Abstract
The complexity of modern in vivo magnetic resonance imaging (MRI) methods in oncology has dramatically changed in the last ten years. The field has long since moved passed its (unparalleled) ability to form images with exquisite soft-tissue contrast and morphology, allowing for the enhanced identification of primary tumors and metastatic disease. Currently, it is not uncommon to acquire images related to blood flow, cellularity, and macromolecular content in the clinical setting. The acquisition of images related to metabolism, hypoxia, pH, and tissue stiffness are also becoming common. All of these techniques have had some component of their invention, development, refinement, validation, and initial applications in the pre-clinical setting using in vivo animal models of cancer. In this review, we discuss the genesis of quantitative MRI methods that have been successfully translated from pre-clinical research and developed into clinical applications. These include methods which interrogate perfusion, diffusion, pH, hypoxia, macromolecular content, and tissue mechanical properties for improving detection, staging and response monitoring of cancer. For each of these techniques, we summarize the 1) underlying biological mechanism(s), 2) pre-clinical applications, 3) available repeatability and reproducibility data, 4) clinical applications, and 5) limitations of the technique. We conclude with a discussion of lessons learned from translating MRI methods from the pre-clinical to clinical setting, and a presentation of four fundamental problems in cancer imaging that, if solved, would result in a profound improvement in the lives of oncology patients.
Keywords: cancer, diffusion, perfusion, CEST, MT, elastography
Introduction
A biomarker is “a biological molecule found in blood, other body fluids, or tissues that is a sign of a normal or abnormal process, or of a condition or disease” (1). An “imaging biomarker” is defined as “a biological feature or biomarker observable in an image” (2). Imaging biomarkers are ubiquitous in clinical oncology for the diagnosis, staging, guidance of therapy delivery, response assessment, and long-term follow-up of patients. These imaging biomarkers have largely been based on gross changes in anatomy and morphology which tend to occur temporally downstream of the underlying physiological, cellular, and molecular changes. Thus, the imaging community has spent tremendous effort attempting to develop more precise biomarkers that can be used to interrogate the underlying physiology thereby improving patient care as well as discovering new biological mechanisms. Central to more specific imaging biomarkers is the notion of quantitative imaging; that is, imaging methods that provide measurements of intrinsic tissue properties (including anatomical, physiological, or biophysical) specified in an appropriate continuous-variable unit. The first examples of quantitative magnetic resonance imaging (MRI) to assess cancer could be considered Damadian’s seminal work in applying relaxometry (3) to distinguish tumor and normal tissues. One year later, Hazelwood (4) investigated both relaxation times and water diffusion in cancer ex vivo. Though relaxation measurements were later shown to lack specificity in vivo, diffusion MRI is now firmly established as a quantitative MRI biomarker for a wide range of oncological and other clinical applications.
While the path can certainly vary, developing an imaging biomarker frequently begins with phantom studies composed of tissue mimicking materials to represent some relevant aspect of anatomy or physiology, or even in vitro studies designed to test the imaging technique’s ability to characterize particular cellular or molecular features commonly encountered in cancer. The prospective imaging biomarker is then investigated using in vivo preclinical studies (typically mice or rats) designed to test the validity of the measurement in a more realistic setting--both in terms of the complexities of mammalian biology, as well as addressing the logistical issues of physiological motion (e.g., cardiac and respiratory) and limited time available in which to make the measurement. Prior to clinical application, validation studies must be performed to ensure that the imaging measure is actually reporting on what it is designed to characterize. Refinement in the clinical setting is typically first explored in healthy subjects, before proceeding to applications in very specific patient populations at a single imaging center. Additionally, the repeatability and reproducibility of the technique must be established for properly powering clinical trials. If those studies are deemed “successful”, then multi-site studies will follow before eventually leading to routine use in the standard-of-care setting. This process is extremely long and resource intensive. To this end, an effort to identify methods to accelerate the process has recently been contributed (2).
In this review, we begin with a description of the technical and logistical considerations that must be addressed when translating an MRI biomarker into clinical application. We then move to a description of several common and emerging MRI methods appropriate for oncology, while also addressing the underlying biological mechanism, pre-clinical applications, available repeatability and reproducibility data, illustrative clinical applications, and limitations of the technique. We conclude the effort with a discussion of lessons learned from our own efforts at translating MRI methods from the pre-clinical to clinical setting, and a presentation of four fundamental problems in cancer imaging that, if solved, would result in a profound improvement in the lives of oncology patients.
Technical considerations
Repeatability, reproducibility, validity
For quantitative imaging biomarkers to be adopted into clinical practice and used to guide precision medicine, their technical performance must be rigorously evaluated. The Quantitative Imaging Biomarkers Alliance (QIBA) of the Radiological Society of North America (RSNA) has established consensus terminology and methods to describe the performance of perspective imaging biomarkers. QIBA recommends that each biomarker be characterized by estimating measurement bias, precision, and linearity between the measurement and underlying biology (5). Measurement bias refers to the difference between the measurement and true value. For quantitative MRI measurements, the true value can be difficult to ascertain, thus bias estimation often relies on pre-established reference values or phantom measurements. While phantom measurements can provide a useful estimate of bias, they may underestimate bias found in vivo. Repeatability is measurement precision under identical conditions, using the same subject, same scanner, and same imaging protocol. Repeatability is usually estimated through test-retest examinations of the same subject performed within a short interval. Reproducibility, in contrast, measures the same subject using different scanners, operators, processing software, and/or image readers. Precision characterizes the variability in measurement of a single sample, and encompasses both repeatability and reproducibility. Linearity describes the proportionality between the true value and measurement over a biologically relevant range. It can also be established using phantom measurements employing (for example) serial dilutions of contrast agents, subject to the same caveat as bias determination.
Historically, there has been a lack of physical phantoms for small animal imaging. Given the variability of coil and animal holder geometries across vendors, as well as a lack of demand for such measurements as compared to clinical systems, this may not be surprising. Furthermore, the design of phantoms appropriate for pre-clinical is not without obstacles. In particular, the limited space available within the smaller diameter coils places physical constraints on the number of features (e.g., spatial resolution, linearity, and slice profile) a single phantom can assess. The recent increase in demand for characterizing quantitative measures in animal studies, though, may lead to increased availability of such phantoms which could improve standardization and quality assurance of MRI in the preclinical setting.
General limitations of MRI techniques
There are a number of logistical issues associated with performing MRI, including the high cost of the scanner and contraindications in patients with metallic implants. Furthermore, MRI requires relatively long acquisition times, both increasing cost and leading to respiratory and cardiac motion artifacts which can tissue of interest. Additionally, although MRI has demonstrated high sensitivity in a number of oncology applications, its specificity can be rather modest. The MRI signal is a complex function of proton density, relaxation processes, and pulse sequence parameters. As mentioned above, seminal MRI work demonstrated tumor visualization due to alterations in the relaxation properties; however, relaxation times are influenced by (for example) oxygenation, water content, and necrosis. These factors can change concurrently in tumors, resulting in MRI measurements that reflect a hybrid of competing effects. Thus, MR images can be difficult to interpret clinically in the face of biological alterations that exhibit similar radiological manifestations. For instance, in glioma, changes in apparent tumor volume may result from changes in the blood–brain barrier after antiangiogenic therapy (known as pseudoresponse), or transient radiation injury may occur after chemoradiation (termed pseudoprogression) (6). The quantitative techniques described below (see Section 3) represent efforts establish more specific measures of particularly pathophysiology to address such limitations.
Challenges in using MRI to characterize cancer in the pre-clinical setting
While pre-clinical MRI is important for the development and validation of novel techniques, there are both technical and biological challenges that can impede translation. Pre-clinical scanners often have technical advantages over clinical scanners with increased field strength (typically from 4.7 to 9.4T, but can be as high as 22T) and high-performance gradients that allow for higher temporal and spatial resolution experiments compared to the clinical setting. However, pre-clinical systems often lack the wide selection of clinically available coils optimized for target geometry or image acceleration (e.g., multi-channel) requiring the development of lab-specific coils. Furthermore, small animal respiratory (30 – 60 breaths per minute under anesthesia) and cardiac motion (400–600 beats per minute) can be a source of substantial image artifacts requiring the use of fast imaging sequences, or respiratory gating to minimize the effect on image quality. Anesthesia and restraints (or bite bars) can be used to minimize animal motion, but some anesthetic agents may unintentionally alter animal physiology thereby potentially affecting the biological process under investigation (7). Finally, while the higher field strengths of pre-clinical systems are attractive from a signal-to-noise perspective, the tissue contrast can be significantly impacted, especially for T1-weighted imaging, confounding translation to typical clinical field strengths of 1.5 and 3.0 T.
In addition to the technical considerations just presented, the interpretation of novel imaging measures and studies hinges on the selection of an appropriate animal model. While not unique to MRI, animal models of cancer should recapitulate the imaging and pathohistological features of human cancers. For example, the widely used rodent glioma models C6 and 9L grow in very different patterns compared with primary human brain tumors; while models based on patient derived xenografts show distinct pathophysiological features that can recapitulate those found in humans. Thus, knowledge gained from imaging a C6 tumor may be of limited value for optimizing an imaging approach for human glioma.
Challenges in using MRI to characterize cancer in the clinical setting
While many of the quantitative techniques listed below in section 3 have been successful applied in the clinic, translating pre-clinical developments directly to the clinical setting can be challenging. Significant resources (personnel, hardware, etc.) must be available to first develop the technique in the pre-clinical setting, and then implement it on clinical systems. In particular, significant technical challenges exist in transferring pre-clinical imaging sequences developed at higher field strengths (4.7 to 15 T) to the lower fields typically employed in the clinic (i.e., 1.5 – 3 T). Furthermore, there exist limited availabilities of particular imaging sequences across vendors which can limit the validation of quantitative techniques across institutions. Image contrast varies considerably due to the field strength dependence of T1, T2, susceptibility, and inherent signal, requiring the optimization of pulse sequence parameters for different field strengths. Unlike the pre-clinical setting, clinical imaging studies often have hardware, safety (e.g., SAR, cardiac limits, peripheral nerve stimulation), and time limitations (e.g., scan duration, breath hold) which can impact imaging protocol design to achieve equivalent imaging measures. High field clinical scanners ( ≥ 7 T) can also aggravate B0 and B1 field inhomogeneities resulting in regional variations in flip angle and signal intensity across the imaging volume (8). Additional challenges which may impede quantitative imaging in the clinical setting include implantable devices (e.g., pacemakers) or patients who suffer claustrophobia or anxiety which may impose additional limitations on field strength or imaging sequences.
Validation of techniques across institutions can be logistically challenging due to site-to-site variations in hardware, acquisition, and analysis methods. Ongoing efforts from the National Cancer Institute’s Quantitative Imaging Network (QIN) (9) and QIBA (10) aim to harmonize protocols and approaches to facilitate clinical decision making based off of standardized quantitative imaging approaches. Validation via large multi-site trials could benefit from multi-institution research networks such as the Eastern Cooperative Oncology Group American College of Radiation Imaging Network (ECOG-ACRIN).
In addition to the above technical challenges, clinical studies require dedicated research staff to attend to important patient considerations as discussed below in section 4.
Quantitatively interrogating biology
Perfusion
Dysregulated angiogenesis is a common feature of many cancers. Imaging methods have been developed to characterize the hemodynamic and structural features of the resulting vasculature and its response to therapy. The three most common MRI methods for interrogating tissue vascular properties are dynamic contrast-enhanced (DCE) MRI, dynamic susceptibility contrast (DSC) MRI, and arterial spin labeling (ASL). Both DCE- and DSC-MRI track the dynamic passage of an intravenously infused contrast agent to derive pharmacokinetic properties; DCE-MRI characterizes the contrast agent extravasation rate (Ktrans; a mixed measure of both perfusion and vessel permeability) and extravascular extracellular volume fraction (ve), while DSC-MRI characterizes blood volume, blood flow, and mean transit time (see Figure 1). With ASL, the difference between images collected with and without the application of a magnetic label of inflowing arterial blood can be used to derive a quantitative measure of perfusion. Perfusion methods are routinely employed for distinguishing tumor types and grades, reliably detecting progression and early response, and predicting overall patient survival.
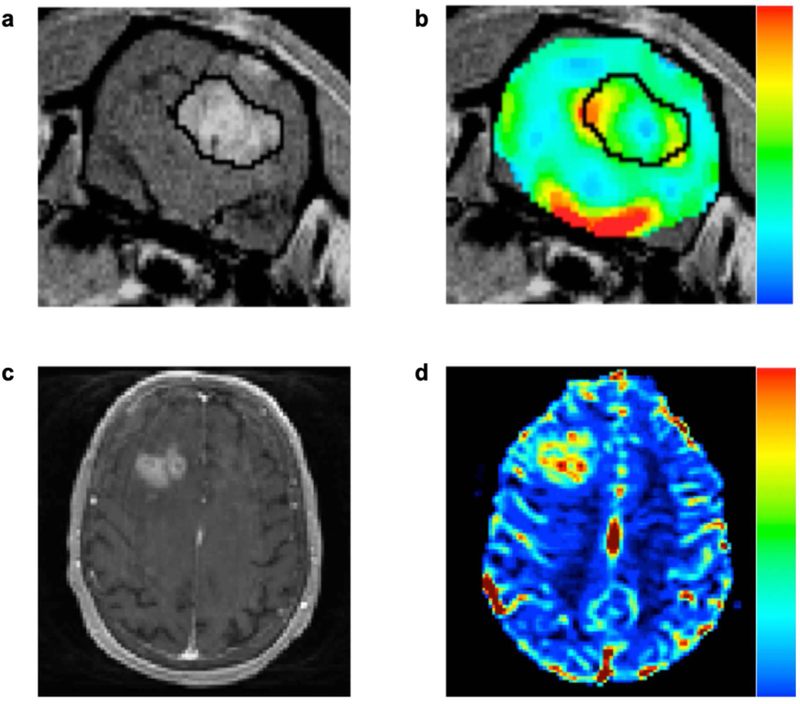
Figure 1.
Dynamic susceptibility contrast MRI methods can be leveraged to assess brain tumor hemodynamics in animals (top row), as well as humans (bottom row). Shown are representative post-contrast T1-weighted images (panels a and c) and DSC-MRI derived cerebral blood volume maps (panels b and d) in an orthotopic C6 rodent glioma model and a patient with a high-grade glioma. Post-contrast T1-weighted images are routinely used to visualize the primary tumor mass due to the accumulation of contrast agent in regions with a disrupted blood brain barrier. In animals and humans, high grade gliomas exhibit CBV values that are markedly higher than that found in normal appearing white matter.
Pre-clinical perfusion imaging has found much utility in evaluating response to anti-vascular therapies. While the most often reported effect of anti-angiogenic therapy is to reduce tumor perfusion, DSC-MRI has also been used to investigate vascular normalization and maturation, where such agents can be used to transiently or permanently enhance perfusion. For example, in a rat model of brain cancer, administration of a multi-targeted receptor tyrosine kinase inhibitor (SU11657), decreased tumor blood volume, vessel size, and mean transit time, while also increasing blood flow and normalizing transit time heterogeneity (11). Similarly, a DCE-MRI study revealed that trastuzumab increased Ktrans in the BT474 mouse model of HER2+ breast cancer (12). Since ASL does not require the use of exogeneous contrast agents, perfusion can be evaluated repeatedly during a scan session. This feature was leveraged to demonstrate that vascular disruption therapy in the SW1222 mouse model of colorectal cancer in the liver yields a significant and rapid decrease in perfusion (13).
Throughout the development of perfusion imaging, pre-clinical and clinical validation efforts have served to verify underlying assumptions, establish accuracy and precision, and identify optimal acquisition and analysis methods. With DSC-MRI, validation efforts have included in vivo comparisons of blood volume and blood flow measures to those derived from PET and SPECT (14), histology (15), and synthetic patient datasets (16). Similarly, numerous studies have validated DCE-MRI methods using physical phantoms (17), and parameters in different organs using autoradiography, histology, and dynamic contrast enhanced CT (18). There is also an extensive literature validating cerebral perfusion measures from ASL using [15O]-water PET (19). Furthermore, the last decade has produced numerous DSC-MRI, DCE-MRI, and ASL standardization and recommendation efforts (20, 21, 22) as well as studies focused on repeatability and reproducibility (23, 24, 25). Such efforts include harmonizing acquisition protocols, post-processing algorithms, and parameter quantification. As a testament to these efforts, all the major MRI vendors have since released commercial products aligned with the recommended protocol.
Cellularity
Diffusion-weighted MRI (DW-MRI or DWI) replies on the mobility of water protons in biological tissues and thus provides a unique means to non-invasively characterize tissue microstructure (26). To remove other contaminating effects such as the T2 shine-through, a quantitative metric termed the apparent diffusion coefficient (ADC) can be obtained with DWI, which reflects the averaged diffusion properties within each voxel. It is typically assumed that, after an effective treatment, the cellularity of a solid tumor will decrease resulting in a reduction of restriction and/or hindrance to water protons which is manifested as an increase in the ADC. In early pre-clinical investigations, Zhao et al. reported a dose-dependent, reversible increase in tumor ADC after treatment using a murine radiation-induced fibrosarcoma model (JS8). Similar findings were reported by Chenevert et al. (27) using a murine brain cancer model. These early pre-clinical investigations provided a new avenue to characterize tumor status and monitor response to treatment (28). DWI has been translated to clinical oncology to differentiate cancer (29), evaluate tumor aggressiveness (30), assess early treatment response (31), and predict treatment efficacy (32). Note that many of these findings were validated with histology, indicating that DWI is sensitive to tumor cellularity. In addition to brain cancer, DWI has been included as either a main or an optional imaging method in the standard-of-care for prostate (33) and liver cancer (34), respectively, and is under investigation for inclusion for breast cancer (35).
Despite the success, there are some remaining challenges that need to be addressed before DWI in can be widely used in large-scale multi-site clinical trials. In particular, DWI typically uses single-shot echo planar imaging that can lead to strong susceptibility artifacts, and gradient coil duty cycle limits the minimal achievable TR and hence increases total scan time. Several approaches have been proposed to overcome the DWI geometric distortion; these include (for example) the segmented EPI (37) and reduced field-of-view (38) acquisitions. The development of multi-band (39) has also been shown to significantly accelerate DWI acquisitions.
Second, efforts on establishing reproducibility and bias across centers should be continued; we note that there have been notable studies on this front using ice-water phantoms for increasing the reproducibility of ADC measurements and identifying vendor- and system-specific bias (39)40 Additionally, DWI may be dominated by perfusion when b values are small (e.g., < 200 sec/mm2) and show non-Gaussian diffusion behavior when b values are large (e.g., > 1200 sec/mm2). Finally, and perhaps most importantly, the ADC has limited specificity to cellularity because it is affected by all microstructural parameters simultaneously. This sometimes causes ambiguous interpretations of tumor status. A possible solution is to develop more advanced diffusion acquisition and analysis methods (37; see Figure 2) which may potentially increase specificity. For example, by including acquisitions with higher diffusion weighting (i.e., higher b values), non-Gaussian diffusion analysis methods have been developed, including bi-exponential (41), kurtosis (42), stretched exponential (43), and restriction spectrum imaging (44), all of which have been applied in oncology and have been suggested to improve sensitivity and/or specificity compared to the ADC. Another approach is to develop biophysical diffusion models (45,46) to extract specific microstructural information such as mean cancer cell size and cell density of tumors.
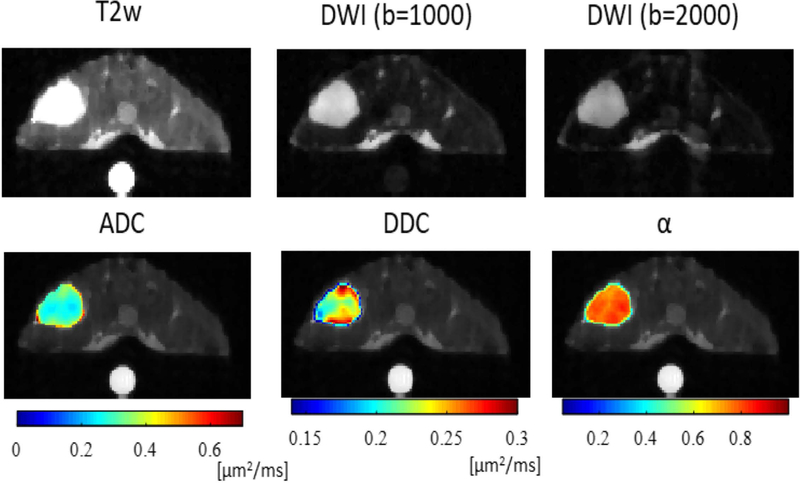
Figure 2.
Multi-parametric maps of a central slice from a representative mouse. The SW620 colon cancer xenograft is the in the upper left. A doped water phantom (5 mM CuSO4) was placed beneath the animal to ensure the accuracy of diffusion measurements. ADC is apparent diffusion coefficient, DDC and α are the distributed diffusion coefficient and heterogeneity index, respectively, obtained using the stretched exponential model. The DDC represents mean intravoxel diffusion rates, while α characterizes the degree of heterogeneity of water diffusion within each voxel.
Oxygen
Tumor hypoxia influences therapy and has been associated with tumor aggressiveness. Oxygen sensitive proton MRI is attractive, since it directly interrogates endogenous tissue characteristics, avoiding the need for exogenous reporter molecules and is entirely non-invasive. Blood oxygen level dependent (BOLD) contrast is based on the transverse relaxation rate of the tissue water proton, which is influenced by the concentration of deoxyhemoglobin generating strong local susceptibility gradients. Correlative trends are reported between BOLD and invasive oximetry (47). Differential BOLD response to an oxygen breathing challenge is related to vascular extent and responses have been reported with respect to diverse pharmacologic interventions, though most studies examine hyperoxic gas (48). Pre-irradiation BOLD response to an oxygen-breathing challenge was related to tumor growth following a single dose of radiation (49). Meanwhile, tissue oxygen level dependent (TOLD) contrast exploits the longitudinal relaxation rate, which is directly sensitive to the concentration of free oxygen molecules, and hence pO2 (50). While the BOLD and TOLD methods show similar enhancement patterns, the changes in longitudinal relaxation tend to be smaller (see Figure 3). In a typical tumor, a hyperoxic gas breathing challenge caused rapid significant response in each parameter, and TOLD lagged BOLD consistent with vascular delivery of oxygen followed by diffusion into the tissue (48). A second tumor, anatomically similar, showed essentially no response and the difference was validated by matching rigorous quantitative 19F MR oximetry. TOLD, but not BOLD, responses enabled tumor stratification with respect to radiation induced growth delay (48). Most studies examine the mean response based on semi-quantitative T1- or T2*-weighted images, or changes in the relaxation rates (R1 and R2*). However, in some tumors, the fraction of tissue showing enhancement was found to be more closely correlated with hypoxia as determined by pimonidazole (51).
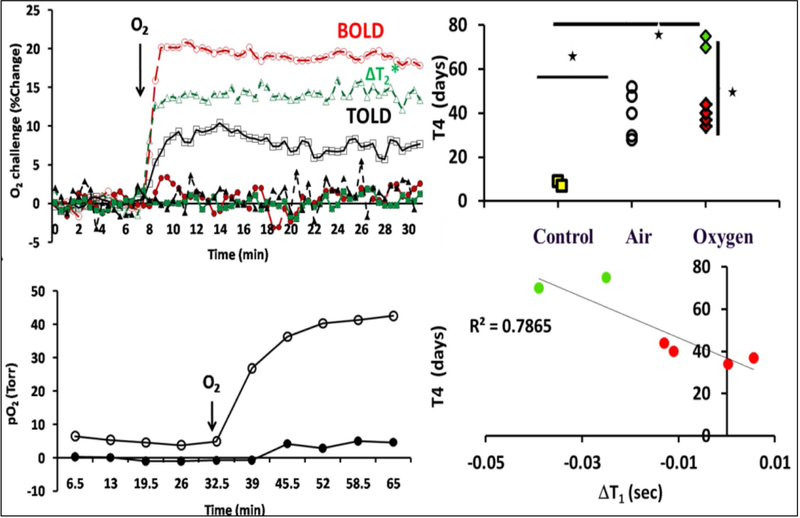
Figure 3.
The left column presents time course data indicates changes in tumor oxygenation in response to oxygen breathing. The upper left panels shows mean changes in BOLD (red O), TOLD (black ▯), %ΔT2* (green Δ) for two similarly sized AT1 tumors exhibiting very different response to 100% O2 breathing with corresponding mean changes in pO2, shown in the bottom left panel. The hypoxic tumor showed little response by all measures, whereas the better oxygenated tumor was rapidly responsive. The upper right panel presents data on tumor growth delay where the effect of breathing air and O2 on radiation response assessed by time for tumor to quadruple in size (T4); * p < 0.01. The lower right panel show the correlation between (T4) and ΔT1 for tumors irradiated during O2 breathing. Modified from Young et al J. Comp. Tomogr. 1981; 5, 543–547.)
Hyperoxic gas breathing interventions are generally reversible (51) and studies have shown that repeated challenges resulted in similar responses (52) allowing effective comparison of hypoxia-modulating interventions (53). Successive imaging on sequential days or over several days showed no significant differences in responses in some tumor types (54). Change in longitudinal relaxation in response to an oxygen breathing challenge has been shown to discriminate tumor tissue from radiation induced necrosis (55). It should be noted that longitudinal relaxation is very sensitive to changes in temperature, and therefore physiological stability is vital. Motivated by these data, clinical applications of BOLD (56) and TOLD (57) have been reported for a range of human cancers. While oxygen sensitive measurements are not included currently in standard-of-care, the required pulse sequences are routine and an oxygen breathing challenge can readily be included in the typical time allotted to a routine radiological examination.
Mechanical properties
Identification of regions of aberrant mechanical properties within soft tissue is a foundational principle for disease detection, with manual palpation serving as an important first-line identification method for the detection of many cancers. Elevations in tumor tissue mechanical stiffness occur through microenvironmental changes in the tissue extracellular matrix that serve to increase the density and organization of structural proteins. The association of mechanical stiffness with cancer progression and treatment response have motivated efforts for imaging-based measurements of tissue mechanical elasticity, termed elastography. There is considerable technological variation among individual elastography methods but, in general, image contrast in elastography is formed through computational processing of tissue deformation response to an applied loading (58).
MR elastography remains an emerging method in oncology and, due to challenges at small length scales, there is a paucity of pre-clinical studies imaging mechanical stiffness for cancer progression or therapy response assessment (59,60). Significant work was reported by Li et al. (61) where MR elastography was used to evaluate response to a therapeutic agent in a murine model of colorectal cancer at early-response time points. Several studies have assessed the repeatability and accuracy of MR elastography; while specific details vary based on individual elastography methods, MR elastography has been found to exhibit similar repeatability and reproducibility (see Figure 4) as other quantitative MR-based methods (62,63). Validating the accuracy of MR elastography is straightforward using gel phantoms with inclusions of known mechanical stiffness assessed with independent mechanical testing, and studies have found accuracy ranging between 10–20% (62). MR elastography measures of tissue stiffness have also been compared to histology, with areas of elevated stiffness correlated to increased collagen and areas of reduced stiffness correlated to tissue necrosis (62,63).
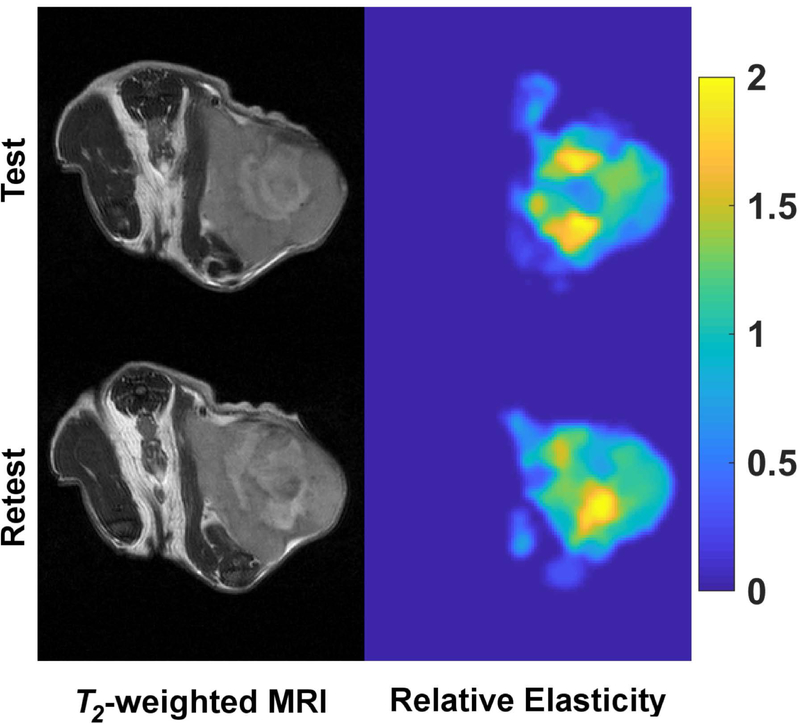
Figure 4.
Pre-clinical mechanical stiffness imaging of a subcutaneous xenograft breast tumor within the context of a test-retest repeatability assessment. T2-weighted MR images (left column) and associated relative mechanical stiffness maps (right column) from quasi-static MR elastography reflect similar mechanical stiffness heterogeneity within the tumor for both the test and retest scans of the same animal.
Clinical applications of MR elastography measurements of tumor mechanical stiffness have shown significant potential for clinical cancer staging and treatment response assessment (64,65). In a recent study by Gordic et al., MR elastography was used to assess tumor stiffness following locoregional therapy for hepatocellular carcinoma (66). Tumor stiffness was found to be significantly reduced in treated tumors, with significant correlations of reduced stiffness to tumor necrosis (66). While MR elastography has prominent emerging clinical implementation in other diseases, most notably in liver fibrosis disease staging (67), its use in cancer quantitative MRI is still emerging without widespread clinical adaptation or inclusion in the standard of care.
While MR elastography imaging for cancer is based on a strong, biological mechanistic foundation, there are a number of limitations. First, small lesions may not be detectable due to resolution limits that preclude sufficient shear wave detection or deformational shape change. Focal lesions may also be confounded and masked by the presence of elevated stiffness of the surrounding parenchyma (e.g., fibrotic organs). Further, mechanical shear wave attenuation can limit resolution depth for vibration-based dynamic elastography. Finally, dedicated hardware and software are often required for MR elastography.
Metabolism
First identified in the 1920’s by Otto Warburg (68), increased tumor glycolysis is one of the metabolic hallmarks of cancers. More recently, several additional metabolic hallmarks have been identified including aberrant glutamine, choline, and lipid metabolism. The three nuclei most commonly used for magnetic resonance spectroscopy (MRS) are 1H, 31P and 13C. Because of its high sensitivity, 1H MRS has been extensively used to characterize lactate, total creatine, and total choline that have concentrations in the mM range. In vivo, the components of the total choline signal that consists of phosphocholine, glycerophosphocholine, and free choline can be resolved, while the total creatine signal consists of phosphocreatine and creatine are not resolved. Although significantly less sensitive than 1H MRS, 31P MRS can provide a wealth of on bioenergetics, phospholipids, and pH (69). 31P MRS studies of cancers have revealed high phosphomonoesters and phosphodiesters (69). 13C MRS has undergone a major renaissance with the development of hyperpolarization. The use of dynamic nuclear polarization of 13C substrates increases sensitivity by ~10,000 allowing the in vivo acquisition of such substrates, with the constraint that the enriched substrate should be water-soluble and have a long longitudinal relaxation time in the liquid state. Pre-clinical applications of dynamic nuclear polarization 13C MRS imaging include detecting lactate dehydrogenase, pyruvate dehydrogenase, and tricarboxcylic acid activity, as well as glutaminolysis, perfusion, necrosis, and extracellular pH (70).
1H MRS is being actively investigated in grading and staging of human cancers. Since brain tumors frequently show decreases of N-acetylaspartate, detection of lactate and lipids, a decrease in total creatine, and an increase of total choline (71), MRS can be used as a companion to MRI, to distinguish malignant tissue from normal tissue or tumor necrosis (72). More generally, studies of many human cancers including breast (73), head and neck (74), and ovarian (75) have identified an increase in total choline. 1H MRS is also being investigated for radiotherapy treatment planning (76) and in predicting therapeutic response (77), and to identify residual or recurrent disease after chemoradiotherapy (78).
Standardization of spectral acquisition that takes into account (for example) metabolite relaxation times and field strengths should allow comparison of metabolites across studies (79); however, because of the variability of parameters, data are frequently presented as metabolite ratios. For example, by using the water signal as an internal reference, total choline levels have been quantitatively measured by using a hybrid time-domain and frequency-domain method for fitting (80). Ongoing efforts are evaluating the repeatability and reproducibility of the technology (70).
Macromolecular content
Tumors comprise a complex milieu of metabolites, proteins, and other large macromolecules. However, the majority of MRI techniques assay protons on free water molecules, and only probe these other tissue components indirectly through their impact on free water relaxation. Often these molecules are present at low concentration and/or their transverse relaxation is too rapid for direct detection. However, magnetization transfer (MT) and chemical exchange transfer (CEST) are two techniques that directly measure macromolecules by targeting their interaction with free water.
MT-MRI interrogates the macromolecular pool by measuring the transfer of energy from free water molecules to water that is bound to macromolecules following an off-resonance saturation pulse. MT can be quantified using a ratio of the signal intensity with and without MT saturation (dubbed the magnetization transfer ratio, MTR), or using quantitative MT (qMT) techniques which separate the MT effects from relaxation (81). An early oncological application performed MT-MRI of excised breast tissue to identify malignant tissue (82). The technique has since been performed in clinical studies, including the use of qMT to assess response of glioblastoma response to chemo-radiation (83). The repeatability and reproducibility of MT-MRI in tumors has not been established, but repeatability has been characterized in healthy breast tissue (84; see Figure 5) and multi-site reproducibility has been assessed in the brain (85). The biological sensitivity of MT-MRI in cancer has not been fully explored. Histological validation of MT in murine pancreatic xenografts demonstrated that MTR correlated with tumor fibrosis, suggesting that MT may be sensitive to tumor desmoplasia (86).
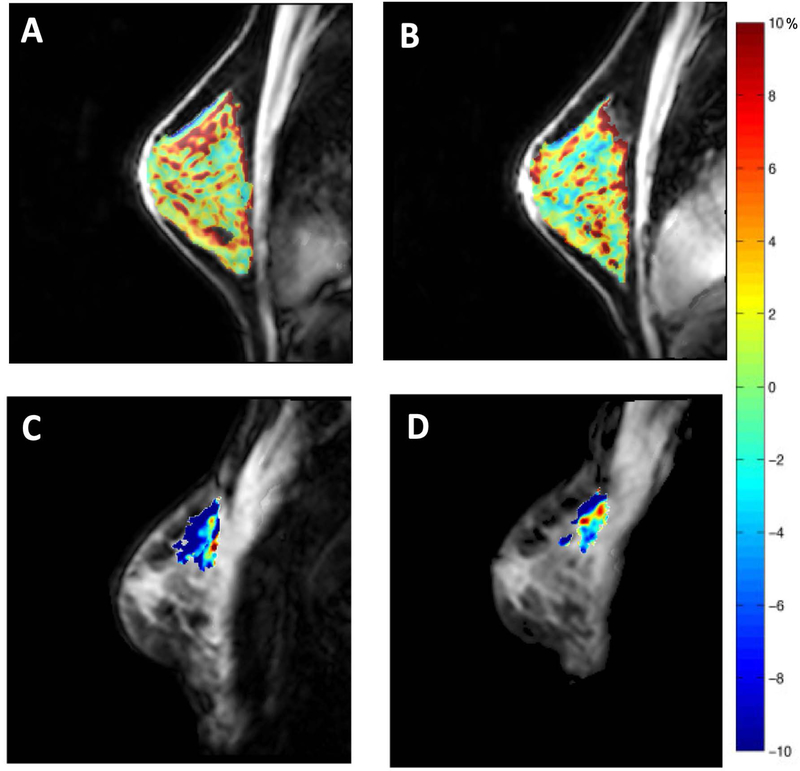
Figure 5.
Panels A and B display the repeatability of CEST measurements in the fibroglandular tissue of the healthy breast. Amide proton transfer residual maps are overlaid on corresponding anatomical images in the same subject before and after repositioning. Tumor amide proton transfer maps of a patient with breast cancer before (panel C) and after (panel D) one cycle of neoadjuvant chemotherapy. The patient achieved partial response to therapy.
While MT-MRI saturates a single frequency, CEST applies off-resonance saturation pulses at a range of frequencies and detects the physical exchange of saturated hydrogen protons with water. Initial applications of CEST to cancer examined amide groups, which may reflect protein concentration. Zhou, et al. were able to detect protein concentrations in the millimolar range in rat gliomas (87) with subsequent translation to imaging of human brain tumors (88). By tuning the saturation frequency to that of known molecules, CEST can also measure the concentration of glutamate, creatine, and lactate. The repeatability of CEST has been examined in healthy breast tissue (89) with repeatability similar to that seen in qMT. The biological correlates of CEST signal in tumors are still under investigation. One study of glioblastoma found that the amide signal from CEST corresponds with proliferation markers on histological sections (90).
pH
Physiological and pathological processes can alter pH homeostasis, resulting in transient or chronic acidic stress. In particular, solid tumors have an acidic extracellular pH that is related to aggressiveness (91); thus, there is a need for robust, reliable, and clinically relevant methods with which to measure these proton dynamics in vivo. The earliest measurements of pH relied on endogenous intracellular signals obtained with 31P MRS, which includes inorganic phosphate (Pi) that has a pH sensitive chemical shift with a pKa ~6.8. Intracellular pH has thus been measured in both animals (92) and humans (93) by single or multi-voxel 31P MRS. It is also possible to measure the extracellular (pHe) pH with 31P MRS in vivo using exogenous indicators, such as 3-aminopropyl phosphonate, and these studies were the first to document that the pHe of tumors was acidic (94). However, such measures have not been translated to the clinically.
While 13C DNP (see section 3.5) is primarily used to study the fates of 13C labeled metabolic substrates, it can also be used to measure tissue pH via polarized 13C-bicarbonate, which establishes a rapid equilibrium with aqueous 13CO2 that can be defined by a Henderson-Hasselbach relationship (95). In cardiac muscle, pH derived from 13C-bicarbonate showed high concordance to that obtained from 31P of Pi, suggesting that the values reflect primarily intracellular pH pHi (96).
Although 1H is very sensitive, it has a small chemical shift dispersion and the presence of a large 110 M signal from 1H2O necessitates sophisticated solvent suppression methods. Nonetheless, with care, moderately high-resolution (1 mm3) 1H MRS images can be obtained to measure pHi and pHe (intracellular and extracellular pH, respectively) in vivo. This was first accomplished using imidazoles in preclinical models (97) and has been extended to measure pHi in human brain following an oral dose of histidine (98). To measure extracellular pH, membrane-impermeant imidazoles have been developed (99) and used to generate the first extracellular pH image of a tumor (100).
Exchange processes are acid- or base-catalyzed and thus CEST imaging can be used to calculate pH in vivo (101; see Figure 6). Although CEST with exogenous contrast agents is offering new opportunities to measure pH in humans, it requires that high doses of contrast be used thereby fundamentally limiting this approach until this shortcoming can be solved. To improve sensitivity and specificity, more powerful CEST contrast agents that contain paramagnetic centers have been developed (PARAmagnetic Chemical Exchange Saturation Transfer; i.e., PARACEST). In this way, the frequency of the contrast agent can be far removed from the water resonance, thus reducing direct magnetization transfer to the water signal, and increasing the sensitivity and precision (102). A number of investigators have been focused on developing PARACEST agents to measure pH of tumors.
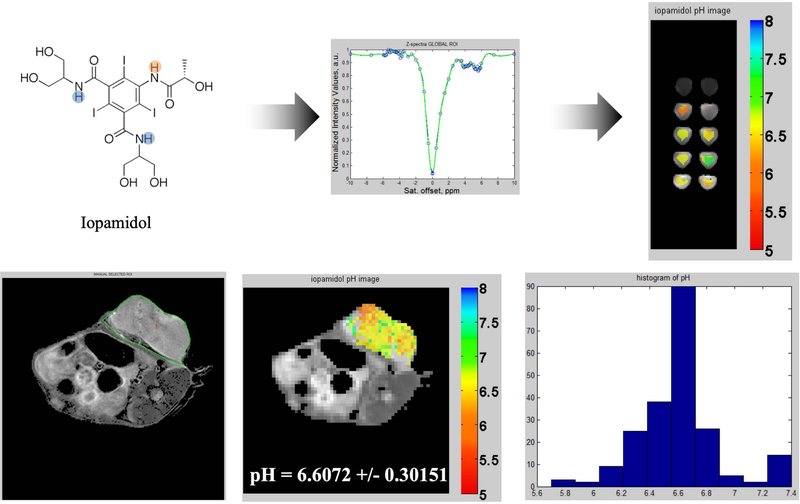
Figure 6.
The top left panel presents the Iopamidol molecule with two pools of 1H highlighted that have different sensitivities to pH. The top middle panel displays the calibration curve z-spectra (blue original, green after B0 correction); note the peaks at 4.2 and 5.5 ppm, corresponding to both pools of protons. The top right panel presents pH maps of 20 mM Iopamidol phantoms. In vivo results are demonstrated in the bottom row of figures via a T2-weighted image of a TUBO tumor in a BALB/c mouse (left panel), a CEST pH map of the tumor (middle panel), and a histogram of the pixels composing the pH map.
Where are we now?
The techniques presented above are at different stages in the translational process from pre-clinical development to clinical application. This section discusses logistical and infrastructure challenges that must be addressed when deploying advanced MRI methods into clinical trials; where appropriate, we share observations and recommendations from our own experience.
Logistical Challenges
Investigators seeking to test MRI methods in human subjects must surmount several logistical challenges, including meeting regulatory requirements, planning for data management and non-standardized acquisition and processing hurdles. To obtain approval from a local Institutional Review Board (IRB; i.e., the committee charged with ensuring that human studies comply with basic ethical principles underling biomedical research), investigators must submit a detailed research protocol describing all study procedures, subject eligibility, risks and benefits to participants, and plans for adverse event management. Examples of specific issues that might arise for an advanced MRI study include drafting an informed consent document describing risks and benefits to participants in lay language, establishing precise inclusion and exclusion criteria for subjects with potential MRI contraindications (e.g., implanted medical devices or borderline renal dysfunction, if subjects are to receive intravenous gadolinium-based contrasts agents), and determining protocols for disclosing incidental findings to participants. One particularly challenging issue involves compensating subjects for participation: IRBs typically look for such compensation to be set at a level whereby the subject is participating free from coercion or undue influence; i.e., any payment should be seen as offsetting the subject’s time and inconvenience rather than as a primary benefit of participation. However, federal regulations offer little guidance in this area and often the appropriate compensation is negotiated between the investigator and the IRB.
Translating preclinical imaging techniques to the clinic sometimes requires approval from specific health authorities, including the FDA. New MRI-based contrast agents for human use may require an Investigational New Drug (IND) application, and new MRI hardware (e.g., a novel RF coil) may be subject to Investigational Device Exemption (IDE) regulations. One particular consideration that may escape notice when translating preclinical MRI methods is the allowed specific absorption rate (SAR) on clinical scanners. To maintain MRI safety, SAR thresholds are set by the manufacturer and the scanners will not execute the pulse sequence if it exceeds those limits.
Adequate data management and storage plans should be developed prior to initiating any clinical imaging study. Specific aspects to consider include local versus cloud-based data storage, standards for image annotation and markup, de-identification requirements, and confidentiality and data security. Local storage is typically sufficient in a single-institution study, but a multicenter trial will require a storage mechanism that can be accessed by individuals at all participating sites, and certain funding mechanisms may also require sharing of de-identified image data. It should be noted that many email servers do not meet data security guidelines governed by the Health Insurance Portability and Accountability Act (HIPAA) for transmitting clinical research data. Most institutions require that investigators err on the side of caution when working with clinical imaging data that may contain patient identifiers. To ensure patient anonymity, a system of identifiers should be used rather than using the full patient name. Depending on the goals of the study, data can be either de-identified (i.e., data can be linked back to the patient using a key known only by the investigator or their designee) or fully anonymized data (i.e., data for which all identifiers are deleted).
For multicenter studies involving collection of data on multiple imaging platforms, data are typically shared via Digital Imaging and Communications in Medicine format (DICOM). However, image annotations, markup, and measurements are saved separately from the DICOMs, and therefore must be transmitted separately if needed for subsequent analyses. When working with advanced analyses of MRI data, clinical workstations may not provide the necessary tools for image analysis and a study-specific image annotation workflow must be designed.
Infrastructure Challenges
Robust mechanisms for screening, referring, consenting, and imaging patients must be established. Even with engaged clinical partners, we have found that a dedicated research nurse responsible for screening and consenting patients has been invaluable for accruing research subjects. Institutional support for a dedicated research nurse is by no means guaranteed, and budgeting for this critical resource should be considered when developing trial budgets.
When imaging trial participants, investigators must often determine whether to use clinical scanners or research-dedicated scanners. Clearly there are certain logistical advantages to obtaining research data as part of a standard-of-care clinical scan, especially if the clinical scan would have been obtained anyway. When attempting to combine research and clinical imaging, investigators must be cognizant of limitations imposed by the available hardware/software on clinical machines. To circumvent this obstacle, we have on occasion designed a research workflow within a standard-of-care exam where the necessary software was uploaded to the scanner to collect research data for each patient; it should be noted, however, that the additional data acquisition did increase the overall time of the clinical exam, and this strategy may not work in certain clinical environments. Alternatively, using research-dedicated scanners may allow investigators to develop and explore novel MRI techniques with fewer limitations associated with both scanner hardware/software and patient throughput demands. Conversely, research facilities may not be located near the clinic and patient transportation issues might limit accrual.
Finally, investigators seeking to deploy advanced MRI techniques within clinical trials must decide whether to develop an isolated and dedicated trial of the imaging technique or whether to “piggyback” an imaging study onto another clinical trial. We have had some success with the latter strategy, attaching evaluations of advanced MRI methods as optional (or in some cases mandatory) correlatives to parent trials of new cancer therapeutic agents. Advantages to this approach include lower costs and faster accrual. An important disadvantage, however, is a potential “two-variable” problem that may arise when evaluating a novel imaging technique for assessing response to novel therapy: if the drug itself has a low response rate, it may be difficult or impossible to assess the accuracy of the imaging technique for predicting response. Hence it is important to match the goal of the imaging correlative with the stage of the clinical trial: early-stage clinical trials are probably best suited for imaging correlatives designed to test feasibility or reproducibility, while later-stage clinical trials (with presumably higher likelihood of drug efficacy) are probably more appropriate for evaluations of predictive power for assessing response.
Where are we going?
Quantitively and specifically imaging the effects of immunotherapy
There are currently hundreds of ongoing clinical trials involving immunotherapy for the treatment of cancer; however, response patterns to these treatments vary greatly from traditional chemotherapeutics and radiation. This variation makes assessing--let alone predicting--response quite challenging. In particular, the Response Evaluation Criteria in Solid Tumors (RECIST; 103) may not be appropriate for characterizing the time-delayed responses frequently seen in patients receiving immunotherapy. Thus, a number of modified response criteria have recently been developed in an attempt to a capture so-called “pseudoprogression” (104); however, these criteria do require additional imaging time points. Furthermore, these modified response criteria are still based strictly on changes in tumor size and not the underlying biology. While there are efforts at using various tracers from positron emission tomography (PET) and different MRI techniques, none have consistently shown to be acceptable for characterizing the biological changes following immunotherapy. Preclinical studies are dominated by radiotracers that are only available at specialized centers, and MRI techniques that are fundamentally limited in their clinical application (e.g., injecting macrophages loaded with iron oxide particles). As new immunotherapies are being developed and show remarkable results in clinical trials (and therefore being fast-tracked into the standard-of-care), there is an immediate need to develop clinically relevant, highly specific imaging biomarkers to assess and predict response to immunotherapies.
Theranostics
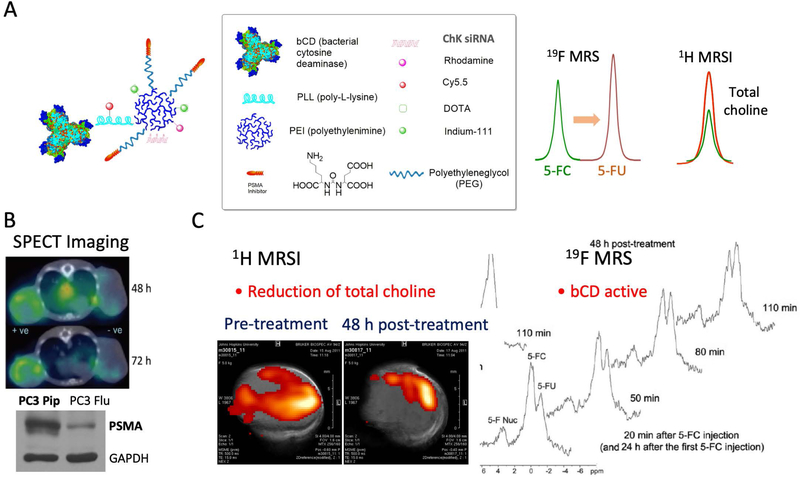
Metabolic characterization of cancers with MRS/I has created new opportunities for ‘metabolotheranostics’ where cancer-cell specific image-guided delivery of a theranostic probe targeting metabolism, and detection of the metabolic effects on the target enzyme can be combined (105). In this approach, cancer-specific aberrant metabolic pathways are detected by MRSI and corrected through the delivery of siRNA or cDNA or pharmacological agents under image-guidance. As an example of a metabolotheranostic, since choline kinase is significantly upregulated in aggressive cancer cells, siRNA to downregulate choline kinase was delivered by a nanoparticle targeted to prostate cancer cells using prostate specific membrane antigen (PSMA) (see Figure 7; (106)). Choline kinase downregulation was detected using 1H MRS and the nanoparticle was detected with SPECT imaging of 111In, since the sensitivity of MRI was not sufficient to detect the PSMA bound nanoparticle (105,107). Normalizing metabolic pathways with siRNA or cDNA delivering probes and detecting their metabolic outcome with MRS provides entirely new opportunities for ‘metabolotheranostic’ strategies, since any gene can be incorporated for cancer treatment strategies. Advances in 13C MRSI hyperpolarized probes (107), combined with image-guided cancer specific delivery of metabolic targeting molecular reagents or pharmacological agents will significantly expand the scope of metabolotheranostic strategies.
Figure 7.
Panel A presents the structure of a PSMA theranostic agent that carries a prodrug enzyme to convert 5-fluorocytosine (5-FC) to 5-fluorouracil (5-FU) that is detected by 19F MRS and siRNA to downregulate choline kinase, thereby resulting in a decrease of total choline detected by 1H MRSI. Panel B shows increased retention of the theranostic agent in a PSMA expressing tumor compared to a non-PSMA expressing tumor. Panel C indicates functional changes in tumor metabolism detected by 1H MRSI and the formation of the cytotoxic drug 5-FU from 5-FC in the tumor detected by 19F MRS. Adapted from (103).
Advanced Analyses
As discussed above, there are an extraordinary number of imaging parameters and techniques that can be used to investigate tumor biology at the region of interest and voxel levels. The quantity and types of measurements on different features can be challenging to analyze and interpret to assess or predict response to therapies. Recently, advanced analysis techniques such as radiomics and imaged-based, mechanistic modeling have been developed which can leverage these large data sets to potentially support clinical decision making.
In radiomics (108), imaging data is converted to higher-dimensional data and analyzed statistically to identify relationships between parameter combinations and treatment outcomes, or to extract unique sub-volumes within tumors (also known as habitats). For example, in oncology, these identified habitats could represent physiologically distinct volumes with variations in cell density, vascularity, necrosis, or extent of edema which could be used to differentiate disease aggressiveness or rate of tumor progression (108). One seminal example by Hawkins et al (109) investigated the use of radiomic techniques to predict malignant lung nodules from screening CT scans. Using 23 stable imaging features, their statistical model could predict nodules which would become cancerous within two years with an accuracy of 79% (area under the curve = 0.75) outperforming the standardized Lung Imaging Reporting and Data System and volume-only approaches.
Imaged-based, mechanistic modeling (110) aims to integrate biophysical models with temporally and spatially resolved, patient-specific data to make patient-specific predictions of tumor growth and treatment response without the need of large training data set. One pre-clinical example by Hormuth et al (111) utilizes cellularity estimates from diffusion weighted MRI acquired in a murine model of glioma before and shortly after single fraction radiation therapy to calibrate an animal-specific model parameters. The animal specific parameters describing tumor cell proliferation, diffusion, and response to radiation therapy, were then used to predict future tumor cell distribution with an error of approximately 10% in tumor volume predictions. Weis et al (112) utilized a similar approach to calibrate patient-specific model parameters for breast cancer patients receiving neoadjuvant therapy. Using one pre- and one post-treatment image, Weis et al was able to more accurately predict complete pathological response as assessed at surgery (area under the curve = 0.87) compared to the standard RECIST criteria approach (area under the curve = 0.71). These promising pre-clinical and clinical studies demonstrate the potential utility of using mechanism based models initialized and calibrated from an individual’s own imaging data predict response to therapy.
From the Ivory Tower to Main street
Even after the technical merits of an MRI method have been demonstrated in the clinical setting, there remain logistical hurdles to integrate it into standard-of-care imaging. The path to widespread clinical dissemination is epitomized by diffusion MRI which was first described in the 1960s, but only recently integrated into standard-of-care imaging for human solid tumors. There exist both hardware and software limitations, the latter of which includes implementing non-standard pulse sequences into FDA-regulated software as well as development of specialized processing tools. There is also a need for additional training, both for the technologist acquiring the image and the radiologist reading the image. As both the image acquisition and read require extra time they incur additional cost. Covering this cost often requires lobbying insurance companies and health payers to justify the additional expense of a new technique. There are a number of techniques under development to speed image acquisition and thus keep costs down. These include hardware advances, progress in parallel imaging techniques, and the development of abbreviated protocols (113). Even after the scientific validity of an MRI technique has been established in human studies, the path from imaging biomarker discovery to widespread clinical implementation requires satisfying of a complex set of requirements (2).
Conclusion
We have presented the salient features of a range of commonly used MR-based imaging techniques that are appropriate for interrogating different features of tumors in the clinical setting. Additionally, we have highlighted several important, but less frequently discussed, challenges related to MRI biomarkers; issues related to repeatability, reproducibility, scientific (technical) obstacles in moving MRI measurements from the bench to the clinic, as well as challenges related to logistics and infrastructure. Finally, we presented three short vignettes of fundamental issues representing areas of critical need. We hope that this contribution both introduces the novice to the field, as well as encourages established investigators to engage in the development of imaging biomarkers for cancer.
Acknowledgments
Grant Support. We thank the National Institutes of Health for funding through R01CA142565, R01CA186193, U01174706, K25CA168936, R01CA158079, K25CA204599, R01CA109106, R35CA209960, P30CA006973 and R01CA193365. We thank the American Cancer Society for RSG-18–006-01-CCE, and the Cancer Prevention and Research Institute of Texas for RR160005, RP140285, and RP120670-P3.
References
- 1.Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89–95. [DOI] [PubMed] [Google Scholar]
- 2.O’Connor JP, Aboagye EO, Adams JE, Aerts HJ, Barrington SF, Beer AJ, Boellaard R, Bohndiek SE, Brady M, Brown G, Buckley DL, Chenevert TL, Clarke LP, Collette S, Cook GJ, deSouza NM, Dickson JC, Dive C, Evelhoch JL, Faivre-Finn C, Gallagher FA, Gilbert FJ, Gillies RJ, Goh V, Griffiths JR, Groves AM, Halligan S, Harris AL, Hawkes DJ, Hoekstra OS, Huang EP, Hutton BF, Jackson EF, Jayson GC, Jones A, Koh DM, Lacombe D, Lambin P27, Lassau N, Leach MO, Lee TY, Leen EL, Lewis JS, Liu Y, Lythgoe MF, Manoharan P, Maxwell RJ, Miles KA, Morgan B, Morris S, Ng T, Padhani AR, Parker GJ, Partridge M, Pathak AP, Peet AC, Punwani S, Reynolds AR, Robinson SP, Shankar LK, Sharma RA, Soloviev D, Stroobants S, Sullivan DC, Taylor SA, Tofts PS, Tozer GM, van Herk M, Walker-Samuel S, Wason J, Williams KJ, Workman P, Yankeelov TE, Brindle KM, McShane LM, Jackson A, Waterton JC. Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol. 2017;14:169–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Damadian R Tumor detection by nuclear magnetic resonance. Science 1971;171:1151–1153. [DOI] [PubMed] [Google Scholar]
- 4.Hazelwood CF, Chang DC, Medina D, Cleveland G, Nichols BL. Distinction between the preneoplastic and neoplastic state of murine mammary glands. Proceedings of the National Academy of Sciences of the United States of America 1972;69:1478–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kessler LG, Barnhart HX, Buckler AJ, Choudhury KR, Kondratovich MV, Toledano A, et al. The emerging science of quantitative imaging biomarkers terminology and definitions for scientific studies and regulatory submissions. Stat Methods Med Res. 2015;24:9–26. [DOI] [PubMed] [Google Scholar]
- 6.Hygino da Cruz LC Jr., Rodriguez I, Domingues RC, Gasparetto EL, Sorensen AG Pseudoprogression and pseudoresponse: imaging challenges in the assessment of posttreatment glioma. AJNR Am J Neuroradiol. 2011;32:1978–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hildebrandt IJ, Su H, Weber WA. Anesthesia and Other Considerations for in Vivo Imaging of Small Animals. ILAR J. 2008;49:17–26. [DOI] [PubMed] [Google Scholar]
- 8.Kraff O, Quick HH. 7T: Physics, safety, and potential clinical applications. J Magn Reson Imaging. 2017;46:1573–89. [DOI] [PubMed] [Google Scholar]
- 9.Yankeelov TE, Mankoff DA, Schwartz LH, Lieberman FS, Buatti JM, Mountz JM, et al. Quantitative Imaging in Cancer Clinical Trials. Clin Cancer Res. 2016;22:284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sullivan DC, Obuchowski NA, Kessler LG, et al. Metrology Standards for Quantitative Imaging Biomarkers. Radiology. 2015;277:813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quarles CC, Schmainda KM: Assessment of the morphological and functional effects of the anti-angiogenic agent SU11657 on 9L gliosarcoma vasculature using dynamic susceptibility contrast MRI. Magn Reson Med 2007;57:680–687. [DOI] [PubMed] [Google Scholar]
- 12.Whisenant JG, Sorace AG, McIntyre JO, et al. Evaluating treatment response using DW-MRI and DCE-MRI in trastuzumab responsive and resistant HER2-overexpressing human breast cancer xenografts. Transl Oncol 2014;7:768–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson SP, Ramasawmy R, Campbell-Washburn AE, et al. Acute changes in liver tumour perfusion measured non-invasively with arterial spin labelling. Br J Cancer 2016;114:897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knutsson L, Börjesson S, Larsson E-M, et al. Absolute quantification of cerebral blood flow in normal volunteers: correlation between Xe-133 SPECT and dynamic susceptibility contrast MRI. J Magn Reson Imaging 2007;26:913–920. [DOI] [PubMed] [Google Scholar]
- 15.Pathak AP, Schmainda KM, Ward BD, Linderman JR, Rebro KJ, Greene AS: MR-derived cerebral blood volume maps: issues regarding histological validation and assessment of tumor angiogenesis. Magn Reson Med 2001;46:735–747. [DOI] [PubMed] [Google Scholar]
- 16.Semmineh NB, Stokes AM, Bell LC, Boxerman JL, Quarles CC: A Population-Based Digital Reference Object (DRO) for Optimizing Dynamic Susceptibility Contrast (DSC)-MRI Methods for Clinical Trials. Tomography 2017;3:41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim H, Mousa M, Schexnailder P, et al. Portable perfusion phantom for quantitative DCE-MRI of the abdomen. Med Phys 2017;44:5198–5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng CS, Waterton JC, Kundra V, et al. Reproducibility and comparison of DCE-MRI and DCE-CT perfusion parameters in a rat tumor model. Technol Cancer Res Treat 2012;11:279–88. [DOI] [PubMed] [Google Scholar]
- 19.Fan AP, Jahanian H, Holdsworth SJ, Zaharchuk G. Comparison of cerebral blood flow measurement with [15O]-water positron emission tomography and arterial spin labeling magnetic resonance imaging: A systematic review. J Cereb Blood Flow Metab 2016;36:842–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alsop DC, Detre JA, Golay X, et al. Recommended implementation of arterial spin-labeled Perfusion mri for clinical applications: A consensus of the ISMRM Perfusion Study group and the European consortium for ASL in dementia. Magn Reson Med 2015;73:102–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welker K, Boxerman J, Kalnin A, et al. ASFNR Recommendations for Clinical Performance of MR Dynamic Susceptibility Contrast Perfusion Imaging of the Brain. Am J Neuroradiol 2015;36:E41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DCE MRI Technical Committee. DCE MRI Quantification Profile, Quantitative Imaging Biomarkers Alliance. Version 1.0. Reviewed Draft. QIBA, July 1, 2012. Available from: http://rsna.org/QIBA.aspx
- 23.Petersen ET, Mouridsen K, Golay X. The QUASAR reproducibility study, Part II: Results from a multi-center Arterial Spin Labeling test-retest study. Neuroimage 2010;49:104–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bane O, Hectors SJ, Wagner M, et al. Accuracy, repeatability, and interplatform reproducibility of T1quantification methods used for DCE-MRI: Results from a multicenter phantom study. Magn Reson Med 2018;79:2564–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prah MA, Stufflebeam SM, Paulson ES, et al. Repeatability of Standardized and Normalized Relative CBV in Patients with Newly Diagnosed Glioblastoma. Am J Neuroradiol 2015;36:1654–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Padhani AR, Liu G, Koh DM, Chenevert TL, Thoeny HC, Takahara T, Dzik-Jurasz A, Ross BD, Van Cauteren M, Collins D, Hammoud DA, Rustin GJ, Taouli B, Choyke PL. Diffusion-weighted magnetic resonance imaging as a cancer biomarker: consensus and recommendations. Neoplasia. 2009;11:102–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chenevert TL, McKeever PE, Ross BD. Monitoring early response of experimental brain tumors to therapy using diffusion magnetic resonance imaging. Clinical Cancer Research 1997;3:1457–1466. [PubMed] [Google Scholar]
- 28.Padhani AR, Liu G, Mu-Koh D, et al. Diffusion-weighted magnetic resonance imaging as a cancer biomarker: consensus and recommendations. Neoplasia 2009;11:102–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kono K, Inoue Y, Nakayama K, et al. The role of diffusion-weighted imaging in patients with brain tumors. American journal of neuroradiology 2001;22:1081–1088. [PMC free article] [PubMed] [Google Scholar]
- 30.Curvo‐Semedo L, Lambregts DM, Maas M, Beets GL, Caseiro‐Alves F, Beets‐Tan RG. Diffusion‐weighted MRI in rectal cancer: Apparent diffusion coefficient as a potential noninvasive marker of tumor aggressiveness. Journal of Magnetic Resonance Imaging 2012;35:1365–1371. [DOI] [PubMed] [Google Scholar]
- 31.Li X, Abramson RG, Arlinghaus LR, Kang H, Chakravarthy AB, Abramson VG, Farley J, Mayer IA, Kelley MC, Meszoely IM, Means-Powell J, Grau AM, Sanders M, Yankeelov TE. Multiparametric magnetic resonance imaging for predicting pathological response after the first cycle of neoadjuvant chemotherapy in breast cancer. Invest Radiol. 2015;50:195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dzik-Jurasz A, Domenig C, George M, et al. Diffusion MRI for prediction of response of rectal cancer to chemoradiation. The Lancet 2002;360:307–308. [DOI] [PubMed] [Google Scholar]
- 33.Barentsz JO, Weinreb JC, Verma S, et al. Synopsis of the PI-RADS v2 guidelines for multiparametric prostate magnetic resonance imaging and recommendations for use. European urology 2016;69:41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchell DG, Bruix J, Sherman M, Sirlin CB. LI‐RADS (Liver Imaging Reporting and Data System): Summary, discussion, and consensus of the LI‐RADS Management Working Group and future directions. Hepatology 2015;61:1056–1065. [DOI] [PubMed] [Google Scholar]
- 35.Partridge S, Zhang Z, Rahbar H, Chenevert T, Kitsch A, Hanna L, Harvey S, Moy L, Demartini W, Schnall M, Lehman C, Comstock C, ACRIN 6702 Trial: A Multi-Center Study Evaluating the Utility of Diffusion Weighted Imaging for Detection and Diagnosis of Breast Cancer. Radiological Society of North America 2017 Scientific Assembly and Annual Meeting, November 26 - December 1, 2017, Chicago IL. [Google Scholar]
- 36.Holdsworth SJ, Skare S, Newbould RD, Guzmann R, Blevins NH, Bammer R. Readout-segmented EPI for rapid high resolution diffusion imaging at 3T. European journal of radiology 2008;65:36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wheeler-Kingshott CA, Hickman SJ, Parker GJ, Ciccarelli O, Symms MR, Miller DH, Barker GJ. Investigating cervical spinal cord structure using axial diffusion tensor imaging. Neuroimage 2002;16:93–102. [DOI] [PubMed] [Google Scholar]
- 38.Feinberg DA, Setsompop K. Ultra-fast MRI of the human brain with simultaneous multi-slice imaging. Journal of magnetic resonance 2013;229:90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malyarenko D, Galbán CJ, Londy FJ, et al. Multi‐system repeatability and reproducibility of apparent diffusion coefficient measurement using an ice‐water phantom. Journal of Magnetic Resonance Imaging 2013;37:1238–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang X, Li H, Xie J, Zhao P, Gore JC, Xu J. Quantification of cell size using temporal diffusion spectroscopy. Magn Reson Med 2016;75:1076–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mulkern RV, Haker SJ, Maier SE. On high b diffusion imaging in the human brain: ruminations and experimental insights. Magn Reson Imaging 2009;27:1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jensen JH, Helpern JA, Ramani A, Lu H, Kaczynski K. Diffusional kurtosis imaging: the quantification of non‐gaussian water diffusion by means of magnetic resonance imaging. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 2005;53:1432–1440. [DOI] [PubMed] [Google Scholar]
- 43.Bennett KM, Schmainda KM, Bennett RT, Rowe DB, Lu H, Hyde JS. Characterization of continuously distributed cortical water diffusion rates with a stretched-exponential model. Magn Reson Med 2003;50:727–734. [DOI] [PubMed] [Google Scholar]
- 44.White NS, McDonald C, Farid N, Kuperman J, Karow D, Schenker-Ahmed NM, Bartsch H, Rakow-Penner R, Holland D, Shabaik A, Bjornerud A, Hope T, Hattangadi-Gluth J, Liss M, Parsons JK, Chen CC, Raman S, Margolis D, Reiter RE, Marks L, Kesari S, Mundt AJ, Kane CJ, Carter BS, Bradley WG, Dale AM. Diffusion-weighted imaging in cancer: physical foundations and applications of restriction spectrum imaging. Cancer Res 2014;74:4638–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang X, Li H, Xie J, Zhao P, Gore JC, Xu J. Quantification of cell size using temporal diffusion spectroscopy. Magn Reson Med 2016;75:1076–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Panagiotaki E, Walker-Samuel S, Siow B, Johnson SP, Rajkumar V, Pedley RB, Lythgoe MF, Alexander DC. Noninvasive quantification of solid tumor microstructure using VERDICT MRI. Cancer Res 2014;74:1902–1912. [DOI] [PubMed] [Google Scholar]
- 47.Thulborn KR, Waterton JC, Matthews PM, and Radda GK Oxygenation Dependence of the Transverse Relaxation-Time of Water Protons in Whole-Blood at High-Field. Biochim. Biophys. Acta 1982;714:265–270. [DOI] [PubMed] [Google Scholar]
- 48.Hallac RR, Zhou H, Pidikiti R, Song K, Stojadinovic S, Zhao D, Solberg T, Peschke P, and Mason RP Correlations of noninvasive BOLD and TOLD MRI with pO2 and relevance to tumor radiation response. Magn. Reson. Med. 2014;71:1863–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodrigues LM, Howe FA, Griffiths JR, and Robinson SP Tumor R-2 * is a prognostic indicator of acute radiotherapeutic response in rodent tumors. J. Magn. Reson. Imaging 2004;19: 482–488. [DOI] [PubMed] [Google Scholar]
- 50.Matsumoto K, Bernardo M, Subramanian S, Choyke P, Mitchell JB, Krishna MC, and Lizak MJ MR assessment of changes of tumor in response to hyperbaric oxygen treatment. Magn. Reson. Med. 2006;56:240–246. [DOI] [PubMed] [Google Scholar]
- 51.O’Connor JPB, Boult JKR, Jamin Y, Babur M, Finegan KG, Williams KJ, Little RA, Jackson A, Parker GJM, Reynolds AR, Waterton JC, and Robinson SP Oxygen-Enhanced MRI Accurately Identifies, Quantifies, and Maps Tumor Hypoxia in Preclinical Cancer Models. Cancer Res. 2016;76:787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang L, Zhao D, Constantinescu A, and Mason RP Comparison of BOLD contrast and Gd-DTPA Dynamic Contrast Enhanced imaging in rat prostate tumor. Magn. Reson. Med. 2004;51:953–960. [DOI] [PubMed] [Google Scholar]
- 53.Rakow-Penner R, Daniel B, and Glover GH Detecting Blood Oxygen Level-Dependent (BOLD) Contrast in the Breast. J. Magn. Reson. Imaging 2010;32:120–129. [DOI] [PubMed] [Google Scholar]
- 54.Zhou H, Zhang Z, Denney R, Williams JS, Gerberich J, Stojadinovic S, Saha D, Shelton JM, and Mason RP Tumor physiological changes during hypofractionated stereotactic body radiation therapy assessed using multi-parametric magnetic resonance imaging. Oncotarget 2017;8:37464–37477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beeman SC, Shui Y-B, Perez-Torres CJ, Engelbach JA, Ackerman JJH, and Garbow JR O2-sensitive MRI distinguishes brain tumor versus radiation necrosis in murine models. Magn. Reson. Med. 2015;75:2442–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Remmele s., Mason r. P., and O’connor j. B. P. (2014) MRI hypoxia measurements. In Functional Imaging in Oncology (Luna A, ed), Springer-Verlag, Heidelberg. [Google Scholar]
- 57.Zhou HL, Hallac RR, Yuan Q, Ding Y, Zhang ZW, Xie XJ, Francis F, Roehrborn CG, Sims RD, Costa DN, Raj GV, and Mason RP Incorporating Oxygen-Enhanced MRI into Multi-Parametric Assessment of Human Prostate Cancer. Diagnostics 2017;7:E48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Glaser KJ, Manduca A, Ehman RL. Review of MR elastography applications and recent developments. J Magn Reson Imaging. 2012;36:757–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jamin Y, Boult JKR, Li J, Popov S, Garteiser P, Ulloa JL, Cummings C, Box G, Eccles SA, Jones C, Waterton JC, Bamber JC, Sinkus R, Robinson SP. Exploring the biomechanical properties of brain malignancies and their pathologic determinants in vivo with magnetic resonance elastography. Cancer Res. 2015;75:1216–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schregel K, Nazari N, Nowicki MO, Palotai M, Lawler SE, Sinkus R, Barbone PE, Patz S. Characterization of glioblastoma in an orthotopic mouse model with magnetic resonance elastography. NMR Biomed. 2018;31:e3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li J, Jamin Y, Boult JK, Cummings C, Waterton JC, Ulloa J, Sinkus R, Bamber JC, Robinson SP. Tumour biomechanical response to the vascular disrupting agent ZD6126 in vivo assessed by magnetic resonance elastography. Br J Cancer. 2014;110:1727–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weis JA, Flint KM, Sanchez V, Yankeelov TE, Miga MI. Assessing the accuracy and reproducibility of modality independent elastography in a murine model of breast cancer. J Med Imaging (Bellingham). 2015;2:036001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sahebjavaher RS, Nir G, Honarvar M, Gagnon LO, Ischia J, Jones EC, Chang SD, Fazli L, Goldenberg SL, Rohling R, Kozlowski P, Sinkus R, Salcudean SE. MR elastography of prostate cancer: quantitative comparison with histopathology and repeatability of methods. NMR Biomed. 2015;28:124–39. [DOI] [PubMed] [Google Scholar]
- 64.Pepin KM, McGee KP, Arani A, Lake DS, Glaser KJ, Manduca A, Parney IF, Ehman RL, Huston J 3rd. MR Elastography Analysis of Glioma Stiffness and IDH1-Mutation Status. AJNR Am J Neuroradiol. 2018;39:31–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shi Y, Gao F, Li Y, Tao S, Yu B, Liu Z, Liu Y, Glaser KJ, Ehman RL, Guo Q. Differentiation of benign and malignant solid pancreatic masses using magnetic resonance elastography with spin-echo echo planar imaging and three-dimensional inversion reconstruction: a prospective study. Eur Radiol. 2018;28:936–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gordic S, Ayache JB, Kennedy P, Besa C, Wagner M, Bane O, Ehman RL, Kim E, Taouli B. Value of tumor stiffness measured with MR elastography for assessment of response of hepatocellular carcinoma to locoregional therapy. Abdom Radiol (NY). 2017;42:1685–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yin M, Glaser KJ, Talwalkar JA, Chen J, Manduca A, Ehman RL. Hepatic MR Elastography: Clinical Performance in a Series of 1377 Consecutive Examinations. Radiology. 2016;278:114–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. The Journal of general physiology. 1927;8:519–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Negendank W Studies of human tumors by MRS: a review. NMR Biomed. 1992;5:303–24. [DOI] [PubMed] [Google Scholar]
- 70.Zaccagna F, Grist JT, Deen SS, Woitek R, Lechermann LM, McLean MA, Basu B, Gallagher FA. Hyperpolarized carbon-13 magnetic resonance spectroscopic imaging: a clinical tool for studying tumour metabolism. Br J Radiol. 2018;91:20170688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Howe FA, Barton SJ, Cudlip SA, Stubbs M, Saunders DE, Murphy M, Wilkins P, Opstad KS, Doyle VL, McLean MA, Bell BA, Griffiths JR. Metabolic profiles of human brain tumors using quantitative in vivo 1H magnetic resonance spectroscopy. Magn Reson Med. 2003;49:223–32. [DOI] [PubMed] [Google Scholar]
- 72.Osorio JA, Ozturk-Isik E, Xu D, Cha S, Chang S, Berger MS, Vigneron DB, Nelson SJ. 3D 1H MRSI of brain tumors at 3.0 Tesla using an eight-channel phased-array head coil. J Magn Reson Imaging. 2007;26:23–30. [DOI] [PubMed] [Google Scholar]
- 73.Gribbestad IS, Singstad TE, Nilsen G, Fjosne HE, Engan T, Haugen OA, Rinck PA. In vivo 1H MRS of normal breast and breast tumors using a dedicated double breast coil. J Magn Reson Imaging. 1998;8:1191–7. [DOI] [PubMed] [Google Scholar]
- 74.Mukherji SK, Schiro S, Castillo M, Kwock L, Muller KE, Blackstock W. Proton MR spectroscopy of squamous cell carcinoma of the extracranial head and neck: in vitro and in vivo studies. AJNR Am J Neuroradiol. 1997;18:1057–72. [PMC free article] [PubMed] [Google Scholar]
- 75.McLean MA, Priest AN, Joubert I, Lomas DJ, Kataoka MY, Earl H, Crawford R, Brenton JD, Griffiths JR, Sala E. Metabolic characterization of primary and metastatic ovarian cancer by 1H-MRS in vivo at 3T. Magn Reson Med. 2009;62:855–61. [DOI] [PubMed] [Google Scholar]
- 76.Nelson SJ, Graves E, Pirzkall A, Li X, Antiniw Chan A, Vigneron DB, McKnight TR. In vivo molecular imaging for planning radiation therapy of gliomas: an application of 1H MRSI. J Magn Reson Imaging. 2002;16:464–76. [DOI] [PubMed] [Google Scholar]
- 77.Saraswathy S, Crawford FW, Lamborn KR, Pirzkall A, Chang S, Cha S, Nelson SJ. Evaluation of MR markers that predict survival in patients with newly diagnosed GBM prior to adjuvant therapy. J Neurooncol. 2009;91:69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.King AD, Yeung DK, Yu KH, Mo FK, Hu CW, Bhatia KS, Tse GM, Vlantis AC, Wong JK, Ahuja AT. Monitoring of treatment response after chemoradiotherapy for head and neck cancer using in vivo (1)H MR spectroscopy. Eur Radiol. 2010;20:165–72. [DOI] [PubMed] [Google Scholar]
- 79.Winfield JM, Payne GS, Weller A, deSouza NM. DCE-MRI, DW-MRI, and MRS in Cancer: Challenges and Advantages of Implementing Qualitative and Quantitative Multi-parametric Imaging in the Clinic. Topics in magnetic resonance imaging: TMRI. 2016;25:245–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bolan PJ, Meisamy S, Baker EH, Lin J, Emory T, Nelson M, Everson LI, Yee D, Garwood M. In vivo quantification of choline compounds in the breast with 1H MR spectroscopy. Magn Reson Med. 2003;50:1134–43. [DOI] [PubMed] [Google Scholar]
- 81.Henkelman RM, Huang X, Xiang QS, Stanisz GJ, Swanson SD, Bronskill MJ. Quantitative interpretation of magnetization transfer. Magn Reson Med. 1993;29:759–66. [DOI] [PubMed] [Google Scholar]
- 82.Callicott C, Thomas JM, Goode AW. The magnetization transfer characteristics of human breast tissues: an in vitro NMR study. Phys Med Biol. 1999;44:1147–54. [DOI] [PubMed] [Google Scholar]
- 83.Mehrabian H, Myrehaug S, Soliman H, Sahgal A, Stanisz GJ. Quantitative Magnetization Transfer in Monitoring Glioblastoma (GBM) Response to Therapy. Sci Rep. 2018;8:2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Arlinghaus LR, Dortch RD, Whisenant JG, Kang H, Abramson RG, Yankeelov TE. Quantitative Magnetization Transfer Imaging of the Breast at 3.0 T: Reproducibility in Healthy Volunteers. Tomography. 2016;2:260–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Berry I, Barker GJ, Barkhof F, Campi A, Dousset V, Franconi JM, et al. A multicenter measurement of magnetization transfer ratio in normal white matter. J Magn Reson Imaging. 1999;9:441–6. [DOI] [PubMed] [Google Scholar]
- 86.Li W, Zhang Z, Nicolai J, Yang GY, Omary RA, Larson AC. Magnetization transfer MRI in pancreatic cancer xenograft models. Magn Reson Med. 2012;68:1291–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou J, Lal B, Wilson DA, Laterra J, van Zijl PC. Amide proton transfer (APT) contrast for imaging of brain tumors. Magn Reson Med. 2003;50:1120–6. [DOI] [PubMed] [Google Scholar]
- 88.Zhou J, Blakeley JO, Hua J, Kim M, Laterra J, Pomper MG, et al. Practical data acquisition method for human brain tumor amide proton transfer (APT) imaging. Magn Reson Med. 2008;60:842–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dula AN, Arlinghaus LR, Dortch RD, Dewey BE, Whisenant JG, Ayers GD, et al. Amide proton transfer imaging of the breast at 3 T: establishing reproducibility and possible feasibility assessing chemotherapy response. Magn Reson Med. 2013;70:216–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sagiyama K, Mashimo T, Togao O, Vemireddy V, Hatanpaa KJ, Maher EA, et al. In vivo chemical exchange saturation transfer imaging allows early detection of a therapeutic response in glioblastoma. Proc Natl Acad Sci U S A. 2014;111:4542–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Swietach P, Vaughan-Jones RD, Harris AL & Hulikova A The chemistry, physiology and pathology of pH in cancer. Philosophical transactions of the Royal Society of London. Series B, Biological sciences 2014;369:20130099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bhujwalla ZM, et al. Combined vascular and extracellular pH imaging of solid tumors. NMR Biomed 2002;15:114–119. [DOI] [PubMed] [Google Scholar]
- 93.Rata M, Giles SL, deSouza NM, Leach MO & Payne GS Comparison of three reference methods for the measurement of intracellular pH using 31P MRS in healthy volunteers and patients with lymphoma. NMR in biomedicine 2014;27:158–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gillies RJ, Liu Z & Bhujwalla Z 31P-MRS measurements of extracellular pH of tumors using 3-aminopropylphosphonate. Am J Physiol 1994;267:C195–203. [DOI] [PubMed] [Google Scholar]
- 95.Gallagher FA, Kettunen MI, Day SE, Hu DE, Ardenkjaer-Larsen JH, Zandt Ri, Jensen PR, Karlsson M, Golman K, Lerche MH, Brindle KM Magnetic resonance imaging of pH in vivo using hyperpolarized 13C-labelled bicarbonate. Nature 2008;453:940–943. [DOI] [PubMed] [Google Scholar]
- 96.Schroeder MA, et al. Measuring intracellular pH in the heart using hyperpolarized carbon dioxide and bicarbonate: a 13C and 31P magnetic resonance spectroscopy study. Cardiovasc Res 2010;86:82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rabenstein DL & Isab AA Determination of the intracellular pH of intact erythrocytes by 1H NMR spectroscopy. Analytical biochemistry 1982;121:423–432. [DOI] [PubMed] [Google Scholar]
- 98.Vermathen P, Capizzano AA & Maudsley AA Administration and (1)H MRS detection of histidine in human brain: application to in vivo pH measurement. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine 2000;43:665–675. [DOI] [PubMed] [Google Scholar]
- 99.Gil S, Zaderenzo P, Cruz F, Cerdan S & Ballesteros P Imidazol-1-ylalkanoic acids as extrinsic 1H NMR probes for the determination of intracellular pH, extracellular pH and cell volume. Bioorganic & medicinal chemistry 1994;2:305–314. [DOI] [PubMed] [Google Scholar]
- 100.van Sluis R Bhujwalla ZM, Raghunand N, Ballesteros P, Alvarez J, Cerdán S, Galons JP, Gillies RJ In vivo imaging of extracellular pH using 1H MRSI. Magn Reson Med 1999;41:743–750. [DOI] [PubMed] [Google Scholar]
- 101.Ward KM & Balaban RS Determination of pH using water protons and chemical exchange dependent saturation transfer (CEST). Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine 2000;44:799–802. [DOI] [PubMed] [Google Scholar]
- 102.Woods M, Woessner DE & Sherry AD Paramagnetic lanthanide complexes as PARACEST agents for medical imaging. Chemical Society reviews 2006;35:500–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schwartz LH, Litière S, de Vries E, Ford R, Gwyther S, Mandrekar S, Shankar L, Bogaerts J, Chen A, Dancey J, Hayes W, Hodi FS, Hoekstra OS, Huang EP, Lin N, Liu Y, Therasse P, Wolchok JD, Seymour L. RECIST 1.1-Update and clarification: From the RECIST committee. Eur J Cancer. 2016;62:132–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Seymour L, Bogaerts J, Perrone A, Ford R, Schwartz LH, Mandrekar S, Lin NU, Litière S, Dancey J, Chen A, Hodi FS, Therasse P, Hoekstra OS, Shankar LK, Wolchok JD, Ballinger M, Caramella C, de Vries EG; RECIST working group. iRECIST: guidelines for response criteria for use in trials testing immunotherapeutics. Lancet Oncol. 2017;18:e143–e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bhujwalla ZM, Kakkad S, Chen Z, Jin J, Hapuarachchige S, Artemov D, Penet MF. Theranostics and metabolotheranostics for precision medicine in oncology. Journal of magnetic resonance. 2018;291:141–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen Z, Penet MF, Nimmagadda S, Li C, Banerjee SR, Winnard PT Jr., Artemov D, Glunde K, Pomper MG, Bhujwalla ZM PSMA-targeted theranostic nanoplex for prostate cancer therapy. ACS nano. 2012;6:7752–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gallagher FA, Kettunen MI, Day SE, Hu DE, Ardenkjaer-Larsen JH, Zandt R, Jensen PR, Karlsson M, Golman K, Lerche MH, Brindle KM. Magnetic resonance imaging of pH in vivo using hyperpolarized 13C-labelled bicarbonate. Nature. 2008;453:940–3. [DOI] [PubMed] [Google Scholar]
- 108.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278:563–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hawkins S, Wang H, Liu Y, Garcia A, Stringfield O, Krewer H, Li Q, Cherezov D, Gatenby RA, Balagurunathan Y, Goldgof D, Schabath MB, Hall L, Gillies RJ. Predicting Malignant Nodules from Screening CT Scans. J Thorac Oncol. 2016;11:2120–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yankeelov TE, Quaranta V, Evans KJ, Rericha EC. Toward a Science of Tumor Forecasting for Clinical Oncology. Cancer Res. 2015;75:918–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hormuth DA II, Weis JA, Barnes SL, Miga MI, Quaranta V, Yankeelov TE. Biophysical modeling of in vivo glioma response following whole brain radiotherapy in a murine model of brain cancer. Int J Radiat Oncol Biol Phys. 2018;100:1270–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Weis JA, Miga MI, Arlinghaus LR, Li X, Abramson V, Chakravarthy AB, Pendyala P, Yankeelov TE. Predicting the Response of Breast Cancer to Neoadjuvant Therapy Using a Mechanically Coupled Reaction-Diffusion Model. Cancer Res. 2015;75:4697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kuhl CK, Schrading S, Strobel K, Schild HH, Hilgers RD, Bieling HB. Abbreviated breast magnetic resonance imaging (MRI): first postcontrast subtracted images and maximum-intensity projection-a novel approach to breast cancer screening with MRI. J Clin Oncol. 2014;32:2304–10. [DOI] [PubMed] [Google Scholar]