Summary
FAT4 mutations lead to several human diseases that disrupt normal development of the kidney. However, the underlying mechanism remains elusive. In studying the duplex kidney phenotypes observed upon deletion of Fat4 in mice, we have uncovered an interaction between the atypical cadherin FAT4 and RET, a tyrosine kinase receptor essential for kidney development. Analysis of kidney development in Fat4−/− kidneys revealed abnormal ureteric budding and excessive RET signaling. Removal of one copy of the RET ligand Gdnf rescues Fat4−/− kidney development, supporting the proposal that loss of Fat4 hyperactivates RET signaling. Conditional knockout analyses revealed a non-autonomous role for Fat4 in regulating RET signaling. Mechanistically, we found FAT4 interacts with RET through extracellular cadherin repeats. Importantly, expression of FAT4 perturbs the assembly of the RET-GFRA1-GDNF complex, reducing RET signaling. Thus FAT4 interacts with RET to fine-tune RET signaling, establishing a juxtacrine mechanism controlling kidney development.
Keywords: Kidney development, FAT4, RET, cadherin, non-autonomous, Van Meldergem syndrome, CAKUT
eTOC blurb
FAT4 mutations occur in humans with kidney disease. Controlling RET tyrosine kinase activity is critical for normal kidney development. Zhang et al. show FAT4 restricts RET signaling activity via binding to RET and reducing RET interactions with its ligand GDNF-GFRA1. Thus FAT4 loss leads to hyperactive RET signaling and disease.
Introduction
Congenital anomalies of the kidney and urinary tract (CAKUT) comprise a wide spectrum of renal and/or urinary tract malformations, and account for ~40–50% of children diagnosed with chronic kidney disease (Vivante et al., 2014). The etiology of CAKUT is influenced by both environmental and genetic factors (Nicolaou et al., 2015; dos Santos et al., 2014). Mouse and human studies have revealed some of the genetic causes behind the anatomical defects in the urinary system, most leading to defects in early kidney development (Vivante et al., 2014).
Mouse kidney development begins at E10.5 with formation of the ureteric bud (UB) from the caudal end of the nephric duct (ND), which invades the neighbouring metanephric mesenchyme (MM) (Short and Smyth, 2016; Dressler, 2009). Reciprocal inductive interactions between the UB and MM lead to repeated branching of the UB and nephrogenesis (Short and Smyth, 2016; Costantini and Kopan, 2010).
The RET receptor tyrosine kinase and its co-receptor GDNF family receptor alpha 1 (GFRA1) are essential to induce the UB and contribute to a program of branching morphogenesis which establishes the collecting duct system. RET and GFRA1 are expressed at low levels in the ND prior to UB invasion and are upregulated in the UB upon its formation, and at the tips of the branching UB upon contact with the MM (Davis et al., 2013; Cacalano et al., 1998; Avantaggiato et al., 1994). The RET/GFRA1 ligand GDNF is expressed in the MM (Durbec et al., 1996; Trupp et al., 1996). Binding of GDNF to RET and GFRA1 induces RET autophosphorylation and activation of multiple downstream signaling cascades, including the RAS-ERK MAP kinase, Phospholipase C gamma (PLCG) and PI3K-AKT pathways (Davis et al., 2013; Costantini, 2011).
Mice lacking Gdnf, Ret or Gfral exhibit impaired UB induction leading to renal agenesis/hypoplasia (Cacalano et al., 1998; Moore et al., 1996; Sanchez et al., 1996; Trupp et al., 1996; Schuchardt et al., 1994). In contrast, over-activating RET by deleting the antagonist Sprouty1 (Spry1), or mutating RET-Y1015 docking tyrosine, induces ectopic UB formation, and subsequently duplex or multiplex kidneys (Hoshi et al., 2012; Michos et al., 2010; Rozen et al., 2009; Jain et al., 2006; Mason et al., 2006; Basson et al., 2005). Most genes required for ureter budding/branching during mammalian kidney development regulate or are regulated by RET signaling (Kobayashi et al., 2007; Grieshammer et al., 2004; Esquela and Lee, 2003; Xu et al., 2003; Bouchard et al., 2002; Wellik et al., 2002; Nishinakamura et al., 2001; Kume et al., 2000; Xu et al., 1999). Yet, despite the pivotal role for RET in kidney development, little is known about the assembly and regulation of the RET-GFRA1-GDNF ternary complex.
Mutations in the atypical cadherin FAT4 cause Van Maldergem syndrome, of which CAKUT is a commonly observed feature (Alders et al., 2014; Cappello et al., 2013). FAT4 variants were also identified in whole-exome sequencing studies of CAKUT patients (van der Ven et al., 2017). In mice, Fat4 deletion leads to kidney defects reminiscent of CAKUT, including multicystic disease (Saburi et al., 2008) and reduced nephron number (Bagherie-Lachidan et al., 2015; Mao et al., 2015; Das et al., 2013). Previously, we noted that a form of CAKUT, termed duplex kidney, occasionally manifests in Fat4 mutants (Saburi et al., 2008).
FAT4 is the mammalian ortholog of Drosophila fat (ft). Two well-defined Ft interacting proteins, Dachsous (Ds) and Four-jointed (Fj), have homologs in mammals (DCHS1, DCHS2, FJX1). Ds acts as a heterophilic binding partner of Ft in trans and activates bidirectional signaling (Blair and McNeill, 2018; Degoutin et al., 2013; Matakatsu and Blair, 2004), and Fj modifies binding between Ft and Ds by phosphorylating their cadherin repeats (Brittle et al., 2010; Simon et al., 2010). Interactions between mammalian FAT4, DCHS1 and FJX1 are conserved (Bagherie-Lachidan et al., 2015; Ishiuchi et al., 2009; Saburi et al., 2008).
While characterizing the duplex kidney phenotype in Fat4 mutants, we discovered that FAT4 is a regulator of RET signaling, and acts via interacting with RET, inhibiting assembly of the RET ternary signaling complex. Our studies identify RET as another extracellular binding partner of Fat cadherins besides Dachsous, and demonstrate a juxtacrine regulation by FAT4 of a central pathway in kidney development.
Results
Loss of Fat4 results in duplex kidneys
Previous studies found duplex kidneys at low penetrance in outbred Fat4−/−;Fjx1−/− mice (Saburi et al., 2008). Backcrossing the Fat4 mutation into a C57BL/6J (B6) background increased this penetrance (60%, n = 10, 9 generations, p=0.004) (Figure S1A–S1B’ and Table S1), whereas backcrossing into a CD1 or 129S1 background did not (Table S1). This background dependency implicates genetic enhancers in the B6 background that promote duplex kidney formation. Co-deletion of Fjx1 in this background somewhat increased the penetrance (80%, n = 15) (Figures 1A–1D’ and Table S1) although this increase was not significant (p=0.28). Deletion of Fjx1 alone did not obviously alter renal development (Figure 1B–1B’ and Table S1). Taken together this implies that Fat4 and Fjx1 act in concert to suppress the formation of duplex kidneys in a manner dependent on genetic modifier(s). Given the high penetrance of the duplex kidney phenotype, subsequent experiments were largely performed in the Fat4−/−;Fjx1−/− mutants.
Figure 1. Fat4 and Fjx1 deletion leads to duplex kidney formation.
(A–D) Macroscopic view of P0 kidneys from wild type, Fjx1−/− or Fat4−/−;Fjx1−/− mice shows both complete (C) and incomplete (D) duplication defects in Fat4−/−;Fjx1−/− kidneys. Pink arrowheads point to two duplicated ureters and the yellow arrowhead to a partial duplicated ureter. (A’–D’) H&E staining of P0 kidneys from wild type, Fjx1−/− or Fat4−/−;Fjx1−/− mice confirms duplex defects in Fat4−/−;Fjx1−/− kidneys. * marks the duplicated pelvis while the black arrows point to the duplicated ureters. Black arrowheads in C’ and D’ point to abnormal nephrogenic zone of the duplex kidneys. (E–F’) PAX2 and WT1 staining on P0 kidneys sections shows abnormal extension of the nephrogenic zone deep into the kidney in Fat4−/−;Fjx1−/− mice. (G–H’) SIX2 and ECAD staining on P0 kidneys sections confirms abnormal nephrogenic zone in Fat4−/−;Fjx1−/− duplex kidneys. Scale bars represent 500μm in A–D’ and 200μm in E–H’. See also Figure S1 and Table S1.
Duplex kidneys can result either from the formation of two UBs from the ND (complete duplication), or precocious branching of the primary UB (incomplete duplication) (Whitten and Wilcox, 2001). Both complete (Figures 1C–1C’) and incomplete duplication (Figures 1D–1D’) were observed in Fat4−/−;Fjx1−/− mice. Histological analyses using hematoxylin and eosin (H&E) staining confirmed the presence of duplex kidneys in Fat4−/−;Fjx1−/− mutants, which often have two pelvises, two ureters and an abnormal indented nephrogenic zone (Figures 1C’ and 1D’) that separates the kidney into two. Staining newborn (P0) kidney sections with antibodies to PAX2 (marker for progenitors and UB), WT1 (progenitors and glomeruli/nephron) and SIX2 (progenitors) confirmed that Fat4−/−;Fjx1−/− kidneys have discontinuous nephrogenic zones consistent with the formation of multiple UB (Figures 1F–1F’ and 1H–1H’), a phenotype not observed in wild type (Figures 1E–1E’ and 1G–1G’) or Fjx1 single mutants (Figures S1C–S1D’).
Duplex kidneys arise from defective kidney induction in Fat4−/−;Fjx1−/− mutants
To determine how duplex kidneys develop in Fat4−/−;Fjx1−/− mutants, we analyzed early kidney induction by whole mount immunostaining. As early as E11.5, when the UB has invaded the MM, abnormal ectopic budding from the ND and abnormal branching of the UB was observed (Figure 2, compare 2A–2A’ with 2B–2B’). Wild type embryos had a single UB at the typical T-stage (E11.5), whereas Fat4−/−;Fjx1−/− mutants frequently had a secondary UB that had already branched from the main UB (Figures 2B–2B’). To confirm that Fat4−/−;Fjx1−/− led to ectopic UBs, urogenital ridge explants were dissected at E11.5 and cultured for 48h. While control explants (from wild type or Fat4 heterozygous animals), predominantly have a single ureteric stalk and normal dividing ureteric tips (Figure 2C), Fat4−/− and Fat4−/−;Fjx1−/− mutant explants frequently have ectopic UBs (Figures 2G and S1E–S1H) arising from either the primary budding site (Figures 2D and S1E–S1F), the ND (Figures 2E and S1G), or the primary UB (Figures 2F and S1H). Some ectopic buds resulted in an extra ureteric stalk with independent branching tree (Figures 2E and 2F); some of the buds did not branch, but could still induce a surrounding MM domain (white arrow in Figures 2I–2I’; compare Figures 2H–2H’ with 2I–2I’). 41% of cultured Fat4 mutant kidneys showed ectopic budding, compared with 72% of Fat4−/−;Fjx1−/− kidneys (Figure 2G), confirming that Fat4 and Fjx1 synergistically regulate budding of the UB to prevent duplex kidney formation. NDs of Fat4−/−;Fjx1−/− mutants were also dissected and stained prior to UB invasion at E10.5 (Figures 2J–2K’, n=5). At E10.5, Fat4−/−;Fjx1−/− mutants were indistinguishable from wild type. The duplex defects observed in Fat4−/− and Fat4−/−;Fjx1−/− P0 kidneys can therefore be explained by ectopic budding and branching of the UB.
Figure 2. Defects in kidney induction in Fat4−/−;Fjx1−/− mice.
(A–B) Whole mount staining of E11.5 Fat4−/−;Fjx1−/− mutants with antibodies against PAX2 (green) and CYTOKERATIN (red) shows abnormal ectopic UB budding (white arrowhead in B), compared with the single normal UB seen in wild type urogenital ridge (A). (C–F) Fat4−/−;Fjx1−/− kidney explants exhibit ectopic budding (white arrows) when cultured ex vivo, as revealed by CALBINDIN staining in red. (G) Summary of the ectopic budding incidence in control (wild type and Fat4+/−), Fat4−/− and Fat4−/−;Fjx1−/− kidney culture. Ectopic budding incidence is shown as the number of kidney with ectopic budding or branching/total number of kidney examined. (H–I’) An ectopic bud in Fat4−/−;Fjx1−/− kidney culture induces the surrounding mesenchyme to condense, as shown by PAX2 staining in green (white arrow). The UB epithelium cells are marked by CALBINDIN in red. (J–K’) No morphological differences were observed between wild type and Fat4−/−;Fjx1−/− urogenital ridge at E10.5 (n=5). The UB epithelium cells are marked by CALBINDIN in red. Scale bars represent 250μm in all panels. See also Figure S1.
RET signaling is up-regulated in Fat4−/−;Fjx1−/− mutants
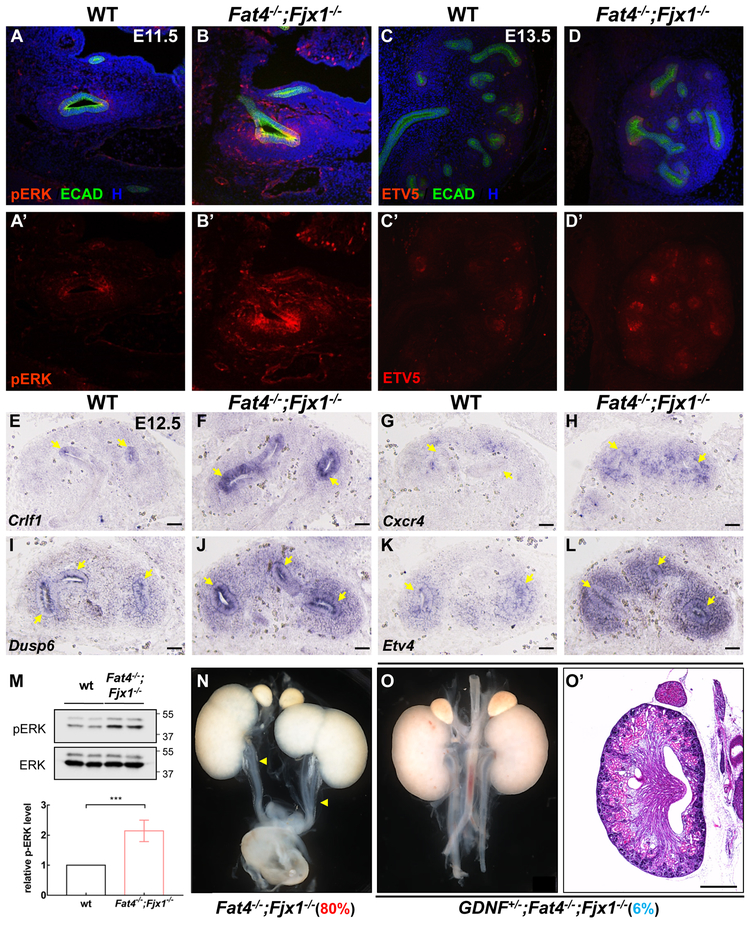
Fat4−/−;Fjx1−/− mutants display several phenotypes caused by increased RET signaling (Hoshi et al., 2012; Jain et al., 2006; Basson et al., 2005). To determine if RET signaling is deregulated in Fat4−/−;Fjx1−/− mutants, we examined the phosphorylation of a downstream effector, ERK, and expression of a transcriptional target Etv5. pERK levels reflect MAPK signaling downstream of RET (Fisher et al., 2001) and ETV5 expression relies heavily on RET signaling (Lu et al., 2009). Significantly, in Fat4−/−;Fjx1−/− E11.5 kidney sections, pERK was increased in the UB (outlined in Figures 3 and S2, compare 3A–3A’ with 3B–3B’ and S2A–S2A’ with S2B–S2B’) and occasionally in surrounding cells (white arrowheads in Figures S2B–2B’). ETV5 expression was also increased at E13.5, when the ureteric buds are undergoing repeated branching events (Figure 3, compare 3C–3C’ with 3D–3D’), although we could not detect alterations in ETV5 at E11.5 (Figures S2C–2D’). To confirm RET signaling is disrupted in Fat4−/−;Fjx1−/− kidneys, we conducted in situ hybridization on other RET signaling targets (Lu et al., 2009). Consistent with increased RET signaling, Crlf1, Cxcr4, Dusp6 and Etv4 were upregulated in the UB of E12.5 Fat4−/−;Fjx1−/− kidney sections, compared with controls (Figures 3E–3L). We also noted increased Cxcr4, Dusp6 and Etv4 expression in cells surrounding the UB.
Figure 3. RET/GDNF signaling is increased in Fat4−/−;Fjx1−/− mice.
(A–B’) Immunostaining with antibodies against phospho-ERK and ECAD on E11.5 kidney sections reveals increased phospho-ERK in Fat4−/−;Fjx1−/− UB tips (outlined) compared with wild type. (C–D’) Immunostaining of ETV5 and ECAD on E13.5 kidney sections shows increased ETV5 signal in Fat4−/−;Fjx1−/− UB tips compared with wild type. (E–L) in situ hybridization on E12.5 kidney sections reveals increased expression of Crlf1, Cxcr4, Dusp6 and Etv4 in Fat4−/−;Fjx1−/− mutants. UBs were labeled by yellow arrows. (M) Western blot on E12.5 kidney lysate confirms increased pERK signal in Fat4−/−;Fjx1−/− mutants. Similar results were obtained in four independent experiments andquantification of pERK signal is represented as mean ± SEM. (N–O’) Haploinsufficiency of GDNF reverts the duplex kidney phenotype in Fat4−/−;Fjx1−/− mutants. The percentage after the genotype shows the incidence for duplex kidney formation. Scale bars represent 50 μm in A–L and 500μm in N–O’.
In line with the immunostaining, western blotting of E12.5 kidney lysates revealed higher pERK levels in Fat4−/−;Fjx1−/− mutants versus wild type controls (Figure 3M). Thus, RET signaling is increased in Fat4−/−;Fjx1−/− mutants. As loss of Fat4 in either a CD1 or 129S1 background does not cause duplex defects (Table S1), we tested whether the variability in the duplex kidney incidence between B6 and CD1 background reflects variability in RET signaling activation, or in the response to this activation, using pERK as a readout. Western blot analysis revealed that Fat4−/− mutants in a CD1 background also have increased levels of pERK (Figures S3A-S3B), implying that the B6 background is more sensitive to altered RET signaling than CD1.
The increase in RET signaling observed in Fat4−/−;Fjx1−/− embryos provides an explanation for the duplex kidney defects observed in the mutants. We reasoned that reducing RET signaling should revert the duplex kidney defects. Strikingly, duplex kidney phenotypes were no longer evident in Gdnf+/−;Fat4−/−;Fjx−/− mutants (n=18, p=1.79E-5) (Table S1 and Figures 3N–3O’) in the B6 background. Thus, Fat4 and Fjx1 regulate RET-GDNF signaling during early kidney induction and their loss increases RET signaling.
The increased RET signaling in Fat4−/−;Fjx−/− kidneys at E12.5 and E13.5 led us to search for morphological defects related to RET over-activation after initial branch formation. Of 53 cultured Fat4−/−;Fjx−/− kidney explants, 12 exhibited UB tip defects such as swollen tip (star in Figure S3E), an expanded tip region (star in Figure S3F) or multiple buddings from one tip associated with a dilated trunk (Figures S3G–S3H), whereas these defects rarely appeared in control kidney explants (1/80) (Figures S3C–S3D). The tip seems to outgrow the trunk in Fat4−/−;Fjx−/− mutants, possibly due to tip region formation at the expense of trunk elongation. These defects resemble Spry1 and Pten mutants (Kim and Dressler, 2007; Basson et al., 2006), and are opposite to the effect of MEK1 inhibition (which increases elongation of the trunk and reduces tip formation) (Watanabe and Costantini, 2004). Both Spry1 and Pten inhibit RET downstream signaling, suggesting the UB tip defects in Fat4−/−;Fjx−/− reflect overactive RET.
We next treated the cultured kidney explants with a low GDNF dose (10ng/ml) to determine if Fat4−/−;Fjx−/− have increased sensitivity. In controls, swollen tip and multiple buds from one UB tip were occasionally induced (5/33, Figure S3I), but most kidney explants had relatively normal morphology (Figures S3I–S3J). Notably, Fat4−/−;Fjx−/− kidney explants were more sensitive to GDNF induction than littermate controls : more kidneys showed tip defects (16/21), and the defects were more severe than controls (compare Figures S3I–S3J with S3K–S3L). UB stalks were sometimes dilated in Fat4−/−;Fjx−/− mutants (Figure S3K). Taken together, these data suggest that FAT4 suppresses RET signaling throughout kidney development.
Fat4 functions non-autonomously to prevent duplex kidney/ectopic bud formation
Our data demonstrate that FAT4 regulates RET. RET is expressed in the ND and UB cells, where it responds to GDNF produced by the MM. We and others (Bagherie-Lachidan et al., 2015; Mao et al., 2015) showed that Fat4 is expressed predominantly in the stroma and mesenchyme at E13.5 and E14.5. Using an EGFP knock-in allele of Fat4 (Wu et al., 2008), we find Fat4 is also highly expressed in the stroma, moderately in the CM and weakly in the UB at E11.5 (Figures 4B–4B’). In addition, we noticed expression of Fat4 in the cells surrounding the ND, ureter and UB (Figures 4A–4A’ and S4A–S4B). RNAscope further confirmed the expression of FAT4 very weakly in the UB epithelia and mainly in surrounding cells (Figures S4C–S4E).
Figure 4. FAT4 non-autonomously regulates kidney induction.
(A–B’) Immunostaining with anti-GFP on E11.5 Fat4-EGFP kidney sections shows expression pattern of FAT4. Anti-PBX1 staining (in red) marks the metanephric blastema and the surrounding mesenchyme, while anti-PAX2 staining (in blue) marks the ND+UB epithelia and the CM. (C–E’) Macroscopic view and H&E staining of the P0 kidneys from Pax3-cre;Fat4flox/+ (C–C’) and Pax3-cre;Fat4flox/− mice show complete duplication (white arrow in D–D’) and incomplete duplication (E–E’) when Fat4 is deleted from the mesenchymal cells. (F–G) Defect in kidney induction was observed in Pax3-cre;Fat4flox/− mouse kidney culture (white arrow in G). Scale bars represent 200μm in A–A’, 50μm in B–B’, 500μm in C–E’ and 250μm in F–G. See also Figure S4 and Table S2.
To explore how Fat4 impacts renal development, we conditionally deleted it from different cell lineages using tissue specific Cre lines which we backcrossed into the B6 background for ≥ 9 generations. As Fat4 is expressed in cells surrounding the ND/UB, we hypothesized that it may suppress ectopic UB growth/branching at these sites. Pax3-cre was used to remove Fat4 from cells surrounding the ND and UB (Saifudeen et al., 2009; Poladia et al., 2006; Engleka et al., 2005). ~ 47% of Pax3-cre;Fat4flox/− mice at P0 presented with, either complete duplication (Figure 4D) or incomplete duplication (Figure 4E) (Table S2, compared with 60% in Fat4−/− mice and 0 in Pax3-cre controls (Figure 4C-C’)). Pax3-cre;Fat4flox/− kidney explants also exhibited duplex defects (Figure 4G), while Pax3-cre;Fat4flox/+ controls did not (Figure 4F). These data suggest a non-autonomous role for Fat4 in regulating UB induction.
Fat4flox/− mice were crossed with Hoxb7-cre to investigate a possible role for Fat4 in UB cells (Zhao et al., 2004). Duplex kidneys at P0 were occasionally observed in Hoxb7-cre;Fat4flox/− mice (4/26), which was not significantly different from controls (1/19)(Table S2)(p=0.29). Hoxb7-cre;Fat4flox/− kidneys were also cultured from explants taken at E11.5 but no ectopic branching or budding were observed (Figures S4F–4I, n=8). Taken together, these data indicate that FAT4 primarily functions non-autonomously in the cells surrounding the ND, ureter and UB to negatively regulate RET signaling in the UB lineage.
RET forms complexes with FAT4 and DCHS1
We sought to identify the mechanism by which FAT4 non-autonomously suppresses RET signaling in the UB cells. Notably, RET is the only receptor tyrosine kinase with cadherin repeats, and FAT4 and DCHS1 are enormous atypical cadherins that interact via their extracellular cadherin repeats. Phylogenetic analyses of repeat one homologies of all cadherins indicate that FAT4, DCHS1 and RET are highly similar (Hulpiau and van Roy, 2009) (see also Figure 5A). Since Fat and Ds function as a ligand-receptor pair during Drosophila development (Matakatsu and Blair, 2004) as do FAT4 and DCHS1 (Bagherie-Lachidan et al., 2015; Ishiuchi et al., 2009), we hypothesized that RET could interact with DCHS1 or FAT4 via cadherin-cadherin interactions, and thereby modulate RET signaling.
Figure 5. Association between FAT4 and RET via cadherin repeats.
(A) Dendrogram of the cadherin superfamily on the basis of cadherin repeat one homologies, modified from that of Hulpiau and van Roy, 2009, shows high relatedness between FAT4, DCHS1 and RET. (B) Citrine-tagged FAT4 and tdTomato-tagged RET can form a complex in HEK cells. (C) mCherry-tagged DCHS1 and EGFP-tagged RET interact in HEK cells. (D) EGFP-tagged RET can pull down Flag-tagged DCHS1-Cad1-5 and Flag-tagged FAT4-Cad1-5 whereas EGFP-tagged LULU cannot. (E) Schematic drawings of the domain composition of RET protein, Cad1-4, 1-2 and 3-4 truncations of RET. (F) RET with deletion of Cad1-4 almost loses the ability to pull down FAT4-Cad1-5, whereas RET-delta-Cad1-2 seems to retain most of its binding ability. RET-delta-Cad3-4 shows less affinity to FAT4-Cad1-5 than RET-delta-Cad1-2. Similar results were obtained in three independent experiments. See also Figure S5.
Since no antibodies are available to immunoprecipitate endogenous FAT4 (>500kDa), we used cultured cells and tagged constructs to determine if FAT4 co-immunoprecipitates with RET. Stable FAT4-Citrine expressing HEK293 cells were transfected with tdTomato-tagged full length RET. GFP-trap beads were used to immunoprecipitate FAT4-Citrine, and western blots were probed for tdTomato to detect RET. These assays showed that full length FAT4 co-immunoprecipitates with RET (Figure 5B). Similarly, we also detected an interaction between DCHS1-mCherry and RET-EGFP (Figure 5C).
The non-cell-autonomous role of Fat4 in regulating RET signaling in vivo prompted us to test if the FAT4-RET interaction is mediated through the homologous cadherin repeats in their extracellular domains. Previous work in Drosophila showed that the first five cadherin repeats (Cad1-5) of Ds or Ft are sufficient for their interaction (Brittle et al., 2010; Simon et al., 2010). We therefore tested whether RET-EGFP could pull down Flag-tagged FAT4-Cad1-5 and DCHS1-Cad1-5 in transfected cells. Indeed, co-immunoprecipitation was readily detected, as was an interaction between DCHS1 and RET (Figure 5D). Since we did not detect duplex kidney defects in Dchs1 mutants (see discussion), we focused on the interaction between FAT4 and RET.
To determine if the RET-FAT4 interaction of requires cadherin domains, RET constructs were generated lacking these regions (schematic in Figure 5E). Immunostaining confirmed identical localization of wild type RET and RET-delta-Cad1-4 (Figures S5A–S5D). Deleting all 4 cadherin repeats of RET abrogated interaction with FAT4 Cad1-5 repeats (Figure 5F). Thus, FAT4-RET interaction involves cadherin repeats in both proteins.
Two additional RET constructs with different cadherin truncations (RET-delta-Cad1-2 and RET-delta-Cad3-4) (schematic in Figure 5E) were also tested. Consistent with phylogenetic studies conducted on the first cadherin repeats of all cadherins (Hulpiau and van Roy, 2009), RET-delta-Cad3-4 (which contains cadherin 1 & 2) formed a complex with FAT4-Cad1-5 (Figure 5F). Intriguingly, however, RET-delta-Cad1-2 more efficiently immunoprecipitated FAT4-Cad1-5 than did RET-delta-Cad3-4. We then conducted additional in silico analysis of the homology of cadherin repeats between FAT4-Cad1-5 and RET-Cad1-2, and between FAT4-Cad1-5 and RET-Cad3-4. The alignment tool EMBOSS Water revealed a better match of FAT4-Cad1-5 with RET-Cad3-4 than RET-Cad1-2 (data not shown). The co-immunoprecipitation of FAT4 with RET and the homology suggests an interaction that involves multiple cadherin repeats in these molecules.
FAT4 interferes with RET-GFRA1-GDNF ternary complex assembly and suppresses RET activity
Our data indicates that FAT4 non-cell-autonomously inhibits RET signaling and that the FAT4 extracellular domain forms a complex with RET. Therefore, we explored how FAT4 influences RET activity. The binding between RET and its ligand GDNF requires the co-receptor GFRA1-4 and studies in mice suggest that RET, GFRA1 and GDNF function as a complex during kidney development (Cacalano et al., 1998; Durbec et al., 1996; Moore et al., 1996; Sanchez et al., 1996; Trupp et al., 1996). Given the binding between RET and FAT4 cadherin repeats and the close proximity between cells that express these molecules during kidney development, we hypothesized that FAT4 affects the RET ternary complex in trans by competing for RET and restricting GDNF from activating ND/UB cells.
To test this hypothesis, we co-transfected RET and Gfra1 into HEK293T cells to establish RET signaling assays. GDNF addition increased pERK (Figure S6A), and immunoprecipitation detected the RET-GFRA1 complex (Figure 6A), proving the system is functional. Addition of FAT4-Cad1-5 markedly reduced RET-GFRA1 binding (Figure 6A, compare IP lane 2 with lane 3), suggesting interaction between RET and FAT4 inhibits binding between RET and GFRA1.
Figure 6. FAT4 interferes with RET-GFRA1 interaction and RET signaling.
(A) EGFP-tagged RET can pull down Flag-tagged GFRA1 in the absence of GDNF and this interaction can be attenuated by FAT4-Cad-1-5. Similar results were obtained in three independent experiments. Quantification of the ratio of co-IPed GFRA1 intensity to IPed RET intensity is shown. R+G is short for RET+GFRA1, and R+G+F for RET+GFRA1+FAT4-Cad-1-5. (B) GDNF enhances RET-GFRA1 interaction, and FAT4-Cad1-5 affects RET-GFRA1 binding even in the presence of GDNF. Note 1x FAT4-Cad1-5 only marginally affects interaction between RET and GFRA1, whereas 2x FAT4-Cad1-5 enhances this effect. Similar results were obtained in three independent experiments. Quantification of the ratio of co-IPed GFRA1 intensity to IPed RET intensity is shown. R+G+G is short for RET+GFRA1+GDNF, and R+G+F+G for RET+GFRA1+FAT4-Cad1-5+GDNF. (C) MG87RET cells were dramatically less stimulated by GDNF and GFRA1 when mixed with FAT4 expressing cells (Tet+) than when mixed with FAT4 non-expressing cells (Tet−) for 48hr. Two time points of stimulation were performed. Quantifications of the relative increase in pERK level after 10min induction with rhGDNF+rhGFRA1 in 48hr co-cultured MG87RET+FAT4(Tet−) cells versus that in 48hr co-cultured MG87RET+FAT4(Tet+) cells are shown. Results are from three independent experiments. Ratio of pERK/RET is included in the quantification to show that decrease in pERK is not due to decreased RET protein. (D) Diagram summarizing FAT4-RET interactions during kidney development. GDNF produced from the MM binds to RET and GFRA1 expressed in the ND to activate RET signaling in the ND and initiate kidney induction. At the molecular level, RET is activated and recruited by GFRA1 and GDNF into lipid rafts, where it binds to effectors (purple oval), such as SRC and FRS2, and thus activates downstream signaling. We propose that FAT4 expressed in cells surrounding the ND/UB binds to RET and restricts its activity in the ND/UB, possibly via preventing its recruitment into lipid rafts by GFRA1 and GDNF. Quantification data are represented as mean ± SEM. See also Figure S6.
Previous studies showed that RET binds weakly to GFRA1 in the absence of GDNF, and that GDNF enhances this interaction (Klein et al., 1997; Treanor et al., 1996). Consistent with those data, we detected enhanced binding between RET and GFRA1 upon addition of GDNF to the medium (Figure 6B). Intriguingly, GFRA1 bands of higher apparent molecular weight were present in the RET-IP lane upon GDNF stimulation, despite no significant change in whole lysates (Figure 6B). GFRA1 in the lysate showed a smeared pattern after electrophoresis reminiscent of protein glycosylation (Figure 6B). By CIP and PNGase treatments, we determined that the emerging bands of GFRA1 that were pulled down by RET in the presence of GDNF were largely due to glycosylation (Figure S6B).
Importantly, FAT4-Cad1-5 expression attenuated RET-GFRA1 binding in the presence of GDNF (Figure 6B). Increased levels of Fat4-Cad1-5 further inhibited binding of GFRA1 to RET (Figure 6B, compare IP lane 5 with lane 6), corroborating the inhibitory effect of FAT4-Cad1-5 on RET and GFRA1 interaction. Of note, FAT4-Cad1-5 differentially influenced the binding between RET and different GFRA1 isoforms in the presence of GDNF (Figure 6B). This may reflect differences in the ability of various GFRA1 isoforms to bind to RET, however the significance of the different isoforms is not yet clear. These data indicate that FAT4 cadherin repeats can interfere with RET-GFRA1-GDNF ternary complex formation. Since FAT4-Cad1-5 forms a complex with RET (Figure 6B), this effect can be explained by competition for RET binding between FAT4 and GFRA1.
Our experiments showing that FAT4 inhibits RET binding were conducted using co-expression of FAT4, RET and GFRA1 in cultured cells. To test if FAT4 affects RET signaling non-cell-autonomously as it does in vivo, we conducted cell mixing experiments with MG87RET and Tet-on-FAT4 cell lines. MG87RET cells stably express low levels of RET and have been used to investigate RET-GFRA1-GDNF signaling in vitro (Paratcha et al., 2001; Eketjall et al., 1999). Tet-on-FAT4 cells do not express RET, are Tetracycline (Tet) inducible, and only express FAT4 in the presence of Tet (Figure 6C). These two cell lines were co-cultured in Tet plus or minus medium for 24hr and 48hr. Recombinant human GDNF and GFRA1 was then added and RET signaling evaluated. Consistent with our hypothesis, RET signaling in MG87RET cells was suppressed by co-cultured FAT4 cells (Figures 6C and S6C–S6E). The suppression was slight but significant with 24hr co-culture (Figures S6C–S6E), and more pronounced after 48hr (Figures 6C), as revealed by decreased levels of pERK upon exposure to FAT4. Tetracycline on its own had no effect on RET signaling in this context (Figure S6F).
Taken together, these data indicate a role for FAT4 in interacting with RET to inhibit its capacity to signal, via inhibition of RET-GDNF-GFRA ternary complex formation, and provide a mechanism for the in vivo role of FAT4 in modulating RET signaling during kidney development.
Discussion
A tightly controlled signaling transduction event not only relies on positive regulators to initiate, amplify and maintain signaling activity, but also depends on negative regulators to impose restrictions for fine-tuning and limiting excessive activity. During kidney development negative regulation of RET signaling is required to ensure the proper budding of the UB, and disrupting such regulation leads to kidney defects. Despite the central role of RET in the signaling, how RET itself is controlled in vivo is still incomplete.
Here, we dissected the mechanism underlying duplex kidney formation in Fat4 mutant mice, and uncovered a previously unknown mode of RET suppression mediated by a physical interaction with FAT4 during kidney development. Our data suggest a simple model in which FAT4 interacts with RET to disrupt the formation of RET-GFRA1-GDNF ternary complex, thereby attenuating RET activation and subsequent signaling activity.
GDNF promotes the recruitment of RET into lipid rafts by GFRA1, where RET binds to intracellular effector proteins, such as SRC and FRS2 to transduce signals from outside of the cells. This recruitment is believed to protect activated RET from degradation, prolonging activation (Pierchala et al., 2006). Binding of FAT4 to RET could simply sequester RET outside lipid rafts to restrict signal amplification (Figure 6D). Proteins with similar functions have been identified: LRIG1 was reported to bind RET and prevent its entry into lipid rafts, thereby limiting RET activity (Ledda et al., 2008). However, the mechanisms differ as FAT4 acts non-cell-autonomously while LRIG1 functions cell-autonomously to suppress RET signaling.
Fat4 and Dchs1/2 mutants have similar kidney phenotypes (e.g. cystic kidneys, expanded CM) (Bagherie-Lachidan et al., 2015; Mao et al., 2011). We found that RET also pulls down DCHS1 in vitro (Figures 5C–5D), although Dchs1 mutants in the B6 (6) background did not have duplex kidneys (data not shown). However, there may be functional redundancy between Dchs1 and Dchs2 in duplex kidney prevention, as seen in restricting CM expansion (Bagherie-Lachidan et al., 2015). The synergistic interaction between FAT4 and FJX1 is also noteworthy. It might involve phosphorylation of another cadherin protein by FJX1—the obvious candidates would be the Dachsous proteins, or RET. According to the Fj consensus phosphorylation sequence (Ishikawa et al., 2008), there are several FJX1 phosphorylation sites on both DCHS1 and DCHS2, but none on RET. Thus, loss of FJX1 phosphorylation of DCHS1/2 could contribute to the duplex kidney phenotype. In addition, there are FJX1 phosphorylation sites on other FAT-like cadherins, such as FAT1 and FAT3. Loss of Fjx1 could also affect their activity.
Inductive and inhibitory signals between cell lineages ensure proper kidney development. Crosstalk between different cell lineages can be mediated in a paracrine manner through secreted molecules (as in the case of receptor RET and ligand GDNF), or through juxtacrine signaling via direct binding of proteins on the cell surface of two cells, especially when in direct contact. So far most of the crosstalk identified between ND/UB and MM occurs through secreted signals and, to a large extent, is related to RET/GDNF pathway. Our in vivo and in vitro analyses suggest a juxtacrine cross-talk model between the ND/UB and mesenchyme as an additional regulatory mechanism. We propose that FAT4 in the mesenchyme/stroma binds to RET in the epithelia during kidney development to restrict RET activity and prevent ectopic UB induction (Figure 6D). Our data suggest that FAT4 sequesters RET in a state that is inaccessible to its co-receptor GFRA1 and ligand GDNF. The presence of FAT4 in cells surrounding the ND/UB could facilitate the establishment of a GDNF threshold to refine the site of UB outgrowth. Normally, only a narrow cluster of ND cells adjacent to the center of the GDNF expressing domain respond to GDNF and sprout (Chi et al., 2009). ND cells outside this region cannot be stimulated because of relatively low local RET signaling activity, possibly due to either lower Ret expression, or limited access to GDNF. When the ND/UB is surrounded by cells that lack FAT4, (e.g. in Fat4 mutants), more ND/UB cells can bud due to a lowered GDNF threshold. This could give rise to duplex kidney formation, as occurs in Spry1 mutants.
According to this model, the epithelial UB and ND cells would have to make contact with the surrounding FAT4 expressing mesenchymal cells through the intervening basement membrane. While several studies have showed that the basement membrane is continuous (Kim and Dressler, 2007; Meyer et al., 2004; Brandenberger et al., 2001), other studies have hinted at a discontinued basement membrane around the UB tips (Barasch et al., 1999; Ekblom et al., 1980). Specifically, some of the basement membrane components, such as laminin and collagen IV, were lacking in the UB tip region (Barasch et al., 1999; Ekblom et al., 1980). UB induction/formation and branching involve the remodeling of basement membrane to allow the UB cells to migrate into the surrounding mesenchymal cells, where matrix metalloproteinases (MMPs) are believed to play roles (Lenz et al., 2000). Inhibiting these proteases impairs branching, as seen in anti-MMP9 and tissue inhibitors of metalloproteinase 1 (TIMP-1) treated mouse kidney culture (Lelongt et al., 1997), TIMP-2 treated rat kidney culture (Barasch et al., 1999) and MMP9 mutants (Arnould et al., 2009). The continual degradation and remodeling of the ECM, may allow FAT4 expressing mesenchymal cells to directly contact UB epithelial cells. In this model the basement membrane is a first line of defense between the UB and the mesenchyme. Once the basement membrane is degraded by MMPs, the FAT4-expressing cells can act as a second line of defense to suppress Ret activation and restrict budding and branching.
An alternative possibility of how FAT4 may affect RET signaling could involve shedding of the extracellular cadherin domains and diffusion to bind to RET in the UB. Notably, Drosophila Fat protein maturation involves an enzymatic cleavage at the extracellular domain (ECD) (Feng and Irvine, 2009; Sopko and McNeill, 2009), which could be released. Additionally, the ECD of FAT1 is shed and released into the secretome of human pancreatic cancer cells (Wojtalewicz et al., 2014). Here we showed that 5 cadherin repeats from the N-terminal interact with RET and affect RET-GFRA1-GDNF formation (Figures 5 and 6). Current antibodies do not recognize the ECD of FAT4, precluding testing this hypothesis. Future super resolution microscopy, and improved antibodies and/or tagging of the endogenous Ret and Fat4 loci will facilitate spatial analysis of the interaction as well as dynamic changes of this interaction during kidney development.
The functional and physical interactions between RET and FAT4 revealed in this study are unlikely to be limited to kidney development and may represent a more general mechanism underlying mesenchymal-epithelial interactions during development, such as in the intestine, another site where both RET and FAT4 are expressed (Lake and Heuckeroth, 2013; Rock et al., 2005). The RET-FAT4 interaction may also influence breast and pancreatic cancer, where both RET activation and FAT4 mutation are found (Mulligan, 2014; Katoh, 2012). Therefore, future investigation of the spatio-temporal regulation of FAT4-RET interaction will deepen our understanding of cell-cell communication and signaling transduction in development and disease.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Please contact the Lead Contact, Helen McNeill (mcneillh@wustl.edu), for reagents and resources generated in this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fat4 ko and Fat4-flox (Saburi et al., 2008), Fat4EGFP (Wu et al., 2008), Fjx1 ko (Probst et al., 2007), Hoxb7-cre (Zhao et al., 2004), and Pax3-cre (Engleka et al., 2005) have been described. Gdnf ko allele was generated by Dr. Arnon Rosenthal’s lab (Moore et al., 1996). Ai14 is a Cre reporter allele that has a loxP-flanked STOP cassette preventing transcription of a CAG promoter-driven tdTomato. All mice used in this study were maintained in a B6 background except for Fat4EGFP. Both male and female animals were used in this study as duplex kidney defects were observed in Fat4 mutants of both. Genotyping was performed using genomic DNA prepared from ear punches. Primer information for genotyping could be found from Table S3. Presence of a vaginal plug on noon of the day was considered embryonic day 0.5. Husbandry and ethical handling of mice were conducted according to guidelines approved by the Canadian Council on Animal Care. All mice were maintained at the Toronto Centre for Phenogenomics (TCP), a disease free environment for breeding of mouse colonies. Mice were given ad libitum a sterile rodent diet from Harlan (2918X) and continuous water supply. The light-dark cycle was kept constant with lights on from 7:00 to 19:00.
METHOD DETAILS
Histological analyses
Hematoxylin and eosin staining was carried out on 7μm paraffin sections. Slides were deparaffinized in xylene, serially rehydrated and stained for 15min in Mayer’s hematoxylin. Tissues were counterstained with freshly made eosin for 1min, dehydrated, mounted and visualized using a Nikon Eclipse 80i microscope with a DS-Ri1 camera.
Immunofluorescence on paraffin sections
Immunofluorescence (IF) was carried out on embryos embedded in paraffin according to standard protocols. Briefly, embryos were fixed overnight in 4% paraformaldehyde, washed in PBS, followed by serial dehydration with ethanol prior to embedding. Tissues were sectioned at 7μm, deparaffinized, rehydrated, and boiled for 22 minutes with Antigen Unmasking Solution (H-3300, Vector Laboratories). Slides were subsequently blocked in blocking buffer (10% NGS, 3% BSA, 0.1% Triton X-100 and 1x PBS) for an hour, probed overnight (O/N) at 4°C with primary antibodies and washed with PBS. FITC, Alexa-488, Cy3, or Cy5-conjugated secondary antibodies were used to visualize the protein of interest via fluorescence. The following antibodies (Abs) were used: anti-SIX2 (ProteinTech, 11562-1-AP; 1:300), anti-PBX1B (Santa Cruz, Sc-101852; 1:300), anti-WT1 (Dako, M3561; 1:200), anti-ECAD (BD Biosciences, 610181; 1:300), anti-PAX2 (BioLegend, 901001; 1:300), anti-phospho-ERK (Cell Signaling, #4370; 1:100), anti-ETV5 (ProteinTech, 13011-1-AP; 1:200), anti-GFP (Abcam, ab13970; 1:500). A Nikon D-Eclipse C1 confocal microscope was used to image all IF samples.
Whole mount immunofluorescence
Embryos were dissected at E10.5 or E11.5 and fixed in 4% PFA/PBS (pH 7.4) at 4°C O/N. Fixed embryos were rinsed with PBS at RT for several times. The urinary tracts were dissected, blocked in PBS-BB (1% BSA, 0.2% skim milk, 0.3% Triton X-100 and 1x PBS) and incubated at 4°C O/N with primary antibodies against CALBINDIN (Calbiochem PC253L, 1:300), PAX2 (BioLegand 901001, 1:300) or CYTOKERATIN (Sigma F3418, 1:300). The tissues were washed twice with PBST (0.3% TritonX-100,1x PBS) at RT for 1–2 h and then 4°C O/N. The tissues were then incubated with secondary antibody O/N at 4°C, washed several times in PBST at RT and imaged on a Nikon C1 confocal system with NIS Elements software (Nikon Instruments Inc., America).
In situ hybridization on paraffin sections
Anti-sense RNA probes labelled with digoxigenin for Crlf1, Cxcr4, Dusp6 and Etv4 were generated and samples were processed as previously described (Reginensi et al., 2016). Briefly, E12.5 embryos were dissected and fixed in 4% PFA/PBS at 4°C O/N, paraffin embedded, sectioned at 7 μm and transferred onto superfrost glass slides. The sections were deparaffinized, rehydrated, fixed and treated with proteinase K, before being hybridized O/N with anti-sense RNA probes (3μg/ml) at 70°C. The sections were washed and blocked, followed by anti-Digoxigenin-AP (Sigma) incubation O/N at 4°C. The colorimetric reaction was performed using BM Purple (Sigma).
RNAscope analyses
RNAscope® 2.5 HD Detection Kit-RED (Advanced Cell Diagnostics, Cat No: 322350) was used to determine Fat4 expression pattern in E11.5 kidney. Briefly, E11.5 embryos were fixed in 10% NBF for 24hr at room temperature, paraffin embedded, cut at 5μm and transferred onto superfrost glass slides. Sample preparation, pretreatment and RNAscope assay were performed according to Formalin-Fixed Paraffin-Embedded (FFPE) Sample Preparation Pretreatment Guide User Manual, Part 1 (Cat No. 322452-USM) and RNAscope® 2.5 HD Detection Kit (RED) User Manual, Part 2 (Cat No. 322360-USM). Probe for mouse Fat4 (Cat No.447511) and positive probe for mouse Ppib (Cat No. 313911) were used.
ex vivo kidney explants
Urogenital ridges from E11.5 embryos were dissected and placed on 0.4μm pore size culture plate inserts (millipore, PICM0RG50), which were in direct contact with DMEM supplemented with 10% FBS, penicillin-streptomycin and GlutaMAX. Cultures were incubated in 5% CO2 and at 37°C for 48h. Kidney rudiments were then fixed in ice-cold methanol at 4°C, washed in PBS and blocked for 1 h in 2% BSA/PBS at RT. Immunostaining was performed using antibodies against CALBINDIN (Calbiochem PC253L, 1:300) and PAX2 (BioLegand 901001, 1: 300), followed by incubation with FITC or Cy3-conjugated secondary antibody (Jackson Laboratories, 1:300) and visualization using a fluorescence microscope.
Cell culture and Transfection
HEK293T and MG87RET cell lines were maintained in DMEM supplemented with 10% FBS, 100 U/ml Penicillin, 100ug/ml Streptomycin and 1% GlutaMAX (complete DMEM). MG87RET cell line is a cell line expressing human c-RET long isoform resulted from retroviral infection (Eketjall et al., 1999). All cells were cultured at 37°C in a humidified atmosphere of 5% CO2. DCHS1-mCherry and FAT4-Citrine cell lines were described recently (Loza et al., 2017) and were kindly provided by Dr. David Sprinzak. Transient transfections into HEK and MG87RET cells and stable transfection into MG87RET cells were performed using Lipofectamine 3000 according to the manufacturer’s instructions. Stable MG87RET+FAT4-EC1-5V5 cell line was selected and maintained in complete DMEM with 500μg/ml Neomycin. Tetracycline-inducible FAT4 expressing cell line (Tet-on-FAT4) was made by cloning FAT4 cDNA into the pcDNA5-FRT/TO-BirA-Flag expression vector (gift from Dr. Brian Raught) by Keyclone Technologies. HEK293 T-REx Flp-In cells (Thermo Fisher Scientific, R78007) were transfected with FAT4-BF and the Flp-recombinase expression vector pOG44 (gift from Dr. Anne-Claude Gingras) and selected with complete DMEM containing Hygromycin B (200μg/ml).
Plasmids used in this study are as follows. pEGFP-C1-Lulu (gift from Dr. Jane McGlade) was used as a negative control for the co-IP experiments using GFP beads. FAT4-Cad1-5-3Flag and DCHS1-Cad1-5-3Flag were cloned into pcDNA3 with triple Flag tag sequence designed to be included in the primer. FAT4-Cad1-5V5 expression vector was made by fusing the secretion signal of Rat TRANSIN protein with FAT4-Cad1-5 coding sequence (aa43-454) and cloning the fused sequence into pcDNA3 using NEB Hifi DNA Assembly. The sequence is in frame with the V5 coding sequence of the vector. PB-T-PAF vector (gift from Dr. James Rini) (Li et al., 2013) was used as the template for Transin PCR amplification.
RET51-EGFP was constructed on the basis of pEGFPN1-RET9, which is a gift from Dr. Jeff Milbrandt, made by cloning an N-terminal HindIII-EcoRI and a C-terminal EcoRI-EcoRI PCR fragments of RET9 into the pEGFPN1 vector (clontech) using HindIII and EcoRI sites of the vector, and an internal EcoRI site of RET9. The C-terminal part of RET9 was swapped for that of RET51 through the two EcoRI sites to make pEGFPN1-RET51-EGFP. EGFP was then swapped for tdTomato to get RET51-tdTomato using the flanking AgeI and NotI sites. pcDNA3-FLAG-ratGFRa1 is an expression vector for rat GFRA1 with a FLAG tag inserted after the signal sequence (gift from Dr. Jeff Milbrandt). RET51-Cad1-4 was deleted by site-directed mutagenesis using the pEGFPN1-RET51-EGFP as a template to get RET51-delta-Cad1-4-EGFP. A SpeI restriction enzyme site was introduced into the deletion site. PCR products of RET51-Cad1-2 and RET51-Cad3-4 were cloned into the SpeI site of RET51-delta-Cad1-4-EGFP through the NEB HiFi DNA Assembly to get the RET51-delta-Cad3-4-EGFP and RET51-delta-Cad1-2-EGFP respectively. For details, please refer to Table S4.
For full-length RET and FAT4 or DCHS1 interaction, stable cells for FAT4 or DCHS1 was transfected with RET. The cell lysate was split into two parts. FAT4 or DCHS1 was pulled down with either tag antibody (test) or IgG (ctrl) to show specific interaction between full-length FAT4 or DCHS1 with full-length RET. For interaction between FAT4- or DCHS1-Cad1-5 and full-length RET, FAT4- or DCHS1-Cad1-5-3Flag was either co-transfected with RET-EGFP, or with Lulu-GFP as a control. For co-immunoprecipitation assays to test the dependency of RET-Cad(s), FAT4-Cad1-5V5 was co-transfected with RET-EGFP, RET-delta-cad1-4-EGFP, RET-delta-Cad1-2-EGFP, RET-delta-Cad3-4-EGFP or Lulu-GFP. To test the effect of FAT4-Cad1-5 on RET-GFRA1 interaction, RET-EGFP was either co-transfected with Flag-GFRA1 or with Flag-GFRA1 and FAT4-Cad1-5V5 together. The transfected HEK cells were then either left untreated or induced with recombinant hGDNF (50–100ng/ml) for 10 min after O/N serum starvation.
Immunoprecipitation, Cell mixing, and Western blot
For all co-immunoprecipitation assays, cells were lysed in IP lysis buffer containing 20 mM Tris, pH7.5, 150 mM NaCl, 2 ml CaCl2, 1% Triton X-100, 10% glycerol and protease and phosphatase inhibitors. The lysate was cleared and incubated with antibody conjugated agarose or magnetic beads O/N at 4°C. For cell mixing experiments, MG87RET cells (1.8×105) were mixed with 3 times more Tet-on-FAT4 cells (5.4×105) per well in 6-well plate. The cells were cultured in complete DMEM containing 10μg/ml tetracycline or not for 48hrs, serum-starved for 5hrs, stimulated with recombinant hGDNF (150ng/ml, R&D system, 212-GD-010) and hGFRA1-Fc Chimera Protein (500ng/ml, R&D system, 714-GR-100) for 10min or 30min as indicated, and then lysed in lysis buffer containing 25mM Tris, pH7.4, 1% NP40, 1% Triton X-100, 1% n-Octyl glucoside, 2mM EDTA and protease and phosphatase inhibitors.
Antibodies used for IP and WB include: anti-mCherry (Abcam, ab167453; 1:1000), anti-GFP (Abcam, ab290; 1:5000), anti-tdTomato (Mybiosource, MBS448092; 1:1000), anti-Flag (Sigma, F1840; 1:1000), anti-V5 (Bethyl, A190-120A; 1:1000), anti-RET (CST, #3223; 1:1000), anti-ERK (CST, #4696; 1:1000), anti-phospho-ERK (CST, #4370; 1:2500), anti-GFRA1 (neuromics, GT15004; 1 :1000), anti-ACTIN (EMD Millipore, MAB1501; 1:1000)
Generation of the dendrogram for the cadherin superfamily
The multiple sequence alignments based on cadherin-repeat one homologies were first performed by ClustalX (NJ approach) and the circular tree was then generated by the Interactive Tree of Life (iTOL) tool. For detailed information on the sequence information, please refer to Hulpiau and van Roy, 2009. Of note, the EC1 of RET they referred to in their paper is actually Cad2 in this study. Sequences for cadherin repeats from Homo sapiens, Mus musculus, Drosophila melanogaster, Gallus gallus, Caenorhabditis elegans, Xenopus laevis, Xenopus tropicalis and Ciona intestinalis were included in this study.
QUANTIFICATION AND STATISTICAL ANALYSIS
Image lab (Bio-Rad) was used to quantify mean value of western blot bands. Briefly, for pERK level in E12.5 kidney lysate in Figures 3 and S3, intensity of pERK was first divided by that of total ERK. The ratio from such calculation for Fat4−/−;Fjx1−/− was then normalized to that for wild type (arbitrarily set as 1) to get the relative pERK level. For the cell mixing experiments in Figures 6 and S6, intensity of pERK was first divided by intensity of either ERK or the upper band of RET-the mature form of RET. The ratio for the Tet+ group was again normalized to that for the Tet− group (arbitrarily set as 1) to get the relative pERK level. For co-IP quantification in Figure 6, intensity of co-IPed GFRA1 was first divided by that of IPed RET. The ratio from such calculation for R+G in Figure 6A and the ratio for R+G+G in Figure 6B were set as 1 and ratios for other groups were then normalized to 1 and compared. Statistical comparisons were performed by un-paired two-tailed Student’s t-test for all butFigure 6B, which was done by one-way ANOVA, using GraphPad Prism 7 software. Statistical significance between groups was indicated as follows: *p<0.05, **p<0.01, ***p<0.001. The statistical significance for duplex phenotype in different background was determined by performing two-proportion test (https://measuringu.com/ab-cal/).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-SIX2 | ProteinTech | 11562-1-AP |
| Mouse anti-PBX1B | SCTB | sc-101852 |
| Mouse anti-WT1 | Dako | M3561 |
| Mouse anti-ECAD | BD Transduction Laboratories | 610181 |
| Rabbit anti-PAX2 | BioLegend | 901001 |
| Rabbit anti-phospho-ERK | Cell Signaling | #4370 |
| Mouse anti-ERK | Cell Signaling | #4696 |
| Rabbit anti-ETV5 | ProteinTech | 13011-1-AP |
| Chicken anti-GFP | Abcam | ab13970 |
| Rabbit anti-GFP | Abcam | Ab290 |
| Rabbit anti-CALBINDIN | Calbiochem | PC253L |
| Mouse anti-CYTOKERATIN | Sigma | F3418 |
| Rabbit anti-mCherry | Abcam | Ab167453 |
| Goat anti-tdTomato | Mybiosource | MBS448092 |
| Mouse anti-Flag | Sigma | F1840 |
| Rabbit anti-V5 | Bethyl | A190-120A |
| Rabbit anti-RET | Cell Signaling | #3223 |
| Goat anti-GFRA1 | Neuromics | GT15004 |
| Mouse anti-ACTIN | EMD Millipore | MAB1501 |
| GFP-Trap_MA | Chromotek | gtma-20 |
| Bacterial and Virus Strains | ||
| TOP10 | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant Human GDNF | R&D system | 212-GD-010 |
| Recombinant Human GFRA1 Fc Chimera protein | R&D system | 714-GR-100 |
| Anti-Digoxigenin-AP | Sigma | 11093274910 |
| BM purple | Sigma | 11442074001 |
| Critical Commercial Assays | ||
| RNAscope® 2.5 HD Detection Kit-RED | Advanced Cell Diagnostics | 322350 |
| RNAscope® Probe- Mm-Fat4 | Advanced Cell Diagnostics | 447511 |
| RNAscope® Probe- Mm-Ppib | Advanced Cell Diagnostics | 313911 |
| Experimental Models: Cell Lines | ||
| HEK293T | Dr. David Sprinzak | N/A |
| FAT4-Citrine | Dr. David Sprinzak | N/A |
| DCHS1-mCherry | Dr. David Sprinzak | N/A |
| MG87RET | This paper | N/A |
| HEK293 T-REx Flp-In | Thermo Fisher Scientific | R78007 |
| Tet-on-FAT4 | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: Fat4tm1.1Hmc | Saburi et al., 2008 | N/A |
| Mouse: Fat4tm1Hmc | Saburi et al., 2008 | N/A |
| Mouse: Fat4EGFP | Wu et al., 2008 | N/A |
| Mouse: Fjx1tm1Awp | Probst et al., 2007 | N/A |
| Mouse: Tg(Hoxb7-cre)5526Cmb | Zhao et al., 2004 | N/A |
| Mouse: Pax3tm1(cre)Joe | Engleka et al., 2005 | N/A |
| Mouse: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | The Jackson Laboratory | #7914 |
| Mouse: Gdnftm1Rosl | Moore et al., 1996 | N/A |
| Mouse: C57BL/6J | The Centre for Phenogenomics | N/A |
| Recombinant DNA | ||
| pcDNA5-FRT/TO-FAT4-BirA-Flag | This paper | N/A |
| pOG44 | Gift from Dr. Anne-Claude Gingras | N/A |
| pEGFP-C1-Lulu | Gift from Dr. Jane McGlade | N/A |
| pcDNA3-FAT4-Cad1-5-3Flag | This paper | N/A |
| pcDNA3-DCHS1-Cad1-5-3Flag | This paper | N/A |
| pcDNA3-TransinSP-FAT4-Cad1-5-V5 | This paper | N/A |
| PB-T-PAF | Gift from Dr. James Rini | N/A |
| pEGFP-N1-RET51 | This paper | N/A |
| RET51-tdTomato | This paper | N/A |
| pcDNA3-FLAG-ratGFRa1 | Gift from Dr. Jeffrey Milbrandt | N/A |
| pEGFP-N1-RET51-delta-Cad1-4 | This paper | N/A |
| pEGFP-N1-RET51-Cad1+2 | This paper | N/A |
| pEGFP-N1-RET51-Cad3+4 | This paper | N/A |
| Oligonucleotides | ||
| Primers for genotyping | This paper, see Table S3 | N/A |
| Primers used for cloning | This paper, see Table S4 | N/A |
| Software and Algorithms | ||
| Image Lab | Bio-Rad | www.bio-rad.com |
| Prism 7 | Graphpad | N/A |
Highlights.
Fat4 mutant mice have duplex kidney defects due to ectopic bud formation
FAT4 functions non-autonomously in the mesenchyme to prevent kidney duplication
RET signaling is overactive in Fat4 mutants
FAT4 interacts with RET, perturbs RET-GFRA1-GDNF assembly and reduces RET signaling
ACKNOWLEDGEMENT
We thank Dr. Mario Cappecchi for the Fat4EGFP/+ mouseline and Dr. Carl Bates for the Hoxb7-cre mice. We also thank Dr. Anne-Claude Gingras, Dr. Brian Raught, Dr. James Rini, Dr. Jane McGlade and Dr. Jeffrey Milbrandt for plasmids. We are grateful to Dr. David Sprinzak for providing cell lines and Dr. Antoine Reginensi for assistance in in situ hybridization. We thank Bendi Gong for help in generating the RET signaling related plasmids.
MBL was funded by OGS and CIHR awards. Grants to HM were provided by a CIHR Foundation award, and start-up funds from Washington University School of Medicine and the BJC investigator program. SJ is supported by R01 DK082531.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests
REFERENCES
- Alders M, Al-Gazali L, Cordeiro I, Dallapiccola B, Garavelli L, Tuysuz B, Salehi F, Haagmans MA, Mook OR, Majoie CB, et al. (2014). Hennekam syndrome can be caused by FAT4 mutations and be allelic to Van Maldergem syndrome. Hum Genet 133, 1161–7. [DOI] [PubMed] [Google Scholar]
- Arnould C, Lelievre-Pegorier M, Ronco P, and Lelongt B (2009). MMP9 limits apoptosis and stimulates branching morphogenesis during kidney development. J Am Soc Nephrol 20, 2171–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avantaggiato V, Dathan NA, Grieco M, Fabien N, Lazzaro D, Fusco A, Simeone A, and Santoro M (1994). Developmental expression of the RET protooncogene. Cell Growth Differ 5, 305–11. [PubMed] [Google Scholar]
- Bagherie-Lachidan M, Reginensi A, Zaveri HP, Scott DA, Helmbacher F, and McNeill H (2015). Stromal Fat4 acts non-autonomously with Dachsous1/2 to restrict the nephron progenitor pool. Development. [DOI] [PubMed] [Google Scholar]
- Barasch J, Yang J, Qiao J, Tempst P, Erdjument-Bromage H, Leung W, and Oliver JA (1999). Tissue inhibitor of metalloproteinase-2 stimulates mesenchymal growth and regulates epithelial branching during morphogenesis of the rat metanephros. J Clin Invest 103, 1299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson MA, Akbulut S, Watson-Johnson J, Simon R, Carroll TJ, Shakya R, Gross I, Martin GR, Lufkin T, McMahon AP, et al. (2005). Sprouty1 is a critical regulator of GDNF/RET-mediated kidney induction. Dev Cell 8, 229–39. [DOI] [PubMed] [Google Scholar]
- Basson MA, Watson-Johnson J, Shakya R, Akbulut S, Hyink D, Costantini FD, Wilson PD, Mason IJ, and Licht JD (2006). Branching morphogenesis of the ureteric epithelium during kidney development is coordinated by the opposing functions of GDNF and Sprouty1. Dev Biol 299, 466–77. [DOI] [PubMed] [Google Scholar]
- Blair S, and McNeill H (2018). Big roles for Fat cadherins. Curr Opin Cell Biol 51, 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard M, Souabni A, Mandler M, Neubuser A, and Busslinger M (2002). Nephric lineage specification by Pax2 and Pax8. Genes Dev 16, 2958–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenberger R, Schmidt A, Linton J, Wang D, Backus C, Denda S, Muller U, and Reichardt LF (2001). Identification and characterization of a novel extracellular matrix protein nephronectin that is associated with integrin alpha8beta1 in the embryonic kidney. J Cell Biol 154, 447–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittle AL, Repiso A, Casal J, Lawrence PA, and Strutt D (2010). Four-jointed modulates growth and planar polarity by reducing the affinity of dachsous for fat. Curr Biol 20, 803–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacalano G, Farinas I, Wang LC, Hagler K, Forgie A, Moore M, Armanini M, Phillips H, Ryan AM, Reichardt LF, et al. (1998). GFRalpha1 is an essential receptor component for GDNF in the developing nervous system and kidney. Neuron 21, 53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappello S, Gray MJ, Badouel C, Lange S, Einsiedler M, Srour M, Chitayat D, Hamdan FF, Jenkins ZA, Morgan T, et al. (2013). Mutations in genes encoding the cadherin receptor-ligand pair DCHS1 and FAT4 disrupt cerebral cortical development. Nat Genet 45, 1300–8. [DOI] [PubMed] [Google Scholar]
- Chi X, Michos O, Shakya R, Riccio P, Enomoto H, Licht JD, Asai N, Takahashi M, Ohgami N, Kato M, et al. (2009). Ret-dependent cell rearrangements in the Wolffian duct epithelium initiate ureteric bud morphogenesis. Dev Cell 17, 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini F (2011). GDNF/Ret signaling and renal branching morphogenesis: From mesenchymal signals to epithelial cell behaviors. Organogenesis 6, 252–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini F, and Kopan R (2010). Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev Cell 18, 698–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Tanigawa S, Karner CM, Xin M, Lum L, Chen C, Olson EN, Perantoni AO, and Carroll TJ (2013). Stromal-epithelial crosstalk regulates kidney progenitor cell differentiation. Nat Cell Biol 15, 1035–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TK, Hoshi M, and Jain S (2013). To bud or not to bud: the RET perspective in CAKUT. Pediatr Nephrol 29, 597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degoutin JL, Milton CC, Yu E, Tipping M, Bosveld F, Yang L, Bellaiche Y, Veraksa A, and Harvey KF (2013). Riquiqui and minibrain are regulators of the hippo pathway downstream of Dachsous. Nat Cell Biol 15, 1176–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos ACS, Marques de Miranda D, and Simoes e Silva AC (2014). Congenital Anomilies of the Kidney and Urinary Tract: An Embryogenetic Review. Birth Defects Research (Part C) 102, 374–381. [DOI] [PubMed] [Google Scholar]
- Dressler GR (2009). Advances in early kidney specification, development and patterning. Development 136, 3863–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbec P, Marcos-Gutierrez CV, Kilkenny C, Grigoriou M, Wartiowaara K, Suvanto P, Smith D, Ponder B, Costantini F, Saarma M, et al. (1996). GDNF signalling through the Ret receptor tyrosine kinase. Nature 381, 789–93. [DOI] [PubMed] [Google Scholar]
- Ekblom P, Alitalo K, Vaheri A, Timpl R, and Saxen L (1980). Induction of a basement membrane glycoprotein in embryonic kidney: possible role of laminin in morphogenesis. Proc Natl Acad Sci U S A 77, 485–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eketjall S, Fainzilber M, Murray-Rust J, and Ibanez CF (1999). Distinct structural elements in GDNF mediate binding to GFRalpha1 and activation of the GFRalpha1-c-Ret receptor complex. EMBO J 18, 5901–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engleka KA, Gitler AD, Zhang M, Zhou DD, High FA, and Epstein JA (2005). Insertion of Cre into the Pax3 locus creates a new allele of Splotch and identifies unexpected Pax3 derivatives. Dev Biol 280, 396–406. [DOI] [PubMed] [Google Scholar]
- Esquela AF, and Lee SJ (2003). Regulation of metanephric kidney development by growth/differentiation factor 11. Dev Biol 257, 356–70. [DOI] [PubMed] [Google Scholar]
- Feng Y, and Irvine KD (2009). Processing and phosphorylation of the Fat receptor. Proc Natl Acad Sci U S A 106, 11989–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher CE, Michael L, Barnett MW, and Davies JA (2001). Erk MAP kinase regulates branching morphogenesis in the developing mouse kidney. Development 128, 4329–38. [DOI] [PubMed] [Google Scholar]
- Grieshammer U, Le M, Plump AS, Wang F, Tessier-Lavigne M, and Martin R (2004). SLIT2-mediated ROBO2 signaling restricts kidney induction to a single site. Dev Cell 6, 709–17. [DOI] [PubMed] [Google Scholar]
- Hoshi M, Batourina E, Mendelsohn C, and Jain S (2012). Novel mechanisms of early upper and lower urinary tract patterning regulated by RetY1015 docking tyrosine in mice. Development 139, 2405–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulpiau P, and van Roy F (2009). Molecular evolution of the cadherin superfamily. Int J Biochem Cell Biol 41, 349–69. [DOI] [PubMed] [Google Scholar]
- Ishikawa HO, Takeuchi H, Haltiwanger RS, and Irvine KD (2008).Four-jointed is a Golgi kinase that phosphorylates a subset of cadherin domains. Science 321, 401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiuchi T, Misaki K, Yonemura S, Takeichi M, and Tanoue T (2009). Mammalian Fat and Dachsous cadherins regulate apical membrane organization in the embryonic cerebral cortex. J Cell Biol 185, 959–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Encinas M, Johnson EM Jr., and Milbrandt J (2006). Critical and distinct roles for key RET tyrosine docking sites in renal development. Genes Dev 20, 321–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M (2012). Function and cancer genomics of FAT family genes (review). Int J Oncol 41, 1913–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, and Dressler GR (2007). PTEN modulates GDNF/RET mediated chemotaxis and branching morphogenesis in the developing kidney. Dev Biol 307, 290–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RD, Sherman D, Ho WH, Stone D, Bennett GL, Moffat B, Vandlen R, Simmons L, Gu Q, Hongo JA, et al. (1997). A GPI-linked protein that interacts with Ret to form a candidate neurturin receptor. Nature 387, 717–21. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Kawakami K, Asashima M, and Nishinakamura R (2007). Six1 and Six4 are essential for Gdnf expression in the metanephric mesenchyme and ureteric bud formation, while Six1 deficiency alone causes mesonephric-tubule defects. Mech Dev 124, 290–303. [DOI] [PubMed] [Google Scholar]
- Kume T, Deng K, and Hogan BL (2000). Murine forkhead/winged helix genes Foxc1 (Mf1) and Foxc2 (Mfh1) are required for the early organogenesis of the kidney and urinary tract. Development 127, 1387–95. [DOI] [PubMed] [Google Scholar]
- Lake JI, and Heuckeroth RO (2013). Enteric nervous system development: migration, differentiation, and disease. Am J Physiol Gastrointest Liver Physiol 305, G1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledda F, Bieraugel O, Fard SS, Vilar M, and Paratcha G (2008). Lrig1 is an endogenous inhibitor of Ret receptor tyrosine kinase activation, downstream signaling, and biological responses to GDNF. J Neurosci 28, 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelongt B, Trugnan G, Murphy G, and Ronco PM (1997). Matrix metalloproteinases MMP2 and MMP9 are produced in early stages of kidney morphogenesis but only MMP9 is required for renal organogenesis in vitro. J Cell Biol 136, 1363–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz O, Elliot SJ, and Stetler-Stevenson WG (2000). Matrix metalloproteinases in renal development and disease. J Am Soc Nephrol 11, 574–81. [DOI] [PubMed] [Google Scholar]
- Li Z, Michael IP, Zhou D, Nagy A, and Rini JM (2013). Simple piggyBac transposon-based mammalian cell expression system for inducible protein production. Proc Natl Acad Sci U S A 110, 5004–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loza O, Heemskerk I, Gordon-Bar N, Amir-Zilberstein L, Jung Y, and Sprinzak D (2017). A synthetic planar cell polarity system reveals localized feedback on Fat4-Ds1 complexes. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu BC, Cebrian C, Chi X, Kuure S, Kuo R, Bates CM, Arber S, Hassell J, MacNeil L, Hoshi M, et al. (2009). Etv4 and Etv5 are required downstream of GDNF and Ret for kidney branching morphogenesis. Nat Genet 41, 1295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Francis-West P, and Irvine KD (2015). A Fat4-Dchs1 signal between stromal and cap mesenchyme cells influences nephrogenesis and ureteric bud branching. Development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Mulvaney J, Zakaria S, Yu T, Morgan KM, Allen S, Basson MA, Francis-West P, and Irvine KD (2011). Characterization of a Dchs1 mutant mouse reveals requirements for Dchs1-Fat4 signaling during mammalian development. Development 138, 947–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JM, Morrison DJ, Basson MA, and Licht JD (2006). Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol 16, 45–54. [DOI] [PubMed] [Google Scholar]
- Matakatsu H, and Blair SS (2004). Interactions between Fat and Dachsous and the regulation of planar cell polarity in the Drosophila wing. Development 131, 3785–94. [DOI] [PubMed] [Google Scholar]
- Meyer TN, Schwesinger C, Bush KT, Stuart RO, Rose DW, Shah MM, Vaughn DA, Steer DL, and Nigam SK (2004). Spatiotemporal regulation of morphogenetic molecules during in vitro branching of the isolated ureteric bud: toward a model of branching through budding in the developing kidney. Dev Biol 275, 44–67. [DOI] [PubMed] [Google Scholar]
- Michos O, Cebrian C, Hyink D, Grieshammer U, Williams L, D'Agati V, Licht JD, Martin GR, and Costantini F (2010). Kidney development in the absence of Gdnf and Spry1 requires Fgf10. PLoS Genet 6, e1000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MW, Klein RD, Farinas I, Sauer H, Armanini M, Phillips H, Reichardt LF, Ryan AM, Carver-Moore K, and Rosenthal A (1996). Renal and neuronal abnormalities in mice lacking GDNF. Nature 382, 76–9. [DOI] [PubMed] [Google Scholar]
- Mulligan LM (2014). RET revisited: expanding the oncogenic portfolio. Nat Rev Cancer 14, 173–86. [DOI] [PubMed] [Google Scholar]
- Nicolaou N, Renkema KY, Bongers EM, Giles RH, and Knoers NV (2015). Genetic, environmental, and epigenetic factors involved in CAKUT. Nat Rev Nephrol 11, 720–31. [DOI] [PubMed] [Google Scholar]
- Nishinakamura R, Matsumoto Y, Nakao K, Nakamura K, Sato A, Copeland NG, Gilbert DJ, Jenkins NA, Scully S, Lacey DL, et al. (2001). Murine homolog of SALL1 is essential for ureteric bud invasion in kidney development. Development 128, 3105–15. [DOI] [PubMed] [Google Scholar]
- Paratcha G, Ledda F, Baars L, Coulpier M, Besset V, Anders J, Scott R, and Ibanez CF (2001). Released GFRalpha1 potentiates downstream signaling, neuronal survival, and differentiation via a novel mechanism of recruitment of c-Ret to lipid rafts. Neuron 29, 171–84. [DOI] [PubMed] [Google Scholar]
- Pierchala BA, Milbrandt J, and Johnson EM Jr. (2006). Glial cell line-derived neurotrophic factor-dependent recruitment of Ret into lipid rafts enhances signaling by partitioning Ret from proteasome-dependent degradation. J Neurosci 26, 2777–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poladia DP, Kish K, Kutay B, Hains D, Kegg H, Zhao H, and Bates CM (2006). Role of fibroblast growth factor receptors 1 and 2 in the metanephric mesenchyme. Dev Biol 291, 325–39. [DOI] [PubMed] [Google Scholar]
- Probst B, Rock R, Gessler M, Vortkamp A, and Puschel AW (2007). The rodent Four-jointed ortholog Fjx1 regulates dendrite extension. Dev Biol 312, 461–70. [DOI] [PubMed] [Google Scholar]
- Reginensi A, Enderle L, Gregorieff A, Johnson RL, Wrana JL, and McNeill H(2016). A critical role for NF2 and the Hippo pathway in branching morphogenesis. Nat Commun 7, 12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock R, Schrauth S, and Gessler M (2005). Expression of mouse dchs1, fjx1, and fat-j suggests conservation of the planar cell polarity pathway identified in Drosophila. Dev Dyn 234, 747–55. [DOI] [PubMed] [Google Scholar]
- Rozen EJ, Schmidt H, Dolcet X, Basson MA, Jain S, and Encinas M (2009). Loss of Sprouty1 rescues renal agenesis caused by Ret mutation. J Am Soc Nephrol 20, 255–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saburi S, Hester I, Fischer E, Pontoglio M, Eremina V, Gessler M, Quaggin SE, Harrison R, Mount R, and McNeill H (2008). Loss of Fat4 disrupts PCP signaling and oriented cell division and leads to cystic kidney disease. Nat Genet 40, 1010–5. [DOI] [PubMed] [Google Scholar]
- Saifudeen Z, Dipp S, Stefkova J, Yao X, Lookabaugh S, and El-Dahr SS (2009). p53 regulates metanephric development. J Am Soc Nephrol 20, 2328–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez MP, Silos-Santiago I, Frisen J, He B, Lira SA, and Barbacid M (1996). Renal agenesis and the absence of enteric neurons in mice lacking GDNF. Nature 382, 70–3. [DOI] [PubMed] [Google Scholar]
- Schuchardt A, D'Agati V, Larsson-Blomberg L, Costantini F, and Pachnis V (1994). Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature 367, 380–3. [DOI] [PubMed] [Google Scholar]
- Short KM, and Smyth IM (2016). The contribution of branching morphogenesis to kidney development and disease. Nat Rev Nephrol 12, 754–767. [DOI] [PubMed] [Google Scholar]
- Simon MA, Xu A, Ishikawa HO, and Irvine KD (2010). Modulation of fat:dachsous binding by the cadherin domain kinase four-jointed. Curr Biol 20, 811–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R, and McNeill H (2009). The skinny on Fat: an enormous cadherin that regulates cell adhesion, tissue growth, and planar cell polarity. Curr Opin Cell Biol 717–23. [DOI] [PubMed] [Google Scholar]
- Treanor JJ, Goodman L, de Sauvage F, Stone DM, Poulsen KT, Beck CD, Gray C, Armanini MP, Pollock RA, Hefti F, et al. (1996). Characterization of a multicomponent receptor for GDNF. Nature 382, 80–3. [DOI] [PubMed] [Google Scholar]
- Trupp M, Arenas E, Fainzilber M, Nilsson AS, Sieber BA, Grigoriou M, Kilkenny C, Salazar-Grueso E, Pachnis V, and Arumae U (1996). Functional receptor for GDNF encoded by the c-ret proto-oncogene. Nature 381, 785–9. [DOI] [PubMed] [Google Scholar]
- van der Ven AT, Shril S, Ityel H, Vivante A, Chen J, Hwang DY, Laricchia KM, Lek M, Tasic V, and Hildebrandt F (2017). Whole-Exome Sequencing Reveals FAT4 Mutations in a Clinically Unrecognizable Patient with Syndromic CAKUT: A Case Report. Mol Syndromol 8, 272–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivante A, Kohl S, Hwang DY, Dworschak GC, and Hildebrandt F (2014). Single-gene causes of congenital anomalies of the kidney and urinary tract (CAKUT) in humans. Pediatr Nephrol 29, 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, and Costantini F (2004). Real-time analysis of ureteric bud branching morphogenesis in vitro. Dev Biol 271, 98–108. [DOI] [PubMed] [Google Scholar]
- Wellik DM, Hawkes PJ, and Capecchi MR (2002). Hox11 paralogous genes are essential for metanephric kidney induction. Genes Dev 16, 1423–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitten SM, and Wilcox DT (2001). Duplex systems. Prenat Diagn 21, 952–7. [DOI] [PubMed] [Google Scholar]
- Wojtalewicz N, Sadeqzadeh E, Weiss JV, Tehrani MM, Klein-Scory S, Hahn S, Schmiegel W, Warnken U, Schnolzer M, de Bock CE, et al. (2014). A soluble form of the giant cadherin Fat1 is released from pancreatic cancer cells by ADAM10 mediated ectodomain shedding. PLoS One 9, e90461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Ying G, Wu Q, and Capecchi MR (2008). A protocol for constructing gene targeting vectors: generating knockout mice for the cadherin family and beyond. Nat Protoc 3, 1056–76. [DOI] [PubMed] [Google Scholar]
- Xu PX, Adams J, Peters H, Brown MC, Heaney S, and Maas R (1999). Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet 23, 113–7. [DOI] [PubMed] [Google Scholar]
- Xu PX, Zheng W, Huang L, Maire P, Laclef C, and Silvius D (2003). Six1 is required for the early organogenesis of mammalian kidney. Development 130, 3085–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Kegg H, Grady S, Truong HT, Robinson ML, Baum M, and Bates CM (2004). Role of fibroblast growth factor receptors 1 and 2 in the ureteric bud. Dev Biol 276, 403–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.