Abstract
Objective
To investigate the role of E26 transformation-specific variant 4 (ETV4) in sorafenib and cisplatin resistance in hepatocellular carcinoma (HCC).
Methods
HCC cell lines SMMC-7721 and HCC-LM3 were transfected with an ETV4- overexpressing plasmid or small interfering RNAs (siRNAs) targeting ETV4. The cells with ETV4 overexpression or ETV4 interference were treated with DMSO, sorafenib (5 μmol/L) or cisplatin (5 μmol/L) for 48 h, and the total protein and total RNA were collected. Western blotting, flow cytometry, EdU proliferation assay were used to analyze the apoptosis and proliferation of the cells. We also obtained clinical specimens of HCC tissues and paired adjacent tissues from 11 patients for detecting ETV4 mRNA expression levels using real-time fluorescence quantitative PCR (q-PCR). The effect of ETV4 interference on the mRNA expression levels of immediate early response gene 3 (IER3) was examined in HCC cells that were treated with DMSO, sorafenib or cisplatin for 48 h.
Results
The expression of ETV4 mRNA was significantly higher in HCC tissues than in the paired adjacent tissues. Overexpression of ETV4 in the HCC cell lines obviously inhibited cell apoptosis induced by sorafenib or cisplatin. Conversely, ETV4 interference significantly enhanced the apoptosis and inhibited the proliferation of the HCC cells following treatments with sorafenib or cisplatin. In addition, ETV4 regulated the mRNA expression levels of IER3 in the cells treatmed with sorafenib and cisplatin.
Conclusion
ETV4 promotes resistance of HCC cells to sorafenib or cisplatin in vitro.
Keywords: hepatocellular carcinoma, E26 transformation-specific variant 4, drug resistance, apoptosis, proliferation
Abstract
目的
研究E26特异性转化变异体4(ETV4)对肝细胞癌(HCC)顺铂和索拉非尼耐药的影响。
方法
顺铂耐药HCC细胞株SMMC-7721(对索拉非尼敏感)和HCC-LM3(对索拉非尼不敏感)经质粒转染诱导ETV4过表达或用siRNA干扰ETV4后, 分别用DMSO、索拉非尼(5μmol/L)或顺铂(5μmol/L)刺激48 h并收集总蛋白或总RNA。采用Western blotting、流式细胞术、EdU增殖检测法检测处理后HCC细胞凋亡水平和增殖能力的变化。利用实时荧光定量PCR(q-PCR)技术检测11例HCC患者肿瘤组织及配对的癌旁组织中ETV4 mRNA的表达水平。肝癌细胞株经干扰ETV4后, 分别用DMSO、索拉非尼或顺铂刺激48 h, 利用q-RCR技术检测早期反应基因3(IER3)的mRNA表达水平。
结果
肝癌组织中ETV4 mRNA的表达水平显著高于对应的癌旁组织。在两株肝癌细胞中过表达ETV4能显著减少索拉非尼或顺铂诱导的细胞凋亡, 而干扰ETV4能显著增加索拉非尼或顺铂诱导的细胞凋亡, 并显著抑制索拉非尼或顺铂刺激下的细胞增殖能力。此外, 在索拉非尼或顺铂的刺激下ETV4能调节IER3的mRNA表达水平。
结论
ETV4过表达能促进HCC细胞对索拉非尼或顺铂产生耐药。
Keywords: 肝细胞癌, E26特异性转化变异体4, 耐药, 细胞凋亡, 细胞增殖
INTRODUCTION
Hepatocellular carcinoma (HCC), the sixth most common tumor and the third leading cause of cancer mortality worldwide[1, 2], poses serious health threat, for which an effective therapy has not been available. Currently HCC is treated primarily by surgical resection, interventional therapy, radiotherapy and drug therapy. Sorafenib has been the first-line drug for advanced HCC since 2007 and as a multi-target tyrosine kinase inhibitor (TKI), it is shown to significantly improve the overall survival (OS) of the patients[3]. In some cases, however, the effectiveness of sorafenib can be compromised by the development of drug resistance. In recent years, researchers have been trying to develop other TKI drugs, such as everolimus and monoclonal antibodies (ramucirumab)[4], but so far they remain only second-line drugs for patients with sorafenib intolerance or resistance.
Sorafenib resistance involves multiple pathways including the Ras/Raf/MEK/ERK, PI3K/AKT and JAK/ STAT pathways[5]. ERK1/2 is one of the many targets of sorafenib, which inhibits ERK1/2 phosphorylation to suppress its activation and the transcription of the related genes. But long-term use of sorafenib can result in reactivation of ERK1/2 and hence drug resistance; some researchers proposed that HCC cells develop drug resistance by activating autophagy [6]. Although the mechanism of sorafenib resistance is becoming clear, effective methods have not been found to solve this problem.
Cisplatin is a widely used chemotherapeutic drug in transcatheter arterial chemoembolization (TACE) for various cancers. Cisplatin has been shown to activate ERK and promote autophagy in ovarian cancer cells to lower its apoptosis-inducing effect and result in drug resistance[7]. Studies have identified an array of molecules that participate in cisplatin resistance, such as BCL-2[8], ERCC1[9], MAST1[10], MRP2/ABCC2[11] and survivin[12], but these studies failed to provide convincing evidence to support the clinical relevance of these molecules.
E26 transformation-specific (ETS) variant 4 (ETV4; also known as PEA3 or E1AF) is a member of the ETS family, whose other members include ETV1 and ETV5[13]. Highly expressed in several cancers including prostate cancer, breast cancer and liver cancer, ETV4 functions as a co-transcription factor and forms a transcription complex[14]; it also directly binds to the promoter regions to promote the transcription of the downstream target genes such as MYC[15], cyclin D1 and MMP1 and play roles in promoting cell proliferation, migration and invasion[16-19]. In addition, ETV4 is a classic downstream of Capicua (CIC), which inhibits the proliferation and metastasis of liver cancer cells by decreasing ETV4 expression[20]. More importantly, ETV4 is also a downstream factor of K-RAS and MET pathways[21], which strongly suggests the association of ETV4 with sorafenib and cisplatin resistance.
Immediate-early response 3 (IER3, also known as IEX-1) is highly expressed in many cancers and widely recognized as an oncogene. IER3 is reported to participate in many biological processes to inhibit the apoptosis and promote the survival of the cancer cells; it is also associated with the development and aggressive behaviors of carious cancers[22-25]. In addition, IER3 is involved in the modulation of responses to DNA damage[26]and the resistance to the chemotherapeutic drugs including tamoxifen[27] and sorafenib[28], suggesting its likely involvement in not only sorafenib resistance but also cisplatin tolerance. In the present study, we aimed to examine the roles of ETV4 and IER3 in sorafenib or cisplatin resistance in HCC cell lines.
MATERIALS AND METHODS
HCC tissue samples
Clinical specimens of HCC tissues and paired adjacent tissues were collected from 11 patients undergoing surgical resection of the tumor in the Department of Hepatobiliary Surgery, Nanfang Hospital of Southern Medical University (Guangzhou, China) between March, 2016 and October, 2017 and stored in liquid nitrogen. All the specimens were confirmed by pathological diagnosis. This study was conducted with approval by the Ethics Committee of Nanfang Hospital. Written informed consent was obtained from all the patients and the anonymity of the samples was carefully maintained.
Cell lines
Human HCC cell lines SMMC-7721 and HCC-LM3 were purchased from the cell bank of Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Science (Shanghai, China). Both the cell lines are resistant to cisplatin and sensitive to sorafenib. The cells were maintained routinely at 37 ℃ with 5% CO2. SMMC-7721 cells were incubated in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS; Gemini). HCC-LM3 cells were cultured in DMEM supplemented with 10% FBS.
Cell treatment
Small interfering RNAs (siRNAs) specific to ETV4 were provided by Guangzhou Ribobio Company, and cell transfection experiments were carried out according to the instructions of RNAimax. Three siRNA constructs were used:
si-ETV4-1: CTCCGATACTATTATGAGA;
si-ETV4-2: GGTGAGCGTTACGTGTACA;
si-ETV4-3: GGCCAGCCATGAATTACGA.
The negative control sequence was synthesized by Ribobio Company, which was also responsible for interpretation of the results. ETV4 overexpression plasmid was provided by Shanghai Jikai Chemical Technology Company, and a Flag blank plasmid was used as the negative control. The plasmids were transfected into the cells via Lipofectamine 3000 (Shanghai Thermo Fisher Company). The cells were transfected with the plasmids or siRNAs for 24 h, followed by treatment with DMSO, sorafenib (5 μmol/L) or cisplatin (5 μmol/L) for 48 h.
Western blotting
After the treatments, the cells were washed with PBS twice and whole cell lysates were prepared in RIPA. BCA method was used to determine the total protein concentration. Following separation of the proteins with 10% sodium dodecyl sulphate-polyacrylamyde gel electrophoresis (SDS-PAGE), the fractionated proteins were transferred to a PVDF membrane (90 V, 120 min), which was blocked with 5% bovine serum albumin (BSA; Whiga, Guangzhou) for 30 min at room temperature and then incubated with ETV4 antibody (1:2000, Abcam), β-actin antibody (1:7500, Ray Antibody Biotech), and PARP antibody (1:1000, Proteintech) at 4 ℃ overnight. The membrane was washed with TBST 5 times (10 min each time) and incubated for 1 h with the secondary anti-rabbit or anti-mouse antibody (1:10 000, ZB-2301 HRP/ZB-2305 HRP, ZSGB-BIO) at room temperature. The membrane was washed again with TBST for 5 times (10 min each time) and illuminated by ECL luminescence method in the dark room. The protein expressions were semi-quantitatively analyzed by measuring the grayscale value using ImageJ software.
Real-time PCR
The cells with ETV4 interference were treated with DMSO, sorafenib (5 μmol/L) or cisplatin (5 μmol/L) for 48 h. The total RNA was extracted from the cells by Trizol method. Reverse transcription was performed using TAKALA reverse transcription kit (RR047A), and real-time PCR was performed using the TB Green PCR Kit (RR420A, TAKALA Biotechnology). The primer sequences used are listed below:
ETV4,
GAAAACCTAGCTTTCCACAGCC (upstream),
CAGGGGACTTGATGGCGATT (downstream);
IER3,
CCCAAGCTTATGTGTCACTCTCGCAGCTG(upstream),
CGCGGATCCGCGAAGGCCGGGTGTTGCT(downstream);
β-actin,
ACAGAGCCTCGCCTTTGCC (upstream),
GATATCATCATCCATGGTGAGCTGG(downstream);
U6,
CTCGCTTCGGCAGCACA (upstream), AACGCTTCACGAATTTGCGT (downstream).
All the primer sequences were designed by PubMed Primer-BLAST and synthesized by Shanghai Thermo Fisher Company. All q-PCR primers were verified as specific and efficient. Gene-specific amplification was performed using LightCycler® 480Ⅱ. The reaction was carried out in a 20 μL reaction volume with initial denaturation at 95 ℃ for 30 s followed by 40 thermal cycles of 95 ℃ for 5 s and 60 ℃ for 20 s. Relative quantification (2-ΔΔCt) method was used for calculating the fold changes of the target mRNA expressions relative to that ofβ-actin.Eachq-PCRassaywasperformedintriplicate.
Detection of cell apoptosis
After treatment with DMSO, sorafenib (5 μmol/L) or cisplatin (5 μmol/L) for 48 h, SMMC-7721 cells were washed with PBS and digested with trypsin containing no EDTA. The cells were resuspended in 500 μL binding buffer, in which 5 μL Annexin V-APC and 5 μL 7-AAD dye were added. Within 30 min, flow cytometry (BD LSRFortessaTM) was performed to detect the proportions of early and late apoptotic cells.
Detection of EdU cell proliferation
SMMC-7721 cells were inoculated in 96-well plates, and after treatment with DMSO, sorafenib (5 μmol/L) or cisplatin (5 μmol/L) for 48 h, 100 μL EdU (1000:1 diluted with culture medium) was added in each well. The plate was incubated at 37 ℃ with 5% CO2 for 2 h, and washed gently with PBS twice. In each well 50 μL 4% paraformaldehyde solution was added at room temperature, and 30 min later the plate was washed once with 2 mg/mL glycine solution. The cells were gently washed with PBS twice and incubated with 100 μL 0.5% Triton X-100 solution at room temperature for 30 min. After washing the cells with PBS once, 100 μL Apollo dye reaction solution was added, and the plate was incubated in darkness at room temperature for 30 min; 100 μL reagent F (1:100 diluted with deionized water) was then added, and 30 min later, the cells were washed with PBS twice and photographed under a fluorescence microscope.
Gene expression database
HCC tissue dataset in TCGA, which contains miRNAs and mRNA expression profiles of 351 liver cancer tissues and 50 paired adjacent tissues, were analyzed for mRNAexpressionsofETV4andIER3andtheircorrelation.
Gene set enrichment analysis (GSEA)
GSEA method was used to confirm which gene sets or characteristics were associated with the expression of ETV4 in the TCGA dataset. We ranked 351 HCC patients in TCGA HCC tissue dataset according to the mRNA level of ETV4, and divided the patients into 4 groups by quartile method. The group with the highest expression was defined as the high ETV4 expression group, and the group with the lowest expression was defined as the low ETV4 expression group. GSEA analysis was conducted for all the mRNAs in these two groups using GSEA V.2.0 software (<a href="http://www.broadinstitute.org/" target="_blank">http://www.broadinstitute.org/</a>).
Statistical methods
GraphPad Prism 5.01 software was used for statistical analysis of the data. Independent sample t-test, one-way ANOVA and Pearson's correlation were used for data analysis. The data was represented as Mean ± SD, and a P value less than 0.05 was considered to indicate a statistically significant difference between the groups.
RESULTS
Expression of ETV4 in HCC
We used the TCGA database for analysis of ETV4 mRNA expression in HCC and paired adjacent tissues (n=50) and also examined the mRNA levels of ETV4 in 11 fresh HCC samples using fluorescence quantitative PCR. The results showed that the expression levels of ETV4 mRNA were significantly higher in HCC tissues than in the paired adjacent tissues (Fig. 1), suggesting that ETV4 is an oncogene highly expressed in HCC.
1.
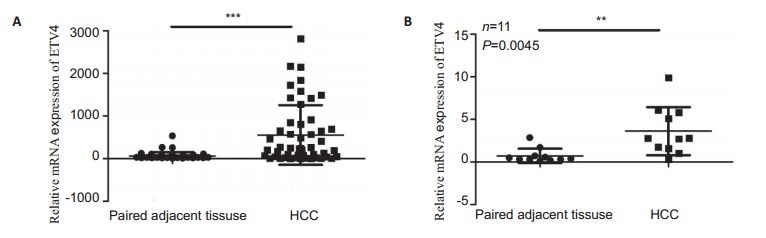
Relative mRNA expression of ETV4 in HCC and paired adjacent tissues. A: TCGA database analysis (n=50) showing high ETV4 mRNA expression in HCC tissues (error bars indicate SD; ***P < 0.0001 by t-test); B: ETV4 mRNA expression is significantly higher in fresh clinical HCC samples than in the adjacent tissues (n=11; error bars indicate SD; ***P < 0.01 by t-test).
Sorafenib or cisplatin enhances ETV4 expression in HCC cells
Western blot analysis showed that ETV4 protein expressions were significantly upregulated in HCC cell lines SMMC-7721 and HCC-LM3 after treatment with sorafenib or cisplatin (Fig. 2A). Gene set enrichment analysis (GSEA) suggested that the high expression of ETV4 was significantly correlated with 46 drug resistance genes in HCC (Fig. 2B). These results suggest the high likeliness that ETV4 is associated with drug resistance in HCC.
2.
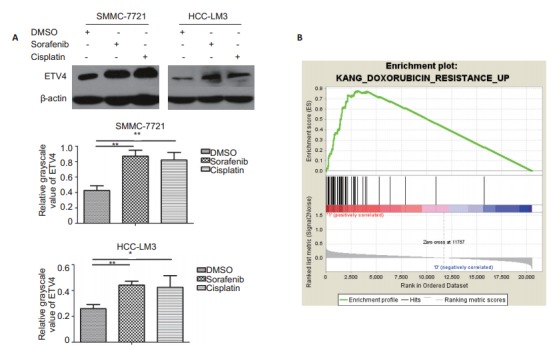
Expression of ETV4 in HCC cell lines SMMC-7721 and HCC-LM3 treated with sorafenib or cisplatin and gene set enrichment analysis (GSEA). A: Western blotting of ETV4 expression in SMMC-7721 and HCC-LM3 cells treated with DMSO, sorafenib (5μmol/L) or cisplatin (5 μmol/L) for 48 h (error bars indicate SD; **P < 0.01 by One-way ANOVA); B: GSEA for analysis of gene enrichment in high and low ETV4 expression groups showing an enrichment score (ES) of 0.7768348 and a normalized enrichment score (NES) of 2.9248652
Effects of ETV4 on sorafenib-or cisplatin-induced apoptosis of HCC cells
Western blot analysis showed that overexpression of ETV4 in SMMC-7721 cells significantly reduced sorafenib-induced apoptosis in SMMC-7721 cells, whereas ETV4 knockdown significantly increased the cell apoptosis induced by sorafenib (Fig. 3A). We found that among the 3 siRNAs, si-ETV4-2 and si-ETV4-3 had better interference efficiencies, and we thus used only si-ETV4-2 and si-ETV4-3 in the subsequent interference experiments. Similarly, overexpression of ETV4 in SMMC-7721 cells significantly reduced cisplatin-induced apoptosis and si-ETV4 significantly increased the cell apoptosis induced by cisplatin (Fig. 3B). Flow cytometry showed consistent results that interference of ETV4 significantly increased apoptosis of SMMC-7721 cells induced by either sorafenib or cisplatin (Fig. 4).
3.
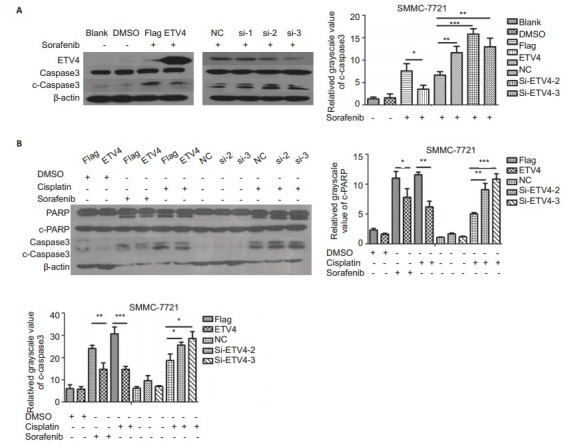
Western blotting of apoptosis-related proteins in sorafenib- or cisplatin-treated SMMC-7721 cells with ETV4 overexpression or interference. A: Western blotting of apoptosis-related proteins in SMMC-7721 cells transfected with Flag, ETV4, NC, si-ETV4-1, si-ETV4-2, or si-ETV4-3 for 24 h followed by treatment with blank, DMSO or sorafenib (5 μmol/L) for 48 h; B: Western blotting of apoptosis-related proteins in SMMC-7721 cells transfected by Flag, ETV4, NC, si-ETV4-2, or si-ETV4-3 for 24 h followed by treatment with DMSO, sorafenib (5 μmol/L), or cisplatin (5μmol/L). Data were analyzed using One-way ANOVA, and the error bars indicate SD (*P < 0.05, **P < 0.01, ***P < 0.0001)
4.
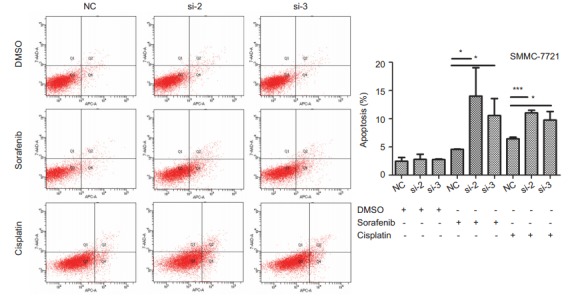
Flow cytometric analysis of SMMC-7721 cell apoptosis induced by sorafenib or cisplatin after ETV4 interference SMMC-7721 cells were transfected by NC, si-ETV4-2 or si-ETV4-3 for 24 h followed by treatment with DMSO, sorafenib (5 μmol/L) or cisplatin (5 μmol/L) for 48 h. Data were analyzed using One-way ANOVA, and the error bars indicate SD (*P < 0.05, ***P < 0.0001)
ETV4 knockdown inhibits proliferation of sorafenib- or cisplatin-treated HCC cells
The results of EdU proliferation assay showed that ETV4 knockdown using si-ETV4 significantly inhibited the proliferation of SMMC-7721 cells treated with sorafenib or cisplatin (Fig. 5). This result confirms the role of ETV4 as a key factor in the regulation of sorafenib or cisplatin resistance in HCC.
5.
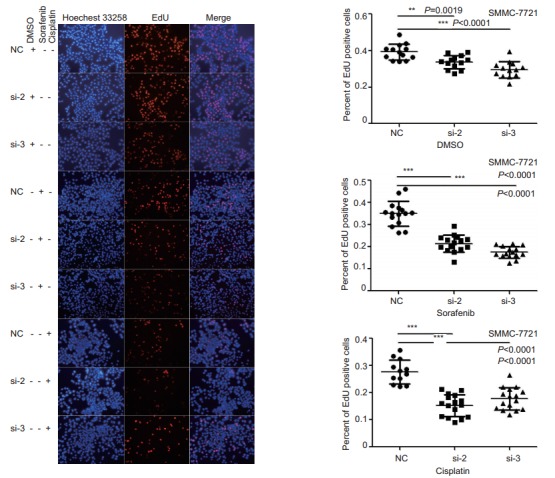
EdU proliferation assay of SMMC-7721 cells with ETV4 interference treated with sorafenib or cisplatin. SMMC-7721 cells were analyzed with EdU cell proliferation analysis after transfection with NC, si-ETV4-2, or si-ETV4-3 for 24 h followed by treatment with DMSO, sorafenib (5 μmol/L) or cisplatin (5 μmol/L) for 48 h. The cells were photographed under an inverted microscope (×200). Data were analyzed using One-way ANOVA, and the error bars indicate SD (**P < 0.01, ***P < 0.0001)
ETV4 knockdown lowers IER3 expression in sorafenib- or cisplatin-treated HCC cells
Analysis of GSE96793 database showed that IER3 was among the oncogenes upregulated by sorafenib treatment in HCC cells. The results of real-time PCR showed that in SMMC-7721 cells transfected with si-ETV4, the mRNA level of IER3 was significantly reduced after sorafenib or cisplatin treatment (<xref ref-type="fig" rid="Figure6">Fig. 6A</xref>). Meanwhile, we found that the mRNA level of IER3 increased in the cells stimulated sorafenib or cisplatin without transfection with si-ETV4, similar to the results in <xref ref-type="fig" rid="Figure2">Fig. 2A</xref>. ETV4 is mainly distributed in the nucleus and has transcriptional factor activity<sup>[<xref ref-type="bibr" rid="b30">30</xref>]</sup>, and consistently, the analysis of the JASPAR database (<a href="http://jaspar.binf.ku.dk/" target="_blank">http://jaspar.binf.ku.dk/</a>) showed that ETV4 could potentially bind to IER3 promoter region at multiple binding sites spanning 2 000 bp (<xref ref-type="table" rid="Table1">Tab. 1</xref>), indicating the very probable role of ETV in regulating IER3. Meanwhile, TCGA data analysis showed that the mRNA level of IER3 was significantly correlated with that of ETV4 (<xref ref-type="fig" rid="Figure6">Fig. 6B</xref>).
6.
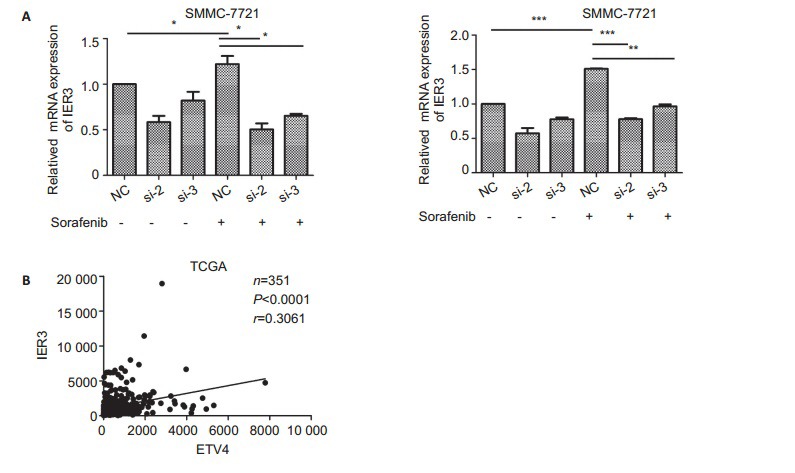
ETV4 knockdown lowers IER3 mRNA expression in HCC cells treated with sorafenib or cisplatin. A: SMMC-7721 cells were treated with DMSO, sorafenib (5 μmol/L) or cisplatin (5μmol/L) for 48 h after transfection with si-ETV4 for 24 h. Data were analyzed using One-way ANOVA, and the error bars indicate SD. *P < 0.05, **P < 0.01, ***P < 0.0001. B: TCGA database was used to analyze the mRNA correlation between IER3 and ETV4. The data were analyzed using Pearson's correlation analysis (n=351, P < 0.0001, r=0.3061)
1.
ETV4 binding sites in IER3 promoter region predicted from website
| Number | Score | Relative score | Start | End | Strand | Predicted site sequence |
| 1 | 6.782 | 0.845 | 148 | 157 | -1 | acaggatctg |
| 2 | 6.264 | 0.835 | 208 | 217 | 1 | taaggaagtt |
| 3 | 5.288 | 0.816 | 1181 | 1190 | -1 | gaaggaaatt |
| 4 | 4.742 | 0.805 | 1261 | 1270 | 1 | gcaggatggc |
| 5 | 7.115 | 0.851 | 1483 | 1492 | 1 | tcaggaaatc |
| 6 | 7.204 | 0.853 | 1586 | 1595 | 1 | tcaggaagct |
| 7 | 4.553 | 0.801 | 1599 | 1608 | 1 | gcaggaaaat |
DISCUSSION
We found that ETV4 was significantly overexpressed in HCC, and GSEA also suggested that a high expression of ETV4 was associated with doxorubicin resistance. In the two human HCC cell lines SMMC-7721 and HCC-LM3, the mRNA expression of ETV4 increased significantly in response to exposure to sorafenib or cisplatin, and exogenous overexpression of ETV4 significantly reduced cell apoptosis induced by sorafenib or cisplatin, while specific interference of ETV4 obviously increased the cell apoptosis. In addition, ETV4 knockdown significantly inhibited the proliferation of SMMC-7721 cells following treatment with sorafenib or cisplatin, indicating that ETV4 plays a role in regulating sorafenib or cisplatin resistance in HCC. This also hints that ETV4 may not only regulate drug resistance of the HCC cells in the presence of a chemotherapeutic agent, but also promote cell survival in the context of radiotherapy or hypoxia.
ETV4, as a transcription factor, was shown to have multiple likely binding sites in the IER3 promoter region to regulate the mRNA level of IER3 in HCC cells exposed to sorafenib or cisplatin. IER3 is a downstream acting factor involved in the regulation of cell clonal formation, cell growth and migration under the stimulation by sorafenib[28]. Similarly, we found that ETV4 regulated the proliferation and apoptosis of HCC cells in the presence of sorafenib or cisplatin treatment, suggesting the high likeliness that ETV4 regulates cell survival and proliferation under the stimulation by sorafenib or cisplatin through IER3. These observations shed light on new strategies for reversing drug resistance of HCC using ETV4 as the therapeutic target.
Biography
陈筱卉,硕士,E-mail: 15625179551@163.com
Funding Statement
国家自然科学基金(81672756)
Contributor Information
Xiaohui CHEN (陈 筱卉), Email: 15625179551@163.com.
Xin LI (李 欣), Email: xinli268@gmail.com.
Dehua WU (吴 德华), Email: 18602062748@163.com.
References
- 1.DaFonseca LG, Marta GN, Braghiroli MIFM, et al. Safety and efficacy of cytotoxic chemotherapy in hepatocellular carcinoma after first-line treatment with sorafenib. BMC Cancer. 2018;18(1):1250. doi: 10.1186/s12885-018-5173-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics. Ca Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Dawkins J, Webster RM. The hepatocellular carcinoma market. Nat Rev Drug Disc. 2018:146. doi: 10.1038/nrd.2018.146. [DOI] [PubMed] [Google Scholar]
- 4.Villanueva A, Llovet JM. Targeted therapies for hepatocellular carcinoma. Gastroenterology. 2011;140(5):1410–26. doi: 10.1053/j.gastro.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yun Z, Yang C, and Aimin A. Research progress on the mechanism of sorafenib resistance in primary hepatocellular carcinoma. Chin Pharmacol Bull. 2013;29(6):752–5. [Google Scholar]
- 6.Lu S, Yao Y, Xu G, et al. CD24 regulates sorafenib resistance via activating autophagy in hepatocellular carcinoma. Cell Death & Dis. 2018;9(6):646. doi: 10.1038/s41419-018-0681-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Wu GS. Role of autophagy in cisplatin resistance in ovarian cancer cells. J Biol Chem. 2014;289(24):17163–73. doi: 10.1074/jbc.M114.558288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han JY, Hong EK, Choi BG, et al. Death receptor 5 and Bcl-2 protein expression as predictors of tumor response to gemcitabine and cisplatin in patients with advanced non-small-cell lung cancer. Med Oncol. 2003;20(4):355–62. doi: 10.1385/MO:20:4:355. [DOI] [PubMed] [Google Scholar]
- 9.Metzger R, Leichman CG, Danenberg KD, et al. ERCC1 mRNA levels complement thymidylate synthase mRNA levels in predicting response and survival for gastric cancer patients receiving combination cisplatin and fluorouracil chemotherapy. J Clin Oncol. 1998;16(1):309–16. doi: 10.1200/JCO.1998.16.1.309. [DOI] [PubMed] [Google Scholar]
- 10.Jin L, Chun J, Pan C, et al. MAST1 drives cisplatin resistance in human cancers by rewiring craf-independent mek activation. Cancer Cell. 2018;34(2):315–30. doi: 10.1016/j.ccell.2018.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liedert B, Materna V, Schadendorf D, et al. Overexpression of cMOAT (MRP2/ABCC2) is associated with decreased formation of platinum-DNA adducts and decreased G2-arrest in melanoma cells resistant to cisplatin. J Invest Dermatol. 2003;121(1):172–6. doi: 10.1046/j.1523-1747.2003.12313.x. [DOI] [PubMed] [Google Scholar]
- 12.Ikeguchi M, Kaibara N. Changes in survivin messenger RNA level during cisplatin treatment in gastric cancer. Int J Mol Med. 2001;8(6):661–6. doi: 10.3892/ijmm.8.6.661. [DOI] [PubMed] [Google Scholar]
- 13.Xiao J, Yang S, Shen P, et al. Phosphorylation of ETV4 at Ser73 by cancer. Biochem Biophys Res Commun. 2017;486(4):1062–8. doi: 10.1016/j.bbrc.2017.03.163. [DOI] [PubMed] [Google Scholar]
- 14.Currie SL, Doane JJ, Evans KS, et al. ETV4 and AP1 transcription factors form multivalent interactions with three sites on the MED25 Activator-Interacting Domain. J Mol Biol. 2017;429(20):2975–95. doi: 10.1016/j.jmb.2017.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollenhorst PC, Paul L, Ferris MW, et al. The ETS gene ETV4 is required for anchorage-independent growth and a cell proliferation gene expression program in PC3 prostate cells. http://www.wanfangdata.com.cn/details/detail.do?_type=perio&id=PubMed000002224357. Genes Cancer. 2011;1(10):1044–52. doi: 10.1177/1947601910395578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyagi N, Deshmukh SK, Srivastava SK, et al. ETV4 facilitates cell-cycle progression in pancreatic cells through transcriptional regulation of cyclin D1. Mol Cancer Res. 2018;16(2):187–96. doi: 10.1158/1541-7786.MCR-17-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu C, Yu T, Huang Y, et al. ETS (E26 transformation-specific) up-regulation of the transcriptional co-activator TAZ promotes cell migration and metastasis in prostate cancer. J Biol Chem. 2017;292(22):9420–30. doi: 10.1074/jbc.M117.783787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dumortier M, Ladam F, Damour I, et al. ETV4 transcription factor and MMP13 metalloprotease are interplaying actors of breast tumorigenesis. Breast Cancer Res. 2018;20(1):73. doi: 10.1186/s13058-018-0992-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singsuksawat E, Thuwajit C, Charngkaew K, et al. Increased ETV4 expression correlates with estrogen-enhanced proliferation and invasiveness of cholangiocarcinoma cells. Cancer Cell Int. 2018;18:25. doi: 10.1186/s12935-018-0525-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim E, Kim D, Lee J, et al. Capicua suppresses hepatocellular carcinoma progression by controlling the ETV4-MMP1 axis. Hepatology. 2018;67(6):2287–301. doi: 10.1002/hep.29738. [DOI] [PubMed] [Google Scholar]
- 21.Deshmukh SK, Singh AP, Singh S. ETV4: an emerging target in pancreatic cancer. http://d.old.wanfangdata.com.cn/Periodical/wfyxyxb201002004. Oncoscience. 2018;5(9-10):260–1. doi: 10.18632/oncoscience.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Q, Hahn J, Neupane B, et al. Dysregulated IER3 expression is associated with enhanced apoptosis in Titin-based dilated cardiomyopathy. Int J Mol Sci. 2017;18(4):723. doi: 10.3390/ijms18040723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin H, Won M, Shin E, et al. EGR2 is a gonadotropin-induced survival factor that controls the expression of IER3 in ovarian granulosa cells. Biochem Biophys Res Com. 2017;482(4):877–82. doi: 10.1016/j.bbrc.2016.11.127. [DOI] [PubMed] [Google Scholar]
- 24.Garcia MN, Grasso D, Lopez-Millan MB, et al. IER3 supports KRASG12D-dependent pancreatic cancer development by sustaining ERK1/2 phosphorylation. J Clin Invest. 2014;124(11):4709–22. doi: 10.1172/JCI76037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye J, Zhang Y, Cai Z, et al. Increased expression of immediate early response gene 3 protein promotes aggressive progression and predicts poor prognosis in human bladder cancer. BMC Urology. 2018;18(1):82. doi: 10.1186/s12894-018-0388-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pawlikowska P, Leray I, DeLaval B, et al. ATM-dependent expression of IEX-1 controls nuclear accumulation of Mcl-1 and the DNA damage response. Cell Death Differ. 2010;17(11):1739–50. doi: 10.1038/cdd.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Semlali A, Oliva J, Badia E, et al. Immediate early gene X-1(IEX-1), a hydroxytamoxifen regulated gene with increased stimulation in MCF-7 derived resistant breast cancer cells. J Steroid Biochem Mol Biol. 2004;88(3):247–59. doi: 10.1016/j.jsbmb.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 28.Emma MR, Iovanna JL, Bachvarov D, et al. NUPR1, a new target in liver cancer: implication in controlling cell growth, migration, invasion and sorafenib resistance. Cell Death Dis. 2016;7(6):e2269. doi: 10.1038/cddis.2016.175. [DOI] [PMC free article] [PubMed] [Google Scholar]


