ABSTRACT
Background
Low–glycemic load dietary patterns, characterized by consumption of whole grains, legumes, fruits, and vegetables, are associated with reduced risk of several chronic diseases.
Methods
Using samples from a randomized, controlled, crossover feeding trial, we evaluated the effects on metabolic profiles of a low-glycemic whole-grain dietary pattern (WG) compared with a dietary pattern high in refined grains and added sugars (RG) for 28 d. LC-MS-based targeted metabolomics analysis was performed on fasting plasma samples from 80 healthy participants (n = 40 men, n = 40 women) aged 18–45 y. Linear mixed models were used to evaluate differences in response between diets for individual metabolites. Kyoto Encyclopedia of Genes and Genomes (KEGG)–defined pathways and 2 novel data-driven analyses were conducted to consider differences at the pathway level.
Results
There were 121 metabolites with detectable signal in >98% of all plasma samples. Eighteen metabolites were significantly different between diets at day 28 [false discovery rate (FDR) < 0.05]. Inositol, hydroxyphenylpyruvate, citrulline, ornithine, 13-hydroxyoctadecadienoic acid, glutamine, and oxaloacetate were higher after the WG diet than after the RG diet, whereas melatonin, betaine, creatine, acetylcholine, aspartate, hydroxyproline, methylhistidine, tryptophan, cystamine, carnitine, and trimethylamine were lower. Analyses using KEGG-defined pathways revealed statistically significant differences in tryptophan metabolism between diets, with kynurenine and melatonin positively associated with serum C-reactive protein concentrations. Novel data-driven methods at the metabolite and network levels found correlations among metabolites involved in branched-chain amino acid (BCAA) degradation, trimethylamine-N-oxide production, and β oxidation of fatty acids (FDR < 0.1) that differed between diets, with more favorable metabolic profiles detected after the WG diet. Higher BCAAs and trimethylamine were positively associated with homeostasis model assessment-insulin resistance.
Conclusions
These exploratory metabolomics results support beneficial effects of a low–glycemic load dietary pattern characterized by whole grains, legumes, fruits, and vegetables, compared with a diet high in refined grains and added sugars on inflammation and energy metabolism pathways. This trial was registered at clinicaltrials.gov as NCT00622661.
Keywords: metabolomics, dietary patterns, whole grains, glycemic load, dietary intervention, crossover, inflammation, insulin resistance
Introduction
Dietary patterns characterized by low-glycemic, minimally processed plant foods, i.e., whole grains, legumes, fruits, vegetables, nuts, and seeds, are associated with reduced risk of several chronic diseases, including many cancers, cardiovascular disease, obesity, and related metabolic diseases (1–4). Conversely, diets high in glycemic load, i.e., refined grains and added sugars, are associated with increased postprandial hyperglycemia and oxidative stress leading to metabolic dysregulation and increased risk of several chronic diseases (5, 6). Although many mechanisms for the protective effects of a low-glycemic, whole-foods dietary pattern have been proposed (7), properties related to higher fiber content are thought to play a leading role. In addition to being rich sources of fiber and providing many essential nutrients, intact plant foods also contain a multitude of phytochemicals, e.g., polyphenols, glucosinolates, omega-3 fatty acids, etc., which may act synergistically (8). These bioactive constituents may exert beneficial effects through modulation of signaling pathways related to inflammation, oxidative stress, cell growth and proliferation, and metabolism (9). Diets higher in fiber have also been shown to favorably alter the composition and functional potential of the gut microbiome (10).
We found previously that among overweight adults controlled feeding of a low-glycemic whole-grain diet resulted in lower serum C-reactive protein (CRP) and higher adiponectin concentrations than a diet high in refined grains and added sugars (11). We also reported that the low-glycemic whole-grain diet was associated with beneficial differences in circulating incretins (12), reduced postprandial glycemic response (13), and improved subjective mood and energy levels (14). While important, these outcomes are based on a limited number of biomarkers known to be altered with diet and metabolic-related diseases. There may be other changes in metabolic pathways that have consequences affecting human physiology and subsequent disease risk that are not detected with a few select measures. With the advent of high-throughput technologies, we can advance our understanding of metabolic response to diet by measuring hundreds of metabolites and characterizing their co-ordinated interactions within and between metabolic pathways. While many studies have used metabolomics to discover biomarkers of dietary intake, few have evaluated metabolic profiles in response to dietary patterns in a controlled feeding study in healthy humans. Using archived plasma samples from this controlled, crossover feeding study(11), our aim in the present analysis was to compare the effects on the targeted plasma metabolome of a low–glycemic load dietary pattern, characterized by whole grains, fruits, vegetables, legumes, and nuts and seeds, with the effects of a diet high in refined grains and added sugars.
Methods
Study design
Data and biologic samples for the present analysis were derived from the Carbohydrates And Related Biomarkers (CARB) study, conducted between June 2006 and July 2009 at the Fred Hutchinson Cancer Research Center (Fred Hutch). The CARB study was a randomized, controlled, crossover feeding study, the primary aims of which were to evaluate the effects of glycemic load on chronic disease susceptibility biomarkers, e.g., markers of systemic inflammation, insulin resistance, and adipokines (11–13). Participants were assigned to blocks based on body mass index and sex to receive 2 eucalorically similar controlled diets in a computer-generated, randomly assigned order, with a 28-d washout period in between diets where participants could eat as desired. The study was double blinded, for both participants and outcome assessors. While participants knew they were consuming 2 different diets, they were unaware of what the specific differences were, i.e., that the diets differed in carbohydrate source and type. To maintain blinding during analyses, laboratory personnel and statisticians were only provided with binary class labels (0,1). The study protocol, including the present ancillary study, was conducted in accordance with the ethical standards of the Declaration of Helsinki and approved by the Fred Hutch Institutional Review Board. All participants gave written informed consent. Based on a fixed sample size of 80 participants in this ancillary study and metabolomics pilot data from this cohort, we estimated that we would have 80% power to detect mean differences in effect sizes of 0.2 for at least 25 metabolites. This trial was registered at clinicaltrials.gov as NCT00622661.
Participants
Details on recruitment and study design have been published previously (11, 15). Briefly, healthy, nonsmoking individuals between the ages of 18 and 45 y were recruited from the Greater Seattle area. Exclusion criteria comprised impaired fasting glucose measured at a study clinic visit (fasting blood glucose ≥5.6 mmol/L), any physician-diagnosed condition requiring a restricted diet, food allergies, regular use of hormones or anti-inflammatory medication, pregnancy or lactation, or heavy use of alcohol (>2 drinks/d). Of the 84 randomly assigned participants, 80 completed all study activities and had complete biospecimen data (Supplemental Figure 1). At baseline, anthropometric data were collected, including height and weight, and body composition using a whole-body DXA scan (GE Lunar DPX-Pro, GE Healthcare), and self-administered questionnaires on demographic characteristics, usual physical activity, and medical history were completed.
Study diets
Participants received 2 controlled diets, a low–glycemic load diet characterized by whole grains, legumes, fruits, vegetables, nuts, and seeds (WG) or a high–glycemic load diet, high in refined grains and added sugars (RG). All food was provided by the Fred Hutch Human Nutrition Laboratory during each intervention period, with weekday dinners consumed under supervision at the study center and the next day's breakfast and lunch taken home for consumption. On Fridays, participants received all weekend meals. Any leftover foods were returned to the study center, weighed, and recorded. Participants were instructed to consume only the foods and beverages provided during both study periods and completed a daily checklist confirming consumption of study foods, with space to record consumption of any nonstudy foods. Adherence to the study diets was ∼98% (13).
Within each intervention diet, a 7-d menu rotation was created using ProNutra (version 3.2, Viocare), with a 2400-kcal reference menu for each day. As part of baseline data collection, participants completed 3-d food records to estimate their habitual energy intake based on their height, weight, sex, and usual activity level. The intake data were used to determine energy needs and food proportions from the reference menu for each participant. The 2 diets were designed to be identical in energy and macronutrient composition (15% energy from protein, 30% energy from fat, and 55% energy from carbohydrate) but to differ in glycemic load (125 compared with 250), and thus, fiber (55 and 28 g/d) for the WG and RG diets, respectively.
Nutrient and phytochemical differences between diets
Although the 2 intervention diets were similar in macronutrient composition, substitutions of more processed foods, i.e., refined grains for whole grains, fruit juices for fruits, etc., resulted in differences in many micronutrients and phytochemicals. To characterize these differences, overall content of accurately captured macro- and micronutrients and other bioactives in each of the diets (over the final 7-d menu rotation, days 21–28) was calculated [Nutrition Data System for Research (NDSR) software v.2016, developed by the Nutrition Coordinating Center; Supplemental Table 1]. Lignans were calculated using the Phenol Explorer database (16).
Specimen collection and metabolomics analysis
Blood was collected at baseline and after each 28-d diet period in the morning after a minimum of 12-h overnight fast and processed and stored at −80°C using a standard protocol. Plasma was collected using two 10-mL EDTA tubes then immediately mixed by inversion to ensure complete mixing with anticoagulant. This was followed by centrifugation at 4°C for 10 min and subsequent aliquoting into 0.1-mL cryovials with O-rings. Samples were stored for a duration of 7–10 y prior to analysis. Serum concentrations of overnight fasting high-sensitivity CRP (11), glucose, and insulin (13) were assayed previously as described. The HOMA-IR was calculated by taking the product of the fasting insulin in microunits per milliliter and glucose in milligrams per deciliter and dividing the result by 405 (13). These outcomes were used in post hoc analyses.
Stored plasma samples were used for metabolite profiling, which was completed at the University of Washington's Northwest Metabolomics Research Center. Targeted metabolomics analysis was carried out using an LC-MS/MS platform in both positive and negative ion modes against 203 standard metabolites (Supplemental Table 2) from more than 25 Kyoto Encyclopedia of Genes and Genomes (KEGG)–defined metabolic pathways (17) (e.g., glycolysis, tricyclic acid cycle, amino acid metabolism, glutathione, etc.) of potential significance to monitor diet effects, along with 24 stable-isotope labeled internal standards for concentration determinations (18). Samples were prepared and analyzed as previously described (15, 19). Briefly, a standard protocol was used, where 25 µL plasma and 150 µL HPLC-grade methanol were combined in an Eppendorf vial and vortexed for 2 min. After 20-min storage at −20°C, the samples were centrifuged at 4ºC for 10 mins at 18,000 × g, and then 250 µL was collected and dried at 30°C in a Speed-Vac for 3 h. Samples were reconstituted with 100 μL 5 mM ammonium acetate in 95% water/5% acetonitrile plus 0.5% acetic acid, and filtered through 0.45-μm polyvinylidene difluoride filters (Phenomenex) prior to analysis. LC-MS/MS experiments were performed on a Waters Acquity I-Class UPLC TQS-micro MS system. Each sample was injected twice, 2 μL and 10 μL, for analysis using positive and negative ionization modes, respectively. Both chromatographic separations were performed in hydrophilic interaction chromatography mode on an AB Sciex 5500 QTrap LC-MS/MS system. The flow rate was 0.3 mL min−1, autosampler temperature was kept at 4°C, and the column compartment was set at 40°C. The mobile phase was composed of solvents A (5 mM ammonium acetate in H2O + 0.5% acetic acid + 0.5% acetonitrile) and B (acetonitrile + 0.5% acetic acid + 0.5% water). The LC gradient conditions were the same for both positive and negative ionization modes. After an initial 1.5-min isocratic elution of 10% A, the percentage of solvent A was increased linearly to 65% at time (t) = 9 min, then remained the same for 5 min (t = 14 min), and then reduced to 10% at t = 15 min to prepare for the next injection. After chromatographic separation, MS ionization and data acquisition was performed using an electrospray ionization source and Sciex Analyst 1.6 software. A pooled study sample was used as the quality control (QC) and run once for every 10 study samples. The intra-assay average CV based on this QC sample was 7.8% across all samples.
Statistical analyses
In total, 121 metabolites were reliably identified and quantitated in >98% of all samples and retained for analysis. Six metabolites had values below the limit of detection for 2% of samples (4 participants at a single time-point). For these 6 metabolites, both time points for an individual were entered as missing in the univariate analyses. Most metabolites tended to be skewed toward higher values and were natural log-transformed to better approximate a normal distribution. Linear mixed models adjusting for batch, diet sequence, period, sex, age, body fat mass, and baseline metabolite concentrations were used to evaluate differences in response between diets for individual metabolites. Potential carryover effects between diet periods were evaluated by adding an interaction term for treatment *period; no evidence of carryover was found. The same models were used for subgroup analyses, i.e., sex, and adiposity group, and to assess post hoc associations between metabolites and CRP and HOMA-IR. Subgroup analyses based on sex and adiposity status were planned a priori. Although we recruited on the basis of BMI, this measure tends to be a poor indicator of body fat (20). Thus, analyses were conducted based on the following adiposity classifications as determined by DXA: high–body fat mass for females >32.0% and males >25.0% (21). This resulted in recategorization of some participants, with n = 29 comprising the normal/low–fat mass group and n = 51 comprising the overweight/high–fat mass group. The Benjamini-Hochberg algorithm was used to control the false discovery rate (FDR < 0.05 for univariate analyses). Univariate analyses were conducted in Stata (v15.1, StataCorp).
To reduce the complexity and consider metabolites that act co-ordinately, pathway analysis was conducted using correlation-adjusted mean rank gene set analysis (CAMERA) (22). Unlike permutation-based methods, such as gene-set enrichment analysis (GSEA), this method directly estimates the correlation among metabolites when assessing pathway enrichment. We initially considered pathways in the KEGG database of metabolic pathways (17). Because a targeted panel of 121 metabolites does not provide full coverage of all pathways, we also employed 2 data-driven methods to identify novel pathways. Specifically, we complemented the 23 KEGG pathways mapped to our targeted metabolomics panel with 1) 17 pathways identified based on clusters of correlated metabolites using the weighted gene coexpression network analysis (WGCNA) method (23); and 2) 12 pathways identified based on highly connected components in an estimated metabolite network using the network gene-set analysis (NetGSA) package in R (24–26). Together, these 2 methods identify data-driven clusters of metabolites either based on correlations among metabolites (WGCNA) or based on a network of connectivities informed by metabolic reactions and refined based on partial correlations among metabolite interactions (NetGSA), the motivation being that groups of metabolites may function similarly in a metabolic pathway. Correlations suggest dependence among the metabolites and the relation may be positive or inverse, suggesting similar or opposing shifts between diets. Pathways were inferred based on biologic function of the metabolites identified in these clusters. These de novo pathways facilitate new discoveries and better reflect the evidence from targeted metabolomics assays. In total, 52 pathways with ≥5 metabolites were included in the 3 pathway enrichment analyses using CAMERA (22). The Benjamini-Hochberg procedure was applied to control the number of false discoveries (FDR < 0.1), conservatively, with all 52 pathways (KEGG and 2 data driven) considered together.
Results
Participant characteristics, overall and stratified by adiposity, are given in Table 1. In the univariate analysis, 18 of the 121 metabolites were statistically significantly different between the WG and RG diets at day 28 (FDR < 0.05; Table 2). Geometric mean ratios of plasma metabolites ranged from 0.61 to 1.18, with melatonin (lower after the WG diet) and inositol (higher after the WG diet) having the greatest fold change between diets. Results were similar when stratified by sex (data not shown) and adiposity (Table 2) for the most significant metabolites, with 2 additional metabolites differing between diets within strata of adiposity groups: after the WG diet, xanthurenate was lower among individuals with a high fat mass, and shikimic acid, a plant phenolic metabolite, was higher among individuals with a normal fat mass (FDR < 0.05; Table 2).
TABLE 1.
Characteristics of participants in the CARB study1
| Characteristics | All participants (n = 80) | Low adiposity2 (n = 29) | High adiposity2 (n = 51) | P value3 |
|---|---|---|---|---|
| Age, y | 29.6 ± 8.1 | 26.4 ± 6.4 | 31.5 ± 8.5 | <0.01 |
| Male sex | 40 (50%) | 19 (66%) | 21 (41%) | <0.05 |
| Weight, kg | 81.1 ± 21.6 | 67.2 ± 9.2 | 89.1 ± 22.8 | <0.001 |
| Body fat, % | 32.8 ± 11.8 | 21.2 ± 6.9 | 39.7 ± 8.3 | <0.001 |
| CRP, mg/L | 1.5 ± 2.7 | 1.4 ± 5.5 | 2.12 ± 2.9 | 0.19 |
| HOMA-IR | 2.6 ± 2.2 | 1.5 ± 0.1 | 3.4 ± 0.3 | <0.001 |
| Race/ethnicity | 0.6 | |||
| Non-Hispanic white | 35 (44%) | 10 (34%) | 25 (49%) | |
| Hispanic | 19 (24%) | 8 (28%) | 11 (22%) | |
| African American | 17 (21%) | 7 (24%) | 10 (20%) | |
| Other4 | 9 (11%) | 4 (20%) | 5 (10%) |
Data presented as means ± SDs or n (%) unless otherwise indicated. CARB, Carbohydrate and Related Biomarkers; CRP, C-reactive protein.
Adiposity classifications as determined by DXA: females: low fat mass <31.9%, high fat mass >32.0%; males: low fat mass <24.9%, high fat mass >25.0%.
Determined by chi-square test or paired t-test.
Other category includes Asian/Pacific Islander/Native American.
TABLE 2.
Plasma metabolites significantly different at day 28 between a low–glycemic load dietary pattern characterized by whole grains, legumes, fruits, and vegetables, compared with a diet high in refined grains and added sugars overall, and stratified by adiposity group1
| All participants (n = 80) | Low adiposity2 (n = 29) | High adiposity2 (n = 51) | ||||
|---|---|---|---|---|---|---|
| Plasma metabolite | Ratio3 | P value4 | Ratio3 | P value4 | Ratio3 | P value4 |
| Inositol | 1.18 (0.18) | 1.46 × 10−23* | 1.21 (0.19) | 1.19 × 10−11* | 1.17 (0.17) | 7.80 × 10−9* |
| Melatonin | 0.61 (0.28) | 3.58 × 10−22* | 0.62 (0.28) | 0.0003* | 0.61 (0.29) | 3.65 × 10−8* |
| Hydroxyphenylpyruvate | 1.16 (0.23) | 3.02 × 10−12* | 1.15 (0.24) | 0.002* | 1.17 (0.23) | 1.54 × 10−5* |
| Betaine | 0.92 (0.12) | 1.75 × 10−10* | 0.90 (0.11) | 0.003* | 0.93 (0.11) | 0.02 |
| Creatine | 0.84 (0.25) | 1.19 × 10−7* | 0.79 (0.20) | 0.0002* | 0.87 (0.27) | 0.001* |
| Acetylcholine | 0.92 (0.15) | 8.54 × 10−7* | 0.90 (0.16) | 0.001* | 0.92 (0.14) | 0.008 |
| Citrulline | 1.06 (0.15) | 1.34 × 10−4* | 1.06 (0.17) | 0.06 | 1.07 (0.14) | 0.007 |
| Ornithine | 1.08 (0.21) | 3.83 × 10−4* | 1.04 (0.19) | 0.27 | 1.11 (0.23) | 0.005 |
| 13-Hydroxyoctadecadienoic acid | 1.14 (0.37) | 4.32 × 10−4* | 1.25 (0.38) | 0.003* | 1.07 (0.35) | 0.11 |
| Aspartic acid | 0.91 (0.23) | 7.25 × 10−4* | 0.93 (0.25) | 0.14 | 0.90 (0.22) | 0.001* |
| Hydroxyproline | 0.87 (0.32) | 7.77 × 10−4* | 0.85 (0.41) | 0.03 | 0.88 (0.26) | 0.001* |
| Methylhistidine | 0.77 (0.55) | 8.18 × 10−4* | 0.80 (0.56) | 0.08 | 0.75 (0.54) | 0.004* |
| Tryptophan | 0.94 (0.14) | 9.19 × 10−4* | 0.95 (0.15) | 0.10 | 0.94 (0.14) | 0.003* |
| Cystamine | 0.87 (0.35) | 0.002* | 0.85 (0.33) | 0.03 | 0.88 (0.36) | 0.02 |
| Glutamine | 1.03 (0.09) | 0.003* | 1.01 (0.07) | 0.47 | 1.04 (0.09) | 0.002* |
| Carnitine | 0.95 (0.14) | 0.004* | 0.92 (0.13) | 0.001* | 0.97 (0.15) | 0.19 |
| Trimethylamine | 0.95 (0.16) | 0.007* | 0.90 (0.17) | 0.004* | 0.98 (0.15) | 0.44 |
| Oxaloacetate | 1.04 (0.15) | 0.007* | 1.03 (0.14) | 0.26 | 1.05 (0.15) | 0.01 |
| Xanthurenic acid | 0.96 (0.15) | 0.03 | 0.99 (0.15) | 0.79 | 0.94 (0.15) | 0.005* |
| Shikimic acid | 1.13 (0.48) | 0.06 | 1.18 (0.37) | 0.003* | 1.10 (0.53) | 0.12 |
FDR, false discovery rate; RG, refined grain and added sugars dietary pattern; WG, low–glycemic load dietary pattern.
Adiposity classifications as determined by DXA: females: low fat mass <31.9%, high fat mass >32.0%; males: low fat mass <24.9%, high fat mass >25.0%.
Geometric mean ratio (geometric SD) of plasma metabolite concentrations comparing WG diet vs. RG diet at day 28; a ratio >1 means that the metabolite was higher in the WG diet relative to the RG diet, whereas a ratio <1 means that the metabolite was lower.
P values derived from linear mixed model adjusting for diet sequence, assay batch, age, sex, body fat percentage, and baseline metabolite concentrations; stratified analyses excluded body fat percentage as a covariate. *Significant with Benjamini-Hochberg FDR <0.05.
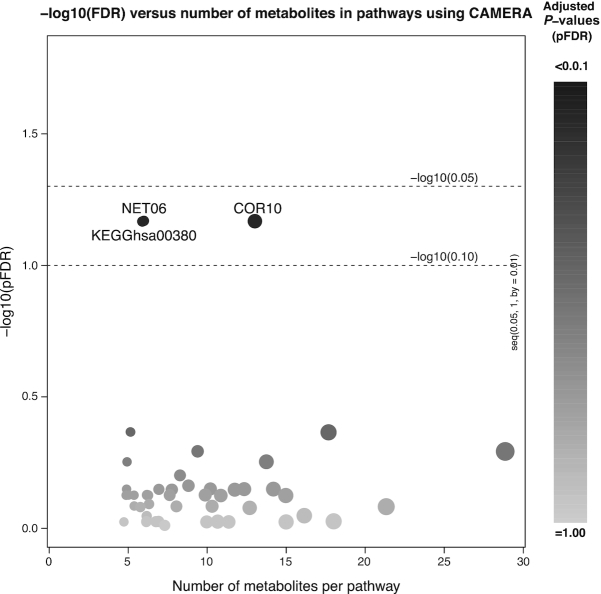
Enriched pathways from KEGG and 2 data-driven methods of identifying metabolic pathways significant at FDR < 0.1 are presented in Figures 1 and 2. A total of 3 pathways were identified, one from each method. Among KEGG metabolic pathways, tryptophan metabolism (KEGGhsa00380) differed between the 2 diets. Of the 6 metabolites measured in our panel, tryptophan and melatonin were statistically significantly lower after the WG diet in the univariate analysis, and xanthurenate was significantly lower after the WG diet among individuals with higher adiposity. Because higher concentrations of tryptophan metabolites have been associated with inflammation (27), we also explored the relation of these six metabolites with CRP. Both kynurenine and melatonin were positively associated with CRP (P = 4.5 × 10−6 and 0.007, respectively; Table 3).
FIGURE 1.
Metabolic pathway analysis comparing a low-glycemic whole-grain dietary pattern to a diet high in refined grains and added sugars conducted using CAMERA. The x-axis shows the pathway size and the y-axis shows the negative logarithm (in base 10) of FDR-adjusted P-values from pathway enrichment analysis. In addition to a KEGG pathway (hsa00380) 2 data-driven metabolic pathways (NET06 and COR10) were enriched at <10% FDR. Metabolites in enriched pathways are shown in Figure 2. CAMERA, correlation-adjusted mean rank gene set analysis; FDR, false discovery rate; KEGG, Kyoto Encyclopedia of Genes and Genomes.
FIGURE 2.
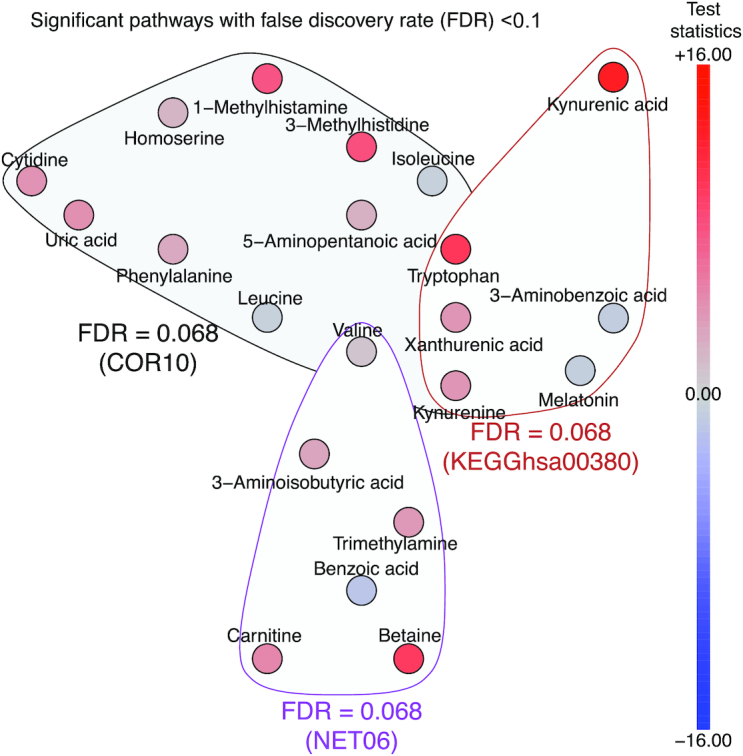
Metabolites in enriched pathways (at 10% FDR). The color of each node corresponds to the test statistic from univariate analysis, as depicted in the color bar. In total, 4 metabolites are included in more than 1 enriched pathway. FDR, false discovery rate; KEGG, Kyoto Encyclopedia of Genes and Genomes.
TABLE 3.
Associations between pathway metabolites and CRP and HOMA-IR1
| All participants (n = 80) | Low adiposity2(n = 29) | High adiposity2 (n = 51) | ||||
|---|---|---|---|---|---|---|
| Metabolite | β ± SE3 | P value3 | β ± SE3 | P value3 | β ± SE3 | P value3 |
| Tryptophan metabolites and CRP | ||||||
| Kynurenine | 1.78 ± 0.39 | 4.5 × 10–6* | 1.79 ± 0.60 | 0.003* | 1.16 ± 0.41 | 0.004* |
| Melatonin | 0.51 ± 0.19 | 0.007* | 0.54 ± 0.32 | 0.09 | 0.40 ± 0.16 | 0.01 |
| Kynurenic acid | −0.14 ± 0.18 | 0.43 | −0.43 ± 0.33 | 0.19 | 0.13 ± 0.15 | 0.37 |
| Tryptophan | 0.35 ± 0.61 | 0.56 | −0.42 ± 1.13 | 0.71 | 0.29 ± 0.52 | 0.58 |
| Xanthurenate | 0.24 ± 0.64 | 0.70 | 0.94 ± 1.33 | 0.48 | −0.67 ± 0.49 | 0.17 |
| 2-Aminobenzoic acid | 0.47 ± 1.13 | 0.95 | 1.81 ± 2.12 | 0.39 | −0.80 ± 0.96 | 0.40 |
| TMAO substrates and CRP | ||||||
| Betaine | 1.63 ± 0.71 | 0.12 | 2.94 ± 1.14 | 0.10 | 0.82 ± 0.64 | 0.20 |
| TMA | −0.36 ± 0.54 | 0.51 | −0.59 ± 0.97 | 0.54 | 0.81 ± 0.47 | 0.09 |
| Carnitine | −0.37 ± 0.59 | 0.53 | −0.02 ± 1.16 | 0.99 | 0.56 ± 0.50 | 0.26 |
| Choline | 0.28 ± 0.59 | 0.64 | 2.89 ± 1.17 | 0.01 | −0.46 ± 0.51 | 0.37 |
| TMAO | 0.07 ± 0.16 | 0.66 | −0.18 ± 0.26 | 0.48 | 0.03 ± 0.15 | 0.87 |
| BCAAs and HOMA-IR | ||||||
| Isoleucine | 0.92 ± 0.26 | 0.0003* | 0.72 ± 0.28 | 0.009* | 1.10 ± 0.38 | 0.004* |
| Valine | 0.84 ± 0.29 | 0.004* | 0.41 ± 0.36 | 0.25 | 0.98 ± 0.41 | 0.02 |
| Leucine | 0.54 ± 0.34 | 0.11 | 0.46 ± 0.38 | 0.23 | 0.52 ± 0.50 | 0.30 |
| TMAO substrates and HOMA-IR | ||||||
| TMA | 0.51 ± 0.24 | 0.03 | 0.88 ± 0.25 | 0.0005* | 0.12 ± 0.36 | 0.73 |
| Betaine | −0.54 ± 0.32 | 0.09 | 0.12 ± 0.37 | 0.74 | −1.03 ± 0.45 | 0.02 |
| Choline | −0.37 ± 0.26 | 0.16 | 0.30 ± 0.35 | 0.40 | −0.70 ± 0.35 | 0.05 |
| Carnitine | 0.24 ± 0.26 | 0.36 | 0.70 ± 0.33 | 0.03 | −0.14 ± 0.37 | 0.70 |
| TMAO | 0.06 ± 0.07 | 0.39 | −0.09 ± 0.07 | 0.25 | 0.25 ± 0.11 | 0.03 |
BCAA, branched-chain amino acid; CRP, C-reactive protein; FDR, false discovery rate; TMA, trimethylamine; TMAO, trimethylamine-N-oxide.
Adiposity classifications as determined by DXA: females: low fat mass <31.9%, high fat mass >32.0%; males: low fat mass <24.9%, high fat mass >25.0%.
β coefficient ± SE and P value derived from linear mixed model adjusting for diet, diet sequence, assay batch, age, sex, and body fat percentage; stratified analyses excluded body fat percentage as a covariate. *Indicates statistical significance with Benjamini-Hochberg FDR <0.05.
Among the pathways identified based on metabolite correlations, a cluster of 13 metabolites was significant at FDR < 0.1 (COR10; Figure 2). These were mainly amino acids involved in tryptophan metabolism, suggesting good correspondence between pathway analysis methods, and valine, leucine, and isoleucine degradation. Given that higher concentrations of branched-chain amino acids (BCAAs) are associated with insulin resistance (28), we conducted a post hoc analysis evaluating the association between BCAAs and HOMA-IR. Valine and isoleucine were statistically significantly associated with plasma HOMA-IR (P < 0.004 and 0.0003, respectively; Table 3). While the direction of the relation between leucine and HOMA-IR was also positive, this association was not statistically significant. The remaining metabolites in this pathway did not fall into overlapping pathways and were primarily amino acid metabolites of protein-rich foods [cytidine, phenylalanine, homoserine, 1 (Nτ)-methylhistamine, 3 (Nπ)-methylhistidine, and 5-aminopentanoic acid] and uric acid, a metabolite of purine degradation. All were higher after the RG diet.
The last significant pathway at FDR < 0.1 was derived from network interactions (NET06; Figure 2), and primarily included metabolites related to trimethylamine-N-oxide (TMAO) production [trimethylamine (TMA), carnitine and betaine] (29), β oxidation of fatty acids [carnitine, and 3-aminoisobutyric acid (30)], and insulin resistance [valine and 3-aminoisobutyric acid (30, 31); Figure 2]. As reported above, plasma valine concentrations were higher after the RG diet and significantly positively associated with HOMA-IR. There was no association between 3-aminoisobutyric acid and HOMA-IR (data not shown). As TMAO has also been associated with inflammation and insulin resistance, we explored the association between TMAO and its substrates with CRP and HOMA-IR. Only TMA, which was positively associated with HOMA-IR among lean individuals, was significant after controlling for multiple testing (Table 3). The sixth metabolite in the network, benzoic acid, a common component of plant foods, was higher after the WG diet.
Discussion
A growing body of epidemiologic literature suggests beneficial effects of dietary patterns emphasizing minimally processed plant foods on various health outcomes (4, 32, 33). To gain mechanistic insight into these protective effects, we evaluated the impact of a low– versus a high–glycemic load dietary pattern on a panel of metabolites using plasma from a completed randomized, controlled feeding study in healthy adults. In addition to highly significant differences in 18 metabolites between the 2 diets, we found differences in several metabolic pathways related to inflammation signaling and energy metabolism.
Of the KEGG-defined metabolic pathways evaluated, the tryptophan metabolism pathway differed, with the majority of metabolites lower in plasma after the WG diet. In addition to its role in protein synthesis, tryptophan is a precursor for many biologically important metabolites involved in neurotransmission, redox reactions, and inflammation and immune responses (34). The majority of dietary tryptophan (∼95%) is converted to kynurenine through the enzymatic action of indoleamine 2,3-dioxygenase (35). From there kynurenine branches to form either kynurenic acid or quinolinic acid, considered anti-inflammatory and proinflammatory, respectively (36). Kynurenine, and xanthurenic acid—an intermediate in the production of quinolinate in the proinflammatory branch, were both nonsignificantly lower overall after the WG diet, despite tryptophan intake being higher. However, xanthurenic acid was significantly lower after the WG diet among individuals in the high adiposity group, corresponding with previous findings of lower CRP in this group (11). Kynurenine, known to be upregulated in response to an inflammatory stimulus (35), was positively correlated with the serum inflammatory marker CRP. These data suggest that the WG dietary pattern may be associated with a shift away from the proinflammatory branch of the kynurenine pathway, while the RG dietary pattern is related to proinflammatory metabolism.
A smaller proportion of dietary tryptophan (∼5%) is converted to serotonin, which can be further metabolized to melatonin. Melatonin is produced mainly by the pineal gland, and regulates circadian rhythms and sleep (37). In addition to human endogenous production, fruits, vegetables, and grain products contain measurable amounts of serotonin and melatonin, which serve to defend the plant against environmental stressors (38, 39). Given that the RG diet was lower in unrefined plant foods, it is unlikely that dietary intake was a major contributor to the observed difference in melatonin between diets. Higher melatonin may be linked to greater postprandial insulin release on the RG diet (13). Insulin lowers plasma concentrations of large neutral amino acids, including tryptophan and BCAAs, by increasing their uptake across the blood–brain barrier (40).
In addition to pathway-specific analyses, we also evaluated data-driven associations between diets based on empirical correlations among the metabolites and their networks. The BCAAs leucine, isoleucine, and valine were differentially clustered between diets in the metabolite correlation analysis. BCAA dysregulation has been strongly associated with insulin resistance and other metabolic risk factors, independent of fat mass (41). Further, plasma valine concentration has been identified as a predictor of future development of type 2 diabetes (31, 42), possibly through increased activation of mammalian target of rapamycin complex 1 (43). In our data, plasma valine concentrations were positively associated with HOMA-IR. Valine was also identified in the network correlation analysis, along with 3-aminoisobutyric acid, a metabolite produced by valine catabolism. This latter metabolite has also recently been associated with altered energy metabolism and cardiometabolic risk. A cross-sectional study of 2000 patients enrolled in the Framingham Heart Study found inverse associations between plasma 3-aminoisobutyric acid and fasting glucose, insulin, HOMA-IR, triglycerides, and total cholesterol (30). In complementary studies in mice, investigators demonstrated that 3-aminoisobutyric acid secreted from myocytes increased the expression of brown adipocyte-specific genes in white adipose tissue, and genes involved in fatty acid β-oxidation in hepatocytes (31). Two other studies have reported beneficial modulation of inflammation and energy metabolism with a dietary pattern higher in minimally processed, low-glycemic foods. Using an untargeted approach, Acar et al. (44) characterized the plasma metabolome after a 26-wk dietary intervention in 146 individuals following either a diet high in fruit, vegetables, whole grains, and fish or a diet higher in processed foods. Similar to our findings, metabolites linked to fatty acid metabolism and glucose utilization exhibited more favorable profiles after the unrefined diet. Another study assessed the plasma metabolome and found that both leucine and valine were lower after a low–glycemic index diet compared to a low-fat diet, with leucine positively related to higher serum IL-6 (45).
Among the network-based correlations were TMA, carnitine, and betaine, substrates for endogenously produced TMAO (29). TMAO contributes to insulin resistance and altered cholesterol metabolism (46) and in animals promotes vascular inflammation through interaction with NF-κB (47). In our study, neither TMAO nor substrates involved in its formation were associated with CRP or HOMA-IR. Intact TMAO can be found in fish or can be synthesized by gut bacteria from betaine, carnitine, and choline to TMA, which can then be oxidized to TMAO by flavin-dependent mono-oxygenase-3 (FMO-3) in the liver (29). While TMA was higher after the RG diet, TMAO was non-significantly higher after the WG diet. In addition to provision of required substrates, dietary components may alter production of TMAO in other ways. Differences in amounts and types of fiber can alter gut microbial composition, altering consortia of bacteria capable of generating TMA (29). There are also data to suggest that some bioactive compounds inhibit FMO-3, i.e., indoles from cruciferous vegetables and resveratrol, a polyphenol in the skins of red grapes and berries, modulating the conversion of TMA to TMAO (48, 49). The extent to which these dietary components contributed to TMAO production is not known, and presumably other phytochemicals would also have the potential to modulate production of TMAO and other metabolites, either through interaction with signaling pathways or through altering gut bacterial structure or activity (50, 51).
There are many strengths of this study, including the randomized, controlled intervention with all individuals consuming the same foods for the 2 dietary patterns being compared. The crossover study design, with each person serving as their own comparison, further reduced the influence of nondietary confounding factors. Although we used a targeted metabolomics panel with metabolites represented in >25 metabolic pathways, not all members in the pathways were measured. For this reason, we employed data-driven analyses evaluating the correlations between both the metabolites and metabolic networks. While we found good correspondence with the pathway-based analyses, it is important to note that these data-driven pathway analyses are only empirical and the relationships between metabolites are not certain. However, differences in the same pathways, i.e., tryptophan and TMAO, were noted in a small subset of these participants using the same metabolomics platform previously, indicating a consistent signal (15). Finally, it is worth noting that the homogeneous population and specific foods used in our study reduce the generalizability of our results. Nevertheless, evaluation of the metabolome in healthy individuals in response to controlled feeding allows for discovery of novel associations and understanding of the mechanisms underlying effects of dietary components in a human system.
In conclusion, in this exploratory analysis using targeted metabolite profiles, we found that a low–glycemic load dietary pattern characterized by whole grains and other minimally processed plant foods compared to a diet high in refined grains and added sugars resulted in differences in several metabolic pathways related to inflammation and energy metabolism, which are likely beneficial. These results illustrate the complexity of dietary patterns and the need to understand the synergistic effects among dietary components, rather than individual compounds, on physiologic and biochemical responses and health outcomes.
Supplementary Material
Acknowledgments
The authors’ responsibilities were as follows—SLN, MK, MLN, MAH, PDL, DR, JWL: designed the research; HG, DD, DR: conducted the research; SLN, AT, AS, TWR, KJO: analyzed the data; SLN: wrote the manuscript; and all authors: read and approved the final manuscript. None of the authors had a personal or financial conflict of interest.
Notes
Funding was provided by the National Institutes of Health (grants R01 CA192222, U54 CA116847, P30 CA015704, P30DK035816, and R01 GM114029).
Supplemental Figure 1 and Supplemental Tables 1 and 2 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/ajcn/.
Abbreviations used: BCAA, branched-chain amino acid; CAMERA, correlation-adjusted mean rank gene set analysis; CARB, Carbohydrates And Related Biomarkers; CRP, C-reactive protein; FDR, false discovery rate; FMO-3, flavin-dependent mono-oxygenase-3; Fred Hutch, Fred Hutchinson Cancer Research Center; KEGG, Kyoto Encyclopedia of Genes and Genomes; NetGSA, network gene-set analysis; QC, quality control; RG, refined grain and added sugars dietary pattern; TMA, trimethylamine; TMAO, trimethylamine-N-oxide; WGCNA, weighted gene coexpression network analysis; WG, low–glycemic load dietary pattern characterized by whole grains, legumes, fruits, vegetables, nuts, and seeds.
References
- 1. McRae MP. Health benefits of dietary whole grains: an umbrella review of meta-analyses. J Chiropr Med. 2017;16(1):10–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bradbury KE, Appleby PN, Key TJ. Fruit, vegetable, and fiber intake in relation to cancer risk: findings from the European Prospective Investigation into Cancer and Nutrition (EPIC). Am J Clin Nutr. 2014;100 Suppl 1:394S–8S. [DOI] [PubMed] [Google Scholar]
- 3. Kahleova H, Levin S, Barnard N. Cardio-metabolic benefits of plant-based diets. Nutrients. 2017;9(8):E848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bamia C. Dietary patterns in association to cancer incidence and survival: concept, current evidence, and suggestions for future research. Eur J Clin Nutr. 2018;72(6):818–25. [DOI] [PubMed] [Google Scholar]
- 5. Barclay AW, Petocz P, McMillan-Price J, Flood VM, Prvan T, Mitchell P, Brand-Miller JC. Glycemic index, glycemic load, and chronic disease risk - a metaanalysis of observational studies. Am J Clin Nutr. 2008;87(3):627–37. [DOI] [PubMed] [Google Scholar]
- 6. Brand-Miller JC. Glycemic load and chronic disease. Nutr Rev. 2003;61(5):S49–55. [DOI] [PubMed] [Google Scholar]
- 7. Satija A, Bhupathiraju SN, Spiegelman D, Chiuve SE, Manson JE, Willett W, Rexrode KM, Rimm EB, Hu FB. Healthful and unhealthful plant-based diets and the risk of coronary heart disease in U.S. adults. J Am Coll Cardiol. 2017;70(4):411–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mozaffarian D, Wu JHY.. Flavonoids, dairy foods, and cardiovascular and metabolic health: a review of emerging biologic pathways. Circ Res. 2018;122(2):369–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu RH. Dietary bioactive compounds and their health implications. J Food Sci. 2013;78 Suppl 1:A18–25. [DOI] [PubMed] [Google Scholar]
- 10. Kerley CP. A review of plant-based diets to prevent and treat heart failure. Card Fail Rev. 2018;4(1):54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Neuhouser ML, Schwarz Y, Wang C, Breymeyer K, Coronado G, Wang CY, Noar K, Song X, Lampe JW. A low-glycemic load diet reduces serum C-reactive protein and modestly increases adiponectin in overweight and obese adults. J Nutr. 2012;142:369–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Runchey SS, Valsta LM, Schwarz Y, Wang C, Song X, Lampe JW, Neuhouser ML. Effect of low- and high-glycemic load on circulating incretins in a randomized clinical trial. Metabolism. 2013;62(2):188–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Runchey SS, Pollak MN, Valsta LM, Coronado GD, Schwarz Y, Breymeyer KL, Wang C, Wang CY, Lampe JW, Neuhouser ML. Glycemic load effect on fasting and post-prandial serum glucose, insulin, IGF-1 and IGFBP-3 in a randomized, controlled feeding study. Eur J Clin Nutr. 2012;66:1146–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Breymeyer KL, Lampe JW, McGregor BA, Neuhouser ML. Subjective mood and energy levels of healthy weight and overweight/obese healthy adults on high- and low-glycemic load experimental diets. Appetite. 2016;107:253–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barton S, Navarro SL, Buas MF, Schwarz Y, Gu H, Djukovic D, Raftery D, Kratz M, Neuhouser ML, Lampe JW. Targeted plasma metabolome response to variations in dietary glycemic load in a randomized, controlled, crossover feeding trial in healthy adults. Food Funct. 2015;6(9):2949–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Neveu V, Perez-Jimenez J, Vos F, Crespy V, du Chaffaut L, Mennen L, Knox C, Eisner R, Cruz J, Wishart D et al.. Phenol-Explorer: an online comprehensive database on polyphenol contents in foods. Database (Oxford). 2010;2010:bap024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kanehisa M, Goto S.. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu J, Djukovic D, Deng L, Gu H, Himmati F, Chiorean EG, Raftery D. Colorectal cancer detection using targeted serum metabolic profiling. J Proteome Res. 2014;13:4120–30. [DOI] [PubMed] [Google Scholar]
- 19. Gu H, Du J, Carnevale Neto F, Carroll PA, Turner SJ, Chiorean EG, Eisenman RN, Raftery D. Metabolomics method to comprehensively analyze amino acids in different domains. Analyst. 2015;140(8):2726–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rothman KJ. BMI-related errors in the measurement of obesity. Int J Obes (Lond). 2008;32 Suppl 3:S56–9. [DOI] [PubMed] [Google Scholar]
- 21. Li C, Ford ES, Zhao G, Balluz LS, Giles WH. Estimates of body composition with dual-energy X-ray absorptiometry in adults. Am J Clin Nutr. 2009;90(6):1457–65. [DOI] [PubMed] [Google Scholar]
- 22. Wu D, Smyth GK.. CAMERA: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 2012;40(17):e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pei G, Chen L, Zhang W. WGCNA application to proteomic and metabolomic data analysis. Methods Enzymol. 2017;585:135–58. [DOI] [PubMed] [Google Scholar]
- 24. Shojaie A, Michailidis G.. Analysis of gene sets based on the underlying regulatory network. J Comput Biol. 2009;16(3):407–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shojaie A, Michailidis G.. Network enrichment analysis in complex experiments. Stat Appl Genet Mol Biol. 2010;9:Article22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ma J, Shojaie A, Michailidis G. Network-based pathway enrichment analysis with incomplete network information. Bioinformatics. 2016;32(20):3165–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Christensen MHE, Fadnes DJ, Rost TH, Pedersen ER, Andersen JR, Vage V, Ulvik A, Midttun O, Ueland PM, Nygard OK et al.. Inflammatory markers, the tryptophan-kynurenine pathway, and vitamin B status after bariatric surgery. PLoS One. 2018;13(2):e0192169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gannon NP, Schnuck JK, Vaughan RA. BCAA metabolism and insulin sensitivity—dysregulated by metabolic status?. Mol Nutr Food Res. 2018;62(6):e1700756. [DOI] [PubMed] [Google Scholar]
- 29. Janeiro MH, Ramirez MJ, Milagro FI, Martinez JA, Solas M. Implication of trimethylamine N-Oxide (TMAO) in disease: potential biomarker or new therapeutic target. Nutrients. 2018;10(10):E1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roberts LD, Bostrom P, O'Sullivan JF, Schinzel RT, Lewis GD, Dejam A, Lee YK, Palma MJ, Calhoun S, Georgiadi A et al.. β-Aminoisobutyric acid induces browning of white fat and hepatic beta-oxidation and is inversely correlated with cardiometabolic risk factors. Cell Metab. 2014;19(1):96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Palomino-Schatzlein M, Simo R, Hernandez C, Ciudin A, Mateos-Gregorio P, Hernandez-Mijares A, Pineda-Lucena A, Herance JR. Metabolic fingerprint of insulin resistance in human polymorphonuclear leucocytes. PLoS One. 2018;13(7):e0199351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mozaffarian D. Dietary and policy priorities for cardiovascular disease, diabetes, and obesity: a comprehensive review. Circulation. 2016;133(2):187–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Neuhouser ML. The importance of healthy dietary patterns in chronic disease prevention. Nutr Res. 2018.doi: 10.1016/j.nutres.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Palego L, Betti L, Rossi A, Giannaccini G. Tryptophan biochemistry: structural, nutritional, metabolic, and medical aspects in humans. J Amino Acids. 2016;2016:8952520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Badawy AA. Kynurenine pathway of tryptophan metabolism: regulatory and functional aspects. Int J Tryptophan Res. 2017;10:1178646917691938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Badawy AA. Hypothesis kynurenic and quinolinic acids: the main players of the kynurenine pathway and opponents in inflammatory disease. Med Hypotheses. 2018;118:129–38. [DOI] [PubMed] [Google Scholar]
- 37. Peuhkuri K, Sihvola N, Korpela R. Dietary factors and fluctuating levels of melatonin. Food Nutr Res. 2012;56.doi: 10.3402/fnr.v56i0.17252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kocadagli T, Yilmaz C, Gokmen V. Determination of melatonin and its isomer in foods by liquid chromatography tandem mass spectrometry. Food Chem. 2014;153:151–6. [DOI] [PubMed] [Google Scholar]
- 39. Feldman JM, Lee EM.. Serotonin content of foods: effect on urinary excretion of 5-hydroxyindoleacetic acid. Am J Clin Nutr. 1985;42(4):639–43. [DOI] [PubMed] [Google Scholar]
- 40. De Montis MG, Olianas MC, Haber B, Tagliamonte A. Increase in large neutral amino acid transport into brain by insulin. J Neurochem. 1978;30(1):121–4. [DOI] [PubMed] [Google Scholar]
- 41. Wiklund PK, Pekkala S, Autio R, Munukka E, Xu L, Saltevo J, Cheng S, Kujala UM, Alen M, Cheng S. Serum metabolic profiles in overweight and obese women with and without metabolic syndrome. Diabetol Metab Syndr. 2014;6(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C et al.. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17(4):448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yoon MS. The emerging role of branched-chain amino acids in insulin resistance and metabolism. Nutrients. 2016;8(7):E405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Acar E, Gurdeniz G, Khakimov B, Savorani F, Korndal SK, Larsen TM, Engelsen SB, Astrup A, Dragsted LO. Biomarkers of individual foods, and separation of diets using untargeted LC-MS-based plasma metabolomics in a randomized controlled trial. Mol Nutr Food Res. 2018;63(1):e1800215. [DOI] [PubMed] [Google Scholar]
- 45. Hernandez-Alonso P, Giardina S, Canueto D, Salas-Salvado J, Canellas N, Bullo M. Changes in plasma metabolite concentrations after a Low-Glycemic Index diet intervention. Mol Nutr Food Res. 2019;63(1):e1700975. [DOI] [PubMed] [Google Scholar]
- 46. Brown JM, Hazen SL.. The gut microbial endocrine organ: bacterially derived signals driving cardiometabolic diseases. Annu Rev Med. 2015;66:343–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Seldin MM, Meng Y, Qi H, Zhu W, Wang Z, Hazen SL, Lusis AJ, Shih DM. Trimethylamine N-Oxide promotes vascular inflammation through signaling of mitogen-activated protein kinase and nuclear factor-kappaB. J Am Heart Assoc. 2016;5(2):e002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cashman JR, Xiong Y, Lin J, Verhagen H, van Poppel G, van Bladeren PJ, Larsen-Su S, Williams DE. In vitro and in vivo inhibition of human flavin-containing monooxygenase form 3 (FMO3) in the presence of dietary indoles. Biochem Pharmacol. 1999;58(6):1047–55. [DOI] [PubMed] [Google Scholar]
- 49. Chen ML, Yi L, Zhang Y, Zhou X, Ran L, Yang J, Zhu JD, Zhang QY, Mi MT. Resveratrol attenuates trimethylamine-N-oxide (TMAO)-induced atherosclerosis by regulating TMAO synthesis and bile acid metabolism via remodeling of the gut microbiota. MBio. 2016;7(2):e02210–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. World Cancer Research Fund/American Institute for Cancer Research. Continuous update project report: diet, nutrition, physical activity and colorectal cancer.[Internet].2017. Available from: wcrf.org/colorectal-cancer-2017. [Google Scholar]
- 51. Koay YC, Wali JA, Luk AWS, Macia L, Cogger VC, Pulpitel TJ, Wahl D, Solon-Biet SM, Holmes A, Simpson SJ et al.. Ingestion of resistant starch by mice markedly increases microbiome-derived metabolites. FASEB J. 2019;33(7):8033–42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.