Significance
During early neurogenesis, progenitors maintain the balance between self-renewal and differentiation. This equilibrium is preserved by the coordinated action of developmental cues and epigenetic mechanisms. We analyzed the role of the H3K9me2 histone demethylase PHF2 in neural stem cells biology in vitro and in early neurogenesis in the chicken neural tube. We found that PHF2 is essential for proliferation of progenitors. Additionally, PHF2 maintains the genome integrity, preventing R-loop accumulation and DNA damage. Our work contributes to the clarification of both the physiological and molecular roles of PHF2 during early neurogenesis. It opens avenues to understand the role of H3K9 methylation not only in development but also in diseases such as autism, cancer, and obesity where PHF2 has been involved.
Keywords: histone demethylation, chromatin transcription, neuronal progenitor proliferation, PHF2
Abstract
Histone H3 lysine 9 methylation (H3K9me) is essential for cellular homeostasis; however, its contribution to development is not well established. Here, we demonstrate that the H3K9me2 demethylase PHF2 is essential for neural progenitor proliferation in vitro and for early neurogenesis in the chicken spinal cord. Using genome-wide analyses and biochemical assays we show that PHF2 controls the expression of critical cell cycle progression genes, particularly those related to DNA replication, by keeping low levels of H3K9me3 at promoters. Accordingly, PHF2 depletion induces R-loop accumulation that leads to extensive DNA damage and cell cycle arrest. These data reveal a role of PHF2 as a guarantor of genome stability that allows proper expansion of neural progenitors during development.
During neural development, multipotent progenitor cells self-renew and ultimately originate specialized neurons and glial cells (1, 2). The chromatin acting factors are essential players in both proliferation and cell differentiation events during embryo development. This epigenetic regulation is achieved by stabilizing chromatin structure that allows establishing heritable gene expression patterns. The epigenetic control is mainly mediated by covalent modifications of histones and DNA (3). Recently, histone methylation/demethylation has received special attention as an essential regulator of gene expression and genome stability during development (4). One critical histone modification during development is dimethylation of histone H3 at lysine 9 (H3K9me2), implicated in the silencing of genes playing important roles in chromatin homeostasis and development (5). In embryonic stem (ES) cells, it has been proposed that H3K9me2 increases across the genome as cells differentiate and acquire lineage specificity (6), although this is contentious (7). In addition, H3K9me2 together with H3K9me3 are essential components of the constitutive heterochromatin (8–10).
Despite the importance of this modification during development, little is known about the role of the enzymes responsible for this mark in neurogenesis. PHF2 is a member of the KDM7 histone demethylase (HDM) family (11). It contains a plant homeodomain (PHD) in the N-terminal and the Jumonji-C (JMJC) domain, which has demethylase enzymatic activity (12). Biochemical studies demonstrated that PHF2 demethylates H3K9me2 upon interaction with H3K4me2/3 through its PHD domain (13). PHF2 was first identified as a candidate gene for hereditary sensory neuropathy type I (14) because it is expressed at high levels in the neural tube and dorsal root ganglia (15). The physiological role of PHF2 in vivo is not yet clear, but it is a coactivator of multiple transcription factors (11). Working with them, PHF2 regulates various differentiation processes (16–18). Alterations in PHF2 have been identified in several cancer types (19–21). Interestingly, PHF2 mutations have been found in patients with autism spectrum disorder (ASD) (22–24). Despite PHF2 high expression in the neural tube and its implication on mental diseases, PHF2 involvement in neural development has not yet been explored.
In this study, we analyzed the role of PHF2 in neural stem cell (NSC) biology. We found that PHF2 is essential for progenitor proliferation in vitro and in vivo, in the chicken spinal cord. PHF2 binds and regulates cell cycle gene promoters, particularly those involved in DNA replication and cell cycle progression. Moreover, PHF2 depletion induces R-loop accumulation, DNA damage, and cell cycle arrest. These data reveal a role for PHF2 as a safeguard of genome stability during development.
Results
PHF2 Binds Promoters and Mediates H3K9me2 Demethylation.
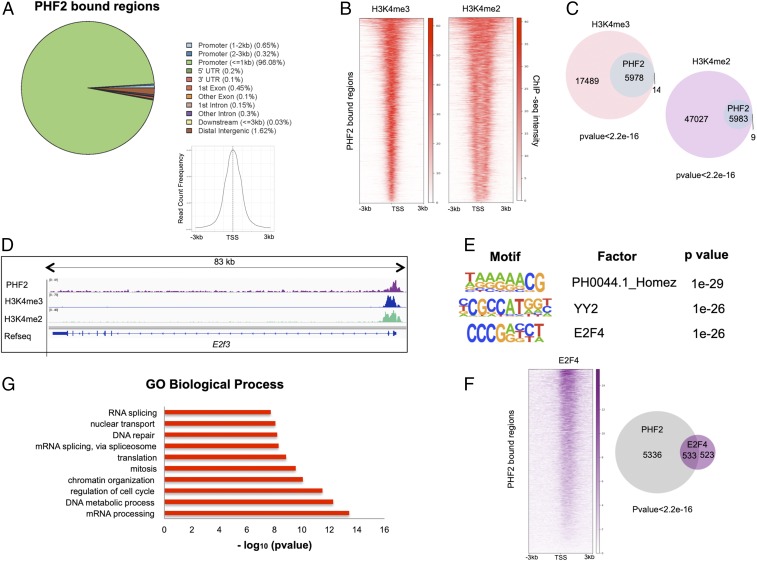
To gain insight into the biological substrate of PHF2 in NSCs, we performed chromatin immunoprecipitation coupled with sequencing (ChIP-seq) experiment. We detected 5,992 peaks normalized to the input (P value of 1e-10) in ChIP data for PHF2. SI Appendix, Fig. S1A shows validation of ChIP-seq results by qPCR for a random set of PHF2 targets. Then, we examined the genomic distribution of the PHF2 peaks. Our results showed that 97% of PHF2 peaks localized on gene promoters, around the transcription start site (TSS) (Fig. 1A), as it has been reported in a different cellular context (25).
Fig. 1.
PHF2 binds promoters in NSCs. (A) Genomic distribution of PHF2 ChIP-seq peaks in NSCs showing that PHF2 mainly binds promoter regions around the TSS. (B) Heatmaps depicting PHF2 binding to H3K4me3- and H3K4me2-marked promoters in NSCs 3 kb around the TSS. Scales indicate ChIP-seq intensities. (C) Venn diagrams showing overlap between PHF2-bound and H3K4me3 and H3K4me2-marked regions. P value is the result of an equal proportions test performed between H3K4me3 and H3K4me2 peaks and a random set. (D) IGV capture showing PHF2, H3K4me3, and H3K4me2 peaks in E2f3 gene in NSCs. (E) Motif enrichment analysis of PHF2 ChIP-seq peaks in NSCs using the Homer de novo motif research tool showing the 3 top enriched motifs. (F) Heatmap representation of PHF2 and E2F4 co-occupied regions (Left) and Venn diagrams (Right) showing peak overlapping between PHF2-bound and E2F4-bound regions in published ChIP-seq in HeLa S3 cells. (G) Gene ontology analysis showing the biological process of the PHF2-bound genes was performed using as a background the whole Mus musculus genome.
It has been proposed that PHF2 through its PHD domain interacts with H3K4me2/3 histone marks; thus, we analyzed the colocalization of PHF2 and H3K4me2/3 at the genome-wide level using previously published H3K4me2/3 ChIP-seq data from NSCs. Doing that, we identified 5,978 (99.7%) (P value <2.2e-16) and 5,983 (99.8%) (P value <2.2e-16) PHF2-bound regions that also contained H3K4me3 or H3K4me2, respectively (Figs. 1 B–D and 2B and SI Appendix, Fig. S1B). The high overlapping suggests a required H3K4me2/3 mark for effective PHF2 binding at promoters as it is described (13, 25). To better understand how PHF2 is targeted to the chromatin, we performed bioinformatics analysis that revealed the most statistically significant predicted PHF2 binding sites using the Homer de novo motif research tool. We identified as the top 3: Homez, YY2, and E2F4 DNA binding motifs (Fig. 1E). These transcription factors, particularly E2F4 and YY2, are essential to control progenitor self-renewal and cell cycle progression (26–28), suggesting a potential role of PHF2 in cell proliferation. To reinforce these data, we analyzed the colocalization of PHF2 and E2F4 using previously published ChIP-seq data in HeLa S3 cells. The results showed that the 50.3% (P value <2.2e-16) of E2F4-bound regions, respectively, also contained PHF2 (Fig. 1F). These results reveal a potential role of PHF2 in regulating E2F-mediated transcription as it has been described for another KDM7 family member, PHF8 (29). Accordingly, gene ontology (GO) analysis of PHF2-bound regions indicated that PHF2 was associated with the promoter region of genes involved in cell cycle regulation, DNA repair, RNA processing, and chromatin organization, among others (Fig. 1G).
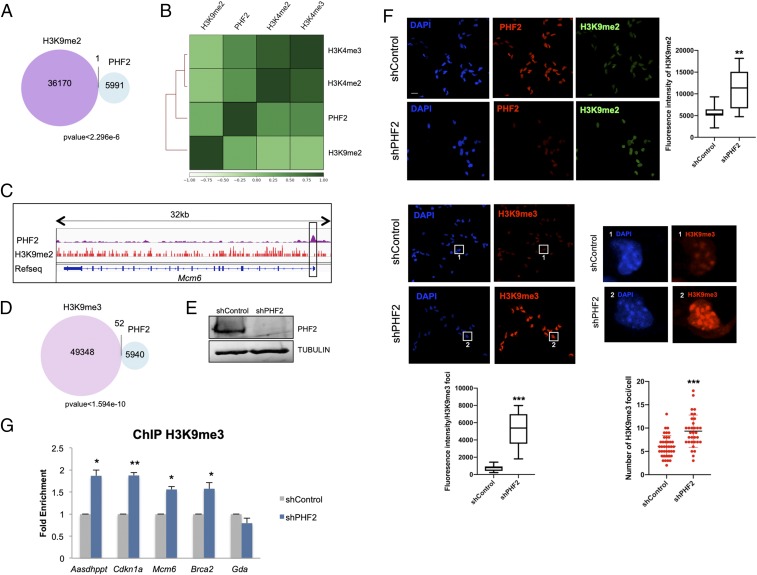
Fig. 2.
PHF2 mediates H3K9me2 demethylation. (A) Venn diagram showing overlap between PHF2-bound and H3K9me2-marked regions in NSCs. (B) Clustered heatmap showing Pearson correlation of H3K9me2, H3K4me2, H3K4me3, and PHF2 ChIP-seq samples based on read coverage within genomic regions. (C) IGV capture showing PHF2 peaks and H3K9me2-marked regions in the Mcm6 gene. The promoter region in the box shows no H3K9me2 enrichment where PHF2 binds. (D) Venn diagram depicting overlap between PHF2-bound and H3K9me3-marked regions in NSCs. (E) NSCs were infected with lentivirus expressing shRNA control (shControl) or shRNA specific for PHF2 (shPHF2). After 48 h, total protein extracts were prepared and the PHF2 and TUBULIN levels were determined by immunoblot. (F) Immunostaining of shControl and shPHF2 cell lines. Cells were fixed and stained with PHF2, H3K9me2, and H3K9me3 antibodies and DAPI (Scale bar: 20 μm.) Zoom in showing H3K9me3 foci in shControl and shPHF2. Cell fluorescence for H3K9me2 staining and H3K9me3 foci was measured using ImageJ. Boxplots represent the quantification of the fluorescence intensities as well as the number of H3K9me3 foci/cell in shControl and shPHF2 cells. **P < 0.01; ***P < 0.001 (Student’s t test). (G) ChIP of H3K9me3 in shControl and shPHF2 cells was analyzed by qPCR at the indicated gene promoters that were identified by PHF2 ChIP-seq as PHF2 targets. An intragenic region of the Gda gene marked by H3K9me2/3 but devoid of PHF2 was used as negative control. Data from qPCR were normalized to the input and expressed as fold enrichment over the data obtained in shControl. Results are the mean of 2 to 3 biological independent experiments. Error bars represent SEM. *P < 0.05; **P < 0.01 (Student’s t test).
Previous studies demonstrated that PHF2 demethylates mainly H3K9me2 (13, 25). We thus analyzed the distribution of H3K9me2 in NSCs by performing ChIP-seq experiments and compared it with PHF2 genomic-associated regions. As expected for a H3K9me2 demethylase, the vast majority (99.99%) (P value <2.3e-6) of PHF2-bound regions were totally excluded from the H3K9me2 positive regions in NSCs (Fig. 2 A–C and SI Appendix, Fig. S2A). Accordingly, clustered heatmaps showed a clear exclusion of the H3K9me2 mark and H3K4me2/3-associated regions (Fig. 2B). Moreover, a strong overlapping of PHF2 peaks with H3K4me2/3-associated regions was observed, in agreement with the data presented in Fig. 1 B and C. Altogether these results suggest that PHF2 binds to genomic regions marked with histone modifications related to transcriptional activation. Similarly to H3K9me2, H3K9me3-enriched regions identified by previously published ChIP-seq were excluded from the PHF2 positive regions in NSCs (Fig. 2D). To evaluate the impact of PHF2 depletion on histone modifications in vivo, NSCs were transduced with lentivirus containing specific PHF2 shRNA that efficiently decreased the PHF2 levels (Fig. 2E and SI Appendix, Fig. S2B) or with a control shRNA (shControl) (see Materials and Methods). The reduction of PHF2 did not affect the expression of its homologous PHF8 (SI Appendix, Fig. S2B). After viral transduction, the levels of H3K9me2 were analyzed by immunofluorescence assays. In Fig. 2F we observed a slight increase in H3K9me2 levels as it has been demonstrated in other cellular contexts (13). As the H3K9me2 mark serves as a substrate for the histone methyltransferase SUV39H, we tested the levels of H3K9me3 in PHF2-depleted NSCs. Results in Fig. 2F clearly showed an increase in H3K9me3. Intriguingly, both the intensity and the number of H3K9me3 foci increased upon PHF2 depletion. Accordingly, a clear accumulation of HP1α was also detected in PHF2-depleted cells (SI Appendix, Fig. S2C) without increase in other histone marks such as H3K4me3 (SI Appendix, Fig. S2D).
These data strongly suggest that depletion of PHF2 led to an increase in heterochromatin-related marks at global levels. Thus, we sought to test whether PHF2 is also important to prevent H3K9me3 increase at a local level. To do so, we chose some PHF2-target regions identified in the ChIP-seq experiment and tested the effect of PHF2 depletion on H3K9me3 levels by ChIP-qPCR assays. A clear increase in the H3K9me3 mark was observed (Fig. 2G), at the analyzed promoters, without affecting the intragenic region of the Gda gene (a non-PHF2 target used as a negative control), in accordance with the global increase noticed in Fig. 2F. These data strongly suggest that PHF2 prevents H3K9me3 accumulation, limiting ectopic heterochromatin formation, particularly at promoter regions.
PHF2 Regulates Cell Cycle Gene Transcription.
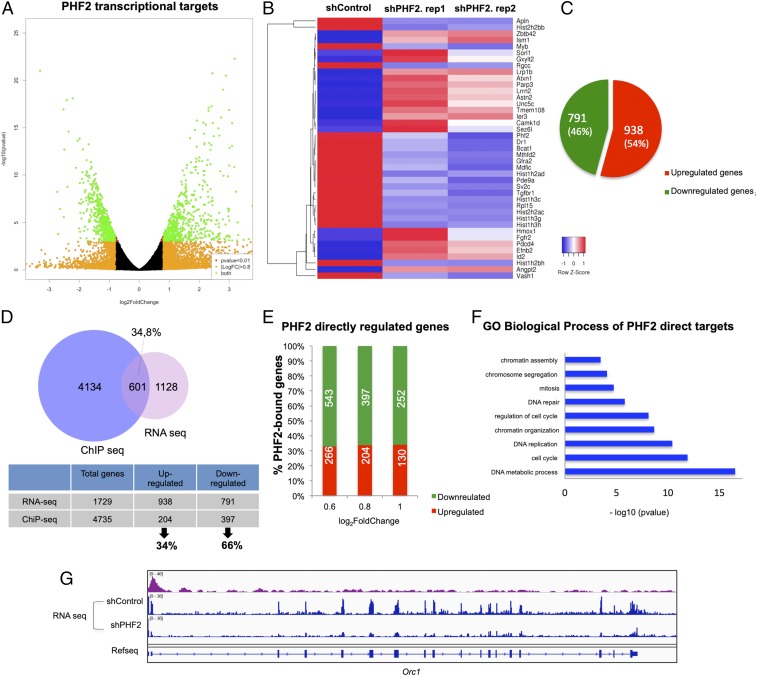
To gain further knowledge into PHF2’s function in NSCs, we identified the PHF2-associated transcriptional profile by RNA-seq. We found 1,729 transcripts that significantly changed their expression (log2 fold change [FC] > 0.8 and FC < −0.8 and P value <0.01), in the 2 biological independent experiments in PHF2-depleted NSCs (shPHF2) compared with control (shControl) cells (Fig. 3 A and B). Among these, 791 (45.8%) were down-regulated and 938 (54.2%) up-regulated upon PHF2 depletion (Fig. 3C). These results were confirmed by depletion of PHF2 by using another shRNA against a distinct region of PHF2 (see Materials and Methods) as we tested by qPCR analysis of some randomly chosen genes (SI Appendix, Fig. S3A). Next, we identified the PHF2 direct transcriptional targets by comparing the PHF2-associated transcriptional profile with the ChIP-seq data. Among the genes that showed a PHF2 dependency for transcriptional regulation (log2 FC > 0.8 and log2 FC < −0.8) in the RNA-seq experiment, 601 (34.8%) were bound by PHF2 (Fig. 3D). Interestingly, the proportion of direct down-regulated transcripts (397, 66%) was higher than the direct up-regulated ones (204, 34%) (Fig. 3D). This percentage was almost maintained at different FCs (Fig. 3E). Moreover, half of the down-regulated genes in the RNA-seq experiment (50.2%) contain PHF2 bound at the promoter region as we determined in the ChIP-seq experiment (SI Appendix, Fig. S3B), while only 21.7% of the up-regulated genes were direct targets of PHF2 (SI Appendix, Fig. S3C). These data are in accordance with the activator role proposed for PHF2 (17). Nevertheless, the fact that 34% of the direct targets became up-regulated upon PHF2 depletion, suggest a potential role of PHF2 contributing to transcriptional repression or preventing activation as it has been demonstrated for another member of the KDM7 family, PHF8, in a different cellular context (30). Enrichment analysis of GO terms over the 601 PHF2 direct target genes showed that the most enriched were associated with cell cycle categories, particularly G1/S transition (E2f2/3/7/8, Cdc7, Cdc25a, Cdk4, and Mcm3/4/8), DNA replication (Orc1/2/6 and Pcna), mitosis (Cdk1, Smc2/3/4, Aurkb, and Topo2a), as well as chromatin activity (Cenpa, Kdm1b, Hat1, Parp1, and Prmt5) (Fig. 3 F and G and SI Appendix, Fig. S4A). Intriguingly, some of them were E2F target genes (Ccnd1, Cdc25a, Pcna, Mcm3/4/6/8, and Smc4, including E2f family genes) (Fig. 3 F and G and SI Appendix, Fig. S4B). To further characterize the differences between control and PHF2 knockdown (KD) NSCs, we performed a GO enrichment analysis of down-regulated, up-regulated, and unregulated direct target genes to identify those biological processes most sensitive to PHF2 depletion. The results revealed that PHF2 down-regulated genes were strongly associated with cell cycle progression, chromatin activity, and DNA repair among others (SI Appendix, Figs. S3D and S4B). The same analysis of the up-regulated genes did show functional categories related to morphogenesis, signal transduction, and developmental process (SI Appendix, Fig. S3E) or to RNA processing and chromatin organization among others in the case of unregulated ones (SI Appendix, Fig. S3F). It is important to notice that the number of genes and the statistically significant values of up-regulated and unregulated gene-associated categories were much lower (P values ranging from −log10 2–6) compared with the down-regulated genes (P values −log10 4–23). This fact, led us to focus on the potential role of PHF2 as a transcriptional activator of cell cycle and DNA repair genes in this particular neural context. Finally, we confirmed by immunoblot the decrease of some proteins whose genes were down-regulated in PHF2-depleted cells (SI Appendix, Fig. S4C). Interestingly, we did not detect any reduction in the histone protein levels, although their mRNAs were down-regulated upon PHF2 depletion (SI Appendix, Fig. S4D), probably due to the tight control of the histone levels inside the cells. Altogether, these data strongly suggest that PHF2 binds to the cell cycle gene promoters to fine tune their chromatin activity and facilitate their transcription.
Fig. 3.
PHF2 regulates cell cycle gene transcription. (A) Volcano plot represents PHF2 transcriptional targets identified by RNA-seq in shControl and shPHF2 NSCs. The green dots represent all of the genes with false discovery rate (FDR) < 0.01 and log2 FC > 0.8 and log2 FC < −0.8. (B) Heatmap showing the top 20 regulated genes identified by RNA-seq in shControl and shPHF2. Two biological replicates of shPHF2 cells were used for RNA-seq. All of the genes showed P value <0.01 and log2 FC > 0.8 and log2 FC < −0.8. (C) Diagram showing the number and percentage of up- and down-regulated genes in the RNA-seq experiment comparing shControl and shPHF2 NSCs. (D) Venn diagram showing overlapping between PHF2-bound genes and PHF2 transcriptional targets. A total of 34.8% of differentially expressed genes identified by RNA-seq with log2 FC > 0.8 and log2 FC < −0.8 were also PHF2 direct targets identified by ChIP-seq. From these, 34% were up-regulated and 66% down-regulated. (E) Graph depicting the percentage and number of up-regulated and down-regulated genes in the shPHF2 according to the RNA-seq that contain PHF2 in their promoter classified by log2 fold change. (F) Gene ontology analysis showing the “biological process” of the PHF2 direct targets was performed using as a background the whole M. musculus genome. (G) IGV capture showing PHF2 peaks and RNA levels in shControl and shPHF2 in the Orc1 gene.
We next sought to analyze the transcriptional consequences of PHF2 overexpression in NSCs. To do that, we established a NSC line that expressed PHF2 wild type (WT) upon addition of doxycycline (see Materials and Methods). We induced the PHF2 expression and the levels of some direct PHF2 targets, identified in the RNA-seq and ChIP-seq experiments, were analyzed by qPCR. No changes or slight alterations of the expression (up and down) were observed upon PHF2 overexpression (SI Appendix, Fig. S4E).
PHF2 Regulates Cell Proliferation.
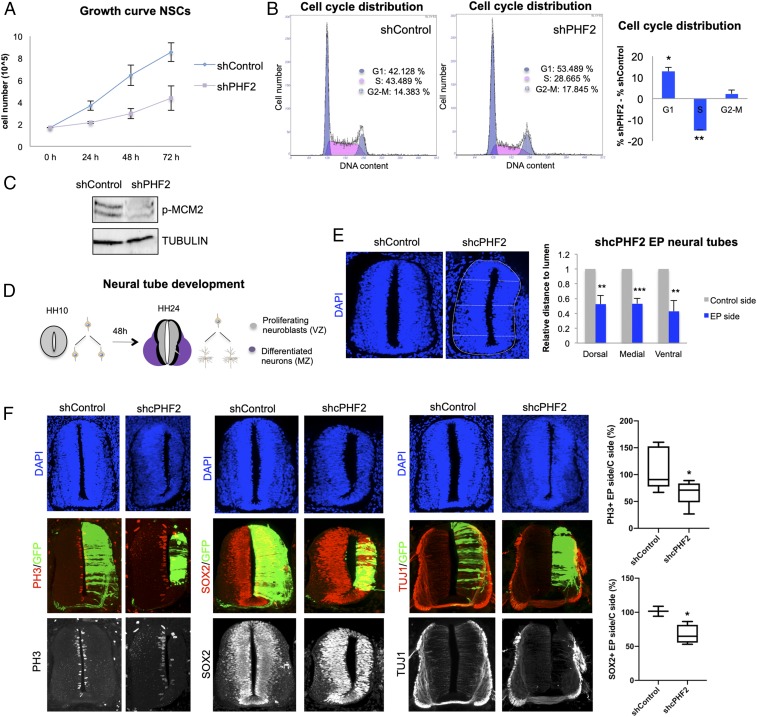
To further analyze the potential role of PHF2 in cell proliferation, we examined the consequences of its reduction. PHF2-depleted NSCs exhibited a striking decrease in cell growth (Fig. 4A). Moreover, flow-cytometry analysis demonstrated a delay in G1/S transition (%G1 shControl 42.2%, shPHF2 53.5%) upon PHF2 depletion (Fig. 4B). Moreover, the levels of the Mcm2 factor (the putative helicase essential for DNA replication initiation and elongation in eukaryotic cells) phosphorylated at S40 required for the initiation of DNA replication were lower in PHF2-depleted cells compared with control progenitors (Fig. 4C). Interestingly, the expression of PHF2 was regulated throughout the cell cycle with higher levels at G1/S transition (SI Appendix, Fig. S5A) and its recruitment to the S-phase-activated gene promoter ORC1 also increased at G1/S transition (SI Appendix, Fig. S5B). These data reinforce the idea that PHF2 contributes to cell cycle progression by facilitating the expression of cell cycle progression genes, particularly those involved in DNA replication.
Fig. 4.
PHF2 depletion blocks neurogenesis in the spinal cord. (A) Growth curve showing the proliferation rate of NSCs infected with lentivirus expressing shRNA control (shControl) or shRNA for PHF2 (shPHF2) from 0 to 72 h. (B) Flow cytometry analysis of NSCs control and PHF2-depleted cells previously stained with propidium iodide. (C) Total protein extracts were prepared from NSCs infected with lentivirus expressing shRNA control (shControl) or shRNA specific for PHF2 (shPHF2) and the phospho-MCM2 (p-MCM2) and TUBULIN levels were determined by immunoblot. (D) Schematic representation indicating the regions occupied by proliferating neural progenitors (VZ) and postmitotic neurons (MZ) in HH10 and HH24 chicken embryo spinal cord. (E) HH10–12 embryos were electroporated with shControl or shRNA for cPHF2 (shcPHF2) cloned into pSUPER vector and GFP-expressing vector. Transversal sections of electroporated neural tubes are indicated above stained with DAPI 48 h postelectroporation (PE). Graphs show the quantification of the size of the control side and shcPHF2-electroporated side. To do so, we measured the dorsal, medial, and ventral distances to the lumen on each side, relative to the length of the central line of the lumen. Data represent the mean of 4 to 5 embryos (from 2 to 4 biological independent experiments). Error bars indicate SD **P < 0.01; ***P < 0.001 (Student’s t test). (F) Transversal sections of neural tubes from HH10–12 embryos electroporated in ovo with shControl or shcPHF2 and stained for H3S10P (PH3), Sox2, or TUJ1 48 PE. Boxplots are showing the quantification of the corresponding immunostaining. Data represent the mean of 4 to 12 embryos (from 3 to 4 biological independent experiments). *P < 0.05 (Student’s t test).
PHF2 Depletion Blocks Neurogenesis in the Spinal Cord.
The findings described above support the idea that PHF2 activates genes essential for neural progenitor proliferation (Fig. 3F and SI Appendix, Fig. S4 A and B). Thus, we sought to test this notion in vivo using the chicken embryo neural tube as a model. Structurally, 2 main zones can be distinguished in a transversal section of the neural tube: the ventricular zone (VZ), where proliferating progenitors reside, and the mantle zone (MZ), where final differentiated neurons accumulate (Fig. 4D). To analyze the function of PHF2 in early neurogenesis, we first cloned 2 shRNAs for chick PHF2 (cPHF2) in a bicistronic vector containing GFP sequence, which reduced PHF2 levels (SI Appendix, Fig. S5C). Then, we knocked down PHF2’s expression in Hamburger and Hamilton stage 10–12 (HH10–12) chicken embryo spinal cords by in ovo electroporation of the shRNAs for cPHF2 or a control shRNA (shControl). Remarkably, cPHF2 KD resulted in a neural tube reduced in size (Fig. 4E and SI Appendix, Fig. S5D). As the 2 cPHF2 shRNAs gave the same phenotype, we chose the shcPHF2, that provided better cPHF2 depletion (SI Appendix, Fig. S5C), to perform the rest of the experiments. The observed reduced size of the neural tube could mainly be due to cell death or to proliferation defects. As we did not observe apoptotic cells in the electroporated neural tubes and our previous results indicated that PHF2 is essential for neural progenitor proliferation in vitro, we hypothesized that cPHF2 plays an active role in maintaining neuroblast proliferating. To explore this idea, HH10–12 neural tubes were electroporated with shcPHF2 or shControl and the effect on neural progenitor proliferation was analyzed after 48 h. We evaluated neural progenitor entry into mitosis by analyzing the presence of H3S10p. Neural tubes electroporated with shcPHF2 showed a reduction in H3S10p (PH3)-positive mitotic cells (ratio of PH3+ cells electroporated [EP] side/control side: shControl 106.5 ± 36.7, shcPHF2 65.3 ± 21; P < 0.05) (Fig. 4F). Accordingly, reduction of the neural progenitor marker SOX2 (ratio of SOX2+ cells EP site/control site: shControl 101.6 ± 10.3, shcPHF2 67.3 ± 14; P < 0.05) (Fig. 4F) indicates that cPHF2 is required for neural progenitor self-renewal in the neural tube. We next explored the possibility that the inhibition of proliferation observed upon cPHF2 reduction would also correspond with a premature differentiation of neuroblasts. Neural tubes electroporated in ovo with shcPHF2 and stained for TUJ1, a neuronal differentiation marker, showed neither premature differentiation nor ectopic localization (Fig. 4F). Only a clear reduction of TUJ1-positive cells was observed, probably reflecting the decrease on the progenitor population. Finally, we investigated whether the neuroblast proliferation impairment affected similarly to all neural subpopulations along the dorsal–ventral axis. To do that, we depleted cPHF2 and analyzed different populations by using different neural markers: MNR2 for ventral neurons (motoneurons) and Pax6 for dorsal progenitors. The results in SI Appendix, Fig. S5E show that PHF2 depletion impairs the generation of all analyzed cell types. Overall, these results point to an essential role of cPHF2 promoting early neurogenesis by controlling progenitor proliferation.
Although we did not observe any effect on neuroblast differentiation upon PHF2 depletion, it has been demonstrated that PHF2 plays an important role in differentiation in other models (16–18). To deeply understand the potential contribution of PHF2 to neuronal commitment, we first analyzed the PHF2 expression during neuronal differentiation. Our results in SI Appendix, Fig. S6A indicated that PHF2 levels slightly increased during differentiation (SI Appendix, Fig. S6A). Next, we analyzed the consequences of overexpressing PHF2 during neuronal differentiation that we measured by the induction of the neuronal marker TUJ1 and the repression of the pluripotency gene NESTIN. Results in SI Appendix, Fig. S6 B and C indicate that PHF2 overexpression did not affect neuronal differentiation in vitro.
As PHF2 depletion led to cell cycle arrest, we finally tested whether PHF2 overexpression overcame G1/S cellular checkpoint imposed by growth factors removal or neuronal differentiation induction. Cell cycle reentry was analyzed by measuring the levels of some cell cycle-related genes by qPCR and cell cycle analysis by flow cytometry. Results in SI Appendix, Fig. S7 A–C suggest that, although we observed the induction of some genes, PHF2 overexpression did not overcome the cell cycle arrest imposed by either growth factor removal or neuronal differentiation induction. Altogether these data strongly indicate that the major role of PHF2 at early neurogenesis is to facilitate neural progenitor proliferation.
PHF2 Depletion Leads to DNA Damage and Genome Instability.
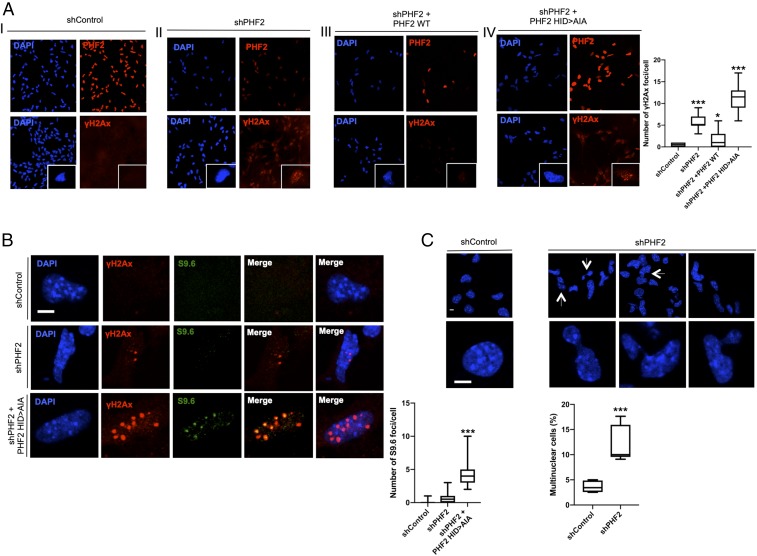
As PHF2 depletion caused profound defects on replication machinery expression (E2f, Cyclin E, Orc1/2, and Mcm2/7) as well as repair components (Brac1/2, Rad51b, and ATR/M) that finally led to cell cycle arrest, we tested whether this fact induced DNA damage that we measured by the γH2Ax content. The histone variant H2AX is phosphorylated at the Ser-139, forming γH2AX as an early cellular response to the DNA double-strand breaks. Then, we quantified the γH2AX content as a measure of DNA damage in control and PHF2-depleted cells. Upon PHF2 KD, a clear accumulation of γH2Ax was detected (Fig. 5A, panel II). This increase was not observed in cells transduced with the shControl RNA (Fig. 5A, panel I). Interestingly, the observed DNA damage was rescued by overexpression of the PHF2 WT (Fig. 5A, panel III) but not by overexpression of the catalytic dead mutant (249H > A) (Fig. 5A, panel IV, and Fig. 5B); indeed, in the latter case, an apparent increase in γH2Ax reactivity, accompanied by a higher decrease of replication-related gene expression (SI Appendix, Fig. S8), was detected, suggesting an important role of PHF2’s enzymatic activity in preventing damage. R-loop (RNA–DNA hybrids formed by reannealing of nascent transcripts to their DNA template, leaving the nontemplated strand as single-stranded DNA) accumulation is associated with DNA replication mistiming that increases collisions with the transcriptional machinery (31), leading to DNA damage (31, 32). Therefore, we investigated whether γH2Ax reactivity observed in PHF2-depleted cells was related to R-loop accumulation. For this purpose, we performed immunofluorescence experiments in control, PHF2 depleted, and PHF2-depleted that overexpressed the PHF2 (249H > A) mutant NSCs (where the γH2Ax signal was higher, see Fig. 5A, panel IV) using the S9.6 antibody that specifically recognizes RNA–DNA hybrids without cross-reacting with single-or double-stranded DNA. The data in Fig. 5B show a clear increase in S9.6 reactivity that colocalized with the γH2Ax-positive regions.
Fig. 5.
PHF2 depletion leads to DNA damage and genome instability. (A) Immunostaining of NSCs expressing shControl (I), shPHF2 (II), shPHF2 together with PHF2 WT (III), and shPHF2 together with PHF2 (249H > A) (IV). Cells were fixed and stained with PHF2 and γH2Ax antibodies and DAPI. Enlarged images are showing individual cells. More than 30 cells were quantified. Data shown are representative of 2 to 3 biological independent experiments (Scale bar: 20 μm.) Boxplots represent the number of γH2Ax foci/cell. *P < 0.05; ***P < 0.001 (Student’s t test). (B) Immunostaining of a representative enlarged cell expressing shControl, shPHF2, and shPHF2 together with PHF2 (249H > A). Cells were fixed and stained with γH2Ax and S9.6 antibodies and DAPI. More than 30 cells were quantified. Data shown are representative of 2 to 3 biological independent experiments (Scale bar: 5 μm.) Boxplots represent the number of S9.6 foci/cell. ***P < 0.001 (Student’s t test). (C) Formation of multinucleated cells and segregation defects are shown for shControl and shPHF2 cells. A total of 50 to 100 cells were quantified. Data shown are representative of 4 independent experiments (Scale bar: 5 μm.) Boxplots represent the percentage of multinuclear cells. ***P < 0.001 (Student’s t test).
DNA damage is often linked with genome instability; accordingly, we noticed an increased frequency of chromosome segregation defects, in particular, anaphase chromatin bridges and multinuclear cells in PHF2-depleted compared with control NSCs (Fig. 5C). Altogether these data demonstrate that PHF2’s deficiency induced DNA damage and R-loop accumulation that led to chromosome segregation defects and ultimately genome instability.
Discusssion
Our data reveal an unforeseen role of PHF2 during development. We demonstrate that this HDM binds to the cell cycle gene promoters facilitating their transcription and preventing genome instability. In that way, PHF2 allows neural stem cell proliferation during progenitor expansion. To do that, PHF2 required its catalytic activity. PHF2 prevents H3K9me3 accumulation, limiting ectopic heterochromatin formation particularly at promoter regions.
Our data indicate that the histone demethylase PHF2 plays a pivotal role in the neural development by promoting the proliferation of neural progenitors. Interestingly, a synergy for regulation of the key proliferation factor E2F was observed. PHF2 directly promotes the expression of several members of the E2F family and at the same time might cooperate with them to facilitate their transcriptional activity. These data are in agreement with previous studies reporting that E2F target genes were repressed by H3K9me2/3 marks and HP1 factor (33, 34). Thus, PHF2 might prevent H3K9me2/3 accumulation at the cell cycle gene promoters before S entrance, and at the same time it facilitates their demethylation to promote cell cycle progression. Interestingly, previous studies have shown that the absence of other member of the KDM7 family, PHF8, also impaired G1/S transition in conjunction with E2F family factors (29).
Our results indicate that PHF2 depletion was associated with the accumulation of RNA:DNA hybrids, DNA damage, and genome instability. The link between R-loops and DNA damage is well established (32). One of the major causes responsible for R-loop accumulation is the replication fork collapse or stall (31, 35–37). Thus, the replication temporal changes due to replication machinery alteration generated by PHF2 depletion might lead to unscheduled collisions of the replication and transcription machineries, inducing DNA breaks (37–40). On the other hand, the observed decrease of expression of factors involved in R-loop resolution and DNA damage repair (ATM/ATR, Rad51, and BRCA1) in PHF2-depleted cells that could contribute to accumulation of double-strand breaks and genome instability. Finally, it has been proposed the demethylation of H3K9 may be an important step in the repair of double-strand breaks. Thus, the global increase in H3K9me3 observed in PHF2-depleted progenitors (Fig. 2F) could delay or impair DNA repair, as it has been described for KDM4B demethylase (41–43). Interestingly, PHF2 associated with p53 and facilitates its activity (44). p53 is an essential tumor suppressor that maintains genomic stability. p53, in addition to the cell cycle checkpoint control, maintains genomic integrity and replication fidelity by preventing DNA topological conflicts between transcription and replication (45). Thus, PHF2 depletion might decrease p53 activity, leading to accumulation of transcription–replication conflicts and genomic instability.
PHF2 has been related to ASD (22–24), but the function of PHF2 in neural development is still poorly understood. Phf2 KD mice showed partial neonatal death, growth retardation, and reduced body weight. Interestingly, the brain weights of Phf2 KD mice were larger than wild-type littermates (46). Macrocephaly is a common phenotype associated with ASD. Intriguingly, we observed that Phf2 KD in cortical progenitors and in the neural tube results in defective neural progenitor proliferation. A similar paradox has been described for another chromatin factor strongly associated with ASD, the remodeling factor CHD8 (47–49). Although at this moment we do not have an explanation for this phenomenon, it would be interesting to elucidate the role of PHF2 in the development of other cell types such as astrocytes. Furthermore, our results raise the possibility that some of the ASD-associated phenotypes in patients carrying PHF2 mutations may be caused by defects during early neural development.
Our work paves the way for investigating the contribution of PHF2 to genomic stability and transcriptional regulation in other cellular contexts. In particular, PHF2 has been widely involved in cancer. Several studies use H3K9me modifier enzymes as targets in cancer treatment (50, 51). Moreover, alteration of the demethylase activity of PHF2 has been suggested as a new target to treat disorders linked to diet-induced obesity, due to its essential role in regulating adipogenesis (46). Our results from PHF2-depleted cells, as well as published data, removing H3K9me2/3 histone methyltransferases (HMTs) (52), indicate that alteration or removal of H3K9 methylation might not be a suitable therapeutic strategy. In both cases, genomic instability might be a drawback in these treatments. Therefore, our work helps to improve our understanding of the multiple cross-talks between epigenetics, development, and diseases.
Materials and Methods
Cell Culture and Differentiation.
Mouse NSCs were prepared from cerebral cortices of C57BL/6J mouse fetal brains (embryonic day [E]12.5). They were cultured in poly-d-lysine (5 μg/mL, 2 h, 37 °C) and laminine (5 μg/mL, 4 h, 37 °C) precoated dishes (53, 54). Cells were maintained in culture as previously described (55) with fibroblast growth factor (FGF) and epidermal growth factor (EGF) to 10 and 20 ng/mL, respectively. Human 293T cells were maintained in culture under standard conditions (56). Chicken UMNSAH/DF1 and HeLa cells were cultured in DMEM supplemented with 10% FBS. NSC differentiation protocol is described in SI Appendix, Materials and Methods.
Antibodies and Reagents.
Antibodies used were anti: PHF2 (Cell Signaling, D45A2), H3K9me3 (Abcam, ab8898), H3K9me2 (Abcam, ab1220), H3K4me3 (Abcam, ab8580), H3Sp10 (Millipore, 06-570), anti-phospho-H2A.X (Millipore, 07-164), β-tubulin III (TUJ1, Covance, MMS-435P), SOX2 (Millipore, AB5603), β-tubulin (Millipore, MAB3408), MNR2 (DSHB, 81.5C10), HP1α (Euromedex), NESTIN (Abcam, ab5968), p-MCM2 (Abcam, ab133243), ATR (Santa Cruz, sc-21848), Rad51 (Santa Cruz, sc8349), histones H4 (Abcam, ab10158), H3 (Abcam, ab1791), H2B (Abcam, ab1790) and PAX6 (DSHB). Mouse monoclonal S9.6 is described in ref. 57. DAPI was obtained from Thermo Fisher (1306). Doxycyclin (Millipore, 324385) was used at 1 µg/mL.
Plasmids.
PHF2 cDNA from p3xFLAG-PHF2 (kindly provided by Jiemin Wong, East China Normal University, Shangai, China) was cloned into pInducer between attB1 and attB2 sites. Both, PHF2 WT and mutant were induced upon doxycycline addition (1 µg/mL). pLKO.1 lentiviral vectors expressing short hairpin RNA against mPHF2 (CGTGGCTATTAAAGTGTTCTA), shPHF2 or control (CAACAAGATGAAGAGCACC) were purchased from Sigma and (CCTTATCCACTCCCACTTGACC) shPHF2_2 was cloned in pLKO.1 lentiviral vector. DNA sequences coding cPHF2 short hairpin RNAs (GGAGCTTCGAAGTCGCACT) shcPHF2 and (CTATGTCGGACCAGAGAGA) shcPHF2_2 were cloned into pSUPER or pSHIN vectors (58), as indicated. pSHIN vector contains the pSUPER and the EGFP expression cassette.
RNA Extraction and qPCR Assays.
RNA was extracted using TRIZOL reagent (Invitrogen). High-capacity cDNA reverse transcription kit (Invitrogen) and 50 ηg of RNA were utilized for reverse transcription. qPCR assays were performed with SYBR Green (Roche) in a LightCycler 480 (Roche) machine using specific primer pairs (SI Appendix, Table S1).
Chick in Ovo Electroporation and Immunostaining.
Eggs from White-Leghorn chickens were used in the in ovo electroporation experiments. They were incubated at 38.5 °C and 70% humidity. Embryo developmental stage was determined following HH (59). Embryos were electroporated with the indicated DNAs at 3 μg/μL with 50 ηg/mL of Fast Green as previously described (54, 60). Expanded protocol is described in SI Appendix, Materials and Methods.
Lentiviral Transduction.
Lentiviral transduction was carried out as previously described (30, 61). Extended protocol is provided in SI Appendix, Materials and Methods.
Indirect Immunofluorescence.
Immunofluorescence assays were performed basically as previously described (61, 62). Cells were fixed in 4% paraformaldehyde for 20 min and permeabilized using PBS-Triton X-100 (0.1%). Then, they were blocked at room temperature for 1 h in 1% BSA (in PBS with 0.1% Triton X-100). Primary antibodies were incubated overnight at 4 °C. Finally, Alexa-conjugated secondary IgG antibodies and DAPI (0.1 ηg/μL) (Sigma) were used for 2 h at room temperature. Images were obtained using a Leica SP5 confocal microscope by LAS-AF software.
Western Blot.
Immunoblotting was performed using standard procedures and visualized using the ECL kit (Amersham).
ChIP Assays.
ChIP assays were performed as previously described (63). See extended protocol in SI Appendix, Materials and Methods.
Cell Cycle Analysis by Flow Cytometry.
Cells were fixed in cold 70% ethanol and stored at −20 °C until they were ready for staining. Cells were washed twice with cold PBS and pelleted by centrifugation. The cell pellet was resuspended in PBS containing 200 µL of 1 mg/mL propidium iodide and 2 mg RNase A and incubated at 37 °C for 15 min.
ChIP-Seq Procedure.
Chromatin immunoprecipitation as well as sample preparation for sequencing from one replicate were done as previously described (54). For details see expanded protocol in SI Appendix, Materials and Methods and Table S2. ChIP-seq data have been deposited in the GEO database under the accession GSE122263 (access token ovizqewwblullyx).
RNA-Seq Procedure.
RNA was extracted using the High Pure RNA isolation kit from Roche followed by DnaseI treatment from 2 biological independent samples. Libraries were prepared using the TruSeq Stranded Total RNA Sample Preparation kit with Ribo-Zero Human/Mouse/Rat Kit (Illumina, RS-122–2201/2) according to the manufacturer’s protocol. Briefly, 500 ηg of total RNA was used for ribosomal RNA depletion. Then, after removing ribosomal RNA, the remaining RNA was fragmented for 4.5 min. The remaining steps of the library preparation were followed according to the manufacturer’s instructions. Final libraries were analyzed using an Agilent DNA 1000 chip to estimate the quantity and check size distribution, and then quantified by qPCR using the KAPA Library Quantification kit (Roche, 07960204001) prior to amplification with Illumina’s cBot. The libraries were sequenced on Illumina High HiSeq 2500 with paired-end 50 base pair long reads (SI Appendix, Table S2). Alignment was performed using HISAT2 (64), assignment of aligned reads to genes was performed using HTSeq (65), and differential expression analysis was performed using DESeq2 (66). RNA-seq data have been deposited in the GEO database under the accession GSE122264 (access token anmrkicwntqjjsp).
ChIP-Seq Data Acquisition and Analysis.
ChIP-seq data were downloaded from Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) (Accessions used in this paper are specified in SI Appendix, Table S3). See further explanations in SI Appendix, Materials and Methods.
Statistical Analysis.
Quantitative data were expressed as mean and SD (for immunofluoresence countings and RNA transcription experiments) and as mean and SEM (for ChIPs). The significance of differences between groups was assessed using the Student’s t test.
Data and Materials Availability.
RNA-seq and ChIP-seq data have been deposited in the GEO database under accessions GSE122264 and GSE122263, respectively. All other materials are available upon request. All the experiments have been approved by the Bioethics Committee of the Consejo Superior de Investigaciones Científicas.
Supplementary Material
Acknowledgments
We thank Drs. Jiemin Wong and Jiwen Li for the PHF2 plasmid; Dr. Borisy for the pSHIN vector; and Drs. A. Jordá, F. Azorín, J. Bernues, L. L. Espinás, S. de la Luna, A. Vaquero, N. Agell, J. Roig, G. Vicent, M. Beato, M. Arbonés, and E. Martí for reagents; and all members of the M.A.M.-B. laboratory for helpful comments and suggestions. This study was supported by grants BFU2012-34261 and BFU2015-69248-P (to M.A.M.-B.) from the Spanish Ministry of Economy and Fondation Jérôme Lejeune (to M.A.M.-B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: RNA-seq and ChIP-seq data have been deposited in the GEO database under accessions GSE122264 and GSE122263, respectively.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903188116/-/DCSupplemental.
References
- 1.Tiberi L., Vanderhaeghen P., van den Ameele J., Cortical neurogenesis and morphogens: Diversity of cues, sources and functions. Curr. Opin. Cell Biol. 24, 269–276 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Temple S., The development of neural stem cells. Nature 414, 112–117 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Kouzarides T., Chromatin modifications and their function. Cell 128, 693–705 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Fueyo R., García M. A., Martínez-Balbás M. A., Jumonji family histone demethylases in neural development. Cell Tissue Res. 359, 87–98 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Epsztejn-Litman S., et al. , De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat. Struct. Mol. Biol. 15, 1176–1183 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wen B., Wu H., Shinkai Y., Irizarry R. A., Feinberg A. P., Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat. Genet. 41, 246–250 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filion G. J., van Steensel B., Reassessing the abundance of H3K9me2 chromatin domains in embryonic stem cells. Nat. Genet. 42, 4; author reply 5-6 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Nakayama J., Rice J. C., Strahl B. D., Allis C. D., Grewal S. I., Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Allshire R. C., Javerzat J. P., Redhead N. J., Cranston G., Position effect variegation at fission yeast centromeres. Cell 76, 157–169 (1994). [DOI] [PubMed] [Google Scholar]
- 10.Saksouk N., Simboeck E., Déjardin J., Constitutive heterochromatin formation and transcription in mammals. Epigenetics Chromatin 8, 3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okuno Y., Inoue K., Imai Y., Novel insights into histone modifiers in adipogenesis. Adipocyte 2, 285–288 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fortschegger K., Shiekhattar R., Plant homeodomain fingers form a helping hand for transcription. Epigenetics 6, 4–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wen H., et al. , Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation. J. Biol. Chem. 285, 9322–9326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicholson G. A., et al. , The gene for hereditary sensory neuropathy type I (HSN-I) maps to chromosome 9q22.1-q22.3. Nat. Genet. 13, 101–104 (1996). [DOI] [PubMed] [Google Scholar]
- 15.Hasenpusch-Theil K., et al. , PHF2, a novel PHD finger gene located on human chromosome 9q22. Mamm. Genome 10, 294–298 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Kim H. J., et al. , Plant homeodomain finger protein 2 promotes bone formation by demethylating and activating Runx2 for osteoblast differentiation. Cell Res. 24, 1231–1249 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee K. H., Ju U. I., Song J. Y., Chun Y. S., The histone demethylase PHF2 promotes fat cell differentiation as an epigenetic activator of both C/EBPα and C/EBPδ. Mol. Cells 37, 734–741 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J., et al. , Epigenetic regulation of megakaryocytic and erythroid differentiation by PHF2 histone demethylase. J. Cell. Physiol. 233, 6841–6852 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Pattabiraman D. R., et al. , Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science 351, aad3680 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh A., et al. , Association of FANCC and PTCH1 with the development of early dysplastic lesions of the head and neck. Ann. Surg. Oncol. 19 (suppl. 3), S528–S538 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Lee C., Kim B., Song B., Moon K. C., Implication of PHF2 expression in clear cell renal cell carcinoma. J. Pathol. Transl. Med. 51, 359–364 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.C Yuen R. K., et al. , Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 20, 602–611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iossifov I., et al. , De novo gene disruptions in children on the autistic spectrum. Neuron 74, 285–299 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cotney J., et al. , The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat. Commun. 6, 6404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bricambert J., et al. , The histone demethylase Phf2 acts as a molecular checkpoint to prevent NAFLD progression during obesity. Nat. Commun. 9, 2092 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu X. N., et al. , Methylation of transcription factor YY2 regulates its transcriptional activity and cell proliferation. Cell Discov. 3, 17035 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lammens T., Li J., Leone G., De Veylder L., Atypical E2Fs: New players in the E2F transcription factor family. Trends Cell Biol. 19, 111–118 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tahmasebi S., et al. , Control of embryonic stem cell self-renewal and differentiation via coordinated alternative splicing and translation of YY2. Proc. Natl. Acad. Sci. U.S.A. 113, 12360–12367 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu W., et al. , PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 466, 508–512 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asensio-Juan E., et al. , The histone demethylase PHF8 is a molecular safeguard of the IFNγ response. Nucleic Acids Res. 45, 3800–3811 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gan W., et al. , R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 25, 2041–2056 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sollier J., Cimprich K. A., Breaking bad: R-Loops and genome integrity. Trends Cell Biol. 25, 514–522 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen S. J., et al. , Rb targets histone H3 methylation and HP1 to promoters. Nature 412, 561–565 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Panteleeva I., et al. , HP1alpha guides neuronal fate by timing E2F-targeted genes silencing during terminal differentiation. EMBO J. 26, 3616–3628 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tuduri S., et al. , Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 11, 1315–1324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helmrich A., Ballarino M., Tora L., Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell 44, 966–977 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Deshpande A. M., Newlon C. S., DNA replication fork pause sites dependent on transcription. Science 272, 1030–1033 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Hoffman E. A., McCulley A., Haarer B., Arnak R., Feng W., Break-seq reveals hydroxyurea-induced chromosome fragility as a result of unscheduled conflict between DNA replication and transcription. Genome Res. 25, 402–412 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merrikh H., Machón C., Grainger W. H., Grossman A. D., Soultanas P., Co-directional replication-transcription conflicts lead to replication restart. Nature 470, 554–557 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bayona-Feliu A., Casas-Lamesa A., Reina O., Bernués J., Azorín F., Linker histone H1 prevents R-loop accumulation and genome instability in heterochromatin. Nat. Commun. 8, 283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colmenares S. U., et al. , Drosophila histone demethylase KDM4A has enzymatic and non-enzymatic roles in controlling heterochromatin integrity. Dev. Cell 42, 156–169.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young L. C., McDonald D. W., Hendzel M. J., Kdm4b histone demethylase is a DNA damage response protein and confers a survival advantage following γ-irradiation. J. Biol. Chem. 288, 21376–21388 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng H., Chen L., Pledger W. J., Fang J., Chen J., p53 promotes repair of heterochromatin DNA by regulating JMJD2b and SUV39H1 expression. Oncogene 33, 734–744 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee K. H., et al. , PHF2 histone demethylase acts as a tumor suppressor in association with p53 in cancer. Oncogene 34, 2897–2909 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Yeo C. Q. X., et al. , p53 maintains genomic stability by preventing interference between transcription and replication. Cell Rep. 15, 132–146 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Okuno Y., et al. , Epigenetic regulation of adipogenesis by PHF2 histone demethylase. Diabetes 62, 1426–1434 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Roak B. J., et al. , Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338, 1619–1622 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Rubeis S., et al. ; DDD Study; Homozygosity Mapping Collaborative for Autism; UK10K Consortium , Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bernier R., et al. , Disruptive CHD8 mutations define a subtype of autism early in development. Cell 158, 263–276 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kondo Y., et al. , Downregulation of histone H3 lysine 9 methyltransferase G9a induces centrosome disruption and chromosome instability in cancer cells. PLoS One 3, e2037 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner T., Jung M., New lysine methyltransferase drug targets in cancer. Nat. Biotechnol. 30, 622–623 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Zeller P., et al. , Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat. Genet. 48, 1385–1395 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Currle D. S., Hu J. S., Kolski-Andreaco A., Monuki E. S., Culture of mouse neural stem cell precursors. J. Vis. Exp., e152 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fueyo R., et al. , Lineage specific transcription factors and epigenetic regulators mediate TGFβ-dependent enhancer activation. Nucleic Acids Res. 46, 3351–3365 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Estarás C., et al. , Genome-wide analysis reveals that Smad3 and JMJD3 HDM co-activate the neural developmental program. Development 139, 2681–2691 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Blanco-García N., Asensio-Juan E., de la Cruz X., Martínez-Balbás M. A., Autoacetylation regulates P/CAF nuclear localization. J. Biol. Chem. 284, 1343–1352 (2009). [DOI] [PubMed] [Google Scholar]
- 57.Boguslawski S. J., et al. , Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J. Immunol. Methods 89, 123–130 (1986). [DOI] [PubMed] [Google Scholar]
- 58.Kojima S., Vignjevic D., Borisy G. G., Improved silencing vector co-expressing GFP and small hairpin RNA. Biotechniques 36, 74–79 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Hamburger V., Hamilton H. L., A series of normal stages in the development of the chick embryo. 1951. Dev. Dyn. 195, 231–272 (1992). [DOI] [PubMed] [Google Scholar]
- 60.Akizu N., et al. , EZH2 regulates neuroepithelium structure and neuroblast proliferation by repressing p21. Open Biol. 6, 150227 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Asensio-Juan E., Gallego C., Martínez-Balbás M. A., The histone demethylase PHF8 is essential for cytoskeleton dynamics. Nucleic Acids Res. 40, 9429–9440 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sánchez-Molina S., et al. , Regulation of CBP and Tip60 coordinates histone acetylation at local and global levels during Ras-induced transformation. Carcinogenesis 35, 2194–2202 (2014). [DOI] [PubMed] [Google Scholar]
- 63.Valls E., et al. , Involvement of chromatin and histone deacetylation in SV40 T antigen transcription regulation. Nucleic Acids Res. 35, 1958–1968 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim D., Langmead B., Salzberg S. L., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anders S., Pyl P. T., Huber W., HTSeq–A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.