Significance
Liver has a remarkable regenerative capacity following injuries. However, the cellular dynamics of how hepatocytes are replenished during homeostasis and upon liver injuries remains largely unclear. By using genetic lineage tracing strategies on rare Lgr5+ hepatocytes surrounding the central veins of the liver lobule, this study shows that Lgr5+ hepatocytes are self-maintained during normal homeostasis and various liver injuries, and contributed to hepatocarcinogesis in two different hepatocellular carcinoma (HCC) models. Our findings provide an insight on hepatocyte regeneration under homeostasis and liver injuries and uncover Lgr5+ hepatocytes as potential cellular origin in HCC development.
Keywords: Lgr5, lineage tracing, hepatocyte, liver regeneration, hepatocellular carcinoma
Abstract
Emerging evidence suggests that hepatocytes are primarily maintained by self-renewal during normal liver homeostasis, as well as in response to a wide variety of hepatic injuries. However, how hepatocytes in distinct anatomic locations within the liver lobule are replenished under homeostasis and injury-induced regeneration remains elusive. Using a newly developed bacterial artificial chromosome (BAC)-transgenic mouse model, we demonstrate that Lgr5 expression in the liver is restricted to a unique subset of hepatocytes most adjacent to the central veins. Genetic lineage tracing revealed that pericentral Lgr5+ hepatocytes have a long lifespan and mainly contribute to their own lineage maintenance during postnatal liver development and homeostasis. Remarkably, these hepatocytes also fuel the regeneration of their own lineage during the massive and rapid regeneration process following two-thirds partial hepatectomy. Moreover, Lgr5+ hepatocytes are found to be the main cellular origin of diethylnitrosamine (DEN)-induced hepatocellular carcinoma (HCC) and are highly susceptible to neoplastic transformation triggered by activation of Erbb pathway. Our findings establish an unexpected self-maintaining mode for a defined subset of hepatocytes during liver homeostasis and regeneration, and identify Lgr5+ pericentral hepatocytes as major cells of origin in HCC development.
The liver is an organ with extraordinary regeneration capacity. The normal liver consists of two main epithelial cell lineages, namely hepatocytes and cholangiocytes. It has long been proposed that facultative stem cells exist in the liver and reside in the “Canal of Hering,” which connects bile canaliculi to the interlobular bile duct in the portal region of the liver lobule. These stem cells produce intermediate (progenitor) cells named “oval cells” that give rise to cholangiocytes and mature hepatocytes, and contribute to normal liver maintenance, acute liver regeneration, and repair upon liver injuries (1, 2). However, this hypothesis remains highly controversial as different experimental settings resulted in discrepancy in conclusions (1, 2). Moreover, there are no validated markers that can be used to distinguish the progenitor cells from the cholangiocytes in healthy and diseased liver for further characterization. Nevertheless, accumulating evidence suggests that adult liver mass is primarily replenished through self-renewal of preexisting hepatocytes during normal homeostasis, as well as in response to different liver injuries (3–7). Cholangiocytes and/or progenitor cells may also give rise to hepatocytes under certain extreme conditions, for instance, when proliferation of hepatocytes is profoundly blocked during liver regeneration associated with either genetic interventions or long-term severe liver injuries (8–10).
The liver lobule is the functional unit of the liver. The murine liver lobule can be roughly separated into three distinct zones along the porto-central axis of the hepatocyte plate. Zones 1 and 3 consist of six to eight layers of periportal and several layers of pericentral hepatocytes, respectively. Zone 2, located in the middle of the lobule, is a less defined region containing 6 to 10 layers of hepatocytes (11). Of note, the human liver lobule is twice in size compared to the rodent liver lobule (12). It has been well recognized that the metabolic functions of hepatocytes vary in different zones (12–14). Moreover, recent reports showed that hepatocytes at different liver lobular zones may also play distinct roles in generating new cells to maintain liver mass during normal homeostasis and fuel regeneration upon liver injuries. The messenger RNA (mRNA) of the Wnt target gene Axin2 is expressed in multiple layers of hepatocytes next to the central vein in the liver lobule (11, 15, 16). Axin2-CreERT2 knock-in lineage tracing mouse model showed that Axin2+ cells may serve as a unipotent hepatic stem cells. These cells can differentiate into other hepatocytes across the liver lobule and contribute significantly in sustaining liver homeostasis (17). However, the central role of Axin2+ cells and their contribution to the regeneration of periportal hepatocytes during homeostasis were not supported by another study (16). Moreover, how the Axin2+ hepatocyte subset regulates liver regeneration in response to injury and liver cancer development remains undefined (17). Interestingly, Sox9, an established marker for cholangiocytes, is expressed at low levels in a small subset of hepatocytes adjacent to the portal veins (18). This subset of Sox9+ periportal hepatocytes, but not the Sox9+ cholangiocytes, are able to differentiate into a significant proportion of newly generated hepatocytes in response to liver injuries (18, 19). It remains elusive, however, whether they are the so-called progenitor cells (also known as “oval cells”) that arise directly from cholangiocytes. Another subset of hepatocytes, the Mfsd2a+ cells that reside in most parts of the periportal region in liver lobules, contribute significantly to the generation of new hepatocytes during injury-induced regeneration. However, they are gradually replaced by other cells in normal liver homeostasis (20). The heterogeneity of the liver epithelium is also more complicated than the three-zones model. For example, glutamine synthetase (GS) only marks the hepatocytes in a thin rim of the central veins in zone 3, while Axin2 and Cyp2e1 appear to label a much larger population of hepatocytes in the same zone (11, 15, 16). In fact, it has been recently proposed that the hepatocyte plate between the central and portal veins can be divided into nine different molecular layers based on the expression of six landmark genes (21). Recently, hepatocytes marked by expression of telomerase are found to be sparsely distributed across different layers of hepatocytes within the lobule. These cells were shown to be the main hepatocyte source for normal liver homeostasis and injury-induced liver regeneration, which further reveals the complexity on self-maintenance of hepatocytes (22).
Using Lgr5 as a genetic marker, we have developed a mouse model with high labeling efficiency to permit cell fate tracing of a unique hepatocyte population that reside in the thin rim surrounding the central vein. We found that these cells are long-lived, but mainly give rise to their own lineage during normal homeostasis and rapid regeneration upon partial hepatectomy. We also showed that Lgr5+ hepatocytes are the cells of origin driving the development of hepatocellular carcinoma (HCC) in both oncogene- and chemical carcinogen-mediated liver cancer models. Our findings unravel a self-maintaining paradigm of hepatocyte regeneration and identify a rare subset of hepatocytes defined by Lgr5 expression that serve as cellular origin in HCC.
Results
Lgr5 Is Selectively Expressed in a Defined Population of Hepatocytes Most Adjacent to the Central Veins.
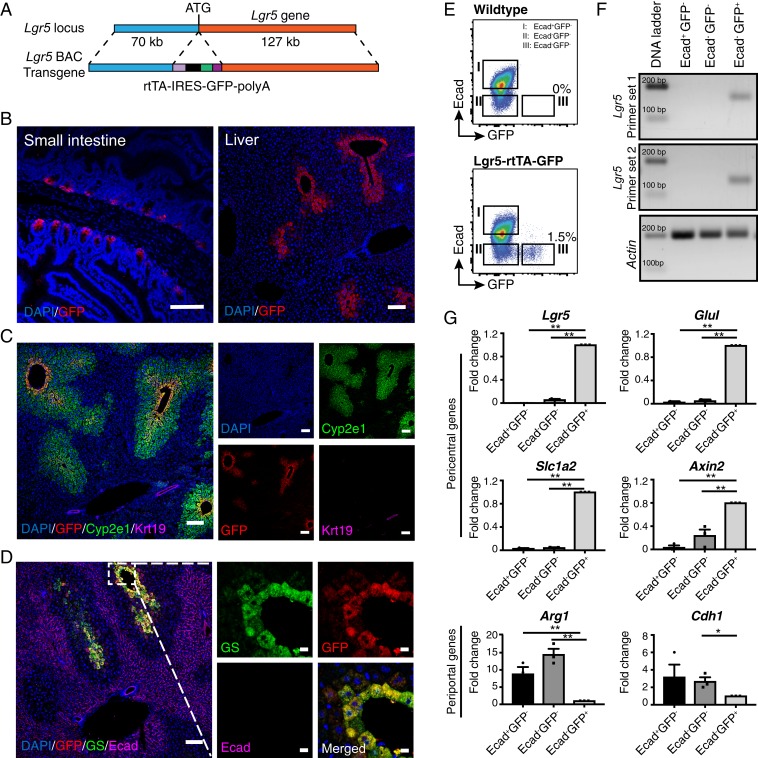
Lgr5 is a stem cell marker for a broad variety of epithelial tissues. While some studies suggested that Lgr5 expression could only be detected in injured livers in Lgr5 knock-in reporter mouse models (23–25), others showed that Lgr5 mRNA was expressed in normal liver (16, 21, 26). In addition, a gene knockout study in mice revealed that Lgr5, together with Lgr4, plays a pivotal role in liver development and zonation (16). However, none of the reporter mouse models in previous studies are able to faithfully mimic the expression pattern of endogenous Lgr5 mRNA in the liver (16, 23–25, 27). We modified a bacterial artificial chromosome (BAC) DNA containing about 200 kb of DNA of the Lgr5 locus and generated a BAC transgenic mouse model harboring the reverse tetracycline transcriptional activator (rtTA)-internal ribosome entry site (IRES)-green fluorescent protein (GFP) to allow cell fate tracking of Lgr5-expressing cells in the liver (Fig. 1A). As previously reported in Lgr5 knock-in mouse models (27, 28), we detected specific expression of GFP in the corpus gland bases of the small intestine of adult Lgr5-rtTA-IRES-GFP BAC transgenic mice (Fig. 1 B, Left). Expression of Lgr5-GFP was exclusively found in a small subset of hepatocytes (Fig. 1 B, Right). Costaining of the Lgr5-rtTA-IRES-GFP liver sections with GFP, Cyp2e1 (marker of hepatocytes in Zone 3), Cytokeratin 19 (Krt19, marker of cholangiocytes), E-cadherin (Ecad, marker of cholangiocytes and periportal hepatocytes), and GS (marker of hepatocytes most adjacent to the central veins) antibodies revealed that Lgr5-GFP+ cells exist within the lobular zone 3 marked by Cyp2e1, but are not present in Ecad-expressing hepatocytes in zone 1. Remarkably, Lgr5-GFP+ and GS+ cells were found to largely overlap with each other in the liver of mice derived from two independent Lgr5-rtTA-IRES-GFP transgenic founders (Fig. 1D and SI Appendix, Fig. S1 A and B). Copy number analysis of the BAC transgene by qPCR on genomic DNA of offsprings derived from the two Lgr5-rtTA-IRES-GFP founders suggested that the two founders harbor 2 copies and 10 copies of the transgene while the intensity and liver lobular zone localization pattern of GFP expression were not significantly different (SI Appendix, Fig. S1C). Taken together, these data clearly indicate that Lgr5-GFP+ cells in the adult liver are mainly restricted to a pericentral hepatocyte subset expressing GS.
Fig. 1.
Lgr5 is expressed in a small subset of hepatocytes mostly adjacent to the central veins. (A) Schematic illustration of the transgene structure for generation of Lgr5-rtTA-IRES-GFP mouse model. The rtTA-IRES-GFP-polyA cassette was inserted in frame to replace the ATG start codon of Lgr5 gene in a BAC DNA clone containing the Lgr5 gene locus. (B) Representative confocal images showing expression of GFP in small intestine (Left) and liver (Right) of adult Lgr5-rtTA-IRES-GFP transgenic mice. (C) Representative confocal images showing costaining of GFP with the pericentral hepatocyte marker Cyp2e1 and bile duct marker Krt19. (D) Representative confocal images depicting that the Lgr5-GFP+ population largely overlaps with the GS-expressing hepatocytes at the thin rim surrounding the central veins and is mutually exclusive to the cells marked by Ecad. (E) Representative FACS plots showing mutual exclusive presence of Lgr5-GFP+ hepatocytes and Ecad+ hepatocytes in adult WT and Lgr5-rtTA-IRES-GFP transgenic mice. (F) Representative gel images showing the expression of endogenous Lgr5 was restricted in the Lgr5-GFP+ hepatocytes. Three subsets of hepatocytes defined by Lgr5-GFP and Ecad expression were FACS sorted and two independent sets of primers targeting different exons of the Lgr5 gene were used for PCR amplification. (G) qPCR analysis of expression of periportal and pericentral genes in the three subsets of hepatocytes defined by Lgr5-GFP and Ecad expression. FACS isolated hepatocytes for each subpopulation were pooled from three mice to procure sufficient total RNA for qPCR analysis. Three independent experiments were conducted for qPCR analyses. *P < 0.05; **P < 0.01. Error bars represent mean ± SEM. (Scale bars: B, C, and D, Left, 100 μm; D, Right, 10 μm.)
To examine whether Lgr5-GFP expression in our BAC transgenic model is able to faithfully mimic the endogenous Lgr5 mRNA expression in hepatocytes, we performed an in vivo two-step collagenase liver perfusion and Percoll gradient centrifugation to obtain hepatocyte-enriched suspension for fluorescence-activated cell sorting (FACS) analysis and sorting (SI Appendix, Fig. S1D). Three subpopulations (i.e., GFP+Ecad–, GFP−Ecad–, and GFP–Ecad+) of hepatocytes defined by Lgr5-GFP and periportal hepatocytes marker Ecad (SI Appendix, Fig. S1D) were isolated. In line with the confocal data (Fig. 1D), FACS analysis also indicated that Lgr5-GFP and Ecad are mutually exclusive in the hepatocytes of Lgr5-rtTA-IRES-GFP mice (Fig. 1E). Notably, Lgr5-GFP only marked ∼2–5% of the total hepatocytes in an adult murine liver (SI Appendix, Fig. S1 E and F). qPCR analysis on the RNAs from the three hepatocyte subsets confirmed that the expression of endogenous Lgr5 mRNA was solely detected in the Lgr5-GFP+Ecad–, but not in other subpopulations using two independent primer sets (Fig. 1 F and G). We then compared expression level of several well-characterized pericentral and periportal hepatocyte marker genes (20, 26) in the three sorted hepatocyte subsets (Fig. 1G). Expression of pericentral hepatocyte genes Glul, Slc1a2, and Axin2 was highly enriched in the Lgr5-GFP+ cells. In contrast, expression of the periportal hepatocyte genes Arg1 and Cdh1 were lower in this cell population. These observations are in concordance with previous studies showing that endogenous Lgr5 mRNA was specifically detected in pericentral hepatocytes of the normal liver by FISH analysis and single cell profiling (16, 21). Altogether, these data suggest that the GFP reporter expression in the Lgr5 BAC transgenic model faithfully reflects the endogenous promoter activity of the Lgr5 gene in the murine liver.
Lgr5+ Hepatocytes Are Long-Lived and Maintain Their Own Lineage during Homeostasis.
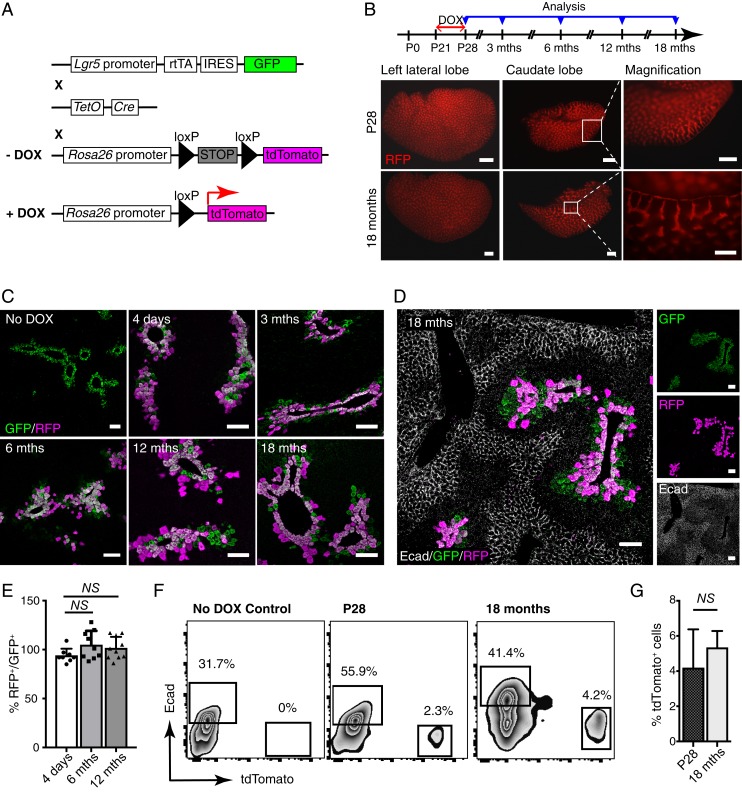
To trace how Lgr5+ hepatocytes contribute to long-term maintenance of the liver mass during homeostasis, we crossed Lgr5-rtTA-IRES-GFP mice with TetO-cre and R26-tdTomato cre reporter lines to generate triple transgenic Lgr5-rtTA/TetO-cre/R26-tdTomato mice. These mice enable doxycycline-inducible, irreversible expression of tdTomato in Lgr5-expressing cells (Fig. 2A). Mice were pulsed at puberty (postnatal day [P]21) and chased for different periods of time. Of note, tdTomato+ cells were not detected in both Lgr5-rtTA/TetO-cre/tdTomato mice without doxycycline induction and in those of control genotypes (i.e., Lgr5-rtTA/tdTomato and TetO-cre/tdTomato) with doxycycline treatment. Whole-mount fluorescent analysis revealed a high labeling efficiency in our lineage tracing system, with similar patterns of tdTomato+ expression evident in the left lateral and caudate lobes from triple transgenic mice immediately postinduction or chased for up to 18 mo (Fig. 2B). Liver sections from mice at 4 d after doxycycline induction and chased for 3, 6, 12, and 18 mo were further analyzed by confocal imaging using GFP and RFP (tdTomato) antibodies (Fig. 2C and SI Appendix, Fig. S2A). The vast majority of hepatocytes immediately adjacent to the central veins showed costaining of GFP and RFP at 4 d after doxycycline induction, further confirming the specificity and high tdTomato labeling efficiency in our model. Interestingly, the pericentral localization pattern of the tdTomato+ hepatocytes within the liver lobules remained unchanged at all time points and persisted through a chase period of up to 18 mo (Fig. 2C). A minute number of tdTomato+GFP– hepatocytes were occasionally found in lobular areas with a few layers of hepatocytes away from central veins, particularly in mice traced for more than 6 mo (Fig. 2C). However, tdTomato+ cells were never detected in the cells lining bile ducts and hepatocytes in zone 1 marked by Ecad (Fig. 2D). To quantify the proportion of tdTomato+ cells during long-term liver homeostasis, percentage of RFP+/GFP+ hepatocytes was calculated from immunofluorescent analysis. More than 80% labeling efficiency was observed among the Lgr5-GFP+ cells in this lineage tracing model at 4 d after doxycycline induction, with no significant increase in proportion of RFP+/GFP+ hepatocytes observed in mice traced up to 12 mo (Fig. 2E). In line with this, tdTomato+ hepatocytes remained at ∼2–5% of total hepatocytes in mice analyzed immediately or 18-mo postinduction by FACS, further suggesting that there is no significant expansion in the progeny of Lgr5-expressing hepatocytes (Fig. 2 F and G). Moreover, FACS data also further confirmed that Lgr5+ cells never gave rise to Ecad+ periportal hepatocytes even after an 18-mo chase (Fig. 2F).
Fig. 2.
Lgr5+ hepatocytes are long-lived and maintain their own lineage under homeostasis condition. (A) Schematic illustration of the genetic Lgr5-rtTA/TetO-Cre/R26-tdTomato mouse model for lineage tracing. Lgr5-rtTA-IRES-GFP mice were crossed with TetO-Cre mice and R26-tdTomato reporter strain to generate doxycycline (DOX)-inducible triple transgenic model. (B) Schematic illustration showing the experimental strategy for doxycycline induction and time points of analysis in lineage tracing of Lgr5-rtTA/TetO-Cre/R26-tdTomato mice. Representative whole-mount fluorescence images of the left lateral and caudate liver lobes showing identical expression pattern of tdTomato in the mice harvested immediately after doxycycline induction and at 18 mo after treatment. (C) Confocal imaging analysis of Lgr5-rtTA/TetO-Cre/R26-tdTomato mice chased for 4 d and 3, 6, 12, and 18 mo. Liver sections were stained with GFP and RFP antibodies. Triple transgenic mice without doxycycline treatment were used as control (No DOX). At least three mice were used for each experimental condition. (D) Representative confocal images for costaining of GFP, RFP, and Ecad on the liver sections of Lgr5-rtTA/TetO-Cre/R26-tdTomato mice chased for 18 mo. The tdTomato+ hepatocytes are mutually exclusive with periportal Ecad+ hepatocytes, indicating zone-restriction of progeny of Lgr5+ cells. (E) Quantification of the percentage of RFP+ in proportion to GFP+ hepatocytes in mice traced for 4 d, 6 mo, and 12 mo after doxycycline induction. An average of 300 cells per site were counted. n = 10 sites on the liver from three mice. Error bars represent mean ± SEM, *P < 0.05 is calculated by using multiple t test. No significant expansion was observed in comparisons of 4 d versus 6 mo (P = 0.07) and 4 d versus 12 mo (P = 0.12). (F) Representative FACS analysis of total hepatocytes from the Lgr5-rtTA/TetO-Cre/R26-tdTomato mice harvested immediately after doxycycline induction and chased for 18 mo. Lgr5-rtTA/TetO-Cre/R26-tdTomato mice without doxycycline treatment (No DOX) showed no tdTomato+ hepatocytes. (G) Bar graph showing no significant changes in percentage of tdTomato+ cells detected by FACS analysis of total hepatocytes from the Lgr5-rtTA/TetO-Cre/R26-tdTomato mice harvested immediately after doxycycline induction at P28 and chased for 18 mo. Error bars represent mean ± SEM, *P < 0.05 is calculated by using multiple t test. (Scale bars: B, 200 μm; C and D, 100 μm.)
Hepatocytes are largely quiescent during homeostasis with a lifespan of about 200–300 d (29). Since the tracing experiments above were started at P21, we asked whether Lgr5-GFP+ hepatocytes proliferate in puberty and adulthood. Lgr5-rtTA-IRES-GFP mice at 3 (puberty) and 8 (adulthood) weeks were treated with drinking water containing BrdU continuously for 3 wk (SI Appendix, Fig. S2B). Costaining of liver sections with GFP and BrdU antibodies showed that a large proportion of Lgr5-GFP+ hepatocytes underwent cell cycling during the liver mass expanding period. BrdU+Lgr5-GFP+ hepatocytes were also observed in the liver of BrdU-treated adult mice, but at an ∼4-fold less frequency compared to adolescents (SI Appendix, Fig. S2 B and C). Liver grows substantially in weight from birth to adulthood, and become constant in adulthood, with threefold increase in weight between 2 wk and 6 mo after birth (SI Appendix, Fig. S2D). We therefore traced the fate of Lgr5-expressing hepatocytes in the perinatal stage by pulsing the pups at P7. Consistent with previous studies (30), staining of GS showed liver zonation has been fully developed in mice at around 5 d (SI Appendix, Fig. S2E). Compared with induction at P21, an expansion of progeny of Lgr5+ hepatocytes labeled at P7 could be detected upon 6-mo tracing. However, most of these cells remained in zone 3 of liver lobules, with only two to three cells away from central veins (SI Appendix, Fig. S2F). Collectively, these observations provide compelling evidence that Lgr5+ hepatocytes are able to proliferate during puberty or adulthood, but they mainly replenish their own lineage for liver mass expansion during early postnatal developmental stage and long-term maintenance under normal homeostasis during adulthood.
Lgr5+ Hepatocytes Generate Their Own Lineage during Acute Liver Regeneration upon Partial Hepatectomy.
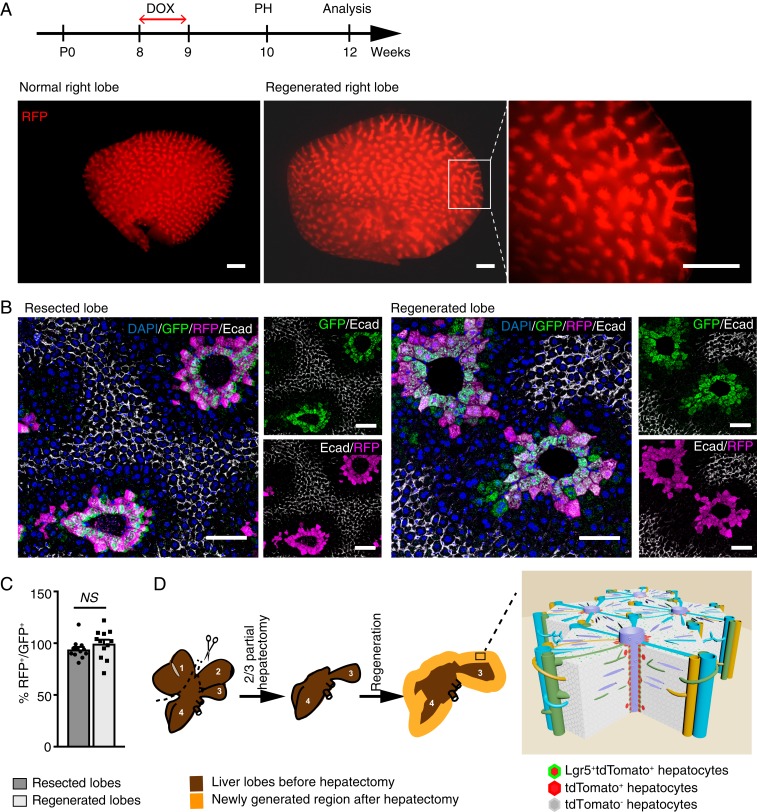
The remarkable regeneration capacity of the liver is evident by complete restoration of the liver mass in adult mice within a few days upon partial hepatectomy (31). We performed two-thirds partial hepatectomy to explore the role of Lgr5+ hepatocytes in acute liver regeneration. Adult Lgr5-rtTA/TetO-Cre/R26-tdTomato mice were pulsed with doxycycline to induce tdTomato expression in Lgr5+ hepatocytes 1 wk prior to the surgery. Interestingly, analysis of whole-mount liver lobes of Lgr5-rtTA/TetO-Cre/R26-tdTomato mice harvested at 14 d postsurgery showed similar tdTomato expression patterns in the regenerated liver and unoperated liver lobes of age-matched mice while significant liver growth was evident by the size increase of the lobe (Fig. 3A). Immunostaining on histology sections of the regenerated liver lobes with RFP, GFP, Ecad, and Cyp2e1 antibodies showed that tdTomato+ hepatocytes remained situated around central veins in the entire lobule, including the newly generated portion at the edges (Fig. 3B and SI Appendix, Fig. S3A). Moreover, lack of detection of GFP+RFP− hepatocytes further indicates that newly regenerated pericentral Lgr5-GFP+ hepatocytes were exclusively derived from preexisting Lgr5+ hepatocytes in the original lobes. No tdTomato+ cells were found in Ecad+ and Sox9+ cells, suggesting that Lgr5+ cells did not give rise to any periportal hepatocytes as well as cholangiocytes during massive regeneration of the liver induced by two-thirds partial hepatectomy (Fig. 3B and SI Appendix, Fig. S3 A and B). The ratio of RFP+ versus GFP+ hepatocytes is similar in resected lobes and regenerated lobes, suggesting that there is no significant expansion of Lgr5 hepatocytes to other hepatocyte lineages after partial hepatectomy (Fig. 3C). Of note, we did not detect any significant difference in the expression patterns of GS, Cyp2e1, Sox9, Krt19, and Ecad in the lobules of resected and regenerated lobues by confocal imaging analysis. It has been well documented that the growth of liver mass after two-thirds partial hepatectomy is primarily due to the proliferation and division of hepatocytes (32, 33). Analysis of Brdu labeling showed that Lgr5+ hepatocytes, similar to other subpopulations of hepatocytes, are highly proliferating after partial hepatectomy (SI Appendix, Fig. S3C). Taken together, our data demonstrate that Lgr5+ hepatocytes mainly take part in generation of their own lineage during rapid liver regeneration following partial hepatectomy (Fig. 3D).
Fig. 3.
Lgr5-derived hepatocytes replenish their own lineage upon partial hepatectomy. (A) Experimental strategy for partial hepatectomy experiments performed on Lgr5-rtTA/TetO-Cre/R26-tdTomato mice. Representative whole-mount images of the right liver lobe from adult littermate control and Lgr5-rtTA/TetO-Cre/R26-tdTomato mice harvested at 2 wk after partial hepatectomy. n = 5 mice. (B) Representative confocal images showing similar immunofluorescence staining pattern of Lgr5-GFP+tdTomato+ population in the resected liver lobe in comparison to the regenerated lobe harvested from the same mouse. Lgr5-GFP+tdTomato+ population remained in the thin-rim surrounding the central veins after two-thirds partial hepatectomy. n = 5. (C) Quantification of the percentage of RFP+ in proportion to GFP+ hepatocytes on resected lobes and regenerated lobes. An average of 200 cells per site were counted. n = 12 sites on the liver from three mice. Error bars represent mean ± SEM, *P < 0.05 is calculated by using multiple t test. (D) Graphic illustration of two-thirds partial hepatectomy procedure and liver regeneration after resection. Left lateral (1) and median (2) lobes are resected, and caudate (3) and right (4) lobes expand to restore the liver mass after surgery. A model illustrates the regeneration of Lgr5+ hepatocytes upon 2/3 partial hepatectomy. Lgr5+ pericentral hepatocytes mainly maintain their own lineage (Lgr5-GFP+tdTomato+) along the central vein upon two-thirds partial hepatectomy and occationally give rise to heptocytes in close pericentral layers (Lgr5-GFP−tdTomato+) within the liver lobule. (Scale bars: A, 200 μm; B, 100 μm.)
Lgr5+ Hepatocytes Can Be Replenished by Lgr5– Cells upon Pericentral Liver Injuries.
Since the Lgr5+ population appears to be restricted to the thin rim surrounding the central vein and is self-maintained during normal liver regeneration, we next asked whether Lgr5+ cells in the adult liver can be replenished by other cells in the event of pericentral liver injuries. The regeneration pattern of Lgr5+ hepatocytes was first tested in a pericentral liver injury model caused by carbon tetrachloride (CCl4) (34). Lgr5-GFP+tdTomato+GS+ hepatocytes were sensitive to the CCl4 insult and were almost completely depleted at 3 d after CCl4 administration (SI Appendix, Fig. S4A). By 6 or 14 d after CCl4 treatment, the Lgr5-GFP+ pericentral area of the liver lobules had recovered with readily detectable GFP and GS expression, but not that of tdTomato (SI Appendix, Fig. S4A). We also investigated the susceptibility of Lgr5+ hepatocytes to another well-established chemical-induced pericentral liver injury in adult mice caused by high dose of diethylnitrosamine (DEN). Adult Lgr5-rtTA/TetO-Cre/R26-tdTomato mice prepulsed with doxycycline were injected with DEN at 100 mg/kg. Analysis of liver sections of mice harvested at 3 d after DEN injection showed that the majority of Lgr5+ hepatocytes have been depleted, with few Lgr5-GFP+tdTomato+ hepatocytes remaining (SI Appendix, Fig. S4B). Expression of Lgr5-GFP and GS surrounding central veins had largely recovered by 14 d after acute DEN injury. However, most of the Lgr5-GFP+GS+ cells were negative for tdTomato expression (SI Appendix, Fig. S4B). Together, these data indicate that pericentral Lgr5+ hepatocytes are vulnerable to both CCl4-and acute DEN-induced pericentral injury but can be regenerated by Lgr5– cells in the process of recovery.
Lgr5+ Hepatocytes Are Highly Susceptible to Neoplastic Transformation.
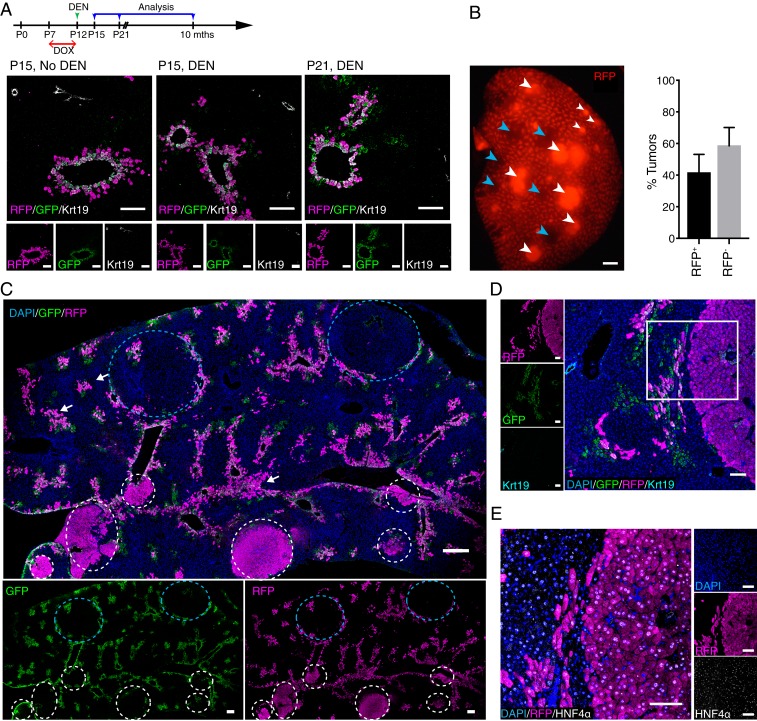
Chronic damage of the liver may lead to hepatocarcinogenesis. This process can be recapitulated in a mouse model induced by administrating a low dose of DEN to male mice at P12–P15 (35). However, it is unclear about the nature of the subset(s) of hepatocytes that are eventually transformed and give rise to HCC in this model. To delineate whether Lgr5+ hepatocytes could be cells of origin in DEN-induced HCC, Lgr5-rtTA/TetO-Cre/R26-tdTomato male pups pulsed with doxycycline at P7 were injected with a single dose of DEN at 25 mg/kg and monitored for tumor formation up to 8 mo. In contrast to depletion of Lgr5-GFP+tdTomato+ cells by acute DEN treatment in adult mice with a high dosage (SI Appendix, Fig. S4B), the majority of Lgr5-GFP+tdTomato+ hepatocytes in prepubertal males treated with a low dose of DEN could survive in this liver cancer model (Fig. 4A). Livers harvested from all of the mice at 8 mo after DEN injection showed multiple tumors (Fig. 4B). Remarkably, while Lgr5+ cells only accounted for about 2% of total hepatocytes, ∼40% of DEN-induced tumors were found to comprise tdTomato+ cells, indicating that these tumors were derived from the small subset of Lgr5+ hepatocytes (Fig. 4B). Notably, GFP+tdTomato+ cells in most areas showing intact liver lobule structure remained in GS+ zone 3 in the DEN-damaged liver, suggesting that nontransformed Lgr5+ cells did not expand significantly in this liver cancer model (SI Appendix, Fig. S5A). GFP was not detected in both tdTomato+ and tdTomato− tumors, suggesting that Lgr5 expression was lost during tumorigenesis even for the tumors derived from Lgr5+ hepatocytes (Fig. 4C and SI Appendix, Fig. S5A). In an attempt to define the difference between tdTomato+ and tdTomato− tumors, tumor samples were stained with a panel of markers, including β-catenin, GS, PCNA, Sox9, Arg1, Krt8, and Krt19, and analyzed by confocal imaging. Of note, no apparent differences were found for these markers between tdTomato+ and tdTomato− tumors in this model. For example, tdTomato+ and tdTomato− tumors were negative for GS, and β-catenin was found to be exclusively expressed in the plasma membrane of tdTomato+ and tdTomato− tumor cells (SI Appendix, Fig. S5B). We hence mainly showed data for the tdTomato+ tumors (Fig. 4 D and E and SI Appendix, Fig. S5). Importantly, tumors are negative for cholangiocyte marker (Krt19) but positive for hepatocyte markers HNF4α and Arg1. This confirmed that both tdTomato− and Lgr5-derived tdTomato+ tumors were HCC (Fig. 4 D and E and SI Appendix, Fig. S5C).
Fig. 4.
Lgr5+ hepatocytes are the primary cellular origin in DEN-induced HCC. (A) Experimental strategy for doxycycline (DOX) and DEN induction for Lgr5-rtTA/TetO-Cre/R26-tdTomato mice. Representative confocal images showing distribution of Lgr5-GFP+tdTomato+ around central veins at P15 without DEN (25 mg/kg) injection, and at P15 and P21, which were 3 d and 9 d after DEN injection, respectively. Dams were treated with doxycycline at P7–P12 of their litters to induce the labeling of tdTomato in Lgr5-expressing hepatocytes of young male pups. (B) Representative whole-mount fluorescence view of a tumorous liver lobe of Lgr5-rtTA/TetO-Cre/R26-tdTomato mouse harvested at 8 mo after DEN induction. tdTomato+ and tdTomato− tumors are depicted by white and blue arrowheads, respectively. Approximately 40% of the tumors detected in the liver of triple transgenic mice showed native tdTomato+ (RFP+) fluorescence. n = 4 mice. (C) Representative confocal tile scan images of a tumorous liver lobe harvested from DEN-treated Lgr5-rtTA/TetO-Cre/R26-tdTomato mouse showing tdTomato+ (white circles) and tdTomato− (blue circles) tumors. White arrows indicate possible tdTomato+ tumor initiation sites. All DEN-induced tumors were negative for GFP. n = 4 mice. (D and E) Representative confocal images showing that tdTomato+ tumors are GFP and Krt19-negative (D), and HNF4α-positive (E). (Scale bars: B and C, 200 μm; D and E, 100 μm.)
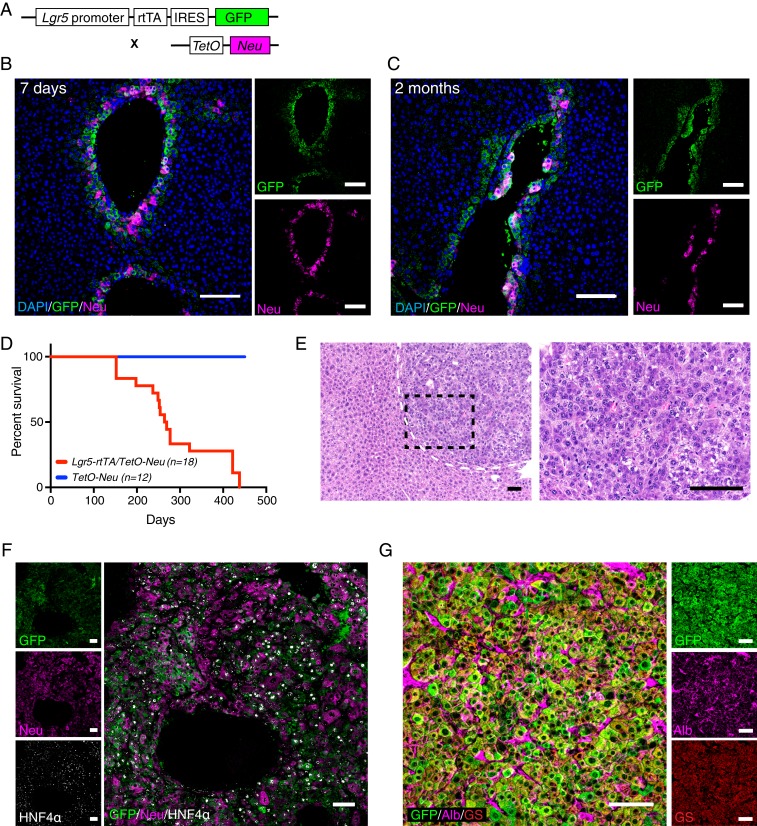
Neu (Her2/ErbB2), together with EGFR (ErbB1/Her1), ErbB3 (Her3), and ErbB4 (Her4), constitutes the EGFR/ErbB/HER family of receptor tyrosine kinases. Among them, EGFR is reported to be overexpressed in about 70% of human HCC incidences, which correlates with aggressive tumors, metastasis, and poor survival (36–38). While Neu/Her2 overexpression is not commonly found in HCCs, different members of the ErbB family share a highly similar downstream signaling cascade. Moreover, many ligands of the ErbB family have been reported to be highly expressed in HCCs (36). Using a Lgr5-rtTA/TetO-Neu mouse model that enables conditional induction of constitutively active Neu/Her2 mutant specifically in Lgr5+ hepatocytes, we tested whether Neu expression is able to transform Lgr5+ hepatocytes to initiate HCC development (Fig. 5A). Expression of exogenous Neu/Her2 in the Lgr5+ cells was confirmed after 7 d of doxycycline induction (Fig. 5B). Small tumor lesions were detected in the mice treated with doxycycline for 2 mo (Fig. 5C) while long-term induction of Neu/Her2 in Lgr5+ hepatocytes from P21 mice was sufficient to drive liver tumor development with a latency of about 8 mo (Fig. 5D). H&E staining showed a solid pattern of HCC with focal area of squamoid appearance (Fig. 5E). The tumor cells are large, polygonal with prominent nucleoli. Moreover, the vast majority of the tumor cells were positive for HNF4α and Albumin (Fig. 5 F and G), confirming the tumors in this model can be classified as HCC. Interestingly, several differences in marker expressions were observed in tumors arised in the Lgr5-rtTA/TetO-Neu model as compared to tumors arised in DEN-damaged liver. Unlike Lgr5+ cell-derived tumors induced by DEN, the tumors arising in the Lgr5-rtTA/TetO-Neu model retained both GS and Lgr5-GFP expression. In addition, DEN-induced tumors lacked cytosolic β-catenin expression while highly expressing Arg1 (SI Appendix, Fig. S5 B and C), whereas tumors in the Lgr5-rtTA/TetO-Neu model showed cytosolic and nuclear β-catenin expression and no expression of Arg1 (SI Appendix, Fig. S6 A and B). Krt8 expression was increased in the tumor cells in the the Lgr5-rtTA/TetO-Neu model, but remained unchanged or down-regulated in the DEN tumors (SI Appendix, Figs. S5D and S7B). Interestingly, the detection of endogenous phophorylated Egfr (p-Egfr) in Lgr5-rtTA/TetO-Neu tumors suggested cross-talk and activation of Egfr by overexpression of constitutively active Neu mutant. In contrast, the expression of p-Egfr and Neu/Her2 was absent in DEN-induced tumors (SI Appendix, Fig. S7E), suggesting that activation of Erbb pathway may not be a main driver for this tumor model. Overall, these data indicate that Lgr5+ hepatocytes are highly susceptible to neoplastic transformation triggered by the activation of Erbb pathway.
Fig. 5.
Expression of constitutively active Neu/Her2 mutant in Lgr5+ hepatocytes induces hepatocellular carcinoma. (A) Schematic illustration of doxycycline-inducible Lgr5-rtTA/TetO-Neu mouse model. Constant treatment of doxycycline allows continuous expression of Neu/Her2 in Lgr5+ hepatocytes. (B and C) Representative confocal images showing expression of GFP and Neu/Her2 on liver sections of mice treated with doxycycline diet for 7 d (B) and 2 mo (C). Neu/Her2 expression was turned on specifically in Lgr5-GFP+ hepatocytes as early as 7 d. (D) Tumor latency in doxycycline-induced Lgr5-rtTA/TetO-Neu transgenic mice. Tumors were detected as early as 4 mo after starting doxycycline treatment. No tumor was found in control TetO-Neu mice. (E) Representative images of H&E staining of liver tumors arisen in Lgr5-rtTA/TetO-Neu mice at low magnification (Left) and high magnification (Right). n = 18 mice. (F) Costaining of GFP, Neu/Her2, an HNF4α revealed that Lgr5-rtTA/TetO-Neu tumors are HNF4α+, and majority of the tumor cells retained expression of Lgr5-GFP and Neu/Her2 (n = 4). (G) Representative confocal image showing Lgr5-rtTA/TetO-Neu tumor cells express hepatocyte markers albumin and GS (n = 4 mice). (Scale bars: 100 μm.)
Discussion
Lgr5 (also known as Gpr49) was identified as a marker for pericentral hepatocytes in genome-wide microarray analysis on isolated periportal and pericentral hepatocytes using digitonin-collagenase perfusion approach (26). By in situ hybridization, the endogenous Lgr5 mRNA was found to be readily detectable in almost all hepatocytes most adjacent to the central vein in the normal adult liver (16). Moreover, Lgr5 was also shown to be expressed exclusively in the hepatocyte layer most adjacent to the central vein in a recent study where single-cell RNA-profiling strategy was used to reconstruct the liver lobule at the molecular level (21). Here, we established a robust BAC transgenic mouse model for studying the roles of Lgr5-expressing hepatocytes in liver regeneration and HCC development. Compared to previously reported Lgr5 knock-in mouse models, the GFP reporter in our transgenic mice is readily detectable in the normal liver. In line with the patterns of endogenous Lgr5 mRNA expression reported from previous studies (16, 21), localization of Lgr5-GFP+ hepatocytes in the liver lobule was restricted to a thin rim of hepatocytes mostly adjacent to the central veins. The fidelity of our model in mimicking endogenous Lgr5 expression in the liver was further confirmed by the exclusive expression of endogenous Lgr5 mRNA in the sorted Lgr5-GFP+ hepatocyte population. Importantly, the high labeling efficiency in the lineage tracing model (up to 85% of total Lgr5-GFP+ hepatocytes) provides a powerful system to track cell fate of the Lgr5-expressing hepatocyte subset in liver homeostasis, injury-induced liver regeneration, and hepatocarcinogenesis. It remains to be determined why our Lgr5 BAC transgenic model is able to recapitulate the endogenous Lgr5 mRNA expression in the liver with high fidelity, compared to other knock-in lines. Similarly, it has been reported that BAC transgenic models can profoundly enhance the accuracy of a reporter of radial glial promoters and lineage tracing of radial glial during neurogenesis (39).
Although hepatocytes in different zones throughout the liver lobule are morphologically similar, they have heterogeneous metabolic functions and regenerative capacity. It was initially proposed that hepatocytes are maintained by a streaming liver model where new hepatocytes are constantly generated during normal homeostasis at the portal vein domain and move forward to replace the hepatocytes in other regions within the lobule (40). Lineage tracing in Axin2-CreERT2 mice, however, has shown that Axin2 expression marks hepatocytes adjacent to the central vein and that these hepatic stem cells play a major role in normal liver homeostasis and give rise to hepatocytes throughout the entire lobule after long-term tracing (17). In contrast, a recent study reported that Tert+ hepatocytes randomly distributed in the liver lobule are responsible for hepatocytes replenishment during both normal liver homeostasis and injury-induced regeneration (22). Accordingly, a local expansion manner of hepatocytes was proposed for liver homeostasis and regeneration in the study. Notably, Tert+ hepatocytes are rarely found in the perivenous zone and the vast majority of them were resident in zones 1 and 2 (22). In this study, we showed that Lgr5+ perivenous hepatocytes persisted throughout the lifespan of the mouse and were mainly replenished by their own duplications during perinatal development, adult homeostasis, and during rapid regeneration induced by two-thirds partial hepatectomy. Genetic lineage tracing on the basis of a dual recombinases approach, however, is necessary to determine in the future whether there are rare Tert+ cells within the Lgr5+ population to repopulate Lgr5+ cells (19). Moreover, new mouse models specific for other hepatocyte layers in the liver lobule are needed to determine whether self-maintenance is a common mode of regeneration for different hepatocyte subsets in normal homeostasis and partial hepatectomy. Our study also revealed plasticity between different subsets of hepatocytes upon severe liver injuries as depletion of Lgr5+ cells by CCl4 or a high dose of DEN in adult liver could be quickly replenished by Lgr5− cells in the liver. We speculate that the cells surrounding the damaged Lgr5+ hepatocytes are likely responsible for this acute damage and repair process. Moreover, the rapid gain of Lgr5 expression in new hepatocytes derived from Lgr5– cells that move to the thin rim surrounding the central vein suggests that a local niche actively defines the property of Lgr5+ hepatocytes. Likely, Wnt signaling-related factors controlled by the endothelial cells of the central veins are actively involved in establishing the pericentral niche for Lgr5+ hepatocytes (17, 41).
It has been reported that hepatocytes are the cellular origin for HCC development in different liver cancer models. However, robust mouse models to define and trace distinct subsets of hepatocytes are unavailable. It is largely unknown which hepatocyte subpopulation represents key cellular targets to initiate HCC development under distinct hepato-oncogenic contexts (42). Sox9/CD133+ progenitor cells were dramatically expanded in a number of different cancer models (43). Liver progenitor cells hence have been proposed as critical cellular targets during liver cancer initiation. However, there is lack of evidence supporting that these cells are cancer initiating cells in multiple mouse liver cancer models (18) while the Sox9+ hepatocytes seemed to show progenitor activity in liver homeostasis and in response to liver injuries. DEN is the most commonly used chemical carcinogen to induce liver cancer in mice for supporting investigation toward understanding of the development of human HCC (42, 44, 45). The sequence of pathophysiological alterations in DEN-induced mouse liver cancer model shows a high degree of similarity with that seen in human HCC development. Notably, DEN needs to be converted into toxic metabolites to cause damage in Cyp2e1+ pericentral hepatocytes. This process also induces inflammation and compensatory proliferation response within the liver, which may, in turn, cause neoplastic transformation of other subsets of hepatocytes. Our data provide compelling evidence that Lgr5+ pericentral hepatocytes that survived DEN-induced damage became the major cells of origin driving the development of DEN-induced liver cancer. Interestingly, expression of Lgr5 and GS were turned off in tumor cells derived from Lgr5+ hepatocytes in this liver cancer model, implying that Lgr5+ hepatocytes lost some critical identities conferred by their niche during tumorigenesis. In contrast, tumors induced by target expression of Neu/Her2 in Lgr5+ hepatocytes maintained high expression levels of Lgr5 and GS, two well-known Wnt target genes. The Wnt pathway is shown to be highly active in the entire zone 3 that is defined by expression of another Wnt target gene Cyp2e1 (15), while expression of Lgr5 and GS is restricted to the thin rim surrounding the central veins. This makes Lgr5 and GS different from other Wnt target genes in the liver while the regulatory mechanism behind remains obscure. It is noteworthy that constitutive activation of Wnt pathway due to somatic mutations of β-catenin or its negative regulator APC and Axin were found in ∼50% of HCC patients. Accordingly, activation of Wnt/β-catenin pathway has been suggested to be a key driver for the development of HCC. Notably, both Lgr5 and GS are among top up-regulated genes in the comparison of Wnt/β-catenin pathway mutation-associated HCCs versus other HCCs (46). The Lgr5-rtTA/TetO-Neu liver cancer model developed in this study may provide a good experimental paradigm for gaining further understanding of Wnt pathway-driven liver cancer and the cross-talk between Wnt and Erbb pathways during the development of HCC.
Materials and Methods
Mice.
The Lgr5-rtTA-IRES-GFP mouse line was generated in the animal facility of the Walter and Eliza Hall Institute of Medical Research, Australia. To generate the Lgr5-rtTA-IRES-GFP transgenic mice, an Lgr5-rtTA-IRES-GFP BAC construct was created using homologous recombination and microinjected into FVB/N zygotes to create the BAC transgenic Lgr5-rtTA-IRES-GFP founder mice. Two independent founders were established (Founders 25 and 28). Of note, descendants from both founders exhibited identical Lgr5-GFP expression evident by immunofluorescent staining of liver sections with various liver zonation markers, and no other difference except for the copy numbers of the transgene was detected in offsprings derived from the two founders. Lgr5-rtTA-IRES-GFP transgenic mice were used to cross with TetO-Cre/R26-tdTomato mice (47, 48) to generate Lgr5/TetO-Cre/R26-tdTomato line for lineage tracing experiments. They were also crossed with TetO-Neu mice (49) to generate Lgr5-rtTA/TetO-Neu mice for conditional exogenous expression of a constitutively active mutant of oncogene Neu/Her2 under the control of Lgr5 promoter. Only male mice were used for DEN-induced liver tumor experiment. A mix of both male and female mice were used for all other experiments and no gender-bias difference was observed. All animals used in this study were bred and housed in individually ventilated cages in specific pathogen-free holdings with temperature and light control (12-h light/dark cycle). All animal experiments were carried out in accordance with protocols approved by SingHealth Institute Animal Care and Use Committee.
Liver Injury Models.
The two-thirds partial hepatectomy procedure was performed to induce liver injury with minimal inflammatory responses (31). Briefly, 8- to 12-wk-old Lgr5-rtTA/TetO-cre/R26-tdTomato mice were pulsed with doxycycline feed for 7 d, and back to normal feed for another 7 d prior to surgery. Mice were first anesthetized with 2% isoflurane and oxygen flow in a Plexiglas chamber and transferred onto a Styrofoam pad. Anesthesia was maintained by isoflurane inhalation with oxygen through a mouthpiece. Following removal of abdominal fur and skin disinfection with iodine, a midline abdominal skin and muscle incision was made to access the liver. A 4-0 silk thread was used to ligate the base of left lateral lobe and the median lobe, and the lobes were resected just above the knots. The peritoneum and skin were then closed with 5-0 suture, and the mouse was placed in individual cage under warming lamp for recovery. Regenerating livers were harvested from the mice at 14 d after partial hepatectomy surgery. Only the right and caudate lobes were used for analysis. Partial hepatectomy procedure for all mice were performed in the morning. For DEN-induced liver injury, male mice at 12 d postnatal were intraperitoneally injected with a single dose of DEN (25 mg/kg) in sterile PBS. Mice were raised normally after DEN injection and euthanized once tumors were detected through abdominal palpation, usually after 8 mo. For CCl4-induced injury, mice at 8 to 12 wk old were intraperitoneally injected with single dose of CCl4 at 1 μL per gram of body weight, diluted in sunflower seed oil at 1:3 dilution. For Neu/Her2 oncogene-induced HCC, Lgr5-rtTA-IRES-GFP transgenic mice were crossed to TetO-Neu transgenic mice to generate Lgr5-rtTA/TetO-Neu transgenic line. Lgr5-rtTA/TetO-Neu transgenic mice were put on continuous doxycycline feed from P21 onwards and harvested when tumors were detected by abdominal palpation at about 8 mo.
Supplementary Material
Acknowledgments
We thank the veterinarians, animal technicians, and staffs from FACS and Advance Bioimaging facilities at Duke-NUS and SingHealth for their technical support. We give special thanks to E. S. Leman for critical reading and insightful comments on the manuscript. C.H.A. is supported by Khoo Postdoctoral Fellowship Award. This work was supported by Duke-NUS Faculty Start-up Fund and Khoo Bridge Funding Award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908099116/-/DCSupplemental.
References
- 1.Kopp J. L., Grompe M., Sander M., Stem cells versus plasticity in liver and pancreas regeneration. Nat. Cell Biol. 18, 238–245 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Miyajima A., Tanaka M., Itoh T., Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell 14, 561–574 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Yanger K., et al. , Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell Stem Cell 15, 340–349 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaub J. R., Malato Y., Gormond C., Willenbring H., Evidence against a stem cell origin of new hepatocytes in a common mouse model of chronic liver injury. Cell Rep. 8, 933–939 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tarlow B. D., et al. , Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell 15, 605–618 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malato Y., et al. , Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. J. Clin. Invest. 121, 4850–4860 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y., et al. , Genetic tracing of hepatocytes in liver homeostasis, injury, and regeneration. J. Biol. Chem. 292, 8594–8604 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raven A., et al. , Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature 547, 350–354 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu W. Y., et al. , Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat. Cell Biol. 17, 971–983 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng X., et al. , Chronic liver injury induces conversion of biliary epithelial cells into hepatocytes. Cell Stem Cell 23, 114–122.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Colnot S., Perret C., “Liver zonation.” in Molecular Pathology of Liver Diseases, Monga S. P. S., Ed. (Springer, New York, 2010), pp. 7–16. [Google Scholar]
- 12.Ben-Moshe S., Itzkovitz S., Spatial heterogeneity in the mammalian liver. Nat. Rev. Gastroenterol. Hepatol. 16, 395–410 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Cheng X., et al. , Glucagon contributes to liver zonation. Proc. Natl. Acad. Sci. U.S.A. 115, E4111–E4119 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gebhardt R., Matz-Soja M., Liver zonation: Novel aspects of its regulation and its impact on homeostasis. World J. Gastroenterol. 20, 8491–8504 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benhamouche S., et al. , Apc tumor suppressor gene is the “zonation-keeper” of mouse liver. Dev. Cell 10, 759–770 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Planas-Paz L., et al. , The RSPO-LGR4/5-ZNRF3/RNF43 module controls liver zonation and size. Nat. Cell Biol. 18, 467–479 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Wang B., Zhao L., Fish M., Logan C. Y., Nusse R., Self-renewing diploid Axin2(+) cells fuel homeostatic renewal of the liver. Nature 524, 180–185 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Font-Burgada J., et al. , Hybrid periportal hepatocytes regenerate the injured liver without giving rise to cancer. Cell 162, 766–779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He L., et al. , Enhancing the precision of genetic lineage tracing using dual recombinases. Nat. Med. 23, 1488–1498 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pu W., et al. , Mfsd2a+ hepatocytes repopulate the liver during injury and regeneration. Nat. Commun. 7, 13369 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halpern K. B., et al. , Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542, 352–356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin S., et al. , Distributed hepatocytes expressing telomerase repopulate the liver in homeostasis and injury. Nature 556, 244–248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huch M., et al. , In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494, 247–250 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin Y., et al. , HGF/R-spondin1 rescues liver dysfunction through the induction of Lgr5+ liver stem cells. Nat. Commun. 8, 1175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao W., et al. , Dynamics of proliferative and quiescent stem cells in liver homeostasis and injury. Gastroenterology 153, 1133–1147 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Braeuning A., et al. , Differential gene expression in periportal and perivenous mouse hepatocytes. FEBS J. 273, 5051–5061 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Barker N., et al. , Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Leushacke M., et al. , Lgr5-expressing chief cells drive epithelial regeneration and cancer in the oxyntic stomach. Nat. Cell Biol. 19, 774–786 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Bucher N. L. R., Malt R. A., Regeneration of Liver and Kidney (Little. Brown, Boston, MA, 1971). [Google Scholar]
- 30.Ober E. A., Lemaigre F. P., Development of the liver: Insights into organ and tissue morphogenesis. J. Hepatol. 68, 1049–1062 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Mitchell C., Willenbring H., A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat. Protoc. 3, 1167–1170 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Caldez M. J., et al. , Metabolic remodeling during liver regeneration. Dev. Cell 47, 425–438.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Miyaoka Y., et al. , Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr. Biol. 22, 1166–1175 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Hoehme S., et al. , Prediction and validation of cell alignment along microvessels as order principle to restore tissue architecture in liver regeneration. Proc. Natl. Acad. Sci. U.S.A. 107, 10371–10376 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santos N. P., Colaço A. A., Oliveira P. A., Animal models as a tool in hepatocellular carcinoma research: A review. Tumour Biol. 39, 1010428317695923 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Komposch K., Sibilia M., EGFR signaling in liver diseases. Int. J. Mol. Sci. 17, E30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.López-Luque J., et al. , Dissecting the role of epidermal growth factor receptor catalytic activity during liver regeneration and hepatocarcinogenesis. Hepatology 63, 604–619 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Lanaya H., et al. , EGFR has a tumour-promoting role in liver macrophages during hepatocellular carcinoma formation. Nat. Cell Biol. 16, 972–977 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anthony T. E., Heintz N., Genetic lineage tracing defines distinct neurogenic and gliogenic stages of ventral telencephalic radial glial development. Neural Dev. 3, 30 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zajicek G., Oren R., Weinreb M. Jr, The streaming liver. Liver 5, 293–300 (1985). [DOI] [PubMed] [Google Scholar]
- 41.Halpern K. B., et al. , Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat. Biotechnol. 36, 962–970 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mu X., et al. , Hepatocellular carcinoma originates from hepatocytes and not from the progenitor/biliary compartment. J. Clin. Invest. 125, 3891–3903 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sia D., Villanueva A., Friedman S. L., Llovet J. M., Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology 152, 745–761 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He G., et al. , Identification of liver cancer progenitors whose malignant progression depends on autocrine IL-6 signaling. Cell 155, 384–396 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen X., et al. , Differential reactivation of fetal/neonatal genes in mouse liver tumors induced in cirrhotic and non-cirrhotic conditions. Cancer Sci. 106, 972–981 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calderaro J., et al. , Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J. Hepatol. 67, 727–738 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Rios A. C., Fu N. Y., Lindeman G. J., Visvader J. E., In situ identification of bipotent stem cells in the mammary gland. Nature 506, 322–327 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Fu N. Y., et al. , Identification of quiescent and spatially restricted mammary stem cells that are hormone responsive. Nat. Cell Biol. 19, 164–176 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Xie W., Chow L. T., Paterson A. J., Chin E., Kudlow J. E., Conditional expression of the ErbB2 oncogene elicits reversible hyperplasia in stratified epithelia and up-regulation of TGFalpha expression in transgenic mice. Oncogene 18, 3593–3607 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.