Significance
Studies of the microbial communities living in association with animals, with plants, and in the environment have revolutionized our understanding of microbial diversity, but the interactions between these microbes remain largely elusive. Here, we characterize pairwise interactions during infection between a human pathogen and 25 diverse microbes from both sympatric (same) and allopatric (different) environments. We demonstrate that allopatric microbes increase survival of the pathogen. Furthermore, we identify the large set of genes (33% of the genome) that are essential for pathogen survival during coinfection, including a core set that are also required in monoinfection and diverse accessory essential genes. This work advances our understanding of how coinfecting microbes modulate the genes required for survival in vivo.
Keywords: Aggregatibacter, CoDE genes, essential, polymicrobial
Abstract
Recent evidence suggests that the genes an organism needs to survive in an environment drastically differ when alone or in a community. However, it is not known if there are universal functions that enable microbes to persist in a community and if there are functions specific to interactions between microbes native to the same (sympatric) or different (allopatric) environments. Here, we ask how the essential functions of the oral pathogen Aggregatibacter actinomycetemcomitans change during pairwise coinfection in a murine abscess with each of 15 microbes commonly found in the oral cavity and 10 microbes that are not. A. actinomycetemcomitans was more abundant when coinfected with allopatric than with sympatric microbes, and this increased fitness correlated with expanded metabolic capacity of the coinfecting microbes. Using transposon sequencing, we discovered that 33% of the A. actinomycetemcomitans genome is required for coinfection fitness. Fifty-nine “core” genes were required across all coinfections and included genes necessary for aerobic respiration. The core genes were also all required in monoinfection, indicating the essentiality of these genes cannot be alleviated by a coinfecting microbe. Furthermore, coinfection with some microbes, predominately sympatric species, induced the requirement for over 100 new community-dependent essential genes. In contrast, in other coinfections, predominately with nonoral species, A. actinomycetemcomitans required 50 fewer genes than in monoinfection, demonstrating that some allopatric microbes can drastically alleviate gene essentialities. These results expand our understanding of how diverse microbes alter growth and gene essentiality within polymicrobial infections.
The interactions between organisms have a significant impact on ecosystem functions. For example, bacterial symbionts in root nodules fix nitrogen, providing ammonia to plants and surrounding microbes (1). While it is important to study interactions to understand community ecology and function, in most cases, interactions are numerous and complex. A community of 25 species has 300 possible pairwise interactions, in addition to any higher-order interactions. As it can be unreasonable to understand all of the interactions within a community, scientists focus on understanding specific key interactions, often using simplified or synthetic communities.
A large body of work shows that interactions are influenced by the nutrients an organism needs and consumes (exploitive competition) and its antagonistic (interference competition) or cooperative functions (2–4). Specifically, antagonism increases with genetic relatedness on some scales but not others, and generalist organisms (i.e., those that thrive in a wide variety of environmental conditions and can use a variety of different resources) are often more antagonistic than specialist organisms (5, 6). Furthermore, organisms’ historical record of interaction—if they are sympatric (from the same environment) or allopatric (found in different environments)—can influence how they interact (5, 7, 8). In addition to coevolution, which can drive specific interactions (9), organisms from the same environment likely fill different niches and are thought to be less likely to compete for nutrients (10, 11).
Here, we define the genetic features required for a focal bacterial pathogen to coexist in vivo with 25 diverse interacting partners, including both allopatric and sympatric species. Specifically, we analyze pairwise coinfections between the human oral pathogen Aggregatibacter actinomycetemcomitans and 15 sympatric oral microbes and 10 allopatric nonoral microbes in a murine abscess. A. actinomycetemcomitans is a gram-negative, facultative anaerobe and opportunistic pathogen associated with aggressive periodontitis (12, 13). Notably, A. actinomycetemcomitans not only infects the oral cavity but can also spread throughout the body to cause abscesses (14). A. actinomycetemcomitans and other oral microbes provide an ideal community for these studies. Beyond their critical importance for human health, their interactions are relatively well studied (15, 16). In addition, most oral microbes including A. actinomycetemcomitans have a highly specialized metabolism and are specialized for growth in the oral cavity (17).
To define the functions required for A. actinomycetemcomitans coinfection with other microbes, we used transposon insertion sequencing (Tn-seq), a method that has proven productive for understanding microbe–microbe interactions in vivo (18–20). This technique combines high-throughput transposon mutant screening with next-generation sequencing to identify genes that are essential for fitness under a condition of interest (21). Tn-seq functions as a single-cell biosensor, reflecting the requirements for fitness for individual mutant bacteria. Using Tn-seq to identify bacterial genes that change in essentiality in the presence of another microbe, recently termed community-dependent essential (CoDE) genes (18), can illustrate how interactions between these microbes alter physiology and can provide insights into the interactions themselves. Recent studies have revealed that coculture generally increases the number of genes required for fitness, although coculture can also alleviate functions necessary during monoculture (18, 20, 22). It is poorly understood, however, whether these functional requirements are generalizable or whether individual organisms require distinct functions with each organism they encounter. In this study, we discovered that A. actinomycetemcomitans requires ∼33% of its genomic loci for fitness during pairwise coinfection. In addition, we identified a set of A. actinomycetemcomitans “core” genes that were required for coinfection with both allopatric and sympatric partners and demonstrated how coinfecting microbes can drastically alleviate or induce the requirement for essential functions.
Results
A. actinomycetemcomitans Coexists with Sympatric (Oral) and Allopatric (Nonoral) Microbes in Murine Abscesses.
We chose 25 diverse microbes to coinfect with A. actinomycetemcomitans in the murine thigh abscess model (Table 1, see Dataset S1 for expanded functional annotations). These microbes include 2 fungi and 23 bacteria. Fifteen of the microbes are commonly found in the oral cavity, and all isolates used here to represent these species were isolated from this environment, except for Candida albicans. The other 10 microbes are not native to the oral cavity; 8 of the nonoral microbes can be host-associated and 2 (Bacillus subtilis, Saccharomyces cerevisiae) are predominately environmental. None of the strains used for these nonoral species were isolated from the oral environment. Together, the microbes chosen are metabolically diverse. For example, they range from strict aerobes to strict anaerobes, and some can use a large range of carbohydrates while others are asaccharolytic. This range in metabolic capacity is reflected in the genome sizes. The oral microbes’ genome sizes range from 1.6 Mb to 4.0 Mb for the bacteria and 14.3 Mb for C. albicans, and the nonoral microbes’ genome sizes range from 1.7 Mb to 8.1 Mb for the bacteria and 11.6 Mb for S. cerevisiae (Table 1 and SI Appendix, Fig. S1). There is no significant difference in genome size between oral and nonoral microbes (Mann–Whitney U test, Padj = 0.16). Similarly, there is no significant difference in the number of protein-coding genes, which range from 1,485 to 6,030 for the oral microbes and 1,548 to 7,116 for the nonoral microbes (Mann–Whitney U test, Padj = 0.16). However, there are significantly more genes annotated with enzyme commission (EC) numbers in the nonoral microbe genomes (SI Appendix, Fig. S1) (Mann–Whitney U test, Padj = 0.0132). The majority of genes with EC numbers encode proteins involved in metabolism, as opposed to structural or regulatory proteins, indicating increased metabolic potential in the nonoral microbes. In addition, the nonoral microbes have significantly more 16S rRNA gene copies (SI Appendix, Fig. S1) (Mann–Whitney U test, Padj = 0.008). High 16S rRNA gene copy number is associated with fast growing microbes and often correlates with success in nutrient-rich environments (23). These microbes allow us to assay coinfection with a diverse set of both sympatric and allopatric organisms.
Table 1.
Broad characterization of A. actinomycetemcomitans and all coinfection species
| Species | Strain | Habitat | Description | Genome size (Mbp) | Protein coding genes | Protein coding genes with EC number | In vivo essential genes |
| Aggregatibacter actinomycetemcomitans | 624 | Oral | Facultative anaerobe | 2.37 | 2,283 | 740 | 199 |
| Actinomyces israelii | F0345 | Oral | Obligate anaerobe | 4.02 | 3,346 | 808 | 370 |
| Actinomyces oris | MG-1 | Oral | Facultative anaerobe | 2.93 | 2,704 | 578 | 207 |
| Campylobacter rectus | 33238 | Oral | Microaerophile | 2.51 | 2,971 | 560 | 283 |
| Candida albicans | SC5314 | Oral | Facultative anaerobe | 14.28 | 6,030 | 1,402 | 182 |
| Filifactor alocis | 35896 | Oral | Obligate anaerobe | 1.93 | 1,641 | 446 | 308 |
| Fusobacterium nucleatum | 25586 | Oral | Obligate anaerobe | 2.17 | 2,067 | 612 | 238 |
| Lactobacillus reuteri | FJ1 | Oral | Aerotolerant anaerobe | 2.15 | 1,945 | 569 | 298 |
| Neisseria mucosa | C102 | Oral | Facultative anaerobe | 2.16 | 2,026 | 706 | 249 |
| Parvimonas micra | 33270 | Oral | Obligate anaerobe | 1.63 | 1,485 | 437 | 263 |
| Porphyromonas gingivalis | W83 | Oral | Obligate anaerobe | 2.34 | 1,909 | 534 | 363 |
| Prevotella intermedia | 25611 | Oral | Obligate anaerobe | 2.67 | 2,191 | 537 | 232 |
| Streptococcus gordonii | DL1.1 | Oral | Facultative anaerobe | 2.20 | 2,076 | 659 | 204 |
| Streptococcus intermedius | 27335 | Oral | Facultative anaerobe | 1.91 | 1,875 | 583 | 294 |
| Streptococcus mutans | UA159 | Oral | Facultative anaerobe | 2.03 | 1,960 | 627 | 193 |
| Streptococcus sanguinis | 10556 | Oral | Facultative anaerobe | 2.39 | 2,272 | 713 | 245 |
| Bacillus subtilis | PY79 | Nonoral | Facultative anaerobe | 4.03 | 4,138 | 1,129 | 257 |
| Burkholderia cenocepacia | J2315 | Nonoral | Obligate aerobe | 8.06 | 7,116 | 1,706 | 326 |
| Enterobacter cloacae | 13047 | Nonoral | Facultative anaerobe | 5.60 | 5,284 | 1,480 | 148 |
| Enterococcus faecalis | V583 | Nonoral | Facultative anaerobe | 3.22 | 3,266 | 789 | 266 |
| Escherichia coli | W3110 | Nonoral | Facultative anaerobe | 4.65 | 4,226 | 1,439 | 191 |
| Francisella sp. | W12-1067 | Nonoral | Obligate aerobe | 1.70 | 1,548 | 533 | 255 |
| Haemophilus influenzae | 86-028NP | Nonoral | Facultative anaerobe | 1.91 | 1,816 | 691 | 146 |
| Saccharomyces cerevisiae | BY4741 | Nonoral | Facultative anaerobe | 11.62 | 5,869 | 1,450 | 115 |
| Serratia marcescens | Db11 | Nonoral | Facultative anaerobe | 5.11 | 4,709 | 1,436 | 246 |
| Streptococcus pyogenes | 950771 | Nonoral | Facultative anaerobe | 1.84 | 1,771 | 584 | 256 |
To study A. actinomycetemcomitans interactions with each microbe during coinfection, we used the murine inner thigh abscess model (19, 24, 25). We chose this persistence model because A. actinomycetemcomitans and other oral pathogens also cause polymicrobial abscess infections both inside and outside the oral cavity (14, 26, 27). In addition, this abscess model allows us to infect the microbes of interest into a sterile environment in the presence of the host immune system. This model has been used to discover molecular interactions between A. actinomycetemcomitans and streptococci that are relevant to the oral cavity (19, 24, 25, 28–30), and recent evidence indicates that A. actinomycetemcomitans gene expression in the murine abscess is similar to that in humans with periodontal disease (25).
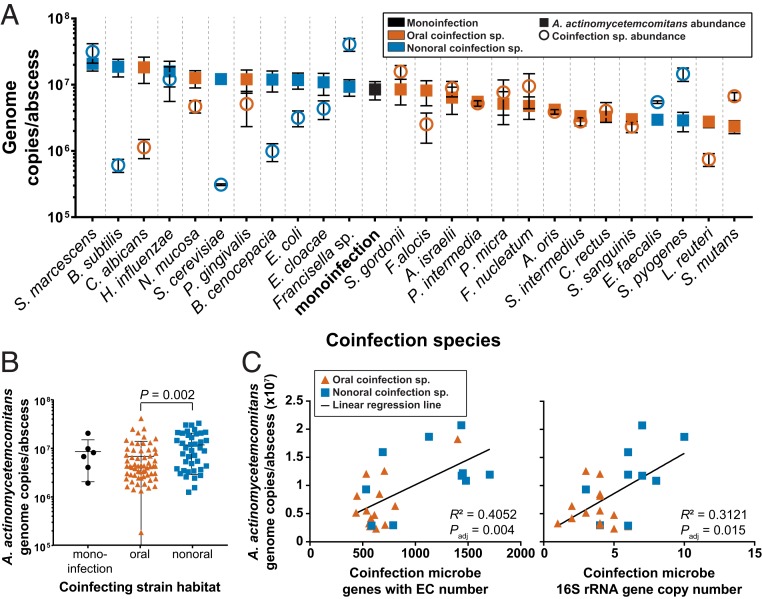
Using this murine inner thigh abscess model, we inoculated 108 colony forming units (CFUs) of A. actinomycetemcomitans in monoinfection and in pairwise coinfection with 108 CFUs of each of the 25 diverse microbes. After 3 d, abscesses were harvested and bacterial loads were assessed using qPCR and 16S rRNA gene amplicon sequencing. A. actinomycetemcomitans and the coinfecting microbes were present in all infections after 3 d (Fig. 1A and Dataset S2). In monoinfection, A. actinomycetemcomitans abundance was 8.5 × 106 genome copies per abscess, while the abundance of A. actinomycetemcomitans varied in coinfection by ∼10-fold between 2.3 × 106 and 2.1 × 107 genome copies per abscess. The coinfecting microbes ranged in abundance from 3.1 × 105 to 4.1 × 107 genome copies per abscess with Serratia marcescens, Francisella sp., and Streptococcus pyogenes representing the highest levels and B. subtilis and S. cerevisiae representing the lowest levels (Fig. 1A). Although the ranges of abundances were similar, A. actinomycetemcomitans was significantly more abundant in the presence of nonoral microbes than oral microbes (Mann–Whitney U test, P = 0.002) (Fig. 1B), indicating that coinfection with allopatric (nonoral) microbes generally increases the fitness of A. actinomycetemcomitans in the murine abscess. No relationship was found between the abundances of A. actinomycetemcomitans and the abundances of the coinfecting microbes (SI Appendix, Fig. S2).
Fig. 1.
(A) Abundance of A. actinomycetemcomitans and coinfection microbes in abscesses. A. actinomycetemcomitans abundance was quantified using qPCR. The abundances of coinfection bacteria were quantified using 16S rRNA gene sequencing and normalized to A. actinomycetemcomitans abundance and 16S rRNA gene copy number. The abundances of coinfecting fungi were determined using qPCR. (B) A. actinomycetemcomitans is significantly more abundant with nonoral microbes than with oral microbes. Each point represents a single abscess. Mann–Whitney U test, P = 0.002. (C) Relationship between A. actinomycetemcomitans abundance in coinfected abscesses and genome metrics of the corresponding coinfection microbe. Linear regression best-fit line is shown with corresponding goodness of fit metric and adjusted P value, calculated using the Benjamini–Hochberg correction. See SI Appendix, Fig. S2 for additional comparisons.
We next assessed whether A. actinomycetemcomitans abundance in the abscess was correlated with the functional capacity of the coinfecting bacterium. As A. actinomycetemcomitans had increased abundance with nonoral microbes, its abundance also positively correlated with characteristics enriched in the nonoral microbe genomes, including, most significantly, the number of genes with an EC number (linear regression, Padj = 0.015) (Fig. 1C). In addition, A. actinomycetemcomitans genome copies per abscess positively correlated with genome size and number of protein-coding genes (Padj = 0.015) (SI Appendix, Fig. S2). We also found that A. actinomycetemcomitans abundance increased with the number of genes in specific cluster of orthologous groups (COG) categories, including: Amino acid, coenzyme, inorganic ion, and lipid transport and metabolism; secondary metabolite biosynthesis, transport, and catabolism; and energy production and conversion (SI Appendix, Fig. S2). Finally, A. actinomycetemcomitans had higher abundance in coinfection with microbes with more 16S rRNA gene copies (Fig. 1C). These data indicate that A. actinomycetemcomitans is more abundant in the murine abscess in the presence of coinfecting microbes with larger functional capacity.
In A. actinomycetemcomitans, 593 Genes Are Essential in the Coinfected Murine Abscess but Not In Vitro.
To identify genes important for A. actinomycetemcomitans fitness during mono- and coinfection, we used a transposon library consisting of 49,423 unique insertion sites across 2,076 (of the total 2,466) annotated genetic loci in the A. actinomycetemcomitans 624 genome (SI Appendix, Table S1). Our analysis focused on “essential” genes, identified by comparing observed transposon distributions following growth in a condition of interest (e.g., mono- or coinfected murine abscess) to transposon distributions under a null model where no gene is important for fitness calculated using a Monte Carlo approach with 1,000 simulations (31). Genes were called essential if they 1) had a Padj < 0.05 and 2) contributed to the left, “reduced” bimodal distribution in the mclust analysis with an uncertainty <0.05 (Datasets S3 and S4) (18, 31). We used a significance cutoff of 0.05 for our analyses here, as we are being additionally stringent by comparing essentialities across samples. Essential genes under the more restrictive cutoff values of Padj < 0.01 and mclust uncertainty <0.01 are available in Dataset S3.
In total, 1,260 genes were identified as essential in the in vitro transposon library or at least 1 murine abscess infection condition (51% of genes). To focus on genes that were only essential in the coinfected murine abscess, the 606 genes identified as essential in the in vitro transposon library and the 1 gene essential only in monoinfection were excluded from all subsequent analyses. We also excluded an additional 60 genes that were < 200 bp in length due to high variability in determining essentiality, likely resulting from poor transposon insertion coverage (SI Appendix, Fig. S3). After excluding these 667 genes, we identified 593 genes that are essential in the murine abscess in coinfection. Of these 593 genes, 199 were essential in monoinfection and at least 1 coinfection, and 394 were essential in at least 1 coinfection but not in monoinfection (coinfection-specific).
To confirm our essentials analysis, we first tested if the fitness of a mutant in isolation replicated its fitness in the Tn-seq mutant library. ACT75_RS08345, a putative molybdenum cofactor guanylyltransferase, was not essential in monoinfection but became essential in coinfection with Actinomyces oris. Similarly, in competition assays with wild-type in the murine abscess, a mutant in this gene was more fit in monoinfection than in coinfection with A. oris (Mann–Whitney U test, P = 0.014) (SI Appendix, Fig. S4). Second, we compared our Tn-seq data to a previous analysis of an ATP synthase mutant in monoinfection and coinfection with Streptococcus gordonii (19). This previous analysis found that an atpB mutant (ACT75_RS06085) had lower fitness than wild-type in both mono- and coinfection, and was less fit in monoinfection than coinfection. Similarly, we detected low read counts for this gene in mono- and coinfection, but atpB was only identified as essential in monoinfection and not in coinfection with S. gordonii (Dataset S3). Finally, we compared the essential genome in monoinfection for A. actinomycetemcomitans 624 (the strain used for these experiments) to A. actinomycetemcomitans VT1169 (SI Appendix, Fig. S5). These 2 strains are different serotypes with a 97.63% 2-way average nucleotide identity and have similar gene fitness profiles in vitro (32). A. actinomycetemcomitans 624 and VT1169 shared 373 essential genes in vivo in monoinfection of their 1,697 orthologs, and had only 182 and 64 unique essential genes, respectively. Thus, while some genes differed in essentiality as expected, overall, their essential genomes were highly similar (2-tailed Fisher’s exact test, P < 0.0001). Therefore, we have confidence in the quality of our sequencing data and analysis.
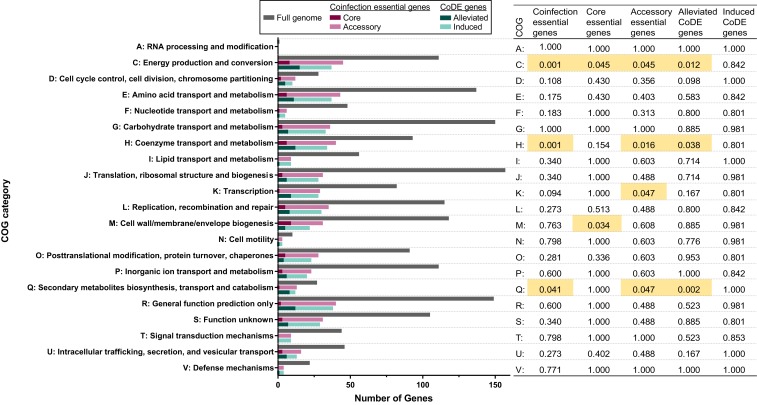
To broadly understand what functions are important for A. actinomycetemcomitans fitness in the murine abscess, we first assessed which COG categories were enriched in the murine coinfection essential gene set. The COG categories energy production and conversion, coenzyme transport and metabolism, and secondary metabolites biosynthesis, transport, and catabolism were all significantly enriched in the essential genes compared to the A. actinomycetemcomitans genome (2-sided Fisher’s exact test, Padj < 0.05) (Fig. 2). We next asked if the variability in the number of coinfection essential genes can be explained by specific traits of the coinfecting microbe. We found no significant correlation between the number of essential genes for each coinfection and broad genome metrics or the number of genes in any COG family of the coinfection microbes (SI Appendix, Fig. S6). Together, these data indicate that ∼25% of the A. actinomycetemcomitans genomic loci are essential for growth in the murine coinfected abscess in the 25 conditions assayed here but not in vitro and that these genes are enriched for specific metabolic functions.
Fig. 2.
Enrichment of COG category genes in essential gene sets relative to the A. actinomycetemcomitans genome. The 593 coinfection specific genes are the sum of the 59 core and 534 accessory essential genes. There are 140 alleviated CoDE genes and 394 induced CoDE genes. Enrichments were performed between each essential gene category and the A. actinomycetemcomitans genome as a whole using a 2-sided Fisher’s exact test. Table shows adjusted P values (Benjamini–Hochberg). Padj < 0.05 are highlighted in yellow.
One-Third of the A. actinomycetemcomitans Genome Is Essential for Fitness in Coinfection.
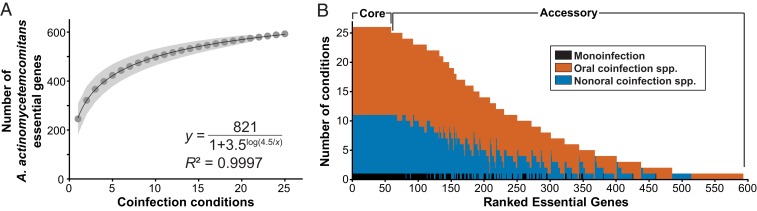
While this pairwise coinfection study is expansive, particularly for assessing fitness determinants, the 593 coinfection specific genes clearly do not encompass all possible genes essential for A. actinomycetemcomitans coinfection fitness in the murine abscess. However, it is possible from these data to estimate the number of A. actinomycetemcomitans essential genes in infinite pairwise coinfections. To accomplish this, we built an essential genes accumulation curve to show the union of A. actinomycetemcomitans essential gene sets across increasing numbers of coinfection conditions (Fig. 3A). This curve is the average of 1,000,000 random iterations and was fit with the Lomolino function, y = asymptote/(1 + slopelog(xmid/x)), where xmid is the number of coinfection conditions where half the maximum number of essential genes is reached, with a high goodness-of-fit (R2 = 0.9997) (33, 34). This analysis was confirmed using Kindt’s exact accumulator (35), which produced curves with best-fit values with high agreement (SI Appendix, Fig. S7). The Lomolino function predicted that A. actinomycetemcomitans would require 821 unique essential genes across infinite pairwise coinfection conditions (95% confidence interval: 801 to 844). Based on this prediction, approximately one-third of the A. actinomycetemcomitans genome is essential for fitness during murine abscess coinfection, and we identified 72% (593 of 821) of these genes in our coinfection experiments. It is worth noting that just 5 coinfection conditions were sufficient to provide over 50% of the predicted total number of coinfection essential genes.
Fig. 3.
(A) Accumulation of unique A. actinomycetemcomitans coinfection essential genes. One million random sampling permutations were used to build the accumulation curve. The gene-accumulation model curve was calculated using the Lomolino function, y = asymptote/(1 + slopelog(xmid/x)), where xmid is the number of coinfection conditions where half the maximum number of essential genes is reached, and the best-fit line and equation are shown. The shaded area shows the 95% confidence interval. (B) Core and accessory coinfection essential genes. Genes are ordered by the number of conditions in which they are essential. The 59 genes essential in all conditions are termed core essential genes, while the remaining genes are termed accessory essential genes.
Respiratory Metabolism Is a Core Essential Function in the Murine Abscess.
To determine if there is a core set of genes essential for A. actinomycetemcomitans coinfection, we calculated the number of coinfection conditions in which each of the 593 coinfection genes was essential (Fig. 3B) and identified 59 genes that were essential across all 25 coinfection conditions. Interestingly, these genes were also essential in monoinfection; thus, the set of genes always essential for coinfection is a subset of the 199 genes essential for monoinfection. We term these 59 genes “core”: Functions always required for fitness in the murine abscess whether in mono- or coinfection (Fig. 3B and Dataset S3). To broadly characterize the functions of the 59 core genes, we first assessed whether COG categories were enriched in the core genes relative to the whole genome. The COG categories energy production and conversion and cell wall/membrane/envelope biogenesis were both significantly enriched (2-sided Fisher’s exact test, Padj < 0.05) (Fig. 2). Further examination revealed that several genes involved in respiratory metabolism were core essential genes. Among these were cydA and cydB, which encode components of the cytochrome bd complex, the sole aerobic respiratory oxidase in A. actinomycetemcomitans. Genes encoding both components of the thiol reductant ABC transporter CydDC, important for assembly of cytochrome bd, were also essential in all datasets. Finally, genes encoding the C and D subunits of the NADH:ubiquinone reductase [Na(+)-transporting], which is involved in respiratory metabolism, were core essential genes. In addition, 3 genes encoding components of the spermidine/putrescine ABC transporter and 3 genes encoding proteins involved in the Tol-Pal system were always essential in coinfection. Thus, the core essential genes required for coinfection are a subset of monoinfection essential genes, and genes encoding proteins critical for aerobic respiration are core fitness determinants in A. actinomycetemcomitans abscess infections regardless of whether A. actinomycetemcomitans is in mono- or coinfection.
“Accessory” A. actinomycetemcomitans Essential Genes in the Murine Abscess.
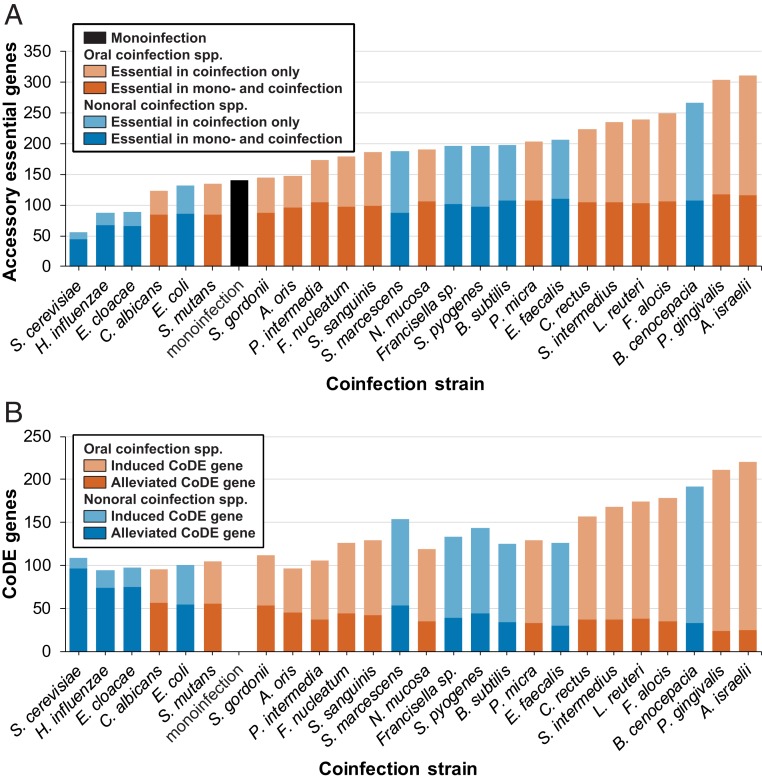
In addition to the 59 core essential genes, there were 534 A. actinomycetemcomitans essential genes termed “accessory” genes, required in at least 1 coinfection but not in all coinfections (Fig. 3B). Of these accessory genes, 140 are essential in mono- and at least 1 coinfection while 394 are coinfection-specific. Furthermore, of the 394 coinfection-specific essential genes, 235 are essential with both sympatric and allopatric microbes, while 124 are unique to coinfection with sympatric microbes and 35 are unique to coinfection with allopatric microbes. Of the sympatric- or allopatric-unique genes, 85% are essential in 2 or fewer coinfection conditions. Overall, ∼20% (108 genes) of the coinfection-specific essential genes were essential in only 1 coinfection, and most of these genes were from coinfections with Porphyromonas gingivalis (23 genes), Actinomyces israelii (22 genes), and Burkholderia cenocepacia (19 genes).
The number of A. actinomycetemcomitans accessory essential genes varied considerably between 56 in coinfection with S. cerevisiae and 311 with A. israelii (Fig. 4A). Although there was no significant difference between the number of accessory essential genes in coinfections with oral and nonoral microbes, it is worth noting that 6 of the 7 conditions with the most accessory essential genes were coinfection with oral bacteria and the 3 conditions with the fewest number of accessory genes were coinfection with nonoral bacteria. In all coinfections, at least a portion of the 140 accessory genes required in monoinfection were also essential (Fig. 4A). However, the number of genes varied widely, between 44 with S. cerevisiae and 117 with P. gingivalis. The number of coinfection-specific accessory genes also varied widely, with as few as 12 required with S. cerevisiae and 195 with A. israelii. These data indicate the number of genes required for A. actinomycetemcomitans fitness during coinfection varies broadly, although at least some functions necessary for fitness during coinfection are also required in monoinfection.
Fig. 4.
(A) Number of accessory essential genes across experimental conditions. Shading indicates if the gene is also essential in monoinfection or only essential in coinfection. (B) Number of CoDE genes across experimental conditions. Shading indicates if the essentiality of the gene is alleviated by coinfection or induced by coinfection.
Conditions Cluster by the Number of A. actinomycetemcomitans Accessory Essential Genes.
We next assessed if the essential genes between coinfection partners were similar by performing a principle coordinate analysis of the Jaccard index between each condition’s accessory essential gene set. This analysis revealed that the coinfection conditions do not cluster based on whether they contained oral or nonoral bacteria (SI Appendix, Fig. S8A). Coinfection with A. oris and Streptococcus mutans clustered most closely to monoinfection, while those with P. gingivalis, B. cenocepacia, A. israelii, and S. cerevisiae clustered furthest from monoinfection. Coinfection with Fusobacterium nucleatum and Neisseria mucosa were the 2 most similar conditions in this analysis. When we extracted the first principle component, it strongly correlated with the number of essential genes in each condition (SI Appendix, Fig. S8B). This analysis shows that coinfections with more accessory genes have similar—not distinct—essential gene sets.
Accessory Essential Genes Are Enriched for Specific Metabolic Functions.
To define what types of functions are sometimes, but not always, important in coinfected abscesses, we determined which COG families are enriched across the accessory essential genes. Similar to the in vivo essential genes and core essential genes, energy production and conversion was enriched in the accessory genes (2-sided Fisher’s exact test, Padj = 0.045) (Fig. 2). The 4 additional components of the NADH:ubiquinone reductase [Na(+)-transporting] were accessory essential genes required in the majority of conditions. Furthermore, in addition to the 1 component of the FOF1 ATP synthase that was a core essential gene, genes encoding 6 additional components of the ATP synthase were accessory essential genes. A. actinomycetemcomitans required all 6 additional ATP synthase genes for fitness in coinfection with A. israelii, A. oris, B. cenocepacia, Enterococcus faecalis, F. nucleatum, Parvimonas micra, Prevotella intermedia, S. pyogenes, and Streptococcus sanguinis, as well as in monoinfection. In contrast, A. actinomycetemcomitans required no additional genes encoding the ATP synthase in coinfection with S. cerevisiae, and only 2 genes with Escherichia coli and Enterobacter cloacae. The requirement for nitrate reduction additionally varied across conditions. Three genes involved in nitrate reduction were essential in coinfection with A. israelii and 2 were essential in coinfection with both B. cenocepacia and P. micra. Finally, the gene encoding ArcA, a regulator activated in anaerobic conditions, was essential with 13 coinfection microbes that range from obligate aerobes to obligate anaerobes, and the gene encoding superoxide dismutase was essential in 11 of 26 conditions, indicating increased oxygen stress during coinfection with some microbes.
In addition to energy production and conversion, COG categories coenzyme transport and metabolism, transcription, and secondary metabolites biosynthesis, transport, and catabolism were also enriched in the accessory essential genes relative to the A. actinomycetemcomitans genome as a whole (2-sided Fisher’s exact test, Padj < 0.05) (Fig. 2). Looking more closely at the accessory genes in these categories, heme synthesis, molybdopterin synthesis, and transcriptional regulation are accessory essential functions.
A. actinomycetemcomitans CoDE Genes.
While the previous analyses focused on all coinfection essential genes regardless of whether they are also required during monoinfection, we are also interested in the subset of genes that are differentially essential in mono- and coinfection. We have previously termed these CoDE genes, and work from our laboratory and others have shown that coinfection can both alleviate the need for genes required during monoinfection as well as necessitate the requirement for new genes (18, 20, 22). The number of A. actinomycetemcomitans CoDE genes varied considerably, between 220 during coinfection with A. israelii and 94 with Haemophilus influenzae (Fig. 4B and Datasets S1 and S3). The coinfection conditions with the most CoDE genes—such as coinfection with A. israelii, P. gingivalis, and B. cenocepacia—have over 100 new genes that become essential during coinfection and fewer than 40 CoDE genes that are alleviated. In contrast, in the conditions in which A. actinomycetemcomitans requires the fewest CoDE genes—such as coinfection with S. cerevisiae, H. influenzae, and E. cloacae—fewer than 25 new genes become essential and over 70 CoDE genes are alleviated.
Alleviated CoDE Gene Functions Are Defined by Functions Essential in Monoinfection.
As 140 of the monoinfection essential genes are alleviated in at least 1 coinfection condition, the alleviated CoDE functions are defined by what is essential in monoinfection. COG enrichment analyses identified that energy production and conversion, coenzyme transport and metabolism, and secondary metabolites biosynthesis, transport, and catabolism were enriched in the alleviated genes (2-sided Fisher’s exact test, Padj < 0.05) (Fig. 2). To further understand functions that are alleviated by coinfection, we identified genes that were essential in monoinfection but in no or few coinfection conditions. A single gene, annotated as a YggU family protein of unknown function, was essential in monoinfection but nonessential in all coinfections. In addition, a gene annotated to encode a toxin-activating lysine-acyltransferase was only essential in monoinfection and in coinfection with Filifactor alocis. The putative product of this gene, LtxC, is an acetyltransferase thought to activate the A. actinomycetemcomitans virulence factor RTX toxin, leukotoxin (LtxA) (36). However, while it has previously been observed that coinfection alleviates the requirement for leukotoxin in the abscess (19), ltxA itself is not essential in any condition, including monoinfection. Therefore, potentially, LtxC has a secondary function for A. actinomycetemcomitans. Finally, 2 genes predicted to encode proteins involved in chromosome partitioning, including cell-division protein ZapB, were each essential in monoinfection and only in 4 coinfection conditions.
We further focused on the 3 conditions with the most alleviated CoDE genes—coinfection with E. cloacae, H. influenzae, and S. cerevisiae—in which 74 to 97 gene essentialities are alleviated. Compared to monoinfection, many of the genetic requirements alleviated across these 3 conditions include: Genes encoding the Flp pilus, which mediates adhesion; genes encoding ATP synthase subunits; and genes encoding proteins involved in deoxyribose synthesis. Therefore, adhesion and ATP synthase are not as critical for A. actinomycetemcomitans survival in the presence of these microbes in the murine abscess model, and A. actinomycetemcomitans has a reduced need to synthesize DNA building blocks.
Induced CoDE Genes Reveal Functions Specific to Coinfection.
To understand how coinfection increases genetic requirements, we analyzed the 394 CoDE genes that are essential in coinfection, but not in monoinfection. No COG categories were enriched in these 394 genes (Fig. 2). We then focused on the 51 CoDE genes that were induced in at least half of the coinfection conditions (13 or more conditions), and these genes revealed a number of functions that are generally important for coinfection. For example, 2 genes encoding subunits of pyruvate dehydrogenase are essential in over 18 coinfection conditions. It is notable that the only coinfection condition where neither identified component of pyruvate dehydrogenase becomes essential is coinfection with B. cenocepacia. A gene encoding a tyrosine transporter was essential in 20 conditions. In addition, tatA and tatC were coinfection-specific essential genes and essential in 15 and 14 conditions, respectively, indicating that the twin-arginine translocase is important in many coinfections. Finally, A. actinomycetemcomitans required a gene encoding a subunit of the MexH family multidrug efflux RND transporter in 21 of 25 coinfection conditions, and a gene encoding the second subunit was essential in 11 coinfection conditions. Therefore, coinfection likely increases the necessity of antimicrobial efflux, as well as flux of pyruvate to acetyl-CoA, tyrosine transport, and secretion of proteins with Tat signal sequences.
Finally, we assessed CoDE genes induced in coinfections in which A. actinomycetemcomitans required the most new essential genes relative to monoinfection: P. gingivalis, A. israelii, and B. cenocepacia. In coinfection with each of these 3 microbes, over 150 A. actinomycetemcomitans genes become essential compared to monoinfection. Analysis of the induced CoDE genes in all 3 conditions show A. actinomycetemcomitans is under stress. A gene encoding a cold-shock protein is essential with these microbes, as well as with 3 other coinfection species, as is a gene annotated as a Fis family transcriptional regulator. In addition, a gene annotated as a stringent starvation protein that has been shown in E. coli to be important to resist acid stress and nutrient starvation is essential in these 3 conditions, but not in monoinfection, and the toxin component of a type II toxin–antitoxin (TA) system is essential in these conditions that is homologous to an A. actinomycetemcomitans D11S toxin activated under acid stress (37, 38). Two other TA toxins are also essential for A. actinomycetemcomitans in coinfection with these 3 strains, but not in monoinfection, and 1 is homologous to A. actinomycetemcomitans D11S_1799, which has been shown to be up-regulated under anaerobic conditions (37). A. actinomycetemcomitans carbon metabolism is likely also altered in the presence of these 3 strains, as a number of genes involved in the TCA cycle and genes encoding a 1-phosphofructokinase and subunits of mannose and fructose PTS transporters are induced CoDE genes in these conditions.
Discussion
Microbes have evolved to survive in complex communities and persist in the presence of both native and invading, unknown microbes. Yet, teasing apart how microbes survive these biological challenges, the underlying microbe–microbe interactions, and the resulting impact on community function is difficult. Previous work has used Tn-seq to identify fitness determinants in mouse models of infections for Staphylococcus aureus, Proteus mirabilis, and A. actinomycetemcomitans, each coinfected with a single organism (18, 20) to identify functional requirements altered by interactions. Here, we present a large-scale dataset of interactions with 25 diverse microbes in vivo. We find that A. actinomycetemcomitans is more abundant with nonoral microbes and microbes with more metabolic capacity. Furthermore, our Tn-seq analysis shows that one-third of the A. actinomycetemcomitans genome is essential for coinfection, A. actinomycetemcomitans always requires aerobic respiration in vivo, and coinfection microbes drastically alter the A. actinomycetemcomitans essential genome.
A. actinomycetemcomitans is a slow-growing, specialized microbe specifically equipped to survive in the oral subgingival crevice and in abscesses (14, 17). While these environments are polymicrobial, only specific microbes are native, and many of the microbes included in this study are absent or possibly transient. However, we found that A. actinomycetemcomitans can compete with both sympatric and allopatric bacteria in the murine abscess, including highly pathogenic, fast-growing microbes (Fig. 1). In addition, on average, A. actinomycetemcomitans was more fit in vivo in the presence of the faster-growing nonoral microbes that it never interacts with, but have larger genomic functional capacity (Fig. 1 and SI Appendix, Fig. S1 and S2). This finding was unexpected as cooperative interactions have been characterized between A. actinomycetemcomitans and other oral microbes. A. actinomycetemcomitans cross-feeds on l-lactate and cross-respires hydrogen peroxide generated by streptococci, and this interaction increases A. actinomycetemcomitans in vivo fitness (19, 29). Therefore, one might have predicted that A. actinomycetemcomitans fitness would be higher in the presence of these oral microbes. In addition, generalist organisms have been shown to be more antagonistic (6). However, our data support a model in which microbes with a more complete metabolic repertoire, whether they be sympatric or allopatric, enhance the fitness of A. actinomycetemcomitans in the murine abscess. We speculate that microbes with more metabolic capacity may synthesize a larger diversity of compounds and more readily cross-feed A. actinomycetemcomitans in vivo, providing nutrients to augment its many auxotrophies. In addition, microbes from other environments or with a larger metabolic capacity may compete less for nutrients with A. actinomycetemcomitans.
We discovered there is a core essential gene set that is universally required for A. actinomycetemcomitans fitness across the breadth of coinfections. As these core genes are also essential in monoinfection, the murine environment is the driving force for selection of core fitness determinants; coinfection cannot alleviate these core fitness determinants. Specifically, aerobic respiration is essential for A. actinomycetemcomitans in the abscess no matter the coinfecting partner. This finding, indicated by the requirements for cytochrome bd, CydDC, and the NADH:ubiquinone reductase, supports data that the abscess, at least in part, is an aerobic environment (19, 39, 40). The essentiality of respiration is also indicated by the requirement for spermidine-putrescine transport, as polyamines protect cells from reactive oxygen stress (41, 42). In addition, as most microbiology experiments are performed in monoinfection, it is an important finding that a proportion of the essential genes identified in monoinfection can never be alleviated by a coinfecting partner. Thus, monoinfection Tn-seq experiments identify genes important for polymicrobial infection and with relevance as drug targets.
Our Tn-seq data demonstrate that all coinfection microbes interacted with A. actinomycetemcomitans, directly or indirectly, as the A. actinomycetemcomitans essential genes in each coinfection differed from monoinfection. There were no conserved essential genes for coinfection specifically with sympatric or allopatric microbes, potentially because of the diversity of the coinfecting species (Fig. 3B and Table 1). However, due to our expansive dataset, we were able to estimate that one-third of the A. actinomycetemcomitans genome is important for mediating survival during coinfection. While this estimate could shift with additional data or alternative modeling, it is clear that coinfection is a pervasive and influential pressure on A. actinomycetemcomitans. Yet, overall, we found it difficult to predict the essential genes in each coinfection condition based on the native environment of the coinfecting microbe, broad physiological metrics, genome metrics, or even phylogeny. The interactions were unique and complex, likely the combination of a range of factors and influenced by the host. A shift in gene essentiality can result from a cooperative or competitive direct interaction with the coinfecting microbe or may reflect a change in the host response during coinfection.
Broadly, we found that coinfection drastically altered A. actinomycetemcomitans gene essentialities. Specifically, in the presence of some nonoral microbes, A. actinomycetemcomitans required 50% fewer genes than in monoinfection and needed few new genes. While there have been very limited studies of coculture Tn-seq, this extreme alleviation of essential genes during coinfection has not been seen before. Although it is hard to pinpoint why a gene is alleviated from our dataset, the alleviation of genes likely occurs because these allopatric coinfecting microbes lessen stress and increase nutrient availability in the host environment. For example, cross-feeding is 1 mechanism through which coinfecting partners could alleviate CoDE genes (43, 44). In other, mainly sympatric, coinfections, A. actinomycetemcomitans required a large set of new CoDE genes and few genes essential in monoinfection were alleviated. Our results indicate the dominant interactions with some sympatric microbes were competitive and exerted increased stress on A. actinomycetemcomitans. However, as specialized interactions have been identified between oral microbes (15, 16, 45), the induction of CoDE genes could also indicate intricate interactions that are important for fitness.
This extensive pairwise dataset provides an expanded context for broadly understanding cross-species interactions. While pairwise interactions might not all scale to more diverse communities or other infection environments, previous studies have found that the majority of essential genes were conserved in higher-order communities (22) and that coinfection had a stronger influence on gene essentiality than infection environment (18). Murine abscesses allow us to study interactions of oral and nonoral microbes in a sterile environment in the presence of the host immune system, but a murine abscess environment with only 2 species is undoubtably different from human abscesses or the human oral cavity. Interestingly, attachment, which is critical for biofilm formation and fitness in the oral cavity, also contributes to fitness in the abscess; genes annotated as pili and pili assembly are essential in many of our conditions, including 1 gene that was essential in all 26 conditions. However, other selective pressures certainly differ in human infections, and interactions are likely more complex. Furthermore, due to the nature of Tn-seq experiments, including the high infectious dose used in this study, the essentiality of secreted public functions is difficult to determine because of complementation from other mutants. Finally, while the 2 A. actinomycetemcomitans strains compared here in monoinfection have highly similar in vivo essential genomes (SI Appendix, Fig. S5), it will be interesting to further explore how the coinfection essential genome varies across strains. Despite these caveats, we have identified a core set of genes whose essentiality cannot be alleviated by coinfection in the host environment and the diversity of genes that can become essential in coinfection. This broad analysis is important for modeling microbe–microbe interactions and for understanding the types of processes that mediate interactions between microbes.
Methods
Genome Functional Annotation.
Genome size and 16S rRNA gene copy number was assigned using the National Center for Biotechnology Information (46). The number of protein-coding genes, protein-coding genes with EC number, and all COG annotations were assigned using the Joint Genome Institute Integrated Microbial Genomes and Microbiomes database (47). If no genome was available for the strain used, metrics were assigned using a closely related species, as indicated in Dataset S1. To confirm that low-quality genomes were not impacting 16S rRNA gene copy number, copy number was compared with closely related finished genomes. The A. actinomycetemcomitans 624 genome (ASM159426v1) was downloaded with assembly accession no. GCF_001594265.1 on September 12, 2017. sRNAs were included as in Jorth et al. (40). For A. actinomycetemcomitans, Kyoto Encyclopedia of Genes and Genomes (KEGG) terms were assigned using the KEGG Automatic Annotation Server and COG terms were assigned using KEGG’s binary relationships to COG in the BRITE database (48, 49). Two-way average nucleotide identity between A. actinomycetemcomitans VT1169 and 624 was calculated as in refs. 50 and 51.
Tn-Seq Murine Abscess Protocol.
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (52). Animal protocols were approved by the Institutional Animal Care and Use Committees of The University of Texas at Austin (protocol no. 00136) and Georgia Institute of Technology (protocol no. A17086).
Stocks of the A. actinomycetemcomitans 624 transposon mutant library (18, 32) were grown both aerobically (with 5% CO2) and anaerobically (gas mix: 5% H2, 10% CO2, 85% N2) overnight at 37 °C as lawns on tryptic soy agar yeast extract medium (TSA + 0.5% YE) to decrease clumping and increase viability. Starter cultures of coinfection microbes and wild-type A. actinomycetemcomitans were grown under the conditions indicated in Dataset S1. Cells were pooled from all starter cultures (including the transposon libraries), washed, and normalized to an OD600 of 2 in PBS. For monoinfection, the A. actinomycetemcomitans transposon library was mixed 1:1 with wild-type A. actinomycetemcomitans. For coinfections, the A. actinomycetemcomitans transposon library was mixed 1:1 with the appropriate strain of interest.
Nair was used to remove hair from both inner thighs of 10-wk-old Swiss Webster mice, and ethanol was used to clean the area before injections. Abscesses were initiated by injecting 100 µL of the mono- or coculture mixture into each murine inner thigh. Animals were euthanized and abscesses were harvested after 3 d. Abscesses were homogenized in PBS for 30 s in BeadBug tubes (Sigma-Aldrich) with 2.8-mm steel beads using a Mini-Beadbeater-16 (BioSpec Products). Next, 500 µL of homogenate was used to inoculate outgrowth cultures in 4.5 mL tryptic soy broth yeast extract + 40 μg/mL kanamycin in 5% CO2 at 37 °C, and the remaining homogenate was frozen at −20 °C. Outgrowths were cultured for 6 h to allow no more than 3 doublings then frozen at −20 °C. Both abscesses per mouse were used for microbial abundance analyses, but only 1 abscess per mouse was used for the Tn-seq analysis.
DNA Extraction for Quantification of Microbe Abundance.
DNA was extracted from homogenized abscesses following enzymatic and chemical lysis, as detailed in SI Appendix, Supplementary Methods.
qPCR for Determination of Microbe Abundance.
A. actinomycetemcomitans, S. cerevisiae, and C. albicans abundances were determined using qPCR. Primers for qPCR of A. actinomycetemcomitans were designed to amplify a 102-bp region from rpoB unique to Aggregatibacter spp.: 5′-TCA CCA AGA AGT GAA TGC AGG CGA AAC C-3′ and 5′-ACG ATG TCC ACG AAT TTC ACG AAA CCG C-3′. S. cerevisiae primers were designed to amplify a 93-bp region from SOD1: 5′-GGT GAC ATG GGT AAC GTA AAG A-3′ and 5′-AAC GGA GGT AGG ACC GAT AA-3′ (modified from ref. 53). C. albicans qPCR primers were designed to amplify a 103-bp region of MLT1: 5′-AAC GAA GGT CTC ATC GGA AAT C-3′ and 5′-CAA ATG CAC CAC CAG CAA TC-3′. To create a standard curve, genomic DNA was extracted from isolates of each microbe using the procedure above and quantified using the Qubit dsDNA HS Assay Kit (Invitrogen). Using this DNA, the standard curve was performed across a 7-log range using 3 separate dilution series in duplicate. Next, 1:10 dilutions of abscess samples were quantified in duplicate, and their Ct values were compared to this standard curve. Each 20-µL qPCR reaction consisted of 2 µL DNA, 1 µL each of forward and reverse primers at 10 µM, 10 µL PowerUP SYBR Green Master Mix (Applied Biosystems), and 6 µL water. qPCR was performed using a Quant Studio 3 (Applied Biosystems) with the following method: 50 °C for 2 min; 95 °C for 2 min; 40 cycles of 95 °C for 15 s and 60 °C for 1 min; and a melting curve consisting of 95 °C for 15 s, 60 °C for 1 min, and 95 °C with a 0.15 °C/s ramping speed then hold for 15 s.
16S rRNA Gene Sequencing and Sequence Analysis for Determination of Microbe Abundance.
The relative abundance of coinfection bacterial strains to A. actinomycetemcomitans was identified using amplicon sequencing with universal primers for the V4 region of the 16S rRNA gene (54). Reads were demultiplexed using the MiSeq system and analyzed in mothur v1.40.3 (55) following a protocol adapted from Kozich et al. (54) and used to calculate genome copies per abscess for each coinfection bacterium. Details are available in SI Appendix, Supplementary Methods and ref. 56.
Construction and Sequencing of Tn-Seq Libraries.
DNA was extracted from 5-mL outgrowth cultures from 1 abscess per mouse using a phenol:chloroform:isoamyl alcohol extraction method as follows. An equal volume of phenol:chloroform:isoamyl alcohol (25:24:1), pH8 was added to each sample. Samples were vortexed for 1 min then centrifuged at 12,000 × g for 5 min. The aqueous phase was precipitated with 0.1 volume 3 M sodium acetate and 0.8 volume 100% isopropanol, mixed, and stored at −20 °C for at least 30 min. DNA was pelleted by centrifuging at 4 °C for 30 min, washed twice with 1 mL 75% ethanol, and dissolved in 500 µL water. To remove RNA, 5 µL 5 M sodium chloride and 5 µL 10 mg/mL RNase A were added to each sample, and samples were incubated for 1 h at 37 °C. Then, samples were reextracted with phenol:chloroform:isoamyl alcohol and ethanol precipitated as above. Final samples were dissolved in 250 µL water.
Samples were prepared for transposon sequencing following previously published protocol (see TdT/2-PCR method for preparing Tn-seq Illumina libraries in supplemental material for ref. 19) with the following modifications: 1) 10 µg of DNA was C-tailed in the TdT tailing reaction; 2) 2 PCR-1 reactions were performed, each with 0.5 to 1 µg DNA and these reactions were pooled in the following purification step; and 3) a mixture of 4 mariner-2 primers were combined with 3N, 5N, 7N, and 9N random bases to increase the sequence diversity of our libraries. Libraries were quantified using qPCR with standard Illumina primers (Kapa Biosystems) and quality controlled using the High Sensitivity DNA Bioanalyzer Analysis (Agilent). Libraries were pooled and sequenced at The University of Texas at Austin Genomic Sequencing and Analysis Facility using a NextSeq 500 sequencer (Illumina) with 1 × 75 v2 chemistry.
Tn-Seq Read Processing, Mapping, and Essentials Analysis.
Sequencing sample metrics are detailed in SI Appendix, Table S1 and ref. 56. For the input, replicates consisted of the 624-pre and 624-amp samples from refs. 18 and 32, which were used along with a replicate combined from 3 aliquots used for the infections (57). All analysis scripts are adapted from previously published protocols (18, 19, 31, 58) and are available at https://github.com/glew8/Tn-seq_co-infection_analyses. Software/language versions used were R v3.4.2, DESeq2 v1.18.1 (59), Bowtie2 v2.3.2 (60), mclust v5.4 (61), and cutadapt v1.8.1 (62).
Briefly, reads were screened to confirm they contained the IR in the correct location (CCAACCTGTTA), allowing 1 mismatch, and mapped to the A. actinomycetemcomitans 624 genome with Bowtie 2 using the TnSeq2.sh script. Insertion site assignments were corrected for polymerase slippage using a custom script to collapse reads mapping within 1 bp onto local maxima using the slippage.sh and slippage.py scripts (19). A. actinomycetemcomitans VT1169 monoinfection data (63) were analyzed as here and as in ref. 19.
Then, the TnSeqDESeq2Essential_mariner.R script was run with 1,000 expected pseudodatasets, to trim = 0 (none of the most abundant sites were removed from the analysis), and no site filtering (all transposon insertion sites were analyzed, no matter the number of replicates they were identified in). This essentials analysis normalized samples for sequencing depth using estimateSizeFactors() in DESeq2. Then, pseudodatsets were constructed with the same number of insertion sites and total reads mapping to those sites as the average of the experimental replicates, randomly distributed across the genome at TA sites. In this analysis, 1,000 pseudodatasets were created for each condition, although no significant difference was seen at 400 pseudodatasets. See ref. 18 for a list of the TA sites in the A. actinomycetemcomitans 624 genome. Next, we binned insertion sites in the experimental and pseudodatasets by gene. A custom gff file was prepared with sRNAs added and the 10% of the sequence trimmed from the 3′ end of each genetic element to remove noise from insertions at the ends of genes using Excel (Dataset S3). DESeq2 was used to compare the mutant abundances in the experimental data to the mutant abundances in pseudodatasets with estimateDispersons() run with fitType=“local”. We ran nbinomWaldTest() with betaPrior = TRUE and betaPrior = FALSE (Dataset S4). The data analyzed for this paper were with betaPrior = TRUE, as this shrinks log2 fold-changes when counts are low, dispersion is high, or degrees-of-freedom are low, and we feel this is the more conservative flag. Finally, we performed bimodal clustering using mclust with modelNames = “V”. Genes analyzed as essential here 1) had a Padj < 0.05 in the DESeq2 analysis and 2) contributed to the left, “reduced” bimodal distribution in the mclust analysis with an uncertainty <0.05.
ACT75_RS08345 Mutant Construction and Essentiality Confirmation.
DNA was transformed into A. actinomycetemcomitans 624 from the A. actinomycetemcomitans VT1169 mutant library following previously published protocols (24, 32, 64), as detailed in SI Appendix, Supplementary Methods.
Statistical Analyses.
Accumulation curves and clustering were performed in R using the vegan package (65). Fisher’s exact test and P value corrections were performed in R. Correlations, Mann–Whitney U tests, and all other statistical analyses were performed in Prism 7 (GraphPad Software).
Data Availability.
Raw 16S rRNA gene sequencing data and raw Tn-seq sequencing files are available in the National Center for Biotechnology Information Sequence Read Archive (SRA) under BioProject accession no. PRJNA540913. A. actinomycetemcomitans input replicates pre (p) and amp (a) are available under SRA accession nos. SRX2426036 and SRX2426058, respectively. Monoinfection A. actinomycetemcomitans VT1169 data are available under SRA accession no. SRP070130.
Supplementary Material
Acknowledgments
We thank the M.W. laboratory for discussion of this manuscript and the Texas Advanced Computing Center at The University of Texas at Austin for providing high performance computing resources that have contributed to the research results reported within this paper. This work was supported by National Institutes of Health Grants R01DE020100 (to M.W.), R01DE023193 (to M.W. and R.J.L.), F32DE027281 (to G.R.L.), and F31DE024931 (to A.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Raw 16S rRNA gene sequencing data and raw transposon insertion sequencing files have been deposited in the NCBI Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (BioProject accession no. PRJNA540913).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907619116/-/DCSupplemental.
References
- 1.Mylona P., Pawlowski K., Bisseling T., Symbiotic nitrogen fixation. Plant Cell 7, 869–885 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hibbing M. E., Fuqua C., Parsek M. R., Peterson S. B., Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birch L., The meanings of competition. Am. Nat. 91, 5–18 (1957). [Google Scholar]
- 4.Nadell C. D., Xavier J. B., Foster K. R., The sociobiology of biofilms. FEMS Microbiol. Rev. 33, 206–224 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Vetsigian K., Jajoo R., Kishony R., Structure and evolution of Streptomyces interaction networks in soil and in silico. PLoS Biol. 9, e1001184 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russel J., Røder H. L., Madsen J. S., Burmølle M., Sørensen S. J., Antagonism correlates with metabolic similarity in diverse bacteria. Proc. Natl. Acad. Sci. U.S.A. 114, 10684–10688 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moné Y., et al. , An example of molecular co-evolution: Reactive oxygen species (ROS) and ROS scavenger levels in Schistosoma mansoni/Biomphalaria glabrata interactions. Int. J. Parasitol. 41, 721–730 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Germain R. M., Weir J. T., Gilbert B., Species coexistence: Macroevolutionary relationships and the contingency of historical interactions. Proc. Biol. Sci. 283, 20160047 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson J., The Coevolutionary Process (University of Chicago Press, Chicago, 1994). [Google Scholar]
- 10.Levine J. M., HilleRisLambers J., The importance of niches for the maintenance of species diversity. Nature 461, 254–257 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Hardin G., The competitive exclusion principle. Science 131, 1292–1297 (1960). [DOI] [PubMed] [Google Scholar]
- 12.Fine D. H., Kaplan J. B., Kachlany S. C., Schreiner H. C., How we got attached to Actinobacillus actinomycetmecomitans: A model for infectious diseases. Periodontol. 2000 42, 114–157 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Fine D. H., et al. , Aggregatibacter actinomycetemcomitans and its relationship to initiation of localized aggressive periodontitis: Longitudinal cohort study of initially healthy adolescents. J. Clin. Microbiol. 45, 3859–3869 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaplan A. H., Weber D. J., Oddone E. Z., Perfect J. R., Infection due to Actinobacillus actinomycetemcomitans: 15 cases and review. Rev. Infect. Dis. 11, 46–63 (1989). [DOI] [PubMed] [Google Scholar]
- 15.Kuramitsu H. K., He X., Lux R., Anderson M. H., Shi W., Interspecies interactions within oral microbial communities. Microbiol. Mol. Biol. Rev. 71, 653–670 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mark Welch J. L., Rossetti B. J., Rieken C. W., Dewhirst F. E., Borisy G. G., Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. U.S.A. 113, E791–E800 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown S. A., Whiteley M., A novel exclusion mechanism for carbon resource partitioning in Aggregatibacter actinomycetemcomitans. J. Bacteriol. 189, 6407–6414 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibberson C. B., et al. , Co-infecting microorganisms dramatically alter pathogen gene essentiality during polymicrobial infection. Nat. Microbiol. 2, 17079 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stacy A., Fleming D., Lamont R. J., Rumbaugh K. P., Whiteley M., A commensal bacterium promotes virulence of an opportunistic pathogen via cross-respiration. MBio 7, 1–10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Armbruster C. E., et al. , Genome-wide transposon mutagenesis of Proteus mirabilis: Essential genes, fitness factors for catheter-associated urinary tract infection, and the impact of polymicrobial infection on fitness requirements. PLoS Pathog. 13, e1006434 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Opijnen T., Camilli A., Transposon insertion sequencing: A new tool for systems-level analysis of microorganisms. Nat. Rev. Microbiol. 11, 435–442 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morin M., Pierce E. C., Dutton R. J., Changes in the genetic requirements for microbial interactions with increasing community complexity. eLife 7, e37072 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klappenbach J. A., Dunbar J. M., Schmidt T. M., rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66, 1328–1333 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stacy A., et al. , Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc. Natl. Acad. Sci. U.S.A. 111, 7819–7824 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stacy A., Abraham N., Jorth P., Whiteley M., Microbial community composition impacts pathogen iron availability during polymicrobial infection. PLoS Pathog. 12, e1006084 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moazzam A. A., Rajagopal S. M., Sedghizadeh P. P., Zada G., Habibian M., Intracranial bacterial infections of oral origin. J. Clin. Neurosci. 22, 800–806 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Robertson D., Smith A. J., The microbiology of the acute dental abscess. J. Med. Microbiol. 58, 155–162 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Liu X., et al. , Real-time mapping of a hydrogen peroxide concentration profile across a polymicrobial bacterial biofilm using scanning electrochemical microscopy. Proc. Natl. Acad. Sci. U.S.A. 108, 2668–2673 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramsey M. M., Rumbaugh K. P., Whiteley M., Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 7, e1002012 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramsey M. M., Whiteley M., Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc. Natl. Acad. Sci. U.S.A. 106, 1578–1583 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner K. H., Wessel A. K., Palmer G. C., Murray J. L., Whiteley M., Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc. Natl. Acad. Sci. U.S.A. 112, 4110–4115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narayanan A. M., Ramsey M. M., Stacy A., Whiteley M., Defining genetic fitness determinants and creating genomic resources for an oral pathogen. Appl. Environ. Microbiol. 83, e00797-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lomolino M. V., Ecology’s most general, yet protean pattern: The species-area relationship. J. Biogeogr. 27, 17–26 (2000). [Google Scholar]
- 34.Dengler J., Which function describes the species-area relationship best? A review and empirical evaluation. J. Biogeogr. 36, 728–744 (2009). [Google Scholar]
- 35.Kindt R., Van Damme P., Simons A. J., Patterns of species richness at varying scales in western Kenya: Planning for agroecosystem diversification. Biodivers. Conserv. 15, 3235–3249 (2006). [Google Scholar]
- 36.Balashova N. V., Shah C., Patel J. K., Megalla S., Kachlany S. C., Aggregatibacter actinomycetemcomitans LtxC is required for leukotoxin activity and initial interaction between toxin and host cells. Gene 443, 42–47 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Schneider B., Weigel W., Sztukowska M., Demuth D. R., Identification and functional characterization of type II toxin/antitoxin systems in Aggregatibacter actinomycetemcomitans. Mol. Oral Microbiol. 33, 224–233 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansen A. M., et al. , SspA is required for acid resistance in stationary phase by downregulation of H-NS in Escherichia coli. Mol. Microbiol. 56, 719–734 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Fetiye K., et al. , Comparison in a rat thigh abscess model of imipenem, meropenem and cefoperazone-sulbactam against Acinetobacter baumannii strains in terms of bactericidal efficacy and resistance selection. Ann. Clin. Microbiol. Antimicrob. 3, 2 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jorth P., Trivedi U., Rumbaugh K., Whiteley M., Probing bacterial metabolism during infection using high-resolution transcriptomics. J. Bacteriol. 195, 4991–4998 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shah P., Swiatlo E., A multifaceted role for polyamines in bacterial pathogens. Mol. Microbiol. 68, 4–16 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Chattopadhyay M. K., Tabor C. W., Tabor H., Polyamines protect Escherichia coli cells from the toxic effect of oxygen. Proc. Natl. Acad. Sci. U.S.A. 100, 2261–2265 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Foster K. R., Bell T., Competition, not cooperation, dominates interactions among culturable microbial species. Curr. Biol. 22, 1845–1850 (2012). [DOI] [PubMed] [Google Scholar]
- 44.D’Souza G., et al. , Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat. Prod. Rep. 35, 455–488 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Kolenbrander P. E., et al. , Bacterial interactions and successions during plaque development. Periodontol. 2000 42, 47–79 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Benson D. A., et al. , GenBank. Nucleic Acids Res. 42, D32–D37 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen I. A., et al. , IMG/M v.5.0: An integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 47, D666–D677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moriya Y., Itoh M. , Okuda S., Yoshizawa A. C., Kanehisa M., KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanehisa M., Goto S., KEGG automatic annotation server. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goris J., et al. , DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez-R L., Konstantinidis K., The enveomics collection: A toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ. Preprints 4, e1900v1 (2016). [Google Scholar]
- 52.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
- 53.Erjavec N., Nyström T., Sir2p-dependent protein segregation gives rise to a superior reactive oxygen species management in the progeny of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 104, 10877–10881 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kozich J. J., Westcott S. L., Baxter N. T., Highlander S. K., Schloss P. D., Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schloss P. D., et al. , Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lewin G. R., Stacy A., Michie K. L., Lamont R. J., Whiteley M., Large-scale identification of pathogen essential genes during co-infection with sympatric and allopatric microbes. Sequence Read Archive. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA540913. Deposited 3 May 2019. [DOI] [PMC free article] [PubMed]
- 57.Stacy A., Whiteley M. , Tn-seq on Aa 624 input pool, 624 abscess, 624+Sg abscess. Sequence Read Archive. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA357601. Accessed 26 September 2017.
- 58.Powell J. E., Leonard S. P., Kwong W. K., Engel P., Moran N. A., Genome-wide screen identifies host colonization determinants in a bacterial gut symbiont. Proc. Natl. Acad. Sci. U.S.A. 113, 13887–13892 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scrucca L., Fop M., Murphy T. B., Raftery A. E., mclust 5: Clustering, classification and density estimation using Gaussian finite mixture models. R J. 8, 289–317 (2016). [PMC free article] [PubMed] [Google Scholar]
- 62.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal 17, 10–12 (2011). [Google Scholar]
- 63.Stacy A., Whiteley M., Tn-seq of A. actinomycetemcomitans: mono-infection of murine thigh abscess. Sequence Read Archive. https://www.ncbi.nlm.nih.gov/sra/SRP070130. Accessed 25 April 2019.
- 64.Wang Y., Goodman S. D., Redfield R. J., Chen C., Natural transformation and DNA uptake signal sequences in Actinobacillus actinomycetemcomitans. J. Bacteriol. 184, 3442–3449 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oksanen J., et al. , vegan: Community ecology package. R Package Version 2.4-4. https://CRAN.R-project.org/package=vegan. Accessed 5 December 2017.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw 16S rRNA gene sequencing data and raw Tn-seq sequencing files are available in the National Center for Biotechnology Information Sequence Read Archive (SRA) under BioProject accession no. PRJNA540913. A. actinomycetemcomitans input replicates pre (p) and amp (a) are available under SRA accession nos. SRX2426036 and SRX2426058, respectively. Monoinfection A. actinomycetemcomitans VT1169 data are available under SRA accession no. SRP070130.