Abstract
We report a method, Methyl-HiC, which captures the chromosome conformation and DNA methylome together in a cell. We use it to reveal coordinated DNA methylation status between distal genomic segments that are in spatial proximity in the nucleus and delineate heterogeneity of both the chromatin architecture and DNA methylome in a mixed population. Methyl-HiC enables simultaneous characterization of cell-type specific chromatin organization and epigenome in complex tissues.
DNA methylation is dynamically regulated1–3 and exhibits characteristic patterns in different cell types4. DNA methylation can be profiled by whole genome bisulfite sequencing (WGBS) at base resolution5, which shows that the methylation status of adjacent CpGs is often correlated6,7. Since DNA is spatially organized into three-dimensional structures, distal genomic regions may be brought into proximity through chromatin folding8. Thus, it is possible that spatially proximal DNA sequences may also exhibit coordinated DNA methylation. However, conventional WGBS have limited power in detecting coordinated DNA methylation across large genomics distances due to short DNA fragments profiled in such assay.
The chromosome conformation capture (3C) technologies have been used to study the chromatin architecture9–11. In 3C experiments, spatial proximity is captured through restriction digestion and in situ ligation of proximal DNA segments12. In principle, methylation status of cytosines on the ligated DNA is preserved in 3C experiments and can be detected through bisulfite sequencing approaches. We therefore combine in situ Hi-C11 and WGBS5 to simultaneously profile chromatin conformation and DNA methylation. We name this method Methyl-HiC, which begins with a standard in situ HiC procedure, followed by an additional step of bisulfite conversion before library construction and paired-end sequencing (Supplementary Fig. 1a). The methylation status of cytosines and the pairwise contact frequencies are then determined using custom software (Supplementary Fig. 1b–e) (see Methods for description).
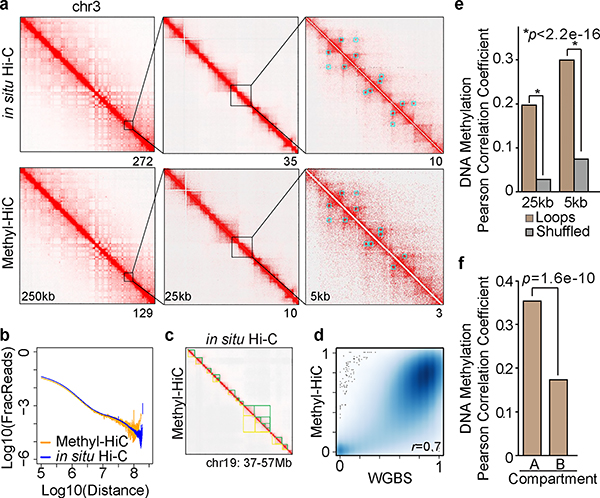
To demonstrate the performance of Methyl-HiC, we applied it to mouse embryonic stem cells (mESCs) (Supplementary Fig. 1f). The resulting contact matrix is highly similar to that from in situ Hi-C (Fig. 1a), with indistinguishable distribution of contact probabilities as a function of genomic distances (Fig. 1b). Similar sets of chromatin loops were detected from both assays (Fig. 1a, Supplementary Fig. 2a, 2b, and Supplementary Table 1). Comparable topologically associating domains (TADs) were also identified from the two datasets (Fig. 1c, Supplementary Fig. 2c and 2d). We also compared the Methyl-HiC results with WGBS data from the same cell type. The Methyl-HiC data captured over 16 million CpGs in the mouse genome and the methylation levels of these CpGs were highly consistent with that from WGBS (Supplementary Fig. 2e and Fig. 1d). Notably, compared to WGBS, Methyl-HiC profiled about 20% fewer CpGs and more CpGs in open chromatin regions such as promoters and enhancers, likely due to reduced access to heterochromatin by the restriction enzyme (Supplementary Fig. 2e and 2f). These results, taken together, demonstrate that Methyl-HiC can simultaneously and accurately profile both the general features of chromosomal architecture and >80% of the methylome in a biological sample.
Fig.1. Methyl-HiC simultaneously profiles long-range chromatin interactions and DNA methylome in mouse embryonic stem cells.
a. Comparison of contact matrix between in situ Hi-C and Methyl-HiC at different resolutions. Blue squares are loops identified from corresponding dataset at 25kb resolution. Numbers below each map show the maximum contact values. b. Comparison of contact frequency distance decay curve obtained from in situ Hi-C and Methyl-HiC data. c. A snapshot on chromosome 19 showing TADs identified from in situ Hi-C and Methyl-HiC (yellow and green triangles, respectively). d. Pearson correlation for DNA methylation in common CpGs with 15X coverage (n=1,161,200) between WGBS and Methyl-HiC datasets (p<2.2e-16). e. Pearson Correlation Coefficient of DNA methylation concordance between chromatin loop anchors identified using HiCCUPS11 at 5kb and 25kb resolutions. Only reads containing 2 or more CpGs on each end were included (n=13,645 for 5kb and n=53,645 for 25kb). Paired reads located within the same loop anchors were re-shuffled and used to calculate the expected values. Significance was assessed using Fisher’s (1925) z in cocor.indep.groups function at cocor package in R21. f. Pearson Correlation Coefficient of DNA methylation on reads from loop anchors in compartment A (n=14,016) compared to those in compartment B (n=12,126). Significance was assessed using Fisher’s (1925) z in cocor.indep.groups function at cocor package in R.
Methyl-HiC allows us to test the hypothesis that cytosines linearly separated but positioned proximally in space may also have coordinated methylation status. We analyzed the CpG methylation on Methyl-HiC reads separated in chromatin loop anchors (Supplementary Fig. 3a–c). Indeed, the methylation status of CpGs in Methyl-HiC read pairs mapping to separated loop anchors showed a significant correlation (Fig. 1e and Supplementary Fig. 3d). Further, such correlation was not restricted to chromatin loops mediated by CTCF13, as loop anchors not occupied by CTCF also showed significant correlation of DNA methylation (Supplementary Fig. 3e). We further tested whether chromatin organizational features influence such coordinated methylation. Indeed, Methyl-HiC read pairs within the same TADs exhibited slightly higher DNA methylation concordance than these from two different TADs (Supplementary Fig. 3f). Although read pairs in either compartment A or B showed similar DNA methylation concordance (Supplementary Fig. 3g), CpGs from loop anchors showed higher methylation concordance in compartment A than those in compartment B (Fig. 1f). We also observed a weaker but significant concordance for inter-chromosomal interactions (Supplementary Fig. 3h). Finally, we observed that the degree of DNA methylation concordance varies depending on the underlying chromatin states (Supplementary Fig 4). Taken together, the above results demonstrate that Methyl-HiC can reveal coordinated DNA methylation between linearly separated but spatially close genomic regions.
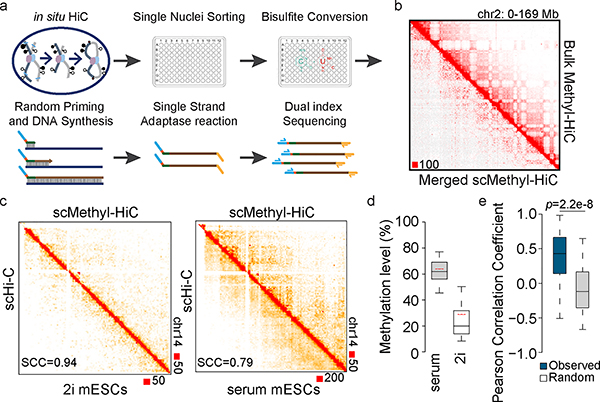
To further demonstrate the utility of Methyl-HiC, we used it to analyze chromatin conformation and DNA methylation together in single cells. Cellular heterogeneity presents a significant challenge for chromatin architecture analysis in complex tissues4,14. Single cell Hi-C has previously been performed to characterize dynamic chromatin organization in early embryogenesis and during cell cycle15,16, and to uncover the heterogeneity of chromatin architecture within a population of cells17. Single cell Methyl-HiC can in principle resolve the heterogeneity of both DNA methylation and chromatin organization from the same cells. To achieve this, we adapted the above Methyl-HiC protocol for single cell analysis. Briefly, after proximity ligation we sorted individual nucleus into 96-well plates where bisulfite conversion was carried out with each nucleus. The resulting DNA was then amplified for sequencing (Fig. 2a). We generated single cell Methyl-HiC (scMethyl-HiC) data for 103 mESCs cultured in serum+LIF condition (serum mESCs) and 47 mESCs cultured in 2i+LIF condition (2i mESCs), obtaining a median number of 80,763 informative contacts per nucleus for serum mESCs and 64,618 for 2i mESCs (Supplementary Table 2). The contact probabilities of the aggregate scMethyl-HiC was comparable to the bulk dataset (Fig. 2b, Supplementary Fig. 5a–c), and to previously published single cell Hi-C data of mESCs15 (Fig. 2c). The scMethyl-HiC experiments also generated DNA methylomes with an average of 567,380 CpGs per nucleus, comparable to previous single cell Methylome datasets4,18 (Supplementary Table 2). The average methylation levels of CpGs in serum and 2i mESCs were 61.9% and 23.4% while the methylation levels of CpH were 1.3% and 0.9% (Fig. 2d and Supplementary Table 2), respectively, which are also consistent with previous observations18. As in bulk Methyl-HiC data, DNA methylation at the chromatin loop anchors was coordinated in individual cells (Fig. 2e). These results showed that Methyl-HiC can simultaneously profile DNA methylation and chromatin architecture in single cells.
Fig.2. Simultaneous analysis of DNA methylome and chromatin architecture in individual cells by single cell Methyl-HiC.
a. Workflow of single cell Methyl-HiC. b. Comparison between ensemble contact matrix of single cell Methyl-HiC from 103 serum mESCs and bulk Methyl-HiC on chromosome 2. Contact matrixes are normalized by sequencing coverage and showed at 1Mb resolution. c. Comparison of contact matrixes on chromosome 14 between same number (n=50) of ensemble single cell Hi-C (scHi-C)15 and single cell Methyl-HiC for mESCs in 2i and serum conditions. Matrix similarity is evaluated by HiCRep22 at 1Mb resolution. SCC means stratum-adjusted correlation coefficient. d. The comparison of methylation levels distribution between serum (n=103) and 2i (n=47) mESCs from scMethyl-HiC. Boxes are median with quartiles and whiskers extend to 5th and 95th percentile. Red dotted lines show the average methylation levels from previously published data18. e. DNA methylation concordance on loop anchors in individual single cell Methyl-HiC datasets. Chromatin loops are identified from bulk Methyl-HiC using HiCCUPS (n=3110). Boxes are median with quartiles and whiskers extend to 5th and 95th percentile.
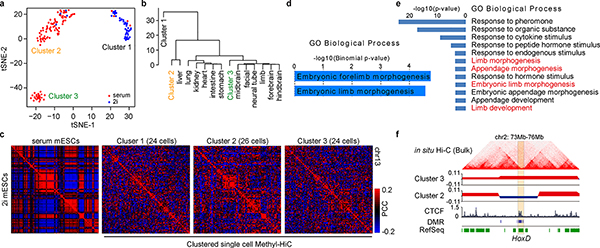
To reveal the heterogeneity of cell population using the above scMethyl-HiC data, we first clustered all the cells based on the DNA methylome of each cell alone. Interestingly, in addition to a clear separation between mESCs grown in serum and 2i conditions, the mESCs grown in serum condition were further divided into two subpopulations, consistent with a previous report18 (Fig. 3a). We compared these subpopulations with DNA methylation profiles from multiple cell lineages and found that Cluster 3 showed potential embryonic limb development trend (Fig. 3b). The aggregate chromatin contact matrixes in each cluster also showed clear cluster-specific chromosome conformation (Fig. 3c), evidenced by differential compartments between clusters (Supplementary Fig. 6a and Table 3) that were accompanied by Differentially Methylated Regions (DMRs) (Supplementary Fig. 6b–d). Similarly, DMRs and genes located in these differential compartments were enriched with GO terms related to embryonic limb development (Fig. 3d and 3e). For example, Epha4 and genes within the HoxD locus19 were differentially methylated and exhibit differential compartment between Cluster 2 and Cluster 3 (Fig. 3f, Supplementary Fig. 6e and 6f). These results suggest that scMethyl-HiC can be used to resolve cell-type specific chromosomal architecture in heterogeneous cell populations.
Fig.3. Single cell Methyl-HiC reveals heterogeneity of cultured mouse embryonic stem cells.
a. t-SNE visualization of unsupervised clustering results according to DNA methylation from individual single cell Methyl-HiC dataset (n=150). Methylation level is calculated in non-overlapping 1Mb bins. b. Unsupervised clustering of aggregated DNA methylome in subgroups of serum mESCs with tissue-specific methylomes in mouse embryonic development. c. Pearson’s correlation matrixes from different cell clusters. Similar numbers of cells are randomly selected to plot the matrixes (n=24, 26, and 24). The left map shows the bulk in situ Hi-C matrixes from serum and 2i mESCs, respectively. Pearson’s correlation is calculated under 1Mb resolution. Color ranges have been set to the same scale. d. GO biological process terms predicted by GREAT for DMRs (n=99) switching from Compartment B in Cluster 2 to Compartment A in Cluster 3. e. GO biological process terms of genes (n=672) switching from Compartment B in Cluster 2 to Compartment A in Cluster 3. P-values are from modified Fisher Exact test for gene-enrichment analysis. f. Snapshot of HoxD cluster genes in differential compartments between Cluster 2 and 3. Hi-C matrix are from bulk in situ Hi-C of the same cell line to show the chromosome organization in nearby region. Two tracks below the HiC matrix are eigenvector decomposition. The positive/negative values correspond to compartment A/B, respectively. DMRs between Cluster 2 and Cluster 3 are also plotted.
In summary, we have developed a method to study the higher-order organization of chromosomal structures and DNA methylomes simultaneously in mixed populations and in single cells. Combining two assays together allowed us to uncover coordinated DNA methylation at genomic regions that are far apart on linear sequence but spatially close in the nucleus. Combined analysis of DNA methylation and chromatin conformation in single cells further enabled us to resolve heterogeneous DNA methylome and chromosome conformation in a mixed cell population. We propose that scMethyl-HiC may be used to study cell-type specific epigenome and chromatin organization in complex tissues. While the current scMethyl-HiC protocol is still limited by data sparsity, this limitation may be overcome in the future by alternative DNA methylation detection strategies20.
METHODS
Cell culture
The F1 Mus musculus castaneus × S129/SvJae mouse ESC line (F123) was obtained from Whitehead Institute as described previously23. Serum mESCs were cultured with irradiated mouse embryonic fibroblasts (Gibco, A34180) in medium with 85% DMEM, 15% Knock-out Serum Replacement (Gibco, 10828–028), 1X penicillin/streptomycin, 1X non-essential amino acids (Gibco, 11140–050), 1X GlutaMax (Gibco, 35050), 0.4 mM β-mercaptoethanol and 1000U/ml LIF (Millipore, ESG1107). 2i mESCs were adapted from serum mESCs by passaging cells in MEF and serum free 2i medium, which contained 50% NEUROBASAL (Gibco, 21103–049), 50% DMEM/F12 (Gibco, 11320–033), 0.5% N2-SUPPLEMENT (Gibco, 17502–048), 1% B27+RA (Gibco, 17504–044), 0.05% BSA (Gibco, 15260–037), 1X penicillin/streptomycin, 2mM GLUTAMINE (Gibco, 25030–081), 150 μ M Monothioglycerol (Sigma, M6145), 1000U/ml LIF (Millipore, ESG1107), 1 μ M MEK inhibitor (Stemgent, 04–0006), and 3 μ M GSK3 inhibitor (Stemgent, 04–0004). Serum mESCs were collected and plated on 0.2% gelatin-coated feeder-free plates for 30 mins before harvesting to remove feeder cell contamination.
Methyl-HiC
In situ Hi-C was performed according to a published protocol11. Briefly, two million cells were cross-linked with 1% formaldehyde for 10 mins at room temperature. Reaction was then quenched with 0.2M Glycine. Cell pellets were washed with cold PBS and lysed with lysis buffer to get nuclei pellets. Nuclei were permeabilized with 0.5% SDS. DNA was in situ digested with 100 units of DpnII (NEB, R0543) overnight. The ends of restriction fragments were filled with biotinylated nucleotides (Invitrogen, 19524016) and in situ ligated with T4 DNA ligase (NEB, M0202). After reversal of crosslinks, ligated DNA was ethanol precipitated and sheared to about 400bp by sonication (Covaris). Sonicated products were pulled down with streptavidin beads. Library construction was then performed on beads. After adapter ligation, beads were suspended in 20ul TE buffer and subjected to bisulfite conversion with EZ DNA Methylation-Gold™ Kit (Zymo, D5005). Unmethylated lambda DNA (Promega, D1501) was sonicated and ligated with the same adapter for Methyl-HiC sample and then was spiked in at 0.5% before bisulfite conversion. Purified bisulfite converted DNA was amplified with HiFi Hotstart Uracil+Ready Mix (KAPA, KK2802).
Single cell Methyl-HiC
In situ Hi-C was performed as same as above bulk Methyl-HiC up to the steps of proximal ligation. After ligation, nuclei pellets were centrifuged and washed with PBS. Pellets were suspended in PBS stained with 1:200 DRAQ7 (CST, 7406S). The nuclei were FACS sorted into 96 well plate, one nucleus a time, containing 9ul PBS in each well. Bisulfite conversion was then performed in each well with EZ-96 DNA Methylation-Direct™ Kit according to the manufactory manual (Zymo, D5020). 0.5% of fragmented lambda DNA was spiked in before bisulfite conversion. Following bisulfite conversion of single nuclei, random priming of bisulfite-converted DNA with high concentration Klenow fragments (Enzymatics, P706L) incorporates an internal indexed P5 adapter to 5’ ends of synthesized fragments, which can be used for downstream multiplexing capability. Exonuclease I (NEB, M0293) and Shrimp Alkaline Phosphatase (NEB, M0371) treatments were then performed to digest unused random primer and inactivate dNTPs, followed by a SPRI bead-based purification step. P7 adapter were then ligated to the 3’ end of single-stranded products by Adaptase module (Swift, 33096). Library amplification was then performed using indexed primers that incorporate dual indexing to enable 96-plex sequencing. Amplified libraries were pooled together and subjected to size selection and library quantification. Please refer to the Supplementary protocol for more details.
Whole-genome bisulfite sequencing
Genomic DNA was first extracted from mESCs (Qiagen, 69504). 1–1.5 μg of genomic DNA was fragmented by sonication (Covaris), end-repaired, dA-tailed, and ligated to cytosine methylated Illumina Truseq adapter. Ligation product was subjected to bisulfite conversion reaction according to the manufacturer’s instructions (Zymo, D5005). 0.5% unmethylated λDNA was spiked-in before the conversion. Bisulfite converted DNA was then PCR amplified and purified.
Sequencing of DNA libraries
The quantification of sequencing libraries was determined by qPCR and TapeStation DNA analyzer (Agilent Technologies). Pooling of multiplexed sequencing samples, clustering, and sequencing were carried out as recommended by the manufacturer on Illumina HiSeq 2500 or HiSeq 4000. For bisulfite-converted libraries, at least 50% of balanced libraries or Phix were multiplexed to overcome the imbalance of GC ratio. All libraries were sequenced in paired-end mode.
Analysis of WGBS data
Raw reads were first trimmed as paired-end reads using Trimmomatic with default parameters to remove the adapters and low quality reads. Trimmed reads were aligned to mm9 using Bismark (v12.5). PCR duplications were removed with Picard (http://broadinstitute.github.io/picard/). CpG methylation level were calculated by Bis-SNP in bissnp_easy_usage.pl with default parameters.
Analysis of in situ Hi-C data
Each end of the raw reads was mapped separately to the mm9 reference genome using BWA-mem. Filtered reads were then paired with in-house script and de-duplicated by Picard. Reads that map to the same fragment were further removed. Contact matrices were generated at different resolution using Juicer pipeline with KR normalization and visualized using Juicebox24. Loops were then called by HiCCUPS11.
Methyl-HiC reads mapping by Bhmem
Raw reads were first trimmed as paired-end reads using Trimmomatic with default parameters to remove the adapters and low-quality reads. The reference genome was in silico converted to make C/T reference (all Cs were converted to Ts) and G/A reference (all Gs were converted to As). Paired-end reads were mapped in single end mode to each converted reference by BWA-MEM. Only uniquely mapped and mapping quality passed reads on both ends were joined. Joint read pairs with more than 20kb insertion size were considered as long-range interactions and subjected to following interaction analysis. CpG methylation level were calculated by Bis-SNP25. Only bases with quality score more than 5 were included in the downstream methylation analysis. More details were implemented in Bhmem.java.
Correlation analysis of Methylation in Methyl-HiC datasets
The reads with incompletely converted cytosine (M-bias) were filtered out as previously described25. Only read pairs with methylation level at both ends were kept for the analysis. For the regions of interests, such as HiCCUPS loops anchor regions, Pearson Correlation Coefficient (PCC) was calculated directly from the methylation level of each read of the read pair, not by the average methylation level at each end of anchor regions. Only read pairs spanning at least 20kb genomic distance were considered for the analysis. As control, the all the read pairs within the same genomic regions no matter whether they have long-range interaction or not were randomly shuffled for 100 times and then subjected to PCC calculation. Fisher’s (1925) z, implemented in “cocor.indep.groups” at R package “cocor”21, was used to assess the significance between observed PCC from interacted read pairs and PCC of the random read pairs. The scripts for this analysis is provided in MethyCorAcrossHiccups.java.
Single cell Methyl-HiC analysis
Reads were mapped by the same Bhmem pipeline with additional parameters to adapt the single cell protocol (G is converted to A in read 1 and C is converted to T in read 2; read 1 and read 2 mapped to different bisulfite converted genomes were also considered as the candidate of best pair when join them). For read pairs map to multiple locations, we first filtered out these with lower mapping quality and higher mismatch number. For these multiple mapped reads with exact mapping quality and mismatches, we then utilized the restriction enzyme cutting sites on the reference genome to help with the reads mapping. Only read pairs with restriction enzyme cutting sites near the reads (+/−50bp nearby alignment start and end of the reads) were kept for the further analysis. Low quality reads, PCR duplicates, and read ends originating from same location but has more than 10 distinct paired ends were removed for the further analysis. Reads originating from distinct restriction fragments and have linear distance larger than 1kb in cis were considered as informative contacts. Contact matrices were generated at different resolution using Juicer pipeline with VC normalization and visualized using Juicebox24. Ensemble matrix similarity was evaluated by HiCRep22.
Methylation density on CpG and CpH were calculated with Bis-SNP. Methylation level in each non-overlapped 1Mb window was further calculated and bin with missing data was replaced by the mean methylation level of bins at the same location across all single cells. Bins with missing data in all of cells were removed from analysis. Then t-Distributed Stochastic Neighbor Embedding (t-SNE) was applied to represent the methylation level structure in single cell level (Rtsne package in R 3.4.0 with plexity=10 and max_iter=5000). Different plexity level and random seed were applied to test the robustness of representation. Methylated count and total count (missing data is not replaced by mean value yet) at each window within each sub-cluster was aggregated and then calculated the methylation level. DNA methylomes of different development lineages were from ENCODE. Methylation density was also calculated in non-overlapped 1Mb windows. Euclidean distance between 3 merged scMethyl-HiC clusters and external ENCODE methylomes was calculated and clustered by “ward.D2” implemented at hclust function in R.
Life Sciences Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Figures show merged data from all replicates. The bulk Methyl-HiC, single cell Methyl-HiC, and WGBS datasets generated in this study have been deposited in the Gene Expression Omnibus (GEO) under the accession number GSE119171. Previously published data used for this study are listed in Supplementary Notes.
Code availability
All the source code for Bhmem software is publicly available at Bitbucket via https://bitbucket.org/dnaase/bisulfitehic/src/master/
Supplementary Material
Acknowledgments
We thank Samantha Kuan and Dr. Bin Li for processing of sequencing. We also thank Dr. Ramya Raviram for kindly revision and comments on the manuscript. The study was supported in part by the NIH 4D Nucleome program (U54DK107977) (to B.R.), 1U01HG007610-01 (to M.K.) and a start-up grant from CCHMC (to Y.L.).
Footnotes
Competing financial interests
B.R. is a cofounder of Arima Genomics, Inc.
References
- 1.Iurlaro M, von Meyenn F & Reik W DNA methylation homeostasis in human and mouse development. Current Opinion in Genetics & Development 43, 101–109 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Lyko F The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet 19, 81–92 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Guo JU, Su Y, Zhong C, Ming G.-l. & Song H Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145, 423–434 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo C et al. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science 357, 600 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lister R et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo S et al. Identification of methylation haplotype blocks aids in deconvolution of heterogeneous tissue samples and tumor tissue-of-origin mapping from plasma DNA. Nature Genetics 49, 635 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shoemaker R, Deng J, Wang W & Zhang K Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Research 20, 883–889 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon Jesse R., Gorkin David U. & Ren B Chromatin Domains: The Unit of Chromosome Organization. Molecular Cell 62, 668–680 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieberman-Aiden E et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 326, 289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon JR et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao Suhas S.P. et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmitt AD, Hu M & Ren B Genome-wide mapping and analysis of chromosome architecture. Nature Reviews Molecular Cell Biology 17, 743 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubo N et al. Preservation of Chromatin Organization after Acute Loss of CTCF in Mouse Embryonic Stem Cells. bioRxiv, 118737 (2017). [Google Scholar]
- 14.Schwartzman O & Tanay A Single-cell epigenomics: techniques and emerging applications. Nature Reviews Genetics 16, 716 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Nagano T et al. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 547, 61–67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flyamer IM et al. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544, 110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramani V et al. Massively multiplex single-cell Hi-C. Nat Methods 14, 263–266 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smallwood SA et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nature Methods 11, 817 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodríguez-Carballo E et al. The HoxD cluster is a dynamic and resilient TAD boundary controlling the segregation of antagonistic regulatory landscapes. Genes & Development 31, 2264–2281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y et al. Bisulfite-free direct detection of 5-methylcytosine and 5-hydroxymethylcytosine at base resolution. Nature Biotechnology 37, 424–429 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Diedenhofen B & Musch J cocor: A Comprehensive Solution for the Statistical Comparison of Correlations. PLOS ONE 10, e0121945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang T et al. HiCRep: assessing the reproducibility of Hi-C data using a stratum- adjusted correlation coefficient. Genome Research (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gribnau J, Hochedlinger K, Hata K, Li E & Jaenisch R Asynchronous replication timing of imprinted loci is independent of DNA methylation, but consistent with differential subnuclear localization. Genes Dev 17, 759–73 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durand NC et al. Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom. Cell systems 3, 99–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Siegmund KD, Laird PW & Berman BP Bis-SNP: Combined DNA methylation and SNP calling for Bisulfite-seq data. Genome Biology 13, R61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Figures show merged data from all replicates. The bulk Methyl-HiC, single cell Methyl-HiC, and WGBS datasets generated in this study have been deposited in the Gene Expression Omnibus (GEO) under the accession number GSE119171. Previously published data used for this study are listed in Supplementary Notes.