Significance
Doxorubicin is a DNA-damaging agent that is highly effective against various types of cancers, but a subset of treated patients develop heart failure for unclear genetic reasons. In the current study using p53 mouse models, low-dose doxorubicin as administered in the clinics surprisingly revealed that the absence of p53 increases susceptibility to doxorubicin cardiotoxicity while a mutant of p53 that retains mitochondrial regulation is protective. Notably, promoting mitochondrial biogenesis with a simple vitamin supplement ameliorated the cardiotoxicity. Furthermore, mitochondrial blood markers observed in association with doxorubicin susceptibility could provide guidance for the safer use of this effective chemotherapy.
Keywords: p53, anthracycline, cardiomyopathy, mitochondria, mtDNA
Abstract
Doxorubicin is a widely used chemotherapeutic agent that causes dose-dependent cardiotoxicity in a subset of treated patients, but the genetic determinants of this susceptibility are poorly understood. Here, we report that a noncanonical tumor suppressor activity of p53 prevents cardiac dysfunction in a mouse model induced by doxorubicin administered in divided low doses as in the clinics. While relatively preserved in wild-type (p53+/+) state, mice deficient in p53 (p53−/−) developed left ventricular (LV) systolic dysfunction after doxorubicin treatment. This functional decline in p53−/− mice was associated with decreases in cardiac oxidative metabolism, mitochondrial mass, and mitochondrial genomic DNA (mtDNA) homeostasis. Notably, mice with homozygous knockin of the p53 R172H (p53172H/H) mutation, which like p53−/− state lacks the prototypical tumor suppressor activities of p53 such as apoptosis but retains its mitochondrial biogenesis capacity, showed preservation of LV function and mitochondria after doxorubicin treatment. In contrast to p53-null state, wild-type and mutant p53 displayed distinct mechanisms of transactivating mitochondrial transcription factor A (TFAM) and p53-inducible ribonucleotide reductase 2 (p53R2), which are involved in mtDNA transcription and maintenance. Importantly, supplementing mice with a precursor of NAD+ prevented the mtDNA depletion and cardiac dysfunction. These findings suggest that loss of mtDNA contributes to cardiomyopathy pathogenesis induced by doxorubicin administered on a schedule simulating that in the clinics. Given a similar mtDNA protection role of p53 in doxorubicin-treated human induced pluripotent stem cell (iPSC)-derived cardiomyocytes, the mitochondrial markers associated with cardiomyopathy development observed in blood and skeletal muscle cells may have prognostic utility.
Despite its well-known cardiotoxicity, the anthracycline doxorubicin (Dox) continues to be widely used in the clinics because it is an effective chemotherapeutic agent against a variety of cancers (1–5). The cardiotoxic side effects of Dox are cumulative-dose dependent and can be either acute or chronic in presentation (6). Acute cardiotoxicity occurs within days of treatment and can manifest with transient arrhythmias and reversible myopericardial inflammation. In contrast, chronic Dox cardiotoxicity can present years after treatment as a cardiomyopathy with significant morbidity and mortality due to its progressive nature. In a prospective analysis of breast and lung cancer patients treated with a cumulative Dox dose of 150 mg/m2, well below the critical heart failure threshold of 550 mg/m2 (1), up to 7% of the patients were estimated to develop on-study evidence of compromised left ventricular (LV) function (7). Given the large number of patients who receive Dox, even a small risk of developing cardiomyopathy represents a significant number of individuals, making it imperative to understand its pathogenesis for developing new preventive and therapeutic strategies.
The mechanism of Dox cardiotoxicity has intrigued both basic and clinical investigators for many years and is thought to be mediated by a number of different cellular processes (6). Dox-induced oxidative stress and cardiomyocyte death are widely believed to play a deleterious role with more recent translational work showing mitochondrial dysfunction and disruption of intracellular iron and calcium homeostasis (6, 8–10). Consistent with its interference of DNA replication and transcription in cancer cells, Dox has been reported to cause DNA double-strand breaks via topoisomerase IIβ (TOP2B), an enzyme expressed in cardiomyocytes, with impairment of mitochondrial biogenesis and increased oxidative stress (11, 12). p53 is a tumor suppressor gene that can maintain genomic integrity by either repairing DNA or causing cell death. p53-regulated cell death pathways have been shown to mediate acute Dox cardiotoxicity using mouse models with bolus injection of high-dose Dox, which can result in global cardiac dysfunction and mortality within days (8, 13, 14). Interestingly, a study using 2-wk-old p53+/+ mice (∼7-y-old human equivalent) chronically treated with low doses of Dox initially showed decreased cardiac function that recovered after 13 wk, while mice expressing a dominant mutant p53 R193P, which were initially resistant to Dox cardiotoxicity, exhibited decreased cardiac function at a later 13-wk time point (15). This study suggested that specific p53 activities may be cardioprotective, especially evident after longer follow-up periods. Thus, p53 appears to have contrasting effects on the development of Dox cardiomyopathy depending on the dosage and timing of the genotoxic stress, a dual nature that is well described for its many regulatory activities in response to cellular stress (16, 17). This also suggested a need to delineate the role of p53 by comparing p53−/− and p53-mutant states that lose or retain specific activities of p53 in an animal model simulating clinical dosing of Dox.
Accumulating evidence points to the mitochondrion as playing a central role in Dox-induced cardiomyopathy. Genetic studies have shown an association between Dox exposure and mitochondrial genomic DNA (mtDNA) depletion in cardiac tissues of both animal models and patients, consistent with the observation of increased susceptibility of mitochondria to Dox in cardiomyocytes derived from patients with the cardiomyopathy (10, 18, 19). The maintenance of mtDNA after Dox exposure has been reported to involve p53, mitochondrial topoisomerase I (TOP1MT), and sirtuin 3 (SIRT3) (20–22). Given these investigations implicating mtDNA in pathogenesis, the characterization of p53-mutant and -null mice with similar loss of apoptosis activity but contrasting mitochondrial effects provided an opportunity to dissect the role of p53 in Dox cardiomyopathy (23, 24). Wild-type and mutant p53 can regulate mitochondrial biogenesis through nuclear transcriptional mechanisms as well as by translocating into mitochondria to maintain mtDNA homeostasis (25). Mice with knockin of the p53 R172H mutation (p53172H/H), homologous to human TP53 R175H that causes the early-onset cancer disorder Li-Fraumeni syndrome (LFS), retains mitochondrial biogenesis and mtDNA maintenance activities while p53-null mice have mitochondrial deficiency (24, 26–28). To dissect the role of p53-regulated cell death and mitochondrial biogenesis on cardiac function after low-dose Dox exposure, p53172H/H mice were compared with wild-type and p53-null (p53−/−) mice. Here, we show that mutant p53 can protect against late-onset Dox cardiotoxicity and that this protective activity is associated with its regulation of the mitochondria but not with the loss of apoptosis or cell-cycle arrest activity. Importantly, treatment of p53−/− mice with nicotinamide mononucleotide (NMN), a vitamin B3 metabolite that promotes mitochondrial biogenesis, prevented mtDNA damage and Dox-induced cardiac dysfunction. Studies using human induced pluripotent stem (iPS) cell-derived cardiomyocytes showed that this p53 activity translates across species with therapeutic implications.
Results
Dox Decreases Cardiac Function and Mitochondrial Metabolism in p53−/− Mice.
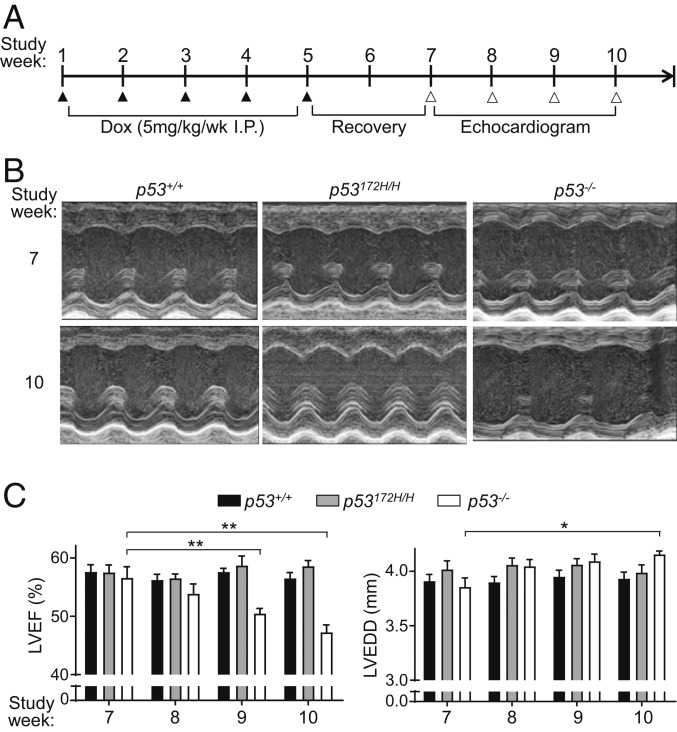
To model chronic Dox cardiomyopathy, wild-type p53+/+, mutant p53172H/H, and null p53−/− mice were treated with low doses of Dox over a 5-wk period beginning at age 5 wk (young adult human equivalent) (Fig. 1A). After a 2-wk recovery period, the cardiac function of these mice was assessed by echocardiography over a 4-wk time course. Dox-treated p53−/− mice showed a progressive decline in LV ejection fraction that became significant 4 wk after treatment (study week 9 vs. week 7), while these changes were not evident in p53172H/H or p53+/+ mice (Fig. 1 B and C). This was consistent with the clinical observation that only a small fraction of Dox-treated patients develop cardiomyopathy (7). The decline in the cardiac function of p53−/− mice was also associated with significant LV dilatation at week 10 (Fig. 1C). Because the R172H mutation of p53 results in the loss of its apoptotic and cell cycle arrest activities (26), the retention of cardiac function in p53172H/H mice after Dox treatment suggested that a nonprototypical p53 tumor suppressor activity, such as mitochondrial regulation, may be conferring the cardiac protection. Therefore, after the final echocardiogram, the mice were killed to assess cardiac mitochondrial function and metabolism. The mitochondrial utilization of fatty acids is known to be impaired in heart failure, so exogenous palmitate was used as energy substrate to measure respiration in isolated cardiac mitochondria (29, 30). The mitochondria of Dox-treated p53−/− mouse hearts showed decreased fatty acid oxidation, while those of both p53+/+ and p53172H/H mice were not significantly affected, reflecting their preserved cardiac function by echocardiogram (Fig. 2A).
Fig. 1.
Both wild-type and mutant p53 prevent cardiac dysfunction after chronic low-dose doxorubicin (Dox) exposure. (A) Schedule of Dox treatment and cardiac function evaluation. Male mice were injected with Dox i.p. weekly over a 5-wk period (study week 1–5). After 2 wk of recovery, LV systolic function was evaluated by weekly echocardiograms for 4 wk prior to killing for tissue analysis (end of study week 10). (B) Representative M-mode echocardiographic images of the indicated p53 genotype hearts at study week 7 and 10. (C) LV ejection fraction (LVEF) and LV end-diastolic dimension (LVEDD) of the indicated p53 genotype mice by echocardiogram (n = 9–12). Statistical testing by 2-way ANOVA in comparison with study week 7 of the corresponding p53 genotype group. Values are mean ± SEM. *P < 0.05; **P < 0.01.
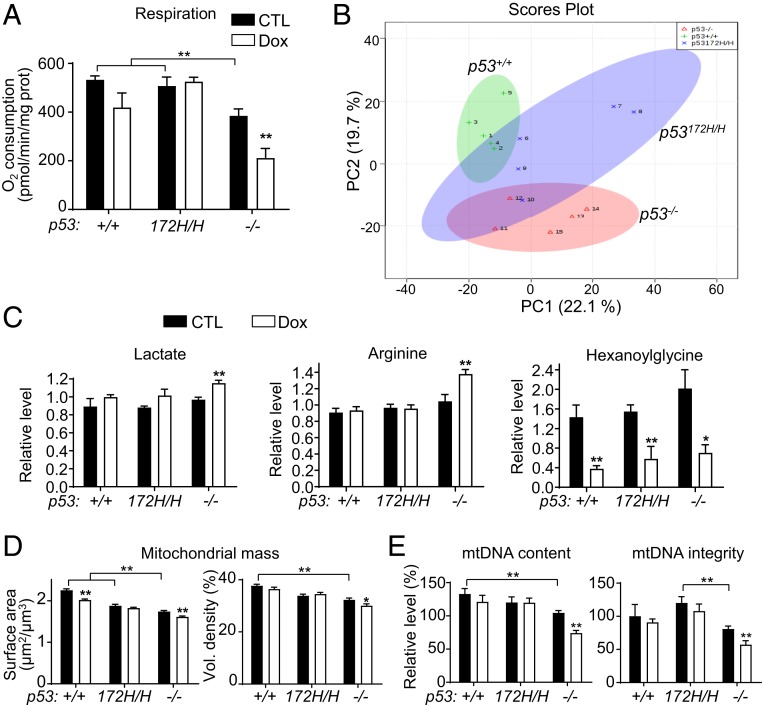
Fig. 2.
Dox-treated p53−/− mouse hearts show decreased mitochondrial function and mass. Dox-treated mice were killed after completion of echocardiograms at the end of study week 10, and heart tissues were used for mitochondrial analysis and metabolomic profiling. (A) Fatty acid oxidation was assessed in isolated mitochondria using palmitoyl-carnitine as substrate during state 3 respiration (n ≥ 4). (B) Principal-component analysis (PCA) of metabolite changes caused by Dox treatment. Heart samples of each p53 genotype are as follows: p53+/+, green plus, +1 to +5; p53172H/H, blue “x,” Χ6 to Χ10; and p53−/−, red triangle, Δ11 to Δ15 (n = 5). (C) Relative levels of some metabolites significantly altered by Dox treatment identified by metabolomic screening (SI Appendix, Fig. S2A; n = 5). (D) Quantification of cardiac mitochondrial mass in TEM images using stereological techniques (>50 TEM images were analyzed for each sample; SI Appendix, Fig. S2). (E) Quantification of mtDNA content and integrity in cardiac tissues at the end of study week 10 (n ≥ 6). Statistical testing by 2-way ANOVA in comparison with untreated control (black bar) of the respective genotype. Values are mean ± SEM. *P < 0.05; **P < 0.01.
By mass spectrometry-based metabolomic profiling, p53−/− heart tissue showed distinct metabolic changes in response to Dox treatment compared with that of p53+/+ and p53172H/H mice, while the p53172H/H tissue displayed partial overlap with that of both p53+/+ and p53−/− mice (Fig. 2B and SI Appendix, Fig. S1A). The heart tissues of the 3 genotypes showed disparate changes in mitochondrial metabolism, such as linoleic acid metabolism in p53+/+, glutamine/glutamate metabolism in p53172H/H, and glyoxylate/dicarboxylate metabolism in p53−/− genotype states (SI Appendix, Fig. S1B). Notably, cardiac tissue lactate levels were increased only in p53−/− mouse hearts, possibly representing a compensatory increase in glycolysis due to decreased oxidative metabolism (Fig. 2C). Like lactate, the steady-state levels of arginine were also increased only in p53−/− mice, consistent with reports of elevated arginine levels in mitochondrial dysfunction and heart failure (Fig. 2C) (31, 32). All 3 genotypes showed significant alterations in the steady-state levels of hexanoylglycine, an intermediate in the branch-chain amino acid catabolism and stress response marker (33), suggesting that low-dose Dox treatment can affect cardiac metabolism regardless of p53 status but that the mitochondria of p53−/− mice are more susceptible to damage (Fig. 2C).
Dox Decreases Mitochondrial Mass and mtDNA Content in p53−/− Hearts.
Given the functional changes in the mitochondria, we examined the morphology of the mitochondria in heart tissue samples by transmission electron microscopy (TEM). Although the mitochondrial morphology appeared qualitatively unchanged by Dox treatment (SI Appendix, Fig. S2), stereologic quantification of the TEM micrographs revealed a modest but significant decrease in both the surface area and volume of the mitochondria in Dox-treated p53−/− mouse hearts while these parameters were unchanged in the mutant p53172H/H state (Fig. 2D). Notably, the p53+/+ cardiac mitochondrial surface area was also decreased by Dox treatment, suggesting some degree of sensitivity to Dox (Fig. 2D). As another estimate of mitochondrial mass, quantification of mtDNA revealed significant decreases in both its quantity and quality in Dox-treated p53−/− mouse hearts (Fig. 2E). There was only a trend of decreased mtDNA content in wild-type hearts after Dox treatment, while the mtDNA level of mutant p53172H/H hearts was higher than that of p53−/− hearts in both quantity and quality (Fig. 2E). Although early cancer development precluded long-term follow-up of p53−/− and p53172H/H mice, some wild-type mice displayed diminished cardiac function in association with lower mtDNA content greater than a year after Dox exposure, a correlation that was also statistically significant in a pooled analysis (SI Appendix, Fig. S3). Besides highlighting the importance of mtDNA maintenance in cardiac function, this observation supported the validity of the cardiomyopathy model utilized in this study as the human disease also occurs sporadically long after Dox exposure. Although p53−/− mice do not have an overt cardiac phenotype, the basal mitochondrial content and function were significantly lower than that of either p53+/+ or p53172H/H mice, which could contribute to their susceptibility to Dox-induced cardiotoxicity (Fig. 2 A, D, and E).
Both Wild-Type and Mutant p53 Regulate Genes Involved in mtDNA Homeostasis.
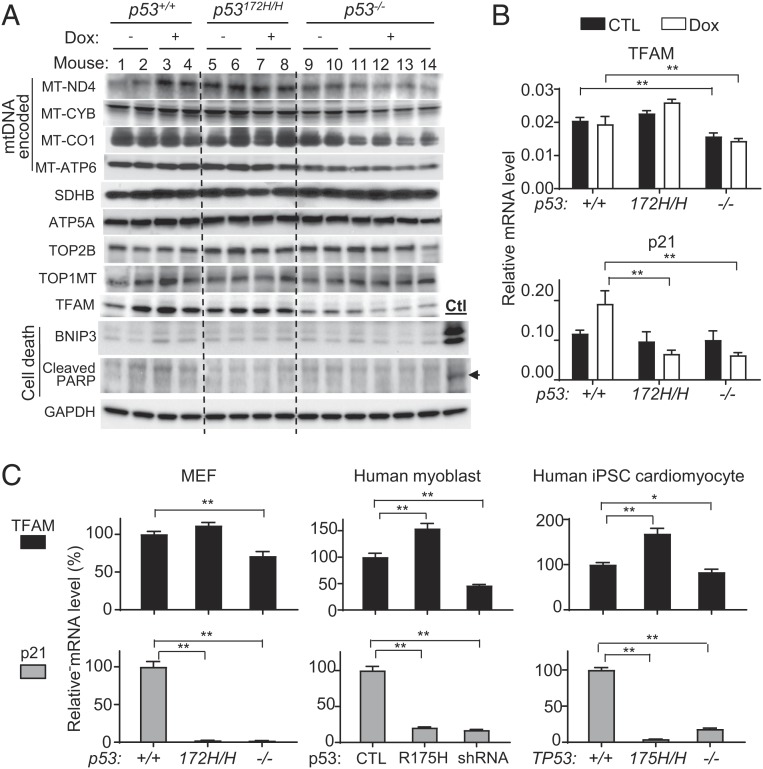
Because the median half-life of mitochondrial proteins in the heart has been estimated at 17 d, the chronic depletion of mtDNA by Dox treatment would be expected to result in a generalized deficiency of mtDNA-encoded respiratory complex subunit proteins over a period of time (34, 35). Indeed, there was decreased protein expression of some representative mtDNA-encoded respiratory complex subunits in the cardiac tissues of p53−/− mice, but not in those of either p53+/+ or p53172H/H mice, 6 wk after Dox treatment (Fig. 3A). In contrast, nuclear genome-encoded succinate dehydrogenase B (SDHB) and ATPase subunit 5 (ATP5A) levels remained unchanged.
Fig. 3.
Dox-treated p53−/− mouse hearts show decreased mtDNA-encoded proteins in association with low TFAM levels. (A) Immunoblots of mtDNA- and nuclear DNA-encoded proteins of cardiac tissues at the end of study week 10 as described in Fig. 1A. Positive control cardiac tissue sample (Ctl) for cell death markers was obtained from p53+/+ mice 4 d after treatment with a bolus dose of Dox (20 mg/kg body weight). GAPDH serves as loading control. (B) To detect the Dox-induced transcriptional activity of p53, TFAM and p21 mRNAs were quantified in cardiac tissues of mice 1 d after final Dox injection per schedule (n = 4–6). (C) Differential effect of p53-mutant and -null states on TFAM and p21 expression in MEF, human myoblasts, and human iPS cell-derived cardiomyocytes (n ≥ 3). Statistical testing by 1- or 2-way ANOVA. Values are mean ± SEM. *P < 0.05; **P < 0.01.
Mechanistically, p53-mediated acute cell death via BNIP3 and ferroptosis have been reported to be involved in Dox cardiotoxicity in the high-dose Dox mouse model (8, 14). Under the divided low-dose Dox cardiotoxicity condition, the levels of apoptosis were similarly low in the cardiac tissue of all p53 genotypes as assessed by both immunoblotting and TUNEL staining, which contrasted with their marked induction by acute high-dose Dox treatment in the p53+/+ state (Fig. 3A and SI Appendix, Fig. S4). Genomic DNA damage by cardiomyocyte-specific topoisomerase 2β (TOP2B) and mitochondrial topoisomerase 1 (TOP1MT) have also been associated with Dox cardiotoxicity, but their protein levels were not altered by either p53 genotype or Dox treatment (Fig. 3A) (12, 21). This suggested the involvement of other factors in the susceptibility of p53−/− cardiac mtDNA to low-dose Dox cardiotoxicity. We examined the level of mitochondrial transcription factor A (TFAM), which can be regulated by both wild-type and mutant p53 and is known to be critical for both mtDNA transcription and maintenance (24, 28). Indeed, TFAM protein levels were relatively lower in p53−/− cardiac tissue at baseline and appeared further reduced after Dox treatment (Fig. 3A). Consistent with its transactivation by p53, TFAM mRNA levels were lower in the cardiac tissue of p53−/− mice compared with that of either p53+/+ or p53172H/H mice (Fig. 3B, Top). As positive control, p53 target gene p21 mRNA expression was induced by Dox only in p53+/+ mice (Fig. 3B, Bottom). Taken together, our data suggested that deficiency in TFAM and mtDNA regulation may contribute to the development of late-onset cardiomyopathy caused by low-dose injections of Dox as administered in the clinics.
The generalizability of the mutant p53 effect on TFAM expression was confirmed in vitro using mouse embryonic fibroblasts (MEFs) and human myoblasts obtained from needle biopsies of skeletal muscle. Both mouse and human mutant p53, either endogenously expressed in MEFs or transduced with lentivirus into myoblasts, showed transactivation of TFAM but not of p21, while both genes were expressed at low levels in p53-null MEFs and p53 short hairpin RNA (shRNA) knockdown myoblasts (Fig. 3C). To determine whether this activity of mutant p53 is preserved in the cell type affected in cardiomyopathy pathogenesis, human iPS cells with homozygous R175H knockin (TP53175H/H, human homolog of the mouse p53 R172H mutation) or disruption (TP53−/−) of the TP53 gene were generated using CRISPR-Cas9 technique. The iPS cells were differentiated into cardiomyocytes with high efficiency in chemically defined medium, and the day 30 cardiomyocytes were confirmed for the expression of cardiac markers before experimental use (SI Appendix, Fig. S5) (36). Ki67 staining of the cardiomyocytes was similarly low for all TP53 genotypes indicating their comparable nonproliferative state (SI Appendix, Fig. S5B). Human TP53175H/H cardiomyocytes showed a similarly disparate pattern of TFAM and p21 expression, confirming the important difference in mtDNA regulation by p53-null and p53-mutant states in the human cell type affected by Dox cardiomyopathy (Fig. 3C).
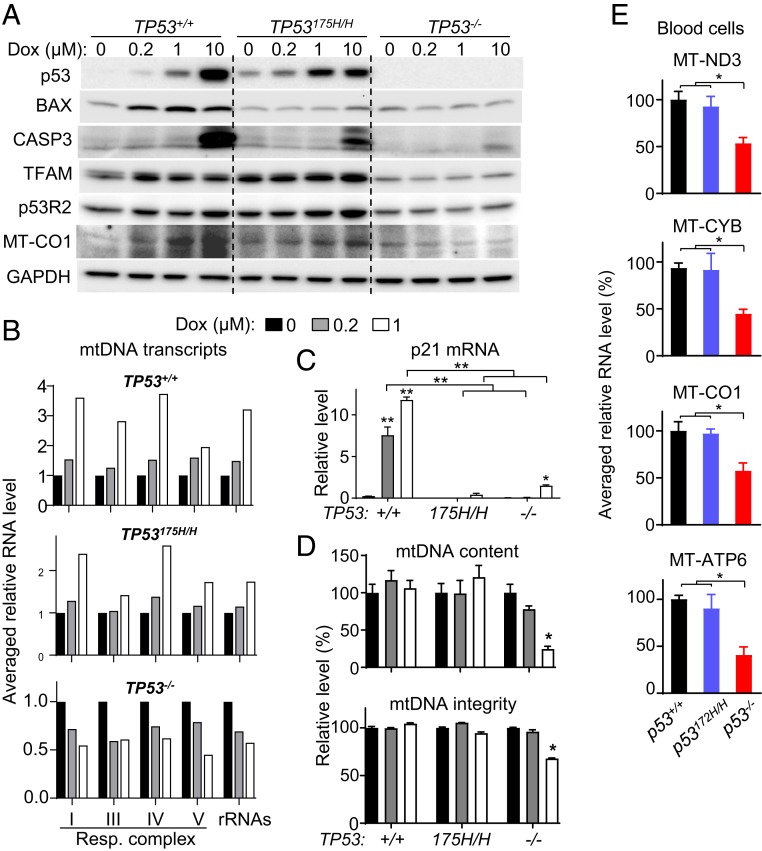
To further delineate the mechanisms underlying pathogenesis, we examined the response of human iPS cell-derived cardiomyocytes with different TP53 genotypes 48 h after Dox exposure. Like TP53−/− cells, TP53175H/H cardiomyocytes expressed lower levels of apoptosis mediator BAX compared with TP53+/+ cells, which showed marked induction of cleaved caspase 3 only at the highest concentration of Dox (Fig. 4A). However, in contrast to the null state, TP53175H/H cardiomyocytes displayed higher basal and Dox-induced protein levels of TFAM, reflective of its mRNA level (Figs. 3C and 4A). Another p53 target gene RRM2B (p53R2) is also known to play an important role in mtDNA replication and repair in quiescent cells (37, 38). Its basal and Dox-induced levels in wild-type and mutant p53 human cardiomyocytes were higher compared with p53-null state (Fig. 4A). This indicated that mutant p53 retains the capacity to promote p53R2 expression, which may also contribute to the repair and maintenance of mtDNA.
Fig. 4.
Absence of p53 impairs mtDNA maintenance and transcription in cardiomyocytes and blood cells. Human iPS cell-derived cardiomyocytes were treated with the indicated concentration of Dox for 48 h (A–D). (A) Immunoblots of whole-cell lysates examining apoptosis and mtDNA regulators. CASP3, cleaved caspase 3. Note that a high dose of Dox (10 μM) was required to elicit p53-dependent cell death. (B) Averaged relative RNA levels of mtDNA-encoded genes grouped by respiratory complexes and rRNAs (individual genes in SI Appendix, Fig. S6). (C) p21 mRNA expression in response to Dox exposure serves as marker of wild-type p53 activity (n = 3). (D) Quantification of mtDNA content and integrity (n ≥ 4). (E) Representative mtDNA transcript levels for the mitochondrial respiratory complex in mouse peripheral blood mononuclear cells of the indicated p53 genotype. Statistical testing by 1- or 2-way ANOVA in comparison with untreated control or between indicated groups. Values are mean ± SEM. *P < 0.05; **P < 0.01.
Dox Has Paradoxical Effects on mtDNA Transcription Dependent on p53.
The decrease in the mtDNA-encoded respiratory subunit proteins of p53-null mouse cardiac tissue and MT-CO1 of human cardiomyocytes in the setting of decreased TFAM levels suggested the possibility of impaired mtDNA transcription after Dox exposure (Fig. 4A). Indeed, the relative transcript levels of mtDNA-encoded genes for both respiratory complex subunits (13 genes) and ribosomal RNAs (2 genes) were decreased by Dox treatment in TP53−/− cardiomyocytes while they were increased in the TP53+/+ and TP53175H/H genotypes (Fig. 4B and SI Appendix, Fig. S6). In TP53175H/H cardiomyocytes, the increase in mtDNA transcripts with concurrent loss of cell cycle inhibitor p21 induction confirmed the dissociation of tumor suppressor and mitochondrial regulatory activities of mutant p53 (Fig. 4C). As observed in mouse heart tissue, mtDNA content and integrity were also preserved in human TP53+/+ and TP53175H/H, but not in TP53−/−, cardiomyocytes with Dox treatment (Fig. 4D). Based on the correlation between decreased mtDNA transcripts and Dox-induced cardiomyopathy, we wondered whether blood cell mtDNA transcripts would show a similar pattern. Peripheral blood mononuclear cells were isolated from the 3 different p53 genotype mice, and their representative mtDNA transcripts were quantified. The mRNA levels for ND3, CYB, CO1, and ATP6, representing respiratory complexes I, III, IV, and V, respectively, were all lower in p53−/− compared with p53+/+ or p53172H/H blood cells, raising the possibility that they could serve as markers of susceptibility to Dox cardiomyopathy (Fig. 4E).
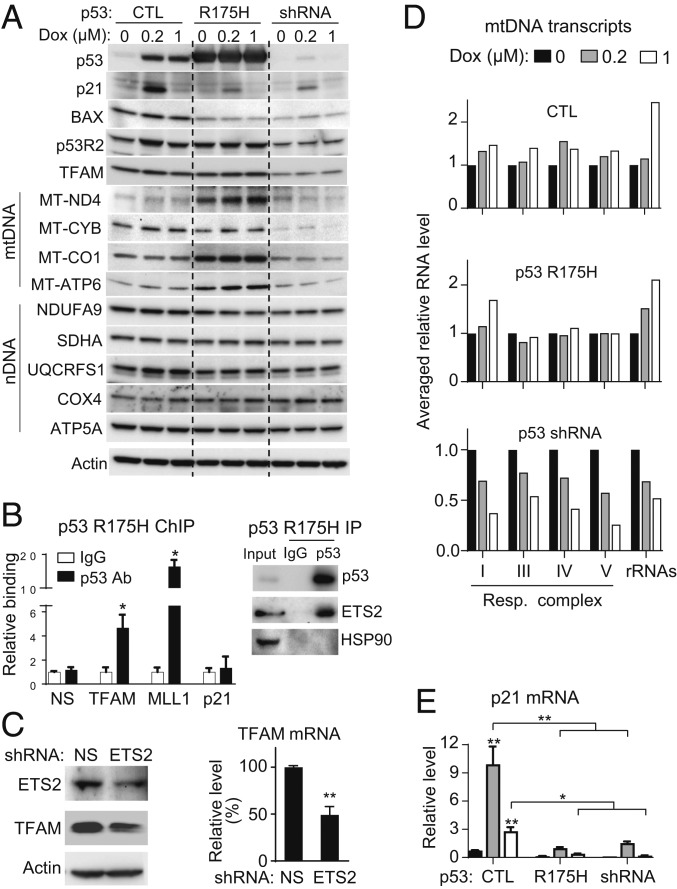
Dox chemotherapy has been associated with skeletal muscle weakness and fatigue in cancer patients (39). Because cells with proliferative potential such as skeletal muscle progenitors may be more sensitive to Dox and given the reduced mtDNA copy number and integrity of Dox-treated p53−/− mouse skeletal muscle (SI Appendix, Fig. S7), we wondered whether the Dox-induced, p53-dependent mtDNA transcriptional response would also be present in human skeletal muscle cells. Human myoblasts were transduced with lentivirus to express mutant p53 R175H or shRNA to knockdown endogenous wild-type p53 and then exposed to Dox. As observed in quiescent human cardiomyocytes, p53 R175H-expressing myoblasts showed higher levels of TFAM and p53R2 protein compared with p53 knockdown cells while concurrently losing p21 and BAX induction (Fig. 5A). The levels of mtDNA-encoded respiratory complex subunit proteins were decreased in p53 depleted human myoblasts by Dox exposure but maintained or increased in mutant p53 R175H-expressing myoblasts (Fig. 5A). In contrast, the levels of nuclear DNA-encoded respiratory complex subunits were generally unaffected by Dox treatment.
Fig. 5.
Mutant p53 can interact with TFAM gene and maintain mtDNA transcription after Dox exposure in proliferating human myoblasts. Primary human skeletal muscle myoblasts were transduced with lentivirus containing control empty vector (CTL), human mutant p53 R175H, or p53 shRNA, and then treated with Dox for 16 h. (A) Whole-cell lysate immunoblots of indicated p53 regulated genes and nuclear DNA (nDNA)/mtDNA-encoded respiratory complex subunits. Actin serves as protein loading control. (B, Left) p53 ChIP of human myoblasts transduced with p53 R175H-expressing lentivirus followed by real-time PCR of the TFAM ETS2-binding sequence. p53 binding is shown relative to nonspecific IgG samples (n = 4). MLL1 (KMT2A) serves as positive control for mutant p53 binding to its ETS2 motif, while the p53-response element of p21 serves as negative control. B2M serves as a nonspecific control gene (NS). (B, Right) ETS2 immunoblot of the p53 R175H immunoprecipitate. HSP90 serves as a negative control. (C) Human myoblasts were transduced with control nonspecific (NS) or ETS2 shRNA lentivirus. ETS2 knockdown was confirmed by immunoblotting and TFAM expression was assessed by real-time PCR (n = 6). (D) Averaged relative RNA levels of mtDNA-encoded genes grouped by respiratory complexes and rRNAs (individual genes in SI Appendix, Fig. S9). (E) p21 mRNA expression in response to Dox exposure serves as marker of wild-type p53 activity (n = 6). Statistical testing by unpaired t test or by 2-way ANOVA in comparison with untreated or between indicated groups. Values are mean ± SEM. *P < 0.05; **P < 0.01.
In support of our finding that mutant p53 can regulate mtDNA homeostasis, a chromatin immunoprecipitation (ChIP) study had reported that mutant p53, including the R175H mutation, enhances p53R2 expression as a gain-of-function activity by interacting with transcription factor ETS2 (40). We hypothesized that such a mechanism could also be involved in the regulation of TFAM expression by mutant p53. Indeed, analysis of published p53 ChIP-seq data revealed potential interaction of mutant p53 with an ETS2-binding sequence in the TFAM gene (SI Appendix, Fig. S8A) (41, 42). p53 ChIP-qPCR of p53 R175H-expressing human myoblasts showed that the mutant p53 binds to the ETS2 site in TFAM, with the previously described ETS2 site of MLL1 serving as a positive control (Fig. 5B) (42). Immunoblotting confirmed protein–protein interaction between mutant p53 R175H and ETS2 by p53 IP (Fig. 5B, Right). As additional controls, mutant p53 failed to bind to the prototypical p53 response element of p21 while endogenous wild-type p53 in control cells showed binding as expected (Fig. 5B and SI Appendix, Fig. S8B). Like the p53R2 gene, TFAM expression also required ETS2 as demonstrated by its suppression with ETS2 knockdown (Fig. 5C). Thus, although our observation could be explained by nontranscriptional effects of mutant p53 on mtDNA homeostasis (43, 44), its transcriptional effects on mtDNA regulators such as p53R2 and TFAM, also known to be transactivated by wild-type p53 through distinct p53 response elements (28, 45), may contribute to its maintenance of mtDNA after Dox exposure.
As in cardiomyocytes, mtDNA-encoded gene transcripts were increased or maintained in wild-type and mutant p53-expressing skeletal muscle myoblasts after Dox exposure but decreased in p53 knockdown cells (Fig. 5D and SI Appendix, Fig. S9). As a common control in the setting of this disparate mtDNA transcriptional response, both p53 R175H-expressing and p53-depleted myoblasts showed loss of p21 induction by Dox compared to control wild-type p53 cells (Fig. 5E). Taken together, these results further substantiated the important effect of p53 as well as its mutant on mtDNA transcription under genotoxic stress, while its deficiency could result in skeletal muscle myopathy after Dox treatment through a similar loss of mtDNA homeostasis.
NAD+ Supplementation Prevents Dox-Induced mtDNA Damage and Cardiac Dysfunction.
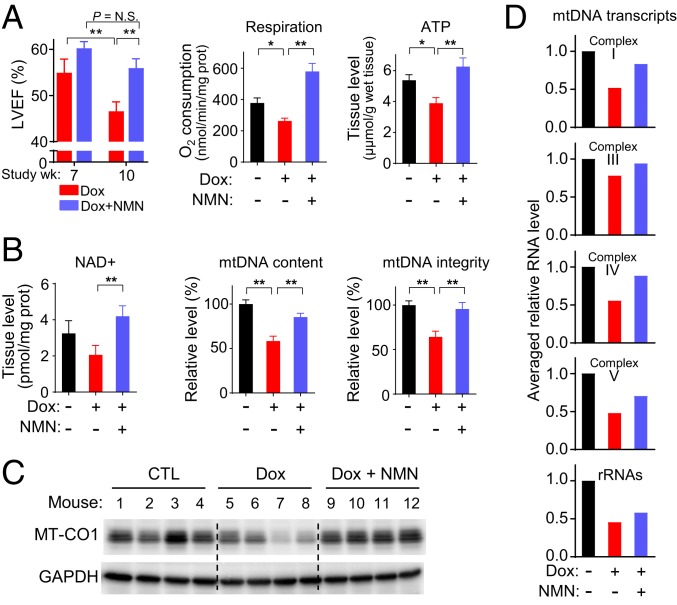
Increasing the cellular levels of nicotinamide adenine dinucleotide (NAD+), involved in DNA repair and mitochondrial homeostasis, has been shown to alleviate a wide variety of pathologic conditions including pressure overload-induced heart failure (46, 47). In a genetic model of mitochondrial myopathy caused by mutated mitochondrial helicase Twinkle, disease progression and mtDNA deletion were improved by NAD+ supplementation (48). Mitochondrial SIRT3, known to be activated by NAD+, appears to protect against Dox-induced mtDNA damage and cardiomyopathy (22), but the effect of direct NAD+ supplementation on Dox-induced cardiomyopathy has not been reported. We therefore examined whether treatment with the NAD+ precursor NMN could ameliorate Dox cardiotoxicity and mtDNA damage in p53−/− mice. Mice undergoing Dox treatment were injected with NMN for the entire duration of the study until the final echocardiogram (study week 10). Remarkably, NMN treatment prevented the significant decline in cardiac function of Dox-treated p53−/− mice (study week 7 versus 10) along with rescuing the decreased mitochondrial respiration and tissue ATP depletion caused by Dox (Fig. 6A). Further analysis showed that NMN supplementation increased heart tissue NAD+ levels in Dox-treated mice (Fig. 6B). In parallel, the Dox-induced reduction in cardiac mtDNA content, mtDNA integrity, and mtDNA gene expression were rescued by NMN to levels comparable to that of untreated control mice (Fig. 6 B–D and SI Appendix, Fig. S10). The decrease in skeletal muscle mtDNA content and integrity caused by Dox exposure was also prevented by NMN supplementation, indicating a common mechanism of mtDNA damage for both Dox-induced cardiomyopathy and skeletal muscle myopathy (SI Appendix, Fig. S11).
Fig. 6.
Nicotinamide mononucleotide (NMN) prevents Dox-induced cardiac and mitochondrial dysfunction. p53−/− mice were treated per divided low-dose Dox protocol and with either vehicle control or NMN i.p. (3 times per wk) over the duration of the entire study. Unless indicated, all analyses were performed at study week 10. (A) LV ejection fraction (LVEF) of p53−/− mice treated with Dox only or Dox plus NMN was measured by echocardiogram at study week 7 and 10 (n = 8). Mitochondrial respiration and ATP levels were measured in cardiac tissue (n ≥ 3). (B) Measurement of NAD+ levels and relative mtDNA content/integrity in cardiac tissue (n ≥ 3). (C) Immunoblot of representative mtDNA-encoded protein MT-CO1 in cardiac tissue. GAPDH is shown as protein loading control. (D) Averaged relative RNA levels of mtDNA-encoded genes grouped by respiratory complexes and rRNAs in cardiac tissue (individual genes in SI Appendix, Fig. S10). Statistical testing by 1- or 2-way ANOVA in comparison with untreated control or between indicated groups. Values are mean ± SEM. *P < 0.05.
Discussion
Although p53 can promote acute cardiotoxicity at high-dose Dox by activating cell death pathways, we report a mechanism whereby it can also confer cardiac protection from a chronic form of Dox cardiotoxicity, which more closely models the human disease. By comparing p53172H/H mutant with p53−/− null genotype states, both of which are similarly defective in cell cycle regulation and apoptosis, our work reveals that p53 regulation of the mitochondrial genome upon Dox exposure plays an important role in preventing Dox cardiotoxicity. These findings are also consistent with emerging evidence that p53 serves to maintain the mitochondrial genome in addition to its well-established role as “guardian” of the nuclear genome (20, 49–51).
In an attempt to understand the disparate effects of p53 on Dox cardiotoxicity and to gain insights into its pathogenesis (13, 15, 52), we utilized p53+/+, p53172H/H, and p53−/− mice and treated them with divided low doses of Dox to induce late-onset cardiomyopathy as observed in patients. No significant cardiac cell death was observed, and LV systolic function was preserved in all p53 genotypes up to 3 wk after treatment, which contrasted with the bolus high-dose Dox-treated mice showing cardiac dysfunction and death within days (8, 13, 14). In the clinics, Dox cardiomyopathy can manifest years after treatment without prior history of acute toxicity so simulating the delayed nature of the cardiac dysfunction in model systems is likely important for understanding its pathogenesis. In a shared setting deficient in p53-dependent apoptosis, p53172HH and p53−/− mice displayed markedly contrasting cardiac function weeks after Dox treatment, suggesting that mtDNA regulation by mutant p53 through both nuclear and nonnuclear mechanisms may confer this protection against late-onset Dox cardiomyopathy. Thus, by comparing the 3 different genotypes of p53, our study suggests that disrupting mtDNA homeostasis, but not cell death, plays a major role in the pathogenesis of chronic Dox cardiomyopathy and may also contribute to the associated peripheral myopathy.
Dox can accumulate in the mitochondria and intercalate between DNA bases, potentially contributing to mtDNA deletions and depletion, which have been reported in human Dox cardiomyopathy tissue samples (19, 53, 54). As demonstrated in a controlled genetic model, the long-term effect of such primary deletions in the mtDNA of mutator Polg257A/A mice is sufficient to drive heart failure pathogenesis and result in LV dilatation (55). Previous studies from a number of different groups have shown that mutant p53, including the p53 R175H mutation, can regulate metabolism and the mitochondria (23, 24, 40, 56). In addition to maintaining mtDNA homeostasis through its transcriptional regulation of genes such as p53R2 in the nucleus, p53 can also interact with TFAM and POLG in the mitochondrial matrix (43, 44). In this regard, both wild-type and mutant p53 have been shown to translocate into the mitochondria via a disulfide relay system to maintain mtDNA content and integrity (25, 51). Our finding that TFAM and p53R2 deficiency in p53−/− mice may contribute to its Dox-induced cardiac dysfunction is further supported by studies showing that increasing either TFAM or p53R2 levels can improve heart function (57, 58). In addition, other DNA regulators such as mitochondrial TOP1MT or TOP2B may also be involved in pathogenesis via interactions with p53 (21, 59).
Besides clarifying the role of p53 in preserving myocardial function against genotoxic stress, the mechanistic insights of the present study may be relevant to the clinics with both prognostic and therapeutic implications. As a clinical translation of the our study, it could be worthwhile examining whether LFS patients who are heterozygous for missense mutations of TP53 that result in mitochondrial “gain of function” are protected from Dox cardiotoxicity and, if so, perhaps can tolerate higher doses of Dox for more effective eradication of cancer (24). In contrast, individuals with LFS who are heterozygous for TP53 nonsense mutations that result in a haploinsufficient state may be predicted to have increased susceptibility to Dox cardiomyopathy. A homozygous TP53-null state is unlikely to exist in humans, but our study suggests that more subtle preexisting mitochondrial deficits may confer increased susceptibility to developing Dox cardiomyopathy in cancer patients. Therefore, measuring mtDNA transcripts in peripheral blood mononuclear cells as suggested by Fig. 4E, for example, may assist in the identification of patients at greater risk of Dox cardiomyopathy who may then benefit from more intensive cardiac management. Based on evidence that NAD+ can promote mitochondrial biogenesis and DNA repair, we also tested whether supplementation with its precursor NMN can prevent Dox cardiomyopathy. Although caution should be exercised in extrapolating our current finding to Dox cardiomyopathy in humans, the marked preservation of cardiac function with maintenance of mitochondrial function and mtDNA by NMN in our p53-null mouse model make it compelling to consider translating this finding to the clinics.
Materials and Methods
Study Approvals.
Human samples were obtained from research participants after informed consent as approved by the National Heart, Lung, and Blood Institute (NHLBI)–NIH Internal Review Board (ClinicalTrials.gov identifier NCT00406445) and as previously reported (24). All animal studies were approved by the Animal Care and Use Committee of the NHLBI–NIH.
Mouse Studies.
Mice were obtained from the following sources: p53 wild-type (p53+/+) and null (p53−/−) mice, The Jackson Laboratory; and p53 R172H knockin mutant (p53172H/H) mice, National Cancer Institute Frederick Mouse Repository. All mice were of the C57BL6 strain or backcrossed at least 5 generations into the C57BL6 background; only male mice were used for all experiments. For the chronic low-dose Dox treatment protocol, 5-wk-old male mice were injected with control normal saline or Dox (5 mg/kg body weight, i.p., doxorubicin HCL dissolved in normal saline; Pfizer) once a week for 5 wk for a total cumulative dose of 25 mg/kg as previously described (15, 60). Mice were allowed to recover for 2 wk prior to echocardiography at the indicated time points. Supplementation with NMN (catalog #HY-F0004; MedChemExpress) was adapted from a previously described protocol (46). Mice concurrently treated with Dox were injected with NMN dissolved in normal saline (500 mg/kg, i.p., 3 times per week on alternating days) 2 d prior to initiation of low-dose Dox treatment and continued for the entire duration of the study. Cardiac function was evaluated by echocardiography as described in SI Appendix, Materials and Methods.
Cells.
Primary human skeletal muscle myoblasts were cultured in DMEM (GlutaMax-I; Invitrogen) containing 20% FBS and 2 μM uridine as previously described (24). MEFs were isolated from 13.5- to ∼14.5-d-old embryos and cultured in DMEM supplemented with 10% FBS as previously described (61). Mouse peripheral blood mononuclear cells were isolated by incubating whole blood in ammonium-chloride-potassium (ACK) lysing buffer, centrifuging, and washing 3 times in PBS as previously described (62).
Generation of Human iPS Cells and Cardiomyocytes.
Human iPS cells with homozygous R175H knockin (TP53175H/H) or disruption (TP53−/−) of the TP53 gene were generated from ND2.0 iPS cell line using the CRISPR-Cas9 technique (63, 64). Briefly, TP53 was knocked out by using high-fidelity eSpCas9 (Addgene; 79145) and gRNA targeting 5′-CAAGCAGTCACAGCACATGA sequence in exon 4. The R175H mutation was knocked in by the same eSpCas9/gRNA and ssODN (5′-aaccagccctgtcgtctctccagccccagctgctcacCATCGCTATCTGAGCAGCGCTCATGGTGGGGGCAGTGCCTCACAACTTCAGTCATGTGCTGTGACTGCTTGTAGATGGCCATGGCGCGGA), which converts CGC to CAC (reverse complementary of GTG) and incorporates a silent mutation for AcuI (underlined) site. Both knockout and knockin iPS cells were confirmed by Sanger sequencing and maintained on Matrigel-precoated plates (catalog #354230; BD Biosciences) at an area concentration of 10 µg/cm2 in Essential 8 Medium (Invitrogen) with daily media changes. A highly efficient method for differentiating human iPS cells into cardiomyocytes (day 30) using heparin in chemically defined medium was utilized as previously described (36). Differentiation into cardiomyocytes was confirmed using cardiac troponin T, α-actinin, and NKX2.5 as cell-specific markers.
Lentiviruses and Cell Transduction.
Human p53 R175H cDNA was subcloned into pLEX-MCS as previously described (65). shRNAs targeting human p53 and ETS2 were obtained from MISSION shRNA Library, TRCN0000003756 and TRCN0000424904, respectively (Sigma-Aldrich), and lentiviruses were prepared using the MISSION lentiviral packaging mix (Sigma-Aldrich). Cells were transduced with lentiviruses for 24 h and selected using 2 µg/mL puromycin for >1 wk.
RNA Quantification by Real-Time RT-PCR.
Total RNA was isolated from cells and mouse tissues using RNeasy Plus Universal Kit (Qiagen). cDNA was synthesized by reverse transcription (Superscript III; Invitrogen), and RT-PCR was performed using a 7900HT Sequence Detection System (Applied Biosystems) as previously described (66). Relative gene expression levels were calculated from cycle threshold (Ct) values normalized to the housekeeping gene eukaryotic translation initiation factor EIF35S (TIF). Primer sequences are listed in SI Appendix, Materials and Methods.
Coimmunoprecipitation Analysis.
Cells were solubilized in lysis buffer (150 mM NaCl, 20 mM Tris, 1% Nonidet P-40, 0.25% sodium deoxycholate, pH 7.4). After preincubating with mouse IgG agarose (Santa Cruz; sc-2343), the cell lysate was incubated with either mouse nonimmune IgG or anti-p53 antibody (DO-1) at 4 °C overnight followed by addition of Protein G PLUS-Agarose (Santa Cruz; sc-2002). The agarose beads were pelleted and washed 3 times with the lysis buffer containing 0.1% Nonidet P-40, and associated proteins were immunoblotted.
ChIP Analysis.
The previously published wild-type/mutant p53 ChIP-seq data (accession number GSE59176) (42) was uploaded into the University of California, Santa Cruz, genome browser and the ETS2 ChIP-seq data (accession number GSE36752) (41) was used for BLAT search to identify mutant p53 interacting ETS2-binding sequences. ChIP assay was performed as described previously with some modifications (67). Cells were cross-linked with 1% formaldehyde for 10 min, and the isolated nuclear extracts were sonicated and immunoprecipitated with anti-p53 antibody (DO-1; sc-126; Santa Cruz) or control mouse IgG (sc-2025; Santa Cruz) at 4 °C overnight. Antibody-bound DNA fragments were purified and quantified by real-time PCR using the primers as listed in SI Appendix, Materials and Methods.
mtDNA Copy Number and Integrity Assay.
DNA was purified from cells or tissues using the DNeasy Blood and Tissue Kit (Qiagen), and mtDNA copy number was measured using real-time PCR as described previously (28). Relative mtDNA copy number was calculated from Ct values of MT-CO2 normalized to the nuclear gene of 18S rRNA. mtDNA integrity was determined by amplifying long and short mtDNA fragments using the LA PCR Kit (TaKaRa) as previously described (68). PCR products were quantified by PicoGreen dye (Molecular Probes) fluorescence, and relative mtDNA integrity was calculated by the ratio of long to short fragments. Primer sequences are listed in SI Appendix, Materials and Methods.
Immunoblotting.
Protein samples were solubilized in cold RIPA buffer supplemented with protease/phosphatase inhibitors (Roche) and centrifuged at 16,000 × g for 15 min. The supernatant was mixed with SDS protein sample buffer, resolved by Tris-glycine SDS/PAGE, and transferred to Immobilon-P membrane (Millipore) for standard ECL immunoblotting. Primary antibodies used are listed in SI Appendix, Materials and Methods.
Mitochondrial Purification and Respiration Studies.
Anesthetized mice were perfused with ice-cold PBS prior to killing and the harvesting of the heart. Mitochondria were isolated from homogenized heart tissues by differential centrifugation per standard protocol (69). The isolated mitochondria were incubated in respiration buffer (100 mM sucrose, 20 mM K+-TES [pH 7.2], 50 mM KCl, 2 mM MgCl2, 1 mM EDTA, 4 mM KH2PO4, 3 mM malate, 0.1% of fatty acid-free BSA) containing 50 μM palmitoyl-carnitine as fatty acid substrate as previously described (70). State 3 respiration was initiated by adding ADP to a final concentration of 4.5 mM and measured using a Clark-type oxygen microelectrode at 37 °C (Instech Laboratories).
Metabolomic Screening.
Heart tissues were dissected and flash-frozen in liquid nitrogen. Small metabolite profiling and semiquantification were performed on 100–150 mg of the tissue using gas chromatography with mass spectrometry and liquid chromatography with tandem mass spectrometry platforms by Metabolon.
Tissue NAD+ and ATP Assays.
NAD+ levels in heart as marker of NMN treatment were determined using the NAD/NADH Quantitation Colorimetric Kit (catalog #K337; BioVision). Cardiac tissue ATP levels were measured using Colorimetric/Fluorometric ATP Assay Kit (catalog #ab83355; Abcam) according to the manufacturer’s protocol. Briefly, ∼10 mg of heart sample was homogenized using perchloric acid/KOH deproteinization followed by absorbance measurement at 550 nm. ATP levels were calculated based on a standard curve and normalized to tissue weight.
TEM and Stereological Measurements.
Mice were perfused, and tissues were fixed and processed for TEM analysis as described in detail in SI Appendix, Materials and Methods.
Statistics.
Statistical analysis was performed using GraphPad Prism software, version 7.02 (GraphPad). The statistical tests used were as follows: 2-tailed Student’s t test for comparison with control condition; 1-way ANOVA followed by Sidak multiple-comparison testing for more than 2 conditions; and 2-way ANOVA with Tukey posttest multiple-comparison testing for serial measurements or multiple conditions within different groups. Values are shown as mean ± SEM. P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Stasia A. Anderson (NHLBI Animal MRI Core), Zu-Xi Yu (NHLBI Pathology Core), Camron Keshavarz and Erin S. Stempinski (NHLBI Electron Microscopy Core Facility), and Matthew F. Starost (NIH Division of Veterinary Resources) for helpful advice and assistance during the course of this study. We also thank Michele D. Allen (NHLBI Murine Phenotyping Core) for excellent coordination and management of mouse experiments, Christian A. Combs (NHLBI Light Microscopy Core) for confocal microscopy, and Matthew P. Donnelly (NHLBI) for critical reading of the manuscript. This work was supported by the NHLBI–NIH Division of Intramural Research (HL006050-09) (to P.M.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904979116/-/DCSupplemental.
References
- 1.Von Hoff D. D., et al. , Risk factors for doxorubicin-induced congestive heart failure. Ann. Intern. Med. 91, 710–717 (1979). [DOI] [PubMed] [Google Scholar]
- 2.Singal P. K., Iliskovic N., Doxorubicin-induced cardiomyopathy. N. Engl. J. Med. 339, 900–905 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Chatterjee K., Zhang J., Honbo N., Karliner J. S., Doxorubicin cardiomyopathy. Cardiology 115, 155–162 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raj S., Franco V. I., Lipshultz S. E., Anthracycline-induced cardiotoxicity: A review of pathophysiology, diagnosis, and treatment. Curr. Treat. Options Cardiovasc. Med. 16, 315 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Moslehi J., Amgalan D., Kitsis R. N., Grounding cardio-oncology in basic and clinical science. Circulation 136, 3–5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carvalho F. S., et al. , Doxorubicin-induced cardiotoxicity: From bioenergetic failure and cell death to cardiomyopathy. Med. Res. Rev. 34, 106–135 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Swain S. M., Whaley F. S., Ewer M. S., Congestive heart failure in patients treated with doxorubicin: A retrospective analysis of three trials. Cancer 97, 2869–2879 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Dhingra R., et al. , Bnip3 mediates doxorubicin-induced cardiac myocyte necrosis and mortality through changes in mitochondrial signaling. Proc. Natl. Acad. Sci. U.S.A. 111, E5537–E5544 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ichikawa Y., et al. , Cardiotoxicity of doxorubicin is mediated through mitochondrial iron accumulation. J. Clin. Invest. 124, 617–630 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burridge P. W., et al. , Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat. Med. 22, 547–556 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyu Y. L., et al. , Topoisomerase IIbeta mediated DNA double-strand breaks: Implications in doxorubicin cardiotoxicity and prevention by dexrazoxane. Cancer Res. 67, 8839–8846 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Zhang S., et al. , Identification of the molecular basis of doxorubicin-induced cardiotoxicity. Nat. Med. 18, 1639–1642 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Zhu W., et al. , Acute doxorubicin cardiotoxicity is associated with p53-induced inhibition of the mammalian target of rapamycin pathway. Circulation 119, 99–106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang X., et al. , Ferroptosis as a target for protection against cardiomyopathy. Proc. Natl. Acad. Sci. U.S.A. 116, 2672–2680 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu W., Zhang W., Shou W., Field L. J., P53 inhibition exacerbates late-stage anthracycline cardiotoxicity. Cardiovasc. Res. 103, 81–89 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhuang J., Ma W., Lago C. U., Hwang P. M., Metabolic regulation of oxygen and redox homeostasis by p53: Lessons from evolutionary biology? Free Radic. Biol. Med. 53, 1279–1285 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kruiswijk F., Labuschagne C. F., Vousden K. H., p53 in survival, death and metabolic health: A lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 16, 393–405 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Lebrecht D., Setzer B., Ketelsen U. P., Haberstroh J., Walker U. A., Time-dependent and tissue-specific accumulation of mtDNA and respiratory chain defects in chronic doxorubicin cardiomyopathy. Circulation 108, 2423–2429 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Lebrecht D., Kokkori A., Ketelsen U. P., Setzer B., Walker U. A., Tissue-specific mtDNA lesions and radical-associated mitochondrial dysfunction in human hearts exposed to doxorubicin. J. Pathol. 207, 436–444 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Nithipongvanitch R., et al. , Evidence for p53 as guardian of the cardiomyocyte mitochondrial genome following acute adriamycin treatment. J. Histochem. Cytochem. 55, 629–639 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Khiati S., et al. , Mitochondrial topoisomerase I (top1mt) is a novel limiting factor of doxorubicin cardiotoxicity. Clin. Cancer Res. 20, 4873–4881 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pillai V. B., et al. , Sirt3 protects mitochondrial DNA damage and blocks the development of doxorubicin-induced cardiomyopathy in mice. Am. J. Physiol. Heart Circ. Physiol. 310, H962–H972 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li T., et al. , Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 149, 1269–1283 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang P. Y., et al. , Increased oxidative metabolism in the Li-Fraumeni syndrome. N. Engl. J. Med. 368, 1027–1032 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhuang J., et al. , Mitochondrial disulfide relay mediates translocation of p53 and partitions its subcellular activity. Proc. Natl. Acad. Sci. U.S.A. 110, 17356–17361 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lang G. A., et al. , Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 119, 861–872 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Saleem A., Adhihetty P. J., Hood D. A., Role of p53 in mitochondrial biogenesis and apoptosis in skeletal muscle. Physiol. Genomics 37, 58–66 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Park J. Y., et al. , p53 improves aerobic exercise capacity and augments skeletal muscle mitochondrial DNA content. Circ. Res. 105, 705–712, 11 p following 712 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taegtmeyer H., Energy metabolism of the heart: From basic concepts to clinical applications. Curr. Probl. Cardiol. 19, 59–113 (1994). [DOI] [PubMed] [Google Scholar]
- 30.Fillmore N., Lopaschuk G. D., Targeting mitochondrial oxidative metabolism as an approach to treat heart failure. Biochim. Biophys. Acta 1833, 857–865 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Williams D., et al. , Abnormal mitochondrial L-arginine transport contributes to the pathogenesis of heart failure and rexoygenation injury. PLoS One 9, e104643 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Q., et al. , Rewiring of glutamine metabolism is a bioenergetic adaptation of human cells with mitochondrial DNA mutations. Cell Metab. 27, 1007–1025.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tyburski J. B., et al. , Radiation metabolomics. 1. Identification of minimally invasive urine biomarkers for gamma-radiation exposure in mice. Radiat. Res. 170, 1–14 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menzies R. A., Gold P. H., The turnover of mitochondria in a variety of tissues of young adult and aged rats. J. Biol. Chem. 246, 2425–2429 (1971). [PubMed] [Google Scholar]
- 35.Kim T. Y., et al. , Metabolic labeling reveals proteome dynamics of mouse mitochondria. Mol. Cell. Proteomics 11, 1586–1594 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Y., et al. , Heparin promotes cardiac differentiation of human pluripotent stem cells in chemically defined albumin-free medium, enabling consistent manufacture of cardiomyocytes. Stem Cells Transl. Med. 6, 527–538 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bourdon A., et al. , Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat. Genet. 39, 776–780 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Pontarin G., Ferraro P., Bee L., Reichard P., Bianchi V., Mammalian ribonucleotide reductase subunit p53R2 is required for mitochondrial DNA replication and DNA repair in quiescent cells. Proc. Natl. Acad. Sci. U.S.A. 109, 13302–13307 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilliam L. A., St Clair D. K., Chemotherapy-induced weakness and fatigue in skeletal muscle: The role of oxidative stress. Antioxid. Redox Signal. 15, 2543–2563 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kollareddy M., et al. , Regulation of nucleotide metabolism by mutant p53 contributes to its gain-of-function activities. Nat. Commun. 6, 7389 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Do P. M., et al. , Mutant p53 cooperates with ETS2 to promote etoposide resistance. Genes Dev. 26, 830–845 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu J., et al. , Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature 525, 206–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshida Y., et al. , P53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer Res. 63, 3729–3734 (2003). [PubMed] [Google Scholar]
- 44.Achanta G., et al. , Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. EMBO J. 24, 3482–3492 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka H., et al. , A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature 404, 42–49 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Lee C. F., et al. , Normalization of NAD+ redox balance as a therapy for heart failure. Circulation 134, 883–894 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katsyuba E., Auwerx J., Modulating NAD+ metabolism, from bench to bedside. EMBO J. 36, 2670–2683 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khan N. A., et al. , Effective treatment of mitochondrial myopathy by nicotinamide riboside, a vitamin B3. EMBO Mol. Med. 6, 721–731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lane D. P., Cancer. p53, guardian of the genome. Nature 358, 15–16 (1992). [DOI] [PubMed] [Google Scholar]
- 50.Bakhanashvili M., et al. , p53 in mitochondria enhances the accuracy of DNA synthesis. Cell Death Differ. 15, 1865–1874 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Park J. H., Zhuang J., Li J., Hwang P. M., p53 as guardian of the mitochondrial genome. FEBS Lett. 590, 924–934 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shizukuda Y., Matoba S., Mian O. Y., Nguyen T., Hwang P. M., Targeted disruption of p53 attenuates doxorubicin-induced cardiac toxicity in mice. Mol. Cell. Biochem. 273, 25–32 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Tokarska-Schlattner M., Zaugg M., Zuppinger C., Wallimann T., Schlattner U., New insights into doxorubicin-induced cardiotoxicity: The critical role of cellular energetics. J. Mol. Cell. Cardiol. 41, 389–405 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Ashley N., Poulton J., Mitochondrial DNA is a direct target of anti-cancer anthracycline drugs. Biochem. Biophys. Res. Commun. 378, 450–455 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Trifunovic A., et al. , Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429, 417–423 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Kolukula V. K., et al. , SLC25A1, or CIC, is a novel transcriptional target of mutant p53 and a negative tumor prognostic marker. Oncotarget 5, 1212–1225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ikeuchi M., et al. , Overexpression of mitochondrial transcription factor a ameliorates mitochondrial deficiencies and cardiac failure after myocardial infarction. Circulation 112, 683–690 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Nowakowski S. G., et al. , Transgenic overexpression of ribonucleotide reductase improves cardiac performance. Proc. Natl. Acad. Sci. U.S.A. 110, 6187–6192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cowell I. G., et al. , Human topoisomerase IIalpha and IIbeta interact with the C-terminal region of p53. Exp. Cell Res. 255, 86–94 (2000). [DOI] [PubMed] [Google Scholar]
- 60.van Acker S. A., et al. , Doxorubicin-induced cardiotoxicity monitored by ECG in freely moving mice. A new model to test potential protectors. Cancer Chemother. Pharmacol. 38, 95–101 (1996). [DOI] [PubMed] [Google Scholar]
- 61.Parrinello S., et al. , Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat. Cell Biol. 5, 741–747 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen J., Kang J. G., Keyvanfar K., Young N. S., Hwang P. M., Long-term adaptation to hypoxia preserves hematopoietic stem cell function. Exp. Hematol. 44, 866–873.e4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen G., et al. , Chemically defined conditions for human iPSC derivation and culture. Nat. Methods 8, 424–429 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ran F. A., et al. , Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang P. Y., et al. , Inhibiting mitochondrial respiration prevents cancer in a mouse model of Li-Fraumeni syndrome. J. Clin. Invest. 127, 132–136 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhuang J., et al. , Forkhead box O3A (FOXO3) and the mitochondrial disulfide relay carrier (CHCHD4) regulate p53 protein nuclear activity in response to exercise. J. Biol. Chem. 291, 24819–24827 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee J. E., et al. , Brd4 binds to active enhancers to control cell identity gene induction in adipogenesis and myogenesis. Nat. Commun. 8, 2217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Furda A., Santos J. H., Meyer J. N., Van Houten B., Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol. Biol. 1105, 419–437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frezza C., Cipolat S., Scorrano L., Organelle isolation: Functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat. Protoc. 2, 287–295 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Timmers S., et al. , Augmenting muscle diacylglycerol and triacylglycerol content by blocking fatty acid oxidation does not impede insulin sensitivity. Proc. Natl. Acad. Sci. U.S.A. 109, 11711–11716 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.