Significance
The ATP-fueled TRiC/CCT acts in the folding of 10% cytosolic proteins. TRiC consists of 8 paralogous subunits, each of which plays special roles in TRiC assembly, allosteric cooperativity, and substrate folding. However, due to lack of a thorough picture of TRiC conformational landscape and atomic-resolution details, the underlying structural mechanisms of TRiC subunit specificity in nucleotide usage and substrate binding, and the allosteric transition during ring closure remain unclear. Here, through cryo-electron microscopy (cryo-EM) analysis, we captured a thorough picture of TRiC conformational landscape from open to closed states and its gradually enhanced allosteric coordination, including the N termini, in unprecedented structural detail. Our study also offers insights into the TRiC subunit specificities in nucleotide usage and ring closure.
Keywords: chaperonin TRiC/CCT, allosteric network, ATPase cycle, conformational landscape, cryo-EM
Abstract
TRiC/CCT assists the folding of ∼10% of cytosolic proteins through an ATP-driven conformational cycle and is essential in maintaining protein homeostasis. Here, we determined an ensemble of cryo-electron microscopy (cryo-EM) structures of yeast TRiC at various nucleotide concentrations, with 4 open-state maps resolved at near-atomic resolutions, and a closed-state map at atomic resolution, revealing an extra layer of an unforeseen N-terminal allosteric network. We found that, during TRiC ring closure, the CCT7 subunit moves first, responding to nucleotide binding; CCT4 is the last to bind ATP, serving as an ATP sensor; and CCT8 remains ADP-bound and is hardly involved in the ATPase-cycle in our experimental conditions; overall, yeast TRiC consumes nucleotide in a 2-ring positively coordinated manner. Our results depict a thorough picture of the TRiC conformational landscape and its allosteric transitions from the open to closed states in more structural detail and offer insights into TRiC subunit specificity in ATP consumption and ring closure, and potentially in substrate processing.
Chaperonins play an essential role in maintaining cellular homeostasis, dysfunction of which is closely related to cancer and neurodegenerative diseases (1–3). The eukaryotic group II chaperonin TRiC/CCT assists the folding of ∼10% of cytosolic proteins, including many key structural and regulatory proteins, such as actin, tubulin, cell cycle regulator CDC20, and cancer-related VHL tumor suppressor and STAT3 (4–10). Moreover, up-regulation or mutations of several individual subunits of TRiC have been suggested to be related to breast cancer, hepatocellular carcinoma, Huntington’s disease, and Leber congenital amaurosis (1, 11–13).
TRiC is an ATPase macromolecular machine that utilizes energy of ATP binding and hydrolysis to drive its conformational transitions to facilitate substrate binding and folding. TRiC has evolved into a heterooligomeric double-ring complex, consisting of 8 homologous but distinct subunits (CCT1 to CCT8 in yeast) in each ring. The heterooligomeric nature of TRiC plays a key role in its functionality by allowing complicated substrate-binding modes in its open form (14, 15) and differentiation of nucleotide usage in its ring-closure process (16–18). However, due to lack of a complete picture of the TRiC conformational landscape and atomic resolution structural details, the underlying structural mechanisms of TRiC subunit specificity in nucleotide usage and substrate binding and the ATP-driven allosteric transitions of the complex remain unclear.
Each TRiC subunit consists of 3 domains: the apical domain (A domain), the intermediate domain (I domain), and the equatorial domain (E domain). The E domain contains the ATP-binding site and is involved in inter- and intra-ring subunit contacts (19). Between 2 adjacent intra-ring subunits, there are contacts linking their E domains via a mixed 4-stranded β-sheet involving the N and C termini of one subunit and the stem loop of the neighboring subunit, forming a “hand-in-hand” intra-ring interaction structural signature, which plays an essential role in intra-ring allosteric network (20).
Extensive efforts have been made to study the structures of archaeal group II chaperonins (21–23), as well as the eukaryotic group II chaperonin TRiC, by crystallography (24, 25) and cryo-electron microscopy (cryo-EM) (4, 5, 19, 26–28). Recently, we resolved 2 cryo-EM structures of Saccharomyces cerevisiae TRiC in a newly identified nucleotide partially preloaded state (NPP-TRiC) and in the ATP-bound state (TRiC-AMP-PNP) (29). The relatively high sequence identity (∼23% to 35% for yeast TRiC or ∼27% to 39% for bovine TRiC) and similar conformations of the subunits, especially in the closed state, have made it challenging to identify the different subunits in the TRiC complex by structure studies. Not long ago, the subunit arrangement was determined by chemical cross-linking and mass spectrometry (30, 31). We have recently identified locations of subunits in the open state TRiC and found the on-axis CCT2 subunit pair forming an unexpected Z-shaped feature and the preloaded nucleotide in the consecutive CCT3, CCT6, and CCT8 subunits in yeast NPP-TRiC (29, 32).
It has been reported that the ATPase activity of TRiC is a function of ATP concentration (33–35): For yeast TRiC, its ATPase reaction velocity reaches the plateau at 0.2 mM ATP, indicating that all TRiC subunits have been activated, while, at lower ATP concentration (0.05 mM), only a fraction of the subunits are activated (35). However, the underlying structural mechanism remains elusive on how the TRiC allostery gradually coordinates with increased ATP concentration. Moreover, ADP-AlFx has been previously used to mimic multiple nucleotide states, including ATP binding and hydrolysis transition states (36); therefore, ADP-AlFx (ADP-aluminium fluoride) could be used as the nucleotide mimic to study TRiC conformational transitions in the gradually increased nucleotide concentration, so as to reveal the initiation of TRiC allosteric cooperativity and its ring closure mechanism.
In the present study, we resolved an ensemble of cryo-EM structures of yeast TRiC at various concentrations of ADP-AlFx from 0.05 mM to 0.5 mM, with 9 of these structures in distinct open conformational states and another 2 in the both-ring-tightly-closed state. Four out of the 9 open-state maps were resolved at near-atomic resolutions, and a closed-state map was determined to a resolution of 2.99 Å, representing so far the best resolved TRiC structure. This closed-state map reveals an unreported extra layer of allosteric network formed by the N termini of multiple subunits, and distinct nucleotide states of TRiC subunits. Overall, we capture a more complete picture of the conformational landscape of TRiC and delineate the trajectory for the nucleotide usage and ring closure of individual subunits. This study facilitates our understanding of the functional specificities of TRiC subunits in nucleotide usage and substrate processing.
Results
Initial Allosteric Transition of TRiC at an Extremely Low Nucleotide Concentration (0.05 mM ADP-AlFx).
To capture the initial allosteric cooperativity of TRiC at an extremely low nucleotide concentration, we carried out cryo-EM investigation of TRiC in the presence of 0.05 mM ADP-AlFx, based on previous biochemical analysis (35). This value is much lower than the physiological nucleotide concentration (1 to 2 mM) and, to some extent, one that may mimic stress conditions (37, 38). In this condition, the molar ratio between TRiC subunits and nucleotide is 1:3. We obtained 4 cryo-EM maps of TRiC, all in distinct but open conformations, termed as the 0.05-C1, 0.05-C2, 0.05-C3, and 0.05-C4 states, to resolutions of 6.80 Å, 4.45 Å, 7.18 Å, and 4.38 Å, respectively (Fig. 1 and SI Appendix, Fig. S1).
Fig. 1.
An ensemble of cryo-EM maps of TRiC in the presence of 0.05 mM ADP-AlFx, and the corresponding nucleotide statuses. (A–D) Four cryo-EM maps of TRiC in distinct open conformations in the presence of 0.05 mM ADP-AlFx, with these conformational states denoted as 0.05-C1 (A), 0.05-C2 (B), 0.05-C3 (C), and 0.05-C4 (D). The cryo-EM map of NPP-TRiC (EMDB: 9540) is also shown in the small Upper Inset of A (different subunits in distinct colors and labeled). A side view (Left) and 2 end-on views (Middle and Right) are shown for each state. The CCT7 and CCT2 subunits displaying observable conformational changes (in one ring or both rings) compared with NPP-TRiC are colored dodger blue and red, respectively. (E–H) Nucleotide occupancy statuses of the 4 maps. Both rings of 0.05-C1 (E), 0.05-C2 (F), 0.05-C3 (G), and 0.05-C4 (H) are shown here. Shown are a slice of the model near the nucleotide pocket region and the nucleotide densities (in red). These rendering styles were followed throughout.
Compared with NPP-TRiC, the cis-ring CCT7 subunit in the 0.05-C1 map shows a movement in the upper portion of the subunit, then resuming the orientation typically observed in the other subunits (Fig. 1A). In the 0.05-C2 map, the A and I domains of CCT2 appear detached from the neighboring CCT5 and show the typical orientation of other subunits (Fig. 1B and SI Appendix, Fig. S4A). In the 0.05-C3 map, both CCT2 and CCT7 appear moved back to the typical orientation (Fig. 1C). However, in all the 3 maps, the entire trans-ring and the other 6 subunits (except CCT2 and CCT7) in the cis-ring remain in the NPP-TRiC conformation (29) (Fig. 1 A–C). Interestingly, in the 0.05-C4 map, CCT7 subunits from both rings simultaneously move back, with other subunits unmoved (Fig. 1D and SI Appendix, Fig. S4B). Altogether, CCT7 and CCT2 could be the first 2 subunits reacting to nucleotide binding.
We then examined the nucleotide occupancies of individual subunits. Overall, in all of the 4 maps, similar nucleotide occupancy patterns were observed for the 2 rings. All these maps display strong nucleotide densities for CCT3, CCT6, CCT8, and CCT1. The nucleotide densities for CCT7, and especially CCT2, were observed to be generally weaker and more variable among the different maps (Fig. 1 E–H). We previously found that CCT3, CCT6, and CCT8 in NPP-TRiC have preloaded nucleotide from the environment of the yeast cells (29), which substantiates our current observation. Furthermore, our results suggest that ATP binding is not sufficient to trigger CCT1 conformational changes.
Interestingly, although the nucleotide densities appear relatively weak in CCT7 and CCT2 in all of these maps, these 2 subunits react relatively sensitively to nucleotide binding, and their conformational changes indeed correlate with their nucleotide binding status. CCT7 shows nucleotide densities in the cis-rings in the 0.05-C1 and 0.05-C3 maps and both rings of the 0.05-C4 map (Fig. 1 E, G, and H); correspondingly, the conformation of these CCT7 subunits differs noticeably from that in the NPP-TRiC map (Fig. 1 A, C, and D).
Taken together, at this rather low nucleotide concentration, CCT1, CCT7, and CCT2 are the subunits involved in ATP binding in the very early stage of the TRiC ATPase cycle, which might be related to their originally solvent-exposed nucleotide pockets (29). Furthermore, the overall similar nucleotide occupancy patterns of the 2 rings in each of these maps indicate a positive inter-ring cooperativity in yeast TRiC, with regard to nucleotide binding, although the movements of CCT7 or CCT2 appear less symmetrical between the 2 rings, due to the extremely low nucleotide occupancy status in the current condition.
TRiC Shows Strengthened Allosteric Cooperativity with Increased Nucleotide Concentration (0.1 mM).
To investigate the further enhanced allosteric cooperativity of TRiC, we increased the ADP-AlFx concentration to 0.1 mM, a value still considered to be a cell stress condition (37, 38). We here resolved 5 cryo-EM maps, all showing TRiC in distinct open conformational states. Most populations of the particles were used to reconstruct 2 of these maps: 0.1-C1 at 4.62 Å resolution (44.4% of the particles) and 0.1-C4 at 4.69 Å resolution (39.4%). The remaining particles (∼15%) were used to reconstruct the other 3 states, including 0.1-C2a, 0.1-C2b, and 0.1-C3 at 12.98 Å, 7.67 Å, and 16.87 Å resolution, respectively (Fig. 2 A–E and SI Appendix, Fig. S2). Inspection of these maps relative to the NPP-TRiC map revealed the ATP-driven conformational variations of up to 4 subunits. We then classified the maps into 4 states (Fig. 2 A–E), which may represent the relatively stable structural intermediates in the TRiC energy landscape toward ring closure: in stage 1, only 1 subunit (CCT7) rotated inward and showed the typical orientation of other subunits (Fig. 2A and SI Appendix, Fig. S4C); on that basis, in stage 2, either CCT2 (0.1-C2a) or CCT7′ (0.1-C2b) moved inwards (Fig. 2 B and C); in stage 3 (0.1-C3), these 2 conformations merged into 1, with either CCT7′ or CCT2 showing additional movement relative to 0.1-C2a or 0.1-C2b, respectively (Fig. 2D); and in stage 4 (0.1-C4), CCT2′ rotated inwards to resume the typical conformation (Fig. 2E and SI Appendix, Fig. S4D). Analysis of these data suggests that, with increased nucleotide concentration that may strengthen allosteric cooperativity, some subunits may react to nucleotide binding in a certain order: specifically, perhaps CCT7 first, and then CCT2.
Fig. 2.
TRiC shows sequential allosteric cooperativity in the presence of 0.1 mM ADP-AlFx. (A–E) Five cryo-EM maps of TRiC at distinct open conformations in the presence of 0.1 mM ADP-AlFx, denoted as 0.1-C1 (A), 0.1-C2a (B), 0.1-C2b (C), 0.1-C3 (D), and 0.1-C4 (E). The CCT7 and CCT2 subunits, displaying observable conformational changes compared with NPP-TRiC, are colored dodger blue and red, respectively. Based on the number of subunits showing conformational changes, we classified the maps into 4 stages. (F and G) Nucleotide occupancy statuses of the relatively better resolved 0.1-C1 (F) and 0.1-C4 (G) maps. In 0.1-C4, only CCT4 in both rings remained completely unoccupied. (H) Visualization of the CCT4 nucleotide pocket region of the 0.1-C4 map (gold surface). Neither density for the β-hairpin motif (indicated by a red dashed line) nor for nucleotide (indicated by a black dashed line) was observed in the 0.1-C4 map. The model is in medium purple, and the nucleotide is rendered as ball-and-stick. We borrowed the model from the 0.2-C1 state (presented later), which has this portion of the map very well resolved, and both structures share overall similar conformation in the CCT4 nucleotide pocket region.
Inspection of the better resolved 0.1-C1 and 0.1-C4 maps also revealed an association between subunit conformational changes and nucleotide occupancy status. In 0.1-C1, relatively strong nucleotide density could be observed in CCT7 but rather weak density in CCT7′ (Fig. 2F), and, correspondingly, only CCT7 appeared in the rotated-back conformation (Fig. 2A). Moreover, in 0.1-C4, we observed strong nucleotide densities in CCT2 and CCT2′, and weak ones in CCT5 and CCT5′ (Fig. 2G). Accordingly, dramatic structural differences in CCT2 (Fig. 2E) and noticeable ones in CCT5 (SI Appendix, Fig. S4I) in both rings were observed. Here, 0.1-C4 adopted a conformation similar to that of TRiC-AMP-PNP (mimicking the ATP-bound state) (SI Appendix, Fig. S4H), indicating that TRiC can bind but not hydrolyze ATP at up to a nucleotide concentration of 0.1 mM because no additional motion beyond the TRiC-AMP-PNP conformation was detected. Moreover, inspection of our 0.1-C1 and 0.1-C4 maps revealed strong nucleotide densities in CCT1, CCT3, CCT6, and CCT8 in both rings although no obvious conformational changes were detected (Fig. 2 A and E–G). In addition, each of these 2 maps revealed similar nucleotide occupancy patterns for the 2 rings (Fig. 2 F and G).
CCT4 Is the Last Subunit That Binds Nucleotide.
Surprisingly, in 0.1-C4 map, CCT4 is the only subunit without bound nucleotide in either ring; additionally, the β-hairpins covering its nucleotide pockets were missing in both rings, similar to that in the NPP-TRiC structure (Fig. 2 G and H) (29). Therefore, at relative low nucleotide concentration, the intrinsically dynamic β-hairpin of CCT4 may be unable to lock nucleotide inside the pocket. The β-hairpin of CCT4 may play a role in the relatively unusual ATP-binding property of CCT4 in yeast.
Atomic-Resolution Map of TRiC in the Closed Conformation in the Presence of 0.2 mM ADP-AlFx.
We then asked at which nucleotide concentration can TRiC initiate nucleotide hydrolysis to shut the rings closed. To address this question, we increased the ADP-AlFx concentration to 0.2 mM and obtained a cryo-EM map of TRiC (named 0.2-C1) (Fig. 3A) at the both-ring-tightly-closed conformation in the dominant population (∼95%). The map was evaluated at a nominal resolution of 2.99 Å, with the resolution ranging from 2.60 Å to 4.20 Å and showing atomic-resolution structural details (SI Appendix, Fig. S3 A–C), which allowed us to build a pseudoatomic model (Fig. 3B). Direct visualization of the protruding affinity CBP (calmodulin binding peptide) tag in the CCT3 subunit in the early stage of the refinement (∼8 Å resolution) allowed us to locate this subunit in both rings (SI Appendix, Fig. S5A). Combined with the available subunit ordering of TRiC (29–32, 39), we were able to identify all of the subunits in this map. Further visualizations of the unique insertions in the CCT1, CCT4, CCT5, and CCT6 subunits (SI Appendix, Fig. S5 D and E) and the specific kink feature in the apical protruding helix of CCT6 (SI Appendix, Fig. S5B) corroborated our subunit assignment, based on the CCT3-CBP location. Altogether, our data suggest that, at the relatively high nucleotide concentration of 0.2 mM, driven by ATP-hydrolysis, TRiC can simultaneously shut both rings closed and form the tightly closed conformation (SI Appendix, Fig. S5C). To further prove that the both-ring-closed conformation is not an artifact caused by ADP-AlFx, we resolved another cryo-EM map of TRiC in the presence of 0.2 mM ATP (termed 0.2-ATP) at 4.00 Å resolution (Fig. 3C and SI Appendix, Fig. S3B). This map is also in the both-rings-closed conformation similar to that of the 0.2-C1 map, demonstrating that TRiC can also close the rings in the presence of 0.2 mM natural nucleotide.
Fig. 3.
Cryo-EM maps of TRiC in the both-rings-tightly-closed conformation. (A) The atomic-resolution cryo-EM map of TRiC in the presence of 0.2 mM ADP-AlFx (0.2-C1). An end-on view (with individual subunits labeled) and 2 side views are shown. The color scheme is followed throughout. (B) Pseudoatomic model of 0.2-C1 (ribbon). (C and D) The cryo-EM maps of TRiC in the presence of 0.2 mM ATP (0.2-ATP, C) and 0.5 mM ADP-AlFx (0.5-C1, D), respectively. Both maps are in the both-rings-tightly-closed conformation similar to that of 0.2-C1.
Asymmetric Nucleotide Occupancy within a Ring in the Closed State of TRiC.
In the 0.2-C1 map, the nucleotide densities for all subunits except CCT8 very well matched ADP-AlFx and a magnesium ion; in addition, the map also revealed a water molecule attacking the gamma phosphate (Fig. 4A), involved in the formation of the ATP hydrolysis reaction center. Surprisingly, for both rings, CCT8 remained in the ADP form without the attacking water molecule (Fig. 4A). Our NADH-coupled enzymatic assay (40) analysis on purified CCT8 monomer showed that CCT8 has a rather weak ATP hydrolysis ability in comparison to the wild-type (WT) TRiC or CCT5 monomer (Fig. 4B and SI Appendix, Fig. S6 A and C and Table S3). Although there was no obvious aggregation in the negative staining EM analysis of CCT8 and CCT5 monomers, indicating very likely that they are in the correctly folded states, we cannot rule out the possibility that part of them may not be in the completely properly folded form (SI Appendix, Fig. S6 B and D). Our results suggest that CCT8 may initially bind ATP from the environment and hydrolyze it to ADP, which may be hardly exchanged in the rest of the ATP cycle in this experimental condition. Moreover, with increased nucleotide concentration to 0.5 mM, we resolved an additional cryo-EM map of TRiC at 3.44 Å resolution also in the closed conformation (Fig. 3D and SI Appendix, Fig. S3B). In this map and the 0.2-ATP map (in the presence of 0.2 mM native ATP) (Fig. 3C), CCT8 was the only subunit remaining bound with ADP in both rings (SI Appendix, Fig. S3 D and E), substantiating the notion that the original ADP in CCT8 may be hardly exchanged in the ATPase cycle of yeast TRiC in our experimental conditions.
Fig. 4.
CCT8 remains in the ADP state, distinct from all of the other subunits in the 0.2-C1 TRiC structure. (A) Portions of the 0.2-C1 map in the nucleotide pocket region showing all of the subunits, except CCT8, bound with ADP-AlFx (stick model) and a magnesium ion (green ball), as well as a water molecule (red ball) in an attacking position, suggesting that these subunits are in the ATP-hydrolysis transition state. However, CCT8 remains bound to ADP, without AlFx or the attacking water molecule, indicating that this subunit is still in the ADP state. The TRiC subunits here are shown following their sequential order within a ring as visualized from the top of the complex. (B) NADH coupled enzymatic assays revealed a lower ATPase activity of the CCT8 monomer than those of the CCT5 monomer and WT TRiC. The ATPase rates (denoted by velocity [V], with the unit of “mole ATP/[mole TRiC • min]”) determined by fitting the liner part of the ATP hydrolysis reaction curve are also provided. The significant difference analysis (Right) also suggests that the CCT8 monomer shows a significantly lower ATPase activity compared to the CCT5 monomer and TRiC complex, with the statistical significance ***P < 0.001 and ****P < 0.0001, respectively.
We have previously shown that CCT6 binds ADP in the NPP state (29). These findings, combined with our current observation, suggest that the originally bound ADP in CCT6 can be replaced by ATP in the nucleotide hydrolysis stage, as a result of the collective motions of other subunits driven by hydrolysis of ATP. Following that, CCT6 could hydrolyze ATP to enter the TRiC ATPase cycle while, for CCT3, after the binding stage, CCT3 does hydrolyze ATP to enter the ATPase cycle. Therefore, for yeast TRiC, not only ATP binding but also ATP hydrolysis occurs in a stepwise manner.
An Extra Layer of N-Terminal Allosteric Network Plays Important Roles in TRiC Ring Closure.
The N-terminal β-strand of each TRiC subunit has been suggested to be involved in the formation of a 4-stranded β-sheet in the hand-in-hand structural signature. However, we did not observe the N-terminal β-strands of all subunits, except CCT1, in any of our open-state TRiC structures resolved here (SI Appendix, Fig. S7 A–H) and our previous NPP-TRiC and TRiC-AMP-PNP maps (29) also in the open state. This suggests that, in the open-state, the N-terminal β-strands and the preceding N termini of most subunits are dynamic and little involved in the allosteric network. In contrast, in the atomic-resolution closed-state 0.2-C1 map, not only the N-terminal β-strands but also the N termini are stabilized and better resolved, compared with the previous X-ray structure at 3.8 Å resolution (24, 30) (Fig. 5 A–E and SI Appendix, Fig. S7 K and L), specifically for the CCT2, CCT1, CCT3, CCT6, CCT7, and CCT5 subunits; and the N-terminal features of CCT4 and CCT8 are similar to those described previously (24, 30). In addition, the stabilized N termini in the closed-state map indicates that they could involve in and enhance the allosteric regulation of TRiC during its ring closure process.
Fig. 5.
The N termini of TRiC subunits may play an essential role in intra- and inter-ring allosteric cooperativity. (A) The locations of the resolved N termini (in color) in the 0.2-C1 TRiC structure. The visualization angle and region are illustrated in the Inset. (B) Magnified view of the interaction between helix H1 of CCT3 and the N terminus of CCT6: specifically, its small 1-turn α-helix before its N-terminal β-sheet. (C) Magnified view of the resolved N-terminal extension of CCT5. (D) Magnified view of the CCT4 N terminus, observed to be bent and extended toward the outside of the ring. The image here is viewed from outside of the TRiC structure. (E) An inter-ring interacting network formed by the N termini of CCT7 and CCT8 from the cis-ring, and the N and C termini of CCT6′ and the stem loop of CCT3′ from the trans-ring. Shown is the model and the surface rendering of the model. The visualization angle and direction are indicated in the Inset. The CCT7 N-terminal tail extends around the E domain of the neighboring CCT8, and to contact the N terminus of the trans-ring CCT6′ (white dashed oval line). The N terminus of CCT8 bends across the interface between the 2 rings, to bundle together with the N and C termini of the trans-ring CCT6′ and to form a contact with the stem loop of the trans-ring CCT3′ (black dashed oval line). A magnified view of the hydrophobic interaction between the N termini of CCT7 and CCT8 is shown on the right. (F) The growth rates of the CCT4-NTD and CCT7-NTD yeast strains were measured to be lower than that of the WT TRiC strain (control). (G) Statistical analyses for doubling time were performed using a 1-way ANOVA, demonstrating the growth defect due to the truncations in the N-terminal tail of CCT4 and CCT7, respectively. The significant difference, ****P < 0.0001, was indicated.
Interestingly, the N terminus of every subunit except CCT4 can be observed in the 0.2-C1 map to contact H1 of the neighboring subunit, potentially enhancing the intra-ring interaction in the closed state (SI Appendix, Fig. S7J), while that of CCT4 is instead bent and extended toward the outside of the ring (Fig. 5 A and D). Our truncation of the CCT4 N terminus (CCT4-NTD) induced an obvious decrease in the yeast growth rate (Fig. 5 F and G and SI Appendix, Table S2), suggesting that this N terminus may contribute to the allosteric cooperativity between the 2 rings. Interestingly, from the open to closed state, the N-terminal region of CCT6 showed a large secondary structural element (SSE) transition from a loop to a β-strand followed by a small 1-turn α-helix, forming clear contact with H1 of the neighboring CCT3 (SI Appendix, Fig. S7E and Fig. 5B) and participating in intra-ring interactions. For CCT5, the close contact between its C terminus and the H1 of CCT7 in the open state was replaced by a contact between its N terminus and CCT7 H1 in the closed state (SI Appendix, Fig. S7I).
Our 0.2-C1 map also showed that the CCT7 N terminus (with F3) extends around the E domain of the neighboring CCT8 and inserts into a hydrophobic pocket composed by a turn of the CCT8 N terminus (with F14 and Y18) (Fig. 5 E, Right), forming a hydrophobic intra-ring interaction network. Interestingly, the N terminus of CCT7 was observed to be also involved in a previously unreported allosteric network by linking the termini of CCT8, CCT6′, and CCT3′ together (Fig. 5E). We further truncated the entire N terminus (N2 to T17) of CCT7 (CCT7-NTD), which induced an obvious growth rate decrease compared with WT TRiC (Fig. 5 F and G and SI Appendix, Table S2). Altogether, our structural and functional analyses suggest that the CCT7 N terminus may play a key role in the intra- and inter-ring allostery of TRiC.
Furthermore, we also found that the N terminus of CCT8 is involved in the unreported interaction network with multiple inter-ring neighboring subunits (Fig. 5E): It bends and crosses the inter-ring interface, to bundle together with the N and C termini of the trans-ring CCT6′ and to form a contact with the stem loop of trans-ring CCT3′ (black dashed ellipsoid line in Fig. 5E). Therefore, instead of participating in the ATPase cycle, CCT8 may play a special role in TRiC ring closure through this allosteric network.
A Unique Cross-Shaped Inter-ring Interaction Pattern in TRiC.
For group II chaperonins, their E domains are involved in inter-ring interactions through a “feet-on-feet” mode: i.e., with the pair of opposing subunits standing on top of each other contacting through corresponding residues. Interestingly, comparison of 0.1-C4 with 0.2-C1 revealed that the E domains, in both the open and closed states, interact through a direct charge–charge pivot interaction pattern in every other subunit pair (specifically CCT2-CCT2′, CCT1-CCT7′, CCT6-CCT6′, and CCT7-CCT1′, which are well conserved) of the 8 inter-ring subunit pairs (SI Appendix, Fig. S8C), together forming a cross-shaped signature in the inter-ring interface (SI Appendix, Fig. S8D). This observation suggests that the 4 pairs of charge–charge pivot contacting subunits spaced out at intervals may be more important in assembly and allosteric regulation between the rings than the other 4 subunits pairs, and this pattern of interaction may also be beneficial for liberating the restraints between the 2 rings for smooth ring closure.
A Detailed Asymmetric Ring Closure Mechanism of TRiC.
Efforts have been made to delineate the ring closure mechanism of TRiC; however, the structures they used to perform the analysis were either with imposed symmetry or at intermediate to low resolutions (5, 15, 27, 28). Therefore, a detailed ATP-driven ring closure mechanism for the asymmetric TRiC complex remains elusive. Here, our near-atomic and atomic-resolution TRiC structures in the open (0.1-C4) and closed (0.2-C1) states allow us to deduce its ring closure mechanism in more structural detail. Unlike the symmetric archaeal group II chaperonin Mm-cpn, which closes the ring through a more rigid-body rocking motion (23), TRiC subunits adopt considerably distinct domain-wise movements in the ring-closing process (SI Appendix, Fig. S9 A–C and Movie S1). Regarding the collective domain motion of individual subunits (Fig. 6B), CCT1 and the neighboring CCT3 display the most dramatic rotations (85.6° and 79.6°, respectively), indicating that they may play a special role in TRiC ring closure or release of substrate into the folding chamber (41). CCT8 exhibits the least movement (29.7°), in line with our finding that CCT8 may be barely involved in the ATPase cycle of TRiC. Overall, the quite different magnitudes of movements of different TRiC subunits could be related to their functional specificities in TRiC ring closure or substrate folding, and this also suggests an asymmetric ring closure mechanism of TRiC.
Fig. 6.
Domain-wise conformational transitions of TRiC from open to closed states, revealing an asymmetric chamber closure mechanism of TRiC. (A) Decomposition of the conformational changes of CCT3 from open 0.1-C4 (gray) to closed 0.2-C1 (colored) state into 3 portions corresponding to the 3 domains. The A domain is shown in red, I domain in yellow, and E domain in sky blue. The rotation axis and angle for each domain are labeled. (B) Rotational angle summation of the 3 domains for each individual subunit, indicating an asymmetric chamber closure of TRiC. (C) Magnified view of the conformational transitions of the key structural elements in the nucleotide pocket of CCT3 from the open 0.1-C4 (gray) to the closed 0.2-C1 (colored) state. Locations of NSL, H11, stem loop, attacking water molecule, and key residues are shown in the Left. The depiction of the active site (Right) illustrates the attacking water nucleophile being held and polarized by hydrogen bonds to the side chains of Asp60 and Asp398, and Lys162 moving to a proximity position to sense the existence of the γ-phosphate (represented by the AlF3 group in our structure).
Furthermore, we take CCT3 as an example, which shows a remarkably large scale of movement in the chamber-closing process, to illustrate the domain-wise conformational transitions of TRiC subunits in more detail (Fig. 6 A and B). We found that, during ATP hydrolysis, the hydrolytic residue Asp60 located adjacent to the stem loop (Fig. 6C) [equivalent to thermosome Asp60 (3)] drags the connecting stem loop by 11.0 Å toward the nucleotide (Movie S2 and SI Appendix, Fig. S9D). This motion, together with similar stem-loop motions of other subunits, could be propagated to the E domains through the hand-in-hand intra-ring allosteric network, leading to an inward 24.2° rotation of the CCT3 E domain toward the central cavity (Fig. 6A, SI Appendix, Fig. S9C, and Movie S1). Surprisingly, analysis of our 0.2-C1 map suggested that the large stem-loop movement in CCT3 (SI Appendix, Fig. S9D) could be captured by the N terminus of CCT8′ from the trans-ring (black dashed ellipsoid in SI Appendix, Fig. S8B); thus, this movement can be propagated to CCT8′, leading to conformational rearrangements in the opposing ring. Noteworthily, this specific CCT3–CCT8′ inter-ring interaction has a symmetric CCT3′–CCT8 interaction. This mechanism could contribute to the inter-ring allosteric coordination during TRiC ring closure. In the meantime, the I domain of CCT3 consists of the key nucleotide-sensing residue Lys162 in the nucleotide sensing loop (NSL) and the key hydrolytic residue Asp398 at H11 (Fig. 6C), equivalent to thermosome Lys161 and Asp386, respectively (3). ATP binding and hydrolysis could bring these 2 critical factors together with closely related elements into proximity of the nucleotide: i.e., bring NSL and H11 of CCT3 closer to the nucleotide by 12.0 Å and 10.5 Å, respectively, facilitating ATP hydrolysis. This motion could lead to conformational transitions of the entire I domain, leading to an anticlockwise rotation of 31.9° downward toward the nucleotide (Fig. 6A and SI Appendix, Fig. S9B). The motion could then be propagated to the connecting A domain, which also showed an overall anticlockwise rotation of 23.5° (Fig. 6A and SI Appendix, Fig. S9A). Collectively, those diversified movements of individual TRiC subunits, however sharing common directions of inward tilting associated with anticlockwise rotation, could be coordinated to shut the ring closed.
Discussion
In our current study, through varying nucleotide concentrations and deep classification, we captured an ensemble of cryo-EM structures of TRiC from open to closed states, with one closed state reaching unprecedented atomic resolution in the TRiC structural study. Analysis of the ensemble of structures revealed a thorough picture of the conformational landscape of TRiC in the ring closure process and its gradually enhanced allosteric regulation in this process. Moreover, of the 8 subunits of TRiC, CCT7 may initiate TRiC ring closure, followed by CCT2; CCT4 is the last subunit to bind nucleotide which may serve as an ATP sensor; and CCT8 remains ADP-bound and is hardly involved in the ATPase-cycle in our experimental conditions. Our atomic resolution closed-state map also depicts an unreported extra layer of allosteric network formed by the N termini of multiple subunits, including CCT4, CCT6, CCT8, and CCT7, which could play an essential role, especially in inter-ring coordination and ring closure. Our findings also delineate the trajectory for the nucleotide usage and ring closure of individual subunits and greatly enhance our understanding of the subunit specificity of TRiC in ATP consumption and ring closure.
CCT4 May Serve as an ATP Sensor and Regulate the ATPase Cycle of TRiC.
Our results suggest that the nucleotide occupancy status of CCT4 is closely related to the nucleotide concentration in the environment. At very low ATP concentrations (up to 0.1 mM, which, to some extent, resembles the cell starvation condition) (37), CCT4 was found to be the only subunit not binding nucleotide. In this situation, TRiC cannot proceed to hydrolyze ATP to close the ring, which may conserve energy for the sustainment of the cells. When the ATP concentration was increased to a level that would allow cells to recover from starvation conditions, CCT4 was observed to bind ATP, and the TRiC complex can resume hydrolyzing ATP and to close the ring for substrate folding. Thus, we conclude that CCT4 is the last subunit to bind nucleotide, and it may serve as a nucleotide sensor of the complex. Taken together, CCT4 appears to play a key role in regulating the ATPase cycle of TRiC and may also be the first subunit in the complex to fire ATP hydrolysis. Substantiating this, previous studies showed that depletion of the ATP binding and hydrolysis ability of CCT4 is lethal for yeast, and CCT4 is the only subunit displaying this phenotype (17, 42).
CCT8 Plays a Special Role in TRiC Ring Assembly and Allosteric Cooperativity.
Surprisingly, our biochemical analysis suggested a fairly weak ability of CCT8 to hydrolyze ATP (Fig. 4B and SI Appendix, Table S3), and we also structurally revealed that CCT8 is the only subunit to remain bound to ADP in our closed-state 0.2-C1, 0.2-ATP, and 0.5-C1 maps, hardly participating in the TRiC ATPase cycle in our experimental conditions (Fig. 4A and SI Appendix, Fig. S3 D and E). Consistent with our results, Chaetomium thermophilum TRiC, but with ATPase-deficient CCT8, has been shown to exhibit an ATPase activity very similar to that of the WT TRiC (43). Also, CCT8 is distinct from other subunits in the ATP binding-related sites, which are well conserved (39). Taken together, we postulate that a nascent CCT8 subunit alone may bind ATP from the environment and hydrolyze it to ADP during TRiC ring assembly; afterward, this ADP in CCT8 is extremely difficult to exchange in the ATPase cycle of TRiC, to some extent comparable to that occurring for GDP in β-tubulin in microtubule assembly process (44). Still, we cannot rule out the possibility that, at much higher ATP concentration and longer reaction time, or in the presence of substrate, the original ADP in CCT8 may gradually be exchanged by ATP. Instead, CCT8 may play an important role in TRiC intra- and inter-ring allosteric cooperativity and ring closure through its N-terminal interaction network (Fig. 5E). Additionally, it has been shown that CCT8 might be a potential activator/modulator of TRiC assembly (45). In summary, CCT8 may be barely involved in the ATP consumption of TRiC; instead, it could play a special role in TRiC ring assembly and allosteric cooperativity.
The Trajectory for the Stepwise Nucleotide Consumption and Ring Closure of TRiC Subunits.
It is noteworthy that our current study additionally disclosed a trajectory for the stepwise nucleotide consumption and ring closure of TRiC subunits, which was not reported before. Based on the sequence of nucleotide binding and spatial locations of individual subunits, we categorize the 8 subunits into 4 classes distributed symmetrically about a “mirror” that is perpendicular to the inter-ring interface (Fig. 7A). The class I subunits, namely CCT3, CCT6, and CCT8, have been indicated to have preloaded ADP from the environment in the very beginning of the nucleotide cycle (29). With gradually increased nucleotide concentration, the class II subunits, CCT7 and CCT1, could bind nucleotide in the early stage of the cycle; however, only CCT7 shows an obvious inward rotation initiating the movements of the entire complex while CCT1 does not move. Then, CCT2, assigned to class III, could bind ATP in a fairly early stage (but with relatively low occupancy) and show a dramatic movement following that of CCT7. The class IV subunits CCT5 and CCT4 are the last 2 subunits to bind nucleotide. CCT5 could subsequently bind ATP after CCT2, showing an observable movement. Therefore, the nucleotide binding is stepwise, and the corresponding subunit conformational changes follow a certain order. Until this stage, TRiC remains in the open state. Eventually, the last subunit CCT4 could bind nucleotide at a nucleotide concentration of about 0.2 mM, and, subsequently, the entire TRiC complex could proceed to the ATP-hydrolysis stage to shut the ring closed. Therefore, CCT4 ATP binding could be the rate-limiting step in the ATPase cycle of TRiC. Moreover, the originally bound ADP in CCT3 and CCT6 could be replaced by ATP, so as to catch up with the hydrolysis process, while the ADP in CCT8 can hardly be exchanged.
Fig. 7.
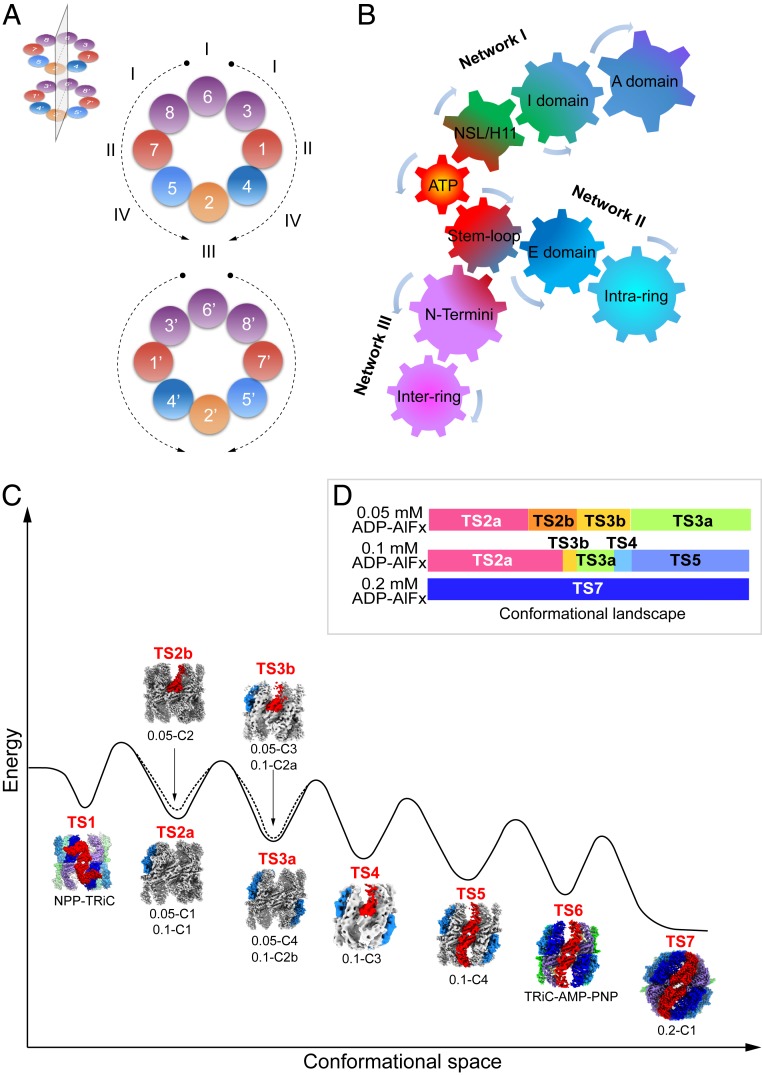
ATP consumption and allosteric regulation mechanisms of TRiC and the conformational landscape of TRiC during ring closure. (A) Ordering of TRiC subunit nucleotide binding. The 8 subunits of TRiC can be categorized into 4 classes, arranged symmetrically about a mirror plane located in the position illustrated in the Inset. (B) Diagram illustrating the proposed 3 sets of gears forming the allosteric networks of TRiC, from interactions within a subunit (Network I) to that of intra-ring (Network II) and inter-ring (Network III) allosteric networks. (C) Hypothetical energy landscape of TRiC, illustrating its conformational transitions during the ring closure process from TS1 to TS7 state. For TS2a, TS2b, TS3a, and TS3b, there are 2 maps in a similar conformation; thus, the names for both maps are listed underneath a representative better resolved map. (D) The conformational landscape of TRiC in the 0.05 mM, 0.1 mM, and 0.2 mM ADP-AlFx conditions.
Three Sets of “Gears” in the Allosteric Transitions of TRiC.
Our results suggest that ATP hydrolysis drives the allosteric transition of TRiC through 3 sets of gears, from interactions within a subunit to intra- and inter-ring interactions (Fig. 7B). According to our proposed model, in network I (allostery within a subunit), during ATP hydrolysis, NSL and the Asp398 located on H11 can be recruited to attach ATP (Movie S2 and Fig. 6C), inducing an inward bending motion of the I domain, which could then be propagated to the A domain, leading it to tilt inward involving in the dome formation to close the ring. In network II (intra-ring allostery), ATP hydrolysis could drag the stem loops of most subunits toward the nucleotide (Fig. 6C), which, through the hand-in-hand signature (SI Appendix, Fig. S8A), could drive E domains to rotate inward toward the central cavity. This leads to a positive cooperativity among subunits within a ring (Fig. 7B). In network III (inter-ring allostery), the N termini of CCT4, CCT6, and CCT7 are directly involved in the inter-ring allosteric network; and the E-domain inward rotation could lead the CCT8 N terminus to approach the stem loop of CCT3′ in the opposite ring, and eventually lock them together (Figs. 5E and 7B). These interactions would add extra anchor points between the 2 rings and lock the complex in a tightly closed conformation. The 3 sets of gears are highly coordinated once the loaded ATP is “fired,” and can form the framework of the allostery of TRiC complex.
Conformational Landscape of TRiC from the Open to Closed States.
Conformational comparison of the 10 maps resolved here, combined with our previous NPP-TRiC and TRiC-AMP-PNP maps (29), showed that the 12 maps can be classified into 9 distinct TRiC states (termed TSs) (Fig. 7C), with 3 pairs (0.05-C1 versus 0.1-C1, 0.05-C3 versus 0.1-C2a, and 0.05-C4 versus 0.1-C2b) in similar conformations, respectively (Fig. 7C and SI Appendix, Fig. S4 E–G). Overall, our ensemble of cryo-EM maps define the conformational landscape of TRiC as it gradually binds more and eventually hydrolyzes nucleotide, suggesting a population shift from open to closed states with increased nucleotide concentration (Fig. 7D). Furthermore, we present a hypothetical energy landscape of the 9 distinct states of TRiC on the basis of ligand (nucleotide) binding status, complex stability, and population distribution (Fig. 7C). NPP-TRiC, with the least bound nucleotide and overall larger B-factors of the model (SI Appendix, Fig. S9E), represents the starting point in the ATPase cycle (TS1) while the 0.2-C1 map with fully loaded nucleotide, in tightly closed conformation and in high population distribution, may stand for the ending point in this process (TS7). Gradually increased nucleotide binding to more subunits could drive TRiC to climb surmountable barriers between the basins of the 2 states and stabilize the later state until the lowest level (Fig. 7C). There are 2 potential basins in TS2, both with 1 subunit showing conformational changes. We then placed the conformer with the larger population (TS2a, Fig. 7D) in the bottom of the basin while the other was in a slightly higher position (TS2b, Fig. 7C). TS3 is in a similar situation. Our study also indicates that TS5 (all subunits bind nucleotide except CCT4) could be a rate-limiting state, and only when the nucleotide concentration increases to about 0.2 mM, the last subunit CCT4 can bind and hydrolyze nucleotide, and then TRiC can climb through this barrier. This proposed TRiC energy landscape from open to closed states could delineate the key components that are needed to trace the mechanism of allosteric events, which may assist in drug discovery.
Positive Cooperativity of Yeast TRiC in Nucleotide Consumption and Ring Closure.
Whether group II chaperonins display positive or negative cooperativity between the 2 rings of a chaperonin has been a long-standing question. Our studies delineate that, for yeast TRiC, the same subunit in both rings appears to bind or hydrolyze nucleotide in a synchronized manner, suggesting an overall positive inter-ring cooperativity in terms of nucleotide binding and hydrolysis. While in some conformers, CCT7 or CCT2 may bind slightly more nucleotide in one ring than in the opposing ring at rather low nucleotide concentrations (0.05 to 0.1 mM), the overall trend for nucleotide binding appears to follow the same order in both rings. Furthermore, once the nucleotide concentration reaches a certain threshold (about 0.2 mM), the TRiC complex initiates hydrolysis of ATP, and both rings can simultaneously shut their chambers closed. Our structure of TRiC at 0.2 mM ATP also showed that both rings of TRiC are closed (Fig. 3C), substantiating our conclusion that the both-rings-closed conformation of TRiC exhibiting the inter-ring positive cooperativity does exist in the presence of natural nucleotide. Still, we could not exclude the possibility that there might be a hybrid conformation of TRiC, with one ring closed and the other ring open, which may transiently exist and is an indication of negative inter-ring cooperativity of chaperonins. However, at 0.2 mM ATP, even by reducing the reaction time to less than 1 min, we did not capture such a hybrid conformation under current technical conditions. Furthermore, we also resolved a negative staining EM map of yeast TRiC bound with yeast prefoldin (PFD) (SI Appendix, Fig. S10), the cochaperone that delivers cytoskeletal proteins to TRiC (46). Our TRiC-PFD map shows that PFD can simultaneously bind both ends of TRiC in yeast (SI Appendix, Fig. S10 C and D); a similar phenomenon was also observed in bovine TRiC-human PFD (47), and human TRiC-PFD (human TRiC can bind 1 or 2 human PFDs depending on the concentration of PFD) (48). In summary, both rings of TRiC could potentially interact with the cochaperone for receiving delivered substrate at the same time. Still, we would like to point out that the TRiC–PFD interaction could be independent of any cooperativity-related signaling. Unlike group I chaperonins that have a detachable lid, group II chaperonins, such as TRiC, evolved to form a built-in lid, statistically beneficial for simultaneous ring closure and efficient substrate folding. In the crowded environment of cells and with the sufficiently high physiological concentration of nucleotide (1 to 2 mM), it is possible that both rings of TRiC could perform substrate folding concurrently for efficient protein quality control.
In summary, our study presents a thorough picture of the conformational landscape of TRiC and delineates the trajectory for the stepwise nucleotide usage and ring closure of individual subunits. Our findings shed light on our understanding of the subunit specificity of TRiC. Furthermore, these data also suggest a positive inter-ring cooperativity of yeast TRiC in nucleotide consumption and ring closure, beneficial for efficient protein folding in the complex environment of eukaryotic cells. Our research demonstrates the power of high-resolution cryo-EM in delineating the allosteric trajectory and potential energy landscape of dynamic macromolecular machines, beneficial for future therapeutic strategy development.
Materials and Methods
Protein Purification.
Yeast TRiC was purified according to our published protocol (29). The supernatant of yeast lysate was incubated with calmodulin resin (GE Healthcare) overnight at 4 °C. Elution of TRiC was achieved by using elution buffer (20 mM Hepes, pH 7.5, 5 mM MgCl2, 0.1 mM EDTA, 50 mM NaCl, 1 mM DTT, 10% glycerol) with 2 mM EGTA. The pooled eluate containing TRiC was concentrated with a Millipore Ultrafree centrifugal filter device (100-kDa cutoff).
Cryo-EM Sample Preparation.
To prepare the sample of TRiC with 0.05 mM ADP-AlFx, the purified yeast TRiC was diluted by dilution buffer (50 mM Tris⋅HCl, pH 7.5, 50 mM KCl) to 1 mg/mL and incubated in the presence of 0.05 mM ADP, 5 mM MgCl2, 0.25 mM Al(NO3)3, and 1.5 mM NaF for 1 h at 30 °C prior to freezing, and a 2.2-μL aliquot of sample was applied to a glow-discharged holey carbon grid (R1.2/1.3, 200 mesh; Quantifoil). The grid was blotted with Vitrobot Mark IV (FEI, now Thermo Fisher Scientific) and then plunged into liquid ethane cooled by liquid nitrogen. To handle the preferred orientation problem usually associated with group II chaperonins, especially in the open state, the grid was pretreated with polylysine (29, 49). The TRiC samples containing 0.1 mM, 0.2 mM, and 0.5 mM ADP-AlFx were prepared, respectively, following similar procedures, but with the concentrations of ADP, Al(NO3)3, and NaF increased accordingly.
To prepare the sample of TRiC with 0.2 mM ATP, the purified yeast TRiC was diluted to 1 mg/mL in the presence of 0.2 mM ATP and 5 mM MgCl2. Then a 2.2-μL aliquot of this sample was applied to a glow-discharged holey carbon grid (R1.2/1.3, 200 mesh; Quantifoil), and the sample was frozen within 1 min after the addition of ATP. The TRiC-PFD sample was made by mixing TRiC with PFD (with a molar ratio of 1:6) at 4 °C for 30 min. The mixed sample was negatively stained with 0.75% UF.
A more detailed description of materials and methods used in this work can be found in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank J. Zhou and Z. Zhou (Institute of Biochemistry and Cell Biology) for providing haploid yeast and plasmids, and D. Li for suggestions on model building of the nucleotide. We thank the cryo-EM facility and the Database and Computing facility of the National Center for Protein Science Shanghai, and also the Protein Expression and Purification Facility for instrument support. This work was supported by National Basic Research Program of China Grant 2017YFA0503503; National Natural Science Foundation of China Grants 31670754, 31872714, and 31861143028; the Chinese Academy of Sciences (CAS) Major Science and Technology Infrastructure Open Research Projects; and CAS Shanghai Science Research Center Grants CAS-SSRC-YH-2015-01 and DSS-WXJZ-2018-0002.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Cryo-EM maps have been deposited in the Electron Microscopy Data Bank, https://www.ebi.ac.uk/pdbe/emdb/ (accession nos. EMD-0756–EMD-0767), and the associated models have been deposited in the Protein Data Bank, www.rcsb.org (accession nos. PDB-6KS6–PDB-6KS8, PDB-6KRE, and PDB-6KRD).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903976116/-/DCSupplemental.
References
- 1.Tam S., et al. , The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat. Struct. Mol. Biol. 16, 1279–1285 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khabirova E., et al. , The TRiC/CCT chaperone is implicated in Alzheimer’s disease based on patient GWAS and an RNAi screen in Aβ-expressing Caenorhabditis elegans. PLoS One 9, e102985 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pereira J. H., et al. , Mechanism of nucleotide sensing in group II chaperonins. EMBO J. 31, 731–740 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Llorca O., et al. , Eukaryotic chaperonin CCT stabilizes actin and tubulin folding intermediates in open quasi-native conformations. EMBO J. 19, 5971–5979 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Llorca O., et al. , Eukaryotic type II chaperonin CCT interacts with actin through specific subunits. Nature 402, 693–696 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Camasses A., Bogdanova A., Shevchenko A., Zachariae W., The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Mol. Cell 12, 87–100 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Plimpton R. L., et al. , Structures of the Gβ-CCT and PhLP1-Gβ-CCT complexes reveal a mechanism for G-protein β-subunit folding and Gβγ dimer assembly. Proc. Natl. Acad. Sci. U.S.A. 112, 2413–2418 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kasembeli M., et al. , Modulation of STAT3 folding and function by TRiC/CCT chaperonin. PLoS Biol. 12, e1001844 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trinidad A. G., et al. , Interaction of p53 with the CCT complex promotes protein folding and wild-type p53 activity. Mol. Cell 50, 805–817 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McClellan A. J., Scott M. D., Frydman J., Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell 121, 739–748 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Minegishi Y., et al. , CCT2 mutations Evoke Leber congenital Amaurosis due to chaperone complex instability. Sci. Rep. 6, 33742 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guest S. T., Kratche Z. R., Bollig-Fischer A., Haddad R., Ethier S. P., Two members of the TRiC chaperonin complex, CCT2 and TCP1 are essential for survival of breast cancer cells and are linked to driving oncogenes. Exp. Cell Res. 332, 223–235 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Huang X., et al. , Chaperonin containing TCP1, subunit 8 (CCT8) is upregulated in hepatocellular carcinoma and promotes HCC proliferation. APMIS 122, 1070–1079 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Spiess C., Miller E. J., McClellan A. J., Frydman J., Identification of the TRiC/CCT substrate binding sites uncovers the function of subunit diversity in eukaryotic chaperonins. Mol. Cell 24, 25–37 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Llorca O., et al. , Analysis of the interaction between the eukaryotic chaperonin CCT and its substrates actin and tubulin. J. Struct. Biol. 135, 205–218 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Rivenzon-Segal D., Wolf S. G., Shimon L., Willison K. R., Horovitz A., Sequential ATP-induced allosteric transitions of the cytoplasmic chaperonin containing TCP-1 revealed by EM analysis. Nat. Struct. Mol. Biol. 12, 233–237 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Amit M., et al. , Equivalent mutations in the eight subunits of the chaperonin CCT produce dramatically different cellular and gene expression phenotypes. J. Mol. Biol. 401, 532–543 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Gruber R., Levitt M., Horovitz A., Sequential allosteric mechanism of ATP hydrolysis by the CCT/TRiC chaperone is revealed through Arrhenius analysis. Proc. Natl. Acad. Sci. U.S.A. 114, 5189–5194 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cong Y., et al. , 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement. Proc. Natl. Acad. Sci. U.S.A. 107, 4967–4972 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pereira J. H., et al. , Crystal structures of a group II chaperonin reveal the open and closed states associated with the protein folding cycle. J. Biol. Chem. 285, 27958–27966 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ditzel L., et al. , Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT. Cell 93, 125–138 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Shomura Y., et al. , Crystal structures of the group II chaperonin from Thermococcus strain KS-1: Steric hindrance by the substituted amino acid, and inter-subunit rearrangement between two crystal forms. J. Mol. Biol. 335, 1265–1278 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Zhang J., et al. , Mechanism of folding chamber closure in a group II chaperonin. Nature 463, 379–383 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dekker C., et al. , The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins. EMBO J. 30, 3078–3090 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muñoz I. G., et al. , Crystal structure of the open conformation of the mammalian chaperonin CCT in complex with tubulin. Nat. Struct. Mol. Biol. 18, 14–19 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Martín-Benito J., et al. , The inter-ring arrangement of the cytosolic chaperonin CCT. EMBO Rep. 8, 252–257 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cong Y., et al. , Symmetry-free cryo-EM structures of the chaperonin TRiC along its ATPase-driven conformational cycle. EMBO J. 31, 720–730 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Booth C. R., et al. , Mechanism of lid closure in the eukaryotic chaperonin TRiC/CCT. Nat. Struct. Mol. Biol. 15, 746–753 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zang Y., et al. , Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM. Nat. Struct. Mol. Biol. 23, 1083–1091 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Leitner A., et al. , The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure 20, 814–825 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalisman N., Adams C. M., Levitt M., Subunit order of eukaryotic TRiC/CCT chaperonin by cross-linking, mass spectrometry, and combinatorial homology modeling. Proc. Natl. Acad. Sci. U.S.A. 109, 2884–2889 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zang Y., et al. , Development of a yeast internal-subunit eGFP labeling strategy and its application in subunit identification in eukaryotic group II chaperonin TRiC/CCT. Sci. Rep. 8, 2374 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reissmann S., Parnot C., Booth C. R., Chiu W., Frydman J., Essential function of the built-in lid in the allosteric regulation of eukaryotic and archaeal chaperonins. Nat. Struct. Mol. Biol. 14, 432–440 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korobko I., Nadler-Holly M., Horovitz A., Transient kinetic analysis of ATP hydrolysis by the CCT/TRiC chaperonin. J. Mol. Biol. 428, 4520–4527 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Shimon L., Hynes G. M., McCormack E. A., Willison K. R., Horovitz A., ATP-induced allostery in the eukaryotic chaperonin CCT is abolished by the mutation G345D in CCT4 that renders yeast temperature-sensitive for growth. J. Mol. Biol. 377, 469–477 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Han H., et al. , Binding of substrates to the central pore of the Vps4 ATPase is autoinhibited by the microtubule interacting and trafficking (MIT) domain and activated by MIT interacting motifs (MIMs). J. Biol. Chem. 290, 13490–13499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang H., et al. , Physiological levels of ATP negatively regulate proteasome function. Cell Res. 20, 1372–1385 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soini J., et al. , Transient increase of ATP as a response to temperature up-shift in Escherichia coli. Microb. Cell Fact. 4, 9 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalisman N., Schröder G. F., Levitt M., The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning. Structure 21, 540–549 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nørby J. G., Coupled assay of Na+,K+-ATPase activity. Methods Enzymol. 156, 116–119 (1988). [DOI] [PubMed] [Google Scholar]
- 41.Joachimiak L. A., Walzthoeni T., Liu C. W., Aebersold R., Frydman J., The structural basis of substrate recognition by the eukaryotic chaperonin TRiC/CCT. Cell 159, 1042–1055 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reissmann S., et al. , A gradient of ATP affinities generates an asymmetric power stroke driving the chaperonin TRIC/CCT folding cycle. Cell Rep. 2, 866–877 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamamoto Y. Y., et al. , Asymmetry in the function and dynamics of the cytosolic group II chaperonin CCT/TRiC. PLoS One 12, e0176054 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou J., Giannakakou P., Targeting microtubules for cancer chemotherapy. Curr. Med. Chem. Anticancer Agents 5, 65–71 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Noormohammadi A., et al. , Somatic increase of CCT8 mimics proteostasis of human pluripotent stem cells and extends C. elegans lifespan. Nat. Commun. 7, 13649 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hansen W. J., Cowan N. J., Welch W. J., Prefoldin-nascent chain complexes in the folding of cytoskeletal proteins. J. Cell Biol. 145, 265–277 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martín-Benito J., et al. , Structure of eukaryotic prefoldin and of its complexes with unfolded actin and the cytosolic chaperonin CCT. EMBO J. 21, 6377–6386 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gestaut D., et al. , The chaperonin TRiC/CCT associates with prefoldin through a conserved electrostatic interface essential for cellular proteostasis. Cell 177, 751–765.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding Z., et al. , High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx. Cell Res. 27, 373–385 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.