Abstract
Spatial and molecular characteristics determine tissue function, yet high-resolution methods to capture both concurrently are lacking. Here, we developed High-Definition Spatial Transcriptomics (HDST), which captures RNA from histological tissue sections on a dense spatially barcoded bead array. Each experiment recovers several hundred thousand transcript-coupled spatial barcodes at 2μm resolution, as demonstrated in mouse brain and primary breast cancer. This opens the way to high-resolution spatial analysis of cells and tissues.
Editorial summary
A dense, spatially-barcoded bead array captures RNA from histological tissue sections for spatially-resolved gene expression analysis.
Charting cells’ spatial organization and molecular features is essential to understand how they interact in both normal and diseased tissues1,2. Massively parallel single-cell RNA-Seq (scRNA-Seq)3,4 can profile hundreds of thousands of dissociated individual cells, but does not retain their spatial position, and can introduce biases in cell recovery5. Conversely, spatial profiling captures detailed positional information in intact tissue, but current methods require pre-selected markers, rely on non-standard instrumentation6–11, or have limited spatial resolution, scalability, or applicability. In particular, Spatial Transcriptomics12 (ST) is a spatially barcoded RNA-Seq method providing transcriptome-wide coverage in many systems, but at a resolution of 100μm (3–30 cells).
To bridge this gap, we developed High-Definition Spatial Transcriptomics (HDST, Fig. 1a) and demonstrate its application to large tissue areas in the mouse brain and human tumors in situ. In HDST, we deposit barcoded poly(d)T oligonucleotides into 2μm wells with a randomly ordered bead array-based fabrication process13 and decode their positions by a sequential hybridization and error-correcting strategy13,14. After a frozen tissue section is placed on the decoded slide, stained and imaged, RNA is captured and then profiled by RNA-Seq.
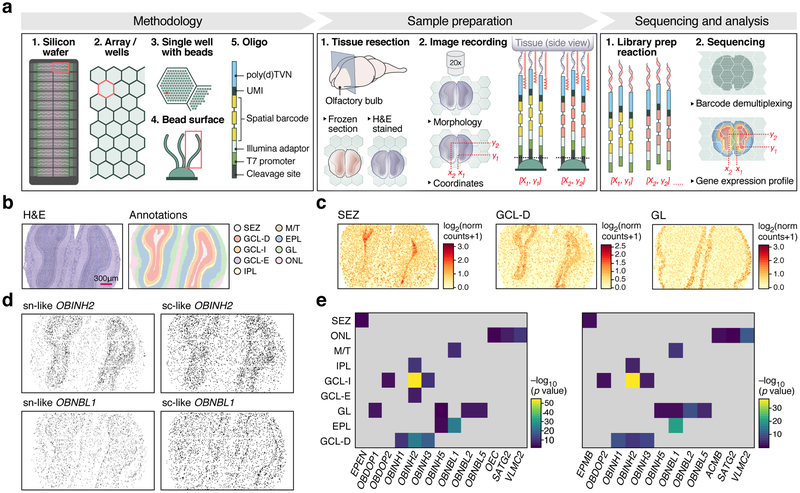
Figure 1. High-definition spatial transcriptomics (HDST).
(a) HDST workflow. (b) Labeling of morphological layers. HDST H&E image of a main olfactory bulb and matching HDST (x,y) barcodes annotated into 9 morphological areas. (c) Layer-specific DE patterns in HDST. Shown is the summed normalized expression of positively enriched signature genes significantly (FDR<0.1, two-sided t-test) associated with each layer as annotated in (b). (d, e) Nuclei segmentation and binning of HDST as in (b). (d) Segmented nuclei (“sn-like”) and lightly binned (“sc-like”) spatial barcodes assigned (black) to each of two cell types as in (b). (e) Enrichment of sn- and sc-like spatial barcodes with assigned cell types (columns) to morphological layers (rows) as in (b). −log10(p-value) (one-sided Fisher’s exact test, Bonferroni adjusted, p-value<0.01) represents the color bar and grey tiles non-significant values.
To produce a high-resolution, high-density bead array, we generated 2,839,865 individual barcoded beads with a split-and-pool approach (Supplementary Fig. 1a), randomly placed them into a hexagonal array of >1.4M 2μm wells, and then decoded each bead’s location (Fig. 1a)13,14 with several hybridization rounds. Each round hybridizes a set of complementary and labeled decoder oligonucleotides (”decoders”) (Methods), records fluorescence across the entire slide area, and then strips the decoders. The process is repeated log3N times (14 times for the array presented here), where N is the number of sequences to be decoded with 3 labels used (Supplementary Fig. 1b). In this way, each bead and barcode receives a unique spatial color address14 creating a HDST array in ~3h total processing time.
To test HDST, we first profiled the main olfactory bulb (MOB) of the mouse brain, whose neurons have traditionally been defined by the presence of neuronal cell bodies across morphological layers15. We assessed whether HDST molecular data can be related to layers and other histological features. We analyzed three replicate sections by HDST and tested its performance in two key tasks: (1) generating high-resolution spatial expression patterns of individual genes, and (2) detecting cell types and assigning them to correct, high-resolution positions.
We confirmed that RNA capture was specific and in overall agreement with bulk RNA-Seq controls, despite the relatively low number of transcripts captured per spatial barcode. We first accounted for barcode redundancy (“clashing”), decoding efficiency and stringent barcode demultiplexing (Supplementary Fig. 2a, Supplementary Table 1). Next, we observed that at saturating sequencing depth (Supplementary Fig. 2b), 85.6±3.3% of all genes detected were located within the area physically covered by the tissue specimen (without using any lower cutoffs), with almost 160,000 barcodes generating spatially-mapped transcripts per assay (n=3 sections, Supplementary Fig. 2c). Although there were few UMIs per barcode location (7.1±6.0 (mean±sd), n=3 sections), a very distinct spatial in situ profile followed the tissue boundary (Supplementary Fig. 2d,e), suggesting detection specificity. Moreover, a combined “bulk” expression profile of each HDST dataset correlated significantly with published MOB bulk RNA-Seq (Supplementary Fig. 3a; Spearman’s ρ = 0.69±0.02; mean±sd) and across the three replicate experiments (Spearman’s ρ = 0.82±0.06; mean±sd). Most detected genes agreed between the bulk and HDST datasets (Supplementary Fig. 3b). Finally, comparing the HDST capture per bead to that observed from smFISH for 3 genes (Penk, Slc17a7 and Fabp7), we estimated HDST capture efficiency at 1.3% per bead (Methods, Supplementary Table 2).
Next, supervised analysis of HDST data correctly identified layer-specific expression signatures. We first annotated morphological layers (Methods) from the H&E stain of each specimen (Fig. 1b). We reasoned that 24 neighboring wells (r ~ 6.5μm) will likely capture transcripts from the same cell. We thus enhanced the signal by light binning, which pooled reads within a short range (e.g., 5X compared to the 5×5 hexagonal wells). On average, each 5X “enhanced” bin had observations from 5.6±2.7 (mean±sd, n=3 sections) (x,y) decoded beads and 44.4±30.6 (mean±sd, n=3 sections) UMIs. Finally, we assigned each “enhanced” bin to a layer, to robustly identify differentially expressed (DE) genes between morphological layers (Methods). Following a smoothing Gaussian filter on the binned data (63.5±38.6 (mean±sd) UMIs per bin, n=3 sections), we performed a two-sided t-test (FDR<0.1), identifying DE signatures specific to morphological layers (Fig. 1c, Supplementary Table 3, Supplementary Fig. 4). Layer-enriched upregulated DE genes (FDR<0.05; log2 fold change>1.5) were correctly assigned, as assessed by comparing their average and individual signatures to their in situ hybridization (ISH) score from the Allen Brain Atlas (ABA)16 (Supplementary Fig. 5).
To test spatial assignment of cell types, we developed a multinomial naïve Bayes classifier to map the sparse high-resolution HDST data to cell type annotations by integration with scRNA-Seq (Methods). We first used scRNA-Seq UMI counts17 to compute the maximum likelihood estimates of the multinomial parameters for each cell type (Supplementary Table 4). We then estimated the likelihood that an expression profile of a given HDST barcode originated from a scRNA-Seq cell type and, using posterior probabilities, assigned cell types to barcode locations (Supplementary Table 5).
Approximately 49.4±15.9% (mean±sd, n=3 sections) of spatially barcoded HDST (1X) profiles were confidently assigned to a single cell type. We then leveraged the matched H&E images in HDST to segment single cell nuclei based on the nuclear stain (Methods), related beads within nuclei and then used this aggregated expression information to also perform cell typing. To estimate our cell assignment’s sensitivity to read depth and spatial resolution, we decreased the resolution using segmenting and binning (Fig. 1d,e) and compared the assigned data to a ST dataset12 (Supplementary Fig. 6a). The posterior probabilities of cell type assignments increased in the aggregated data (Supplementary Fig. 6b,c, Supplementary Table 6), with a cell type confidently predicted in 58.1±5.3% (mean±sd, n=3 sections) of segmented and 61.3±3.7% (mean±sd, n=3 sections) of all (x,y) positions (Supplementary Fig. 6d), compared to 0.4% of (x,y) positions in ST data. DE markers drove the assignment task (Supplementary Fig. 6e, Supplementary Table 7).
Collecting H&E stains jointly with HDST data allowed us to further relate high-resolution barcodes to sub-cellular features. To demonstrate this, we performed nuclear segmentation and identified transcripts with preferential nuclear localization, by comparing RNAs associated with barcodes within or outside of segmented nuclei (Supplementary Fig. 6e,f, Supplementary Table 8). Most of the 186 genes identified as nucleus specific by both HDST and single-nucleus RNA-seq (Methods) were protein-coding. Furthermore, HDST barcodes overlapping within segmented nuclei showed significantly higher (p<0.05, one-sided unpaired Welch’s t-test) ratios of intronic vs. exonic reads. This analysis can be extended to other sub-cellular features imaged with dedicated stains (e.g., dendrites).
We related spatially assigned cell types to morphological layers (Fig. 1d,e, Methods), finding layer specific patterns for 15 of 63 tested cell types, typically consistent between segments and lightly binned data. For example, a neuroblast population (OBNLB1) was enriched in the mitral (M/T) and external (EPL) plexiform layers, inhibitory neurons (OBINH1–3) in the deep granular zone (GCL-D), astroependymal cells (EPMB and EPEN) in the subependymal zone (SEZ), and olfactory ensheathing cells (OEC), vascular cells (VLMC2) and satellite glia (SATG2) in the olfactory nerve layer (ONL). GABAergic (OBNBL5), dopaminergic periglomerular (OBDOP1) and VGLUT1/2 (OBNLB2) neuroblasts were found in the glomerular layer (GL). Many of these associations and classifications have previously been reported15,17, with inhibitory neurons (OBINH2) dominating the granular (GCL-E, GCL-I and GCL-D) and internal plexiform (IPL) layers.
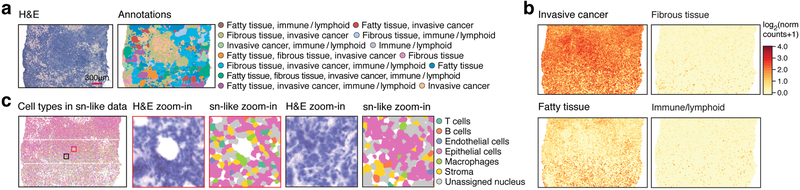
Relating histopathology and transcriptional profiles could help improve our understanding of disease biology and patient diagnosis and treatment. We assessed HDST’s clinical potential in a tumor section from a histological grade 3 breast HER2+ cancer patient (Fig. 2a, Methods). We annotated clinically relevant morphological features in an H&E stain, and performed segmentation, differential expression analysis and cell typing, leveraging published auxiliary breast cancer scRNA-seq18 (Supplementary Table 9). DE genes between morphological areas (Fig. 2b, Supplementary Fig. 7a,b) using smoothed and binned data (38.03±23.6 UMIs per bin containing 6.1±3.1 beads at 91% library saturation) revealed that invasive cancer-specific areas were high in KRT19 and ERBB2, as expected, but also in TMSB10, a marker promoting migration of breast cancer cells19 (Supplementary Tables 10–11). A single cell type could be assigned to 59.8% of segments and 75.5% of bins driven by DE genes (Fig. 2c, Supplementary Fig. 7c, Supplementary Tables 12–13).
Figure 2. HDST distinguishes cell types and niches in a breast cancer resection.
(a) Labeling of morphological layers. HDST H&E image (left) of a breast cancer section and matching HDST (x,y) barcodes annotated into 13 morphological areas (right, color code). (b) Layer-specific spatial DE patterns in HDST. Summed normalized expression of positively enriched signature genes significantly (FDR<0.1, two-sided t-test) associated with each layer as in (a). (c) Cell type assignments by single nuclei as in (a). Two zoomed in regions (black and red squares) with H&E and color coded segments.
In conclusion, HDST is a high-definition method to measure in situ spatial information, at 1,400-fold higher resolution than ST, in healthy and pathological tissue. HDST is readily deployable as it relies on robust and commoditized tissue, molecular, bead-array and imaging modular tasks. Recently, Slide-Seq, a spatial RNAseq method with comparably low capture rates was developed20 with related features. However, Slide-Seq does not include histology, provides 25X lower resolution than HDST and has a higher rate of measurements confounded by signals from multiple cells. While HDST data is currently relatively sparse, signals are highly specific and interpretable by computational integration with morphological features and single-cell profiles. Further HDST development will improve understanding of tissue organization and function in health and disease.
Online Methods
Bead production
We used a split-and-pool approach to generate a total of 2,893,865 different quality controlled 2 μm silica beads. A primer precursor containing (1) the T7 promoter, (2) an Illumina sequencing handle, (3) a 15 bp “Spatial barcode pool” and (4) a 7 bp “bridge” oligonucleotide sequence (/AmC6/UUUUUGACTCGTAATACGACTCACTATAGGGACACGACGCTCTTCCGATCT-Spatial_barcode_Pool1-Bridge1) (IDT) were linked to the bead surface using amine chemistry14. To increase the bead pool size, we pursued two additional ligation steps adding 14 bp and 15 bp pools of spatial barcode sequences eg. /Pho/Bridge2-Spatial_barcode_Pool2-Bridge3 was ligated to Bridge1 through a complementary Bridge1’Bridge2’ 14 bp helper sequence. T4 DNA ligase (NEB) following the manufacturer’s protocol was used to couple the second spatial oligonucleotide construct, which was added in a 2:1 ratio to the precursor oligonucleotide. In the second ligation, the ligated sequence ending with the Bridge3 sequence acted as the precursor for the next spatial barcode pool (/Pho/Bridge4-Spatial_barcode_Pool3 ligated through Bridge3’Bridge4’). Spatial_barcode_Pool3 was followed by a 5 bp unique molecular identifier and 20 poly(d)TVN. The ligated Bridge1Bridge2 sequences read GACTTGTCTAGAGC and Bridge3Bridge4 TGATGCCACACTACTC. All sequences used in the split-and-pool ligation steps (except the first precursor oligonucleotide containing the Spatial_barcode_Pool1) were synthesized on Illumina’s “Big Bird” high-throughput oligonucleotide synthesis platform using phosphoramidite synthesis chemistry.
Array generation
The complete bead pool was used to load a total of 1,467,270 individual hexagonal wells covering a 13.7mm2 area (5.7mm × 2.4mm). The wells were etched using a weak acid in a planar silica slide and polished to 1μm height. A total of 24 such areas were made on each slide. The bead pool (~120mg) was loaded in ethanol onto the planar slides with shaking. To ensure only one 2μm bead would fit one well, the wells were etched at a diameter of 2.05μm yielding a single bead per well coverage in over 99% of the wells.
Array decoding
Two sets of complementary and fluorescently labeled (FAM and Cy3) oligonucleotides were synthesized, deprotected and purified. An additional set of unlabeled but still complementary probes was made. Each decoder set represented an individual decoder pool (10nM). A total of 14 different color-coded (red; green and dark) pools were made. For example, if decoding 65 spatial oligonucleotide barcodes (Spatial_barcode_Pool1), the first decoder pool contained oligonucleotides complementary to spatial barcodes 1–27 labeled with FAM, barcodes 28–54 labeled with Cy3 and finally oligonucleotides 55–65 with no color label attached. In the second decoding cycle, oligonucleotides complementary to spatial barcodes 1–9; 28–36 and 55–63 were labeled with FAM, oligonucleotides complementary to barcodes 10–18; 37–45 and 64–65 were labeled with Cy3, and the rest were unlabeled. The color scheme was cycled for another 2 cycles for decoding 65 oligonucleotides in a total of 4 cycles. The same approach was then repeated to decode 211 barcoded oligonucleotides (Spatial_barcode_Pool2) with 5 cycles and another 5 cycles to decode 211 barcoded oligonucleotides in Spatial_barcode_Pool3 for a total of 14 cycles. This decoding approach was conducted as previously published14 resulting in each (x,y) well position encoded with a combination of three colors (FAM; Cy3 and “dark”). Decoded arrays and (x,y) files were shared by Illumina. The decoding scheme and oligonucleotide sequences are proprietary as offered in the Illumina array product line.
Tissue samples
Adult C57BL/6J mice (at 12 weeks age) were euthanized and their main olfactory bulb (MOB) dissected. The samples were then frozen in an isopentane (Sigma-Aldrich) bath at −40°C, and transferred to −80°C for storage until sectioning. The frozen bulbs were embedded at −20°C in Tissue-Tek OCT (Sakura). Cryosections were taken at 10μm thickness. Breast cancer biopsies were snap frozen and embedded into OCT. Cryosections were taken at 16μm thickness. This study complied with all relevant ethical regulations regarding experiments involving animal and human tissue samples.
Tissue staining and imaging
Tissue sections were first adhered to the surface by keeping the slide at 37°C for 1 min. A fixation step on the slide surface was performed using 4% formaldehyde (Sigma-Aldrich) in 1x phosphate buffered saline (PBS, pH 7.4) for 10 min at room temperature (RT). The sections were stained using standard hematoxylin and eosin (H&E) staining, as previously described12, and 20x imaged with a Ti-7 Nikon Eclipse.
RNA-Seq library preparation and sequencing
We followed a protocol as described in Stahl et al12and Salmén et al21. Briefly, tissue sections were permeabilized using exonuclease I buffer (NEB) for 30 min at 37°C and 0.1X pepsin (pH 1) for 10 min at 37°C, followed by in situ cDNA synthesis overnight at 42°C using Superscript III supplemented with RnaseOUT in 1XFS buffer, 5 mM DTT, 0.5 mM dNTP mix (all from ThermoFisher Scientific), 50 μg/ml actinomycin D in 1% DMSO (Sigma-Aldrich) with 0.19 mg/ml BSA (NEB). For breast cancer samples, the tissue permeabilization steps included a 20 min tissue incubation with 14 U collagenase I in Hank’s balanced salt solution (ThermoFisher Scientific) followed by digestion with 0.1X pepsin (pH 1) for 10 min at 37°C. Tissue sections were digested after cDNA synthesis using proteinase K (Qiagen) for 1h at 56°C and the barcode-transcript information cleaved using a USER (NEB) (for 2h at 37°C). The material was processed into libraries as described in Jemt et al22 and 1.08 pM sequenced on an Illumina Nextseq 500 instrument with v2 chemistry using paired-end 300 bp reads (R1 125 bp and R2 175 bp for MOB; R1 150 bp with R2 150 bp for the breast cancer samples).
HDST data pre-processing
FASTQ files were processed using the ST Pipeline v1.5.1 software23. The forward read contained both the barcode the bridge sequences and were trimmed retaining the following bases: 1–15, 31– 45 and 61–76. This created a forward read containing only a spatial barcode followed by a UMI. R2 transcripts were mapped with STAR24 to the mm10 mouse or GRCh38.79 human reference genomes. Mapped reads were counted using the HTseq count tool25 “gene” feature. Spatial barcodes were demultiplexed using an implementation of TagGD26 as described23. We allowed a 2 bp “padding” overhang on the total length of the spatial barcode enabling correction for either a 1 bp insertion or deletion error at the beginning or end of the barcode sequence. Demultiplexing was based on building a hashmap of 11 bp k-mers. All barcodes were then compared allowing a Hamming distance of 4 mismatches. UMI (77–82 bp in R1) duplicated sequences paired to the annotated reads were collapsed using a hierarchical clustering approach. All UMIs, with allowing for 1 mismatch, were clustered using the spatial barcode, mapped gene locus (with a window offset of 250 bp25,27) and strand information.
Histological image processing
To relate the histological image and the counts matrix, we assigned image pixel coordinates to the centroid of each bead well28. First, we detected the arrays’ boundaries and corners, and assumed a perfect hexagonal well matrix distribution. We then translated pixel coordinates into fixed centroid (x,y) coordinates using the total detected area of the array. Well coordinates detected under the tissue boundaries were used in further analysis.
Image annotation
Images used in the study were annotated with an interactive user interface for selecting spatial barcodes and their (x,y) coordinates based on the tissue morphology. Each (x,y) barcode position could be assigned to one or more of the 9 distinct regions in the mouse olfactory bulb: Olfactory Nerve Layer (ONL), Granular Cell Layer External (GCL-E), Granular Cell Layer Internal (GCL-I), Deep Granular Zone (GCL-D), External Plexiform Layer (EPL), Mitral Layer (M/T), Internal Plexiform Layer (IPL), Subependymal Zone (SEZ) and the Granular Cell Layer (GL). For HDST MOB annotation, exactly one regional tags was assigned to one (x,y) spatial barcode. For ST, more than one tag could be assigned per (x,y) spatial spot location. For breast cancer histological sections, we used 6 areas: Invasive cancer, Fatty tissue, Fibrous tissue, Normal glands, Vascular space and Immune/lymphoid cells with allowing for multiple region assignments. If a barcode position was not covered by an annotation polygon, the position was assigned to the closest polygon in cases the unassigned barcode was within 5 pixels distance.
H&E image segmentation for single cell identification
Single cell segmentation based on the H&E image was done by combining Ilastik 1.3.229 and CellProfiler 3.1.830. In Ilastik, we trained a random forest classifier to identify two distinct classes (nuclei and background). Based on this, we were able to predict and export probability maps for the nuclei. We used CellProfiler30 to segment the probability maps and identify single cell masks for downstream analysis.
smFISH data processing
Previously published and processed MOB smFISH data12 was used in the analysis. Briefly, a 10 μm section was attached to a cover slip and the tissue was stained with 250 nM fluorescent label probes (LGC Biosearch Technologies) for three of the genes (Penk, Slc17a7 and Fabp7) diluted in staining buffer31 and a counterstain with Hoescht applied. z-stacks were imaged at 0.3 μm distance on a Nikon Ti-E. The images were stitched in Fiji32. Three replicate ROIs corresponding to 100 μm (in diameter) were randomly placed in three layers of the olfactory bulb: granular layer, glomerular layer and olfactory nerve layer. To estimate the average number of nuclei present per one ROI region in each layer, we used a grid approach on our histological image and counted detected nuclei. We then estimated the number of RNA spots per nucleus for each region and layer. These averaged RNA counts were compared to UMI counts in the HDST data averaged again per nucleus for each region and layer. Data was adjusted for the effective bead packing density and average number of beads lost at demultiplexing after sequencing. The average number of RNAs per one HDST bead were finally compared to smFISH counts for the same sized area.
Binning of HDST data
We divided the total area of each HDST array into non-overlapping bins, each covering an area of X×X beads, where X∈{5,38}, and summed the UMIs of beads within each spatial bin. In order to ensure appropriate bin sizes, we first considered all manufactured wells as a 1918×765 matrix. On average, around 1,370 (x,y) wells filled with beads would size up to one ST spot (100 μm; X=38) when taking into account the center-to-center distance between two wells. We took the binned data containing 1,370 wells per bin and took every second bin into account in both x and y directions. This was to ensure space between two ST spots would be accounted for. This bin size was called “ST-like” in all further analyses. We did not take into consideration that this bin actually represents 63% of the transcriptome profiled per ST spot due to space between two hexagonal wells. Second, we made bins with fewer wells per bin in a logarithmic manner until reaching the smallest bin (5X). The 5X bin was referred to as single cell like of “sc-like”.
Spatial differential expression analysis
Binned 5X data was smoothed using a Gaussian kernel (5×5) with 0.5 standard deviations equally in both x and y directions. The smoothed binned data was then scaled for purposes of visualization such that the maximum expression value stayed the same. We performed a two-sided t-test to identify DE genes for each HDST morphological region (one vs. rest). The genes with a log2 fold change>1.5 and FDR<10% were identified as differentially expressed and used in further analyses. Scanpy package was used for visualization and differential expression analysis33.
Validation of differentially expressed genes
To validate layer-specific gene expression in the HDST data, we performed enrichment analysis using layer specific gene sets from the Allen Brain Atlas as reference. Layers annotated in both datasets were used in the analysis with all HDST granular layers merged into one instance to be comparable to the data provided in the Allen Brain Atlas. Genes with a layer specific log2 fold change of greater than 1.5 (implying upregulation) and FDR<10% were tested for enrichment in the layer-specific gene sets (“expression fold” change greater than 1.5) from the Allen Brain Atlas. Only genes passing the respective fold-change thresholds in both data-sets were included in the analysis. Images for the top gene present in each layer were downloaded from ABA’s High Resolution Image Viewer and stitched using Fiji32.
Assessing nuclear RNAs in HDST data
Single cell and single nucleus data from the mouse (10x Chromium 3 v2 sequencing) from the M1 region on the mouse brain was downloaded from www.biccn.org. BICCN data, tools, and resources are released under the Creative Commons Attribution 4.0 International (CC BY 4.0, https://creativecommons.org/licenses/by/4.0/legalcode) License. The single cell dataset was published from the U19 Zeng team (1U19MH114830–01) and the single nucleus dataset from the U19 Huang team (1U19MH114821–01). 50,000 randomly selected single nuclei and cells were used in downstream analysis. HDST data were split into two sets, based on whether the respective (x,y) coordinate overlapped or not the segmented nucleus. We then identified those genes in each of the subsets that are present in either nuclei or cells. We observed 186 genes (128 protein coding, 58 non-coding) that are expressed exclusively in single-nucleus data and whose HDST barcodes are present in segmented nuclei. Furthermore, we calculated the ratio of intronic and exonic reads using a reference which contains both introns and exons in the alignment and an exon-only reference, respectively. The barcodes that overlap with nucleus segmentations (n=36722) showed significantly higher (intron+exon)/exon log-ratios than rest of the barcodes (n=75887) (log-transformed total UMIs in nuclear barcodes: 0.04807±0.1507 (mean±sd), non-nuclear barcodes: 0.0461±0.1475 (mean±sd), p-value: 0.017, one-sided unpaired Welch’s t-test).
Cell type assignment to HDST barcodes
For analysis of MOB data, we downloaded the matrix containing mean expression values x per cell type j from Zeisel et al17 using the loompy package (https://github.com/linnarsson-lab/loompy). We subseted the matrix to contain only cell types annotated in the olfactory bulb (OB) and non-neuronal cell types for a total of 63 cell types. For analysis of HDST breast cancer data, we downloaded the expression matrix for a triple negative breast cancer single-cell RNA-seq data set18 from GEO (GSE118389) and calculated mean expression values x per cell type j for all genes contained in the matrix. Cell type annotations were obtained from the study’s github repository (https://github.com/Michorlab/tnbc_scrnaseq/blob/master/data/cell_types_tab_S9.txt).
The vector of probabilities Θj of each gene being captured in a cell type j is defined as follows:
i = gene, j = cell type, k = total number of genes, x = mean expression
We calculate the likelihood L of the cell type j specified by Θj, given the observed UMIs b per gene for a HDST (x,y) barcode as follows:
b = vector of UMI counts per gene for an individual HDST (x, y) profile
n = total number of UMIs for an individual HDST (x, y) profile
For each HDST (x,y) and cell type j, we calculated the ratio between the likelihood of that cell type Lj and the likelihood of the most likely cell type Lmax as a measure to assess how good the secondary cell type assignments are compared to the most likely cell type (primary assignment):
For each HDST (x,y) and cell type j, we calculated the posterior probability i.e. normalized likelihood LN as the ratio between the likelihood of that cell type Lj and the sum of all likelihoods for that barcode Ltot as a measure to assess how distinct each assignment is compared to all others:
where P(cj) denotes the uniform prior for cell type j. P(cj|b) and P(b|cj) represent the posterior probability and the likelihood, respectively. P(b), the evidence term, is defined as and used for the normalization.
Finally, to test against the null-hypothesis that HDST (x,y) expression profiles originate from random expression profiles for each cell type j and respective each Θj, we retained only non-zero elements of each Θj, and shuffled them in 1000 iterations while keeping the distribution of UMIs b as in the corresponding HDST (x,y) expression profile. We then calculated the randomized likelihood Lrand for each HDST (x,y), cell type and iteration. Finally, an empirical p-value pemp was calculated for each HDST (x,y) and cell type assignment as the fraction of Lrand that yie6lded a likelihood higher or equal to the cell type likelihood Lj multiplied by the probability of drawing only non-zero values from Θj given b, with correction for multiple testing (Benjamini-Hochberg). For each HDST (x,y) the cell type with the highest likelihood (LR = 1) was considered the primary assignment and all cell types with LR ≥ 0.8 were considered secondary assignments. Finally, for a cell type assignment to be considered valid, we required LN ≥ 0.1 and Pemp ≤ 0.01 for MOB and LN ≥ 0.7 and Pemp ≤ 0.05 for breast cancer. For further analysis only HDST (x,y) with exactly one valid cell type assignment were considered. Cell type assignments, ratios and the empirical p-values for all HDST (x,y), segmented and binned MOB profiles have been reported in Supplementary Tables 5–6. Differential expression analysis was performed between the cell types using two-sided t-test (log2 fold change>1.5 and FDR<10%).
Auxiliary data pre-processing
Public bulk RNA-Seq datasets12 were downloaded from NCBI’s SRA with accession PRJNA316587, mapped to the mm10 reference and UMI filtered using the ST pipeline v1.3.1. Averaged and naively adjusted34 bulk gene expression signatures were compared to those of the three replicates created with HDST and normalized the same way. Allen Brain Atlas (ABA) gene lists were downloaded from the ABA API using the ConnectedServices module of the allensdk Python package version 0.16.0. The standard ST data as a counts matrix was downloaded from http://www.spatialtranscriptomicsresearch.org/.
Supplementary Material
Acknowledgments:
We thank the NGI Stockholm, SciLifeLab, Flatiron Institute and Simons Foundation for providing infrastructure support. We thank Leslie Gaffney, Ania Hupalowska and Jennifer Rood for help with manuscript preparation. Work was supported by the Knut and Alice Wallenberg Foundation, Swedish Foundation for Strategic Research, Swedish Cancer Society and the Swedish Research Council (to S.V., J.L. and P.L.S.), the EMBO long-term fellowship (ALTF 738–2017) to J.K., Early Postdoc Mobility fellowship (P2ZHP3_181475) to D.S., the NIH HuBMAP HIVE project, the Klarman Cell Observatory and HHMI (to A.R.). S.V is supported as a Wallenberg Fellow at the Broad Institute of MIT and Harvard.
Footnotes
Competing interests
F.S., J.F., J.L. and P.L.S. are authors on patents PCT/EP2012/056823 (WO2012/140224), PCT/EP2013/071645 (WO2014/060483) and PCT/EP2016/057355 applied for by Spatial Transcriptomics AB (10x Genomics) covering the described technology. M.R. is employed by Illumina Inc. A.R. is a founder and equity holder of Celsius Therapeutics and an SAB member of Syros Pharmaceuticals and Thermo Fisher Scientific.
Reporting summary: Further information on research design is available in the Life Sciences Reporting Summary linked to this article.
Data availability: The raw mouse data have been deposited to NCBI’s GEO archive GSE130682. Raw files for the breast cancer sample are available through an MTA with Åke Borg (ake.borg@med.lu.se). All processed data is available at the Single Cell Portal (https://portals.broadinstitute.org/single_cell/study/SCP420).
Code availability: All code has been deposited on GitHub at https://github.com/klarman-cell-observatory/hdst.
References
- 1.Oh SW et al. Nature 508, 207–214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lein E, Borm LE & Linnarsson S Science 358, 64–69 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Macosko EZ et al. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng GXY et al. Nat. Commun 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van den Brink SC et al. Nat. Methods 14, 935–936 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Chen KH, Boettiger AN, Moffitt JR, Wang S & Zhuang X Science 348, aaa6090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X et al. Science 361, eaat5691 (2018).29930089 [Google Scholar]
- 8.Coskun AF & Cai L Nat. Methods 13, 657–660 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beliveau BJ et al. Nat. Commun 6, 7147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JH et al. Science 343, 1360–1363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ke R et al. Nat. Methods 10, 857–860 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Ståhl PL et al. Science 353, 78–82 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Michael KL, Taylor LC, Schultz SL & Walt DR Anal. Chem 70, 1242–1248 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Gunderson KL et al. Genome Res. 14, 870–877 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagayama S, Homma R & Imamura F Front. Neural Circuits 8, 98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lein ES et al. Nature 445, 168–176 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Zeisel A et al. Cell 174, 999–1014.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karaayvaz M et al. Nature Communications 9, 3588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X et al. Breast Cancer Research 19, 15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriques SG et al. Science 363, 1463–1467 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salmén F et al. Nat. Protoc 13, 2501–2534 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Jemt A et al. Sci. Rep 6, 37137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Navarro JF, Sjöstrand J, Salmén F, Lundeberg J & Ståhl PL Bioinformatics 33, 2591–2593 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Dobin A et al. Bioinformatics 29, 15–21 (2012).23104886 [Google Scholar]
- 25.Anders S, Pyl PT & Huber W Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costea PI, Lundeberg J & Akan P PLoS One 8, e57521 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gautheret D, Poirot O, Lopez F, Audic S & Claverie JM Genome Res. 8, 524–530 (1998). [DOI] [PubMed] [Google Scholar]
- 28.Wong K, Navarro JF, Bergenstråhle L, Ståhl PL & Lundeberg J Bioinformatics 34, 1966–1968 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Sommer C, Straehle C, Kothe U & Hamprecht FA 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro 230–233 (2011). [Google Scholar]
- 30.Lamprecht MR, Sabatini DM & Carpenter AE Biotechniques 42, 71–75 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Lyubimova A et al. Nat. Protoc 8, 1743–1758 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Schindelin J et al. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolf FA, Angerer P & Theis FJ Genome Biol. 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Svensson V, Teichmann SA & Stegle O Nat. Methods 15, 343–346 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.