Significance
Genetic analysis of Rickettsia has been difficult. We developed a transposon and selection scheme to facilitate the isolation of Rickettsia conorii mutants with insertional lesions. Here, we demonstrate that the R. conorii polysaccharide synthesis operon (pso) encompasses genetic determinants for biosynthesis of the O antigen, which also affect the composition of outer-membrane proteins, invasion of host cells, and pathogenesis. The O antigen provides essential barrier functions and plays a major role in host–pathogen interactions. Our findings suggest that infected hosts develop protective immunity against R. conorii via the production of antibodies targeting the O antigen. Conservation of pso among rickettsial species suggests that it may play a universal role in O-antigen synthesis, disease pathogenesis, and the development of immunity.
Keywords: transposon mutagenesis, lipopolysaccharide, O antigen, Weil–Felix reaction, polysaccharide synthesis operon
Abstract
Rickettsial diseases have long been diagnosed with serum antibodies cross-reactive against Proteus vulgaris (Weil–Felix reaction). Although Weil–Felix antibodies are associated with the development of immunity, their rickettsial target and contribution to disease pathogenesis are not established. Here, we developed a transposon for insertional mutagenesis of Rickettsia conorii, isolating variants defective for replication in cultured cells and in spotted fever pathogenesis. Mutations in the polysaccharide synthesis operon (pso) abolish lipopolysaccharide O-antigen synthesis and Weil–Felix serology and alter outer-membrane protein assembly. Unlike wild-type R. conorii, pso mutants cannot elicit bactericidal antibodies that bind O antigen. The pso operon is conserved among rickettsial pathogens, suggesting that bactericidal antibodies targeting O antigen may generate universal immunity that could be exploited to develop vaccines against rickettsial diseases.
Antibodies directed against bacterial surface carbohydrates, i.e., the capsular polysaccharide or the O antigen of lipopolysaccharide (LPS), activate, complement, and promote microbial killing (1). Bactericidal antibodies are acquired during colonization or invasive disease with pathogens such as Escherichia coli, Haemophilus influenzae, and Neisseria meningitidis or following immunization with carbohydrate-conjugate vaccines (2–4). Bactericidal antibodies represent a correlate for immunity; however, disease protection is limited to specific pathogen serotypes and countered by the selection of variants with distinct carbohydrate antigens (5). Rickettsial pathogens rely on hematophagous arthropods for host transmission and disease pathogenesis (6). Due to their obligate requirement for intracellular replication, Rickettsia spp. cannot be propagated on laboratory media (7). Rickettsial diseases have therefore been diagnosed with the Weil–Felix serology, the detection of IgG or IgM cross-reactive with Proteus vulgaris (8). Increased Weil–Felix serology has been associated with positive clinical outcome and with protective immunity (9). For survivors of epidemic typhus, waning Weil–Felix serology has been observed in patients with recrudescent typhus (Brill–Zinsser disease) (10). Administration of epidemic typhus vaccines, for example extracts from lice infected with Rickettsia prowazekii, elicits Weil–Felix antibodies and disease protection (11). Nevertheless, the rickettsial target of Weil–Felix antibodies and its contribution to rickettsial disease pathogenesis are not established.
Here, we developed a transposon mutagenesis technology enabling facile isolation of insertional mutants defective in the obligate intracellular life cycle of Rickettsia conorii. We describe that R. conorii variants harboring transposon insertions in the conserved polysaccharide synthesis operon cannot produce O-antigen polysaccharides and fail to induce Weil–Felix serology with a significant virulence defect in the murine infection model. The results suggest that humans may develop protective immunity against pathogenic rickettsiae by targeting the conserved carbohydrate epitope present in the O antigen of LPS.
Results
kkaebi Transposon Mutagenesis Identifies R. conorii Variants Defective in Intracellular Replication.
Earlier work developed transposon mutagenesis for the selection of antibiotic-resistant Rickettsia variants. However, the selection for transposon mutants was hindered by rickettsial variants with spontaneous resistance to antibiotic selection (12, 13). To overcome this obstacle, we measured the frequency of spontaneous resistance in R. conorii replicating in Vero cells. R. conorii rifampin-resistant plaques appeared at a frequency of 1 × 10−7 plaque-forming unit (PFU), whereas chloramphenicol-resistant plaques were not isolated (mutational frequency <1 × 10−8 PFU). Using whole-genome sequencing, we identified a missense mutation in the rpoB gene of rifampin-resistant R. conorii (amino acid substitution D533G [accession no. SRR8404401 [wild-type R. conorii] and SRR8404402 [rifampin-resistant R. conorii]; refs. 14 and 15). These findings corroborate previous work, demonstrating that rpoB missense mutations (R546K) provide for rifampin resistance in R. prowazekii (16). However, chloramphenicol does not appear to select for spontaneously resistant R. conorii variants and was therefore used for the selection of transposon mutants.
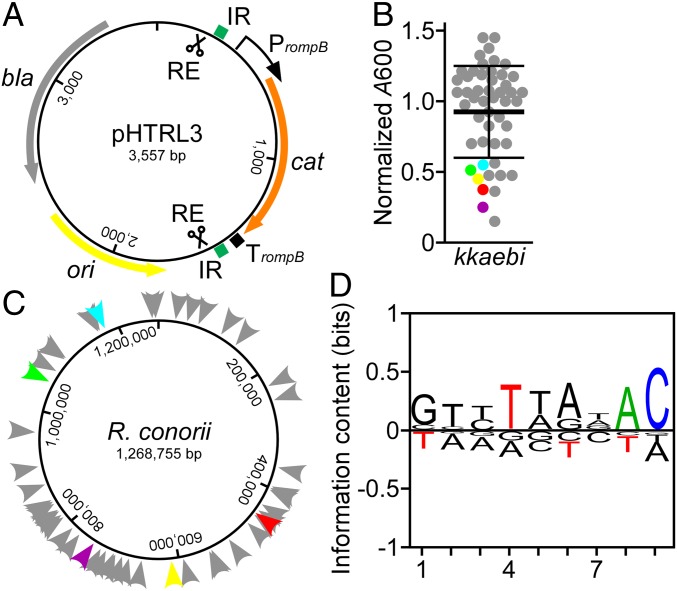
For insertional mutagenesis, we generated pHTRL3, which carries a codon-optimized gene for chloramphenicol acetyltransferase (cat) flanked by the inverted repeats (IR) of the Tn5 transposon under control of the R. rickettsii rompB promoter (PrompB) (Fig. 1A). The minitransposon was named kkaebi. kkaebi DNA was PCR amplified from pHTRL3 template DNA, cleaved with restriction enzyme (RE), and incubated with Tn5 transposase to generate transposome complexes that, when electroporated into R. conorii and selected on Vero cell cultures in the presence of chloramphenicol, generated insertional mutations at a frequency of 5 × 10−8/µg kkaebi. Isolated variants were cataloged with HK numbers in the R. conorii mutant library database (SI Appendix, Table S1). After 4 d of plaque expansion in Vero cells, R. conorii HK mutants were purified via gradient density centrifugation and absorption at 600 nm (A600) was determined. Most of the kkaebi mutants replicated to the same level as wild-type R. conorii (Fig. 1B). However, A600 and PFU for the HK2, HK24, HK27, HK28, and HK50 variants were lower than those of wild-type Rickettsia (Fig. 1B). Sequence analysis of the transposon insertion sites revealed that all R. conorii variants harbored single insertions of kkaebi distributed randomly across the rickettsial genome (Fig. 1C and SI Appendix, Table S1). Alignment of the 9-nucleotide region for each recorded insertion revealed that transposition of kkaebi is not biased by specific nucleotide sequences (Fig. 1D).
Fig. 1.
Transposon mutagenesis in R. conorii. (A) Map of pHTRL3 with the kkaebi minitransposon. (B) A600 was measured to determine the yield of R. conorii kkaebi variants after 4 d growth on Vero cells. Five variants (HK2, HK24, HK27, HK28, and HK50) with defective growth are colored in red, yellow, green, purple, and light blue, respectively. (C) Diagram illustrating the position of kkaebi insertion sites on the R. conorii chromosome (color code same as in B). (D) BLogo sequence logo plot generated with 53 insertion events of kkaebi. Nine bases were used to generate the sequence logo. Nucleotide frequency at each position is indicated by the relative height of each letter; significantly increased or reduced frequency (P < 0.05) at a given position is shown in color.
Genes Involved in the Biosynthesis of the O-Antigen Polysaccharide and Weil–Felix Serology.
Sequence analysis identified 2 variants, HK2 and HK15, with kkaebi insertions in Rc0457 and Rc0459. Rc0457 and Rc0459 are located in a gene cluster here designated as the rickettsial polysaccharide synthesis operon (pso) (Fig. 2A and SI Appendix, Fig. S1). The first 3 genes (Rc0456 to Rc0458) of pso are conserved among all pathogenic Rickettsia and exhibit sequence homology with 3 gene products from P. vulgaris (3074 to 3076) and Vibrio cholerae (wbvB, wbvR, and wbvD) (SI Appendix, Fig. S2). When expressed and purified from E. coli, WbvB/WbvR/WbvD catalyze synthesis of UDP-l-QuiNAc (N-acetyl-l-quinovosamine), a key constituent of the O antigen of V. cholerae, P. vulgaris, and Rickettsia (SI Appendix, Fig. S2) (17–19). The Rickettsia typhi O antigen comprises the polysaccharide repeat [→4)-α-d-Glc-(1→3)-α-l-QuiNAc-(1→4)-[α-d-Glc-(1→3)-α-l-QuiNAc-(1→4)]n-α-d-Glc-(1→] with a short side chain [d-GlcNAc-(1→3)-α-l-QuiNAc-(1→3)-α-d-GlcNAc→] linked to l-QuiNAc (20). Of note, the disaccharide motif, [α-l-QuiNAc-(1→3)-α-d-GlcNAc], is also present in the O antigen of P. vulgaris OX19 (21). However, in P. vulgaris OX2, which does not generate cross-reactive Weil–Felix antibodies with R. typhi, the disaccharide unit is linked to the O-antigen repeats via a β-glycosidic bond (21).
Fig. 2.
The Rickettsia pso is the determinant of Weil–Felix serology. (A) Diagram of the pso in pathogenic Rickettsia. Arrowheads indicate transposon insertion sites. (B) Silver-stained polyacrylamide gel of affinity-purified LPS from R. conorii wild type (WT) or the pso variants HK2 and HK15. (C) Immunoblot of R. conorii LPS with R. conorii (α-RcHu) or O. tsutsugamushi (α-OtHu) convalescent human sera. (D) Rabbit antisera raised against affinity-purified LPS from R. conorii wild type (α-LPSWT) or the pso variants HK2 (α-LPSHK2) and HK15 (α-LPSHK15) were examined for antibodies against WT, HK2, and HK15 LPS and compared with naive rabbit serum. (E) Silver-stained polyacrylamide gel of affinity-purified LPS from P. vulgaris OX2 and OX19 and P. mirabilis OXK. (F) Immunoblot of P. vulgaris OX2 and OX19 and P. mirabilis OXK LPS with α-RcHu or α-OtHu. (G) α-LPSWT, α-LPSHK2, or α-LPSHK15 was examined for antibodies cross-reactive against P. vulgaris OX2 and OX19 and P. mirabilis OXK LPS.
To determine whether the R. conorii variants HK2 and HK15 exhibit O-antigen synthesis defects, we purified R. conorii LPS and analyzed molecules by acrylamide gel electrophoresis and silver staining. As expected, LPS from wild-type R. conorii migrated as a spectrum of molecules with tethered O-antigen repeats and ladder-like appearance on polyacrylamide gels (Fig. 2B). LPS isolated from the HK15 mutant exhibited a similar migration pattern (Fig. 2B). In contrast, a single LPS species was purified from the HK2 variant, which migrated faster on the polyacrylamide gel than wild-type and HK15 LPS (Fig. 2B). Immunoblot analysis with R. conorii-specific human antisera (α-RcHu) revealed antibodies that bound LPS from wild-type and HK15 R. conorii, but not LPS from the HK2 variant (Fig. 2C). Rabbit IgG against R. conorii LPS (raised via immunization with purified LPS, α-LPSWT) bound LPS from wild-type and HK15 R. conorii, but displayed only weak binding for HK2 LPS (Fig. 2D). Rabbit IgG raised against purified HK2 and HK15 LPS (α-LPSHK2 and α-LPSHK15) bound to the cognate LPS antigens but did not exhibit cross-reactivity with LPS from wild type or the other mutant strain (Fig. 2D). The LPS O-antigen biosynthesis defect in R. conorii HK2 was in part restored by transformation with pHTRL8. This plasmid carries Rc0457 and upstream promoter sequences as well as Rc0458 to Rc0460 (SI Appendix, Fig. S3). Rc0457 encodes UDP-GlcNAc 4,6-dehydratase/3,5-epimerase, an enzyme that is essential for QuiNAc and O-antigen polysaccharide synthesis in V. cholerae. Next, we performed immunoblot analyses with affinity-purified LPS from P. vulgaris OX2 and OX19 and Proteus mirabilis OXK (Fig. 2E). As expected, α-RcHu, but not α-OtHu (human immune serum from individuals infected with Orientia tsutsugamushi) harbored Weil–Felix antibodies against OX2 and OX19 LPS (Fig. 2F). Finally, rabbit α-LPSWT, but not rabbit α-LPSHK2 or α-LPSHK15, recognized P. vulgaris OX2 LPS (Fig. 2G). As previously reported, we detected OX19 LPS-specific antibodies in naive rabbit serum, which prohibited further cross-reactivity analysis (22). Together these data indicate that R. conorii pso encodes genes for the synthesis of the O antigen of LPS, which represents the rickettsial target of Weil–Felix antibodies.
Without O-Antigen Synthesis, R. conorii Displays Altered Outer-Membrane Protein Content and Reduced Host Cell Invasion Activity.
We wondered whether the O-antigen synthesis defects of pso mutants interfere with the assembly of rickettsial outer-membrane proteins. Outer-membrane extracts of wild-type R. conorii and the pso mutant strains HK2 and HK15 were analyzed by Coomassie-stained sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), which revealed increased abundance of proteins migrating at 190, 120, and 32 kDa from the outer membrane of R. conorii HK2 (Fig. 3A). Corresponding gel slices were excised and tryptic peptides of proteins analyzed by mass spectrometry (MS) with in silico comparison of tryptic peptides derived from the R. conorii genome (SI Appendix, Table S2). The data identified rOmpA (190 kDa), rOmpB (120 kDa), and rOmpB (32 kDa) as the most abundant species. Of note, the 32-kDa gel slice harbored peptides from 2 additional outer-membrane proteins, rOmpA and Sca1. rOmpA and rOmpB are members of the autotransporter superfamily, forming surface (S) layers that contribute to rickettsial invasion of host cells (23–25). Autotransporters are synthesized as large precursor species with N-terminal signal peptides for secretion via the Sec pathway. The Bam complex subsequently translocates and assembles autotransporters in the outer membrane (26). During assembly, the N-terminal passenger domain of autotransporters is cleaved and displayed on the bacterial surface, while the C-terminal β-barrel domain functions to anchor the passenger domain in the outer membrane. Outer-membrane samples of R. conorii strains were subjected to immunoblotting with monoclonal antibodies specific for the 190-kDa passenger domain of rOmpA or polyclonal antipeptide antibodies specific for the 32-kDa β-barrel domain of rOmpA. Compared to wild-type R. conorii and the HK15 mutant strain, the abundance of the rOmpA passenger and β-barrel domains was increased in R. conorii HK2 (Fig. 3B). Of note, the β-barrel domain exhibited increased mobility in R. conorii HK2, suggesting that the O-antigen synthesis defect altered not only the abundance of the autotransporter but also its proteolytic cleavage. Immunoblotting with polyclonal antibodies against rOmpB also revealed an increased abundance of this autotransporter in R. conorii HK2 (Fig. 3B). Electroporation of R. conorii HK2 with pHTRL8 restored the abundance of rOmpA and rOmpB and of the overall outer-membrane protein content to levels closer to those observed for wild-type R. conorii (SI Appendix, Fig. S3).
Fig. 3.
R. conorii HK2 is defective for outer-membrane protein display and attachment to host cells. (A and B) Outer-membrane fractions isolated from R. conorii wild-type (WT) or the pso variants HK2 and HK15 were analyzed by (A) Coomassie-stained PAGE or (B) immunoblotting with monoclonal antibodies against the passenger domain of rOmpA (α-rOmpAP), polyclonal antipeptide antibodies against the β-barrel domain of rOmpA (α-rOmpAβ), or polyclonal antibodies against rOmpB (α-rOmpB). (C) Rickettsia replication in Vero cells was quantified with the plaque assay (n = 3). (D) Quantification of cytopathic areas of Vero cells infected with R. conorii variants at day 3 postinfection (n = 24 for WT, n = 11 for HK2, and n = 23 for HK15). Data are the mean (±SEM) of 3 independent determinations. Statistically significant differences were analyzed with (C) 2-way ANOVA with Bonferroni posttests and (D) 1-way ANOVA with Dunnett’s posttest. *P < 0.05; **P < 0.001; ***P < 0.0001. (E) Representative microscopic images of cytopathic areas were identified by differential interference contrast microscopy (DIC) and colored yellow. (Scale bar, 50 µm.) (F) Samples were thin-sectioned, stained with uranyl-acetate, and viewed via transmission electron microscopy (TEM). Arrowheads identify intracellular R. conorii. (Scale bar, 200 nm.)
Defects in O-antigen synthesis and autotransporter display in R. conorii HK2 were also associated with reduced attachment to Vero cells (WT, 4.0 log10 PFU; HK2, 2.4 log10 PFU, P < 0.0001 at 1-h postinoculation; Fig. 3C). In pairwise comparisons of rickettsial replication at timed intervals, wild-type and HK15 R. conorii expanded at similar rates (WT, 7.5 log10 PFU; HK15, 7.1 log10 PFU at 6 d postinoculation, P > 0.05; Fig. 3C). In contrast, R. conorii HK2 replicated at a slower rate (HK2, 5.8 log10 PFU at 6 d postinoculation; WT vs. HK2, P < 0.0001; Fig. 3C). The defects in host cell attachment and intracellular growth were restored when R. conorii HK2 was transformed with pHTRL8 [WT, 7.3 log10 PFU; HK2 (pHTRL7), 5.8 log10 PFU; HK2 (pHTRL8), 7.0 log10 PFU at 6 d postinoculation; WT vs. HK2 (pHTRL7), P < 0.0001; WT vs. HK2 (pHTRL8), P > 0.05; SI Appendix, Fig. S3]. In addition to the attachment defect, the HK2 mutant produced a significantly reduced cytopathic area on Vero cell cultures after 3 d of infection [WT, 7.5 (±0.6) × 104 µm2; HK2, 0.8 (±0.1) × 104 µm2; HK15, 4.0 (±0.3) × 104 µm2; WT vs. HK2, P < 0.001; WT vs. HK15, P < 0.001; Fig. 3 D and E]. Electron microscopy analysis of cytopathic Vero cells identified R. conorii, mostly within the cytoplasm of host cells, without significant changes in bacterial size and shape (Fig. 3F and SI Appendix, Fig. S4).
R. conorii O-Antigen Synthesis Is Required for Spotted Fever Pathogenesis.
To investigate whether R. conorii HK2 or HK15 exhibits virulence defects in the mouse model for acute disease, cohorts of mice were inoculated i.v. with 1 × 106 PFU wild-type R. conorii or its HK2 or HK15 variants. Animals infected with wild-type R. conorii exhibited disseminated vascular disease with severe body weight loss and lethal outcome within 3 d of inoculation (Fig. 4 A and B). The HK15 mutant also caused lethal outcome, whereas inoculation of mice with R. conorii HK2 did not produce disease (Fig. 4 A and B). Animals infected with 1 × 103 PFU of wild-type R. conorii exhibited body weight loss (18.5%, P < 0.0001) during the first 7 d of infection followed by a slow recovery for the next 7 d (returning to 96.5% of the original body weight). Animals infected with R. conorii HK2 or HK15 exhibited only modest weight losses (Fig. 4C). After 14 d, infection with wild-type R. conorii (α-WT) elicited IgG antibody responses against rickettsial LPS that were not observed in animals infected with R. conorii HK2 or HK15 (Fig. 4D). However, preexisting antibodies recognizing OX2 and OX19 LPS in naive mice prevented further correlation between R. conorii infection and positive Weil–Felix serology (Fig. 4E). Importantly, α-WT, but neither α-HK2 nor α-HK15, promoted complement-mediated killing of R. conorii in mouse plasma (Fig. 4F).
Fig. 4.
R. conorii O-antigen synthesis is required for pathogenesis. (A and B) Kaplan–Meier analysis for survival (A) and body-weight analysis (B) of C3H/HeN mice (n = 10) infected with 1 × 106 PFU R. conorii WT or the pso mutants HK2 and HK15 or mock infected (Mock). (C) Body-weight analysis of C3H/HeN mice (n = 10) infected with 1 × 103 PFU of R. conorii WT and pso mutant strains. Data are representative of 2 independent experiments. The proportion of survival animals was analyzed using the 2-tailed log-rank test. Two-way ANOVA with Bonferroni posttests were performed to analyze the statistical significance of body-weight change. (D) Immunoblotting of affinity-purified LPS from R. conorii WT, HK2, or HK15 with α-WT, α-HK2, or α-HK15 mouse immune serum. (E) Immunoblotting of affinity-purified LPS from P. vulgaris OX2 or OX19 with α-WT or naive mouse sera. (F) Survival of R. conorii in mouse plasma mixed with naive, α-WT, α-HK2, or α-HK15 mouse immune serum (n = 3). Data are the mean (±SEM) of 3 independent determinations. Statistically significant differences were analyzed with 1-way ANOVA with Dunnett’s posttest. *P < 0.05; **P < 0.001; ***P < 0.0001.
Discussion
Our results demonstrate that the R. conorii pso locus is responsible for O-antigen biosynthesis, contributes to the pathogenesis, and is essential for the development of bactericidal Weil–Felix antibodies. In light of these findings, bactericidal Weil–Felix antibodies can be assigned a role in protective immunity, supporting earlier clinical observations associating increased Weil–Felix antibodies with survival and convalescence and bactericidal Weil–Felix antibodies with protective immunity in individuals receiving whole-cell Rocky Mountain spotted fever or epidemic typhus vaccines (27, 28). Although the pso locus is conserved among spotted fever- or typhus-causing Rickettsia, there exist variations in pso gene content among the 2 groups. We hypothesize that the genetic differences of the pso locus are responsible for differences in Weil–Felix serology as, for example, spotted fever agents, but not typhus agents, elicit antibodies that are cross-reactive with P. vulgaris OX2 (29). Nevertheless, similar to carbohydrate-specific bactericidal antibodies in established bacterial vaccines (5), the discovery of the pso locus may be exploited to generate O-antigen specific subunit vaccines against spotted fever and typhus agents whose bactericidal antibodies are likely to provide a correlate for protective immunity against the corresponding rickettsial diseases.
We show here that chloramphenicol is a suitable antibiotic to select for R. conorii variants with insertional kkaebi lesions. Even if the insertional lesion disrupts a gene that contributes to tissue cell invasion or intracellular replication, some of these mutants will exert partial phenotypes of delayed replication, diminished invasion, or reduced cell-to-cell spread that should allow their isolation and phenotypic characterization. Thus, a library of several thousand R. conorii mutants with mapped insertional lesions may be tremendously useful for the field of rickettsial biology in assigning function to the more than 800 genes that remain as of yet uncharacterized. Others have initiated similar insertional mutagenesis studies focusing on Rickettsia parkeri, Rickettsia rickettsii, and R. prowazekii (30–33). Together, the results from all of these studies should enable comparative genetic analyses of rickettsial species in the near future, which would be a tremendous boost for a field that for many years was hampered by the lack of tools for genetic studies.
We think it is likely that the insertional disruption of Rc0457 in R. conorii HK2 abolishes the synthesis of the O-antigen repeats within LPS, allowing the mutant to synthesize a rudimentary lipid A core molecule that cannot be further modified. LPS is a major outer-membrane component essential for the growth of many gram-negative bacteria. The biosynthesis and transport of LPS are tightly controlled and coupled to the synthesis and assembly of other cell envelope components, such as peptidoglycan and S-layer proteins, to prevent loss of outer-membrane integrity (34, 35). We envision that the altered composition of the outer membrane, notably increased surface proteins rOmpA and rOmpB, contributes to the survival of the HK2 variant under high osmolarity pressure. It would be interesting to learn the detailed structure of R. conorii O antigen and to compare its structure with relevant O antigens from spotted fever- and typhus-Rickettsia as well as P. vulgaris and V. cholerae. Such analyses, combined with the study of antibodies that cross-react with specific LPS molecules, as is provided here, should resolve remaining questions on how certain bacteria elicit antibodies cross-reactive to rickettsial LPS, where these antibodies bind, and how they may be exploited for the design of immune therapeutics and vaccines against Rickettsia.
Materials and Methods
Detailed information describing materials and methods is provided in SI Appendix, Materials and Methods.
kkaebi Transposon Mutagenesis.
The kkaebi minitransposon DNA was PCR amplified (PCRF, 5′-AAAGACAGCTGTCTCTTATACACATCTCAACCATCATC-3′; PCRR, 5′-GACAGCTGTCTCTTATACACATCTCAACCCTGAAG-3′), digested with PshAI, purified, and incubated with Tn5 transposase (Lucigen; 10 µg kkaebi mixed with 2 units Tn5) for 24 h at room temperature to generate transposome complexes. The transposome complexes were dialyzed against 250 mM sucrose prior to electroporation (3 kV·cm−1, 200 Ω, 25 µF, 5.0 ms; Gene Pulser Xcell, Bio-Rad) into electrocompetent R. conorii prepared by washing 3 times in cold 250-mM sucrose. kkaebi variants were immediately recovered with DMEM supplemented with 5% HI-FBS and inoculated on 6-well tissue culture plates of confluent Vero cells. After 1 h infection at 34 °C in 5% CO2 atmosphere, 6-well plates were overlaid with 0.5% agarose in DMEM supplemented with 5% HI-FBS. Infected cells were incubated at 34 °C, 5% CO2 for an additional 5 h and treated with chloramphenicol (MP Biomedicals) at a final concentration of 0.3 µg·ml−1 to select for kkaebi variants. Mutants were isolated via plaque formation after 4 to 15 d of incubation in Vero cell cultures. To expand mutant strains, isolated plaques were resuspended in 2 mL DMEM with 5% HI-FBS and inoculated on each well of 6-well plates of confluent Vero cells. After 1 h infection at 34 °C in a 5% CO2 humidified chamber, medium was aspirated and replaced with fresh DMEM with 5% HI-FBS and 0.3 µg·ml−1 chloramphenicol. At 4 d postinfection, when monolayers of Vero cells were fully infected, Vero cells were mechanically disrupted with 3-mm glass beads, releasing intracellular R. conorii. After host cell debris was removed by centrifugation (1,000 × g, 4 °C, 5 min), the supernatant containing R. conorii was transferred to 225-cm2 flasks of confluent Vero cells to expand at 34 °C, 5% CO2 for 4 d. Rickettsiae were purified from Vero cells by differential centrifugation through 33% MD-76R solution (21,000 × g, 4 °C, 20 min), washed in Sucrose–Phosphate–Glutamate (SPG) buffer (218 mM sucrose, 3.8 mM KH2PO4, 7.2 mM K2HPO4, 4.9 mM l-glutamate, pH 7.2), and suspended in 1 mL SPG buffer. A600 was measured with bacterial samples diluted in SPG buffer.
Nucleotide Sequence Analysis.
BLogo sequence analysis was conducted with type 2 logos and base representation calculated from the 9-bp nucleotide sequences flanking the kkaebi insertion sites (36). The background frequencies of A, C, G, and T used for the BLogo analysis were 0.35, 0.19, 0.17, and 0.29, respectively.
Outer-Membrane Fractionation.
Outer-membrane fractionation was conducted based on a previously published protocol (37). The centrifugation sediments of R. conorii wild-type and pso variants or overnight cultures of P. vulgaris were suspended in 500 µL of buffer A (200 mM Tris⋅HCl, 1 M sucrose, 1 mM EDTA, pH 8.0) and mixed with 100 µL of lysozyme (5 mg·mL−1 in dH2O; Sigma). After 5 min incubation at room temperature, 2 mL of dH2O was added and incubated for 20 min at room temperature. Then, 3 mL buffer B (50 mM Tris⋅HCl, 2% Triton X-100, 10 mM MgCl2, pH 8.0) and 50 µL of DNase I (1 mg·mL−1 in dH2O; Sigma) were added and incubated for 20 min at room temperature. The mixture was ultracentrifuged at 160,000 × g for 60 min at 4 °C. The sediment was suspended in 500 µL of buffer C (200 mM Tris⋅HCl, 2% SDS, 10 mM EDTA, pH 8.8) and used for subsequent analyses.
Affinity Purification of Lipopolysaccharide.
LPS was affinity purified using polymyxin B-agarose (38). Specifically, a fractionated outer-membrane sample was dialyzed (2-kDa molecular weight cutoff; Thermo Scientific) twice against 4 L dH2O at room temperature. This solution was brought to 50 mM Tris⋅HCl, pH 7.5, mixed with 20 µL of proteinase K (10 mg·mL−1 in dH2O; Sigma), and incubated at 55 °C for 5 h. The crude polysaccharide solution was dialyzed (2-kDa molecular weight cutoff; Thermo Scientific) against 4 L dH2O at 4 °C overnight and brought to 100 mM NH4HCO3, pH 8.0, 0.9% NaCl. The crude polysaccharide sample was applied to a 2-mL polymyxin B-agarose (Sigma) column and incubated at 4 °C for 16 h, followed by washing with 10 mL of wash buffer (100 mM NH4HCO3, pH 8.0). LPS was eluted from the column with 10 mL of elution buffer (1% deoxycholic acid in 100 mM NH4HCO3, pH 8.0) and extensively dialyzed against 4 L of deoxycholic acid removal buffer (4 mM Tris⋅HCl, pH 8.8, 0.25% NaCl, 10% EtOH), followed by dialysis against dH2O. LPS samples were concentrated using a Speed-Vac and stored at 4 °C.
SDS-PAGE and Immunoblotting.
Samples were mixed with sample buffer (125 mM Tris⋅HCl, 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.01% bromophenol blue, pH 6.8) and boiled at 95 °C for 10 min. Samples were separated on 15% SDS-PAGE gels and stained with Coomassie Brilliant Blue R-250 for detecting outer-membrane proteins (Amsbio). A silver-staining kit was used for detection of LPS (Bio-Rad). For immunoblot analyses, samples were electrophoretically transferred from the gel onto a 0.22-µm PVDF membrane (GE Healthcare). The membrane was immersed in blocking buffer (TBS-T [100 mM Tris⋅HCl, 150 mM NaCl, pH 7.5, 0.1% Tween-20] with 5% milk) for 1 h at room temperature. The membrane was washed and incubated in a solution containing primary antibodies or antisera for 1 h at room temperature. rOmpA-specific mouse monoclonal antibody 13-3 (1:5,000 dilution), rabbit polyclonal antipeptide antibody c240 targeting the transmembrane domain of rOmpA (1:2,000 dilution), or rabbit polyclonal antibody recognizing rOmpB (1:1,000 dilution) was used as previously described (generously provided by Ted Hackstadt, Rocky Mountain Laboratory, Hamilton, MT) (39, 40). Rabbit antisera specific to LPS purified from wild-type R. conorii, HK2, and HK15 variants were used at a 1:10,000 (against R. conorii) or 1:1,000 (against P. vulgaris) dilution in TBS-T. Mouse hyperimmune sera collected from R. conorii-infected mice were used at a 1:10,000 (against R. conorii) or 1:625 (against P. vulgaris) dilution in TBS-T. Human antisera collected from patients with confirmed diagnoses of R. conorii or O. tsutsugamushi infections were used at a 1:10,000 dilution in TBS-T (kindly provided by Ranjan Premaratna, University of Kelaniya) (41). Of note, consents to store and use human antisera for research and diagnosis purposes were obtained at the time of sample collection. This study was approved by the ethics review committee, Faculty of Medicine, University of Kelaniya (IRB reference no. P/106/04/2018). The membrane was washed 3 times and incubated with peroxidase-conjugated secondary antibodies (anti-mouse IgG and anti-rabbit IgG [Cell Signaling] and anti-human IgG [Abcam]) at a 1:10,000 dilution in TBS-T for 1 h at room temperature. After a final wash, the membrane was developed using SuperSignal West Pico PLUS (Thermo Scientific) and exposed to Amersham Hyperfilm ECL (GE Healthcare).
R. conorii Survival in Mouse Plasma.
Whole blood was collected by cardiac puncture of C3H/HeN mice (Charles River Laboratories) and anticoagulated with 10 µg·mL−1 desirudin (Marathon Pharmaceuticals). Plasma was generated by centrifugation of desirudin-treated blood (1,000 × g for 5 min at 4 °C, followed by 10,000 × g for 3 min at 4 °C) for removal of blood cells. The hyperimmune sera samples were heat inactivated at 56 °C for 30 min, followed by incubation on ice for 5 min. Aliquots (50 µL) of 5 × 106 PFU R. conorii were opsonized with 50 µL of hyperimmune sera on ice for 10 min and mixed with freshly prepared or heat-inactivated plasma (200 µL). The infected samples were incubated at 37 °C with rotation for 60 min, at which time all plasma samples were incubated on ice and brought to 1-mL volume with ice-cold DMEM with 5% HI-FBS. Infectious R. conorii titers were determined by plaque assay. R. conorii survival was calculated as the percentage of the average R. conorii initial inoculum at 60 min.
Supplementary Material
Acknowledgments
We thank T. Hackstadt, T. Clark, and N. Noriea for reagents and technical advice; C. Emolo and X. Chen for experimental assistance; and members of the Howard Taylor Ricketts Laboratory for comments and discussion. This project has been supported by funds from the University of Chicago to the Howard Taylor Ricketts Laboratory and National Institutes of Health (AI144136).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Whole-genome sequencing data for wild-type R. conorii Malish 7 (accession no. SRR8404402) and rifampin-resistant R. conorii (accession no. SRR8404401) are deposited in the Sequence Read Archive.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911922116/-/DCSupplemental.
References
- 1.Robbins J. B., Schneerson R., Polysaccharide-protein conjugates: A new generation of vaccines. J. Infect. Dis. 161, 821–832 (1990). [DOI] [PubMed] [Google Scholar]
- 2.Goldschneider I., Gotschlich E. C., Artenstein M. S., Human immunity to the meningococcus. II. Development of natural immunity. J. Exp. Med. 129, 1327–1348 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneerson R., Barrera O., Sutton A., Robbins J. B., Preparation, characterization, and immunogenicity of Haemophilus influenzae type b polysaccharide-protein conjugates. J. Exp. Med. 152, 361–376 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed A., Li J., Shiloach Y., Robbins J. B., Szu S. C., Safety and immunogenicity of Escherichia coli O157 O-specific polysaccharide conjugate vaccine in 2-5-year-old children. J. Infect. Dis. 193, 515–521 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Micoli F., Costantino P., Adamo R., Potential targets for next generation antimicrobial glycoconjugate vaccines. FEMS Microbiol. Rev. 42, 388–423 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merhej V., Angelakis E., Socolovschi C., Raoult D., Genotyping, evolution and epidemiological findings of Rickettsia species. Infect. Genet. Evol. 25, 122–137 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Sahni A., Fang R., Sahni S. K., Walker D. H., Pathogenesis of rickettsial diseases: Pathogenic and immune mechanisms of an endotheliotropic infection. Annu. Rev. Pathol. 14, 127–152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weil E., Felix A., Zur serologischen diagnose the fleckfiebers. Wien. Med. Wochenschr. 29, 33–35 (1916). [Google Scholar]
- 9.Zinsser H., Castaneda M. R., Active and passive immunization in typhus fever. Proc. Natl. Acad. Sci. U.S.A. 20, 9–11 (1934). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zinsser H., Varieties of typhus fever and the epidemiology of the American form of European typhus fever (Brill’s disease). Am. J. Hyg. 20, 513–532 (1934). [Google Scholar]
- 11.Weigl R., Die Methoden der activen Fleckfieber-Immunisierung. Bull. l’Acad. Polonaise Sci. et Lett. 7, 25–62 (1930). [Google Scholar]
- 12.Qin A., Tucker A. M., Hines A., Wood D. O., Transposon mutagenesis of the obligate intracellular pathogen Rickettsia prowazekii. Appl. Environ. Microbiol. 70, 2816–2822 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baldridge G. D., Burkhardt N., Herron M. J., Kurtti T. J., Munderloh U. G., Analysis of fluorescent protein expression in transformants of Rickettsia monacensis, an obligate intracellular tick symbiont. Appl. Environ. Microbiol. 71, 2095–2105 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim H. K., Premaratna R., Missiakas D. M., Schneewind O., Whole genome sequencing of Rickettsia conorii Malish 7. Sequence Read Archive. https://www.ncbi.nlm.nih.gov/sra/?term=SRR8404402. Deposited 31 July 2019. [Google Scholar]
- 15.Kim H. K., Premaratna Ranjan, Missiakas D. M., Schneewind O., Whole genome sequencing of rifampin-resistant Rickettsia conorii Malish 7. Sequence Read Archive. https://www.ncbi.nlm.nih.gov/sra/?term=SRR8404401. Deposited 31 July 2019. [Google Scholar]
- 16.Rachek L. I., Tucker A. M., Winkler H. H., Wood D. O., Transformation of Rickettsia prowazekii to rifampin resistance. J. Bacteriol. 180, 2118–2124 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amano K. I., Williams J. C., Dasch G. A., Structural properties of lipopolysaccharides from Rickettsia typhi and Rickettsia prowazekii and their chemical similarity to the lipopolysaccharide from Proteus vulgaris OX19 used in the Weil-Felix test. Infect. Immun. 66, 923–926 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kneidinger B., Larocque S., Brisson J. R., Cadotte N., Lam J. S., Biosynthesis of 2-acetamido-2,6-dideoxy-L-hexoses in bacteria follows a pattern distinct from those of the pathways of 6-deoxy-L-hexoses. Biochem. J. 371, 989–995 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amano K., Fujita M., Suto T., Chemical properties of lipopolysaccharides from spotted fever group rickettsiae and their common antigenicity with lipopolysaccharides from Proteus species. Infect. Immun. 61, 4350–4355 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peturova M., Vitiazeva V., Toman R., Structural features of the O-antigen of Rickettsia typhi, the etiological agent of endemic typhus. Acta Virol. 59, 228–233 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Ziolkowski A., et al. , Structures of the O-antigens of Proteus bacilli belonging to OX group (serogroups O1-O3) used in Weil-Felix test. FEBS Lett. 411, 221–224 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Felix A., The rabbit as experimental animal in the study of the typhus group of viruses. Trans. R. Soc. Trop. Med. Hyg. 26, 365–378 (1933). [Google Scholar]
- 23.Martinez J. J., Seveau S., Veiga E., Matsuyama S., Cossart P., Ku70, a component of DNA-dependent protein kinase, is a mammalian receptor for Rickettsia conorii. Cell 123, 1013–1023 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Li H., Walker D. H., rOmpA is a critical protein for the adhesion of Rickettsia rickettsii to host cells. Microb. Pathog. 24, 289–298 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Palmer E. L., Mallavia L. P., Tzianabos T., Obijeski J. F., Electron microscopy of the cell wall of Rickettsia prowazeki. J. Bacteriol. 118, 1158–1166 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernstein H. D., Looks can be deceiving: Recent insights into the mechanism of protein secretion by the autotransporter pathway. Mol. Microbiol. 97, 205–215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker R. R., Rocky mountain spotted fever: Results of ten years’ prophylactic vaccination. J. Infect. Dis. 57, 78–93 (1935). [Google Scholar]
- 28.Malcomson M. E., Wishart F. O., Studies of the serology of typhus fever; complement-fixing antibody response to vaccination and to infection. Can. J. Public Health 37, 411–421 (1946). [PubMed] [Google Scholar]
- 29.Cedzynski M., et al. , Structural and serological studies of the O-antigen of the bacterium Proteus vulgaris OX2 (serogroup O2) used in the Weil-Felix test. Biochemistry (Mosc.) 62, 15–20 (1997). [PubMed] [Google Scholar]
- 30.Lamason R. L., et al. , Rickettsia Sca4 reduces vinculin-mediated intercellular tension to promote spread. Cell 167, 670–683.e10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamason R. L., Kafai N. M., Welch M. D., A streamlined method for transposon mutagenesis of Rickettsia parkeri yields numerous mutations that impact infection. PLoS One 13, e0197012 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark T. R., et al. , Transformation frequency of a mariner-based transposon in Rickettsia rickettsii. J. Bacteriol. 193, 4993–4995 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Z. M., Tucker A. M., Driskell L. O., Wood D. O., Mariner-based transposon mutagenesis of Rickettsia prowazekii. Appl. Environ. Microbiol. 73, 6644–6649 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein G., Raina S., Regulated assembly of LPS, its structural alterations and cellular response to LPS defects. Int. J. Mol. Sci. 20, E356 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morè N., et al. , Peptidoglycan remodeling enables Escherichia coli to survive severe outer membrane assembly defect. MBio 10, e02729-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green B., Bouchier C., Fairhead C., Craig N. L., Cormack B. P., Insertion site preference of Mu, Tn5, and Tn7 transposons. Mob. DNA 3, 3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thein M., Sauer G., Paramasivam N., Grin I., Linke D., Efficient subfractionation of gram-negative bacteria for proteomics studies. J. Proteome Res. 9, 6135–6147 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Laus M. C., et al. , A novel polar surface polysaccharide from Rhizobium leguminosarum binds host plant lectin. Mol. Microbiol. 59, 1704–1713 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Noriea N. F., Clark T. R., Mead D., Hackstadt T., Proteolytic cleavage of the immunodominant outer membrane protein rOmpA in Rickettsia rickettsii. J. Bacteriol. 199, e00826-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anacker R. L., Mann R. E., Gonzales C., Reactivity of monoclonal antibodies to Rickettsia rickettsii with spotted fever and typhus group rickettsiae. J. Clin. Microbiol. 25, 167–171 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Premaratna R., Loftis A. D., Chandrasena T. G., Dasch G. A., de Silva H. J., Rickettsial infections and their clinical presentations in the western province of Sri Lanka: A hospital-based study. Int. J. Infect. Dis. 12, 198–202 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.