Significance
Meristems are highly regulated groups of stem cells that are ultimately responsible for the formation of all branches, lateral organs, and stems in plants, and thus directly affect plant architecture and crop yield. We have identified a highly conserved member of a family of mitochondria-localized proteases that regulates maize inflorescence architecture. Unlike in Arabidopsis, in which its function appears dispensable, we have discovered that this protein is required for maize growth and productivity in field conditions and in high temperatures, and we provide evidence that it maintains reproductive meristem redox status and auxin homeostasis. These data highlight the importance of meristem redox status for sustaining maize organogenesis in challenging environments.
Keywords: maize, meristems, FTSH, ROS, auxin
Abstract
Meristems are highly regulated structures ultimately responsible for the formation of branches, lateral organs, and stems, and thus directly affect plant architecture and crop yield. In meristems, genetic networks, hormones, and signaling molecules are tightly integrated to establish robust systems that can adapt growth to continuous inputs from the environment. Here we characterized needle1 (ndl1), a temperature-sensitive maize mutant that displays severe reproductive defects and strong genetic interactions with known mutants affected in the regulation of the plant hormone auxin. NDL1 encodes a mitochondria-localized ATP-dependent metalloprotease belonging to the FILAMENTATION TEMPERATURE-SENSITIVE H (FTSH) family. Together with the hyperaccumulation of reactive oxygen species (ROS), ndl1 inflorescences show up-regulation of a plethora of stress-response genes. We provide evidence that these conditions alter endogenous auxin levels and disrupt primordia initiation in meristems. These findings connect meristem redox status and auxin in the control of maize growth.
Plant developmental plasticity depends on the maintenance of various groups of stem cells called meristems. In particular, shoot architecture in higher plants is established by apical meristems, such as shoot apical meristems (SAMs) and inflorescence meristems (IMs), and axillary meristems (AMs), initiating at the axils of true or modified leaves. In crop species such as maize, IM and AM activity determines inflorescence architecture, thus directly impacting reproductive potential and yield.
Several plant hormones such as auxin and cytokinin, as well as environmental factors, influence meristem function, which entails initiating lateral primordia at the peripheral zone (PZ) while maintaining a stem cell core in the central zone (CZ) of the meristem (1). While cytokinins are necessary for meristem maintenance, auxins are required for primordia initiation, and the action of both is key to meristem activity (2). Recently, the redox status of the SAM, and in particular the balance between superoxide (O2−) and hydrogen peroxide (H2O2) in the CZ and PZ, have been proposed to integrate stress signals with core meristematic functions (3). These findings complement previous studies whereby mutants affected in meristem redox status show developmental defects (4–9). Understanding how meristem redox regulation is integrated into both transcriptional and hormonal networks is crucial for comprehending plant stress adaptation (10, 11).
Auxins play many roles in plant development. Local auxin concentration gradients are necessary for organ formation, and the dynamic regulation of auxin transport establishes the patterning of inflorescences and reproductive structures (2). In maize, mutants defective in AM initiation and development show a reduction in inflorescence branching and spikelet formation (grass-specific structures containing florets), and often are caused by mutations in auxin-related genes. For example, SPARSE INFLORESCENCE1 (SPI1) encodes a maize ortholog of Arabidopsis YUCCAs, enzymes that function in the main auxin biosynthetic pathway in plants (12, 13), and BARREN INFLORESCENCE1 and 4 (BIF1 and BIF4) encode AUX/IAAs, negative regulators of auxin signaling (14). Here we show that NDL1 encodes a mitochondrial-localized m-AAA protease of the FTSH family (15), and that ndl1 mutants condition high ROS levels that affect auxin homeostasis and consequently reproductive organogenesis.
Results
needle1 Is a Temperature-Sensitive Mutant Affected in Reproductive Organogenesis.
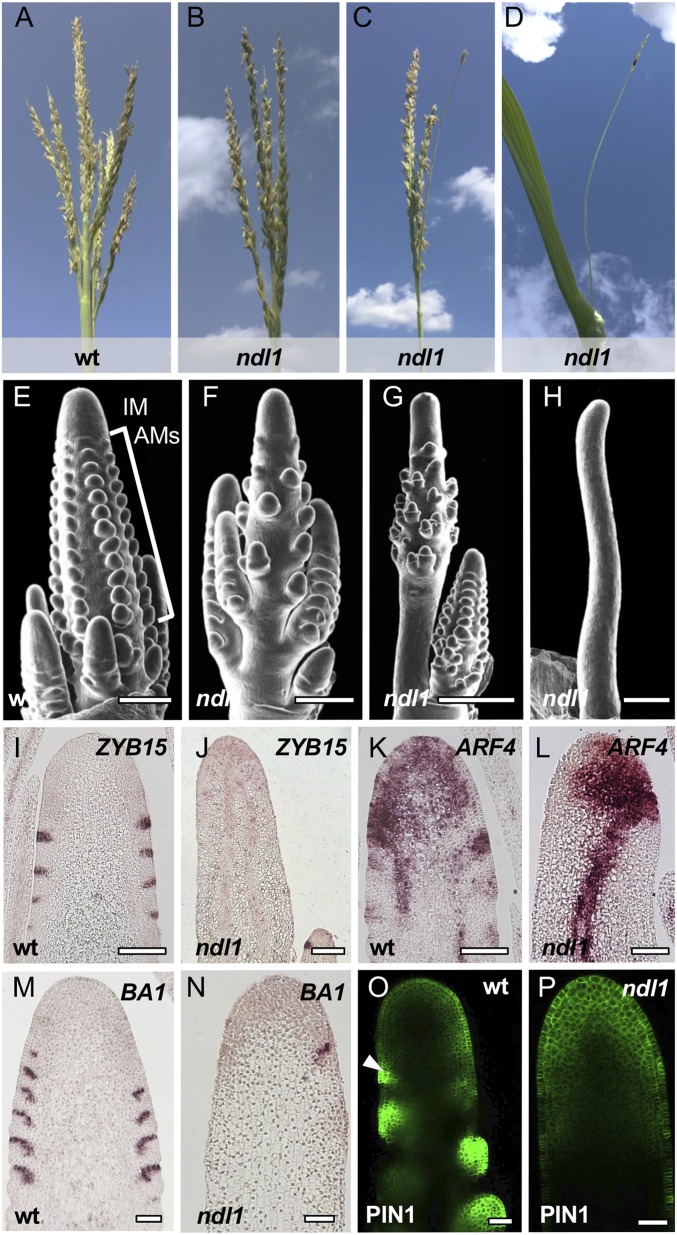
A single allele of the ndl1 mutant (ndl1-ref) was originally identified in an EMS mutagenesis screen for maize inflorescence mutants. Segregation in M2 populations indicated that the phenotype was caused by a single recessive mutation. After introgression into a homogeneous B73 background, homozygous ndl1 plants showed a range of inflorescence phenotypes, with the strongest defects observed in the tassel, where phenotypes ranged from tassels with a slightly reduced number of branches and spikelets to those with no branches and spikelets on the main spike; ears of ndl1 plants were marginally shorter than wild type and occasionally showed disorganized rows of kernels (Fig. 1 A–D and SI Appendix, Fig. S1).
Fig. 1.
ndl1 is defective in reproductive organogenesis. (A–D) The ndl1 tassel phenotype is very variable, from mild (B) to severe (D). (E–H) SEMs of immature tassels in normal and mutant plants also show different expressivity of the ndl1 mutation. AMs, axillary meristems; IM, inflorescence meristem. (Scale bars, 500 μm.) (I–N) mRNA in situ hybridizations of immature tassels with ZYB15, ARF4, and BA1 antisense probes. (Scale bars, 250 μm.) (O and P) Confocal images of normal and ndl1 immature tassels expressing ZmPIN1a-YFP; arrowhead points to PIN1-YFP up-regulation at the site of primordia initiation. (Scale bars, 100 μm.)
The variable expressivity of the ndl1 tassel phenotype was observed in both field and greenhouse conditions; however, the appearance of a strong phenotype in field-grown plants was more likely to occur in late planting fields, i.e., June instead of May, with ∼10 °F higher temperatures (SI Appendix, Fig. S2). To test the hypothesis that temperature was influencing the expressivity of the ndl1 phenotype, we grew ndl1 plants in a growth chamber at mild (24 °C day/20 °C night) and high temperatures (32 °C day/28 °C night). At high temperatures, over 30% of ndl1 plants arrested growth after producing a few leaves, while the remaining ndl1 plants transitioned to flowering but produced tassels with significantly fewer branches than wild type plants (SI Appendix, Fig. S2). None of these phenotypes were observed at mild temperature. We also determined that ndl1 mutants displayed significantly shorter primary roots when grown at increasingly higher temperatures than wild type plants grown in the same conditions (SI Appendix, Fig. S1). These results indicate that ndl1-ref is a temperature-sensitive mutant that shows vegetative and reproductive defects at high temperatures.
To characterize ndl1 inflorescence defects, we used SEM in the early stages of tassel and ear development. In wild type plants, the IM gives rise to a series of reproductive AMs, which subsequently produce spikelets and florets (Fig. 1E). In severe ndl1 tassels, no AMs were visible, whereas in tassels with a weak phenotype, the regular initiation and arrangements of AMs along the inflorescence axis was partially disrupted (Fig. 1 F–H). In ndl1 ears, we observed milder defects in AM initiation compared with tassels, and IMs occasionally showed slight fasciation (SI Appendix, Fig. S1). Altogether, these results suggest that NDL1 functions in AMs initiation as well as in IM maintenance.
In maize inflorescences, the early steps of primordia initiation occur within the PZ of the IM, where suppressed bracts (SBs), the first visible primordia, subtend newly initiating AMs. We used antisense probes for the maize ZYB15, BA1, and ARF4 genes whose expression marks SBs, boundary domains, and meristems, respectively (Fig. 1 I, K, and M), to analyze the early steps of reproductive organogenesis (14, 16). In ndl1 immature tassels, the regular expression patterns of ZYB15 and BA1 were absent in the PZ of the IM (Fig. 1 I, J, M, and N). Similarly, ARF4, whose expression is typically observed in both the IM and the PZ in wild type (14), was expressed normally in the ndl1 IM but absent in the PZ (Fig. 1 K and L).
Polar auxin transport is necessary for maize AM initiation and inflorescence patterning (17, 18), and ZmPIN1a is a membrane-localized auxin efflux transporter whose up-regulation at the PZ of IMs marks newly initiating primordia. We therefore monitored the expression of ZmPIN1a-YFP in wild type and ndl1 immature tassels. A strong up-regulation of ZmPIN1a-YFP was detected on the flank of wild type IMs, which overlapped with SB and AM initiation sites as previously reported (Fig. 1O) (18). This patterning was absent in phenotypically severe ndl1 tassels (Fig. 1P), while, in mild ndl1 tassels, ZmPIN1a-YFP localization resembled the wild type pattern of expression (SI Appendix, Fig. S3). Overall, our results indicate that NDL1 is essential for the early stages of reproductive organogenesis leading to the initiation of SBs and AMs.
ndl1 inflorescence defects were reminiscent of previously described maize mutants affected in auxin biology (12, 14, 17, 19). This observation together with the disruption of ZmPIN1a-YFP expression in ndl1 tassels suggested that NDL1 may affect auxin-related processes. We therefore examined the genetic interaction between ndl1 and auxin-related mutants defective in signaling and biosynthesis. We observed a strong genetic interaction between ndl1 and Bif1, a semidominant auxin signaling mutant caused by a stabilizing mutation in ZmIAA27 (14). A similarly strong interaction was seen between ndl1 and spi1, an auxin biosynthetic mutant (12). In both cases, ndl1 enhanced the inflorescence phenotype of +/Bif1 and spi1/spi1 mutants (SI Appendix, Fig. S4). Double-mutant tassels showed a significant reduction in branch and spikelet-pair number compared with single mutants, while long barren tips in the ears of double mutants were observed (SI Appendix, Fig. S4). A similar ear phenotype was also seen when ndl1 was crossed to semidominant auxin signaling mutant Bif4 (14) (SI Appendix, Fig. S4). Overall, these genetic interactions suggest a functional link between NDL1 and auxin in regulating maize inflorescence development.
NDL1 Encodes a Mitochondrial Localized FTSH Protein.
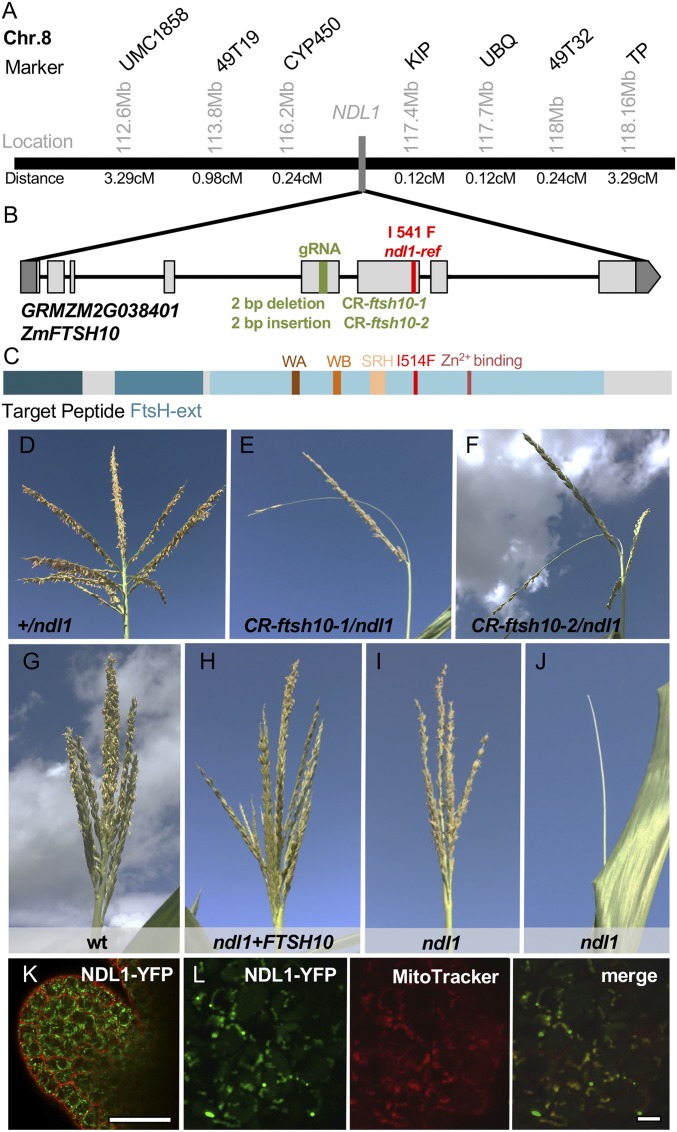
By positional cloning, the ndl1-ref allele was mapped to a 1.2-Mb window on chromosome 8, which contained 27 predicted genes (Fig. 2A). We then performed bulked segregant RNA-seq analysis (20) and identified an A-to-T transversion that was present only in the ndl1 mutant within the coding region of GRMZM2G038401, a gene encoding a protein homologous to FTSH, a highly conserved mitochondrial localized protein present in all organisms from bacteria to humans. The resulting amino acid substitution in ndl1 (I541F) corresponded to a residue that was highly conserved (invariably I or L) in both prokaryotic and eukaryotic organisms (SI Appendix, Fig. S5). We then sequenced the candidate gene in 29 inbred lines and examined 900 lines from the maize HapMap 3.1 for SNPs in GRMZM2G038401 (21) and determined that the A>T SNP was present only in the ndl1-ref mutant, suggesting that it was a mutation rather than an extant polymorphism.
Fig. 2.
NDL1 encodes ZmFTSH10. (A) Positional cloning of NDL1. Molecular markers used and physical (Mb) and genetic distances (cM) are indicated. (B) Schematic representation of the ZmFTSH10 gene and the position of the mutant alleles. Exons and UTRs are depicted as light gray and dark gray rectangles, respectively. The green bar indicates the guide RNA targeting site. The red bar positions the missense mutation. (C) Schematic representation of the ZmFTSH10 protein. FtsH-ext, FtsH-extracellular domain (Pfam); SRH, Second Region of Homology motif; WA, Walker A motif; WB, Walker B motif. (D–F) The tassel phenotype of wild type and ndl1 mutants generated by CRISPR-Cas9. (G–J) The variable tassel phenotype of ndl1 (I and J) is rescued by the pZmFTSH10::FTSH10-YFP construct. (K) Confocal images of an ear AM expressing FTSH10-YFP, counterstained with propidium iodide. (Scale bar, 50 μm.) (L) Confocal images of immature ear AMs expressing NDL1-YFP stained with MitoTracker Red CMXRos. (Scale bars, 10 μm.)
We pursued 2 complementary approaches to confirm that GRMZM2G038401, hereafter referred to as ZmFTSH10, corresponded to NDL1. First, we used a CRISPR-Cas9–based strategy with a gRNA targeting exon 5 of ZmFTSH10 to create additional mutant alleles (Fig. 2B). T1 transgenic plants containing the CRISPR-Cas9 vector were crossed to ndl1-ref homozygous mutants. A barren tassel phenotype was observed in several individual plants in the resulting F1s heterozygous for the ndl1-ref mutation (Fig. 2 D–F and SI Appendix, Fig. S6). Sequencing of ZmFTSH10 in these plants identified 4 different Cas9-induced frame-shift insertions and deletions (SI Appendix, Fig. S6), confirming that mutations in ZmFTSH10 caused the aberrant tassel phenotype. We also generated transgenic plants containing a pZmFTSH10::FTSH10:YFP construct and crossed it to ndl1-ref homozygous mutants. Four independent events expressing the ZmFTSH10-YFP fusion protein were capable of fully complementing the ndl1-ref phenotype (Fig. 2G and SI Appendix, Fig. S7), further confirming that NDL1 corresponds to ZmFTSH10/GRMZM2G038401.
Based on publicly available RNA-seq data and qRT-PCR in different tissues, NDL1 is expressed ubiquitously in all tissues (SI Appendix, Fig. S8), and confocal analysis of pZmFTSH10::FTSH10:YFP transgenic lines showed that NDL1-YFP was present in all tissues examined, appearing as multiple punctate dots within each cell (Fig. 2K and SI Appendix, Fig. S8). Simultaneous imaging of NDL1-YFP with a mitochondrial marker in maize transgenic lines showed colocalization of the 2 signals, indicating that NDL1 was targeted to mitochondria as predicted (Fig. 2L).
FTSHs are ATP-dependent metalloproteases belonging to the AAA protein super family and ensure membrane protein quality control via proteolytic and chaperone-like activities (15). In eukaryotes, FTSHs are nuclear-encoded proteins targeted to chloroplasts and/or mitochondria. Mitochondrial FTSHs are grouped into the m-AAA and i-AAA classes, both localized in the inner membrane system: the m-AAA class has the active site facing the mitochondria matrix, whereas the i-AAA class exposes its catalytic sites to the inner membrane space. Arabidopsis contains 12 FTSH proteins, 4 of which are targeted to mitochondria via an N-terminal target peptide, including 2 m-AAA proteases, AtFTSH3 and AtFTSH10, and 2 i-AAA proteases, AtFTSH4 and AtFTSH11 (22, 23). The maize B73v3 reference genome contains 12 FTSH members. Neighbor-joining analysis placed NDL1 within the clade of m-AAA proteases that includes AtFTSH3 and AtFTSH10 and yeast YTA10 and YTA12 (SI Appendix, Fig. S9). Overall, NDL1 shares 74% and 71% identity and 82% and 78% similarity with AtFTSH3 and AtFTSH10, respectively. Two additional partially truncated genes, LOC103629414 and LOC109941663, encoding proteins with ∼300 amino acids similar with the NDL1 N terminus, were also identified (SI Appendix, Fig. S10). However, both genes lacked sequence encoding all C-terminal domains, and they are likely nonfunctional. Therefore, NDL1 appears to be the only functional maize m-AAA protease.
To test whether the role of NDL1 in reproductive development was conserved, we analyzed Arabidopsis Atftsh3 and Atftsh10 mutants. We first obtained a T-DNA insertion line in AtFTSH3 (SALK_037144) that knocked out AtFTSH3 expression (SI Appendix, Fig. S11). Subsequently, we used CRISPR-Cas9 to edit AtFTSH10 in the ftsh3 insertion line background. A single T2 line, CR-ftsh10, carrying a 4-bp deletion in the coding region of AtFTSH10, was selected for further analysis (SI Appendix, Fig. S11). No specific inflorescence phenotype was observed in Atftsh3;Atftsh10 double mutant relative to wild type plants, even when grown at elevated temperatures. However, double-mutant Atftsh3;Atftsh10 plants showed a reduction in primary root length when compared with wild type, a result consistent with a recent report (23), and, similar to ndl1, the root phenotype was enhanced at higher temperatures (SI Appendix, Fig. S11). These results suggest that maize reproductive development may be more sensitive to the absence of ZmFTSH10 activity than Arabidopsis.
Like other eukaryotic FTSH proteins, NDL1 contains an N-terminal mitochondrial target peptide and several conserved domains (Fig. 2C). These include an N-terminal FTSH extracellular domain (Pfam) flanked by 2 transmembrane regions and a middle region comprising an AAA+ domain containing Walker A and B motifs and an SRH motif, which are crucial for ATPase function. The C terminus contains a peptidase domain containing a Zn2+ binding HEXXH motif. The I541F mutation of the ndl1-ref mutant corresponds to a highly conserved residue located within the AAA+ lid domain (Pfam), which is important for ATPase activity (24) (SI Appendix, Fig. S10). We therefore measured the ATPase activity of recombinant NDL1 and ndl1-ref proteins and determined that the I541F mutation caused ∼50% decrease in NDL1 ATPase activity independently of temperature (SI Appendix, Fig. S5), indicating that the temperature sensitivity of ndl1 mutants was not specific to the ndl1-ref allele, but a common feature of mutants in this protein family (5, 23).
Alternative Respiration Complex and ROS-Related Genes Are Up-Regulated in ndl1.
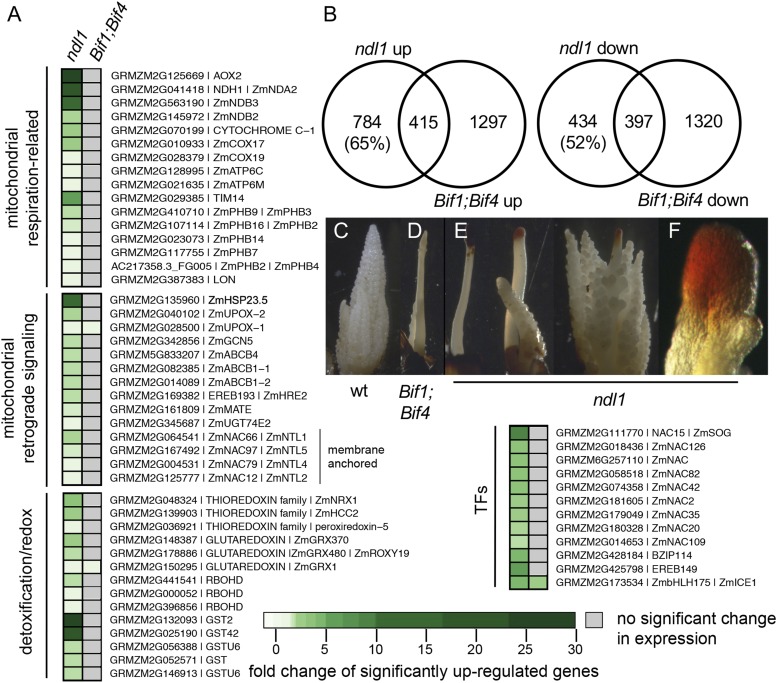
To better understand the molecular phenotype of ndl1, we performed RNA-seq on immature tassels and identified 1,199 up- and 831 down-regulated genes in ndl1 mutants compared with wild type. In agreement with the role of plant m-AAA proteases in the stability and assembly of mitochondrial oxidative phosphorylation complexes, up-regulated genes in ndl1 included many encoding mitochondrial-localized proteins such as those involved in mitochondria proficiency (i.e., PROHIBITINS; PHBs), as well as components of the electron transport chain (OXPHOS pathway) such as AOX (ALTERNATIVE OXIDASE), type II NADPH dehydrogenases (NDBs/NDHs), COX17/COX19, and CYTC-1 (Fig. 3A) (25). AOX and NDBs are involved in alternative respiration pathways that serve as plant-specific bypasses of the electron transport chain and were among the highest up-regulated genes (SI Appendix, Table S1). Strong up-regulation was also observed in genes involved in oxidative stress and redox/detoxification such as RESPIRATORY BURST OXYGEN (RBOHD), GLUTAREDOXINS (GRXs), and several GST-encoding genes (Fig. 3A and SI Appendix, Table S1).
Fig. 3.
ndl1 inflorescences are stressed. (A) Genes significantly up-regulated in ndl1 and Bif1;Bif4 RNA-seq tassel datasets. Gray boxes indicate no significant expression difference observed relative to wild type (FDR < 0.05). (B) Overlap between differentially expressed genes (FDR < 0.05) in ndl1 dataset and Bif1;Bif4 dataset. (C–F) DAB staining of immature tassels to measure H2O2 accumulation. Strong DAB stain is observed only in IMs showing a very severe phenotype, but not in BMs where AMs are forming. Close-up view of IM in F.
Arabidopsis homologs of these genes are implicated in mitochondrial retrograde signaling, which communicates suboptimal mitochondria status with the nucleus to restore cellular homeostasis (26, 27). ndl1 tassels showed 26-fold up-regulation of the AOX2 gene (AOX1a in Arabidopsis), which is known to counteract excess ROS caused by stress or mitochondrial defects in complex I (28, 29) (Fig. 3A). Only AOX2, but not other AOX members that act in different complexes (29), was up-regulated, indicating that NDL1 may be specifically involved in complex I assembly (SI Appendix, Fig. S12). Furthermore, many genes homologous to other retrograde signaling marker genes (Fig. 3A) (30, 31) were significantly up-regulated, including small heat shock protein ZmHSP23.5, UP-REGULATED BY OXIDATIVE STRESS (UPOX) genes, and several PHBs, which physically interact with m-AAA proteases (32) (Fig. 3A). Comparison of transcriptional profiles of Arabidopsis phb3 mutant roots (33) and maize ndl1 tassels showed over 100 orthologous genes that were up-regulated in both mutants (SI Appendix, Table S1 and Dataset S1), indicating that compromised NDL1 function results in a similar transcriptional response to that seen in Arabidopsis under various mitochondrial perturbations, and suggesting conserved retrograde signaling components between these 2 distantly related species. Additional genes that may be involved in mitochondrial stress response include ZmNAC15, a homolog of an Arabidopsis DNA damage response regulator (34), and ZmEREB149, a close homolog of AtERF114 that mediates ROS signaling (33) (Fig. 3A and SI Appendix, Table S1).
To understand whether the transcriptional changes observed in ndl1 tassels were a consequence of the severe inflorescence phenotype, we also performed RNA-seq on immature tassels of the Bif1;Bif4 auxin signaling mutant, which displays a similar phenotype to ndl1 (14), and then carried out comparative transcriptomic analysis. Over 65% (784 of 1,199) of up-regulated genes and 52% (434 of 8,31) of down-regulated genes in the ndl1 dataset were not found in Bif1;Bif4, reflecting largely distinct molecular signatures for each mutant (Fig. 3B). Genes specifically up-regulated in ndl1 but not Bif1;Bif4 included all members of the OXPHOS pathway, certain GSTs, NACs, and EREB149 (Fig. 3A). GO analysis of genes differentially expressed in both ndl1 and Bif1;Bif4, ndl1-only, or Bif1;Bif4-only mutants showed enrichment for the terms “response to chemical, biotic and abiotic stimulus” and “response to stress” for most subsets (SI Appendix, Fig. S12). Down-regulated genes found in both the shared and Bif1;Bif4 datasets included those involved in early reproductive development such as BA1 (16), likely absent due to lack of AMs in both mutants (SI Appendix, Fig. S12). On the contrary, down-regulated genes found only in Bif1;Bif4 showed strong enrichment for “chromatin assembly” and “protein folding” supporting a link between auxin signaling and changes in chromatin state (35) (SI Appendix, Fig. S12).
Since mitochondria are a main source of ROS, and OXPHOS complex defects increase ROS production (36), we performed DAB staining to examine H2O2 levels in immature tassels (Fig. 3 C–F). No staining was observed in wild type or Bif1;Bif4 tassels (Fig. 3 C and D), but strong accumulation was detected at the tip of ndl1 IMs displaying a strong barren phenotype but not in those with a less severe phenotype (Fig. 3E and SI Appendix, Fig. S13). DAB staining was observed only in the IM of the main spike and not in the middle, base, or in secondary branches, suggesting that the up-regulation of AOX2 or scavenging enzymes (28) may quickly restore the redox status in these organs. These results are consistent with the hypothesis that NDL1 functions in maintaining OXPHOS complex activity.
ndl1 Alters Endogenous Auxin Levels and Responses.
Given the role of auxin in the formation of lateral primordia and the synergistic interaction between ndl1 and several auxin mutants, NDL1 may regulate inflorescence development via cross-talk with auxin-related pathways. We therefore examined differentially expressed genes involved in auxin signaling, transport, and homeostasis in ndl1 and Bif1;Bif4 RNA-seq datasets, and observed that many genes including PINs, ARFs, AUX/IAAs, and GH3s were differentially expressed in both mutants. In general, genes within this subset of auxin-related genes showed similar transcriptional responses. For example, both mutants showed significant up-regulation of the auxin transporter gene ZmPIN10 as well as mis-regulation of several GH3 genes involved in auxin inactivation, indicating that auxin transport and homeostasis are similarly perturbed. On the contrary, several Aux/IAA genes were differentially expressed only in one mutant but not the other, suggesting that Aux/IAAs may function in specific contexts (SI Appendix, Fig. S13).
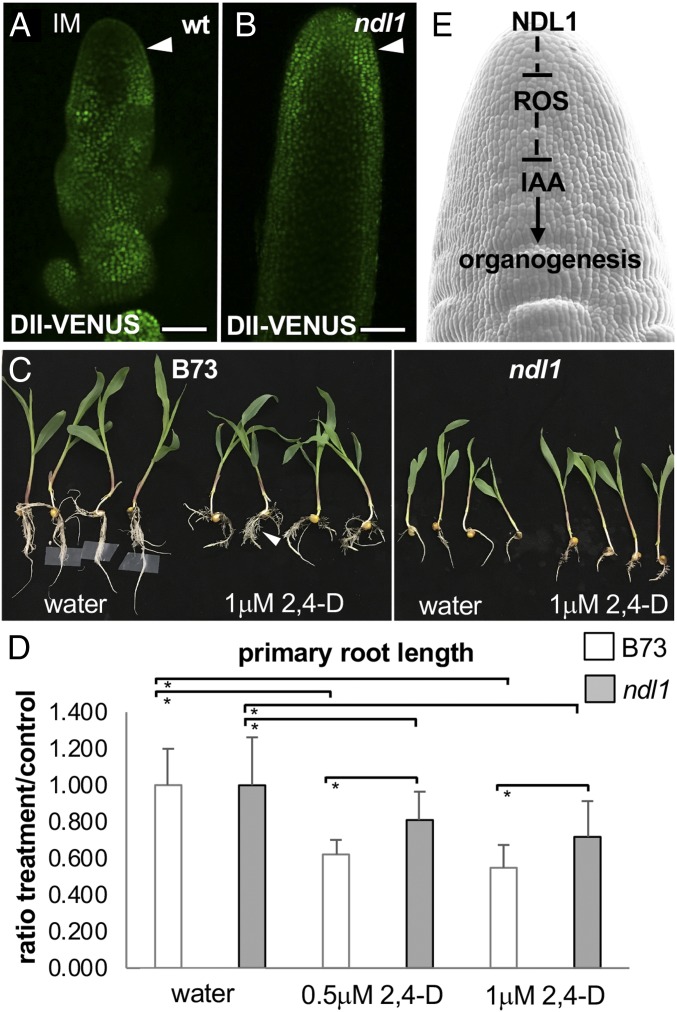
The strong genetic interactions observed with Bif1, which encodes a stabilized AUX/IAA protein insensitive to auxin-induced degradation (14), and with spi1, an auxin biosynthetic mutant (SI Appendix, Fig. S4), suggested that, in ndl1 IMs, the levels of free auxin may be decreased and thus cause early defects in organogenesis. To test this hypothesis, we crossed ndl1 to DII-VENUS, a marker line based on the degron domain of AUX/IAA proteins, whose degradation is controlled by auxin levels in cells (37, 38). In wild type tassels, low expression of DII-VENUS was detected in the IM, indicating relatively high concentration of endogenous auxin. In ndl1 tassels with a severe phenotype, however, the DII-VENUS signal was stronger and uniformly distributed, indicating lower levels of auxin in ndl1 IMs and in particular in the PZ (Fig. 4 A and B and SI Appendix, Fig. S13). This was not observed in ndl1 tassels with a mild phenotype that resembled wild type DII-VENUS expression (SI Appendix, Fig. S13). We also investigated the response of ndl1 roots to applications of 2,4D, a synthetic auxin analog, and measured the primary root length of treated and untreated wild type and ndl1 mutants. 2,4D applications caused a significant reduction in primary root length and a proliferation of lateral roots in wild type as expected (39), while ndl1 roots were less sensitive to the treatments (Fig. 4 C and D). These findings indicate that ndl1 is partially buffered from exogenous auxin applications, suggesting that more auxin is needed to achieve a similar response to wild type roots and that fully functional mitochondria are necessary for a normal auxin response.
Fig. 4.
Auxin-related defects in ndl1 mutants. (A and B) Maximum projection of DII-VENUS signals in wild type (A) and ndl1 mutant (B) immature tassels. IM, inflorescence meristem. Arrowheads point to the PZ where high DII-VENUS signal is observed in ndl1 but not in wild type. (Scale bars, 100 μm.) (C and D) ndl1 roots are less sensitive to exogenous 2,4D applications (n ≥ 34 per treatment; data from at least 2 biological replicates; error bars represent SD; *P < 0.0001, Student’s t test). (E) Schematic model of NDL1 function in organogenesis.
Discussion
Given their role in providing energy to cells and sensing environmental stresses, mitochondria are essential for plant growth (25). How mitochondrial status is integrated with developmental programs such as those controlled by auxin, however, remains unclear. Here we demonstrate that NDL1, a mitochondria-localized m-AAA protease, is necessary for thermotolerant growth of maize. Mitochondrial FTSH proteins are highly conserved across taxa at the sequence and functional levels, form oligomeric complexes, and, together with PHBs, are required for proper assembly and maintenance of mitochondrial oxidative phosphorylation complexes (15, 40). In ndl1 mutants, ROS accumulate to high levels in IMs, mitochondria switch to the alternative oxidase pathway, and meristem defects lead to altered inflorescence architecture. These phenotypes resemble those reported in related Arabidopsis mutants such as ftsh4 and the type-I prohibitin phb3, which show elevated ROS and AOX levels as well as SAM defects and increased shoot branching (5, 7, 33, 41). It is therefore surprising that Arabidopsis ftsh3;ftsh10 mutants did not show an obvious and specific reproductive phenotype but rather displayed only root growth defects (here and in ref. 23). This difference could reflect the fact that high-temperature field conditions are sufficient to trigger the ndl1 phenotype, while the relatively cool standard growth conditions of Arabidopsis are not. Indeed, ndl1 mutants grown at mild temperatures showed no phenotype. Only when grown at constant high temperatures in a controlled environment were ndl1 mutants also impaired in vegetative development, suggesting that the apparent inflorescence-specific phenotype of ndl1 mutants is due to the seasonal maize growth (i.e., germinating in mild temperatures and transitioning to reproductive development in warmer temperatures).
The effect on maize organogenesis caused by the ndl1 mitochondria-related defects resembled that seen in auxin-related mutants, and our analysis showed a synergistic interaction at the genetic level among mutants, a decrease in endogenous auxin levels and auxin transport in ndl1 inflorescences, and attenuated response to exogenous auxin applications in roots. Transcriptional analysis of ndl1 tassels showed the mis-regulation of many auxin-related genes, similar to those observed in the maize Bif1;Bif4 auxin signaling mutant (14). However, ndl1 displayed several unique phenotypes such as elevated accumulation of H2O2 in IMs and a large number of strongly up-regulated ROS-scavenger and redox-related genes, such as those belonging to the PEROXIDASE, GST, and the THIOREDOXIN families (Fig. 3A and SI Appendix, Fig. S12). ROS contribute to plant development and stress responses and may directly influence auxin homeostasis, transport, and signaling (10, 26), i.e., H2O2 was proposed to mediate auxin oxidative degradation via peroxidase activities (7, 42). Such a scenario agrees with lower auxin levels in ndl1 IMs with high ROS. However, we also observed several genes involved in auxin conjugation, such as ZmGH3.2 and ZmGH3.8 and UDP-glucosyl transferase genes (43), that were up-regulated in ndl1. One of the latter genes, UGT74E2, is often up-regulated in mitochondrial stress responses (44). Furthermore, high temperatures directly influence auxin levels (45), highlighting the inherent complexity of these interactions. We propose that NDL1 is required for maintaining the redox status of meristems to sustain maize growth at high temperatures (Fig. 4E). When impaired, stress signals unbalance auxin regulation at the PZ of IMs, which results in defective organogenesis. While heat stress enhances the ndl1 phenotype, it is likely that other stresses also influence it. Indeed, RNA-seq of ndl1 tassels showed up-regulation of many genes whose Arabidopsis homologs have been associated with mitochondrial stresses, dysfunction, and retrograde signaling (46), uncovering many genes likely to regulate maize growth under duress. Understanding how NDL1 and other mitochondrial stress-related genes function is essential to devise strategies for enhancing maize yield in suboptimal environments and harsher climates.
Materials and Methods
The ndl1 reference (ndl1-ref) allele was generated by EMS mutagenesis in the OH43 background by Gerald Neuffer and obtained from the Maize Genetics Cooperation Stock Center (stock 04HI-A632XOH43GN-173; original M2 population). Complete details regarding plant materials, experimental methods, and data analyses are provided in the SI Appendix.
Supplementary Material
Acknowledgments
The authors thank Dr. Gerald Neuffer for generating the ndl1-ref allele, the Maize Genetics Cooperation Stock Center for seeds, Renyta Moses and Gabriel Koslow for help with genotyping, Paula McSteen and the University of Missouri Columbia Plant Transformation Core Facility for transformation, Robert Schmitz and the Georgia University Genetic Department for RNA-seq sequencing, Yaping Feng for assistance with BSA RNA-seq, and Carolyn Rasmussen for DII-VENUS seeds and comments on the manuscript. Q.L. was supported by the China Scholarship Council and by the Waksman Charles and Johanna Busch Fellowship. This research was supported by grants from the National Science Foundation (IOS 1546873 and 1456950 to A.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The NDL1 coding sequence was deposited in GenBank, https://www.ncbi.nlm.nih.gov/genbank/ (accession no. MK674049) and corresponds to GRMZM2G038401/Zm00001d010522_T001 (B73 v3/v4). RNA-seq datasets were deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession nos. GSE129684 and GSE135468).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907071116/-/DCSupplemental.
References
- 1.Pfeiffer A., Wenzl C., Lohmann J. U., Beyond flexibility: Controlling stem cells in an ever changing environment. Curr. Opin. Plant Biol. 35, 117–123 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Shi B., Vernoux T., Patterning at the shoot apical meristem and phyllotaxis. Curr. Top. Dev. Biol. 131, 81–107 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Zeng J., Dong Z., Wu H., Tian Z., Zhao Z., Redox regulation of plant stem cell fate. EMBO J. 36, 2844–2855 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bashandy T., et al. , Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. Plant Cell 22, 376–391 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dolzblasz A., et al. , The mitochondrial protease AtFTSH4 safeguards Arabidopsis shoot apical meristem function. Sci. Rep. 6, 28315 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang F., et al. , A maize glutaredoxin gene, abphyl2, regulates shoot meristem size and phyllotaxy. Plant Cell 27, 121–131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang S., et al. , Perturbation of auxin homeostasis caused by mitochondrial FtSH4 gene-mediated peroxidase accumulation regulates arabidopsis architecture. Mol. Plant 7, 856–873 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Knuesting J., et al. , Arabidopsis glutaredoxin S17 and its partner, the nuclear factor Y subunit C11/negative cofactor 2α, contribute to maintenance of the shoot apical meristem under long-day photoperiod. Plant Physiol. 167, 1643–1658 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong L., et al. , Variable cell growth yields reproducible organ development through spatiotemporal averaging. Dev. Cell 38, 15–32 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Tognetti V. B., Bielach A., Hrtyan M., Redox regulation at the site of primary growth: Auxin, cytokinin and ROS crosstalk. Plant Cell Environ. 40, 2586–2605 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Schippers J. H., Foyer C. H., van Dongen J. T., Redox regulation in shoot growth, SAM maintenance and flowering. Curr. Opin. Plant Biol. 29, 121–128 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Gallavotti A., et al. , Sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc. Natl. Acad. Sci. U.S.A. 105, 15196–15201 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y., Auxin biosynthesis. Arabidopsis Book 12, e0173 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galli M., et al. , Auxin signaling modules regulate maize inflorescence architecture. Proc. Natl. Acad. Sci. U.S.A. 112, 13372–13377 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogura T., Wilkinson A. J., AAA+ superfamily ATPases: Common structure–Diverse function. Genes Cells 6, 575–597 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Gallavotti A., et al. , The role of barren stalk1 in the architecture of maize. Nature 432, 630–635 (2004). [DOI] [PubMed] [Google Scholar]
- 17.McSteen P., et al. , Barren inflorescence2 Encodes a co-ortholog of the PINOID serine/threonine kinase and is required for organogenesis during inflorescence and vegetative development in maize. Plant Physiol. 144, 1000–1011 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallavotti A., Yang Y., Schmidt R. J., Jackson D., The Relationship between auxin transport and maize branching. Plant Physiol. 147, 1913–1923 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phillips K. A., et al. , Vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 23, 550–566 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu S., Yeh C. T., Tang H. M., Nettleton D., Schnable P. S., Gene mapping via bulked segregant RNA-Seq (BSR-Seq). PLoS One 7, e36406 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bukowski R., et al. , Construction of the third-generation Zea mays haplotype map. Gigascience 7, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urantowka A., Knorpp C., Olczak T., Kolodziejczak M., Janska H., Plant mitochondria contain at least two i-AAA-like complexes. Plant Mol. Biol. 59, 239–252 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Kolodziejczak M., Skibior-Blaszczyk R., Janska H., m-AAA complexes are not crucial for the survival of arabidopsis under optimal growth conditions despite their importance for mitochondrial translation. Plant Cell Physiol. 59, 1006–1016 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Miller J. M., Enemark E. J., Fundamental characteristics of AAA+ protein family structure and function. Archaea 2016, 9294307 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Millar A. H., Whelan J., Soole K. L., Day D. A., Organization and regulation of mitochondrial respiration in plants. Annu. Rev. Plant Biol. 62, 79–104 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Kerchev P. I., et al. , Mitochondrial perturbation negatively affects auxin signaling. Mol. Plant 7, 1138–1150 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Woodson J. D., Chory J., Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9, 383–395 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vanlerberghe G. C., Alternative oxidase: A mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int. J. Mol. Sci. 14, 6805–6847 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karpova O. V., Kuzmin E. V., Elthon T. E., Newton K. J., Differential expression of alternative oxidase genes in maize mitochondrial mutants. Plant Cell 14, 3271–3284 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Clercq I., et al. , The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 25, 3472–3490 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Aken O., et al. , Defining the mitochondrial stress response in Arabidopsis thaliana. Mol. Plant 2, 1310–1324 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Steglich G., Neupert W., Langer T., Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria. Mol. Cell. Biol. 19, 3435–3442 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong X., et al. , PHB3 maintains root stem cell niche identity through ROS-Responsive AP2/ERF transcription factors in Arabidopsis. Cell Rep. 22, 1350–1363 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Bourbousse C., Vegesna N., Law J. A., SOG1 activator and MYB3R repressors regulate a complex DNA damage network in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 115, E12453–E12462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weijers D., Wagner D., Transcriptional responses to the auxin hormone. Annu. Rev. Plant Biol. 67, 539–574 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Kirkinezos I. G., Moraes C. T., Reactive oxygen species and mitochondrial diseases. Semin. Cell Dev. Biol. 12, 449–457 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Mir R., et al. , A DII domain-based auxin reporter uncovers low auxin signaling during telophase and early G1. Plant Physiol. 173, 863–871 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brunoud G., et al. , A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482, 103–106 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Alarcón M. V., Salguero J., Lloret P. G., Auxin modulated initiation of lateral roots is linked to pericycle cell length in maize. Front. Plant Sci. 10, 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piechota J., Kolodziejczak M., Juszczak I., Sakamoto W., Janska H., Identification and characterization of high molecular weight complexes formed by matrix AAA proteases and prohibitins in mitochondria of Arabidopsis thaliana. J. Biol. Chem. 285, 12512–12521 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Aken O., et al. , Mitochondrial type-I prohibitins of Arabidopsis thaliana are required for supporting proficient meristem development. Plant J. 52, 850–864 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Kawano T., Roles of the reactive oxygen species-generating peroxidase reactions in plant defense and growth induction. Plant Cell Rep. 21, 829–837 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Tanaka K., et al. , UGT74D1 catalyzes the glucosylation of 2-oxindole-3-acetic acid in the auxin metabolic pathway in Arabidopsis. Plant Cell Physiol. 55, 218–228 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tognetti V. B., et al. , Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22, 2660–2679 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Franklin K. A., et al. , Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. U.S.A. 108, 20231–20235 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y., et al. , Stress responsive mitochondrial proteins in Arabidopsis thaliana. Free Radic. Biol. Med. 122, 28–39 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.