Abstract
Background
Hepatocellular carcinoma is a common malignant cancer and the second most common cause of cancer-related deaths worldwide. Collagen triple helix repeat containing 1 (CTHRC1) has been increasingly reported to be involved in tumorigenesis and/or tumor progression. However, limited data are available regarding the role of CTHRC1 in hepatocellular carcinoma.
Methods
Paraffin-embedded specimens from a total of 29 patients with HCC were collected in our study. The expression of CTHRC1 in hepatocellular carcinoma was evaluated using immunohistochemistry and bioinformatics analysis. Furthermore, Spearman analysis was performed to identify factors of correlation between CTHRC1 and clinicopathological features. Survival curves for hepatocellular carcinoma were produced using the Kaplan-Meier method and the log rank test.
Results
In this study, we confirmed that CTHRC1 is highly expressed in tissues and hepatoma cell lines. The statistical analysis revealed that the levels of CTHRC1 were significantly correlated with cirrhosis (P=0.024), tumor size (P=0.006), vascular invasion (P<0.001), TNM stage (P<0.001), and BCLC stage (P<0.001). High expression of CTHRC1 in hepatocellular carcinoma tissues is significantly associated with poor survival.
Conclusion
CTHRC1 may serve as a prognostic biomarker for hepatocellular carcinoma.
Keywords: hepatocellular carcinoma, CTHRC1, biomarker, bioinformatics
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors in China, and associated with high rates of morbidity and mortality.1 It is prone to postoperative intrahepatic recurrence, as well as intrahepatic or extrahepatic metastasis.2 Early detection of HCC may result in satisfactory prognosis through surgical therapy, transcatheter arterial chemoembolization and use of various kinase inhibitors (eg, sorafenib and regorafenib).3 However, most patients with HCC exhibit an insidious onset, combined with chronic hepatopathy, early symptoms are atypical or not obvious, and the appropriate timing of surgery is often missed by the time of the diagnosis.4,5 This situation has markedly affected the prognosis of patients with HCC.
The detection of biomarkers is a relatively simple and noninvasive method used to evaluate the prognosis of diseases, and thus improve the survival rate of patients. Recently, the expression of biomarkers, such as alpha-fetoprotein, ki-67, tumor protein p53, microRNAs and circular RNAs, has been associated with poor prognosis in patients. Among them, the false negative cases and false positive cases challenge the clinical prognostic assessment. Thus, the identification of molecules associated with the progression of HCC is warranted to achieve better prognosis.
CTHRC1 is a secreted protein involved in a variety of processes, including vascular remodeling, anti-fibrosis, metabolism, and carcinogenesis. CTHRC1 may affect the occurrence and development of tumors by participating in the process of cell proliferation and migration, synthesis of type I collagen, and repair of damage to blood vessels in colorectal cancer, pancreatic cancer, HCC, etc.6–9 CTHRC1 plays an important role in the pathogenesis of systemic lupus erythematosus.10 However, the clinical significance of CTHRC1 expression in HCC remains unclear. The present study was designed to elucidate the relationship between the expression of CTHRC1 and prognosis in patients with HCC.
Materials And Methods
Analysis Of Oncomine Data
Datasets in the Oncomine database (https://www.oncomine.org) were used to determine the expression pattern of CTHRC1 in HCC. Briefly, the CTHRC1 gene was queried in the database and the results were filtered by selecting HCC and HCC versus Normal.
Collection Of Paraffin-Embedded Specimens
Paraffin-embedded specimens of patients with HCC were collected from September 2015 to September 2018. These specimens were stored in the pathology department of the 980th Hospital of PLA Joint Logistics Support Force (Shijiazhuang, China). The clinicopathological characteristics, including the gender, age, tumor size, tumor-node-metastasis (TNM) stage,11 Barcelona Clinic Liver Cancer (BCLC) staging system,12 etc., are summarized. Five specimens were removed due to loss at follow-up, and specimens from a total of 29 patients were collected. The deadline for the calculation of patient survival was March 20, 2019. This study was approved by the Ethics Committee of the 980th Hospital of PLA Joint Logistics Support Force. Written informed consent was provided by all patients.
Immunohistochemical And Hematoxylin Staining And Evaluation
A traditional immunohistochemical staining protocol was used in this study. Briefly, the tissue section was deparaffinized in xylene and rehydrated using different concentrations of alcohol. Subsequently, the section was treated with 3% hydrogen peroxide, followed by antigen retrieval with 10 mM citrate buffer (pH 6.0). After blockage with 10% goat serum for 30 min, the section was incubated with anti-CTHRC1 antibody (rabbit polyclonal antibody, 1:25 dilution, 16,534-1-AP; Proteintech) overnight at 4°C, followed by incubation with a peroxidase-labeled secondary antibody. Subsequently, hematoxylin was used to stain the nuclei.
CTHRC1 staining was semiquantitatively assessed using a grade scoring system according to the intensity of staining.13 The expression of CTHRC1 was classified into two categories: high level (grades 4–9) and low level (grades 0–3). CTHRC1 staining was scored by a pathologist in a blinded manner.
Cell Lines And Culture
HepG2 cells were deposited in the Hebei Key Lab of Laboratory Animal Science, Hebei Medical University (Shijiazhuang, China). LO2 cells were donated by the Department of Traditional and Western Medical Hepatology, Third Hospital of Hebei Medical University (Shijiazhuang, China). Huh7, SMMC7721, and QSG7701 cells were deposited in the Liver Disease Center of PLA, the 980th Hospital of PLA Joint Logistics Support Force. The use of the cell lines was approved by the Ethics Committee of the 980th Hospital of PLA Joint Logistics Support Force. All cell lines were cultured in Dulbecco’s modified Eagle’s medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Gibco) and penicillin/streptomycin, and incubated in 5% carbon dioxide at 37°C.
RNA Isolation And Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
The TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used to extract total RNA from cells according to the protocol provided by the manufacturer. The reverse transcription of mRNA was completed using a mRNA First-Strand cDNA Synthesis kit (Tiangen, Beijing, China). The qPCR analysis was performed using a standard SYBR Green PCR kit (Takara, Japan). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as endogenous control for the detection of mRNA expression. Relative quantification analysis was performed using the comparative CT (2-△△CT) method. Each experiment was performed in triplicate. The primer sequences were as follows: CTHRC1 forward, 5ʹ-CAATGGCATTCCGGGTACAC-3ʹ; CTHRC1 reverse, 5ʹ-GTACACTCCGCAATTTTCCCAA-3ʹ. GAPDH forward, 5ʹ-CAGGAGGCATTGCTGATGAT-3ʹ; GAPDH reverse, 5ʹ-GAAGGCTGGGGCTCATTT-3ʹ.
Statistical Analysis
The SPSS Version 21.0 software package (IBM Corp., Armonk, NY, USA) was used for the statistical analysis. All analyses were performed using the χ2-test or independent sample t-test or Spearman correlation analysis, as appropriate. Survival curves for HCC were produced using the Kaplan-Meier method and the log rank test. All comparisons were two-tailed, and a P<0.05 denoted statistical significance.
Results
High Expression Of CTHRC1 In The Oncomine Database
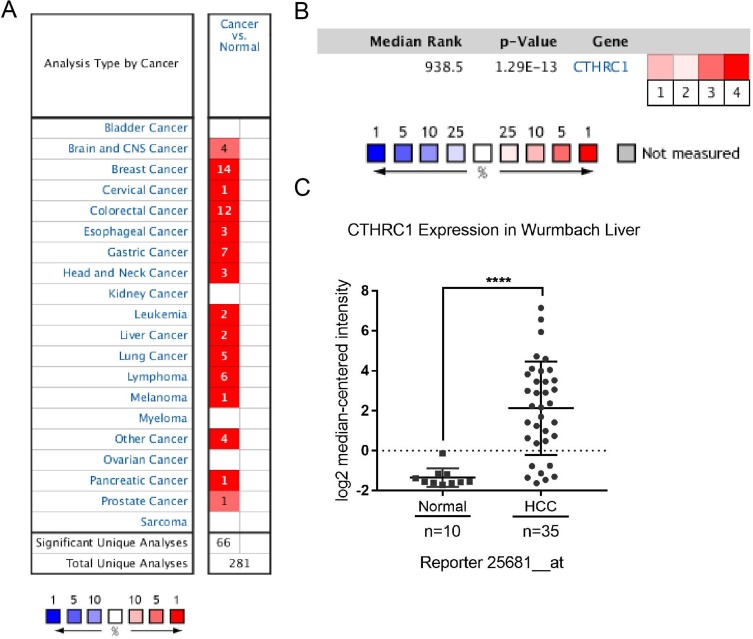
We analyzed the expression of CTHRC1 in cancer tissues and normal tissues from a total of 281 unique analyses in the Oncomine database. Of those, 66 analyses met the following conditions. The gene expression in tumor tissues was two-fold higher than that observed in normal tissues, gene rank was in the top 10%, and P= 0.0001. In HCC, two analyses meet these conditions (Figure 1A).
Figure 1.
Expression of CTHRC1 in the Oncomine database. (A) Expression of the CTHRC1 gene in hepatocellular carcinoma vs normal. (B) Comparisons of the expression of CTHRC1 in HCC in four independent analyses from the Oncomine database. Analysis comparison 1: Guichard liver; 2: Guichard liver 2; 3: TCGA liver; 4: Wurmbach liver. (C) Expression of CTHRC1 in wurmbach liver.
Notes: Compared with normal group, ****P<0.001.
The next part of the study was concerned with the expression of CTHRC1 in HCC tissues versus normal tissues in four independent analyses of data obtained from the Oncomine database. According to Guichard Liver14 (analysis comparison 1), Guichard Liver214 (analysis comparison 2), The Cancer Genome Atlas Liver (analysis comparison 3), and Wurmbach Liver15 (analysis comparison 4), the median rank for CTHRC1 across the afore mentioned analyses was 938.5. According to the median-ranked analysis, the P-value for CTHRC1 was 1.29E-13. Among the four analyses, the Wurmbach Liver demonstrated the highest expression of the CTHRC1 gene (Figure 1B). The 25681_at probe was also observed in the Wurmbach Liver analysis15 (Figure 1C). There was a statistically significant difference between the HCC and the normal group (P<0.001).
Concerns have been expressed regarding the gene copy number. According to The Cancer Genome Atlas and Guichard Liver analyses, the CTHRC1 gene copy number was increased in HCC tissues versus normal liver tissues. Moreover, in the Wurmbach Liver analysis, we observed an upregulation in the mRNA levels of CTHRC1 in HCC tissues versus normal tissues. This upregulation was reflected by a 11.156-fold change in mRNA levels, and the high expression gene rank was 67 (top 1%) (Table 1). Collectively, these observations suggested that CTHRC1 was highly expressed in HCC tissues.
Table 1.
Expression of CTHRC1 In The Oncomine Database
| Datasets (Sample Size) | Comparison Groups | Fold Change | P value | OverexpressionGene Rank |
|---|---|---|---|---|
| Guichard Liver (185) | Hepatocellular carcinoma vs Normal | 1.124 | 2.26E-13 | 948(top 6%) |
| Guichard Liver2 (52) | Hepatocellular carcinoma vs Normal | 1.057 | 0.001 | 2,288(top13%) |
| TCGA Liver (212) | Hepatocellular carcinoma vs Normal | 1.261 | 3.32E-14 | 929(top 5%) |
| Wurmbach Liver (75) | Hepatocellular carcinoma vs Normal | 11.156 | 1.50E-10 | 67(top 1%) |
Abbreviation: TCGA, The Cancer Genome Atlas.
High Expression Of CTHRC1 In HCC Tissues
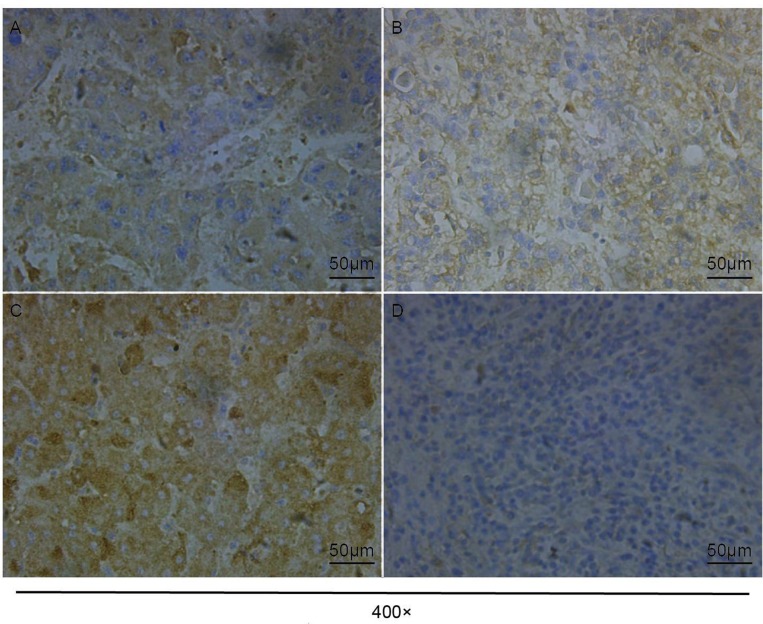
We examined the clinical relevance of CTHRC1 in HCC by evaluating its levels and distribution in human HCC tissue arrays through immunohistochemistry. The results showed that all 29 patients expressed CTHRC1, including 7, 11, and 11 patients with weakly (Figure 2A), moderately (Figure 2B), and strongly (Figure 2C) positive expression, respectively.
Figure 2.
Expression of CTHRC1 in hepatocellular carcinoma and paracancerous tissues. (A) Weakly positive expression (+) in HCC tissues. (B) Moderately positive expression (++) in HCC tissues. (C) Strongly positive expression (+++) in HCC tissues. (D) Negative expression in paracancerous tissues.
Among the 29 paracancerous tissues examined, 12 were negative (Figure 2D), while 13, 1, and 3 cases were weakly, moderately, and strongly positive, respectively. The expression of CTHRC1 in HCC tissues was significantly higher than that measured in paracancerous tissues (P<0.001) (Table 2).
Table 2.
Expression of CTHRC1 in Hepatocellular Carcinoma and Paracancerous Tissues
| Immunohistochemical Grade | HCC Tissue (n=29,100%) | Paracancerous Tissues (n=29,100%) | P value |
|---|---|---|---|
| Negative (-) | 0 | 12 | <0.001 |
| Weakly positive (+) | 7 | 13 | |
| Moderately positive (++) | 11 | 1 | |
| Strongly positive (+++) | 11 | 3 |
High Expression Of CTHRC1 In HCC Cell Lines
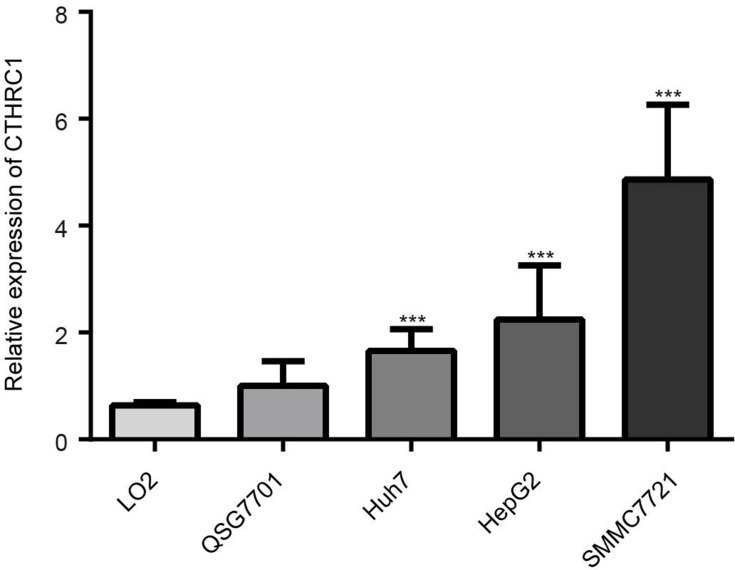
The results from the qRT-PCR showed a higher expression level of CTHRC1 in Huh7, HepG2, and SMMC7721 cell lines versus LO2 and QSG7701 cell lines (Figure 3). The results indicated that the expression of CTHRC1 mRNA in cancer cell lines was significantly higher than that reported in normal cell lines (P<0.01).
Figure 3.
The relative expression of CTHRC1 in normal hepatocyte LO2, QSG7701, and HCC cell lines ***P<0.01 compared with LO2, QSG7701.
High Expression Of CTHRC1 Correlated With Poor Prognosis In HCC
The statistical analysis revealed that the levels of CTHRC1 were significantly correlated with cirrhosis (P=0.024), tumor size (P=0.006), vascular invasion (P<0.001), TNM stage (P<0.001), and BCLC stage (P<0.001). However, there were no statistically significant correlations identified between the expression of CTHRC1 and other clinicopathological characteristics, including gender, age, alpha-fetoprotein, gamma-glutamyl transferase, alanine aminotransferase, hepatitis B virus infection, and number of tumors (Table 3). Furthermore, Spearman analysis of correlation between CTHRC1 and clinicopathological features demonstrated that cirrhosis, tumor size, vascular invasion, TNM stage, and BCLC stage were closely associated with the level of CTHRC1 expression (P≤0.05) (Table 4).
Table 3.
Correlation Between The Expression Of CTHRC1 And Clinicopathological Characteristics In 29 Patients With HCC
| Clinicopathological Variables | Number Of Cases (%) | Low Expression | High Expression | P value |
|---|---|---|---|---|
| Gender | ||||
| Male | 25 (86.2) | 13 | 12 | 0.052 |
| Female | 4 (13.8) | 0 | 4 | |
| Age (years) | ||||
| ≤60 | 11 (37.9) | 7 | 4 | 0.111 |
| >60 | 18 (62.1) | 6 | 12 | |
| AFP(µg/L) | ||||
| ≤400 | 20 (69.0) | 11 | 9 | 0.101 |
| >400 | 9 (31.0) | 2 | 7 | |
| GGT(U/L) | ||||
| ≤54 | 17 (58.6) | 9 | 8 | 0.296 |
| >54 | 12 (41.1) | 4 | 8 | |
| ALT(U/L) | ||||
| ≤45 | 19 (65.5) | 8 | 11 | 0.684 |
| >45 | 10 (34.5) | 5 | 5 | |
| HBV | ||||
| Negative | 6 (20.7) | 3 | 3 | 0.775 |
| Positive | 23 (79.3) | 10 | 13 | |
| Cirrhosis | ||||
| No | 11 (37.9) | 2 | 9 | 0.024 |
| Yes | 18 (62.1) | 11 | 7 | |
| Tumor size(cm) | ||||
| ≤5 | 19 (65.5) | 12 | 7 | 0.006 |
| >5 | 10 (34.5) | 1 | 9 | |
| Tumor number | ||||
| Single | 23 (79.3) | 12 | 11 | 0.119 |
| Multiple | 6 (20.7) | 1 | 5 | |
| Vascular invasion | ||||
| No | 14 (48.3) | 13 | 1 | <0.001 |
| Yes | 15 (51.7) | 0 | 15 | |
| TNM stage | ||||
| I–II | 13 (44.8) | 12 | 1 | <0.001 |
| III–IV | 16 (55.2) | 1 | 15 | |
| BCLC stage | ||||
| 0+A | 12 (41.4) | 12 | 0 | <0.001 |
| B+C | 17 (58.6) | 1 | 16 |
Abbreviations: AFP, alpha-fetoprotein; GGT, gamma-glutamyl transferase; ALT, alanine aminotransferase; HBV, hepatitis B virus; TNM, tumor-node-metastasis; BCLC, Barcelona Clinic Liver Cancer.
Table 4.
Spearman Analysis of Correlation between the Expression of CTHRC1 and Clinicopathological Variables
| Variables | CTHRC1 Expression | |
|---|---|---|
| Spearman Correlation | P value | |
| Gender | 0.361 | 0.055 |
| Age | 0.296 | 0.119 |
| AFP | 0.305 | 0.108 |
| GGT | 0.194 | 0.313 |
| ALT | −0.075 | 0.697 |
| HBV | 0.053 | 0.784 |
| Cirrhosis | −0.419 | 0.024 |
| Tumor size | 0.508 | 0.005 |
| Tumor number | 0.289 | 0.128 |
| Vascular invasion | 0.933 | <0.001 |
| TNM stage | 0.861 | <0.001 |
| BCLC stage | 0.932 | <0.001 |
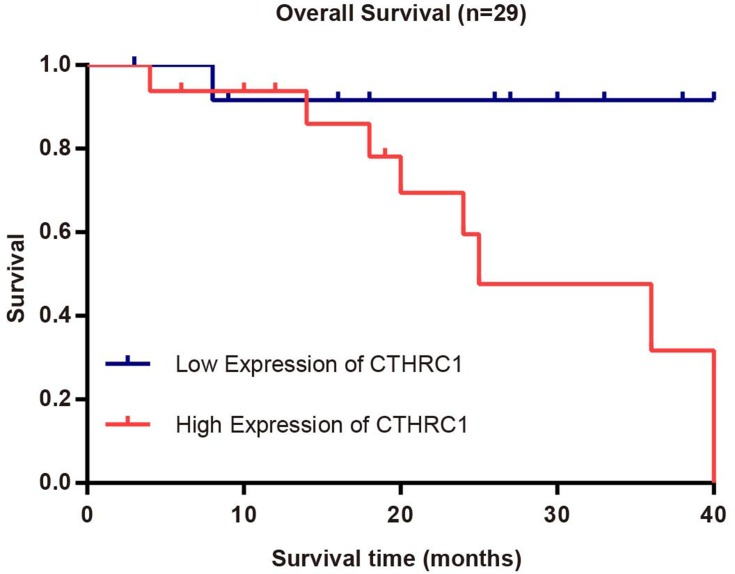
Survival curves for HCC patients using the Kaplan-Meier method and the log rank test indicated that high expression of CTHRC1 may be associated with poor prognosis (Figure 4).
Figure 4.
Survival curves for 29 HCC patients with low/high expression of CTHRC1. n(high)=16, n(low)=13, log rank test: P=0.049.
The Prognostic Value Of CTHRC1 As A Biomarker For HCC
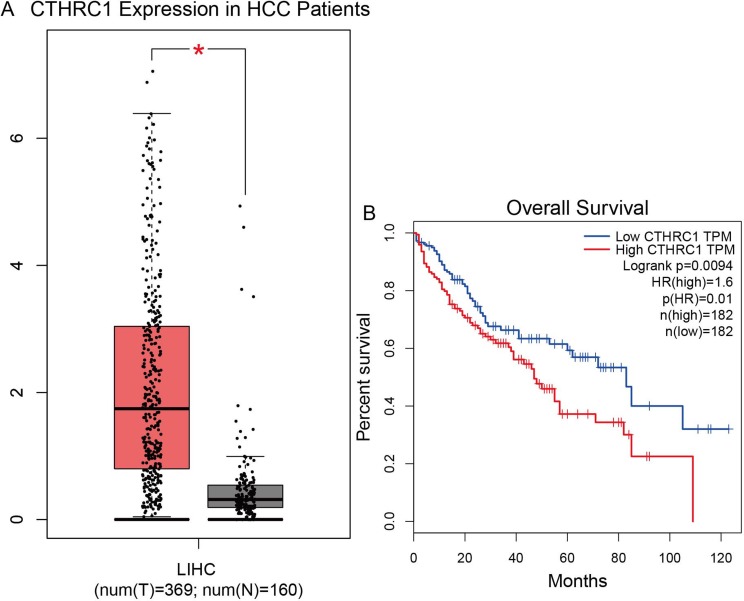
Data from the Gene Expression Profiling Interactive Analysis database and further analysis showed that the expression of CTHRC1 was significantly higher in 369 patients with HCC versus 160 normal tissues (Zefang Tang, Chenwei Li, Boxi Kang. Zhang’s Lab). Further analysis showed that high expression of CTHRC1 may be associated with poor prognosis (Figure 5). These results showed that CTHRC1 may be used as a biomarker for HCC.
Figure 5.
High expression of CTHRC1 in patients with LIHC and overall survival curves. (A) Expression levels of CTHRC1 in healthy controls (n=160) and patients with hepatocellular carcinoma (n=369) from the gene expression profiling interactive analysis database. (B) Overall survival curves for patients with low/high CTHRC1 expression from the Gene Expression Profiling Interactive Analysis database.
Notes: Compared with normal group, *P<0.05.
Abbreviation: LIHC, Liver hepatocellular carcinoma.
Discussion
The identification of molecules associated with HCC progression and prediction of prognosis is of crucial importance. In this study, we investigated the effect of CTHRC1 on HCC. Through bioinformatics analyses and experiments, we confirmed that CTHRC1 is highly expressed in tissues and hepatoma cell lines. High expression of the CTHRC1 gene is associated with poor prognosis. Collectively, the results of the present study indicated that CTHRC1 may serve as a prognostic biomarker for HCC.
CTHRC1 is an oncogene involved in numerous types of human cancers, including HCC.16–21 However, there is limited information regarding the prognostic value of CTHRC1 in HCC. Previous research showed that CTHRC1 promotes tumor invasion and metastasis.6,22,23 More recently, studies reported that CTHRC1 is an important microenvironment factor24 and regulated by DNA hyper-methylation, which may be closely associated with HCC.25 Studies revealed that CTHRC1 induces epithelial-mesenchymal transition and promotes movement of cancer cells.26,27
A recent immunohistochemical study examined 130 tissues with cervical squamous cell carcinoma.28 The relationships between the expression of CTHRC1, clinicopathological variables, and the prognosis of cervical squamous cell carcinomas were investigated. The expression of the CTHRC1 protein in tumors was significantly higher than that reported in normal tissues (P<0.001). The multivariate proportional hazards model revealed that high expression of CTHRC1 was an independent predictor of overall survival and disease-free survival. In addition, the immunohistochemical analysis of the present study indicated that the levels of CTHRC1 expression were significantly elevated in HCC tissues, and closely associated with tumor progression. This finding was consistent with those reported in previous studies.28
Despite the potential implication of CTHRC1 in the progression of cancer, few previous studies have examined its expression levels and clinical significance in HCC. Chen et al found that elevated expression of CTHRC1 was more frequently observed in larger and advanced HCC tumors.6 Furthermore, the expression of CTHRC1 was an indicator of poor prognosis. A total of 1,065 patients with cancer from eight studies were included in the meta-analysis. The results revealed that increased expression of CTHRC1 is associated with advanced TNM stage, increased lymph node metastasis, and tumor size, as well as decreased overall survival and disease-free survival. These findings indicated that CTHRC1 may be a biomarker for the prognosis of patients with cancer.29 There are subtle differences between the present results and those of the meta-analysis. In this study, we confirmed that CTHRC1 is highly expressed in HCC tissues at both the RNA and DNA levels using the publicly available gene expression datasets in the Oncomine and Gene Expression Profiling Interactive Analysis databases. The results obtained from the qRT-PCR analysis also showed higher mRNA levels of CTHRC1 in the Huh7, HepG2, and SMMC7721 cell lines versus the LO2 and QSG7701 cell lines. The results showed that the expression of CTHRC1 in hepatoma cell lines was higher than that observed in the normal cell lines. The clinical relevance analysis showed that positive expression of CTHRC1 in HCC tissues was significantly correlated with cirrhosis, tumor size, vascular invasion, TNM stage, and BCLC stage. This conclusion is partially consistent with the result of previous research. In addition, we evaluated the value of CTHRC1, revealing that high expression of CTHRC1 was associated with poor prognosis in HCC patients.
The present study had several limitations. Firstly, the sample size was small (29 patients). Larger studies are warranted to confirm the results. Secondly, the conclusions were based on data produced using five cell lines, and may not accurately reflect the processes occurring in an intact organism. A study is currently ongoing, aiming to confirm the present results in mice.
In conclusion, the present study demonstrated that high expression of CTHRC1 in HCC tissues is significantly associated with poor survival. This finding suggests that CTHRC1 may act as a novel oncogene in HCC, and may be a therapeutic target.
Acknowledgments
The authors gratefully acknowledge all the staff of the Department of Pathology, The 980th Hospital of PLA Joint Logistics Support Force, who generously provided invaluable assistance in the collection of data throughout the duration of the study. This study was supported by grants from the National Natural Science Foundation of China (No.30571667, No.31702076), the National Key R&D Program of China (No.2016YFD0501000, No.2017YFD0501705), and the Hebei Province Science and Technology Support Program, China (No.15967777D).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 2.Orcutt ST, Anaya DA. Liver resection and surgical strategies for management of primary liver cancer. Cancer Control. 2018;25(1):1073274817744621. doi: 10.1177/1073274817744621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ueshima K, Nishida N, Kudo M. Sorafenib-regorafenib sequential therapy in advanced hepatocellular carcinoma: a single-institute experience. Dig Dis. 2017;35(6):611–617. doi: 10.1159/000480257 [DOI] [PubMed] [Google Scholar]
- 4.Yuan J, Ji H, Xiao F, et al. MicroRNA-340 inhibits the proliferation and invasion of hepatocellular carcinoma cells by targeting JAK1. Biochem Biophys Res Commun. 2017;483(1):578–584. doi: 10.1016/j.bbrc.2016.12.102 [DOI] [PubMed] [Google Scholar]
- 5.Zhao L, Cao J, Hu K, et al. RNA-binding protein RPS3 contributes to hepatocarcinogenesis by post-transcriptionally up-regulating SIRT1. Nucleic Acids Res. 2019;47(4):2011–2028. doi: 10.1093/nar/gky1209 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Chen YL, Wang TH, Hsu HC, Yuan RH, Jeng YM. Overexpression of CTHRC1 in hepatocellular carcinoma promotes tumor invasion and predicts poor prognosis. PLoS One. 2013;8(7):e70324. doi: 10.1371/journal.pone.0070324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim HC, Kim YS, Oh HW, et al. Collagen triple helix repeat containing 1 (CTHRC1) acts via ERK-dependent induction of MMP9 to promote invasion of colorectal cancer cells. Oncotarget. 2014;5(2):519–529. doi: 10.18632/oncotarget.1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pitule P, Vycital O, Bruha J, et al. Differential expression and prognostic role of selected genes in colorectal cancer patients. Anticancer Res. 2013;33(11):4855–4865. [PubMed] [Google Scholar]
- 9.Tang SC, Chen YC. Novel therapeutic targets for pancreatic cancer. World J Gastroenterol. 2014;20(31):10825–10844. doi: 10.3748/wjg.v20.i31.10825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Q, Yang Q, Sun H. Collagen triple helix repeat containing-1: a novel biomarker associated with disease activity in systemic lupus erythematosus. Lupus. 2018;27(13):2076–2085. doi: 10.1177/0961203318804877 [DOI] [PubMed] [Google Scholar]
- 11.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17(6):1471–1474. doi: 10.1245/s10434-010-0985-4 [DOI] [PubMed] [Google Scholar]
- 12.European Association for the Study of the Liver, European Organisation for Research and Treatment of Cancer. Clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56(4):908–943. doi: 10.1016/j.jhep.2011.12.001 [DOI] [PubMed] [Google Scholar]
- 13.Lin XL, Yang L, Fu SW, et al. Overexpression of NOX4 predicts poor prognosis and promotes tumor progression in human colorectal cancer. Oncotarget. 2017;8(20):33586–33600. doi: 10.18632/oncotarget.16829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guichard C, Amaddeo G, Imbeaud S, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44(6):694–698. doi: 10.1038/ng.2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wurmbach E, Chen YB, Khitrov G, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45(4):938–947. doi: 10.1002/hep.21622 [DOI] [PubMed] [Google Scholar]
- 16.Tameda M, Sugimoto K, Shiraki K, et al. Collagen triple helix repeat containing 1 is overexpressed in hepatocellular carcinoma and promotes cell proliferation and motility. Int J Oncol. 2014;45(2):541–548. doi: 10.3892/ijo.2014.2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JH, Baek TH, Yim HS, et al. Collagen triple helix repeat containing-1 (CTHRC1) expression in invasive ductal carcinoma of the breast: the impact on prognosis and correlation to clinicopathologic features. Pathol Oncol Res. 2013;19(4):731–737. doi: 10.1007/s12253-013-9636-y [DOI] [PubMed] [Google Scholar]
- 18.Tan F, Liu F, Liu H, Hu Y, Liu D, Li G. CTHRC1 is associated with peritoneal carcinomatosis in colorectal cancer: a new predictor for prognosis. Med Oncol. 2013;30(1):473. doi: 10.1007/s12032-013-0473-3 [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Abulimiti N, Wang C. Collagen triple helix repeat containing 1 expression in osteosarcoma: a new predictor of prognosis. Ann Clin Lab Sci. 2018;48(3):338–344. [PubMed] [Google Scholar]
- 20.Liu W, Fu XL, Yang JY, et al. Elevated expression of CTHRC1 predicts unfavorable prognosis in patients with pancreatic ductal adenocarcinoma. Am J Cancer Res. 2016;6(8):1820–1827. [PMC free article] [PubMed] [Google Scholar]
- 21.Yang C, Liu T, Xia B, Gu L, Lou G. Correlation of collagen triple helix repeat containing 1 overexpression with lymph node and peritoneal metastasis in epithelial ovarian cancer. Int J Gynecol Cancer. 2017;27(1):22–27. doi: 10.1097/IGC.0000000000000850 [DOI] [PubMed] [Google Scholar]
- 22.Chen G, Wang D, Zhao X, et al. miR-155-5p modulates malignant behaviors of hepatocellular carcinoma by directly targeting CTHRC1 and indirectly regulating GSK-3beta-involved Wnt/beta-catenin signaling. Cancer Cell Int. 2017;17:118. doi: 10.1186/s12935-017-0469-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Lee M, Yu G, Lee H, Han X, Kim D. CTHRC1 activates pro-tumorigenic signaling pathways in hepatocellular carcinoma. Oncotarget. 2017;8(62):105238–105250. doi: 10.18632/oncotarget.22164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Wang Y, Ma M, et al. Autocrine CTHRC1 activates hepatic stellate cells and promotes liver fibrosis by activating TGF-beta signaling. EBioMedicine. 2019;40:43–55. doi: 10.1016/j.ebiom.2019.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun XJ, Wang MC, Zhang FH, Kong X. An integrated analysis of genome-wide DNA methylation and gene expression data in hepatocellular carcinoma. FEBS Open Bio. 2018;8(7):1093–1103. doi: 10.1002/2211-5463.12433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ni S, Ren F, Xu M, et al. CTHRC1 overexpression predicts poor survival and enhances epithelial-mesenchymal transition in colorectal cancer. Cancer Med. 2018;7(11):5643–5654. doi: 10.1002/cam4.1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenburg G, Hay ED. Epithelia suspended in collagen gels can lose polarity and express characteristics of migrating mesenchymal cells. J Cell Biol. 1982;95(1):333–339. doi: 10.1083/jcb.95.1.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li N, Chen L, Liu C, Jiang Y, Rong J. Elevated CTHRC1 expression is an indicator for poor prognosis and lymph node metastasis in cervical squamous cell carcinoma. Hum Pathol. 2019;85:235–241. doi: 10.1016/j.humpath.2018.10.015 [DOI] [PubMed] [Google Scholar]
- 29.Wang SF, Yin Z, Yin JJ, Zhang W, Dong CG. Clinical significance of CTHRC1 protein expression in human cancers: a meta-analysis. Genet Mol Res. 2016;15:2. [DOI] [PubMed] [Google Scholar]