Abstract
Dynamic 3D chromatin conformation is a critical mechanism for gene regulation during development and disease. Despite this, profiling of 3D genome structure from complex tissues with cell-type specific resolution remains challenging. Recent efforts have demonstrated that cell-type specific epigenomic features can be resolved in complex tissues using single-cell assays. However, it remains unclear whether single-cell Chromatin Conformation Capture (3C) or Hi-C profiles can effectively identify cell types and reconstruct cell-type specific chromatin conformation maps. To address these challenges, we have developed single-nucleus methyl-3C sequencing (sn-m3C-seq) to capture chromatin organization and DNA methylation information and robustly separate heterogeneous cell types. Applying this method to >4,200 single human brain prefrontal cortex cells, we reconstruct cell-type specific chromatin conformation maps from 14 cortical cell types. These datasets reveal the genome-wide association between cell-type specific chromatin conformation and differential DNA methylation, suggesting pervasive interactions between epigenetic processes regulating gene expression.
Introduction
Three-dimensional genome architecture is a critical feature of gene regulation in metazoan organisms 1-3. Chromatin conformation profiling has revealed the existence of features such as Topologically Associated Domains (TADs) and enhancer-promoter interactions 4-9. Despite the increasing utility of these datasets, most existing chromatin conformation maps are generated from cell lines or from bulk tissues 4,8-11. While these data has helped to elucidate general principles of chromatin organization, it cannot fully represent the diversity of cell types that arise in vivo. Single-cell 3C or Hi-C represent attractive strategies to resolve cell-type heterogeneity 12-14. However, current single-cell Hi-C profiles from cultured cells primarily capture cell-cycle patterns 12,15. It remains unclear whether single-cell Hi-C profiles are suitable for partitioning constituent cell types.
In contrast to single-cell Hi-C data, single-cell DNA methylome datasets enable high-resolution cell-type classification from cell types in primary human tissues 16,17. DNA methylation (mC) is unaltered in the basic protocol of 3C or HiC, therefore it may be feasible to detect both long-range ligation junctions and mC by combining 3C or HiC with bisulfite sequencing.
Here we describe a method, single-nucleus methyl-3C sequencing (sn-m3C-seq), to jointly profile chromatin conformation and mC from the same cell. Bulk and single-cell m3C-seq profiles accurately recapitulate chromatin architectures of mouse embryonic stem cells (mESCs). Furthermore, we show that sc-m3C-seq can distinguish cultured mouse cell types as well as highly heterogeneous human brain cell populations. Using 4,238 sn-m3C-seq profiles, we identify 14 cell types from human frontal cortex by clustering of mC profiles and from these clusters identify cell-type specific 3D chromatin structures. We observe a strong, cell-type specific relationship between mC and 3D genome structure , suggesting significant crosstalk between these epigenomic features.
Results
Joint profiling of chromatin conformation and DNA methylation from the same DNA molecule
In sn-m3C-seq, we first perform restriction enzyme digestion and ligation on fixed nuclei, as is typically performed in an in situ 3C experiments 8, 18, 19. The ligated 3C nuclei are dispensed into 384 well PCR plates using Fluorescence-activated nuclei sorting (FANS) and subject to proteinase digestion and bisulfite conversion , and libraries are constructed similar to our previous snmC-seq2 method (Fig. 1) 16,20 . When performed as a bulk assay (m3C-seq) ligated nuclei are not sorted into well but treated in bulk.
Figure 1. Outline of the single-nucleus methyl-3C sequencing (sn-m3C-seq) method.
Samples are processed with a typical in situ 3C/Hi-C procedure following by single-cell DNA methylome library preparation using snmC-seq2.
To evaluate the quality of chromatin contact maps generated by m3C-seq, we compared bulk m3C-seq data to conventional bulk in situ 3C-seq and Hi-C profiles in mESCs. Both Hi-C/3C and bisulfite conversion present challenges for read alignment due to the presence of chimeric reads and the conversion of unmethylated cytosines to uracils, respectively. We developed TAURUS-MH (Two-step Alignment with Unmapped Reads Using read Splitting for Methyl-HiC), a mapping pipeline for m3C-seq data using a hybrid of ungapped and read splitting alignments (Supplementary Fig. 1a). Sequencing reads were first mapped to an in silico bisulfite converted genome using Bismark calling an ungapped aligner (bowtie1)21, and unmapped reads are split into 3 segments followed by ungapped alignment. We compared the performance of TAURUS-MH to BWA-METH22, which is designed for bisulfite sequencing data alignment using BWA-MEM. This comparison was performed using typical Hi-C data with in silico simulated bisulfite conversion. We use the alignment of conventional Hi-C data 23 as our gold standard. When compared with BWA-METH, our pipeline showed 19.43% higher in mappability (86.12% vs. 66.69%, Fig. 2a), 3.64% higher in accuracy (97.86% vs. 94.22%, Fig. 2b), and 13.41% higher long-range cis contacts (42.79% vs. 29.38% from total fragments and 49.68% vs. 44.06% from mapped fragments, Fig. 2c).
Figure 2. Data processing and analysis of m3C-seq sequencing reads.
Reads derived from non-bisulfite treated regular Hi-C sequencing are converted C to T (read1) and G to A (read2) in silico and aligned using BWA-meth, Bismark (bowtie1), and Bismark (bowtie1) followed by split-read alignment. Alignment of non-converted reads using conventional alignment pipeline is used as a standard (Conventional, Non-converted). For (a-d), the mapping algorithms were applied to a common test dataset (n=1) to make a fair comparison of their performance. (a) Percent of aligned reads as a pair. (b) Alignment accuracy of different alignment strategies compared with conventional Hi-C alignment using in silico converted reads. (c) Fraction of read pairs with cis short-range reads (cis < 1kb), cis long-range interactions (cis > 1kb), and trans interactions (trans) using different alignment strategies. (d) Similar to panel (c), but for 3C-seq (without conversion), bulk m3C-seq (with conversion, from the same sample as bulk 3C-seq), and combined 192 single-nucleus m3C-seq results. (e) Contact maps from chromosome 17 for conventional bulk Hi-C and bulk m3C data. (f) mC profiles near the Pou5f1 gene for conventional bulk MethylC-seq as well bulk m3C-seq. The experiment was repeated twice independently with similar results.
We then analyzed chromatin contact data quality comparing bulk m3C-seq with a matched 3C-seq library. Bulk m3C-seq libraries showed a comparable fraction of long-range (>1kb) intra-chromosomal ligation events compared to the control 3C-seq library (26.6% in 3C-seq and 19.0% in m3C-seq) (Fig. 2d). Surprisingly, we observed more frequent inter-chromosomal “contacts” in m3C-seq (30.0% in m3C-seq and 12.24% in 3C-seq) (Fig. 2d). Since snmC-seq2 involves random-primed DNA synthesis 20, we speculate that the inter-chromosomal “contacts” are an artifact caused by spurious hybridization and polymerase extension during random-primed DNA synthesis. We further hypothesized that spurious inter-chromosomal ligation is dependent on DNA concentration and the frequency of intermolecular interaction. Consistent with this hypothesis, in sn-m3C-seq, where the random-primed DNA synthesis reaction contains a much lower DNA concentration, we found a similar background level (15.11% in sn-m3C-seq and 12.24% in 3C-seq) of inter-chromosomal ligation.
To assess the methylome quality of bulk m3C-seq, we compared bulk m3C-seq with published WGBS profiles generated from mESC24. With comparable sequencing depth, the m3C-seq library showed more uniform genomic coverage compared to the WGBS library, covering more cytosines and more CpG sites and showed a narrower distribution of coverage at CpG sites (Supplementary Fig. 1b,c).
Finally, we compared contact maps and mC profiles generated by bulk m3C-seq with conventional Hi-C and MethylC-seq data generated from mESC (Fig. 2e,f)25. We observed strong agreement between bulk m3C-seq and Hi-C (Fig. 2e, stratum adjusted correlation coefficient, SCC = 0.91 26). Similarly, we observed strong concordance of methylation profiles from bulk m3C-seq with existing MethylC-seq datasets for mESC (Fig. 2g , Pearson correlation = 0.82). We further compared bulk m3C-seq to multiple published bulk Hi-C and MethylC-seq datasets of mESC and found strong correlations for both types of profiles (Supplementary Fig. 2).
Fluorescence-activated nuclei sorting excludes nuclei multiplets
To generate sn-m3C-seq profiles, in situ 3C treated nuclei were sorted using fluorescence-activated nuclei sorting (FANS) into 384 well PCR plates followed by snmC-seq2 single-cell methylome library preparation. In control species mixture experiments (Supplementary Table 1), we found surprisingly frequent (23.2%) inter-species nuclei multiplets (Supplementary Fig. 3a) due to formaldehyde cross-linking, whereas multiples were eliminated when crosslinking was performed separately for each species (Supplementary Fig. 3b). We found that performing crosslinking with a 10-fold diluted nuclei preparation or stringently selecting nuclei with a 2n genomic DNA content could largely eliminate inter-species nuclei multiplets (7.4% for dilution, 1% for 2n gating) (Supplementary Fig. 3c-e).
sn-m3C-seq generates high-quality single-cell DNA methylation profiles
We systematically compared the technical characteristics of sn-m3C-seq with published single-cell methylome datasets generated using Zymo Pico Methyl-seq 27,28 and scNMT-seq 29. sn-m3C-seq showed superior read mapping rate (72.4±3.6%) than Pico Methyl-seq (33±12.3%) or snNMT-seq (32.2±9.4%, Supplementary Fig. 4a). The library complexity of sn-m3C-seq (maximumly 27.5±9.9% of the mouse genome) is similar to that of scNMT-seq (22.8±11%), and is greater than Pico Methyl-seq (10±4.7%, Supplementary Fig. 4b). scBS-seq and its derivative scNMT-seq show bias (2.48±0.82 fold enriched) towards CpG islands 30, whereas sn-m3C-seq (1.21±0.15), snmC-seq2 (1.14±0.09) and Pico Methyl-seq (1.57±0.1) showed modest enrichment of CpG islands (Supplementary Fig. 4c). Lastly, sn-m3C-seq and Pico Methyl-seq show comparable coverage uniformity (Supplementary Fig. 4d). At a coverage of 1 million non-clonal reads, sn-m3C-seq covers 28.9% of 1kb genomic bins and 89% of 10kb bins, while Pico Methyl-seq covers 29.1% of 1kb bins and 90.5% of 10kb bins. Both assays are less biased than scNMT-seq, which covers 23.5% of 1kb bins and 78% of 10kb bins with 1 million non-clonal reads. We observed a high correlation of the mC profiles between pooled sn-m3C-seq and previously generated bulk WGBS experiments with mESC specific hypomethylation at the promoter regions of pluripotent genes such as Dppa4 and Dppa2 (Pearson r = 0.89, sn-m3C-seq vs. mESC Lee 2014) (Fig. 3d and Supplementary Fig. 2) 35-38.
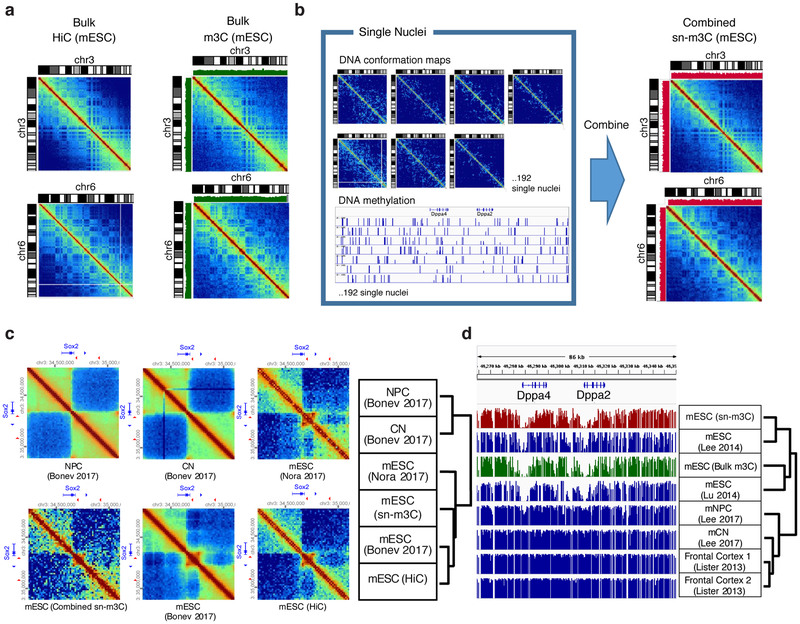
Figure 3. Bulk and single-nucleus m3C-seq of mouse embryonic stem cells.
(a) Comparison of HiC and bulk m3C-seq chromatin contact profiles . Green bar plot shows CpG methylation level from m3C-seq. (b) Reconstructed mESC chromatin conformation map from sn-m3C-seq profiles compared to Hi-C or bulk m3C-seq. Red bar plot shows CpG methylation level from sn-m3C-seq. (c) Bulk and single-nucleus m3C-seq chromatin contact profiles of the Sox2 locus in mESC compared to published HiC data generated from mESC, cortical neurons (CN) and neural progenitor cells (NPC). . (d) Bulk and single-nucleus m3C-seq DNA methylation profiles at Dppa2/4 locus compared to published methylome data generated from mESC, mouse CN and frontal cortex. The experiment was repeated twice independently with similar results.
sn-m3C-seq profiles recapitulate chromatin conformation contact maps
We have compared our data with previous single-cell Hi-C studies. We profiled comparable numbers of cells as the published single-cell Hi-C datasets with the largest numbers of profiled cells to date (Ramani et al., Nagano et al. 12,31; Ramani=10,696; Nagano=3,413 This study=6,200 total cells between mouse ESC, NMuMg, and human brain data), yet we obtain 1.7 fold more contacts than Nagano et al. and 49.27 fold more contacts than Ramani et al. Other studies (Flyamer et al 2017; Tan et al. 2018)32,33 profiled fewer numbers of cells (Tan=35, Flyamer=246) but with higher numbers of contacts (Tan=1,165,296; Flyamer=2,416,802, Supplementary Fig. 5a,b, see supplemental methods for details of analysis). These data indicate that our method generates single-cell chromatin conformation data of comparable quality to existing unimodal methodologies.
Using hierarchical clustering on a matrix of SCC, we observed that contact maps from our mESC sn-m3C-seq clustered with Hi-C data from mESCs 34, while the cortical neurons (CN) and neural progenitor cells (NPC) datasets clustered separately (Fig. 3a-c) 9. In both Hi-C and pooled sn-m3C-seq data, we observed mESC specific contacts, such as enhancer-promoter contacts at the Sox2 locus (Fig. 3c). We have observed additional cell-type specific hypomethylated regions in association with chromatin interaction differences in Tbx5 and Tfap2d (Supplementary Fig. 6-7).
sn-m3C-seq profiles separate mouse cell types
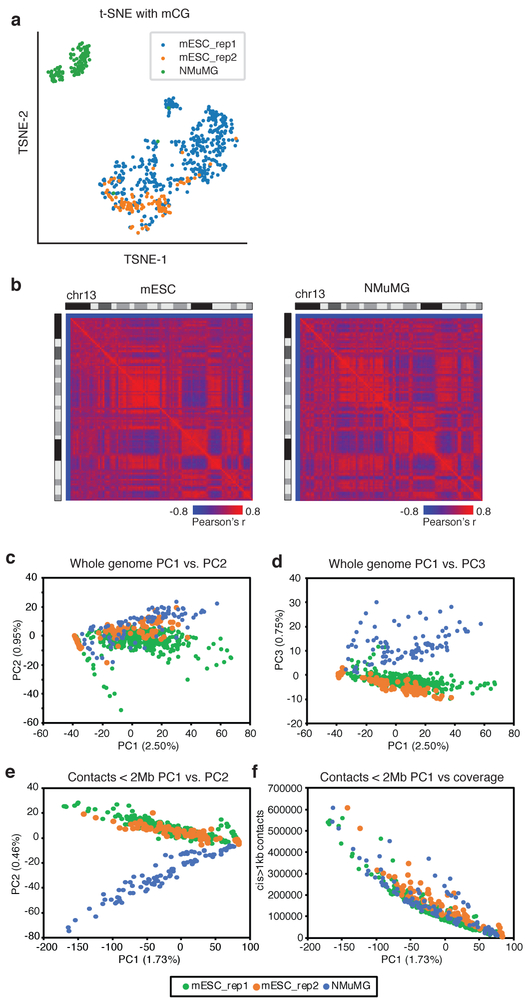
To test the robustness of cell-type identification using sn-m3C-seq profiles, we performed t-distributed stochastic neighbor (tSNE) embedding with CpG methylation levels in non-overlapping 100kb bins from single cells, which shows a clear separation of mESC and NMuMG cells (Fig. 4a). Using pooled sc-m3C-seq data, we identified distinct A/B compartment signatures between the two cell types (Fig. 4b) as well as local differences in Hi-C contacts (arrows in Supplementary Fig. 8a,b).
Figure 4. Single-nucleus m3C-seq reconstructs cell-type specific chromatin conformation maps.
(a) tSNE of single-cell mC profiles of mouse ES cells and NMuMG cells . (b) Chromosome wide Pearson correlation matrix from pooled sc-m3C-seq maps for ES cells and NMuMG cells . (c-d) Principal component analysis (PCA) of whole genome contact matrices from sc-m3C-seq from ES from NMuMG cells (Percentage of variance are marked on the axis). PC1 and PC2 are shown in (c); PC1 and PC3 are shown in (d). (e) PCA of local interactions (<2Mb) from sc-m3C-seq data from NMuMG cells showing PC1 and PC2. (f) Correlation of PC1 and per cell contacts . For (a) and (c-f), n=2 independently prepared mouse ES cell cultures were analyzed. The two mESC replicates each contained 379 and 93 cells. One (n=1) replicate of NMuMG cells containing 96 cells was analyzed.
We also compared the ability of mC or Hi-C contacts to partition sc-m3C-seq into the relevant cell types. DNA methylation profiles could easily distinguish between ES and NMuMG cells using the first principal component (PC) alone, which explains 33.7 % of total variance (Supplementary Fig. 8c,d). In contrast, PCA using whole genome Hi-C contacts at 100kb could not distinguish between ES and NMuMG cells using the first two PCs (Fig. 4c), but the third PC did clearly separate these two cell types (Fig. 4d). PCA using local contacts (< 2Mb) was able to distinguish the two cell types using the second PC (Fig. 4e). We observed that the first PC was highly correlated with per cell sequencing depth (Fig. 4f), suggesting the power for cell type identification using Hi-C contacts is highly dependent on sequencing coverage. In contrast, the ability to distinguish the two cell types using mC profiles is not sensitive to sequencing coverage (Supplementary Fig. 8e,f), indicating the robustness of cell type classification from mC profiles. These results underscore the importance of jointly profiling mC along with chromatin conformation to reliably distinguish cell types in single-cell experiments.
sn-m3C-seq identifies cell-type specific chromatin interaction in human prefrontal cortex
To test whether sn-m3C-seq can be applied to complex human primary tissues, we generated 4,238 sn-m3C-seq profiles from the prefrontal cortex (PFC) region of two post-mortem adult human brains (Supplementary Table 2). We first identified non-neuronal cell types using CG methylation (mCG) signature followed by fine clustering of neuronal subtypes using non-CG methylation (mCH), resulting in the identification of 14 major cell types in human PFC (Fig. 5a,b). We annotated the clusters based on the depletion of mCG and mCH at the gene body of known cell type markers (Supplementary Fig. 9). The methylation profile is highly correlated in each cell type between the two individuals (Supplementary Fig. 10). Brain neuron subtypes (excitatory neuron subtypes: L2/3, L4, L5 and L6; inhibitory neuron subtypes: Pvalb, Sst, Ndnf and Vip) can be identified with much greater resolution using mCH or CG methylation (mCG) signatures, compared to only using chromatin interactions (Fig. 5a-c). However, clustering analysis using chromatin interactions alone or jointly with mCH can robustly resolve non-neuronal brain cell types (Fig. 5c and Supplementary Fig. 11).
Figure 5. Single-nucleus m3C-seq in human brain prefrontal cortex (PFC).
(a-c) Dimension reduction (t-Distributed Stochastic Neighbor Embedding, tSNE) visualization of single human PFC cells using mCH (a) and mCG (b) of non-overlapping 100kb genomic bins, or chromatin interaction at 1Mb resolution (c). L2/3, L4, L5 and L6: excitatory neuron subtypes located in different cortical layers. Ndnf and Vip: CGE derived inhibitory sub-types. Pvalb and Sst: MGE derived inhibitory sub-types. Astro: astrocyte. ODC: oligodendrocyte. OPC: oligodendrocyte progenitor cell. MG: microglia. NN1: non-neuronal cell type 1. Endo: endothelial cell. (d) looping between the SATB2 and LINC01923 locus in excitatory neuron (L2/3, L4, L5 and L6) (e) chromatin looping between PROX1 and RPS6KC1 region in CGE derived inhibitory cell types - Vip and Ndnf. (f-g) mCH (f) and mCG (g) levels at SATB2 locus in excitatory neuron clusters. (h-i) mCH (h) and mCG (i) levels at PROX1 locus in CGE derived inhibitory neuron clusters. All analyses were performed with 4,238 sn-m3C-seq profiles generated from n=2 independent human specimen.
Guided by the cell-type identification using mC signatures, we reconstructed brain cell-type chromatin interaction maps using sn-m3C-seq reads. We further identified 36,559 cell-type specific chromatin interactions using a negative-binomial test based method (edgeR39, FDR=0.1%, Supplementary Table 3), and 6,161 differential domain boundaries using the recently described HiCluster method (Supplementary Table 4)40. We found drastic chromatin interaction dynamics at cell-type signature genes (Fig. 5d-e, Supplementary Figs. 12,13). For example, SATB2 is a marker gene for excitatory neurons and shows reduced mCH and mCG in excitatory neuron clusters (Fig. 5f,g). A distinct chromatin loop between SATB2 promoter region and the adjacent LINC01923 locus located 1.15Mb away is only found in excitatory, but not inhibitory neuron types (Fig. 5d). A specific pattern of increased domain boundary probability at the SATB2 locus was also observed only in excitatory cells. Similarly, PROX1 is a marker gene for inhibitory neuron subtypes (Vip and Ndnf) derived from caudal ganglionic eminence41. PROX1 locus shows reduced mCH and mCG in Vip and Ndnf clusters (Fig 5h,i). Chromatin loops specific to Vip and Ndnf neurons were found between promoters of PROX1 and RPS6KC1. Higher domain boundary probabilities were also observed at the promoter of PROX1 in CGE-derived neurons.
Cell-type specific chromatin interactions are associated with differential DNA methylation signatures
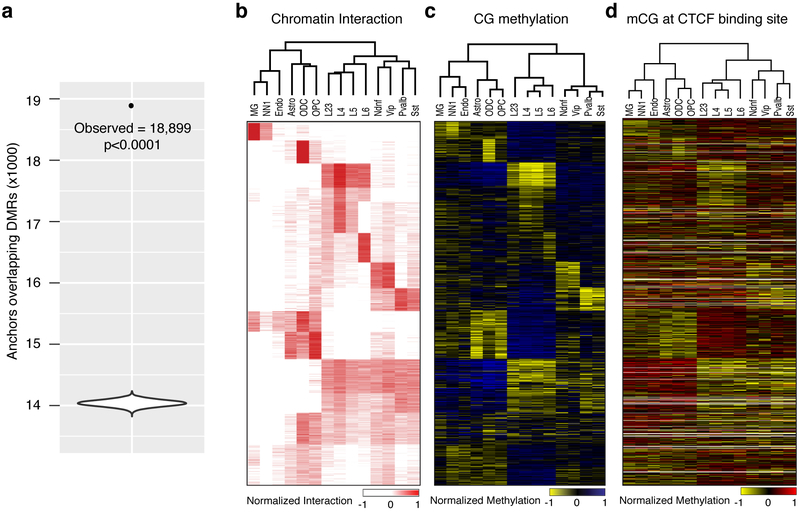
The PFC sn-m3C-seq dataset allowed us to explore the relationship between chromatin architecture and mC across brain cell types. We found a significant overlap between cell-type specific chromatin interactions and the 115,137 differentially methylated regions (DMRs) identified across brain cell types (p<0.0001, two-sided permutation test, Fig. 6a, Supplementary Table 5). Examining the mC profiles over the anchor regions (k-means clustered, k=15) revealed a striking hypo-methylation pattern at the sites of differential interacting regions in the cell types showing enriched interaction frequencies (Fig.6b,c). Therefore, on a global scale, cell-type specific chromatin interactions are associated with differential methylation patterns with matched cell type specificity.
Figure 6. Differential mC signature associated with cell-type specific chromatin interactions .
(a The violin plot of the distribution of the overlap between permuted differential interacting region anchor sites and DMRs ; the labelled point indicates the observed overlap. Violin plot elements: maximum=14,234; minimum=13,822; mean=14038.7. (DMRs, p<0.0001, two-sided permutation test, n=10,000 permutations). (b-d) Heatmap visualization of cell-type specific chromatin interaction (b), CG methylation at anchor regions (c) and CG methylation at CTCF binding sites overlapping with the anchor regions (d).
Higher-order chromatin structure is regulated by an interplay of genomic architectural proteins 42, including the methylation sensitive DNA binding protein CTCF 43,44. We examined whether differential interacting sites also showed variable methylation of the CTCF motif within CTCF binding sites defined by neuronal ChIP-seq 45. Within each cluster of differential interacting regions, CTCF binding sites were generally hypomethylated in the corresponding cell types showing increased chromatin interaction frequency (Fig. 6d).
We also investigated whether differential methylation of the methyl sensitive base at position 4 43,44 in the CTCF motif is associated with differential chromatin interactions. We identified CTCF motifs where position 4 showed variable cytosine methylation across the 14 cell types (highest methylation >80%, lowest <20%). Only a small minority (1,141/57,740) of CTCF motifs showed variable methylation, indicating that a minority of CTCF binding sites may be subject to regulation by variable DNA methylation. One possible reason that such a small portion of CTCF motifs show variable methylation is that variably methylated CTCF motifs are more likely to contain a CpG dinucleotide at position 4-5 relative to the genome wide occurrence of CTCF motifs (Supplementary Fig. 14a), despite the fact that such CpG containing CTCF motifs represent the minority total CTCF motif occurrences in the genome (Supplementary Fig. 14b, 10.99%). We observed that motifs that have variable methylation of position 4 are more likely to be found in variable interacting regions of the genome (Supplementary Fig. 14c, p=1.7e-6, two-sided Fisher’s exact test). These results indicate that a portion of variable interacting regions may be regulated by differential methylation of the CTCF motif, and underscore the importance of multi-omic profiling mC and chromatin conformation.
Finally, we examined the relationship between differential domain boundaries and mC. 73% of the differential domain boundaries colocalize with differential interaction anchors (p<1×10−300, two-sided hypergeometric test), and 46% of the differential domain boundaries overlap with DMRs (p<1×1e−50, two-sided hypergeometric test). Within a given cell type, we found the mC levels of CTCF motifs located at domain boundaries has significantly lower mC levels compared with non-boundary sites (Supplementary Fig. 15a-c). Genes whose transcription start sites (TSS) locate to within 2kb of the boundaries also showed depletion of methylation at their gene bodies compared to the genes at non-boundary sites (Supplementary Fig. 15d), indicating that the gene is more likely to be active when a TAD boundary is identified at their promoters. Taken these together, we have observed strong correlations between 3D genome interaction with mC.
Discussion
Cell-type specific chromatin conformation maps can potentially provide a valuable addition to other single-cell modalities for the creation of cell type atlases 46. This information complements single-cell transcriptomes and the annotation of regulatory elements using single-cell epigenomic profiles, to provide a more comprehensive description of gene regulatory activities. However, it is currently unclear how well single-cell Hi-C/3C methods alone can distinguish unique cell-types in a heterogeneous population. To enhance the cell-type signature in single-cell chromatin conformation data, we devised a method to allow jointly profiling of chromatin interaction and mC from a single nucleus. Consistent with previous single-cell methylome studies, sn-m3C-seq allows unequivocal clustering of cell types, which can then guide the reconstruction of high-quality cell-type specific chromatin conformation maps.
Our results indicate that single cell contact profiles alone can distinguish between drastically different cell types such as mESC and NMuMG. However, the confidence in cell type separation is highly dependent on sample coverage and downstream processing. Indeed in the human prefrontal cortex sn-m3C-seq dataset, It is possible to use contact maps to distinguish between non-neuronal cells and neurons, but not neuronal subtypes beyond coarsely separating excitatory and inhibitory cells. Our strategy of using mC signatures to define 14 cell types from human prefrontal cortex followed by the identification of cell-type specific chromatin interactions clearly demonstrated the advantage of our multi-omic approach.
Supplemental Methods
Cell culture
Mouse ES cells (E14TG2a) were purchased from American Type Culture Collection (ATCC CRL-1821). ES cells were grown in DMEM media (Corning 10-013-CV) supplemented with 10% HyClone FBS (Fisher SH3007003E), 1X MEM Non-essential amino acids (ThermoFisher 11140050), 1X Glutamax supplement (ThermoFisher 35050061), 1X ß-mercaptoethanol (Millipore ES-007-E), 100U/mL Penicillin-Streptomycin (ThermoFisher 15140122), and 1000U/mL Leukemia Inhibitory Factor (Millipore ESG1107). ES cells were cultured in feeder free conditions on 0.5% gelatin coated plates.
GM12878 cells were obtained from Coriell Institute for Medical Research. GM12878 cells were grown in RPMI-1640 medium (ThermoFisher 11875093) supplemented with 15% Fetal Bovine Serum (Corning 35-010-CV) and 100U/mL Penicillin-Streptomycin (ThermoFisher 15140122).
NMuMg cells (RBRC-RCB2868) were obtained from the RIKEN BioResource Center. NMuMg cells were grown in DMEM (Corning 10-013-CV) with 10% Fetal Bovine Serum (Corning 35-010-CV), 10µg/mL Insulin (ThermoFisher 12585014), and 100U/mL Penicillin-Streptomycin (ThermoFisher 15140122).
All cell lines were routinely tested for mycoplasma contamination and tested negative.
Human brain tissue
Postmortem human brain biospecimens were obtained from NIH NeuroBioBank at University of Maryland Brain and Tissue Bank. sn-m3C-seq was applied to BA10 cortical tissues of a 21-year-old Caucasian male (UMB5577) with a postmortem interval (PMI) = 19 h, as well as a 29-year-old Caucasian male (UMB5580) with a PMI = 8 h.
Hi-C and 3C
in situ Hi-C was performed as previously described using the MboI restriction enzyme 8. in situ 3C experiments were performed based on the in situ Hi-C protocol with minor modifications. Briefly, prior to fixation, adherent cells were trypsinized, counted, and collected by centrifugation; suspension cells were counted and collected by centrifugation. Cells were resuspended in culture media at a concentration of 1×106 cells per mL of media and fixed in 1% formaldehyde for 10 minutes at room temperature with shaking. For standard species mixture experiments, equal numbers of mouse and human cells were combined into a single tube prior to fixation. For the 1:10 dilution species mixture experiment, cells were resuspended at a concentration of 1×105 cells per mL of media prior to fixation. For the species mixture experiments where samples were mixed after fixation, each cell type was fixed independently as described above and combined at later stages in the protocol. in situ Hi-C samples were digested with the MboI restriction enzyme and processed as described previously. For in situ 3C experiments, samples were digested with DpnII enzyme overnight at 37ºC with gentle mixing. The following day, the sample was incubated at 62ºC for 10 minutes to inactivate the restriction enzyme. The typical biotin fill in step in the Hi-C protocol was omitted. The sample was then ligated for 4 hours at room temperature with T4 DNA ligase in the same manner as in in situ Hi-C experiments. The sample was then stained with Hoechst (0.1μg/μL) for the final 30 minutes of the ligation step. The sample was then passed through a 40 μM nylon cell strainer (Corning 431750) into a FACS tube prior to sorting. As a quality control step, 10% of the sample was taken for conventional library preparation and sequenced using shallow sequencing on a MiSeq.
Fluorescence-activated nuclei sorting (FANS)
FANS was performed at the Salk Institute Flow Cytometry Core Facility using a BD Influx cell sorter. A 100 micron nozzle tip was used, with 1 × PBS as sheath fluid (sheath pressure was set to 18.5 PSI) with sample and collection cooling set to 4 degrees. The gating strategy for selecting intact, single, Hoechst labelled nuclei from debris was as follows: nuclei were first gated based on Forward Scatter (FSC) and Side Scatter (SSC) pulse height, then multiplet exclusion gating was applied (forward scatter and side scatter pulse width). Finally, nuclei of specific DNA content were selected (e.g. 2N) by virtue of Hoechst fluorescence intensity. Individual nuclei were deposited into wells of 384-well plate using the Single Cell (1-drop single) mode. In preparation for 384-well plate deposition, 20-30 particles (e.g. calibration beads) were sorted onto a transparent plastic plate cover for alignment calibration. 20-30 particles are then directly sorted into the wells for final visual confirmation of alignment precision.
Bulk and single-cell methylome library preparation
Libraries for bulk and single-cell methylomes were generated using snmC-seq2. A detailed step-by-step bench protocol for snmC-seq2 is provided as Supplement Methods in Luo et al. (2018) 20. Bulk methylome libraries were prepared manually using individual tubes. Single-cell methylome libraries were prepared using a Tecan Evo 100 robotic platform as described in Luo et al. (2018) 20. Libraries for mESC and NMuMG samples were sequenced using Illumina MiSeq and HiSeq 4000 instruments in PE150 mode. Libraries for human prefrontal cortex sample were sequenced using Illumina HiSeq 4000 and Novaseq 6000 instruments in PE150 mode.
Data Processing
mESC and NMuMG datasets were mapped to mm10 reference genome. GM12878 data was mapped to hg38 reference genome. Human prefrontal cortex data were mapped to hg19 reference genome. C to T converted and G to A converted reference genomes were prepared for each reference genome using bismark_genome_preparation. The first (upstream) 25bp and last (downstream) 3bp were trimmed from both read1 and read2 to remove random primer sequence and the low complexity tail introduced by Adaptase. Read1 and read2 were mapped separately using Bismark with Bowtie1 with read1 as complementary (always G to A converted) and read2 (always C to T converted) as original strand 21,47. After the initial ungapped alignment, unmapped reads were split into 3 subreads by 40bp, 32bp, and 40bp after removing 5bp of both ends results in having 6 reads (The resulting subread IDs were converted to 1-1,1-2,1-3,2-1,2-2,2-3 for the later steps). Six subreads derived from unmapped reads were mapped separately using Bismark with Bowtie1. All aligned reads were merged into a BAM file using picard and were sorted by query name. The fragments with all the mapped reads aligned to the same positions were considered as duplicates and removed before allc files were generated. For each fragment, the outermost aligned reads were chosen for the chromatin conformation map generation.
Contact matrix generation and visualization
From the contact files, Cooler was used for generating the contact matrix for different sized bins and Higlass was used for visualization 48,49.
-
Generate fixed-width genomic bins:
cooler makebins reference_chrNameLength_file BINSIZE > Genomic_bin_file
-
Sort and index a contact list:
cooler csort --nproc 2 -c1 2 -p1 3 -c2 4 -p2 5 -o output_file input_file reference_chrNameLength_file
-
Cool file generation:
cooler cload pairix Genomic_bin_file input_file output_file
Comparison of m3C CpG methylation data with published WGBS data
We downloaded 6 publicly available WGBS datasets of different cell types including mESC, mCN, mNPC, and fetal mouse brain tissues 35,36,38,50. The methylation level of CpG sites was computed after merging coverage from strands. Average methylation level between 2kb upstream and 2kb downstream of known gene transcription start sites were computed (n=63759). Complete linkage hierarchical clustering with Euclidean distance was performed based on Pearson correlation coefficients (Fig. 3d, Supplementary Fig. 2b).
Comparison of m3C chromatin conformation data with published Hi-C data
We downloaded 4 publicly available Hi-C datasets of different cell types including mNPC, mCN, mESC9,34. Genomic contact matrix at 1Mb resolution was generated for each dataset. Stratum adjusted correlation coefficients (SCC) were calculated using HiCRep for intrachromosomal interactions across the whole genome26 (Fig. 3c, Supplementary Fig. 2a).
Comparison of bulk m3C-seq and sn-m3C-seq data with published methylome datasets
The technical characteristics of bulk m3C-seq were compared to a mESC WGBS dataset (SRX202087)24. Fastq files downloaded from SRX202087 were mapped to mm10 reference genome using Bismark with Bowtie2 aligner. The resulted BAM file was downsampled to match the coverage of bulk m3C-seq data. Published single-cell methylome datasets were downloaded from SRP069120, SRP062328 and SRP13102427-29. The fastq files were mapped to mm10 reference genome using Bismark with Bowtie2 aligner. Preseq51 was used to estimate library complexity using forward reads with Preseq gc_extrap function with options −e 5e + 09 −s 1e + 0712. Library complexity values shown in this study were estimated for the sequencing depth of 50 million read pairs. To determine the enrichment of CpG islands (CGI) in single-cell methylome data, the fraction of CGI on mouse chromosome 1 covered by a single-cell methylome was compared to shuffled regions with matching sizes. The shuffling was carried out using bedtools shuffle and was repeated five times and the average fraction of regions covered by reads was used. Bulk MethylC-seq data downsampled to 1 million non-clonal reads for this analysis. For computing the number of genomic regions covered by reads at different sequencing coverage, 1kb and 10kb bins were generated using bedtools makewindows across the mouse genome. The bins were intersected with bulk MethylC-seq and single cell methylomes downsampled to 100,000 to 1.5 million reads.
Comparison of sn-m3C-seq chromatin conformation data with published single-cell Hi-C data
We downloaded multiple previous single-cell Hi-C datasets 12,14,31-33,52 to compare with our snm3C-seq method. To allow for an unbiased comparison across methods, we processed each dataset uniformly using previously described alignment pipeline 53. For each dataset, we quantified multiple metrics of data quality, including the number of reads sequenced per cell, the number of mapped pairs per cell, the PCR duplication rate per cell, and the fraction of reads that align as short cis fragments (<1kb), long-range (>1kb) cis fragments, and inter-chromosomal pairs. We use a threshold of 1kb as a cut-off for defining cis contacts to eliminate any possibility that two paired reads align to different restriction fragments as a result of either failed digestion or re-ligation. For the Ramani et al. dataset, we only used data deposited in GEO, specifically the ML3 dataset (GSM2254217). Also since the Ramani dataset is a species mixture experiment, we aligned reads to both the human and mouse genomes and only considered reads if they aligned to either the human or mouse genome, but not both. We also noted that Flyamer et al. perform multiple additional filtration steps after alignment due to the use of whole-genome amplification (WGA) to limit the possibility that a given ligation fragment is represented twice in a single cell due to WGA. We did not perform similar filtration steps, as we believed that using a single analytical pipeline as opposed to bespoke sample specific filtration was the least biased approach to compare across datasets and methods. Therefore our pipeline reports more contacts per cell (2,416,802 long range cis contacts) than is reported by Flyamer et al. (1,900,000) in their manuscript.
Cell type identification using DNA methylation signature
CG methylation levels (mCG) are computed for every non-overlapping 100kb bins across the genome in each single cell. The bins with more than 20 CG basecalls in more than 90% of cells were selected for further analysis. Bin-level mCG levels were normalized by global mCG of each cell. Similar to our prior analysis using snmC-seq, we imputed the mCG in each bin with less than 20 CG basecalls by using the mean mCG of that bin across all the cells having more than 20 CG basecalls in the bin 16.
Cell type identification using 3D genome structure
We generated a contact map using 100kb bin in each cell. The interaction frequency of each bin is normalized by dividing the average interaction frequency of the bins at the same distance interactions. The bins that are covered with more than 100 cells (n=19357) were used for the PCA analysis shown in Fig. 4c,d. The bins with an interaction distance of less than 2Mb (n=18004) were used for the PCA analysis shown in Fig. 4e,f.
Quality control for human cortical sn-m3C-seq profiles.
We filtered the cells by total non-clonal reads > 500k, global mCCC < 3%, total autosomal cytosines covered < 100M and total long-range (>10k) cis contacts > 5000. We also required total long-range cis contacts of each chromosome > x, for a chromosome of length × Mb.
Visualization and clustering of human cortical cells based on methylation.
Both CG and non-CG methylation level (mCG and mCH) were computed for every non-overlapping autosomal 100kb bins. Bin-level mCG levels were normalized by global mCG of each cell. For each individual and each sequencing batch, the bins with more than 20 CG basecalls in more than 90% of cells were selected for further analysis. We imputed the mCG in each bin with less than 20 CG basecalls by using the mean mCG of that bin across all the cells having more than 20 CG basecalls in the bin. The mCG matrices of different individuals and batches were integrated together using Scanorama54 using all the bins with default parameters. The first 50 dimensions of Scanorama embedding were used for t-SNE visualization and clustering. For clustering, we used the euclidean distance of the embedding to generate the binary k-nearest neighbor graph A of all the cells with k = 20, where Aij is 1 if cell j is one of the 20 nearest neighbors of cell i. Then A was weighted by the jaccard similarity matrix of A55. Specifically, the weight of the edge between cell i and j in the final graph was the jaccard similarity between Ai and Aj. Louvain clustering was performed on the weighted graph with resolution 1.6. We used mCG to cluster all the 4238 cells, and then selected all the neurons (MEF2C+) to perform another round of clustering using mCH. The mCH matrices were processed in the same way except the basecall cutoff was set to be 100 in 99% of cells. We merged some of the clusters in order to have enough cells in each cluster, but still separating the known cell types. The clusters were annotated as known cell types based on the gene body depletion of mCG and mCH.
Visualization of human cortical cells based on chromosome interactions.
The contact matrices were generated at 1Mb resolution. We used scHiCluster for embedding the single-cell intra-chromosomal contact matrices with default parameters56. The first 20 dimensions of the embedding were used for t-SNE visualization.
Identification of differentially methylated regions (DMRs).
The single-cell methylation profiles at base resolution were merged for each cluster. CG sites from the two strands were merged to enhancer the statistic power. DMRs were identified using methylpy DMRfind50.
Identification of differential interaction regions
Single-cell contact maps were binned at a resolution of 50kb. We retained bins if they contain non-zero values in at least 10% of single cells. Differential interactions were called using edgeR. Specifically, each single cell was treated as a replicate for each corresponding cluster. The data were normalized using calcNormFactors and dispersions were estimated using estimateDisp. We performed quasi-likelihood F-tests identifying differentially interacting regions across all samples using glmQLFit and glmQLFTest. Benjamini-Hochberg corrections were applied for multiple testing, and we retained bins with FDR <0.1%. Finally, we applied additional filters such that we required the maximum cluster-wide average interaction frequency to be at least 2-fold higher than the minimum cluster-wide average interaction frequency, and that the percentage of single cells with contacts had to be least 3 fold greater in the highest cluster than the lowest.
Identification of differential domain boundaries.
For this analysis, we only used cells with more than 50k contacts. The TAD-like structures (TLSs) in single cells were identified using TopDom after scHiCluster imputation at 25kb resolution 56. For the j-th bins, we counted the number of cells where the bin was identified as TLS boundaries in cluster i, denoted as cij. Then we computed the boundary probability by , where ti is the number of cells in cluster i. For each bin, we used the contingency table O to compute the p-value by chi-square test, where Oi0 = cij and Oi1 = ti – cij, and performed multiple test correction using Benjamini-Hochberg procedure. For differential domain boundaries, we required FDR < 0.01, minjpij < 0.05 and maxjpij – minjpij > 0.1. To eliminate the effect of limited resolution for TAD identification (which could shift for 1 or 2 bins), we expanded 50kb on both sides of the selected differential boundaries and repeated the test. We used c'ij to denote the number of cells in which one of the 25kb bins in the 125kb region was called as TLS boundaries. To filter for the significant differential boundaries, we required FDR < 0.01, minjp'ij < 0.3 and maxjp'ij – minjp'ij > 0.1.
Comparison of methylation levels at differential domain boundaries.
In each cell type i, we separated 25kb bins into boundaries (pij > 0.15) and non-boundaries (pij < 0.05). We compared the mCG or mCH level between the boundaries and non-boundaries at 1) those bins, 2) CTCF motifs that overlapping with those bins, 3) the gene bodies whose TSS are within 2kb of those bins.
CTCF Methylation analysis
For analysis of CTCF ChIP-seq binding site methylation, we downloaded data from in vitro differentiated neurons generated by ENCODE (ENCSR822CEA)45,57. Methylation levels were calculated within peak regions. For comparison with differential interacting regions, the CTCF binding site methylation levels were averaged across all CTCF sites within the pair of interacting bins.
For CTCF motif analysis, we used position weight matrices generated by SELEX58. The reason we use SELEX defined motifs for this analysis is that we wanted to limit any biases that may result from using ChIP-seq defined motifs due to the possibility of CpG methylation may change the relative likelihood of observing specific motifs due to the known sensitivity of CTCF to bind to methylated CpGs at the 4th position in the motif. Motifs were identified in the genome using Homer scanMotifGenomeWide using a log-odds detection threshold of 659. We identified variably methylated sites motifs as those that showed at least 1 cell type with methylation of position 4 >80% and one cell type <20%, where at least 10 reads cover the cytosine.
Statistics.
The following statistical tests were used in this manuscript: Fisher’s Exact test and Hypergeometric tests were used to compute associations in contingency tables. Wilcoxon rank-sum tests were used to test for differences between groups and does not make any assumptions regarding the distribution of the underlying data. Pearson correlations were used to evaluate the linear relationship between samples, in particular related to replicate experiments for reproducibility. The Stratum Adjusted Correlation Coefficient was used to compare Hi-C datasets for reproducibility 26. EdgeR was used to analyze differential count data between groups39, namely Hi-C contact frequencies. It assumes the data follows an underlying Negative Binomial distribution. All statistical tests were two-sided.
Reporting Summary.
Further information regarding the experimental design, key resources, statistical analysis, and software used in this study can be found in the Nature Research Reporting Summary linked to this article.
Data Availability Statement
Raw data and processed data for culture mouse cells mESC and NMuMG are available from NCBI GEO accession GSE124391. Raw data and processed data for human prefrontal cortex are available from GEO accession GSE130711. Intermediate files for DNA methylation and chromatin contacts can be downloaded from https://github.com/dixonlab/scm3C-seq
Code Availability Statement
The source code used is publicly available at https://github.com/dixonlab/Taurus-MH and https://github.com/dixonlab/scm3C-seq
Supplementary Material
Acknowledgments
This work was supported by NIH grant 5R21HG009274 to J.R.E and DP5OD023071 to J.R.D. J.R.E is a Howard Hughes Medical Institute investigator. J.R.D is also supported by the Leona M. and Harry B. Helmsley Charitable Trust grant No. 2017-PG-MED001 and a grant from the Salk Institute Innovation Research Fund. This work was also supported by the Flow Cytometry Core Facility of the Salk Institute with funding from NIH-NCI CCSG: P30 014195. We would like to thank the ENCODE consortium and the laboratory of Dr. Michael Snyder from Department of Genetics, Stanford University for the generation of CTCF ChIP-seq data used in this manuscript (GSE127577, ENCODE accession ENCSR822CEA).
Footnotes
Competing Financial Interests Statement
The authors declare no competing interests.
References
- 1.Dixon JR, Gorkin DU & Ren B Chromatin Domains: The Unit of Chromosome Organization. Mol. Cell 62, 668–680 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rowley MJ & Corces VG The three-dimensional genome: principles and roles of long-distance interactions. Curr. Opin. Cell Biol. 40, 8–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dekker J & Heard E Structural and functional diversity of Topologically Associating Domains. FEBS Lett. 589, 2877–2884 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixon JR et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nora EP et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sexton T et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Phillips-Cremins JE et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153, 1281–1295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rao SSP et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonev B et al. Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell 171, 557–572.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon JR et al. Chromatin architecture reorganization during stem cell differentiation. Nature 518, 331–336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitt AD et al. A Compendium of Chromatin Contact Maps Reveals Spatially Active Regions in the Human Genome. Cell Rep. 17, 2042–2059 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagano T et al. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 547, 61–67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagano T et al. Single-cell Hi-C for genome-wide detection of chromatin interactions that occur simultaneously in a single cell. Nat. Protoc. 10, 1986–2003 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Nagano T et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502, 59–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Lin D, Yardimci GG & Noble WS Unsupervised embedding of single-cell Hi-C data. Bioinformatics 34, i96–i104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo C et al. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science 357, 600–604 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hui T et al. High-Resolution Single-Cell DNA Methylation Measurements Reveal Epigenetically Distinct Hematopoietic Stem Cell Subpopulations. Stem Cell Reports 11, 578–592 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee DS, Luo C, Zhou J, Chandran S, Rivkin A, Bartlett A, Nery JR, Fitzpatrick C, O’Connor C, Dixon JR, Ecker JR. Single-cell multi-omic profiling of chromatin conformation and DNA methylation. Protocol Exchange doi: 10.21203/rs.2.11454/v1 [DOI] [Google Scholar]
- 19.Sajan SA & Hawkins RD Methods for identifying higher-order chromatin structure. Annu. Rev. Genomics Hum. Genet. 13, 59–82 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Luo C et al. Robust single-cell DNA methylome profiling with snmC-seq2. Nat. Commun 9, 3824 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krueger F & Andrews SR Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pedersen BS, Eyring K, De S, Yang IV & Schwartz DA Fast and accurate alignment of long bisulfite-seq reads. arXiv [q-bio.GN] (2014). [Google Scholar]
- 23.Li H Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [q-bio.GN] (2013). [Google Scholar]
- 24.Habibi E et al. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell 13, 360–369 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Lee D-S et al. An epigenomic roadmap to induced pluripotency reveals DNA methylation as a reprogramming modulator. Nat. Commun. 5, 5619 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang T et al. HiCRep: assessing the reproducibility of Hi-C data using a stratum-adjusted correlation coefficient. Genome Res. 27, 1939–1949 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gravina S, Dong X, Yu B & Vijg J Single-cell genome-wide bisulfite sequencing uncovers extensive heterogeneity in the mouse liver methylome. Genome Biol. 17, 150 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu B et al. Genome-wide, Single-Cell DNA Methylomics Reveals Increased Non-CpG Methylation during Human Oocyte Maturation. Stem Cell Reports 9, 397–407 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark SJ et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Commun. 9, 781 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smallwood SA et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods 11, 817–820 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramani V et al. Massively multiplex single-cell Hi-C. Nat. Methods 14, 263–266 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flyamer IM et al. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544, 110–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan L, Xing D, Chang C-H, Li H & Xie XS Three-dimensional genome structures of single diploid human cells. Science 361, 924–928 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nora EP et al. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 169, 930–944.e22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee D-S et al. An epigenomic roadmap to induced pluripotency reveals DNA methylation as a reprogramming modulator. Nat. Commun. 5, 5619 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu F, Liu Y, Jiang L, Yamaguchi S & Zhang Y Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev. 28, 2103–2119 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee S-M et al. Intragenic CpG islands play important roles in bivalent chromatin assembly of developmental genes. Proc. Natl. Acad. Sci. U. S. A. 114, E1885–E1894 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lister R et al. Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou J et al. Robust single-cell Hi-C clustering by convolution- and random-walk–based imputation. Proc. Natl. Acad. Sci. U. S. A. 116, 14011–14018 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyoshi G et al. Prox1 Regulates the Subtype-Specific Development of Caudal Ganglionic Eminence-Derived GABAergic Cortical Interneurons. J. Neurosci. 35, 12869–12889 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merkenschlager M & Nora EP CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annu. Rev. Genomics Hum. Genet. 17, 17–43 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Wang H et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 22, 1680–1688 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zimmermann B, Bilusic I, Lorenz C & Schroeder R Genomic SELEX: a discovery tool for genomic aptamers. Methods 52, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Regev A et al. Science forum: the human cell atlas. Elife 6, e27041 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods Only References
- 47.Langmead B Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformatics 32, 11–17 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abdennur N & Mirny L Cooler: scalable storage for Hi-C data and other genomically-labeled arrays. doi: 10.1101/557660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kerpedjiev P et al. HiGlass: web-based visual exploration and analysis of genome interaction maps. Genome Biol. 19, 125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schultz MD et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 523, 212–216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daley T & Smith AD Predicting the molecular complexity of sequencing libraries. Nat. Methods 10, 325–327 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stevens TJ et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544, 59–64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dixon JR et al. Integrative detection and analysis of structural variation in cancer genomes. Nat. Genet. (2018). doi: 10.1038/s41588-018-0195-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hie B, Bryson B & Berger B Efficient integration of heterogeneous single-cell transcriptomes using Scanorama. Nat. Biotechnol. (2019). doi: 10.1038/s41587-019-0113-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Levine JH et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 162, 184–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou J et al. HiCluster: A Robust Single-Cell Hi-C Clustering Method Based on Convolution and Random Walk. bioRxiv 506717 (2018). doi: 10.1101/506717 [DOI] [Google Scholar]
- 57.Davis CA et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 46, D794–D801 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jolma A et al. DNA-binding specificities of human transcription factors. Cell 152, 327–339 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data and processed data for culture mouse cells mESC and NMuMG are available from NCBI GEO accession GSE124391. Raw data and processed data for human prefrontal cortex are available from GEO accession GSE130711. Intermediate files for DNA methylation and chromatin contacts can be downloaded from https://github.com/dixonlab/scm3C-seq