Abstract
目的
分析人类与小鼠lncRNA中的同源与种系特异性lncRNA, 揭示人类与小鼠表观遗传调控的种系差异。
方法
根据同源lncRNA序列搜索和人类/小鼠全基因组双序列比对确定GENCODE项目首期报道的13562个人类lncRNA和10481个小鼠lncRNA中的同源lncRNA基因, 用lncRNA/DNA结合分析软件LongTarget预测lncRNA的DNA结合位点和表观遗传调控靶基因, 根据Gene Ontology数据库分析靶基因的功能。
结果
仅158对人类和小鼠lncRNA被识别为同源lncRNA, 这些同源lncRNA在人类与小鼠基因组有种系特异性的DNA结合位点和靶基因, Gene Ontology分析提示这些同源lncRNA对靶基因的种系特异性调控可能对人类和小鼠的表型差异有重要影响。
结论
仅极少数人类和小鼠lncRNA是同源的, 这些同源lncRNA有种系特异性的表观遗传调控靶基因, lncRNA和表观遗传调控的种系特异性对人类和小鼠的差异(包括疾病模型的差异)有重要影响。
Keywords: 长链非编码RNA, 直系同源基因, 表观遗传调控, 人类, 小鼠
Abstract
Objective
To identify orthologous lncRNAs in human and mice and the species specificity of their epigenetic regulatory functions.
Methods
The human/mouse whole- genome pairwise alignment (hg19/mm10, genome.UCSC.edu) was used to identify the orthologues in 13 562 and 10 481 GENCODE-annotated human and mouse lncRNAs.The Infernal program was used to search the orthologous sequences of all the exons of the 13562 human lncRNAs in mouse genome (mm10) to identify the highly conserved orthologues in mice.LongTarget program was used to predict the DNA binding sites of the orthologous lncRNAs in their local genomic regions.Gene Ontology analysis was carried out to examine the functions of genes.
Results
Only 158 orthologous lncRNAs were identified in humans and mice, and many of these orthologues had species- specific DNA binding sites and epigenetic target genes.Some of the epigenetic target genes executed important functions in determining human and mouse phenotypes.
Conclusions
Only a few human and mouse lncRNAs are orthologues, and most of lncRNAs are species-specific.The orthologous lncRNAs have species-specific epigenetic target genes, and species-specific epigenetic regulation greatly contributes to the differences between humans and mice.
Keywords: long non-coding RNA, orthologous genes, epigenetic regulation, human, mice
大量研究者利用小鼠建立人类疾病的模型[1-2],尤其是许多肿瘤的模型[3-5],来研究人类疾病的发病机制和开发治疗疾病的药物。但是,尽管基因组测序揭示平均85%(60%~99%)的人类与小鼠蛋白质编码基因序列是高度一致的,依据小鼠模型开发的肿瘤治疗药物仅有约5%在人类具有安全而肯定的疗效[6]。
所有哺乳动物都存在大量lncRNA基因[7-8],DNA和组蛋白修饰酶是由lncRNA携带到特定基因组位点的[4, 9-10],lncRNA和基因组修饰对基因表达具有重要调控作用。这些发现促使研究者从表观遗传学角度研究基因表达调控的种系差异[11]。
GENCODE项目首批报道了13562个人类lncRNA和10481个小鼠lncRNA[7-8],但尚无报道揭示人类与小鼠lncRNA中哪些是同源的和哪些是种系特异的,亦无报道系统分析同源与种系特异lncRNA的表观遗传调控靶基因。为此,本研究分析这13562个人类lncRNA和10481个小鼠lncRNA,分析它们的同源/种系特异性和同源基因的表观遗传调控靶基因。
1. 数据和方法
1.1. 同源lncRNA基因识别
我们识别人类与小鼠同源lncRNA基因的方法如下:首先,同源lncRNA基因应位于人类/小鼠全基因组双序列比对中的比上部分。据此我们分析了GENCODE报道的人类与小鼠lncRNA在人类/小鼠全基因组双序列比对(hg19/mm10,genome.UCSC.edu)中的位置,然后按四个条件评判比对块是否构成同源基因:(a)比对块覆盖某个人类lncRNA基因大于50%的序列,(b)比对块覆盖某个小鼠lncRNA基因大于50%的序列,(c)比对上的人类/小鼠lncRNA基因的交集大于50%人类lncRNA序列,(d)比对上的人类/小鼠lncRNA基因的交集大于50%小鼠lncRNA序列。其次,同源lncRNA基因的序列应比较保守。为了获得序列与结构保守的同源lncRNA基因,我们用RNA序列与结构搜索软件Infernal在小鼠基因组(mm10)搜索13562个人类lncRNA的每个外显子[12-13],获得外显子的同源序列。第三,许多同源lncRNA基因包含同源转座子。为此我们根据人类和小鼠转座子注释文件(rmsk.txt,genome. UCSC.edu)分析由第一步识别的同源lncRNA基因哪些含有同源转座子(转座子的class、subclass、name均相同)。
1.2. 同源lncRNA的DNA结合位点与表观遗传调控靶基因预测
许多lncRNA能与DNA结合形成三链结构并将基因组修饰酶招募到特定的基因组位点,调控位点附近的基因组修饰和基因表达[9-10]。LongTarget软件根据lncRNA/DNA结合所遵循的碱基配对规则预测lncRNA中的DNA结合域(TFO,Triplex- Forming Oligonucleotides)和基因组中的DNA结合位点(TTS,Triplex Target Sites)[14]。我们用LongTarget (默认参数)预测了序列与结构保守的同源lncRNA在其200万bp上下游区域的DNA结合位点[15],将启动子区域有TTS的基因当做lncRNA的表观遗传调控靶基因。
1.3. 基因功能注释
我们主要使用DAVID软件对lncRNA的表观遗传调控靶基因进行功能注释<sup>[<xref ref-type="bibr" rid="b16">16</xref>]</sup>(<a href="https://david.ncifcrf.gov/" target="_blank">https://david.ncifcrf.gov/</a>)。
2. 结果
2.1. 大多数人类与小鼠lncRNA是种系特异的
根据人类/小鼠全基因组双序列比对的结果,13562个人类lncRNA中有11729个在双序列比对中有一到多个比对块,根据四个评判条件,仅158对lncRNA是同源lncRNA。进一步,根据用Infernal进行的RNA序列搜索的结果,其中仅40对同源lncRNA序列与结构均保守。再者,根据转座子分析,在158对同源lncRNA中,28对包含同源转座子。这些结果首次具体地揭示哪些人类与小鼠lncRNA是同源的,提示绝大多数人类与小鼠lncRNA是种系特异性的。
在这些同源lncRNA中,大部分在人类与小鼠均尚未被研究与注释(如RP11-329N22.1/Gm37667、RP11- 5P4.1/Gm37556),一些仅在人类(如GLYCTK-AS1/ D030055H07RIK、SNORA71A/Gm23925、SNORA58/ Gm29362)或小鼠(如RP11- 89K21.1/Six3os1、RP4- 794H19.2/Junos)被研究与注释,仅极少数在人类和小鼠中均被研究和注释(包括ZEB2-AS1/Zeb2os、LPPAS2/Lppos、HOTAIRM1/Hotairm1、HOXA11- AS/ Hoxallos、H19/H19、MALAT1/Malat1、NEAT1/Neat1、TUG1/Tug1)。这些说明目前对同源lncRNA的功能缺乏了解。
2.2. 许多人类与小鼠同源lncRNA基因含有同源转座子
我们分析了158对同源lncRNA基因中的转座子,发现114对lncRNA含有转座子,28对同源lncRNA包含同源转座子(转座子在hg19/mm10双序列比对中位于同源位置,且转座子的class、subclass、name均相同),这些结果提示同源转座子对同源lncRNA的形成与进化有重要作用。
2.3. 同源lncRNA在人类与小鼠基因组有种系特异性的DNA结合位点和靶基因
我们预测了40对序列与结构保守的同源lncRNA在它们所在基因组区域(200万bp)的DNA结合位点[15]。首先,若干同源lncRNA在人类与小鼠基因组中均有清晰的TTS,提示它们在此区域有保守的表观遗传调控功能。例如,GLYCTK-AS1/D030055H07RIK的最大TTS信号是248/232,NEAT1/Neat1的TTS信号高达681/381。其次,若干同源lncRNA仅在人类或小鼠中有清晰的TTS,提示这些同源lncRNA的种系特异性表观遗传调控功能。例如,GLYCTK-AS1在人类GLYCTK的启动子区域有清晰的TTS(图 1),但D030055H07RIK在小鼠Glyctk的启动子区域却没有。仔细分析揭示GLYCTK-AS1可能有人类特异性的DNA结合位点和靶基因,这些基因包括PARP3、ABHD14B、ACY1、RPL29、POC1A、ALAS1、PPM1M、WDR82、GLYCTK等。此外,同源lncRNA的种系特异性DNA结合位点还位于种系特异性转座子,小鼠lncRNA 5730585A16Rik是一个典型例子(图 2),揭示了种系特异性DNA结合位点与种系特异性体细胞基因组防御的联系。
1.
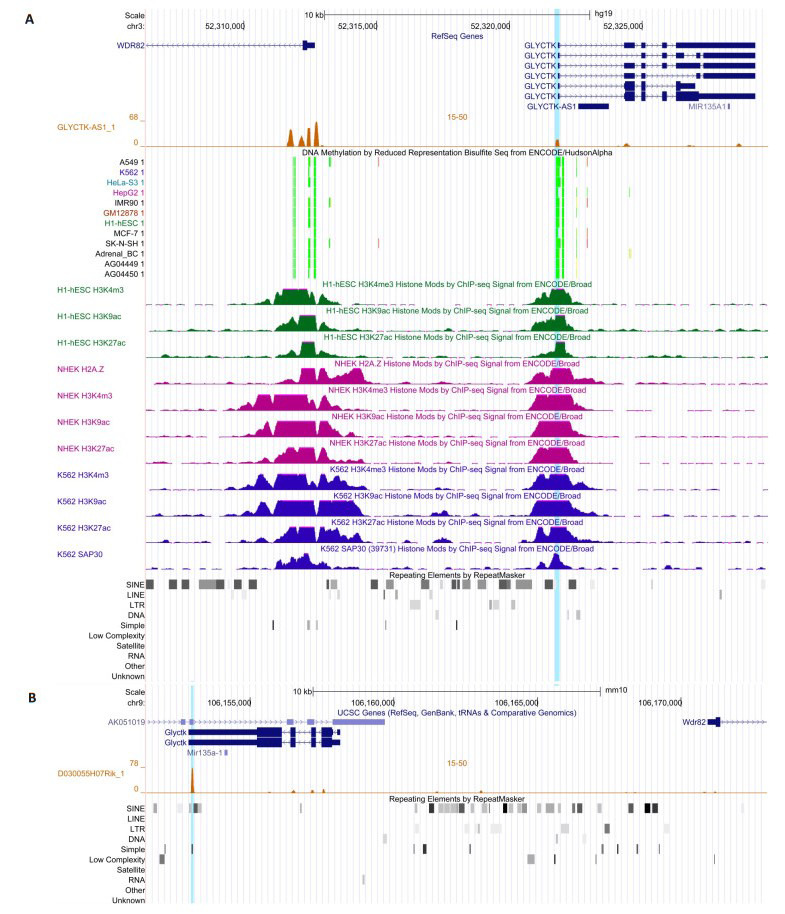
同源lncRNA在其局部基因组区域的DNA结合位点
2.
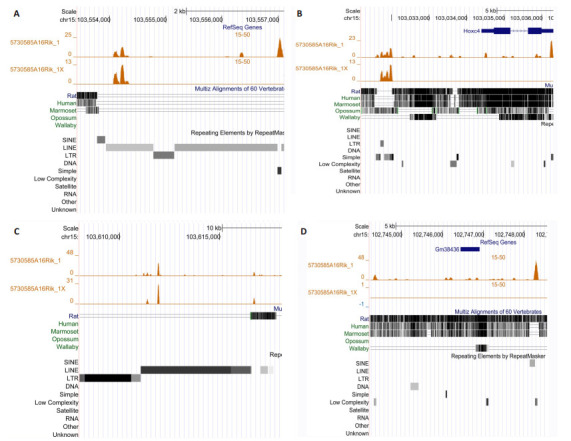
小鼠lncRNA 5730585A16Rik的许多TTS位于小鼠特异性或啮齿类特异性转座子
2.4. 种系特异性表观遗传调控可能显著影响人类与小鼠的表型差异
我们用DAVID软件(版本6.8)分析了上述种系特异性靶基因的功能[16],发现POC1A(一个携带WD40功能域的蛋白质)等3个与GLYCTK-AS1相关的靶基因与一个特殊疾病的表型有关,据此推断GLYCTK-AS1对POC1A的种系特异性表观遗传调控可能对人类与小鼠表型差异具有重要影响。
3. 讨论
基因组测序揭示平均85%的蛋白质编码基因在人类和小鼠高度一致,但对lncRNA基因的保守性尚少有具体的分析。LncRNA介导的表观遗传调控的种系特异性重要但富有争议[17-18]。本研究分析了GENCODE报道的13562个人类lncRNA和10481个小鼠lncRNA,首次具体揭示其中可能仅158个lncRNA是同源基因,这些同源lncRNA在人类和小鼠有种系特异性的DNA结合位点和靶基因,揭示了表观遗传调控的种系特异性。根据这些结果可推断,人类疾病和人类疾病的小鼠模型在表观遗传调控方面存在显著的差异。一个突出例子是许多肿瘤相关的重要分子p53,它既负责DNA修复也激活许多lncRNA表达[19-21],包括同源lncRNA TUG1/Tug1和NEAT1/Neat1[22-24]。人类TUG1能与PRC2结合抑制许多细胞周期基因,而小鼠Neat1与许多染色质结合蛋白在小鼠胚胎干细胞中相互作用。同源lncRNA TUG1/Tug1和NEAT1/Neat1所具有的与肿瘤相关的功能提示[25-26],许多人类肿瘤的小鼠模型与人类肿瘤在基因表达的表观遗传调控方面有不可忽视的差别。
本研究主要分析了序列较保守的40对同源lncRNA。由于人类与小鼠的种系距离接近9千万年且lncRNA基因可累积补偿性突变,其它同源lncRNA的序列和功能差异更大。近几年GENCODE项目以及其它研究报道了更多人类与小鼠lncRNA,但lncRNA数量的增加并不影响本研究的主要结果与结论。与此同时,我们的分析以及相关研究也揭示许多lncRNA基因有趋同进化现象,即功能相同的lncRNA基因独立起源在基因组的相近位置,典型例子包括在真哺乳类控制Kcnq1印迹区基因印迹的Kcnq1ot1和控制Igf2r印迹区基因印迹的Airn[27-28]。趋同进化和种系特异性是lncRNA分析的重要方面。
Biography
蒋洋洋, 在读硕士研究生, E-mail:1775812424@qq.com
Funding Statement
国家自然科学基金(31571348);广州市科技创新局(201607010067)
Supported by National Natural Science Foundation of China (31571348)
Contributor Information
蒋 洋洋 (Yangyang JIANG), Email: 1775812424@qq.com.
张 海 (Hai ZHANG), Email: zhangh@smu.edu.cn.
References
- 1.Koentgen F, Suess G, Naf D. Engineering the mouse genome to model human disease for drug discovery. Methods Mol Biol. 2010;602:55–77. doi: 10.1007/978-1-60761-058-8. [Koentgen F, Suess G, Naf D.Engineering the mouse genome to model human disease for drug discovery[J].Methods Mol Biol, 2010, 602:55-77.] [DOI] [PubMed] [Google Scholar]
- 2.Roper J, Tammela T, Akkad A, et al. Colonoscopy-based colorectal cancer modeling in mice with CRISPR-Cas9 genome editing and organoid transplantation. Nat Protoc. 2018;13(2):217–34. doi: 10.1038/nprot.2017.136. [Roper J, Tammela T, Akkad A, et al.Colonoscopy-based colorectal cancer modeling in mice with CRISPR-Cas9 genome editing and organoid transplantation[J].Nat Protoc, 2018, 13(2):217-34.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frese KK, Tuveson DA. Maximizing mouse cancer models. http://industry.wanfangdata.com.cn/dl/Detail/NSTLQK?id=NSTLQK_NSTL_QKJJ023516520. Nat Rev Cancer. 2007;7(9):645–58. doi: 10.1038/nrc2192. [Frese KK, Tuveson DA.Maximizing mouse cancer models[J].Nat Rev Cancer, 2007, 7(9):645-58.] [DOI] [PubMed] [Google Scholar]
- 4.Khaled WT, Liu P. Cancer mouse models:past, present and future. Semin Cell Dev Biol. 2014;27:54–60. doi: 10.1016/j.semcdb.2014.04.003. [Khaled WT, Liu P.Cancer mouse models:past, present and future [J].Semin Cell Dev Biol, 2014, 27:54-60.] [DOI] [PubMed] [Google Scholar]
- 5.Morin PJ, Weeraratna AT. Genetically-defined ovarian cancer mouse models. J Pathol. 2016;238(2):180–4. doi: 10.1002/path.4663. [Morin PJ, Weeraratna AT.Genetically-defined ovarian cancer mouse models[J].J Pathol, 2016, 238(2):180-4.] [DOI] [PubMed] [Google Scholar]
- 6.Day CP, Merlino G, Van Dyke T. Preclinical mouse cancer models:a maze of opportunities and challenges. Cell. 2015;163(1):39–53. doi: 10.1016/j.cell.2015.08.068. [Day CP, Merlino G, Van Dyke T.Preclinical mouse cancer models:a maze of opportunities and challenges[J].Cell, 2015, 163(1):39-53.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs:analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–89. doi: 10.1101/gr.132159.111. [Derrien T, Johnson R, Bussotti G, et al.The GENCODE v7 catalog of human long noncoding RNAs:analysis of their gene structure, evolution, and expression[J].Genome Res, 2012, 22(9):1775-89.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yue F, Cheng Y, Breschi A, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515(7527):355–64. doi: 10.1038/nature13992. [Yue F, Cheng Y, Breschi A, et al.A comparative encyclopedia of DNA elements in the mouse genome[J].Nature, 2014, 515(7527):355-64.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JT. Lessons from X-chromosome inactivation:long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23(16):1831–42. doi: 10.1101/gad.1811209. [Lee JT.Lessons from X-chromosome inactivation:long ncRNA as guides and tethers to the epigenome[J].Genes Dev, 2009, 23(16):1831-42.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–93. doi: 10.1126/science.1192002. [Tsai MC, Manor O, Wan Y, et al.Long noncoding RNA as modular scaffold of histone modification complexes[J].Science, 2010, 329 (5992):689-93.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brien GL, Valerio DG, Armstrong SA. Exploiting the Epigenome to control cancer-promoting gene-expression programs. Cancer Cell. 2016;29(4):464–76. doi: 10.1016/j.ccell.2016.03.007. [Brien GL, Valerio DG, Armstrong SA.Exploiting the Epigenome to control cancer-promoting gene-expression programs[J].Cancer Cell, 2016, 29(4):464-76.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardner PP. The use of covariance models to annotate RNAs in whole genomes. Brief Funct Genomic Proteomic. 2009;8(6):444–50. doi: 10.1093/bfgp/elp042. [Gardner PP.The use of covariance models to annotate RNAs in whole genomes[J].Brief Funct Genomic Proteomic, 2009, 8(6):444-50.] [DOI] [PubMed] [Google Scholar]
- 13.Nawrocki EP. Annotating functional RNAs in genomes using Infernal. Methods Mol Biol. 2014;1097:163–97. doi: 10.1007/978-1-62703-709-9. [Nawrocki EP.Annotating functional RNAs in genomes using Infernal[J].Methods Mol Biol, 2014, 1097:163-97.] [DOI] [PubMed] [Google Scholar]
- 14.Abu AA, Petrov AI, Stombaugh J, et al. Comprehensive survey and geometric classification of base triples in RNA structures. Nucleic Acids Res. 2012;40(4):1407–23. doi: 10.1093/nar/gkr810. [Abu AA, Petrov AI, Stombaugh J, et al.Comprehensive survey and geometric classification of base triples in RNA structures[J].Nucleic Acids Res, 2012, 40(4):1407-23.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He S, Zhang H, Liu H, et al. LongTarget:a tool to predict lncRNA DNA-binding motifs and binding sites via Hoogsteen base-pairing analysis. Bioinformatics. 2015;31(2):178–86. doi: 10.1093/bioinformatics/btu643. [He S, Zhang H, Liu H, et al.LongTarget:a tool to predict lncRNA DNA-binding motifs and binding sites via Hoogsteen base-pairing analysis[J].Bioinformatics, 2015, 31(2):178-86.] [DOI] [PubMed] [Google Scholar]
- 16.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [Huang DW, Sherman BT, Lempicki RA.Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources [J].Nat Protoc, 2009, 4(1):44-57.] [DOI] [PubMed] [Google Scholar]
- 17.Amandio AR, Necsulea A, Joye E, et al. Hotair is dispensible for mouse development. PLoS Genet. 2016;12(12):e1006232. doi: 10.1371/journal.pgen.1006232. [Amandio AR, Necsulea A, Joye E, et al.Hotair is dispensible for mouse development[J].PLoS Genet, 2016, 12(12):e1006232.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, Helms JA, Chang HY. Comment on "Hotair is dispensable for mouse development". PLoS Genet. 2016;12(12):e1006406. doi: 10.1371/journal.pgen.1006406. [Li L, Helms JA, Chang HY.Comment on "Hotair is dispensable for mouse development"[J].PLoS Genet, 2016, 12(12):e1006406.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan MR, Xiang S, Song Z, et al. The p53-inducible long noncoding RNA TRINGS protects cancer cells from necrosis under glucose starvation. EMBO J. 2017;36(23):3483–500. doi: 10.15252/embj.201696239. [Khan MR, Xiang S, Song Z, et al.The p53-inducible long noncoding RNA TRINGS protects cancer cells from necrosis under glucose starvation[J].EMBO J, 2017, 36(23):3483-500.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michelini F, Pitchiaya S, Vitelli V, et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat Cell Biol. 2017;19(12):1400–11. doi: 10.1038/ncb3643. [Michelini F, Pitchiaya S, Vitelli V, et al.Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks[J].Nat Cell Biol, 2017, 19(12):1400-11.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez Y, Segura V, Marin-Bejar O, et al. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat Commun. 2014;5:5812. doi: 10.1038/ncomms6812. [Sanchez Y, Segura V, Marin-Bejar O, et al.Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature[J].Nat Commun, 2014, 5:5812.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blume CJ, Hotz-Wagenblatt A, Hullein J, et al. p53-dependent noncoding RNA networks in chronic lymphocytic leukemia. Leukemia. 2015;29(10):2015–23. doi: 10.1038/leu.2015.119. [Blume CJ, Hotz-Wagenblatt A, Hullein J, et al.p53-dependent noncoding RNA networks in chronic lymphocytic leukemia[J].Leukemia, 2015, 29(10):2015-23.] [DOI] [PubMed] [Google Scholar]
- 23.Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106(28):11667–72. doi: 10.1073/pnas.0904715106. [Khalil AM, Guttman M, Huarte M, et al.Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression[J].Proc Natl Acad Sci USA, 2009, 106(28):11667-72.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mello SS, Sinow C, Raj N, et al. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017;31(11):1095–108. doi: 10.1101/gad.284661.116. [Mello SS, Sinow C, Raj N, et al.Neat1 is a p53-inducible lincRNA essential for transformation suppression[J].Genes Dev, 2017, 31 (11):1095-108.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanzos A, Carlevaro-Fita J, Mularoni L, et al. Discovery of cancer driver longnoncoding RNAs across 1112 tumour genomes:New candidates and cistinguishing features. Sci Rep. 2017;7:41544. doi: 10.1038/srep41544. [Lanzos A, Carlevaro-Fita J, Mularoni L, et al.Discovery of cancer driver longnoncoding RNAs across 1112 tumour genomes:New candidates and cistinguishing features[J].Sci Rep, 2017, 7:41544.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y, Ge Z, Zhang E, et al. The lncRNA TUG1 modulates proliferation in trophoblast cells via epigenetic suppression of RND3. Cell Death Dis. 2017;8(10):e3104. doi: 10.1038/cddis.2017.503. [Xu Y, Ge Z, Zhang E, et al.The lncRNA TUG1 modulates proliferation in trophoblast cells via epigenetic suppression of RND3[J].Cell Death Dis, 2017, 8(10):e3104.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grant J, Mahadevaiah SK, Khil P, et al. Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature. 2012;487(7406):254–8. doi: 10.1038/nature11171. [Grant J, Mahadevaiah SK, Khil P, et al.Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation[J].Nature, 2012, 487(7406):254-8.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H, Shang X, Zhu H. LncRNA/DNA binding analysis reveals losses and gains and lineage specificity of genomic imprinting in mammals. https://academic.oup.com/bioinformatics/article/33/10/1431/2801463. Bioinformatics. 2017;33(10):1431–6. doi: 10.1093/bioinformatics/btw818. [Liu H, Shang X, Zhu H.LncRNA/DNA binding analysis reveals losses and gains and lineage specificity of genomic imprinting in mammals[J].Bioinformatics, 2017, 33(10):1431-6.] [DOI] [PubMed] [Google Scholar]


