Abstract
The inheritance of monoallelic germline mutations affecting BRCA1 or BRCA2 predisposes with a high penetrance to several forms of epithelial malignancy. The large, nuclear-localized BRCA proteins act as custodians of chromosome integrity through distinct functions in the assembly and activity of macromolecular complexes that mediate DNA repair, replication reactivation and mitotic progression. The loss of these tumour suppressive functions following biallelic BRCA gene inactivation has long been thought to provoke genomic instability and carcinogenesis. However, recent studies not only identify new functions for BRCA1 and BRCA2 in the regulation of transcription and RNA processing potentially relevant to their tumour suppressive activity, but also suggest that monoallelic BRCA2 gene mutations suffice for carcinogenesis. This emerging evidence opens fresh lines of enquiry concerning tissue-specific cancer evolution in BRCA mutation carriers. Collectively, these insights engender new models to explain how BRCA gene mutations cause cancer susceptibility in specific tissues.
Keywords: BRCA1, BRCA2, chromosome stability, carcinogenesis, DNA repair, DNA replication, transcription, R-loops
Cancer susceptibility associated with mutations affecting BRCA1 or BRCA2
Germline mutations affecting a single copy of the breast cancer genes BRCA1 or BRCA2 significantly increase the risk of breast and ovarian cancer (reviewed in Nathanson et al., 2001; Rahman and Stratton, 1998; Welcsh and King, 2001). A recent prospective study (Kuchenbaecker et al., 2017) indicates that the cumulative breast cancer risk to age 80 years is 72% versus 69% for BRCA1 or BRCA2 mutation carriers respectively, whereas the corresponding cumulative ovarian cancer risk is 44% versus 17%. However, the BRCA genes are not only breast and ovarian cancer suppressors. Germline heterozygous mutations affecting BRCA2 also significantly elevate the risk of cancers in the pancreas, male breast, prostate, and other tissues (Breast Cancer Linkage Consortium, 1999; Hout et al., 2019; Huang et al., 2018; Thompson and Easton, 2002). Moreover, inherited bi-allelic mutations affecting BRCA1 as well as BRCA2 may cause congenital syndromes associated with developmental anomalies, chromosome fragility and cancers at various sites (for eg., Howlett et al., 2002; Sawyer et al., 2015; Seo et al., 2018). Collectively, these clinical genetic studies indicate that the BRCA genes operate to suppress carcinogenesis in several different human tissues, and that the inheritance of monoallelic versus biallelic mutations causes distinct clinical manifestations.
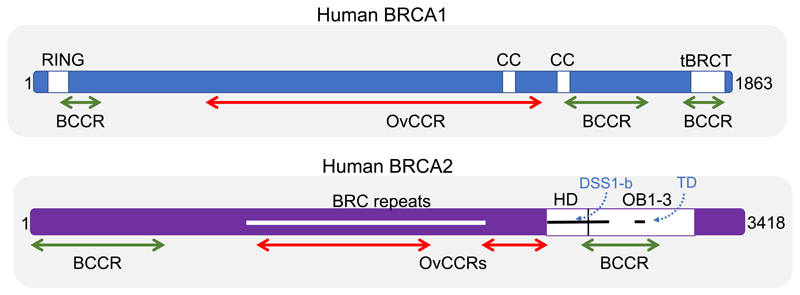
Human BRCA1 encodes an 1863 amino acid protein (Figure 1) containing an amino (N)-terminal RING domain, which heterodimerizes with the partner protein BARD1 to form an E3 ubiquitin ligase. At its carboxyl (C)-terminus, BRCA1 contains tandem repeats of the so-called BRCA1 carboxyl-terminal (BRCT) domains, each comprising about 100 amino acids, which engage phosphopeptide motifs in partner proteins to form functional macromolecular complexes, in turn assisting substrate selection for BRCA1-BARD1 ubiquitin ligase activity. Human BRCA2, a 3418 amino acid protein (Figure 1), also contains distinct motifs that mediate its interaction with partner proteins. The evolutionarily conserved segment of BRCA2 encoded by exon 11 contains 8 repeats of a 35-40 residue motif termed the BRC repeat, which binds to the recombination enzyme RAD51 (Wong et al., 1997), as well as other partners (Lee et al., 2011; Martinez et al., 2016; Rajagopalan et al., 2010). A C-terminally positioned region of BRCA2 interacts with the small, charged protein DSS1 to assume a fold that contains single-stranded (ss)DNA-binding OB domains as well as a putative double-stranded (ds)DNA-binding domain (von Nicolai et al., 2016; Yang et al., 2002). Notably, both BRCA1 and BRCA2 are characterized by extended unstructured regions that likely assume stable conformations only upon engagement with partner proteins. These features have led to the proposal that BRCA1 and BRCA2 represent intrinsically disordered proteins that act as hubs for the formation of macromolecular complexes serving distinct cellular functions (Venkitaraman, 2014).
Figure 1. The human BRCA1 and BRCA2 proteins.
The schematics representing human BRCA1 (1863 amino acids) and BRCA2 (3418 amino acids) are not to the same scale. Regions depicted as white boxes in BRCA1 include the N-terminal RING domain, the C-terminal tandem BRCT (tBRCT) domains, and a region predicted to encode coiled coil (CC) motifs. Regions depicted in BRCA2 include the conserved region of exon 11 encoding 8 repeats of the 30-40 amino acid BRC motif (white line), as well as the adjoining helical domain (HD, white box), and OB1-3 folds (white box), which together form a DNA-binding domain. The segments binding DSS1 (DSS1-b) and encoding the tower domain (TD) are represented as black lines. Proposed mutation cluster regions predisposing preferentially to breast (BCCR, green) or ovarian (OvCCR, red) are shown, based on data from Rebbeck et al., 2015. Caveats concerning the nature and mechanism of the proposed cluster regions are summarized in the main text.
BRCA gene mutations and cancer susceptibility
Thousands of variants affecting the BRCA genes occur in the human genome, but only a fraction of them are reliably known to cause cancer susceptibility (Cline et al., 2018; ClinVar, https://www.ncbi.nlm.nih.gov/clinvar). Founder mutations inducing cancer susceptibility have been identified amongst the Ashkenazim, as well as in Iceland and other regions. The great majority of pathogenic or likely pathogenic mutations (~80%) generate a premature termination codon, truncating the encoded protein and potentially decreasing its expression through nonsense-mediated mRNA decay (Anczuków et al., 2008). A minority are missense variants encoding a stable mutant protein (~10%). In addition, pathogenic or likely pathogenic BRCA1 and BRCA2 alleles may arise through large in-frame deletions that span one or more exons, or alterations that disrupt transcriptional regulatory regions to inhibit expression of the mutant allele. Notably, what is summarized here represents an approximation, since the proportion of truncating versus mis-sense variants, and their likely pathogenicity, vary considerably between the BRCA1 versus BRCA2 genes.
Truncating mutations in BRCA1 and BRCA2 are distributed throughout the protein, with those that substantially shorten the encoded proteins largely being pathogenic. In contrast, most missense variants identified through genome sequencing have been classified using multifactorial likelihood approaches either as being benign or of unknown significance, and probably represent polymorphic variants even when detected in cancer genomes (ClinVar, https://www.ncbi.nlm.nih.gov/clinvar). Mis-sense mutations known to be pathogenic tend to cluster in limited regions of the BRCA proteins, including the N-terminal RING domain and C-terminal tandem BRCT domain in BRCA1, or in the region of BRCA2 that spans the OB folds and helical domain.
A large number of missense alleles are classified as being variants of unknown significance (VUS), particularly for BRCA2, posing a challenge for accurate risk assessment and clinical management in mutation carriers. It can be difficult purely from clinical genetic information to reliably determine the likely pathogenicity of missense variants, owing in part to the small number of cases bearing any given variant. Recent work to address this challenge has therefore attempted to assess the effects of such variants on protein function using in vitro assays. In one recent example, saturation genome editing in haploid Hap1 cells, coupled with an assay for cell viability, was used to “scan” the RING and BRCT regions of BRCA1 for functionally deleterious variants (Findlay et al., 2018). However, not all functionally deleterious BRCA1 variants may affect cell viability, and so it will be important to extend such an approach to other cellular phenotypes. Similar studies for the BRCA2 protein are also warranted, given its larger size and the greater frequency of missense variants still classified as VUS.
It has been proposed from epidemiological studies that truncating mutations affecting different regions of BRCA1 and BRCA2 may preferentially confer risks of breast versus ovarian cancer (Rebbeck et al., 2015). BRCA1 is thought to contain three “breast-cancer cluster” regions (BCCRs) and a single “ovarian cancer cluster” region (OCCR), the latter spanning much of the evolutionarily conserved exon 11. Similar regions have been designated in BRCA2 (Figure 1). The proposed existence of such cluster regions raises the possibility that different BRCA gene mutations may affect functions relevant to tissue-specific carcinogenesis. However, certain caveats warrant attention. Truncating mutations within the proposed cluster regions will not only delete downstream regions of the protein, but also probably suppress protein expression through nonsense-mediated mRNA decay. Thus, such mutations – particularly those that fall within the first few hundred nucleotides of the BRCA1 or BRCA2 coding sequence - seem likely to create functionally “null” alleles. How such alleles might exert tissue-specific effects is puzzling, and so the mechanistic basis for the proposed clustering is unclear.
Cellular functions of BRCA1 and BRCA2 implicated in cancer susceptibility
The ‘chromosomal instability’ model for cancer susceptibility
Current understanding of the tumour suppressive roles of BRCA1 and BRCA2 has largely been founded upon a series of studies that quickly followed their discovery, which defined their essential function in preserving chromosome integrity during cell division. Bi-allelic inactivation of BRCA1 or BRCA2 in transgenic mice was found to cause early embryonal lethality and impaired cell proliferation (Connor et al., 1997; Friedman et al., 1998; Hakem et al., 1996; Liu et al., 1996; Patel et al., 1998; Sharan et al., 1997), as well as hypersensitivity to genotoxins (Chen et al., 1998b; Connor et al., 1997; Patel et al., 1998; Sharan et al., 1997). BRCA1 and BRCA2 were shown to accumulate in nuclear foci induced by DNA damage (Chen et al., 1998a; Scully et al., 1997a), and to interact with different proteins implicated in the DNA damage response (Mizuta et al., 1997; Sharan et al., 1997; Wong et al., 1997). Notably, BRCA2-deficient cells were shown to spontaneously accumulate aberrations in chromosome structure and number during their division (Patel et al., 1998), characterized by breaks affecting a single sister chromatid, as well as abnormal “radial” chromosomes pathognomonic of defects in homologous DNA recombination previously linked to Bloom syndrome and Fanconi anemia (Patel et al., 1998). Cells lacking murine BRCA1 were soon shown to exhibit similar anomalies (Shen et al., 1998). Taken together, these results not only established a critical biological role for BRCA1 and BRCA2 as custodians of chromosome integrity during the cell cycle, but also led to the proposal (Scully and Livingston, 2000; Venkitaraman, 2002) that pathogenic mutations inactivating the BRCA genes cause cancer susceptibility by inducing chromosomal instability and mutagenesis. These foundational ideas, verified and extended in later studies, remain the chief pillars for current paradigms concerning the tumour suppressive activities of BRCA1 and BRCA2.
DNA repair by homologous recombination
BRCA1 and BRCA2 are essential for the repair of dsDNA breaks (DSBs) by homologous recombination (HR) (for eg., Moynahan et al., 1999, 2001). They undertake distinct functions during this process that are well studied (reviewed in for eg., Venkitaraman, 2002, 2014), and therefore only briefly recounted here. Notably, BRCA1 and BRCA2 form a tri-molecular complex with a partner protein, PALB2, that assists in their localization and function at sites of DNA damage (Xia et al., 2006; Sy et al., 2009; Zhang et al., 2009). Put simply, BRCA1 acts first to assemble macromolecular complexes that signal the presence of DNA damage, and subsequently helps to initiate the repair of DSBs by recruiting proteins that process broken ends. By contrast, BRCA2 participates directly in controlling the activity and assembly of the key recombination enzyme, RAD51, on ss- and dsDNA substrates to execute DNA repair by HR.
Stabilization of stalled DNA replication forks
A role for BRCA2 and associated proteins in the protection of stalled DNA replication forks has attracted increasing attention. It was shown in 2003 that BRCA2 deficiency de-stabilizes the structure of DNA intermediates formed at stalled replication forks induced by exposure to hydroxyurea, subsequently triggering their collapse into DSBs, leading to the proposal that BRCA2 stabilizes the structure of arrested forks to allow their error-free resolution (Lomonosov et al., 2003). In 2011, it was found that a C-terminal segment of BRCA2 dispensable for HR protects nascent DNA strands at stalled forks from degradation by the MRE11 nuclease (Schlacher et al., 2011), outlining how this function might be exerted.
Subsequent studies provide further insight into how BRCA1 and BRCA2 act to protect stalled DNA replication forks. This emerging picture envisions a two-step mechanism wherein replication forks first undergo remodeling and reversal promoted by a BRCA-independent function of RAD51, followed by a second step involving BRCA1, BRCA2 and RAD51 that protects the remodeled, reversed fork from degradation by nucleases including MRE11, EXO1, CtIP and possibly, Dna2 (Kolinjivadi et al., 2017; Lemaçon et al., 2017; Mijic et al., 2017). The sequential and dual functions of RAD51 in these events - initially BRCA-independent, and later BRCA-dependent - are yet to be directly established but have instead been indirectly inferred from multiple experiments. For instance, the observed decrease in the frequency of reversed replication forks visualized by electron microscopy in RAD51-depleted cells cannot be ameliorated by MRE11 inhibition, suggesting that RAD51 is necessary for fork reversal in a step that precedes MRE11-induced degradation (Lemaçon et al., 2017; Mijic et al., 2017). By contrast, MRE11 inhibition restores the decrease in reversed forks detected in BRCA2-deficient cells (Mijic et al., 2017), suggesting that fork reversal occurs even in the absence of BRCA2, but that fork protection from MRE11 mandates a second, BRCA2-dependent step. This indirect evidence supporting the proposed two-step mechanism - fork reversal followed by fork protection - remains open to reinterpretation.
Both fork reversal and fork protection likely require multiple activities. Besides RAD51, fork remodeling and reversal involves proteins such as the DNA translocases SMARCAL1, ZRANB3 and HTLF (Kolinjivadi et al., 2017; Lemaçon et al., 2017; Mijic et al., 2017; Vujanovic et al., 2017), and factors such as RADX and FBH1 that regulate RAD51 activity at stalled forks (Dungrawala et al., 2017; Fugger et al., 2015). Moreover, BRCA1/BARD1 and the Fanconi anemia (FA) proteins FANCA, FANCD2 and FANCJ (Schlacher et al., 2012; Billing et al., 2018; Peng et al., 2018) have been implicated along with BRCA2 and RAD51 in fork protection.
Intriguingly, several studies suggest distinct roles for BRCA1 versus BRCA2 at stalled replication forks that may account for distinct patterns of genomic instability in their absence. For example, BRCA1 but not BRCA2 is required to prevent tandem duplications when replication forks stall at Tus/Ter replication fork barriers (Willis et al., 2017). Furthermore, the methyltransferase EZH2 promotes the cleavage of stalled replication forks by the MUS81 nuclease in BRCA2-deficient - but not BRCA1-deficient – cells to augment sensitivity to poly-ADP ribose polymerase (PARP1) inhibitors (Rondinelli et al., 2017). These observations add to the evidence that BRCA1 and BRCA2 play complementary but distinct roles both in HR and in the reactivation of stalled replication forks.
RNA-DNA hybrid (R loop) processing and transcription
A rising number of recent studies implicate BRCA1 and BRCA2 in the turnover of R-loops, hybrids between RNA and ssDNA that are physiological intermediates during gene transcription, but whose unscheduled accumulation may trigger genome instability (Aguilera and García-Muse, 2012). The first clues implicating the BRCA proteins in R-loop turnover came from the concurrent observations that BRCA2 depletion in cells was found to increase R loop accumulation (Bhatia et al., 2014), and that BRCA1 was shown to bind multiple proteins involved in transcription, localize to sites of transcription arrest, and exhibit a genetic interaction with senataxin (SETX), a helicase implicated in R-loop resolution (Hill et al., 2014).
These clues soon led (Hatchi et al., 2015) to the demonstration that BRCA1 engages R-loops that arise physiologically at sites of transcription termination throughout the genome, to which it recruits SETX. Disruption of the BRCA1/SETX complex led to R-loop accumulation, ssDNA breakage in the untranscribed R-loop strand, and DNA damage marked by γ-H2AX. Interestingly, insertion/deletion mutations were found in BRCA1-deficient breast cancers at the transcription termination regions of certain genes adjacent to R-loop mediated BRCA1/SETX binding sites.
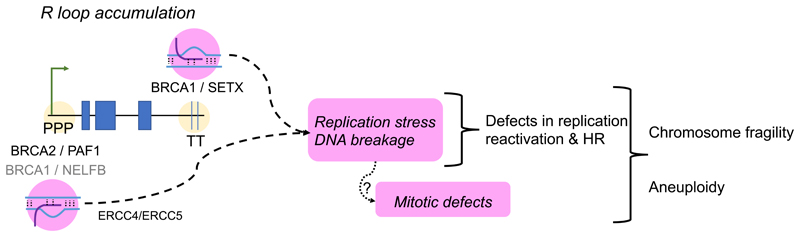
Strikingly, replication fork instability and the frequency of structural chromosomal aberrations like chromatid breaks or radial chromosomes characteristic of BRCA2-deficient cells was reduced (Tan et al., 2017) by Ribonuclease H1, an enzyme that selectively digests R-loops. This observation suggests that R-loop accumulation is a proximal cause of spontaneous genomic instability during DNA replication following BRCA2 inactivation (Tan et al., 2017). Notably, many DNA breaks that occur in cells lacking FANCA or FANCD2 are also R loop-dependent, as is the recruitment of FANCD2 to microscopic nuclear foci at sites of DNA damage (García-Rubio et al., 2015). Collectively, these observations indicate that unscheduled R-loop accumulation may be a major source of the endogenous DNA damage that leads to spontaneous chromosomal instability following the inactivation of BRCA1 or BRCA2 (Figure 2).
Figure 2. R-loop accumulation and chromosome instability in BRCA-deficient cells.
The schematic depicts how transcription-associated R loop accumulation in BRCA2-deficient cells may be a major source of replication stress and DNA damage leading to chromosome fragility. Roles are shown for BRCA1 and BRCA2 in R-loop turnover at the promoter-proximal pausing (PPP) sites, or at transcription termination (TT) sequences, of expressed genes. The RNAPII regulatory factors PAF1 and NELFB work with BRCA2 and BRCA1 respectively, in the switch from pausing to elongation. Senataxin (SETX) is recruited by BRCA1 to TT sites. The ERCC4/ERCC5 nuclease has been implicated in R-loop cleavage to form DNA breaks.
Whereas BRCA1 has been implicated in R-loop turnover at transcription termination sites, the origin of R-loop accumulation following BRCA2 inactivation has been less clear. One idea (Bhatia et al., 2014) is that BRCA2 might participate in mRNP biogenesis through the interaction of its partner protein, DSS1, with PCID2 component of the mRNA transcription and export (TREX) complex, but functional evidence to support such a function is quite limited. Instead, recent attention has turned to a role for BRCA2 in controlling the release of RNA polymerase II (RNAPII) from promoter-proximal pausing (PPP) sites, which has emerged as a fundamental regulatory event during transcription elongation in metazoan cells.
Human BRCA2 has recently been shown to bind to the RNAPII holoenzyme (Shivji et al., 2018). BRCA2 inactivation by depletion or cancer-causing mutations was found to trigger unscheduled RNAPII accumulation and R-loop accrual at PPP sites in actively transcribed genes. DNA breaks marked by γH2AX formation was increased at these genomic sites in BRCA2-deficient cells, suggesting that unscheduled R-loops formed at PPP sites may be processed into DSBs. Indeed, depletion of the ERCC4 endonuclease, which was shown previously to cleave R-loops (Sollier et al., 2014), decreases γH2AX formation at PPP sites in BRCA2-deficient cells. Clues to the mechanism by which BRCA2 may control promoter proximal pausing of RNAPII have also been found (Shivji et al., 2018). BRCA2 inactivation was shown to decrease RNAPII-associated factor 1 (PAF1) recruitment to the holoenzyme, which normally promotes RNAPII release from PPP sites. PAF1 depletion was found to phenocopy R-loop accumulation at PPP sites in wild-type cells, while its overexpression could ameliorate R-loop accumulation and RNAPII pausing after BRCA2 inactivation. Intriguingly, BRCA2 inactivation could diminish H2B Lys120 ubiquitination, a chromatin mark implicated in transcription elongation. Collectively, these observations provide evidence that BRCA2 performs a novel function in the “switching” of RNAPII from promoter-proximal pausing to productive elongation via augmented PAF1 recruitment, and that disruption of this function by cancer-causing BRCA2 mutations triggers R-loop-mediated DNA breakage preferentially at PPP sites in actively transcribed genes (Shivji et al., 2018).
Notably, emerging evidence links R-loop accumulation following BRCA1 inactivation to the pathogenesis of breast cancers. BRCA1 was previously reported to co-immunoprecipitate with RNAPII (Scully et al., 1997b), and specifically to bind (Aiyar et al., 2004) to negative elongation factor B (NELF-B, also termed COBRA1), whose release from RNAPII promotes the transition from RNAPII pausing to elongation. Recent work (Zhang et al., 2017) shows that R-loops accumulate preferentially at PPP sites in luminal epithelial cells – but not in other cell types – of non-malignant mammary tissues derived from female carriers of pathogenic BRCA1 mutations. In a murine model for Brca1-associated mammary carcinogenesis, targeted inactivation of NELF-B/COBRA1 was shown to decrease R-loop accumulation and significantly suppress mammary tumourigenesis. Thus, this study reports a first link between the control by BRCA1 of RNAPII transcription elongation and R-loop formation at PPP sites with mammary carcinogenesis.
RNA-DNA hybrid processing in DSB repair
Additional roles ascribed to BRCA1 and BRCA2 in RNA processing may enable DSB repair by HR. Prior studies suggest that DNA breakage triggers the synthesis by RNAPII of damage-induced long non-coding RNAs (dilncRNAs) (Francia et al., 2016; Michelini et al., 2017) or shorter transcripts (Wei et al., 2012) from broken DNA ends, which promote the sensing, signaling and repair of these lesions. Recent evidence (D’Alessandro et al., 2018) suggests that the dilncRNAs anneal with resected ends at DSBs to form RNA-DNA hybrids that are recognized by BRCA1, in turn to assist in the recruitment of BRCA2 and RAD51. Interestingly, BRCA2 engages RNase H2 and recruits this enzyme to DSBs, where it regulates the turnover of RNA-DNA hybrids at the damage sites. Thus, these studies collectively suggest that BRCA1 and BRCA2 may - through mechanisms that remain to be firmly established - promote the repair of DSBs by HR through the processing of DNA-RNA hybrids formed at the lesions.
BRCA1 and oncogenic transcription
Interestingly, emerging evidence suggests that the regulation of RNAPII transcription elongation by BRCA1 may provide insight into the long-standing conundrum as to why BRCA gene inactivation causes cancer susceptibility in particular tissues like the breast. BRCA1 inactivation was found to alter the transcriptional activation of the estrogen receptor (ER)α subunit-encoding ESR1 gene via R-loop formation in the ESR1 enhancer region, altering enhancer-promoter interactions, and decreasing expression of ESR1 and neighbouring gene loci (Chiang et al., 2019). These findings suggest that the normal transcription of genes like ESR1 implicated in the differentiation of mammary luminal epithelial cells requires R-loop suppression by BRCA1, and thus, that pathogenic mutations inactivating BRCA1 may subvert normal differentiation to promote tissue-specific carcinogenesis.
Further new evidence (Herold et al., 2019) speaks to a different role for BRCA1 in tissue-specific carcinogenesis driven by overexpression of the N-MYC oncogenic transcription factor in neuronal and neuroendocrine cells. Recent work indicates that N-MYC-driven transcriptional activation is assisted by the recruitment of BRCA1 to PPP sites in MYC-induced genes, which serves to prevent RNAPII stalling, and enhance transcriptional activation. Interestingly, BRCA1 works in the context of N-MYC-driven transcription to stabilize mRNA de-capping complexes, and suppress R-loop accumulation at PPP sites. These findings raise the possibility that BRCA1 may cooperate more generally with transcription-activating oncogenic factors to drive carcinogenesis in different tissues.
Consistent with this idea, Ewing sarcoma cells also exhibit a dependence on BRCA1 activity to maintain aberrant transcription (Gorthi et al., 2018). These cells express an oncogenic fusion protein in which the N-terminal transactivation domain of the constitutively expressed EWSR1 protein is translocated to the C-terminal DNA binding domain of the rarely expressed FLI1 protein, forming an oncogenic transcription factor. Ewing sarcoma cells were found not only to accumulate R-loops accompanied by enhanced replication stress, but also display impaired HR and sensitivity to the topoisomerase inhibitor, etoposide. Indirect evidence was found to suggest that the increased engagement of BRCA1 by the transcriptional machinery in Ewing sarcoma cells might comprise HR, raising the idea that the differential distribution of BRCA1 to different macromolecular complexes may be a key facet of its physiology.
How do mutations affecting BRCA1 and BRCA2 promote cancer susceptibility?
“Two-hit” tumour suppression by BRCA1 and BRCA2
BRCA1 or BRCA2 mutation carriers typically inherit a single mutant copy in their germline. Early work following the discovery of the BRCA genes suggested that the second, wild-type copy was lost in evolving malignant cells during carcinogenesis (Collins et al., 1995; Gudmundsson et al., 1995; Merajver et al., 1995), such that tumours from mutation carriers consistently exhibit “loss-of-heterozygosity” or LOH, marking genetic inactivation of the wild-type allele. Furthermore, the expression of wild-type BRCA1 in cancers arising in mutation carriers was sometimes found to be silenced by methylation or other epigenetic alterations (Press et al., 2008), even when the wild-type allele was intact. These observations have led to the prevailing belief that the BRCA genes conform to the so-called “two-hit” paradigm for tumour suppression articulated by Knudson (Knudson, 1971), wherein germline inheritance of a monoallelic or heterozygous mutant tumour suppressor gene allele causes tumour formation only when the second copy is inactivated during carcinogenesis through a somatic mutational or epigenetic event.
Niggling but inconclusive evidence has long raised the possibility that heterozygous mutations affecting BRCA2, in particular, might suffice for carcinogenesis, even when the remaining wild-type allele remains actively expressed. For instance, heterogeneous loss of the second allele has been reported in a small group of human BRCA2-linked breast cancer cases (King et al., 2007), and studies on samples from three human pancreatic ductal adenocarcinomas suggested that BRCA2 LOH was dispensable for tumour formation (Goggins et al., 2000). However, the significance and generality of such findings has until recently been difficult to establish.
Mono-allelic BRCA inactivation suffices for carcinogenesis
A clear indication that mono-allelic BRCA2 inactivation suffices for carcinogenesis came from studies on a genetically engineered murine model for tissue-specific carcinogenesis in the pancreas, which were then extended to human samples (Skoulidis et al., 2010). Pancreatic ductal adenocarcinomas arising in mice engineered to express mutant oncogenic KRAS selectively in the pancreas alongside a germline, heterozygous, truncating mutation in Brca2 that recapitulates human pathogenic mutations were found to consistently retain and express a functional, wild-type Brca2 allele. Similar findings were reported in a small collection of pancreatic ductal adenocarcinomas from Icelandic carriers of the human pathogenic truncating mutation, BRCA2999delC (Skoulidis et al., 2010).
Recent results from genomic studies on large collections of cancers arising in human BRCA1 and BRCA2 mutation carriers have provided supporting evidence that a significant fraction of cancers arising in mutation carriers do not undergo LOH, and therefore retain a functional, expressed wild-type copy. For example, in a group of 160 familial breast and ovarian tumors arising in germline carriers of BRCA1 or BRCA2 mutations, retention of the wild-type allele was observed in as many as 46% of BRCA2-mutant breast cancers (Maxwell et al., 2017). Retention of wild-type BRCA2 occurred less frequently - but in a significant fraction (16%) - of BRCA2-mutant ovarian cancers, whereas wild-type BRCA1 retention was overall less frequent, occurring in only 7-10% of ovarian or breast cancers. Notably, cancers retaining the wild-type BRCA allele also retained the capacity for HR as indicated by the lack of a characteristic mutational signature, and were less likely to respond to platinum chemotherapy, together suggesting that the retained wild-type BRCA gene allele remained functional and active (Maxwell et al., 2017).
Consistent with these studies, recent analyses of >10,000 genomes from cancers arising in over 30 different tissues report that as many as 8% of these cancers harbor a pathogenic or likely pathogenic germline mutation affecting one of 21 different genes (Huang et al., 2018). In cancers from different tissues that bear germline BRCA gene mutations, there was no evidence for LOH (either through genetic alterations or at the level of RNA expression) in ~37% of BRCA1-mutant tumors, and in ~53% of BRCA2-mutant tumors, suggesting that a functional copy of the wild-type allele has been retained in a significant fraction. Again, tissue-specific differences were evident, in that the absence of LOH was more frequent in breast than ovarian cancer genomes. Collectively therefore, these studies provide several lines of clinical genetic evidence that the BRCA genes need not always conform to the Knudson “two-hit” paradigm, in that mono-allelic heterozygous mutations affecting BRCA1 or BRCA2 suffice for tumour formation in varying contexts. One implication is that a significant fraction of BRCA-mutant cancers may exhibit primary resistance to targeted therapies such as poly-ADP polymerase inhibitors owing to retention of a wild-type allele. Moreover, secondary resistance to such therapies could also occur through the outgrowth of tumourigenic cells that bear monoallelic BRCA mutations.
Heterozygous BRCA1 or BRCA2 mutations in cancer pathogenesis
How, then, might monoallelic, heterozygous BRCA1 or BRCA2 mutations drive cancer pathogenesis? Chromosomal instability, increased genotoxin sensitivity and defective HR are typically caused by bi-allelic BRCA gene inactivation in murine or human cells, and not by heterozygous truncating mutations (Connor et al., 1997; Patel et al., 1998; Skoulidis et al., 2010). Moreover, organ development and function is grossly normal in genetically engineered mice heterozygous for several mutant BRCA truncating alleles (Connor et al., 1997; Friedman et al., 1998; Ludwig et al., 1997; Sharan et al., 1997; Suzuki et al., 1997), as is homology-directed DNA repair in multiple tissues from heterozygous Brca2-mutant mice (Kass et al., 2016). These observations suggest that many pathogenic BRCA gene truncating alleles induce neither haplo-insufficiency nor trans-dominant loss-of-function, at least for several cellular functions of BRCA1 and BRCA2.
Indeed, primary human mammary epithelial cells or fibroblasts bearing heterozygous BRCA1 gene mutations were found (Pathania et al., 2014) to exhibit normal functions in mediating DSB repair by HR, in enforcing damage-induced cell cycle checkpoints, and in mitotic spindle pole formation. However, when exposed to replication-stressing agents, these cells were defective in the reactivation of stalled forks and in their protection from collapse, accompanied by defective DSB repair by HR. These observations raise the possibility that replication stress may induce genomic instability in heterozygous BRCA1-mutant mammary cells to promote carcinogenesis in BRCA1 mutation carriers.
By contrast (Tan et al., 2017), cell lines engineered to harbor pathogenic monoallelic, heterozygous BRCA2 truncating mutations, as well as mammary epithelial cells from heterozygous carriers of the Icelandic BRCA2999delC founder mutation, exhibit no defect in replication tract stability compared to wild-type cells when challenged with the replication-stressing agent hydroxyurea. Unexpectedly however, in such heterozygous BRCA2 mutant cells, transient exposure to a cellular metabolite and ubiquitous environmental toxin, formaldehyde, was shown to stall and destabilize DNA replication forks, and provoke structural chromosomal aberrations. Formaldehyde was found to selectively deplete BRCA2 protein via proteasomal degradation, through a mechanism that may be independent of ubiquitin recognition by the 19S proteosomal subunit. Only about 30-50 other cellular proteins detected by SWATH-MS were similarly affected, speaking to the selectivity of the mechanism. Heterozygous BRCA2 truncating mutations, by lowering pre-existing BRCA2 protein expression, rendered cells vulnerable to BRCA2 haploinsufficiency induced by transient exposure to formaldehyde or acetaldehyde, an alcohol catabolite. These observations are consistent with a two-step mechanism wherein aldehydes form DNA adducts to induce genome damage and replication stress - whilst also impairing DNA repair and replication reactivation through the selective proteolytic depletion of BRCA2 (Tan et al., 2017). Thus, the genome-damaging and BRCA2-depleting effects of aldehydes may reinforce one another to amplify genome instability.
Intriguingly, emerging evidence suggests that certain forms of genome damage may alter the differentiation of mammary epithelial cells. Human mammary epithelial cells depleted of BRCA1, or exposed to inter-strand DNA-cross linking agents, undergo an epithelial-to-mesenchymal transition implicated in malignant transformation, and exhibit signs of altered differentiation (Wang et al., 2016). BRCA1 has very recently been found to engage NUMB and HES1 in a protein complex that regulates mammary epithelial cell differentiation after DNA damage induced by interstand cross-links (Wang et al., 2019). These findings raise the possibility that spontaneously arising forms of DNA damage induced by endogenous mutagens like aldehydes, or lesions encountered during replication, may initiate tissue-specific carcinogenesis.
Tissue-specific carcinogenesis in BRCA mutation carriers
Why BRCA gene mutations should predispose to cancers in specific tissues, when their physiological functions relevant to tumour suppression seem ubiquitous, remains a major unresolved question. Several recent lines of evidence touched upon in the foregoing open new lines of investigation that may help to address this issue. For example, one important new insight indubitably comes from the accumulating evidence implicating BRCA1 and BRCA2 in transcriptional regulation and R-loop accumulation, since derangements in gene expression are a plausible driver of tissue-specific carcinogenesis because they can induce changes in cell fate and tissue differentiation. Fresh evidence also implicates the genotoxic effects of tissue-specific hormones such as oestrogen active especially in the breast or ovary. Evidence that oestrogen metabolites induce DNA damage is longstanding (reviewed for eg. in, Liehr, 1990); recent evidence now suggests that R-loop formation is a major source of oestrogen-induced genotoxicity (Stork et al., 2016). Another line of enquiry arises from the recent observation that endogenous metabolites like aldehydes, which are turned over differentially through cellular metabolism in different tissues, are able to trigger DNA damage, induce BRCA2 haploinsufficiency accompanied by the outgrowth of genomically unstable progeny (Tan et al., 2017), or even cell death (Tacconi et al., 2017) in BRCA-mutant cells. Aldehyde accumulation may in this way modify tissue-specific cancer progression in BRCA mutation carriers. Finally, results indicating that certain forms of DNA damage – perhaps arising from endogenous metabolites? – may induce altered differentiation in certain BRCA-mutant tissues like the mammary gland (for eg., Wang et al., 2016) again suggest that the intersecting roles of the BRCA proteins in DNA repair, replication reactivation and transcription may conspire to force tissue-specific changes that promote carcinogenesis.
Concluding remarks
In conclusion, I have endeavoured here to briefly summarize emerging evidence that prompts reconsideration of our understanding of how germline mutations affecting BRCA1 or BRCA2 cause tissue-specific carcinogenesis. The essential roles of the BRCA proteins as custodians of chromosome integrity through distinct functions in the assembly and activity of macromolecular complexes that mediate DNA repair, replication reactivation and mitotic progression remain centre-stage, but emerging evidence indicates that new functions for BRCA1 and BRCA2 in the regulation of transcription and RNA processing may powerfully suppress transcription-associated DNA damage to abort events inducing replication stalling or DNA breakage (Figure 2). Moreover, whilst biallelic BRCA gene inactivation has long been thought to induce carcinogenesis in mutation carriers by triggering genomic instability, growing evidence suggests that monoallelic BRCA gene mutations suffice for carcinogenesis in several settings. A significant fraction of cancers arising in BRCA-mutation carriers may therefore exhibit resistance to new therapies whose therapeutic index depends on the idea that biallelic BRCA inactivation is necessary for carcinogenesis. Finally, emerging evidence raises several fresh lines of enquiry concerning tissue-specific cancer evolution in BRCA mutation carriers. Collectively, these insights engender new models summarized here which help to explain how BRCA gene mutations cause cancer susceptibility in specific tissues.
Acknowledgements
I thank members of my laboratory for stimulating discussions, and Drs. Miyoung Lee and Xavier Renaudin for critical comments on this manuscript. Work in my laboratory is funded by Medical Research Council (MRC) Programme grants MC_UU_12022/1 and MC_UU_12022/8.
Abbreviations
- BRCA1
Breast cancer susceptibility gene 1
- BRCA2
Breast cancer susceptibility gene 2
- RING
domainreally interesting new gene domain
- OB fold
oligonucleotide / oligosaccharide binding fold
- BARD1
BRCA1-associated RING domain protein 1
Footnotes
Conflict of interest statement
I declare no conflict of interest concerning the material discussed in this paper.
References
- Aguilera A, García-Muse T. R Loops: From Transcription Byproducts to Threats to Genome Stability. Mol Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Aiyar SE, Sun J, Blair AL, Moskaluk CA, Lu Y, Ye Q-N, Yamaguchi Y, Mukherjee A, Ren D, Handa H, et al. Attenuation of estrogen receptor alpha-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes Dev. 2004;18:2134–2146. doi: 10.1101/gad.1214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anczuków O, Ware MD, Buisson M, Zetoune AB, Stoppa-Lyonnet D, Sinilnikova OM, Mazoyer S. Does the nonsense-mediated mRNA decay mechanism prevent the synthesis of truncated BRCA1, CHK2, and p53 proteins? Hum Mutat. 2008;29:65–73. doi: 10.1002/humu.20590. [DOI] [PubMed] [Google Scholar]
- Bhatia V, Barroso SI, Garcia-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014;511:362–365. doi: 10.1038/nature13374. [DOI] [PubMed] [Google Scholar]
- Billing D, Horiguchi M, Wu-Baer F, Taglialatela A, Leuzzi G, Nanez SA, Jiang W, Zha S, Szabolcs M, Lin C-S, et al. The BRCT Domains of the BRCA1 and BARD1 Tumor Suppressors Differentially Regulate Homology-Directed Repair and Stalled Fork Protection. Mol Cell. 2018;72:127–139.e8. doi: 10.1016/j.molcel.2018.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breast Cancer Linkage Consortium. Cancer risks in BRCA2 mutation carriers. J Natl Cancer Inst. 1999;91:1310–1316. doi: 10.1093/jnci/91.15.1310. [DOI] [PubMed] [Google Scholar]
- Chen J, Silver DP, Walpita D, Cantor SB, Gazdar AF, Tomlinson G, Couch FJ, Weber BL, Ashley T, Livingston DM, et al. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell. 1998a;2:317–328. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- Chen PL, Chen CF, Chen Y, Xiao J, Sharp ZD, Lee WH. The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc Natl Acad Sci U A. 1998b;95:5287–5292. doi: 10.1073/pnas.95.9.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang H-C, Zhang X, Li J, Zhao X, Chen J, Wang HT-H, Jatoi I, Brenner A, Hu Y, Li R. BRCA1-associated R-loop affects transcription and differentiation in breast luminal epithelial cells. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Liao RG, Parsons MT, Paten B, Alquaddoomi F, Antoniou A, Baxter S, Brody L, Cook-Deegan R, Coffin A, et al. BRCA Challenge: BRCA Exchange as a global resource for variants in BRCA1 and BRCA2. PLoS Genet. 2018;14:e1007752. doi: 10.1371/journal.pgen.1007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins N, McManus R, Wooster R, Mangion J, Seal S, Lakhani SR, Ormiston W, Daly PA, Ford D, Easton DF, et al. Consistent loss of the wild type allele in breast cancers from a family linked to the BRCA2 gene on chromosome 13q12-13. Oncogene. 1995;10:1673–1675. [PubMed] [Google Scholar]
- Connor F, Bertwistle D, Mee PJ, Ross GM, Swift S, Grigorieva E, Tybulewicz VL, Ashworth A. Tumorigenesis and a DNA repair defect in mice with a truncating Brca2 mutation. Nat Genet. 1997;17:423–430. doi: 10.1038/ng1297-423. [DOI] [PubMed] [Google Scholar]
- D’Alessandro G, Whelan DR, Howard SM, Vitelli V, Renaudin X, Adamowicz M, Iannelli F, Jones-Weinert CW, Lee M, Matti V, et al. BRCA2 controls DNA:RNA hybrid level at DSBs by mediating RNase H2 recruitment. Nat Commun. 2018;9:5376. doi: 10.1038/s41467-018-07799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dungrawala H, Bhat KP, Le Meur R, Chazin WJ, Ding X, Sharan SK, Wessel SR, Sathe AA, Zhao R, Cortez D. RADX Promotes Genome Stability and Modulates Chemosensitivity by Regulating RAD51 at Replication Forks. Mol Cell. 2017;67:374–386.e5. doi: 10.1016/j.molcel.2017.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay GM, Daza RM, Martin B, Zhang MD, Leith AP, Gasperini M, Janizek JD, Huang X, Starita LM, Shendure J. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562:217. doi: 10.1038/s41586-018-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia S, Cabrini M, Matti V, Oldani A, d’Adda di Fagagna F. DICER, DROSHA and DNA damage response RNAs are necessary for the secondary recruitment of DNA damage response factors. J Cell Sci. 2016;129:1468–1476. doi: 10.1242/jcs.182188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman LS, Thistlethwaite FC, Patel KJ, Yu VP, Lee H, Venkitaraman AR, Abel KJ, Carlton MB, Hunter SM, Colledge WH, et al. Thymic lymphomas in mice with a truncating mutation in Brca2. Cancer Res. 1998;58:1338–1343. [PubMed] [Google Scholar]
- Fugger K, Mistrik M, Neelsen KJ, Yao Q, Zellweger R, Kousholt AN, Haahr P, Chu WK, Bartek J, Lopes M, et al. FBH1 Catalyzes Regression of Stalled Replication Forks. Cell Rep. 2015;10:1749–1757. doi: 10.1016/j.celrep.2015.02.028. [DOI] [PubMed] [Google Scholar]
- García-Rubio ML, Pérez-Calero C, Barroso SI, Tumini E, Herrera-Moyano E, Rosado IV, Aguilera A. The Fanconi Anemia Pathway Protects Genome Integrity from R-loops. PLoS Genet. 2015;11:e1005674. doi: 10.1371/journal.pgen.1005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goggins M, Hruban RH, Kern SE. BRCA2 is inactivated late in the development of pancreatic intraepithelial neoplasia: evidence and implications. Am J Pathol. 2000;156:1767–1771. doi: 10.1016/S0002-9440(10)65047-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorthi A, Romero JC, Loranc E, Cao L, Lawrence LA, Goodale E, Iniguez AB, Bernard X, Masamsetti VP, Roston S, et al. EWS-FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma. Nature. 2018;555:387–391. doi: 10.1038/nature25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson J, Johannesdottir G, Bergthorsson JT, Arason A, Ingvarsson S, Egilsson V, Barkardottir RB. Different tumor types from BRCA2 carriers show wild-type chromosome deletions on 13q12-q13. Cancer Res. 1995;55:4830–4832. [PubMed] [Google Scholar]
- Hakem R, de la Pompa JL, Sirard C, Mo R, Woo M, Hakem A, Wakeham A, Potter J, Reitmair A, Billia F, et al. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell. 1996;85:1009–1023. doi: 10.1016/s0092-8674(00)81302-1. [DOI] [PubMed] [Google Scholar]
- Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, Dimitrov S, Pathania S, McKinney KM, Eaton ML, et al. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol Cell. 2015;57:636–647. doi: 10.1016/j.molcel.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S, Kalb J, Büchel G, Ade CP, Baluapuri A, Xu J, Koster J, Solvie D, Carstensen A, Klotz C, et al. Recruitment of BRCA1 limits MYCN-driven accumulation of stalled RNA polymerase. Nature. 2019;567:545–549. doi: 10.1038/s41586-019-1030-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SJ, Rolland T, Adelmant G, Xia X, Owen MS, Dricot A, Zack TI, Sahni N, Jacob Y, Hao T, et al. Systematic screening reveals a role for BRCA1 in the response to transcription-associated DNA damage. Genes Dev. 2014;28:1957–1975. doi: 10.1101/gad.241620.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hout CVV, Tachmazidou I, Backman JD, Hoffman JX, Ye B, Pandey AK, Gonzaga-Jauregui C, Khalid S, Liu D, Banerjee N, et al. Whole exome sequencing and characterization of coding variation in 49,960 individuals in the UK Biobank. BioRxiv. 2019 doi: 10.1038/s41586-020-2853-0. 572347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–9. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- Huang K-L, Mashl RJ, Wu Y, Ritter DI, Wang J, Oh C, Paczkowska M, Reynolds S, Wyczalkowski MA, Oak N, et al. Pathogenic Germline Variants in 10,389 Adult Cancers. Cell. 2018;173:355–370.e14. doi: 10.1016/j.cell.2018.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TA, Li W, Brogi E, Yee CJ, Gemignani ML, Olvera N, Levine DA, Norton L, Robson ME, Offit K, et al. Heterogenic loss of the wild-type. In BRCA allele in human breast tumorigenesis. Ann Surg Oncol. 2007;14:2510. doi: 10.1245/s10434-007-9372-1. [DOI] [PubMed] [Google Scholar]
- Knudson AG. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolinjivadi AM, Sannino V, De Antoni A, Zadorozhny K, Kilkenny M, Técher H, Baldi G, Shen R, Ciccia A, Pellegrini L, et al. Smarcal1-Mediated Fork Reversal Triggers Mre11-Dependent Degradation of Nascent DNA in the Absence of Brca2 and Stable Rad51 Nucleofilaments. Mol Cell. 2017;67:867–881.e7. doi: 10.1016/j.molcel.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips K-A, Mooij TM, Roos-Blom M-J, Jervis S, van Leeuwen FE, Milne RL, Andrieu N, et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA. 2017;317:2402–2416. doi: 10.1001/jama.2017.7112. [DOI] [PubMed] [Google Scholar]
- Lee M, Daniels MJ, Garnett MJ, Venkitaraman AR. A mitotic function for the high-mobility group protein HMG20b regulated by its interaction with the BRC repeats of the BRCA2 tumor suppressor. Oncogene. 2011;30:3360. doi: 10.1038/onc.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaçon D, Jackson J, Quinet A, Brickner JR, Li S, Yazinski S, You Z, Ira G, Zou L, Mosammaparast N, et al. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat Commun. 2017;8:860. doi: 10.1038/s41467-017-01180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liehr JG. Genotoxic effects of estrogens. Mutat Res. 1990;238:269–276. doi: 10.1016/0165-1110(90)90018-7. [DOI] [PubMed] [Google Scholar]
- Liu CY, Flesken-Nikitin A, Li S, Zeng Y, Lee WH. Inactivation of the mouse Brca1 gene leads to failure in the morphogenesis of the egg cylinder in early postimplantation development. Genes Dev. 1996;10:1835–1843. doi: 10.1101/gad.10.14.1835. [DOI] [PubMed] [Google Scholar]
- Lomonosov M, Anand S, Sangrithi M, Davies R, Venkitaraman AR. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev. 2003;17:3017–3022. doi: 10.1101/gad.279003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez JS, von Nicolai C, Kim T, Ehlén Å, Mazin AV, Kowalczykowski SC, Carreira A. BRCA2 regulates DMC1-mediated recombination through the BRC repeats. Proc Natl Acad Sci U S A. 2016;113:3515–3520. doi: 10.1073/pnas.1601691113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell KN, Wubbenhorst B, Wenz BM, De Sloover D, Pluta J, Emery L, Barrett A, Kraya AA, Anastopoulos IN, Yu S, et al. BRCA locus-specific loss of heterozygosity in germline BRCA1 and BRCA2 carriers. Nat Commun. 2017;8:319. doi: 10.1038/s41467-017-00388-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merajver SD, Frank TS, Xu J, Pham TM, Calzone KA, Bennett-Baker P, Chamberlain J, Boyd J, Garber JE, Collins FS. Germline BRCA1 mutations and loss of the wild-type allele in tumors from families with early onset breast and ovarian cancer. Clin Cancer Res Off J Am Assoc Cancer Res. 1995;1:539–544. [PubMed] [Google Scholar]
- Michelini F, Pitchiaya S, Vitelli V, Sharma S, Gioia U, Pessina F, Cabrini M, Wang Y, Capozzo I, Iannelli F, et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat Cell Biol. 2017;19:1400–1411. doi: 10.1038/ncb3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijic S, Zellweger R, Chappidi N, Berti M, Jacobs K, Mutreja K, Ursich S, Chaudhuri AR, Nussenzweig A, Janscak P, et al. Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat Commun. 2017;8:859. doi: 10.1038/s41467-017-01164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta R, LaSalle JM, Cheng HL, Shinohara A, Ogawa H, Copeland N, Jenkins NA, Lalande M, Alt FW. RAB22 and RAB163/mouse BRCA2: proteins that specifically interact with the RAD51 protein. Proc Natl Acad Sci U A. 1997;94:6927–6932. doi: 10.1073/pnas.94.13.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- Nathanson KN, Wooster R, Weber BL. Breast cancer genetics: what we know and what we need. Nat Med. 2001;7:552–6. doi: 10.1038/87876. [DOI] [PubMed] [Google Scholar]
- von Nicolai C, Ehlén Å, Martin C, Zhang X, Carreira A. A second DNA binding site in human BRCA2 promotes homologous recombination. Nat Commun. 2016;7 doi: 10.1038/ncomms12813. 12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel KJ, Yu VP, Lee H, Corcoran A, Thistlethwaite FC, Evans MJ, Colledge WH, Friedman LS, Ponder BA, Venkitaraman AR. Involvement of Brca2 in DNA repair. Mol Cell. 1998;1:347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- Pathania S, Bade S, Le Guillou M, Burke K, Reed R, Bowman-Colin C, Su Y, Ting DT, Polyak K, Richardson AL, et al. BRCA1 haploinsufficiency for replication stress suppression in primary cells. Nat Commun. 2014;5:5496–5496. doi: 10.1038/ncomms6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Cong K, Panzarino NJ, Nayak S, Calvo J, Deng B, Zhu LJ, Morocz M, Hegedus L, Haracska L, et al. Opposing Roles of FANCJ and HLTF Protect Forks and Restrain Replication during Stress. Cell Rep. 2018;24:3251–3261. doi: 10.1016/j.celrep.2018.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press JZ, De Luca A, Boyd N, Young S, Troussard A, Ridge Y, Kaurah P, Kalloger SE, Blood KA, Smith M, et al. Ovarian carcinomas with genetic and epigenetic BRCA1 loss have distinct molecular abnormalities. BMC Cancer. 2008;8:17. doi: 10.1186/1471-2407-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman N, Stratton MR. The genetics of breast cancer susceptibility. Annu Rev Genet. 1998;32:95–121. doi: 10.1146/annurev.genet.32.1.95. [DOI] [PubMed] [Google Scholar]
- Rajagopalan S, Andreeva A, Rutherford TJ, Fersht AR. Mapping the physical and functional interactions between the tumor suppressors p53 and BRCA2. Proc Natl Acad Sci U S A. 2010;107:8587–8592. doi: 10.1073/pnas.1003689107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbeck TR, Mitra N, Wan F, Sinilnikova OM, Healey S, McGuffog L, Mazoyer S, Chenevix-Trench G, Easton DF, Antoniou AC, et al. Association of Type and Location of BRCA1 and BRCA2 Mutations With Risk of Breast and Ovarian Cancer. JAMA. 2015;313:1347–1361. doi: 10.1001/jama.2014.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondinelli B, Gogola E, Yücel H, Duarte AA, van de Ven M, van der Sluijs R, Konstantinopoulos PA, Jonkers J, Ceccaldi R, Rottenberg S, et al. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat Cell Biol. 2017;19:1371–1378. doi: 10.1038/ncb3626. [DOI] [PubMed] [Google Scholar]
- Sawyer SL, Tian L, Kähkönen M, Schwartzentruber J, Kircher M, University of Washington Centre for Mendelian Genomics, FORGE Canada Consortium. Majewski J, Dyment DA, Innes AM, et al. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov. 2015;5:135–142. doi: 10.1158/2159-8290.CD-14-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. A Distinct Replication Fork Protection Pathway Connects Fanconi Anemia Tumor Suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R, Livingston DM. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 2000;408:429–432. doi: 10.1038/35044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997a;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- Scully R, Anderson SF, Chao DM, Wei W, Ye L, Young RA, Livingston DM, Parvin JD. BRCA1 is a component of the RNA polymerase II holoenzyme. Proc Natl Acad Sci U S A. 1997b;94:5605–5610. doi: 10.1073/pnas.94.11.5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo A, Steinberg-Shemer O, Unal S, Casadei S, Walsh T, Gumruk F, Shalev S, Shimamura A, Akarsu NA, Tamary H, et al. Mechanism for survival of homozygous nonsense mutations in the tumor suppressor gene BRCA1. Proc Natl Acad Sci U S A. 2018;115:5241–5246. doi: 10.1073/pnas.1801796115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- Shen SX, Weaver Z, Xu X, Li C, Weinstein M, Chen L, Guan XY, Ried T, Deng CX. A targeted disruption of the murine Brca1 gene causes gamma-irradiation hypersensitivity and genetic instability. Oncogene. 1998;17:3115. doi: 10.1038/sj.onc.1202243. [DOI] [PubMed] [Google Scholar]
- Shivji MKK, Renaudin X, Williams CH, Venkitaraman AR. BRCA2 Regulates Transcription Elongation by RNA Polymerase II to Prevent R-Loop Accumulation. Cell Rep. 2018;22:1031–1039. doi: 10.1016/j.celrep.2017.12.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoulidis F, Cassidy LD, Pisupati V, Jonasson JG, Bjarnason H, Eyfjord JE, Karreth FA, Lim M, Barber LM, Clatworthy SA, et al. Brca2 heterozygosity promotes Kras(G12D) -driven carcinogenesis in a murine model of familial pancreatic cancer. Cancer Cell. 2010;18:499. doi: 10.1016/j.ccr.2010.10.015. [DOI] [PubMed] [Google Scholar]
- Sollier J, Stork CT, Garcia-Rubio MaL, Paulsen RD, Aguilera As, Cimprich Ka. Transcription-Coupled Nucleotide Excision Repair Factors Promote R-Loop-Induced Genome Instability. Mol Cell. 2014;56:777–785. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork CT, Bocek M, Crossley MP, Sollier J, Sanz La, Chédin F, Swigut T, Cimprich Ka. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. ELife. 2016;5:e17548–e17548. doi: 10.7554/eLife.17548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci U A. 2009;106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacconi EM, Lai X, Folio C, Porru M, Zonderland G, Badie S, Michl J, Sechi I, Rogier M, Matía García V, et al. BRCA1 and BRCA2 tumor suppressors protect against endogenous acetaldehyde toxicity. EMBO Mol Med. 2017;9:1398–1414. doi: 10.15252/emmm.201607446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taglialatela A, Alvarez S, Leuzzi G, Sannino V, Ranjha L, Huang J-W, Madubata C, Anand R, Levy B, Rabadan R, et al. Restoration of Replication Fork Stability in BRCA1- and BRCA2-Deficient Cells by Inactivation of SNF2-Family Fork Remodelers. Mol Cell. 2017;68:414–430.e8. doi: 10.1016/j.molcel.2017.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SLW, Chadha S, Liu Y, Gabasova E, Perera D, Ahmed K, Constantinou S, Renaudin X, Lee M, Aebersold R, et al. A Class of Environmental and Endogenous Toxins Induces BRCA2 Haploinsufficiency and Genome Instability. Cell. 2017;169:1105–1118.e15. doi: 10.1016/j.cell.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D, Easton DF. Cancer Incidence in BRCA1 mutation carriers. J Natl Cancer Inst. 2002;94:1358–1365. doi: 10.1093/jnci/94.18.1358. [DOI] [PubMed] [Google Scholar]
- Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- Venkitaraman AR. Cancer suppression by the chromosome custodians, BRCA1 and BRCA2. Science. 2014;343:1470–1475. doi: 10.1126/science.1252230. [DOI] [PubMed] [Google Scholar]
- Vujanovic M, Krietsch J, Raso MC, Terraneo N, Zellweger R, Schmid JA, Taglialatela A, Huang J-W, Holland CL, Zwicky K, et al. Replication Fork Slowing and Reversal upon DNA Damage Require PCNA Polyubiquitination and ZRANB3 DNA Translocase Activity. Mol Cell. 2017;67:882–890.e5. doi: 10.1016/j.molcel.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Bierie B, Li AG, Pathania S, Toomire K, Dimitrov SD, Liu B, Gelman R, Giobbie-Hurder A, Feunteun J, et al. BRCA1/FANCD2/BRG1-Driven DNA Repair Stabilizes the Differentiation State of Human Mammary Epithelial Cells. Mol Cell. 2016;63:277–292. doi: 10.1016/j.molcel.2016.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Xiang Liu DB, He A, Randle HJ, Zhang KX, Dongre A, Sachs N, Clar AP, Tao L, et al. Inadequate DNA damage repair promotes mammary transdifferentiation leading to BRCA1 breast cancer. Cell. 2019;178:135–151. doi: 10.1016/j.cell.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Rendtlew Danielsen JM, Yang Y-G, Qi Y. A Role for Small RNAs in DNA Double-Strand Break Repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Welcsh PL, King MC. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum Mol Genet. 2001;10:705–713. doi: 10.1093/hmg/10.7.705. [DOI] [PubMed] [Google Scholar]
- Willis NA, Frock RL, Menghi F, Duffey EE, Panday A, Camacho V, Hasty EP, Liu ET, Alt FW, Scully R. Mechanism of tandem duplication formation in BRCA1-mutant cells. Nature. 2017;551:590–595. doi: 10.1038/nature24477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J Biol Chem. 1997;272:31941–31944. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thomä NH, Zheng N, Chen P-L, Lee W-H, Pavletich NP. BRCA2 Function in DNA Binding and Recombination from a BRCA2-DSS1-ssDNA Structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- Zhang F, Ma J, Wu J, Ye L, Cai H, Xia B, Yu X. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19:524. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chiang H-C, Wang Y, Zhang C, Smith S, Zhao X, Nair SJ, Michalek J, Jatoi I, Lautner M, et al. Attenuation of RNA polymerase II pausing mitigates BRCA1-associated R-loop accumulation and tumorigenesis. Nat Commun. 2017;8 doi: 10.1038/ncomms15908. 15908. [DOI] [PMC free article] [PubMed] [Google Scholar]