Abstract
Cell-to-cell variation in the regenerative potential of mesenchymal stromal cells (MSCs) impedes the translation of MSC therapies into clinical practice. Cellular heterogeneity is ubiquitous across MSC cultures from different species and tissues. This review highlights advances to elucidate molecular profiles that identify cell subsets with specific regenerative properties in heterogeneous MSC cultures. Cell surface markers and global signatures are presented for proliferation and differentiation potential, as well as immunomodulation and trophic properties. Key knowledge gaps are discussed as potential areas of future research. Molecular profiles of MSC heterogeneity have the potential to enable unprecedented control over the regenerative potential of MSC therapies through the discovery of new molecular targets and as quality attributes to develop robust and reproducible biomanufacturing processes. These advances would have a positive impact on the nascent field of MSC therapeutics by accelerating the development of therapies with more consistent and effective treatment outcomes.
1. Introduction
Mesenchymal stromal cells (MSCs) are the most common stem cell therapy in clinical trials [1]. This popularity traces back to the groundbreaking research of Friedenstein et al. who identified colony-forming unit fibroblasts (now known as MSCs) in bone marrow [2]. This early research demonstrated that MSCs have a remarkable capacity to regenerate osseous tissue in vivo [3]. MSCs have been given several names over the years, including marrow stromal cells and multipotent stromal cells [4, 5], and have been isolated from many tissues, such as adipose and the umbilical cord [6, 7]. The current popularity of MSCs as a stem cell therapy reflects their broad regenerative properties to home to the site of injury [8], undergo extensive proliferation [9], exhibit multipotency [10], regulate the immune system [11], and secrete trophic factors [12]. The therapeutic applications of these pleiotropic cells are vast. Clinical trials with MSCs are underway to treat skeletal defects, graft-vs.-host disease, and cardiovascular disorders, to name a few [13].
A barrier to realize the therapeutic potential of MSCs is their intrinsic heterogeneity. MSCs are a composite of cell progenitors at different states of lineage commitment [14, 15] and cellular aging [16, 17]. Cellular heterogeneity is ubiquitous across MSC cultures harvested from different species and tissues [18–20]. Cell-to-cell variation in MSC function initiates in vivo in the stem cell niche [21], is evident within single-cell-derived MSC colonies [22], and is exacerbated by replicative stress during ex vivo cultivation [16]. Cell subsets within heterogeneous MSC cultures vary in their regenerative potential, including proliferation potential [23, 24] and potency [10, 14]. Cellular heterogeneity has impacted the effectiveness of MSC therapies in animal models to repair bone, cartilage, and the heart, among other tissues [25–27]. This heterogeneity has been cited as a possible factor contributing to the variability in treatment outcomes of MSC therapies in clinical trials [13, 28, 29]. Variation in the regenerative potential among cell subsets in MSC cultures may confound trial results and slow, if not arrest, the translation of an MSC therapy into clinical practice.
There is a critical need for molecular profiles of MSC heterogeneity to manufacture effective MSC therapies. This review highlights advances to elucidate cell surface markers and global signatures that identify cell subsets with specific regenerative properties in heterogeneous MSC cultures. Molecular profiles of MSC heterogeneity will enable cell enrichment and quality control assessment during the manufacturing of MSC therapies to standardize cell composition. In addition, they will help identify new molecular targets to regulate the regenerative potential of MSCs. Molecular profiles of MSC heterogeneity are expected to make a positive impact on the nascent field of MSC therapeutics by accelerating the development of therapies with more consistent and effective treatment outcomes.
2. Proliferation Potential
MSCs are a rare population of progenitors in adult tissue [10] and are expanded ex vivo to obtain a sufficient amount of cells for clinical applications [30]. Cell-to-cell variation in the proliferation potential of MSCs gives rise to cell population dynamics during ex vivo expansion that alters the composition of cell subsets in culture and, in turn, may impact the efficacy of MSC therapies [31]. Heterogeneity in the proliferation potential of MSC cultures was first reported in morphologically distinct subsets of small, rapidly dividing cells and large, slowly dividing cells [23, 24]. We and others have validated this functional heterogeneity in proliferation potential with single-cell-derived colonies that originated from a common, parental MSC culture [15, 32, 33].
2.1. Cell Surface Markers of Proliferation Potential
A focus of ongoing research on MSC heterogeneity is to elucidate an immunophenotype of proliferation potential. Cell surface markers enable noninvasive and nondestructive isolation of specific cell subsets from MSC cultures for research and clinical applications. The International Society for Cellular Therapy has specified that human MSCs must express CD73, CD90, and CD105 [34]. We and others observed little to no variation in surface expression of these biomarkers between rapidly and slowly dividing cells in cultures of human bone marrow-derived MSCs (hBM-MSCs) [17, 32, 35]. The inability of the standard MSC immunophenotype to detect specific cell subsets in MSC cultures demonstrates the need for new surface markers of MSC heterogeneity.
Several cell surface markers have been identified to isolate MSCs with high colony-forming efficiency from tissues (Table 1). Of these, the 75 kDa endothelial antigen STRO-1 [36, 37], heterotypic intercellular adhesion molecule CD146 [38], and the low-affinity nerve growth factor receptor CD271 [39] are among the most investigated isolation markers. STRO-1 was named for its ability to isolate the stromal fraction of human bone marrow [37]. CD146 is a pericyte marker [40], and its expression by MSCs is consistent with a perivascular origin for the postnatal MSC niche [41]. CD271+ stroma emerges in fetal bone marrow before the onset of hematopoietic activity [42], suggesting that CD271 detects primitive mesenchymal cells. Positive selection with any of these surface markers recovers most, if not all, of the colony-forming unit fibroblasts present in human bone marrow aspirates [37–39]. MSC surface markers exhibit tissue-specific expression. Consider CD271, which is an effective marker to isolate hBM-MSCs and human adipose-derived (hAD) MSCs [39, 43]; however, it is weakly expressed on MSCs from human Wharton's jelly (hWJ) [44]. Once isolated, primary MSC cultures are functionally heterogeneous. As an example, Simmons and Torok-Storb reported that colony-forming unit fibroblasts accounted for only 1% of the STRO-l+/glycophorin A− fraction of human bone marrow [37]. When MSCs are expanded ex vivo, some isolation markers are rapidly downregulated [45], as is the case for CD271 [46].
Table 1.
Representative surface markers of cell subsets for heterogeneous MSCs.
| Cell subset | Surface marker | Comments | References |
|---|---|---|---|
| Fast growing/multipotent MSCs | Low-affinity nerve growth factor receptor (CD271) | Isolation marker that is downregulated in expanded MSCs | [39, 43, 44, 46, 94] |
| Melanoma cell adhesion molecule (CD146) | A pericyte marker expressed in primary and expanded MSCs | [35, 38, 40, 95, 106] | |
| Neuron-glial antigen 2 (NG2) | Upregulated in expanded MSCs. Similar expression in tri- and bipotent MSCs | [35, 47] | |
| Stage-specific embryonic antigen-4 (SSEA-4) | An embryonic stem cell marker expressed on primitive MSCs | [17, 48, 49] | |
| STRO-1 | Clonogenic MSCs constitute a small fraction of the isolated stromal cells | [36, 37, 107] | |
|
| |||
| Osteogenic MSCs | Tissue nonspecific alkaline phosphatase (TNAP) | Selects for MSCs with increased mineralization and expression of osteogenic-related genes | [67] |
|
| |||
| Chondrogenic MSCs | Neural cell adhesion molecule (CD56) | Isolates chrondroprogenitors but is downregulated in expanded MSCs | [70, 71] |
| Receptor tyrosine kinase-like orphan receptor 2 (ROR2) | Isolates chrondroprogenitors from confluent, undifferentiated MSCs | [65] | |
|
| |||
| Triploblastic MUSE cells | Stage-specific embryonic antigen-3 (SSEA-3) | Selected cells exhibit triploblastic differentiation at the single-cell level | [74–79] |
|
| |||
| Immunoregulatory MSCs | Tetherin (bone marrow stromal antigen 2, CD317) | MSCs isolated for tetherin expression have proinflammatory properties and may participate in pathogen clearance | [96] |
| Vascular cell adhesion molecule-1 (CD106) | Selects MSCs that suppress inflammatory cytokine and stimulate regulatory T cells | [98–100] | |
|
| |||
| Aging MSCs | Decoy TRAIL receptor (CD264) | Upregulated concurrently with p21 and remains elevated through senescence | [16] |
MSCs: mesenchymal stem cells. MUSE: multilineage-differentiating stress enduring.
Surface markers that detect cellular heterogeneity in cultured MSCs enable quality control of MSC therapies during ex vivo expansion. One category of markers for cultured MSCs is a subset of isolation markers, including CD146 and stage-specific embryonic antigen-4 (SSEA-4), that is expressed in ex vivo cultures of hBM-MSCs [17, 35]. Surface markers, like neuron-glial antigen 2 (NG2), are another category that is upregulated in hBM-MSCs upon cultivation [47]. While CD146 is a pericyte marker [40], SSEA-4 is an embryonic stem cell marker [48] that is used to isolate induced pluripotent stem cells [49]. NG2 potentiates the activity of β1 integrins and growth factor receptors in regulating cell proliferation, motility, and survival [50]. hBM-MSCs with high levels of SSEA-4 surface expression proliferate at a faster rate than MSCs with low expression levels for this marker [17]. We observed that the surface expression of CD146 and NG2 is inversely correlated to doubling time during the serial passage of single-cell-derived hBM-MSC cultures [35]. In addition, we observed that the fraction of MSCs with high expression of NG2 and low scatter properties is more clonogenic than the parental MSC culture from which it was derived [35].
2.2. Global Molecular Signatures of Proliferation Potential
The global scope of transcriptomic and proteomic profiling can detect differences in cell populations not evident with immunophenotyping [51, 52]. A global molecular signature of MSC heterogeneity may enable greater control over cell composition in MSC therapies than can be achieved with an immunophenotype alone. Microarray analysis of differential gene expression in high- and low-growth populations of hBM-MSCs and human dental tissue-derived (hDT) MSCs identified a common gene signature associated with immature MSCs [19] (Table 2). High-growth MSCs derived from these different tissues exhibited increased expression of genes with critical roles in cell growth and survival: E2F2, PTTG1, and TWIST1 are representative of this common signature [19]. The E2F family of transcription factors regulates the G1/S transition of the cell cycle and DNA synthesis, as well as the DNA damage and repair checkpoint response [53]. PTTG1 (aka pituitary tumor-transforming gene 1) encodes securin, which participates in synchrony of chromosome separation in the anaphase of mitosis [54]. The transcription factor TWIST1 mediates lineage commitment of MSCs and may mediate their self-renewal [55]. hBM-MSCs that were stably transduced with the TWIST1 gene maintained an immature phenotype and exhibited an increased proliferation rate relative to a mock control [55].
Table 2.
Global molecular signatures of cell subsets in heterogeneous MSC cultures.
| Cell subset | Molecular signature | MSC source | Reference |
|---|---|---|---|
| Fast growing/multipotent relative to slow-growing MSCs | Upregulated genes: ASPM, AURKB, CCNB2, CDC2, CDC20, CENPF, CEP55, CHEK1, CIT, CKS2, DLG7, E2F2, GINS2, LDB2, MAD2L1, NCAPG, PBK, POLQ, PTTG1, RPA3, RRM2, TOP2A, TWIST1, UBE2C | Human bone marrow and dental tissue | [19] |
|
| |||
| Fast growing/multipotent MSCs | Upregulated genes: ACAN, ALP1, BMP2, CDC2, CDH1, COL1A1, COL2A1, DLL3, DVL1, FGF2, FOXA2, GDF2, IGF1, JAG1, NEUROG2, NOTCH1, SOX2 | Human bone marrow | [56] |
|
| |||
| Slow-growing MSCs | Upregulated genes: ALDH1A1, CCND2, CD44, DTX1, FGF1, HSPA9, MSX1, TUBB3 | ||
|
| |||
| Fast growing/multipotent MSCs | Upregulated proteins: CALM1, POMC, TPM4 | Human bone marrow | [60] |
|
| |||
| Slow-growing MSCs | Upregulated proteins: ANXA1, CALD1, ENO1, GAPDH, HSP27, LMNA, PKM | ||
|
| |||
| Osteogenic MSCs | Upregulated genes: Col1a1, Col1a2, Comp, Dlx3, Dlx5, Fgfr3, Fmod, Gli1, Hey1, Ibsp, Pitx1, Prrx2, Ptch1, Pth1r, Ror2, Sp7, Tbx3 | Mouse bone marrow | [20] |
|
| |||
| Adipogenic MSCs | Upregulated genes: Abca1, Abcg1, Cebpa, Ctgf, Cxcl12, Dlk1, Foxc2, Inhbb, Lpl, Nr1h3, Pgf, Plin4, Pparg, Prdm16, Sox5, Ucp2 | ||
|
| |||
| Marrow-forming MSCs relative to MSCs that form fibrous tissue | Upregulated genes: ASPN, BMP2, BMP4, CXCL1, DCN, EYA1, GNAS, ICAM1, IGF1, IL8, MEOX2, MN1, MSX2, OGN, OMD, PRRX1, SFRP2, WISP1 | Human bone marrow | [84] |
| Downregulated gene: CNN1 | |||
|
| |||
| Immunomodulatory, VCAM-1+ relative to VCAM-1− MSCs | Upregulated genes: COX2, IDO1, IL1A, IL1B, IL6, IL8 | Human term placental chorionic villi | [98] |
| Upregulated secreted protein: PGE2 | |||
|
| |||
| Immunomodulatory, tetherin+ relative to tetherin− MSCs | Upregulated genes: ADAR, BST2, EIF2AK2, IL18, ISG15, ISG20, MX1, MX2, OAS1, OAS2, OAS3, OASL | Human bone marrow | [96] |
|
| |||
| Trophic activity, ALDHBright MSCs | Uniquely secreted proteins: ACVR1, ANG, GREM1, IGF1, METRN, PDGFA, PLXND1, SPON1, VEGFB, WNT5A | Human bone marrow | [116] |
|
| |||
| Trophic activity, ALDHDim MSCs | Uniquely secreted proteins: ANGPTL3, APOH, BMP2, MMP19, PF4, PLG, PTPRM, PTPRU, TIE1 | ||
MSCs: mesenchymal stem cells. Nomenclature for global molecular signatures is provided in the supplementary material ().
In another study of fast- and slow-growing subsets of hBM-MSCs from osteoarthritis patients, genes encoding sex-determining region Y-box 2 (SOX2), notch homolog 1 (NOTCH1), and the notch ligand delta-like 3 (DLL3) were among the upregulated genes in fast-growing MSCs [56]. These genes are associated with embryonic and postembryonic stem cell renewal [57, 58]. Their expression in fast-growing MSCs is suggestive of an immature phenotype with developmental plasticity [56]. The gene for heat shock 70 kDa protein 9 (aka mortalin or HSPA9) was the most overexpressed, by nearly 10-fold, among the genes upregulated in the slow-growing MSC subset. Overexpression of mortalin in senescent normal human lung fibroblasts increased cumulative population doublings, induced a younger cell morphology, and lowered senescence-associated β-galactosidase activity relative to controls [59]. Perhaps mortalin upregulation by slow-growing MSCs is an attempt to extend their lifespan.
Proteomic profiling of hBM-MSCs from osteoarthritis patients revealed an overexpression of calcium-binding and actin-binding proteins involved in cytokinesis, such as calmodulin 1 (CALM1) and tropomyosin (TPM4), in fast-growing MSC populations relative to their slow-growing counterparts [60]. Overexpressed proteins in the slow-growing MSCs included heat shock protein 27 (HSP27) and annexin A1 (ANXA1) [60], indicative of cellular stress [61]. Slow-growing MSCs overexpressed caldesmon 1 (CALD1) as well [60]. Caldesmon 1 binds to calmodulin 1 and tropomyosin to inhibit cell division by regulating actin activation of myosin ATPase [62]. This suggests a scenario whereby caldesmon 1 and its targets, calmodulin 1 and tropomyosin, act in a coordinated manner to regulate cell division in MSCs.
3. Differentiation Potential
Trilineage potential to exhibit adipo-, chondro-, and osteogenesis is one of the minimal criteria for hMSCs established by the International Society for Cellular Therapy [34]. This criterion pertains to the MSC culture as a whole, which is an ensemble of individual cells with different differentiation potentials. Cellular heterogeneity in the differentiation potential of MSC cultures arises, in part, from progenitors at different stages of lineage commitment, ranging from multi- to unipotent [10, 14, 15]. Initially, lineage commitment was attributed to a sequential loss in differentiation potential [14, 63]. We expanded on this work by developing an in vitro high-capacity assay to quantify the clonal heterogeneity in the trilineage potential of hBM-MSCs [15]. By analyzing large numbers of single-cell-derived colonies, we revealed a more complex hierarchy of lineage commitment that results in heterogeneous MSC cultures containing cells with all possible combinations of adipo-, chondro-, and osteogenic potential. RNA sequencing of single cells and single-cell-derived colonies of murine bone marrow-derived (mBM) MSCs validated the inherent complexity of lineage commitment and revealed multiple lineage-specific transcriptional profiles in individual cells and colonies [20, 64].
3.1. Cell Surface Markers of Differentiation Potential
Surface markers of proliferation potential can isolate cells that are both rapidly dividing and multipotent from heterogeneous MSC cultures [17, 35, 43]; however, there are limitations to the use of a proliferation biomarker to predict the differentiation potential of MSCs. For instance, a cell subset of hBM-MSCs with an enhanced capacity for chondrogenic differentiation had a similar expression profile for the standard MSC markers (CD73, CD90, and CD105) and proliferation markers (STRO-1, CD146, and CD271) as compared with a cell subset that exhibited low chondrogenic differentiation from the same MSC culture [65]. Likewise, we observed that NG2 surface expression was correlated to the proliferation potential for single-cell-derived hBM-MSC colonies; however, the correlation did not extend to the differentiation potential because tri- and bipotent MSC colonies had similar proliferation potentials [35, 66]. The limitation of a proliferation biomarker to detect some changes in lineage commitment necessitates identifying surface markers whose expression in undifferentiated MSCs is predictive of their differentiation potential.
Much of the research in this area has focused on surface markers of osteogenic and chondrogenic potential given that bone repair and cartilage repair account for nearly 20% of MSC-based clinical trials [13]. Positive activity for tissue nonspecific alkaline phosphatase (TNAP, aka mesenchymal stem cell antigen-1) identified single-cell-derived colonies and sorted groups of undifferentiated hBM-MSCs that had a higher degree of calcium mineralization and greater levels of osteogenic-related genes during in vitro osteogenesis than TNAP− MSCs from the same culture [67]. TNAP plays a critical role in the maintenance of bone mineralization by regulating phosphate levels [68, 69]. Expression of TNAP is tissue specific: undifferentiated MSCs from human cord blood (hCB-MSCs) are negative for this marker [67]. As another example, CD271+ hBM-MSCs can be divided into a CD56+ fraction that is enriched in chondroprogenitors relative to the CD56− fraction [70]. CD56 is a cell adhesion molecule that is coexpressed with CD271 on the bone surface that lines the marrow cavity [70]. Like CD271, CD56 is rapidly downregulated in cultured MSCs [71].
Recently, the receptor tyrosine kinase-like orphan receptor 2 (ROR2) was identified as a predictive marker of chondrogenic potential for hBM-MSCs [65]. Positive selection for ROR2 in confluent cultures of undifferentiated hBM-MSCs isolates a cell subset with an enhanced capacity for chondrogenic differentiation in vitro and in vivo [65]. Possibly, ROR2 mediates Wnt5a regulation of chondrogenesis by differential use of the nuclear factor of activated T cells (NFAT) and nuclear factor-κB (NF-κB) pathways [72, 73]. ROR2+ cells were more prevalent in expanded hBM-MSC cultures from osteoarthritis patients than in control cultures from healthy donors [65]. Consistent with this observation, MSC cultures from the patients produced more cartilage tissue in vitro relative to the controls [65].
There is a growing body of evidence that MSCs contain a rare population of pluripotent cells, which have been named multilineage-differentiating stress-enduring (MUSE) cells [74]. MUSE cells have been isolated from mesenchymal tissues by their positive expression of stage-specific embryonic antigen-3 (SSEA-3), a pluripotency marker, and endoglin (CD105), an MSC marker [74]. SSEA-3+ CD105+ cells account for as little as 1% of hBM-MSCs and 3-9% of hAD-MSCs [74, 75]. MUSE cells express pluripotency factors, such as octamer-binding transcription factor 3/4 (OCT3/4) and SOX2 [74, 76], and exhibit triploblastic differentiation at the single-cell level [77]. This pluripotent phenotype is characteristic of MUSE cells isolated from various tissues, including human bone marrow, skin, and adipose [74, 77, 78]. As their name suggests, MUSE cells are capable of surviving extreme cellular stress [78, 79], which has been exploited to enrich adipose-derived MUSE cells [78]. In addition, MUSE cells from different tissues are nontumorigenic in vivo [74, 75]. Given concerns about induced pluripotent stem cells forming teratomas [80], MUSE cells may potentially provide a safer source of pluripotent cells for clinical applications.
3.2. Global Molecular Signatures of Differentiation Potential
RNA sequencing elucidated transcriptional signatures that distinguish between single cell-derived colonies of mBM-MSCs with osteogenic and adipogenic potential [20]. The transcriptional signature for undifferentiated colonies with osteogenic potential included genes for osterix (Sp7) and its mediator, distal-less homeobox 5 (Dlx5), that are master transcription factors in the bone morphogenetic protein pathway [81]. In contrast, the signature for adipogenic potential contained genes encoding the transcription factors peroxisome proliferator-activated receptor γ (Pparg) and CCAAT/enhancer-binding protein α (Cebpa) that regulate each other in a positive feedback loop [20, 82]. Undifferentiated mBM-MSC colonies with both osteogenic and adipogenic potential shared these two distinct gene signatures [20]. Coexistence of distinct transcriptional profiles in MSCs resembles the lineage priming mechanism during early commitment of hematopoietic progenitors [83].
In another study, microarray analysis revealed a transcriptional signature of hBM-MSCs that forms an ectopic ossicle containing both bone and marrow when implanted in immunodeficient mice [84]. Single colony-derived strains of hBM-MSCs are highly variable in their capacity to form this bone/marrow organ [84]. Genes for secreted frizzled-related protein 2 (SFRP2) and calponin 1 (CNN1) were the most upregulated and downregulated, respectively, in MSC strains that formed a bone/marrow organ relative to those that formed only fibrous tissue [84]. SFRP2 encodes a soluble inhibitor of Wnt signaling [85], but its involvement in the formation of a bone/marrow organ could extend to other signaling pathways. For example, SFRP2 binds to the fibronectin- (FN-) integrin-α5β1 complex to inhibit apoptosis [86], which may promote MSC survival in ectopic implants. Calponin 1 is a matrix-binding protein that induces actin polymerization [87]. In regulating cytoskeletal structure, calponin 1 may affect cell proliferation and/or differentiation in MSC implants. Consistent with the downregulation of the CNN1 gene in bone/marrow-forming MSC strains, CNN1-null mice exhibit increased bone formation [88].
4. Immunomodulatory and Trophic Properties
MSCs possess extraordinary immunomodulatory and trophic properties to orchestrate endogenous tissue repair. To resolve inflammation in damaged tissue, MSCs suppress inflammation and may promote clearance of inflammatory stimuli [89]. To promote healing, MSCs produce trophic factors to increase angiogenesis and stimulate endogenous cell growth and differentiation [90]. The majority of MSC-based clinical trials utilize immunomodulatory and trophic properties of MSCs to repair damaged tissue of nonmesenchymal origin and tissue injured by inflammation [13]. The first commercial stem cell drug was an MSC therapy—Osiris Therapeutics' Prochymal brand of Remestemcel-L—which is used to treat acute inflammation in graft-versus-host disease [91]. Potential therapeutic benefits of immunomodulation and trophic effects extend to MSC repair of myocardial tissue and other mesenchymal tissues [92, 93]. Given the breadth of these applications, it is critically important to identify molecular profiles of cell-to-cell variation in the immunomodulatory and trophic properties of heterogeneous MSC cultures.
4.1. Cell Surface Markers of Immunomodulatory and Trophic Properties
Surface markers that select for colony-forming MSCs, such as CD271 and STRO-1, have been used to enrich MSCs with increased immunosuppressive capacity [94, 95]. In addition to these biomarkers, other antigens have been identified that detect cell-to-cell variation in the immunoregulatory activity of MSCs [96–98]. Of these, the expression of vascular cell adhesion molecule-1 (VCAM-1, CD106) in MSCs has been extensively studied [98–100]. Cell-cell adhesion via VCAM-1 plays a key role in the MSC regulation of T cells: MSC-induced suppression of T-cell proliferation correlates with T-cell adhesion to MSCs and is reversed with blocking VCAM-1 antibody [100]. When MSCs derived from human term placental chorionic villi (hCV-MSCs) were divided into VCAM-1− and VCAM-1+ subsets, the latter was far more effective in (1) suppressing the secretion of inflammatory cytokines by activated peripheral blood mononuclear cells and (2) stimulating the formation of anti-inflammatory regulatory T cells [98]. The content of VCAM-1+ cells in MSC cultures is tissue specific: more in hCV-MSCs than in hBM-MSCs and none in hAD-MSCs [98]. The high levels of immunosuppressive VCAM-1+ cells in hCV-MSCs may be associated with fetomaternal tolerance. The placenta provides an immune-privileged environment for the fetus [101]. VCAM-1+ MSCs within chorionic villi may help suppress an immune response at the border between maternal and fetal blood.
A growing body of data suggests that MSCs may exhibit proinflammatory properties in certain instances, perhaps to stimulate pathogen clearance [89, 102]. Recently, the surface marker tetherin (aka bone marrow stromal antigen 2 and CD317) identified a subset (~1%-3%) of hBM-MSCs with elevated secretion of interleukin-7 (IL-7) [96], which mediates B and T cell development [103]. As its name suggests, tetherin inhibits the spread of viral infection by tethering budding viruses to infected cells [104]. In addition, tetherin activates NF-κB to induce an inflammatory response [105]. Tetherin+ MSCs could potentially have a role in the clearance of viral infections. It is noteworthy that the differentiation potential of tetherin+ hBM-MSCs was compromised relative to tetherin− MSCs from the same culture [96], suggesting that there are limitations to using differentiation markers to select for MSCs with specific immunomodulatory properties.
A few surface markers have been examined for their ability to isolate MSCs with enhanced trophic activity to increase angiogenesis and promote endogenous tissue repair. For instance, murine gastrocnemius muscle injected with CD146+ hCV-MSCs had more blood vessels at the injection site than controls receiving the CD146− cell fraction [106]. Cell-to-cell variation in the trophic activity of STRO-1+ hBM-MSCs was evident when they were divided into STRO-1Bright and STRO-1Dim subsets: conditioned medium from the STRO-1Bright MSCs elicited greater cardiac cell proliferation and endothelial tube formation [107]. Inhibition of stromal cell-derived factor 1 (SDF1, CXCL12) and hepatocyte growth factor (HGF) in the conditioned medium attenuated these trophic effects [107].
4.2. Global Molecular Signatures of Immunomodulatory and Trophic Properties
Microarray and qPCR analysis of VCAM-1+ hCV-MSCs provided insight into the enhanced anti-inflammatory potential of this cell subset [98]. Relative to VCAM-1− hCV-MSCs, the VCAM-1+ subset had elevated mRNA levels of cyclooxygenase-2 (COX2), indoleamine 2,3-dioxygenase 1 (IDO1), and other key immune modulators, as well as secreted more prostaglandin E2 (PGE2) protein [98]. While COX-2 is typically associated with inflammation [108], its increased expression in VCAM-1+ MSCs may have an anti-inflammatory function. Specifically, COX-2 may increase the production of the immunosuppressors PGE2 [109] and IDO-1 [110] in MSCs. In support of this possibility, COX-2 is the rate-limiting enzyme in PGE2 synthesis [111] and regulates IDO-1 expression in animal models of cancer [112]. Overexpression of COX-2 increases the immunosuppressive activity of MSCs derived from human umbilical cord [113].
Interrogation of the transcriptome of tetherin+ hBM-MSCs revealed a gene signature that is consistent with a possible role for this cell subset in viral clearance [96]. Single colony-derived strains of tetherin+ hBM-MSCs expressed several antiviral genes, including interferon-stimulated gene 20 kDa protein (ISG20) and oligoadenylate synthetase (OAS), at higher levels than the parental culture and tetherin− strains of hBM-MSCs [96]. Both gene products are induced by interferon and involved in the destruction of RNA viruses: ISG20 is a secreted RNase that directly attacks the viruses [114], and OAS acts indirectly by activating intercellular RNase L [115]. The antiviral activity of ISG20 and OAS complements the ability of tetherin to sequester virus particles [104] and induce an inflammatory response [105].
Proteomic analysis of conditioned medium from hBM-MSCs detected uniquely secreted trophic factors for angiogenesis in cell subsets with elevated expression of aldehyde dehydrogenase (ALDH) [116]. A fluorescent substrate enables cell enrichment based on intercellular ALDH activity [117]. Conditioned medium from ALDHBright MSCs stimulated in vitro endothelial cell proliferation and tube formation and in vivo angiogenesis to a greater extent than ALDHDim MSCs from the same culture [116]. While the ALDHBright and ALDHDim subsets had similar gene signatures, their secretome was different [116]. ALDHBright MSCs uniquely secreted several proangiogenic cytokines, which included vascular endothelial growth factor β (VEGFB) and platelet derived growth factor α (PDGFA); whereas, ALDHDim MSCs secreted inhibitors of angiogenesis, such as platelet factor 4 (PF4) and plasminogen (PLG) [116]. The coexistence of pro- and antiangiogenic MSC subsets in the same culture could be akin to an angiogenic on/off switch evident during tumorigenesis [118]. MSC subsets that induce angiogenesis could promote vessel sprouting; whereas, subsets that are inhibitory could cause vessel trimming during vascular remodeling [119].
5. Cellular Aging
Cellular aging and replicative senescence compromises stem cell fitness and is an obstacle to the production of effective MSC therapies. Replicative stress caused by rapid MSC expansion during biomanufacturing can induce DNA damage and cellular aging [120]. During serial passage, MSCs experience a continuous and organized aging process [121] that diminishes their regenerative potential and culminates in replicative senescence [122, 123]. Cellular aging also can occur in vivo prior to harvesting MSCs from a donor. Repeated cell division during tissue maintenance and repair can cause cellular aging over the life of the donor, particularly in the elderly [120]. The negative impact of cellular aging on MSC therapies is twofold. First, the loss of stem cell fitness limits, if not prevents, ex vivo expansion to produce clinically relevant quantities of MSCs. Second, it may impair the effectiveness of MSC therapies to regenerate damaged tissue. We and others have observed that MSC cultures contain a heterogeneous mixture of cells at different stages of aging, starting at early passage [16, 17, 124]. Molecular profiles of this heterogeneity would be useful to develop expansion conditions that mitigate cellular aging, as well as to monitor and control the cell composition of MSC therapies during biomanufacturing.
5.1. Cell Surface Markers of Cellular Aging
Surface markers for rapidly dividing, multipotent MSCs (e.g., CD146, NG2, and SSEA-4) are downregulated in aging MSCs during ex vivo expansion [17, 35]. For example, Rosu-Myler et al. [17] described early passage hBM-MSCs as a heterogeneous mixture of cells with high and low surface expression of SSEA-4. Serial passage on tissue culture plastic depletes clonal, multipotent SEEA-4hi MSCs from culture and causes an accumulation of SSEA-4lo cells with diminished regenerative potential [17]. Block et al. [124] reported that small, SSEA-4+ cells accounted for less than 10% of hBM-MSCs harvested from elderly donors ages 65 and older. They were able to isolate these high-quality cells and expand them on the extracellular matrix from young-donor MSCs to quantities required for clinical applications [124].
A surface marker expressed on aging cells would enable the enrichment of rapidly dividing MSCs by negative selection. Recently, we reported on decoy TRAIL receptor CD264 as the first surface marker of cellular aging for MSCs [16]. The content of CD264+ cells accumulates during serial passaging of hBM-MSC cultures: CD264 is initially upregulated at an intermediate passage concurrently with p21 and remains upregulated at late passage as aging progresses to senescence [16]. There is a strong inverse correlation of CD264+ cell content with multiple metrics of stem cell fitness, including colony-forming efficiency as a measure of proliferation potential [16]. MSCs may increase CD264 expression to promote cell survival during aging possibly via the protein kinase B (aka Akt) signaling pathway [125].
5.2. Global Molecular Signatures of Cellular Aging
Gene and epigenetic signatures have been identified for molecular changes in heterogeneous MSCs associated with replicative senescence [126, 127], but to date there are no senescence signatures for cell subsets in MSC cultures. It should be noted that senescence signatures developed for heterogeneous MSCs represent an ensemble average of molecular changes for individual cells within the bulk culture. As such, they may not be predictive of the properties of specific cell subsets. To emphasize this point, the epigenetic senescence signatures for heterogeneous hBM-MSC cultures and their subclones were compared recently [128]. The signature consists of DNA methylation changes at six CpG sites, which predicts the average passage number of heterogeneous hBM- and hAD-MSC cultures [127]. While the DNA methylation pattern provided a good correlation between predicted and real passage number of the bulk hBM-MSC cultures, the predictions did not correlate at all with the real passage number of the subclones [128]. Moreover, the differentiation potential of the subclones did not correlate with their senescence signature [128]. This example illustrates the need for global senescence signatures that are relevant to cell subsets, not just the bulk culture.
6. Applications
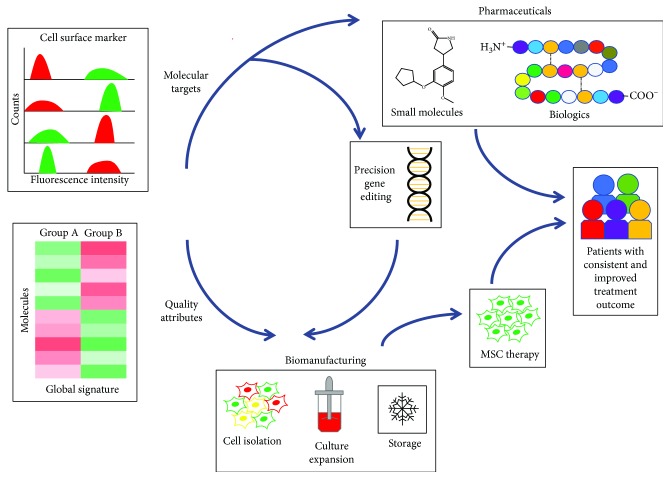
The molecular profiles described here provide new insight into MSC biology. This knowledge can be exploited to identify molecular targets to regulate the regenerative potential of MSCs (Figure 1). Targeted molecules and pathways can be regulated by chemical and biologic agents administered as adjuvants with MSC therapies. For example, we demonstrated that a small-molecule antagonist of macrophage migration inhibitory factor enhances the migratory response of hBM-MSCs to injured bronchial epithelial cells [129]. Matrix proteins when administered as an adjuvant with hBM-MSCs increase in vivo survival of the stem cells by attenuating anoikis [130]. Recent improvements in the nonviral delivery of CRISPR/Cas9 components have enabled safe and effective gene editing of hard-to-transfect MSCs [131]. Adjuvant therapies and precision gene editing inspired from MSC molecular profiles have the potential to improve the effectiveness of MSC therapies by providing unprecedented control over their regenerative potential.
Figure 1.
Applications of molecular profiles of MSC heterogeneity. Surface markers and global signatures identify cell subsets with specific regenerative properties in heterogeneous MSC cultures. Molecular profiles of MSC heterogeneity have application as molecular targets and quality attributes in the production of MSC therapeutics. Targeted molecules can be regulated by small-molecule and biologic pharmaceuticals, as well as by precision gene editing. Quality attributes enable enrichment of a MSC population and its assessment during all stages of biomanufacturing of MSC therapies from cell isolation to culture expansion to storage. This control over the composition and function of MSC therapies has the potential to improve treatment outcomes for patients.
Surface markers and global signatures of MSC heterogeneity have the potential to be effective in predicting treatment outcome in MSC-based clinical trials. Predictive molecular profiles of the regenerative potential of MSCs have utility as quantifiable attributes of cell quality during the manufacturing of MSC therapies [132]. These quality attributes can enable enrichment of a MSC population and its assessment during all stages of manufacturing from the selection of the source stem cell to the preparation of the final clinical-grade product. Most likely, MSC quality attributes will be multivariate and specific to the tissue of origin, with a unique combination of surface markers and global signatures for each application. For example, they could include CD271 and other tissue-specific markers of multipotency [39]; negative markers of an undesirable phenotype, such as cellular aging [16]; and global signatures that can classify MSCs based on a specific function, like immunomodulation [116]. When measured in real time, these quality attributes can enable feedforward and feedback control of the manufacturing process [133] to improve the consistency and quality of an MSC therapy.
7. Future Directions
Molecular profiles of MSC heterogeneity are far from complete. More research is needed in all categories of MSC function and, in particular, homing. Upon remote delivery of MSC therapies to a patient, the stem cells are capable of homing to the site of injury. Despite its importance to tissue repair, little is known about the cell-to-cell variation in the ability of MSCs to home to injured tissue [134]. To date, most of the global profiles of MSC heterogeneity have been gene signatures, with limited analysis of the proteome and secretome. In the future, more comprehensive profiling is warranted that includes miRNA, lncRNA, and the epigenome, given their roles in the regulation of stem cell function [135–137].
There is limited information on how the molecular profiles of MSC heterogeneity are influenced by variables encountered during the manufacturing of MSC therapies. MSC heterogeneity can be profoundly affected by the choice of stem cell donor [65], tissue source [98], and process conditions [16]. Donor variables include age, BMI, sex, and health status. As reflected in this review, popular tissue sources for MSCs are bone marrow and adipose, and there is a growing interest in perinatal tissues as a source of primitive MSCs that can be harvested noninvasively. Process conditions, such as passage number and confluency, determine the extent of cell expansion during the manufacturing of MSC therapies. An understanding of the influence of production variables on the surface markers and global signatures described in this review is essential for these molecular profiles to be adopted as quality attributes during the manufacturing of MSC therapies. This knowledge can guide the selection of donor and tissue source and the design of process conditions that optimize MSC performance in the clinic.
Many of the molecular profiles discussed here have yet to be validated in an animal model. In vitro behavior of MSCs is not a reliable predictor of their in vivo performance. As a case in point, only a fraction of clonal mBM-MSCs, which exhibit in vitro osteogenesis, are capable of in vivo bone formation [20]. Promising surface markers and global signatures that have been identified based on in vitro function of MSCs need to be validated first in animal models and then in clinical trials. Molecular profiles that are predictive of clinical outcome are candidates to use as quality attributes for robust and reproducible manufacturing of MSC therapies.
8. Conclusions
Inconsistencies in the composition of MSC cultures hinder their use in regenerative medicine. There is a critical need for molecular profiles of this heterogeneity to manufacture effective MSC therapies. This review presented cell surface markers and global signatures that isolate cell subsets with specific regenerative properties from heterogeneous MSC cultures. More research is required to ascertain how these molecular profiles are influenced by biomanufacturing conditions. Other areas to be explored include noncoding RNA and epigenetic signatures, in addition to the in vivo validation of many of the molecular profiles. Predictive surface markers and global signatures of regenerative potential will facilitate robust and reproducible manufacturing of MSC therapies, as well as identify new molecular targets to regulate MSC function. This control over MSC composition and function will accelerate the translation of MSC therapeutics into clinical practice.
Acknowledgments
The author would like to thank Ms. Margaret Giler for formatting the references and editing the figure for this article. This work was supported with funding from the Division of Chemical, Bioengineering, Environmental, and Transport Systems of the National Science Foundation (NSF) (grant number 1604129), and a Carol Lavin Bernick Tulane Faculty Grant.
Disclosure
The funding agencies that supported this study were not involved in manuscript writing, editing, approval, or decision to publish.
Conflicts of Interest
The author is an inventor listed on patents and patent applications entitled “Method for Identification and Culture of Multipotent Mesenchymal Stem Cells with High Proliferation Potential” issued in Austria, China, and Germany; pending issuance in France, Switzerland, and UK; and under review in Canada, India, Japan, and the US. In addition, the author is the inventor listed on a patent application entitled “Cell-Surface Marker of Early MSC Aging” pending issuance in the US and under review in Australia, Canada, and Europe.
Supplementary Materials
Supplementary Table: nomenclature for global molecular signatures.
References
- 1.Trounson A., McDonald C. Stem cell therapies in clinical trials: progress and challenges. Cell Stem Cell. 2015;17(1):11–22. doi: 10.1016/j.stem.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Friedenstein A. J., Chailakhjan R. K., Lalykina K. S. The development of fibroblast colonies in monolayer cultures of guinea-pig bone marrow and spleen cells. Cell and Tissue Kinetics. 1970;3(4):393–403. doi: 10.1111/j.1365-2184.1970.tb00347.x. [DOI] [PubMed] [Google Scholar]
- 3.Friedenstein A. J., Chailakhyan R. K., Latsinik N. V., Panasyuk A. F., Keiliss-Borok I. V. Stromal cells responsible for transferring the microenvironment of the hemopoietic tissues: Cloning in vitro and retransplantation in vivo. Transplantation. 1974;17(4):331–340. doi: 10.1097/00007890-197404000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Bianco P., Robey P. G., Simmons P. J. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell. 2008;2(4):313–319. doi: 10.1016/j.stem.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caplan A. I. Mesenchymal stem cells. Journal of Orthopaedic Research. 1991;9(5):641–650. doi: 10.1002/jor.1100090504. [DOI] [PubMed] [Google Scholar]
- 6.da Silva Meirelles L., Chagastelles P. C., Nardi N. B. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. Journal of Cell Science. 2006;119(11):2204–2213. doi: 10.1242/jcs.02932. [DOI] [PubMed] [Google Scholar]
- 7.Kern S., Eichler H., Stoeve J., Klüter H., Bieback K. Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood, or adipose tissue. Stem Cells. 2006;24(5):1294–1301. doi: 10.1634/stemcells.2005-0342. [DOI] [PubMed] [Google Scholar]
- 8.Ponte A. L., Marais E., Gallay N., et al. The in vitro migration capacity of human bone marrow mesenchymal stem cells: comparison of chemokine and growth factor chemotactic activities. Stem Cells. 2007;25(7):1737–1745. doi: 10.1634/stemcells.2007-0054. [DOI] [PubMed] [Google Scholar]
- 9.Sekiya I., Larson B. L., Smith J. R., Pochampally R., Cui J.&.;. G., Prockop D. J. Expansion of human adult stem cells from bone marrow stroma: conditions that maximize the yields of early progenitors and evaluate their quality. Stem Cells. 2002;20(6):530–541. doi: 10.1634/stemcells.20-6-530. [DOI] [PubMed] [Google Scholar]
- 10.Pittenger M. F., Mackay A. M., Beck S. C., et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 11.Selmani Z., Naji A., Zidi I., et al. Human leukocyte antigen-G5 secretion by human mesenchymal stem cells is required to suppress T lymphocyte and natural killer function and to induce CD4+CD25highFOXP3+ regulatory T cells. Stem Cells. 2008;26(1):212–222. doi: 10.1634/stemcells.2007-0554. [DOI] [PubMed] [Google Scholar]
- 12.Park K. S., Kim Y. S., Kim J. H., et al. Trophic molecules derived from human mesenchymal stem cells enhance survival, function, and angiogenesis of isolated islets after transplantation. Transplantation. 2010;101(8, article e290):701. doi: 10.1097/tp.0000000000001859. [DOI] [PubMed] [Google Scholar]
- 13.Squillaro T., Peluso G., Galderisi U. Clinical trials with mesenchymal stem cells: an update. Cell Transplantation. 2016;25(5):829–848. doi: 10.3727/096368915X689622. [DOI] [PubMed] [Google Scholar]
- 14.Muraglia A., Cancedda R., Quarto R. Clonal mesenchymal progenitors from human bone marrow differentiate in vitro according to a hierarchical model. Journal of Cell Science. 2000;113:1161–1166. doi: 10.1242/jcs.113.7.1161. [DOI] [PubMed] [Google Scholar]
- 15.Russell K. C., Phinney D. G., Lacey M. R., Barrilleaux B. L., Meyertholen K. E., O'Connor K. C. In vitro high-capacity assay to quantify the clonal heterogeneity in trilineage potential of mesenchymal stem cells reveals a complex hierarchy of lineage commitment. Stem Cells. 2010;28(4):788–798. doi: 10.1002/stem.312. [DOI] [PubMed] [Google Scholar]
- 16.Madsen S. D., Russell K. C., Tucker H. A., Glowacki J., Bunnell B. A., O’Connor K. C. Decoy TRAIL receptor CD264: a cell surface marker of cellular aging for human bone marrow-derived mesenchymal stem cells. Stem Cell Research & Therapy. 2017;8(1):p. 201. doi: 10.1186/s13287-017-0649-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosu-Myles M., McCully J., Fair J., et al. The globoseries glycosphingolipid SSEA-4 is a marker of bone marrow-derived clonal multipotent stromal cells in vitro and in vivo. Stem Cells and Development. 2013;22(9):1387–1397. doi: 10.1089/scd.2012.0547. [DOI] [PubMed] [Google Scholar]
- 18.Gullo F., De Bari C. Prospective purification of a subpopulation of human synovial mesenchymal stem cells with enhanced chondro-osteogenic potency. Rheumatology. 2013;52(10):1758–1768. doi: 10.1093/rheumatology/ket205. [DOI] [PubMed] [Google Scholar]
- 19.Menicanin D., Bartold P. M., Zannettino A. C. W., Gronthos S. Identification of a common gene expression signature associated with immature clonal mesenchymal cell populations derived from bone marrow and dental tissues. Stem Cells and Development. 2010;19(10):1501–1510. doi: 10.1089/scd.2009.0492. [DOI] [PubMed] [Google Scholar]
- 20.Rostovskaya M., Donsante S., Sacchetti B., et al. Clonal analysis delineates transcriptional programs of osteogenic and adipogenic lineages of adult mouse skeletal progenitors. Stem Cell Reports. 2018;11(1):212–227. doi: 10.1016/j.stemcr.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu V. W. C., Scadden D. T. Heterogeneity of the bone marrow niche. Current Opinion in Hematology. 2016;23(4):331–338. doi: 10.1097/MOH.0000000000000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rennerfeldt D. A., Van Vliet K. J. Concise review: when colonies are not clones: evidence and implications of intracolony heterogeneity in mesenchymal stem cells. Stem Cells. 2016;34(5):1135–1141. doi: 10.1002/stem.2296. [DOI] [PubMed] [Google Scholar]
- 23.Mets T., Verdonk G. In vitro aging of human bone marrow derived stromal cells. Mechanisms of Ageing and Development. 1981;16(1):81–89. doi: 10.1016/0047-6374(81)90035-X. [DOI] [PubMed] [Google Scholar]
- 24.Colter D. C., Sekiya I., Prockop D. J. Identification of a subpopulation of rapidly self-renewing and multipotential adult stem cells in colonies of human marrow stromal cells. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(14):7841–7845. doi: 10.1073/pnas.141221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang L., Ma A., Song L., et al. Cartilage regeneration by selected chondrogenic clonal mesenchymal stem cells in the collagenase-induced monkey osteoarthritis model. Journal of Tissue Engineering and Regenerative Medicine. 2014;8(11):896–905. doi: 10.1002/term.1676. [DOI] [PubMed] [Google Scholar]
- 26.Rada T., Santos T. C., Marques A. P., et al. Osteogenic differentiation of two distinct subpopulations of human adipose-derived stem cells: an in vitro and in vivo study. Journal of Tissue Engineering and Regenerative Medicine. 2012;6(1):1–11. doi: 10.1002/term.388. [DOI] [PubMed] [Google Scholar]
- 27.Zhang S., Ge J., Sun A., et al. Comparison of various kinds of bone marrow stem cells for the repair of infarcted myocardium: single clonally purified non-hematopoietic mesenchymal stem cells serve as a superior source. Journal of Cellular Biochemistry. 2006;99(4):1132–1147. doi: 10.1002/jcb.20949. [DOI] [PubMed] [Google Scholar]
- 28.Dimmeler S., Ding S., Rando T. A., Trounson A. Translational strategies and challenges in regenerative medicine. Nature Medicine. 2014;20(8):814–821. doi: 10.1038/nm.3627. [DOI] [PubMed] [Google Scholar]
- 29.Prockop D. J., Prockop S. E., Bertoncello I. Are clinical trials with mesenchymal stem/progenitor cells too far ahead of the science? Lessons from experimental hematology. Stem Cells. 2014;32(12):3055–3061. doi: 10.1002/stem.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoch A. I., Leach J. K. Concise review: optimizing expansion of bone marrow mesenchymal stem/stromal cells for clinical applications. Stem Cells Translational Medicine. 2014;3(5):643–652. doi: 10.5966/sctm.2013-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selich A., Daudert J., Hass R., et al. Massive clonal selection and transiently contributing clones during expansion of mesenchymal stem cell cultures revealed by lentiviral RGB-barcode technology. Stem Cells Translational Medicine. 2016;5(5):591–601. doi: 10.5966/sctm.2015-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mareddy S., Crawford R., Brooke G., Xiao Y. Clonal isolation and characterization of bone marrow stromal cells from patients with osteoarthritis. Tissue Engineering. 2007;13(4):819–829. doi: 10.1089/ten.2006.0180. [DOI] [PubMed] [Google Scholar]
- 33.Bigildeev A. E., Zhironkina O. A., Shipounova I. N., Sats N. V., Kotyashova S. Y., Drize N. I. Clonal composition of human multipotent mesenchymal stromal cells. Experimental Hematology. 2012;40(10):847–856.e4. doi: 10.1016/j.exphem.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Dominici M., le Blanc K., Mueller I., et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8(4):315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 35.Russell K. C., Tucker H. A., Bunnell B. A., et al. Cell-surface expression of neuron-glial antigen 2 (NG2) and melanoma cell adhesion molecule (CD146) in heterogeneous cultures of marrow-derived mesenchymal stem cells. Tissue Engineering Part A. 2013;19(19-20):2253–2266. doi: 10.1089/ten.tea.2012.0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ning H., Lin G., Lue T. F., Lin C.-S. Mesenchymal stem cell marker Stro-1 is a 75 kd endothelial antigen. Biochemical and Biophysical Research Communications. 2011;413(2):353–357. doi: 10.1016/j.bbrc.2011.08.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simmons P. J., Torok-Storb B. Identification of stromal cell precursors in human bone marrow by a novel monoclonal antibody, STRO-1. Blood. 1991;78:55–62. [PubMed] [Google Scholar]
- 38.Sacchetti B., Funari A., Michienzi S., et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131(2):324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 39.Quirici N., Soligo D., Bossolasco P., Servida F., Lumini C., Deliliers G. L. Isolation of bone marrow mesenchymal stem cells by anti-nerve growth factor receptor antibodies. Experimental Hematology. 2002;30(7):783–791. doi: 10.1016/S0301-472X(02)00812-3. [DOI] [PubMed] [Google Scholar]
- 40.Covas D. T., Panepucci R. A., Fontes A. M., et al. Multipotent mesenchymal stromal cells obtained from diverse human tissues share functional properties and gene-expression profile with CD146+ perivascular cells and fibroblasts. Experimental Hematology. 2008;36(5):642–654. doi: 10.1016/j.exphem.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 41.Crisan M., Yap S., Casteilla L., et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3(3):301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 42.Cattoretti G., Schiró R., Orazi A., Soligo D., Colombo M. P. Bone marrow stroma in humans: anti-nerve growth factor receptor antibodies selectively stain reticular cells in vivo and in vitro. Blood. 1993;81(7):1726–1738. [PubMed] [Google Scholar]
- 43.Quirici N., Scavullo C., de Girolamo L., et al. Anti-L-NGFR and -CD34 monoclonal antibodies identify multipotent mesenchymal stem cells in human adipose tissue. Stem Cells and Development. 2010;19(6):915–925. doi: 10.1089/scd.2009.0408. [DOI] [PubMed] [Google Scholar]
- 44.Margossian T., Reppel L., Makdissy N., Stoltz J. F., Bensoussan D., Huselstein C. Mesenchymal stem cells derived from Wharton’s jelly: comparative phenotype analysis between tissue and in vitro expansion. Bio-medical Materials and Engineering. 2012;22(4):243–254. doi: 10.3233/BME-2012-0714. [DOI] [PubMed] [Google Scholar]
- 45.Bara J. J., Richards R. G., Alini M., Stoddart M. J. Concise review: bone marrow-derived mesenchymal stem cells change phenotype following in vitro culture: implications for basic research and the clinic. Stem Cells. 2014;32(7):1713–1723. doi: 10.1002/stem.1649. [DOI] [PubMed] [Google Scholar]
- 46.Jones E. A., Kinsey S. E., English A., et al. Isolation and characterization of bone marrow multipotential mesenchymal progenitor cells. Arthritis and Rheumatism. 2002;46(12):3349–3360. doi: 10.1002/art.10696. [DOI] [PubMed] [Google Scholar]
- 47.Kozanoglu I., Boga C., Ozdogu H., et al. Human bone marrow mesenchymal cells express NG2: possible increase in discriminative ability of flow cytometry during mesenchymal stromal cell identification. Cytotherapy. 2009;11(5):527–533. doi: 10.1080/14653240902923153. [DOI] [PubMed] [Google Scholar]
- 48.Zhao W., Ji X., Zhang F., Li L., Ma L. Embryonic stem cell markers. Molecules. 2012;17(6):6196–6236. doi: 10.3390/molecules17066196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abujarour R., Valamehr B., Robinson M., Rezner B., Vranceanu F., Flynn P. Optimized surface markers for the prospective isolation of high-quality hiPSCs using flow cytometry selection. Scientific Reports. 2013;3(1) doi: 10.1038/srep01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stallcup W. B., Huang F.-J. A role for the NG2 proteoglycan in glioma progression. Cell Adhesion & Migration. 2014;2(3):192–201. doi: 10.4161/cam.2.3.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vega F., Chang C.-C., Medeiros L. J., et al. Plasmablastic lymphomas and plasmablastic plasma cell myelomas have nearly identical immunophenotypic profiles. Modern Pathology. 2005;18(6):806–815. doi: 10.1038/modpathol.3800355. [DOI] [PubMed] [Google Scholar]
- 52.Gutierrez-Garcia G., Cardesa-Salzmann T., Climent F., et al. Gene-expression profiling and not immunophenotypic algorithms predicts prognosis in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Blood. 2011;117(18):4836–4843. doi: 10.1182/blood-2010-12-322362. [DOI] [PubMed] [Google Scholar]
- 53.Cam H., Dynlacht B. D. Emerging roles for E2F: beyond the G1/S transition and DNA replication. Cancer Cell. 2003;3(4):311–316. doi: 10.1016/S1535-6108(03)00080-1. [DOI] [PubMed] [Google Scholar]
- 54.Holt L. J., Krutchinsky A. N., Morgan D. O. Positive feedback sharpens the anaphase switch. Nature. 2008;454(7202):353–357. doi: 10.1038/nature07050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Isenmann S., Arthur A., Zannettino A. C., et al. TWIST family of basic helix-loop-helix transcription factors mediate human mesenchymal stem cell growth and commitment. Stem Cells. 2009;27(10):2457–2468. doi: 10.1002/stem.181. [DOI] [PubMed] [Google Scholar]
- 56.Mareddy S., Dhaliwal N., Crawford R., Xiao Y. Stem cell-related gene expression in clonal populations of mesenchymal stromal cells from bone marrow. Tissue Engineering Part A. 2010;16(2):749–758. doi: 10.1089/ten.tea.2009.0307. [DOI] [PubMed] [Google Scholar]
- 57.Sarkar A., Hochedlinger K. The Sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12(1):15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chiba S. Concise review: notch signaling in stem cell systems. Stem Cells. 2006;24(11):2437–2447. doi: 10.1634/stemcells.2005-0661. [DOI] [PubMed] [Google Scholar]
- 59.Kaul S. C., Reddel R. R., Sugihara T., Mitsui Y., Wadhwa R. Inactivation of p53 and life span extension of human diploid fibroblasts by mot-2. FEBS Letters. 2000;474(2-3):159–164. doi: 10.1016/S0014-5793(00)01594-5. [DOI] [PubMed] [Google Scholar]
- 60.Mareddy S., Broadbent J., Crawford R., Xiao Y. Proteomic profiling of distinct clonal populations of bone marrow mesenchymal stem cells. Journal of Cellular Biochemistry. 2009;106(5):776–786. doi: 10.1002/jcb.22088. [DOI] [PubMed] [Google Scholar]
- 61.Rhee H. J., Kim G.-Y., Huh J. W., Kim S.-W., Na D. S. Annexin I is a stress protein induced by heat, oxidative stress and a sulfhydryl-reactive agent. European Journal of Biochemistry. 2000;267(11):3220–3225. doi: 10.1046/j.1432-1327.2000.01345.x. [DOI] [PubMed] [Google Scholar]
- 62.Eppinga R. D., Li Y., Lin J. L.-C., Lin J. J.-C. Tropomyosin and caldesmon regulate cytokinesis speed and membrane stability during cell division. Archives of Biochemistry and Biophysics. 2006;456(2):161–174. doi: 10.1016/j.abb.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 63.Banfi A., Muraglia A., Dozin B., Mastrogiacomo M., Cancedda R., Quarto R. Proliferation kinetics and differentiation potential of ex vivo expanded human bone marrow stromal cells. Experimental Hematology. 2000;28(6):707–715. doi: 10.1016/S0301-472X(00)00160-0. [DOI] [PubMed] [Google Scholar]
- 64.Freeman B. T., Jung J. P., Ogle B. M. Single-cell RNA-Seq of bone marrow-derived mesenchymal stem cells reveals unique profiles of lineage priming. PLoS One. 2015;10(9):p. e0136199. doi: 10.1371/journal.pone.0136199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dickinson S. C., Sutton C. A., Brady K., et al. The Wnt5a receptor, receptor tyrosine kinase-like orphan receptor 2, is a predictive cell surface marker of human mesenchymal stem cells with an enhanced capacity for chondrogenic differentiation. Stem Cells. 2017;35(11):2280–2291. doi: 10.1002/stem.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Russell K. C., Lacey M. R., Gilliam J. K., Tucker H. A., Phinney D. G., O'Connor K. C. Clonal analysis of the proliferation potential of human bone marrow mesenchymal stem cells as a function of potency. Biotechnology and Bioengineering. 2011;108(11):2716–2726. doi: 10.1002/bit.23193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim Y. H., Yoon D. S., Kim H. O., Lee J. W. Characterization of different subpopulations from bone marrow-derived mesenchymal stromal cells by alkaline phosphatase expression. Stem Cells and Development. 2012;21(16):2958–2968. doi: 10.1089/scd.2011.0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Golub E. E., Boesze-Battaglia K. The role of alkaline phosphatase in mineralization. Current Opinion in Orthopaedics. 2007;18(5):444–448. doi: 10.1097/BCO.0b013e3282630851. [DOI] [Google Scholar]
- 69.Narisawa S., Fröhlander N., Millán J. L. Inactivation of two mouse alkaline phosphatase genes and establishment of a model of infantile hypophosphatasia. Developmental Dynamics. 1997;208(3):432–446. doi: 10.1002/(SICI)1097-0177(199703)208:3<432::AID-AJA13>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 70.Sivasubramaniyan K., Ilas D. C., Harichandan A., et al. Bone marrow-harvesting technique influences functional heterogeneity of mesenchymal stem/stromal cells and cartilage regeneration. The American Journal of Sports Medicine. 2018;46(14):3521–3531. doi: 10.1177/0363546518804807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Battula V. L., Treml S., Bareiss P. M., et al. Isolation of functionally distinct mesenchymal stem cell subsets using antibodies against CD56, CD271, and mesenchymal stem cell antigen-1. Haematologica. 2009;94(2):173–184. doi: 10.3324/haematol.13740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu Y., Rubin B., Bodine P. V. N., Billiard J. Wnt5a induces homodimerization and activation of Ror2 receptor tyrosine kinase. Journal of Cellular Biochemistry. 2008;105(2):497–502. doi: 10.1002/jcb.21848. [DOI] [PubMed] [Google Scholar]
- 73.Bradley E. W., Drissi M. H. WNT5A regulates chondrocyte differentiation through differential use of the CaN/NFAT and IKK/NF-κB pathways. Molecular Endocrinology. 2010;24(8):1581–1593. doi: 10.1210/me.2010-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuroda Y., Kitada M., Wakao S., et al. Unique multipotent cells in adult human mesenchymal cell populations. PNAS. 2010;107(19):8639–8643. doi: 10.1073/pnas.0911647107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ogura F., Wakao S., Kuroda Y., et al. Human adipose tissue possesses a unique population of pluripotent stem cells with nontumorigenic and low telomerase activities: potential implications in regenerative medicine. Stem Cells and Development. 2014;23(7):717–728. doi: 10.1089/scd.2013.0473. [DOI] [PubMed] [Google Scholar]
- 76.Nitobe Y., Nagaoki T., Kumagai G., et al. Neurotrophic factor secretion and neural differentiation potential of multilineage-differentiating stress-enduring (MUSE) cells derived from mouse adipose tissue. Cell Transplantation. 2019;28 doi: 10.1177/0963689719863809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wakao S., Kitada M., Kuroda Y., et al. Multilineage-differentiating stress-enduring (MUSE) cells are a primary source of induced pluripotent stem cells in human fibroblasts. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(24):9875–9880. doi: 10.1073/pnas.1100816108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heneidi S., Simerman A. A., Keller E., et al. Correction: Awakened by cellular stress: isolation and characterization of a novel population of pluripotent stem cells derived from human adipose tissue. PLoS One. 2013;8(7) doi: 10.1371/annotation/190d4d01-a63c-4adc-a123-e519ee40a03e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alessio N., Squillaro T., Özcan S., et al. Stress and stem cells: adult Muse cells tolerate extensive genotoxic stimuli better than mesenchymal stromal cells. Oncotarget. 2018;9(27):19328–19341. doi: 10.18632/oncotarget.25039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gutierrez-Aranda I., Ramos-Mejia V., Bueno C., et al. Human induced pluripotent stem cells develop teratoma more efficiently and faster than human embryonic stem cells regardless the site of injection. Stem Cells. 2010;28(9):1568–1570. doi: 10.1002/stem.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee M.-H., Kwon T.-G., Park H.-S., Wozney J. M., Ryoo H.-M. BMP-2-induced Osterix expression is mediated by Dlx5 but is independent of Runx2. Biochemical and Biophysical Research Communications. 2003;309(3):689–694. doi: 10.1016/j.bbrc.2003.08.058. [DOI] [PubMed] [Google Scholar]
- 82.Rosen E. D., Hsu C.-H., Wang X., et al. C/EBPα induces adipogenesis through PPARγ: a unified pathway. Genes & Development. 2002;16(1):22–26. doi: 10.1101/gad.948702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ng S. Y.-M., Yoshida T., Zhang J., Georgopoulos K. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity. 2009;30(4):493–507. doi: 10.1016/j.immuni.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sworder B. J., Yoshizawa S., Mishra P. J., et al. Molecular profile of clonal strains of human skeletal stem/progenitor cells with different potencies. Stem Cell Research. 2015;14(3):297–306. doi: 10.1016/j.scr.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rattner A., Hsieh J.-C., Smallwood P. M., et al. A family of secreted proteins contains homology to the cysteine-rich ligand-binding domain of frizzled receptors. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(7):2859–2863. doi: 10.1073/pnas.94.7.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bovolenta P., Esteve P., Ruiz J. M., Cisneros E., Lopez-Rios J. Beyond Wnt inhibition: new functions of secreted Frizzled-related proteins in development and disease. Journal of Cell Science. 2008;121(6):737–746. doi: 10.1242/jcs.026096. [DOI] [PubMed] [Google Scholar]
- 87.Kake T., Kimura S., Takahashi K., Maruyama K. Calponin induces actin polymerization at low ionic strength and inhibits depolymerization of actin filaments. Biochemical Journal. 1995;312(2):587–592. doi: 10.1042/bj3120587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yoshikawa H., Taniguchi S. I., Yamamura H., et al. Mice lacking smooth muscle calponin display increased bone formation that is associated with enhancement of bone morphogenetic protein responses. Genes to Cells. 1998;3(10):685–695. doi: 10.1046/j.1365-2443.1998.00214.x. [DOI] [PubMed] [Google Scholar]
- 89.English K. Mechanisms of mesenchymal stromal cell immunomodulation. Immunology and Cell Biology. 2013;91(1):19–26. doi: 10.1038/icb.2012.56. [DOI] [PubMed] [Google Scholar]
- 90.Caplan A. I., Dennis J. E. Mesenchymal stem cells as trophic mediators. Journal of Cellular Biochemistry. 2006;98(5):1076–1084. doi: 10.1002/jcb.20886. [DOI] [PubMed] [Google Scholar]
- 91.Kurtzberg J., Prockop S., Teira P., et al. Allogeneic human mesenchymal stem cell therapy (Remestemcel-L, Prochymal) as a rescue agent for severe refractory acute graft-versus-host disease in pediatric patients. Biology of Blood and Marrow Transplantation. 2014;20(2):229–235. doi: 10.1016/j.bbmt.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 92.Ranganath S. H., Levy O., Inamdar M. S., Karp J. M. Harnessing the mesenchymal stem cell secretome for the treatment of cardiovascular disease. Cell Stem Cell. 2012;10(3):244–258. doi: 10.1016/j.stem.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu L., Leijten J. C. H., Georgi N., Post J. N., van Blitterswijk C. A., Karperien M. Trophic effects of mesenchymal stem cells increase chondrocyte proliferation and matrix formation. Tissue Engineering Part A. 2011;17(9-10):1425–1436. doi: 10.1089/ten.tea.2010.0517. [DOI] [PubMed] [Google Scholar]
- 94.Kuci S., Kuci Z., Kreyenberg H., et al. CD271 antigen defines a subset of multipotent stromal cells with immunosuppressive and lymphohematopoietic engraftment-promoting properties. Haematologica. 2010;95(4):651–659. doi: 10.3324/haematol.2009.015065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nasef A., Zhang Y. Z., Mazurier C., et al. Selected Stro-1-enriched bone marrow stromal cells display a major suppressive effect on lymphocyte proliferation. International Journal of Laboratory Hematology. 2009;31(1):9–19. doi: 10.1111/j.1751-553X.2007.00997.x. [DOI] [PubMed] [Google Scholar]
- 96.James S., Fox J., Afsari F., et al. Multiparameter analysis of human bone marrow stromal cells identifies distinct immunomodulatory and differentiation-competent subtypes. Stem Cell Reports. 2015;4(6):1004–1015. doi: 10.1016/j.stemcr.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ortiz L. A., DuTreil M., Fattman C., et al. Interleukin 1 receptor antagonist mediates the antiinflammatory and antifibrotic effect of mesenchymal stem cells during lung injury. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(26):11002–11007. doi: 10.1073/pnas.0704421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang Z. X., Han Z.-B., Ji Y. R., et al. CD106 identifies a subpopulation of mesenchymal stem cells with unique immunomodulatory properties. PLoS One. 2013;8(3):p. e59354. doi: 10.1371/journal.pone.0059354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mabuchi Y., Morikawa S., Harada S., et al. LNGFR+THY-1+VCAM-1hi+ cells reveal functionally distinct subpopulations in mesenchymal stem cells. Stem Cell Reports. 2013;1(2):152–165. doi: 10.1016/j.stemcr.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ren G., Zhao X., Zhang L., et al. Inflammatory cytokine-induced intercellular adhesion molecule-1 and vascular cell adhesion molecule-1 in mesenchymal stem cells are critical for immunosuppression. The Journal of Immunology. 2010;184(5):2321–2328. doi: 10.4049/jimmunol.0902023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Trowsdale J., Betz A. G. Mother’s little helpers: mechanisms of maternal-fetal tolerance. Nature Immunology. 2006;7(3):241–246. doi: 10.1038/ni1317. [DOI] [PubMed] [Google Scholar]
- 102.Yang R., Liu Y., Kelk P., et al. A subset of IL-17+ mesenchymal stem cells possesses anti-Candida albicans effect. Cell Research. 2013;23(1):107–121. doi: 10.1038/cr.2012.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ceredig R., Rolink A. G. The key role of IL-7 in lymphopoiesis. Seminars in Immunology. 2012;24(3):159–164. doi: 10.1016/j.smim.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 104.Jouvenet N., Neil S. J. D., Zhadina M., et al. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. Journal of Virology. 2009;83(4):1837–1844. doi: 10.1128/JVI.02211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hotter D., Sauter D., Kirchhoff F. Emerging role of the host restriction factor tetherin in viral immune sensing. Journal of Molecular Biology. 2013;425(24):4956–4964. doi: 10.1016/j.jmb.2013.09.029. [DOI] [PubMed] [Google Scholar]
- 106.Park T. S., Gavina M., Chen C. W., et al. Placental perivascular cells for human muscle regeneration. Stem Cells and Development. 2011;20(3):451–463. doi: 10.1089/scd.2010.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Psaltis P. J., Paton S., See F., et al. Enrichment for STRO-1 expression enhances the cardiovascular paracrine activity of human bone marrow-derived mesenchymal cell populations. Journal of Cellular Physiology. 2010;223:530–540. doi: 10.1002/jcp.22081. [DOI] [PubMed] [Google Scholar]
- 108.Anderson G. D., Hauser S. D., McGarity K. L., Bremer M. E., Isakson P. C., Gregory S. A. Selective inhibition of cyclooxygenase (COX)-2 reverses inflammation and expression of COX-2 and interleukin 6 in rat adjuvant arthritis. The Journal of Clinical Investigation. 1996;97(11):2672–2679. doi: 10.1172/jci118717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yañez R., Oviedo A., Aldea M., Bueren J. A., Lamana M. L. Prostaglandin E2 plays a key role in the immunosuppressive properties of adipose and bone marrow tissue-derived mesenchymal stromal cells. Experimental Cell Research. 2010;316(19):3109–3123. doi: 10.1016/j.yexcr.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 110.Munn D. H., Mellor A. L. Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends in Immunology. 2013;34(3):137–143. doi: 10.1016/j.it.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pettus B. J., Bielawski J., Porcelli A. M., et al. The sphingosine kinase 1/sphingosine-1-phosphate pathway mediates COX-2 induction and PGE2 production in response to TNF-α. The FASEB Journal. 2003;17(11):1411–1421. doi: 10.1096/fj.02-1038com. [DOI] [PubMed] [Google Scholar]
- 112.Cesario A., Rocca B., Rutella S. The interplay between indoleamine 2,3-dioxygenase 1 (IDO1) and cyclooxygenase (COX)-2 in chronic inflammation and cancer. Current Medicinal Chemistry. 2011;18(15):2263–2271. doi: 10.2174/092986711795656063. [DOI] [PubMed] [Google Scholar]
- 113.Li D., Han Y., Zhuang Y., et al. Overexpression of COX-2 but not indoleamine 2,3-dioxygenase-1 enhances the immunosuppressive ability of human umbilical cord-derived mesenchymal stem cells. International Journal of Molecular Medicine. 2015;35(5):1309–1316. doi: 10.3892/ijmm.2015.2137. [DOI] [PubMed] [Google Scholar]
- 114.Espert L., Degols G., Gongora C., et al. ISG20, a new interferon-induced RNase specific for single-stranded RNA, defines an alternative antiviral pathway against RNA genomic viruses. Journal of Biological Chemistry. 2003;278(18):16151–16158. doi: 10.1074/jbc.m209628200. [DOI] [PubMed] [Google Scholar]
- 115.Silverman R. H. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. Journal of Virology. 2007;81:p. 12720. doi: 10.1128/jvi.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sherman S. E., Kuljanin M., Cooper T. T., Putman D. M., Lajoie G. A., Hess D. A. High aldehyde dehydrogenase activity identifies a subset of human mesenchymal stromal cells with vascular regenerative potential. Stem Cells. 2017;35(6):1542–1553. doi: 10.1002/stem.2612. [DOI] [PubMed] [Google Scholar]
- 117.Storms R. W., Trujillo A. P., Springer J. B., et al. Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(16):9118–9123. doi: 10.1073/pnas.96.16.9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hanahan D., Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86(3):353–364. doi: 10.1016/S0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 119.Korn C., Augustin H. G. Mechanisms of vessel pruning and regression. Developmental Cell. 2015;34(1):5–17. doi: 10.1016/j.devcel.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 120.Rando T. A. Stem cells, ageing and the quest for immortality. Nature. 2006;441(7097):1080–1086. doi: 10.1038/nature04958. [DOI] [PubMed] [Google Scholar]
- 121.Wagner W., Horn P., Castoldi M., et al. Replicative senescence of mesenchymal stem cells: a continuous and organized process. PLoS One. 2008;3(5) doi: 10.1371/journal.pone.0002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Galderisi U., Helmbold H., Squillaro T., et al. In vitro senescence of rat mesenchymal stem cells is accompanied by downregulation of stemness-related and DNA damage repair genes. Stem Cells and Development. 2009;18(7):1033–1042. doi: 10.1089/scd.2008.0324. [DOI] [PubMed] [Google Scholar]
- 123.Yang Y.-H. K., Ogando C. R., Wang See C., Chang T. Y., Barabino G. A. Changes in phenotype and differentiation potential of human mesenchymal stem cells aging in vitro. Stem Cell Research & Therapy. 2018;9(1):p. 131. doi: 10.1186/s13287-018-0876-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Block T. J., Marinkovic M., Tran O. N., et al. Restoring the quantity and quality of elderly human mesenchymal stem cells for autologous cell-based therapies. Stem Cell Research & Therapy. 2017;8(1):p. 239. doi: 10.1186/s13287-017-0688-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lalaoui N., Morlé A., Mérino D., et al. TRAIL-R4 promotes tumor growth and resistance to apoptosis in cervical carcinoma HeLa cells through AKT. PLoS One. 2011;6(5):p. e19679. doi: 10.1371/journal.pone.0019679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ren J., Stroncek D. F., Zhao Y., et al. Intra-subject variability in human bone marrow stromal cell (BMSC) replicative senescence: molecular changes associated with BMSC senescence. Stem Cell Research. 2013;11(3):1060–1073. doi: 10.1016/j.scr.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Koch C. M., Joussen S., Schellenberg A., Lin Q., Zenke M., Wagner W. Monitoring of cellular senescence by DNA-methylation at specific CpG sites. Aging Cell. 2012;11(2):366–369. doi: 10.1111/j.1474-9726.2011.00784.x. [DOI] [PubMed] [Google Scholar]
- 128.Franzen J., Zirkel A., Blake J., et al. Senescence-associated DNA methylation is stochastically acquired in subpopulations of mesenchymal stem cells. Aging Cell. 2017;16(1):183–191. doi: 10.1111/acel.12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Barrilleaux B. L., Phinney D. G., Fischer-Valuck B. W., et al. Small-molecule antagonist of macrophage migration inhibitory factor enhances migratory response of mesenchymal stem cells to bronchial epithelial cells. Tissue Engineering Part A. 2009;15(9):2335–2346. doi: 10.1089/ten.tea.2008.0434. [DOI] [PubMed] [Google Scholar]
- 130.Karoubi G., Ormiston M. L., Stewart D. J., Courtman D. W. Single-cell hydrogel encapsulation for enhanced survival of human marrow stromal cells. Biomaterials. 2009;30(29):5445–5455. doi: 10.1016/j.biomaterials.2009.06.035. [DOI] [PubMed] [Google Scholar]
- 131.Xu X., Gao D., Wang P., et al. Efficient homology-directed gene editing by CRISPR/Cas9 in human stem and primary cells using tube electroporation. Scientific Reports. 2018;8(1):p. 11649. doi: 10.1038/s41598-018-30227-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lipsitz Y. Y., Timmins N. E., Zandstra P. W. Quality cell therapy manufacturing by design. Nature Biotechnology. 2016;34(4):393–400. doi: 10.1038/nbt.3525. [DOI] [PubMed] [Google Scholar]
- 133.Kirouac D. C., Zandstra P. W. The systematic production of cells for cell therapies. Cell Stem Cell. 2008;3(4):369–381. doi: 10.1016/j.stem.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 134.Isakova I. A., Baker K., DuTreil M., Dufour J., Gaupp D., Phinney D. G. Age- and dose-related effects on MSC engraftment levels and anatomical distribution in the central nervous systems of nonhuman primates: identification of novel MSC subpopulations that respond to guidance cues in brain. Stem Cells. 2007;25(12):3261–3270. doi: 10.1634/stemcells.2007-0543. [DOI] [PubMed] [Google Scholar]
- 135.Davis C., Dukes A., Drewry M., et al. MicroRNA-183-5p increases with age in bone-derived extracellular vesicles, suppresses bone marrow stromal (stem) cell proliferation, and induces stem cell senescence. Tissue Engineering Part A. 2017;23(21-22):1231–1240. doi: 10.1089/ten.tea.2016.0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tye C. E., Gordon J. A. R., Martin-Buley L. A., Stein J. L., Lian J. B., Stein G. S. Could lncRNAs be the missing links in control of mesenchymal stem cell differentiation? Journal of Cellular Physiology. 2015;230(3):526–534. doi: 10.1002/jcp.24834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ozkul Y., Galderisi U. The impact of epigenetics on mesenchymal stem cell biology. Journal of Cellular Physiology. 2016;231(11):2393–2401. doi: 10.1002/jcp.25371. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table: nomenclature for global molecular signatures.