol. Next, we performed MIC experiments using a fully drug-susceptible HN878-SC strain isolated from our mouse study described above. An LC HN878 colony (HN878-LC) was used as a control after we had confirmed that this strain contained a WT glpK gene. Both strains were isolated from the same lung of an infected mouse that had not been treated with MXF. These HN878 strains showed similar growth in media containing various concentrations of oleic acid (free fatty acid) or glyceryl trioleate (triglyceride derived from glycerol and 3 units of oleic acid, TAG), except in the presence of free glycerol where HN878-LC showed a more rapid growth rate (SI Appendix, Fig. S2 A and B). HN878-SC and HN878-LC also had the same MICs for INH, RIF, EMB, and MXF (Table 2). However, HN878-SC exhibited a 3- to 100-fold growth advantage, as measured by CFU counts, on sub-MIC of each of these drugs in the presence of glycerol, compared to HN878-LC (Fig. 3B). Finally, we created clean glpK knockout mutants in M. tuberculosis strain H37Rv (H37RvΔglpK). The parental H37Rv and H37RvΔglpK complemented with M. tuberculosis glpK inserted at the M. tuberculosis attP site and expressed under its native promoter served as controls. As with the HN878 SC strain, H37RvΔglpK grew more slowly in liquid media supplemented with glycerol (SI Appendix, Fig. S2C), and produced smaller colonies on solid media supplemented with glycerol compared to H37Rv and the glpK complemented H37RvΔglpK controls (SI Appendix, Fig. S2D). H37RvΔglpK had identical INH, RIF, and MXF MICs to both controls (Table 2). However, deletion of glpK in H37Rv enhanced strain survival in the presence of sub-MIC of all 3 antibiotics, similar to the studies performed with the in vivo selected HN878 SCs. This increased survival to antituberculars was apparent both in the relative optical density of each strain cultured in various concentrations of INH, RIF, and MXF compared to the same strain cultured in the absence of drug (Fig. 3 C–E), and the relative survival of each strain plated on solid media containing 0.5-fold the MIC of INH, RIF, and MXF compared to the same strains plated on solid media without drugs (Fig. 3F). The reversible SC phenotype that we observed in 3 different M. tuberculosis strains was not limited to the M. tuberculosis sensu stricto but was also observable in Mycobacterium bovis bacillus Calmette–Guérin (bacillus Calmette–Guérin) (SI Appendix, Text S1 and Fig. S3).
Table 2.
MIC of M. tuberculosis LC and SC variants
| Colony variant | MIC (µg/mL) | Resistance | |||
| MXF | INH | RIF | EMB | ||
| TDR193-LC | 0.2 | 2.5 | >100 | 16 | MDR |
| TDR193-SC | 0.2 | 5 | >100 | 16 | MDR |
| TDR193-SC-RevLC9 | 0.2 | 2.5 | >100 | 16 | MDR |
| HN878-LC | 0.1 | 0.1 | 0.05 | 2 | Susceptible |
| HN878-SC | 0.1 | 0.1 | 0.05 | 2 | Susceptible |
| H37Rv | 0.063 | 0.042 | 0.048 | 1.28 | Susceptible |
| H37RvΔglpK | 0.063 | 0.042 | 0.048 | 1.28 | Susceptible |
| H37RvΔglpK::glpK | 0.063 | 0.042 | 0.048 | 1.28 | Susceptible |
MDR, multiple-drug resistance.
Fig. 3.
Decreased susceptibility of glpK mutants to various antituberculosis drugs. Differential susceptibility of (A) TDR193 and (B) HN878 LC and SC variants to sub-MIC concentrations of MXF, EMB, INH, and RIF were determined by percent survival of CFU on 7H10 medium containing the antibiotic at the indicated concentrations versus no antibiotic control CFU. Differential susceptibilities of H37Rv WT, H37RvΔglpK, and H37RvΔglpK::glpK to sub-MIC concentrations of (C) INH, (D) RIF, and (E) MXF compared to no antibiotic control were determined by OD600 measurements. (F) Susceptibility of H37Rv WT, H37RvΔglpK, and H37RvΔglpK::glpK to MXF, INH, and RIF concentrations were determined by percent survival of CFU at the indicated concentrations compared to no antibiotic control. The values of 3 independent experiments are shown for each graph. Significant differences of survival frequencies were calculated using 2-tailed Student’s t test or 2-way ANOVA analysis, *P < 0.05.
SC glpK Frameshift Mutants Are Tolerant to Supra-MIC Concentrations of Antituberculosis Drugs and Hydrogen Peroxide.
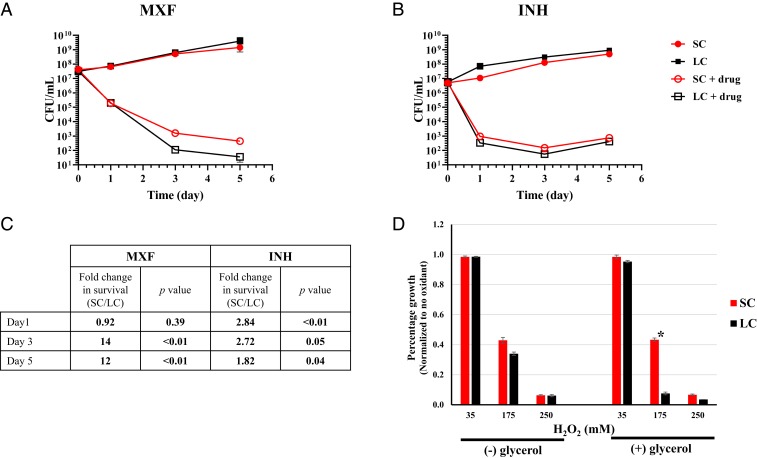
We tested the extent to which SC glpK mutants exhibit a typical tolerant-like phenotype (26, 27). Performing time-kill studies, we found that the HN878-SC (glpK mutant) had delayed killing compared to HN878-LC cultures incubated with either 0.4 µg/mL of MXF (4× the MIC) or 1 µg/mL of INH (20× the MIC) (Fig. 4 A–C). When incubated in MXF, the SC showed a 14-fold increase in survival on day 3 (P < 0.01) and a 12-fold increase on day 5 (P < 0.01). When incubated with INH, the SC showed a 2.84-fold increase in survival on day 1 (P < 0.01) and a 2.72-fold increase on day 3 (P = 0.05), compared to HN878-LC (glpK WT), in the presence of glycerol. We also tested the susceptibility of HN878-SC to the oxidative stress-inducing agent hydrogen peroxide. The HN878-SC was significantly more resistant to hydrogen peroxide (175 mM) in the presence of glycerol, compared to HN878-LC (Fig. 4D).
Fig. 4.
Differential tolerance of HN878 SC and LC variants to supra-MIC concentrations of antibiotics and to hydrogen peroxide. M. tuberculosis HN878 SC and LC strains grown in 7H9 supplemented with OADC and Tween 80 to midlog phase (OD600 = 0.6 to 0.7) were diluted to ∼5 × 106 or 5 × 107 CFU/mL and treated in the presence of glycerol with (A) 0.4 µg/mL MXF (4× MIC) or (B) 1 µg/mL INH (20× MIC). Viability was determined by CFU counts. (C) Fold-change with P value of survival differences between SC and LC identified in A and B. (D) Differential susceptibility of HN878 SC and LC strains to H2O2 compared to control was determined by OD600 measurements. Both strains were grown in 7H9 medium containing OADC and Tween 80 with (+) or without (−) glycerol to midlog phase (OD600 = 0.6 to 0.7). The cultures were diluted 1/20 and treated with different amounts of H2O2. The treated and control cultures were incubated for 6 d at 37 °C. OD600 were recorded and normalized to the corresponding control without oxidant treatment. The values of 3 independent experiments are shown for each graph. Significant differences of survival frequencies were calculated using 1-tailed Student’s t test, *P < 0.05.
Transcriptional Profiling of M. tuberculosis glpK 7C Frameshift Mutants.
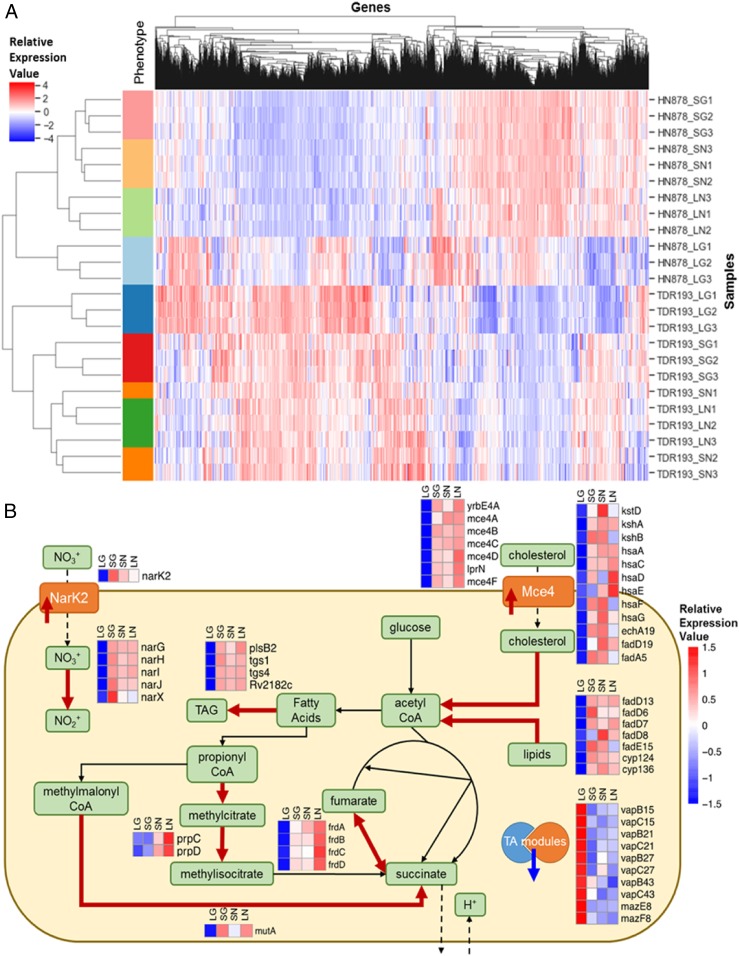
To identify altered transcriptomic pathways highly associated to glpK mutation, we performed RNA-sequencing (RNA-seq) analysis of SC and LC variants with different genetic M. tuberculosis backgrounds, namely the SC strain TDR193-SC and the LC strain TDR193-SC-RevLC9 (lineage 4, Euro/American family), and the HN878-SC and HN878-LC strains (lineage 2, Beijing family), both in the absence and presence of glycerol. In both M. tuberculosis strain backgrounds, the SCs showed gene-expression patterns similar to that of the LCs in the absence of glycerol (Fig. 5A). In contrast, the LCs cultured in the presence of glycerol differed by coordinated expression changes associated with responses typically associated with adaptation to diverse stressors. For example, several stress response regulons, including SigH (28) and DosR (29), were up-regulated in the SCs with or without glycerol, relative to the LC strains in glycerol, while KstR (30) was among regulons down-regulated in the SCs. Conversely, transcriptions of the genes involved in triacylglyceride synthesis were elevated in SC strains compared to their LC controls (Fig. 5B and SI Appendix, RNAseq SI). Collectively, these transcriptional changes are consistent with the possibility that glpK gene expression promotes M. tuberculosis growth under glycerol-replete conditions, whereas loss of functional glpK results in M. tuberculosis general stress responses associated with an adaptive tolerant state in the presence of glycerol.
Fig. 5.
Gene-expression differences appeared between the LCs and SCs in presence and absence of glycerol. (A) Heatmap of ranked gene-expression values for the different conditions profiled by RNA sequencing. Individual samples are arrayed in the rows, and genes are arrayed along the columns. The different conditions profiled are: SG = SC in glycerol (red bars); SN = SC without glycerol (orange bars); LG = LC in glycerol (blue bars); LN = LC without glycerol (green bars). The HN878 strain background is represented in the light color bars, and the TDR193 strain background is represented in the dark color bars. Values reported are ranked RPKM values, scaled such that the mean across samples is 0 and the SD is 1. The hierarchical clustering by sample shows clear separation by condition within each strain background. The largest separation between conditions is the LG vs. the other conditions. (B) Schematic summarizing main bacterial cellular processes impacted by glpK phase variation, as implied by gene expression changes in both TDR193 and HN878 backgrounds. The heatmaps visualize the scaled means of ranked RPKM values for the corresponding genes in each condition. Thick red arrows indicate gene associated proteins or processes that would be up-regulated in SCs and in absence of glycerol, and the thick blue arrow indicates down-regulated proteins.
Lung Glycerol in M. tuberculosis-Infected Mice.
To assess whether M. tuberculosis may encounter glycerol in human-like TB lesions, we tested the lungs of M. tuberculosis-infected C3HeB/FeJ mice for the presence of glycerol using LC-MS. We used this infection model because C3HeB/FeJ mice develop much more human-like lung granulomas exhibiting caseous necrosis following aerosol infection with M. tuberculosis (31) than is observed in C57BL/6 mice (32). Glycerol was detectable in normally appearing lung tissue (61.6 µg/mL), lung tissue with cellular infiltrates (44.13 µg/mL), as well as in lung caseum (10.4 µg/mL), with ∼10 to 30% of the glycerol-extracted from triglycerides. These results suggest in vivo M. tuberculosis infections may be exposed to glycerol that may both stimulate increased growth in WT glpK variants, and also select for glpK mutants where slower growth or drug tolerance may improve survival.
Discussion
Drug-resistant M. tuberculosis has long been thought of as a stable phenotype that occurs through acquisition of heritable mutations. In contrast, drug tolerance and drug persistence have been viewed as reversible phenomena associated with transient phenotypic states. Here, we show in clinical strains of M. tuberculosis a type of drug tolerance that is caused by a rapidly reversible glpK frameshift mutation in a 7C HT sequence, which produces a phenotype that is similar to the traditional definition of drug tolerance (26, 27). These tolerance-conferring frameshifts also slow bacterial growth and produce SCs. The glpK mutant SCs showed small but significant increases in survival following exposure to various antituberculosis agents, compared to glpK wild-type LCs. Although these increases are small, such differences were consistent across different genetic M. tuberculosis backgrounds.
The association between genotypic drug tolerance and slow growth, combined with a rapid reversal rate has potentially far-reaching consequences, in that this process can mimic transient phenotypic drug tolerance. For example, in the case of TB relapse the development of reversible drug tolerance during TB treatment could permit a slow-growing bacterial subpopulation to survive drug treatment while causing minimal clinical symptoms. With the lifting of drug pressure at the end of treatment, the slow-growing population could rapidly revert to a WT population with the capacity to overgrow any drug-tolerant SC cells, making the drug-tolerant subpopulation difficult to detect by conventional microbiological methods unless specifically assessed as in this study. Relapse in such a patient would thus be attributed to regrowth of a (drug-susceptible) “persister” population rather than genotypic drug-tolerant mycobacteria. Our findings are supported by a previous study that identified glpK frameshift mutations in a screen for mutations that increase tolerance to streptomycin and RIF in M. tuberculosis (13). Also demonstrating the potential clinical impact of even small changes in MIC, a recent study showed that increases as little as 0.01 µg/mL INH or RIF MIC in fully drug-susceptible M. tuberculosis isolates substantially raised the odds ratio for clinical relapse after TB treatment (33).
Our discovery that drug tolerance is linked to slow growth and glycerol metabolism suggests an important role for nutritionally controlled differences in growth rates in TB pathogenesis. Earlier studies have shown that M. tuberculosis growth is strongly enhanced by adding glycerol to the culture medium compared to carbon sources, such as glucose or free fatty acids. This has led to the use of glycerol in all standard mycobacterial media (34). Upon uptake, glycerol is phosphorylated by GlpK to glycerol-3-phosphate, which is either utilized for the synthesis of glycerophospholipids (35) or alternatively is converted by glycerol-3-phosphate dehydrogenase to dihydroxyacetone phosphate, which then enters glycolysis and gluconeogenesis (36). Furthermore, GlpK belongs to the ROK (repressor, open-reading frame, kinase) protein family (37); therefore, it is also possible that GlpK [as shown for other sugar kinases (38)], might also moonlight as a transcription regulator. When M. tuberculosis is starved for glycerol by either frameshifting glpK or by depleting glycerol from the medium, we suggest that a general stress response is activated, which phenotypically renders M. tuberculosis broadly stress-resistant and antibiotic-tolerant. The RNA expression pattern that we noted to be part of this nutrient-limited stress response appears to be similar to that reported for WT M. tuberculosis cultured under conditions that have been associated with a “tolerant” “persistent” state (29). Culture of the LC but not the SC in rich glycerol media caused LCs to increase their growth rate. This increased growth and the accompanying down-regulation of stress regulons typified by DosR and SigH may have made the LC cultures more broadly susceptible to drugs and hydrogen peroxide. Therefore, M. tuberculosis appears to induce a general stress response by toggling on and off the expression of the glpK gene via reversible frameshifts within a 7C HT sequence.
We noted that all of the SC glpK mutants were caused by frameshift mutations occurring at a 7C HT located within the coding region of glpK. HTs in MMR-deficient bacteria are known to present slippage sites for DNA polymerases (39, 40), which produce frameshift mutations at high frequency (41, 42). M. tuberculosis also lacks a recognizable MMR system (21, 22). Furthermore, the M. tuberculosis H37Rv reference sequence (GenBank ID AL123456.3) (21), includes 126 homopolymeric sequences (poly-C:G and poly-A:T ≥ 7-mer) located within ORFs and 18 located within intergenic regions. A search of 5,604 M. tuberculosis genomes in the National Center for Biotechnology Information database found at least 1 frameshift mutation in 74% of these homopolymeric sequences (SI Appendix, Table S2). However, this mutational mechanism, to our knowledge, has not been previously reported as a cause of reversible drug tolerance. Previous studies of numerous pathogenic bacteria, such as Neisseria species, Mycoplasma species, and Mycobacterium abscessus have described the occurrence of frameshift mutations via slipped-strand mispairing in regions even without homopolymeric sequences (18, 43, 44). These findings suggest that reversible frameshift mutations as a means of genetically regulating stress responses, including antibiotic tolerance, may be widespread in multiple organisms including M. tuberculosis.
We showed that glpK frameshifted SCs develop during murine infections and that selection increases further with drug treatment. Our study was performed in mice receiving a single drug (MXF), because drug-tolerant M. tuberculosis in human TB is likely to develop in diseased lesions where poor drug penetration often results in effective monotherapy (45). We found that glpK 7C HT variants emerged at a high rate under this circumstance. Pethe et al. (46) showed that pyrimidine-imidazoles (PIs), whose mechanism of action is linked to glycerol metabolism do not inhibit M. tuberculosis infection in a murine model. However, the relative proportion of LC and SC generated in PI-treated mice was not noted or recorded (46). We suspect that it is possible that PI treatment provided a strong selection for the emergence of glpK mutants in these mice. Variants with glpK frameshift mutations are detectable in sputum samples from many human TB patients and, furthermore, these glpK mutants were unstable (47, 48). Human plasma and mouse lung tissue contain significant amounts of free-glycerol (49–52) and our results show that free-glycerol is also present in diseased mouse lung and caseum. The levels of glycerol we detected in vivo are substantially lower than those used in M. tuberculosis culture medium. While the level of glycerol needed to promote a growth difference between WT and glpK mutants in vivo is unknown, we have shown that the in vivo environment provides strong selection for glpK mutations. Overall, our results strongly suggest that SC emergence and drug tolerance caused by GlpK phase variation is relevant in human TB. We also suggest that a larger repertoire of SCs caused by reversible frameshift mutations elsewhere in the M. tuberculosis genome may provide this pathogen with multiple biological mechanisms to adapt to drugs and changing environments.
Methods
Bacterial Strains and Culture Conditions.
Clinical M. tuberculosis strains were randomly selected from a collection of isolates obtained from the Tuberculosis Trials Consortium of the Centers for Disease Control and Prevention-conducted Study 22 (23), and from TDR-TB strain bank established by the World Health Organization Special Program for Research and Training in Tropical Disease using geographic, phylogenetic, and drug-resistance diversity as selection criteria (24). Unless otherwise stated, the M. tuberculosis strains were cultivated at 37 °C either in Middlebrook 7H9 broth (Difco) containing 0.05% Tween 80 or on Middlebrook 7H10 agar supplemented with 0.5% glycerol, both enriched with 10% oleic acid-albumin-dextrose-catalase (Difco). Broth cultures were incubated under gentle shaking. M. bovis bacillus Calmette–Guérin (ATCC 35734) WT and mutant strains were maintained in complete Middlebrook 7H9 medium (BD Difco) supplemented with 0.05% (vol/vol) Tween 80, 0.5% (vol/vol) glycerol, 0.5% albumin, 0.2% glucose, 0.085% sodium chloride, and 0.0003% catalase at 37 °C with agitation at 80 rpm. Pyrazinoic acid (POA) was purchased from Sigma-Aldrich and was freshly dissolved in 90% DMSO at a concentration of 0.5 M and sterilized using 0.2-μm PTFE membrane filters (Acrodisc PALL). The POA-resistant M. bovis bacillus Calmette–Guérin strains used in this study were isolated previously as described in Gopal et al. (53). For all plasmid construction, Escherichia coli strains Top10 (Invitrogen) were grown in Luria-Bertani broth or agar (both from Sigma Aldrich) at 37 °C, supplemented with 50 µg/mL kanamycin (Sigma Aldrich) or 150 µg/mL Hygromycin B (Invitrogen), where appropriate.
Deletion of glpK Gene from M. tuberculosis H37Rv Strain.
The glpK gene was deleted using allelic exchange techniques, as described previously (54). Briefly, 1,500 base pairs upstream and downstream of glpK were cloned into p2NIL suicide vector containing lacZ-sacB selection cassette. All cloning was done in E. coli XL10 Gold (Aglient) and the final mutant construct was confirmed by Sanger sequencing. The H37Rv strain was transformed with the final mutant construct followed by a 2-step selection process, as described previously (54). The glpK deletion was screened by PCR and confirmed by WGS, as described below.
To complement the glpK deletion with the gene transcribed from its own promoter, a fragment containing the glpK gene and 200-bp upstream and 50-bp downstream sequences was cloned into pMV306 integrative plasmid. The resulting construct was electroporated into the glpK mutant and hydromycin-resistant transformants were selected.
MIC Determination.
M. tuberculosis strains were grown in 7H9 medium to midlog phase optical density (OD580 = 0.5 to 0.7). The cultures were then diluted to ∼2 × 104 CFU per starting inoculum and spotted on 7H10 agar plates containing the following drug concentrations: EMB (2, 4, 8, 16, or 32 µg/mL), INH (0.025, 0.05, 0.1, 0.2, 0.625, 1.25, 2.5, 5 µg/mL), MXF (0.05, 0.1, 0.2, 0.4, 0.5, 1, 2, or 4 µg/mL), and RIF (0.0125, 0.025, 0.05, 0.1, 1, 10, or 100 µg/mL). Antibiotic-free 7H10 agar plates spotted with the primary inoculum or a 1:100 dilution were used as controls. The plates were incubated at 37 °C for 2 to 3 wk. The first antibiotic concentration that inhibited growth compared to growth of the 1:100 dilution defined the MIC. All MICs were determined in triplicate. Because each value within a triplicate MIC test was almost always identical to the other values within the same triplicate set, a single MIC value is shown without SD for each test. Susceptibility of H37Rv strains to RIF, INH, and MXF in 7H9 containing 10% OADC and 0.2% glycerol was determined by the microdilution method, as described previously (55, 56). Briefly, the first column of wells of a 96-well plate (Costar) was filled with 200 µL of 7H9 containing the drug at maximum concentration to be tested. The remaining wells were filled with 100 µL 7H9 medium. This was used to perform 2-fold serial dilution of the drugs, RIF 0.82 → 0.003 µg/mL, INH 0.34 → 0.0027 µg/mL, and MXF 1 → 0.008 µg/mL. Each well was inoculated with ∼5 × 105 CFU of M. tuberculosis. Plates were sealed with a Breathe-Easy membrane (Sigma) and incubated at 37 °C and shaking for 10 d. The percentage of growth inhibition in each well was determined by measuring OD600 in a Cytation 3 Imaging plate Reader (BioTek Instruments) and comparing it to a well with no drug. The susceptibility tests were done in triplicate and statistical significance was analyzed using a 2-way ANOVA. Susceptibility of M. bovis bacillus Calmette–Guérin strains to POA in 7H9 broth at near-neutral pH 6.5 (with 0.5% [vol/vol] or without glycerol) was determined as described previously (53). POA MIC50 values reported here are those concentrations of drug that inhibit 50% of growth as compared to a drug-free control after 5 d of incubation. The WT strain was included as a control in all experiments. All MICs were performed in 3 technical and biological replicates.
Reversion of the SC Phenotype to WT Colony Phenotype.
Three independent colonies from SC phenotype cultures were randomly picked and grown in 7H9 liquid medium at 37 °C to late phase (OD580 = 1.6), which was considered passage 0. Each culture was plated on 7H10 agar medium and the frequency of LCs was determined. The cultures were then diluted to 8.69 × 107 ± 2.20 × 107 CFU per starting inoculum, and flasks of 12 mL 7H9 liquid medium were inoculated. The culture flasks were incubated at 37 °C for 1 wk. Serial dilutions of the cultures were plated on 7H10 agar medium to determine the CFU count and to examine the emergence of LC phenotypes. The culture passages were repeated up to 12 times.
Growth and Colony Morphology of POA-Resistant M. Bovis Bacillus Calmette–Guérin.
In order to determine if the mutants had any specific growth-related phenotypes, midlog cultures of M. bovis bacillus Calmette–Guérin were pelleted at 3,200 rpm for 10 min and resuspended in 7H9 broth (with 0.5% [vol/vol] or without glycerol) by adjusting to a fixed starting inoculum (OD600 = 0.03) in T25 culture flasks (SPL Life Sciences) and growth was monitored by measuring OD600 at each time point (Ultrospec 10 cell density meter, Amersham Biosciences). Pictures of M. bovis bacillus Calmette–Guérin colonies were taken under the fixed magnification of a stereomicroscope (Nikon SMZ-Z45T) by a camera (Nikon DS-Fi2) mounted to its eyepiece.
DNA Isolation, PCR, and Bidirectional DNA Sequencing.
Genomic DNA was extracted using a CTAB protocol with slight modification, as described previously (57, 58). To amplify DNA fragments for DNA sequencing, PCR was performed using a mix containing 1 ng of genomic DNA, 5 pmol of each primer, 200 µM dNTPs, 1× PCR buffer, and 1 U of high-fidelity pfx Taq polymerase (Invitrogen) or Phusion DNA polymerase (Thermo Scientific, as per the manufacturer’s protocol) per 50-µL reaction. All PCR products were examined on an ethidium bromide-stained agarose gel and purified using a gel-extraction kit (Qiagen). Direct Sanger sequencing of PCR products was performed with a BigDye Terminator kit and analyzed with an ABI3100 Genetic Analyzer (Applied Biosystems).
WGS and Mutation Detection.
Total genomic DNA was extracted as described above and purified by using MagAttract HMW DNA kit (Qiagen). The DNA of TDR193-SC, TDR193-LC, TDR193-SC-RevLC1, TDR193-SC-RevLC9, C71-SC, C71-LC, C71-SC-RevLC, HN878-XM4-LC1, HN878-XM4-LC2, HN878-XM4-LC3, HN878-XM4-SC4, HN878-XM4-SC5, HN878-XM4-SC6, and HN878-SC strains were submitted to The Genomics Center, Rutgers University, Newark, NJ (http://research.njms.rutgers.edu/genomics/). DNA libraries were constructed using the Nextera XT DNA Library Preparation Kit (Illumina) and samples were sequenced on Illumina MiSeq (Illumina) to produce more than 100× coverage. The quality-trimmed paired-end reads were mapped to M. tuberculosis CCDC5079 (GenBank ID CP002884.1) or to M. tuberculosis H37Rv strain (GenBank ID AL123456.3), and the variants (single nucleotide changes and small deletion up to 50 nucleotides) were detected with the Fixed Ploidy Variant Detection tool in CLC Genomics Workbench v9 (CLC, Bio-Qiagen). Variants falling in PE/PPE family genes were excluded from the analysis.
RNA Extraction, Library Preparation, and Sequencing.
M. tuberculosis strains were grown to midlog phase (OD600 = 0.6 –0.7) at 37 °C in 7H9 medium with or without 1% glycerol. Total RNA was extracted by using TRIzol reagents (Invitrogen), as described previously (59). The extracted RNAs were submitted for transcriptome analysis to The Genomics Center, Rutgers University, Newark, NJ (http://research.njms.rutgers.edu/genomics/). The quality of RNA was checked for integrity on an Agilent 2100 Bioanalyzer using RNA pico6000 kit; samples with RNA integrity number >7.0 were used for subsequent processing. Illumina Ribo-Zero rRNA Removal Kit (Bacteria) was used for the removal of ribosomal RNA according to the manufacturer’s protocol. The Illumina compatible RNA-seq library was prepared using New England Biolabs next ultra RNA-seq library preparation kit. The cDNA libraries were purified using AmpureXP beads and quantified on an Agilent 2100 Bioanalyzer and on Qubit 2.0 Fluorometer (Life Technologies). An equimolar amount of barcoded libraries were pooled and sequenced on Illumina NextSeq platform (Illumina) with 1 × 75 configuration. CLC Genomics Workbench v10.0.1 (CLC, Bio-Qiagen) was used for RNA-seq analysis. Demultiplexed FASTQ files from RNA-Seq libraries were imported into the CLC software. Bases with low quality were trimmed using the following setting: Quality trim limit = 0.05, ambiguity trim maximum value = 2. The trimmed reads were mapped to the reference genome, M. tuberculosis H37Rv (GenBank ID NC_000962.3). The aligned reads were obtained using the following parameters: maximum number of allowed mismatches was 2, minimum length and similarity fraction was set at 0.8, and minimum number of hits per read was 10. For each strain 3 independent RNA extractions and RNA-seq analysis were performed; and the statistical analysis of differentially expressed genes compared to the control strain is carried out based on a negative binomial general linear mixed model influenced by the multifactorial EdgeR method (60), a tool in CLC Genomic Workbench.
We ranked reads per kilobase and million mapped reads (RPKM) expression values for each sample, such that the lowest RPKM value was designated 1, the second lowest value was designated 2, and so forth. We used these ranked values as input into the gene set enrichment analysis (GSEA) Pipeline (http://software.broadinstitute.org/gsea/index.jsp) (61). Using the GSEA analysis software, we tested for enrichment in gene sets based on regulons [information from TFOE (62)]. The genes were ranked ordered by the GSEA software according to their ranked RPKM values using the Student’s t statistic metric assessing difference between conditions compared. Gene sets compromising activated and repressed targets genes were analyzed separately, sets consisting of greater than 10 genes were included in the analysis, and nominal P values calculated from the analyses of the activated and repressed genes were combined using Fisher’s method (63). A “regulon activity change score” was calculated by weighting the gene-set size-normalized enrichment score (NES) values of the activated and repressed gene sets with the relative proportions of activated vs. repressed target genes in the regulon. Regulons were considered enriched if GSEA reported enrichment scores (ES) in opposite directions for the activated vs. repressed target gene sets, and if the overall regulon activity change score had an absolute value greater than 1. Among enriched gene sets, those with higher ES, NES, and leading-edge statistics were prioritized [see GSEA (61) for more information about the individual statistics].
Mouse Infection, MXF Treatment, and Isolation of glpK Mutants.
Pathogen-free female BALB/c mice aged 8 wk were purchased from Charles River Laboratories. Mice were group-housed in a biosafety level III animal facility and maintained with sterile bedding, water, and mouse chow. All animal experiments were conducted in compliance with and approved by the Investigational Animal Care and Use Committee of the New Jersey Medical School, Rutgers University. Nine-week-old female BALB/c mice (weight range 18 to 20 g) were infected with an inoculum of 3 × 106 CFU/mL M. tuberculosis HN878 using a Glas-Col whole-body aerosol unit. This resulted in a lung implantation of 1.56 log CFU per mouse 3 h postinfection. Groups of 4 to 5 mice were killed via cervical dislocation 2 and 4 wk postinfection. At 4 wk postinfection, 2 groups of 5 mice were treated daily with 100 mg/kg MXF for 1 wk. One group was analyzed immediately at the end of the 1-wk treatment period, and the other group was analyzed 2 wk later. Control groups received vehicle only for 1 wk. Whole lungs were homogenized in 8 mL of PBS supplemented with 0.05% Tween 80 and serial dilutions were plated on Middlebrook 7H11 agar supplemented with 10% OADC and 0.5% glycerol to score the bacterial burden. Colonies were counted after 6 wk of incubation at 37 °C.
Glycerol Quantitation in Infected Lung Tissue.
Lung tissues were cryosectioned to 25-µm thick and 3,000,000 µm2 of sectioned tissue was collected by laser-capture microdissection using a Leica LMD 6500 (64). Extraction was performed by adding 50 µL of acetonitrile to 3,000,000 µm2 of 25-µm-thick laser-captured tissue. Extracts were sonicated in a water batch for 10 min and centrifuged at 4,000 rpm for 5 min. Derivatization of glycerol was performed to improve the HPLC retention of glycerol and to reduce spectral background noise. Glycerol derivatization was performed by combining 15 µL of 1% benzoyl chloride derivatization agent in acetonitrile, 15 µL of glycerol standard in acetonitrile, or 15 µL of tissue extract, 15 µL of 1 µg/mL glycerol-d5 in methanol and 15 μL of 100 mM sodium carbonate. An additional sample of 15 µL TAG mixture in acetonitrile was placed in the derivatization reaction conditions to demonstrate that glycerol does not deconjugate from TAG species during the reaction and contribute to glycerol levels. The TAG standard contained a total of 10 different species in equal parts and a total concentration of 500 µg/mL TAGs. The reaction mixture was heated to 50 °C for 1 h to complete derivatization. Sixty microliters of 2% formic acid in water was added to the samples to quench the reaction. Five microliters of the standards and study samples was injection on to the AB Sciex 6500+ Qtrap. Chromatography was performed on an Agilent SB-C8 4.6 × 100 mm 3.5-µm HPLC column using a reverse-phase gradient. The mobile phases were 0.1% formic acid in Milli-Q deionized water and 0.1% formic acid in acetonitrile. The monobenyzl derivative MRM transitions were monitored for both glycerol (197.08/105.03) and glyderol-d5 (202.08/105.03) internal standard.
Statistical Analysis.
All of the data were analyzed using a statistical tool of CLC Genomic Workbench or Student’s t test, as appropriate. The CLC tool was used to analyze RNA-seq data and differentially expressed mRNAs were defined using false-discovery rate-adjusted P ≤ 0.05. Microsoft Excel was used to perform Student’s t test and a P ≤ 0.05 was considered significant.
Data Availability.
Genome and transcriptome data were deposited in the NCBI BioProject database (ID PRJNA478476). The BioSample accession numbers for genomes and transcriptomes analyzed in this study are described in SI Appendix, Table S3.
Supplementary Material
Acknowledgments
We thank Dr. Anne Lenaerts for providing Kramnic mouse samples; Matthew Zimmerman and Yan Pan for finalizing the glycerol data; Claudia Setzer for help in the initial phase of the project; and Drs. Chris Sassetti and Martin Gengenbacher for their helpful discussions. This work was supported by the National Institute of Allergy and Infectious Diseases, Awards U19AI11276, T32AI125185, and R01AI111967; the Singapore Ministry of Health’s National Medical Research Council under its Translational and Clinical Research Flagship Grant NMRC/TCR/011-NUHS/2014 and the Center Grant ‘MINE’ Core #4 BSL-3 NMRC/CG/013/2013 (to T.D.), which is part of Singapore Programme of Research Investigating New Approaches to Treatment of Tuberculosis (SPRINT-TB, http://www.sprinttb.org).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.B. is a guest editor invited by the Editorial Board.
Data deposition: Genome and transcriptome data have been deposited in the NCBI BioProject database (accession no. PRJNA478476). Genomes and transcriptomes analyzed in this study have been deposited under BioSample accession nos. SAMN09517834–SAMN09517846, SAMN10742622–SAMN10742634, and SAMN09517865–SAMN09517876, and are described in SI Appendix, Table S3.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907631116/-/DCSupplemental.
References
- 1.WHO , Global Tuberculosis Report 2017 (World Health Organization, Geneva, 2017). [Google Scholar]
- 2.American Thoracic Society; CDC; Infectious Diseases Society of America , Treatment of tuberculosis. MMWR Recomm. Rep. 52, 1–77 (2003). Erratum in: MMWR Recomm. Rep.53, 1203 (2005). [PubMed] [Google Scholar]
- 3.WHO , Treatment of Tuberculosis Guidelines (World Health Organization, ed. 4, 2010). [PubMed] [Google Scholar]
- 4.Quy H. T. W., et al. , Drug resistance among failure and relapse cases of tuberculosis: Is the standard re-treatment regimen adequate? Int. J. Tuberc. Lung Dis. 7, 631–636 (2003). [PubMed] [Google Scholar]
- 5.Yoshiyama T., Shrestha B., Maharjan B., Risk of relapse and failure after retreatment with the Category II regimen in Nepal. Int. J. Tuberc. Lung Dis. 14, 1418–1423 (2010). [PubMed] [Google Scholar]
- 6.Guerra-Assunção J. A., et al. , Recurrence due to relapse or reinfection with Mycobacterium tuberculosis: A whole-genome sequencing approach in a large, population-based cohort with a high HIV infection prevalence and active follow-up. J. Infect. Dis. 211, 1154–1163 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parmar M. M., et al. , Unacceptable treatment outcomes and associated factors among India’s initial cohorts of multidrug-resistant tuberculosis (MDR-TB) patients under the revised national TB control programme (2007-2011): Evidence leading to policy enhancement. PLoS One 13, e0193903 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voskuil M. I., et al. , Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198, 705–713 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warner D. F., Mizrahi V., Tuberculosis chemotherapy: The influence of bacillary stress and damage response pathways on drug efficacy. Clin. Microbiol. Rev. 19, 558–570 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connolly L. E., Edelstein P. H., Ramakrishnan L., Why is long-term therapy required to cure tuberculosis? PLoS Med. 4, e120 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baek S.-H., Li A. H., Sassetti C. M., Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol. 9, e1001065 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thayil S. M., Morrison N., Schechter N., Rubin H., Karakousis P. C., The role of the novel exopolyphosphatase MT0516 in Mycobacterium tuberculosis drug tolerance and persistence. PLoS One 6, e28076 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torrey H. L., Keren I., Via L. E., Lee J. S., Lewis K., High persister mutants in Mycobacterium tuberculosis. PLoS One 11, e0155127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallis R. S., et al. , Drug tolerance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 43, 2600–2606 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bryant J. M., et al. , Whole-genome sequencing to establish relapse or re-infection with Mycobacterium tuberculosis: A retrospective observational study. Lancet Respir. Med. 1, 786–792 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Witney A. A., et al. ; RIFAQUIN Study Team , Use of whole-genome sequencing to distinguish relapse from reinfection in a completed tuberculosis clinical trial. BMC Med. 15, 71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vandiviere H. M., Loring W. E., Melvin I., Willis S., The treated pulmonary lesion and its tubercle bacillus. II. The death and resurrection. Am. J. Med. Sci. 232, 30–37; passim (1956). [DOI] [PubMed] [Google Scholar]
- 18.van der Woude M. W., Bäumler A. J., Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17, 581–611 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orsi R. H., Bowen B. M., Wiedmann M., Homopolymeric tracts represent a general regulatory mechanism in prokaryotes. BMC Genomics 11, 102 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkhill J., et al. , The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Cole S. T., et al. , Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Mizrahi V., Andersen S. J., DNA repair in Mycobacterium tuberculosis. What have we learnt from the genome sequence? Mol. Microbiol. 29, 1331–1339 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Benator D., et al. ; Tuberculosis Trials Consortium , Rifapentine and isoniazid once a week versus rifampicin and isoniazid twice a week for treatment of drug-susceptible pulmonary tuberculosis in HIV-negative patients: A randomised clinical trial. Lancet 360, 528–534 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Vincent V., et al. , The TDR tuberculosis strain bank: A resource for basic science, tool development and diagnostic services. Int. J. Tuberc. Lung Dis. 16, 24–31 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Comas I., et al. , Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat. Genet. 44, 106–110 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brauner A., Fridman O., Gefen O., Balaban N. Q., Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 14, 320–330 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Kester J. C., Fortune S. M., Persisters and beyond: Mechanisms of phenotypic drug resistance and drug tolerance in bacteria. Crit. Rev. Biochem. Mol. Biol. 49, 91–101 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Mehra S., Kaushal D., Functional genomics reveals extended roles of the Mycobacterium tuberculosis stress response factor sigmaH. J. Bacteriol. 191, 3965–3980 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park H.-D., et al. , Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48, 833–843 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kendall S. L., et al. , A highly conserved transcriptional repressor controls a large regulon involved in lipid degradation in Mycobacterium smegmatis and Mycobacterium tuberculosis. Mol. Microbiol. 65, 684–699 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kramnik I., Demant P., Bloom B. B., Susceptibility to tuberculosis as a complex genetic trait: Analysis using recombinant congenic strains of mice. Novartis Found. Symp. 217, 120–131; discussion 132–137 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Kramnik I., Dietrich W. F., Demant P., Bloom B. R., Genetic control of resistance to experimental infection with virulent Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 97, 8560–8565 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colangeli R., et al. ; DMID 01-009/Tuberculosis Trials Consortium Study 22 Teams , Bacterial factors that predict relapse after tuberculosis therapy. N. Engl. J. Med. 379, 823–833 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franzblau S. G., et al. , Comprehensive analysis of methods used for the evaluation of compounds against Mycobacterium tuberculosis. Tuberculosis (Edinb.) 92, 453–488 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Larrouy-Maumus G., et al. , Discovery of a glycerol 3-phosphate phosphatase reveals glycerophospholipid polar head recycling in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 110, 11320–11325 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ehrt S., Rhee K., Mycobacterium tuberculosis metabolism and host interaction: Mysteries and paradoxes in Pathogenesis of Mycobacterium tuberculosis and its Interaction with the Host Organism, Pieters J., McKinney J. D., Eds. (Springer Berlin Heidelberg, Berlin, Heidelberg, 2013), pp. 163–188. [DOI] [PubMed] [Google Scholar]
- 37.Titgemeyer F., Reizer J., Reizer A., Saier M. H. Jr, Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology 140, 2349–2354 (1994). [DOI] [PubMed] [Google Scholar]
- 38.Flores C.-L., Gancedo C., Unraveling moonlighting functions with yeasts. IUBMB Life 63, 457–462 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Streisinger G., Owen J., Mechanisms of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics 109, 633–659 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Canceill D., Viguera E., Ehrlich S. D., Replication slippage of different DNA polymerases is inversely related to their strand displacement efficiency. J. Biol. Chem. 274, 27481–27490 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Bayliss C. D., et al. , Neisseria meningitidis escape from the bactericidal activity of a monoclonal antibody is mediated by phase variation of lgtG and enhanced by a mutator phenotype. Infect. Immun. 76, 5038–5048 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bayliss C. D., Determinants of phase variation rate and the fitness implications of differing rates for bacterial pathogens and commensals. FEMS Microbiol. Rev. 33, 504–520 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Meyer T. F., van Putten J. P., Genetic mechanisms and biological implications of phase variation in pathogenic neisseriae. Clin. Microbiol. Rev. 2 (April suppl), S139–S145 (1989). [DOI] [PMC free article] [PubMed]
- 44.Pawlik A., et al. , Identification and characterization of the genetic changes responsible for the characteristic smooth-to-rough morphotype alterations of clinically persistent Mycobacterium abscessus. Mol. Microbiol. 90, 612–629 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Dheda K., et al. , Drug-penetration gradients associated with acquired drug resistance in patients with tuberculosis. Am. J. Respir. Crit. Care Med. 198, 1208–1219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pethe K., et al. , A chemical genetic screen in Mycobacterium tuberculosis identifies carbon-source-dependent growth inhibitors devoid of in vivo efficacy. Nat. Commun. 1, 57 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trauner A., et al. , The within-host population dynamics of Mycobacterium tuberculosis vary with treatment efficacy. Genome Biol. 18, 71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Black P. A., et al. , Whole genome sequencing reveals genomic heterogeneity and antibiotic purification in Mycobacterium tuberculosis isolates. BMC Genomics 16, 857 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hagström-Toft E., et al. , Marked heterogeneity of human skeletal muscle lipolysis at rest. Diabetes 51, 3376–3383 (2002). [DOI] [PubMed] [Google Scholar]
- 50.van der Kolk B. W., et al. , Effect of diet-induced weight loss on angiopoietin-like protein 4 and adipose tissue lipid metabolism in overweight and obese humans. Physiol. Rep. 6, e13735 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen Y., Xu Z., An improved GC-MS method in determining glycerol in different types of biological samples. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 930, 36–40 (2013). [DOI] [PubMed] [Google Scholar]
- 52.van der Beek C. M., et al. , The prebiotic inulin improves substrate metabolism and promotes short-chain fatty acid production in overweight to obese men. Metabolism 87, 25–35 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Gopal P., et al. , Pyrazinamide resistance is caused by two distinct mechanisms: Prevention of coenzyme a depletion and loss of virulence factor synthesis. ACS Infect. Dis. 2, 616–626 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parish T., Stoker N. G., Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146, 1969–1975 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Wallace R. J. Jr, Nash D. R., Steele L. C., Steingrube V., Susceptibility testing of slowly growing mycobacteria by a microdilution MIC method with 7H9 broth. J. Clin. Microbiol. 24, 976–981 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiegand I., Hilpert K., Hancock R. E. W., Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175 (2008). [DOI] [PubMed] [Google Scholar]
- 57.van Embden J. D., et al. , Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: Recommendations for a standardized methodology. J. Clin. Microbiol. 31, 406–409 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Safi H., Aznar J., Palomares J. C., Molecular epidemiology of Mycobacterium tuberculosis strains isolated during a 3-year period (1993 to 1995) in Seville, Spain. J. Clin. Microbiol. 35, 2472–2476 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Safi H., et al. , IS6110 functions as a mobile, monocyte-activated promoter in Mycobacterium tuberculosis. Mol. Microbiol. 52, 999–1012 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rustad T. R., et al. , Mapping and manipulating the Mycobacterium tuberculosis transcriptome using a transcription factor overexpression-derived regulatory network. Genome Biol. 15, 502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fisher R. A., Statistical Methods for Research Workers (Oliver and Boyd, London, England, 1925). [Google Scholar]
- 64.Zimmerman M., et al. , Ethambutol partitioning in tuberculous pulmonary lesions explains its clinical efficacy. Antimicrob. Agents Chemother. 61, e00924-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome and transcriptome data were deposited in the NCBI BioProject database (ID PRJNA478476). The BioSample accession numbers for genomes and transcriptomes analyzed in this study are described in SI Appendix, Table S3.